Abstract
We report here the structural determination of the N-linked glycans in the 66-kDa glycoprotein, part of the unique sulfated complex cell wall polysaccharide of the red microalga Porphyridium sp. Structures were elucidated by a combination of normal phase/reverse phase HPLC, positive ion MALDI-TOF MS, negative ion electrospray ionization, and MS/MS. The sugar moieties of the glycoprotein consisted of at least four fractions of N-linked glycans, each composed of the same four monosaccharides, GlcNAc, Man, 6-O-MeMan, and Xyl, with compositions Man8–9Xyl1–2Me3GlcNAc2. The present study is the first report of N-glycans with the terminal Xyl attached to the 6-mannose branch of the 6-antenna and to the 3-oxygen of the penultimate (core) GlcNAc. Another novel finding was that all four glycans contain three O-methylmannose residues in positions that have never been reported before. Although it is known that some lower organisms are able to methylate terminal monosaccharides in glycans, the present study on Porphyridium sp. is the first describing an organism that is able to methylate non-terminal mannose residues. This study will thus contribute to understanding of N-glycosylation in algae and might shed light on the evolutionary development from prokaryotes to multicellular organisms. It also may contribute to our understanding of the red algae polysaccharide formation. The additional importance of this research lies in its potential for biotechnological applications, especially in evaluating the use of microalgae as cell factories for the production of therapeutic proteins.
Keywords: Carbohydrate Structure, Glycoprotein, Glycoprotein Structure, Glycosylation, Mass Spectrometry (MS), N-Glycans, Cell Wall Polysaccharide, Methylated Sugars, Red Microalgae
Introduction
N-Glycosylation is one of the most fundamental protein post-translational modifications in eukaryotes, influencing the physicochemical and biological properties of proteins (1–5). To date, N-glycosylation patterns and N-glycan structures have been studied mainly in mammals, insects, yeasts, and plants (6), whereas seaweeds and microalgae have received very little attention aside from a few studies conducted on green algae. These studies generally revealed the presence of similar glycans to those found in the other species. For example, a lectin blot analysis in combination with N-glycosidase F (PNGase F)3 or endoglycosidase H (Endo H) treatment on the glycoproteins of the flagellar scale of Scherffelia dubia, a member of the Chlamydomonas family, revealed the presence of both high mannose and processed N-glycans (hybrid and complex types) (7, 8). Application of a similar protocol to the N-glycan structures of different flagellar strains of Tetraselmis showed that the scale-associated Tetraselmis striata glycoproteins are composed largely of high mannose glycans. On the other hand, those of Tetraselmis chui consist of many unknown complex glycans (9–11). A deeper insight into the glycan structures associated with the glycoprotein pheromone of the chlorophyte Volvox carteri f. nagariensis was obtained by applying a combination of exoglycosidases digestion, gas chromatographic sugar analysis, and two-dimensional HPLC separation (12). This experimental protocol showed that the N-glycans contain a chitobiose core with one to four additional Man residues with or without an additional Xyl residue attached to the 2-position of the core branching Man.
The current study focuses on the structural characterization of N-glycans in the 66-kDa glycoprotein associated with the cell wall polysaccharide of Porphyridium sp., the most abundant species of red microalga of the division Rhodophyta. Porphyridium sp. has been the subject of intensive study by our group for a number of years (13–17). The cells of the red microalga are encapsulated within a cell wall complex of a polysaccharide, which includes glycoproteins. This polysaccharide complex was found to possess unique characteristics and bioactivities, which offer a vast range of potential applications (13–17). Chemical characterization of red microalgae polysaccharide revealed that it is an anionic heteropolymer (molecular mass 3–5 × 106 Da) composed of about 10 different sugars, the main ones being Xyl, Glc, and Gal, with sulfate groups located on the Glc and Gal moieties (16–21). A primary disaccharide building block, 3-O-(α-d-glucopyranosyluronic acid)-l-galactopyranose has been isolated and characterized from the polysaccharides of various red microalgae species (21–23), and the chemical structure of different fractions separated from the polysaccharide of Porphyridium sp. have been determined (24, 25).
Among the proteins of the Porphyridium sp. cell wall complex that have been detected, the most prominent is a 66-kDa glycoprotein (27, 66). This glycoprotein is tightly bound, but not covalently linked, to the cell wall polysaccharide and consists of a polypeptide of ∼58 kDa and sugar moieties of ∼8 kDa. Sequencing of a cDNA clone encoding the 66-kDa glycoprotein revealed that this is a novel protein with four potential N-glycan sites that does not show similarity to any protein in the public domain databases. However, it does show some structural similarities to protein superfamilies within the carbohydrate binding domain, namely, glycosyltransferases, pectin lyase-like, sialidases, and concanavalin A-like lectins/glucanases in the SCOP databases, indicating a possible role of the 66-kDa glycoprotein in synthesis/modification of the cell wall polysaccharide (27). Moreover, this protein was found at the early stages of the cell wall cycle intermediate (28, 67) and in all cell wall-modified mutants. In addition, the glycoprotein was shown to play a role in biorecognition (30). Initial characterization of the sugar moieties by lectin blot analysis and PNGase F treatment suggested the presence of terminal Man (27).4 The glycoprotein was also detected by a xylose-specific antibody, indicating the presence of Xyl.
In the current study we characterized for the first time the N-linked glycans of the 66-kDa sugar moiety within the cell wall polysaccharide of Porphyridium sp. This study of the N-linked glycosylation patterns in Porphyridium sp. constitutes the first step in the search for enzymes involved in glycosylation-related pathways in this microalga. Because the cell wall glycoprotein was suggested to be involved in polysaccharide biosynthesis, unraveling its structure may contribute to our understanding of the biosynthesis of this polysaccharide. In addition, knowledge of the N-linked glycosylation might be of value for biotechnological applications, especially for production of therapeutic proteins, using the algae as a “cell factory,” as well as for studying the evolution of glycosylation processes.
EXPERIMENTAL PROCEDURES
Overall Experimental Protocol
Isolation and identification of the N-glycans of the 66-kDa glycoprotein bound to the cell wall-sulfated polysaccharide of Porphyridium sp. were performed as follows. The glycoprotein was isolated by SDS-PAGE. Gel bands containing the glycoprotein were subjected to the action of the following endoglycosidases to release the N-glycans: PNGase F, which cleaves between the reducing terminal GlcNAc residue and Asn residues, except those containing a 3-linked Fuc attached to the reducing terminal GlcNAc residue (31); Endo H, which cleaves between the GlcNAc residues of the chitobiose core in oligomannose and hybrid type N-linked glycans (32, 33); N-glycosidase A, which is suitable for releasing N-glycans with a core α 1,3-linked Fuc (34, 35). Samples of the released glycans were either directly derivatized with 2-aminobenzamide (2AB) or left underivatized. The non-labeled PNGase F-released glycans were also hydrolyzed to monosaccharides followed by 2AB-labeling. Both the 2AB-labeled intact glycans and monosaccharides were analyzed by HPLC and mass spectrometry (MS) as described below. In addition, samples of derivatized glycans released by PNGase F and Endo H were submitted to digestion by an array of exoglycosidases and then analyzed by normal phase (NP)-HPLC.
Isolation of the 66-kDa Glycoprotein
The microalga Porphyridium sp. (UTEX 637), obtained from a University of Texas Culture Collection, was grown in artificial seawater as previously described by Arad (18). To collect the extracellular polysaccharide, 12-day-old algae cultures were centrifuged (5000 × g, 10 min), and the supernatant containing the dissolved polysaccharide complex was dialyzed against double-distilled water (DDW) at 4 °C and then freeze-dried. The freeze-dried polysaccharide complex was redissolved in DDW to a concentration of 0.3% w/v. An aliquot (about 1.2 ml) of this solution was boiled in Laemmli sample buffer (0.5 ml) containing SDS and β-mercaptoethanol and loaded onto a funnel-shaped polyacrylamide gel under conditions described previously (27). The 66-kDa glycoprotein (ca. 30 μg) was detected by Coomassie Blue staining.
In-gel Enzymatic Release of N-Linked Glycans
N-Glycans were released by PNGase F (the gene from Flavobacterium meningosepticum was cloned and expressed in Escherichia coli, Roche Applied Science) using in-gel digestion according to Küster et al. (31) or by in-gel Endo H (recombinant from E. coli, Calbiochem) digestion. Briefly, each band containing the 66-kDa glycoprotein was excised from reducing SDS-PAGE gels and washed with 20 mm NaHCO3 (Sigma), pH 7, for PNGase F digestion or with 50 mm sodium phosphate buffer, pH 5.5, for Endo H digestion. The washed gel bands were dried in a vacuum centrifuge and then rehydrated with 200 μl of 20 mm NaHCO3, pH 7, containing 4 units of PNGase F or with 200 μl of 50 mm sodium phosphate buffer, pH 5.5, containing 20 milliunits of Endo H. After incubation for 24 h at 37 °C, the released glycans were extracted from the gel with 200 μl of H2O (×5) with sonication for 20 min. Salts were removed by incubation at room temperature (5 min) with 50 μl of an acid-activated AG-50W (50–100 mesh) slurry (Bio-Rad) followed by filtration through a 0.45-μm filter (Millex-LH, hydrophobic polytetrafluoroethylene, Millipore, Bedford, MA). As required, N-glycans released from bovine fetuin (from fetal calf serum, Sigma) (31) was used as the standard.
Alternatively, N-glycans were released by N-glycosidase A using the method described by Navazio et al. (36). Briefly, each band containing the 66-kDa glycoprotein from reducing SDS-PAGE gels was washed with 50 mm NH4HCO3 (Fluka, Buchs, Switzerland). The washed gel bands were dried in a vacuum centrifuge and then rehydrated with 200 μl of 50 mm NH4HCO3, pH 5, containing 2.5 μg of trypsin (Roche Applied Science). To inactivate any residual trypsin, the samples were heat-treated for 10 min at 100 °C. The samples were dried in a vacuum centrifuge and then incubated with 20 μl of N-glycosidase A dialyzed with 100 mm citrate-phosphate buffer (5 milliunits/250 μl) overnight at 37 °C. After incubation, the released N-linked glycans were eluted 5 times with 0.1% formic acid (Sigma). Salts were removed by using a mixed bed resin column of AG-50W (50 μl) and AG4-X4 (20 μl) (100-200 mesh; Bio-Rad) packed into a gel loading pipette tip.
Hydrolysis of N-Glycans and Monosaccharide Standards after PNGase F Digestion
Monosaccharides were obtained from the N-glycans (500 pmol) by acid hydrolysis in 500 μl of 2 m trifluoroacetic acid (Sigma) in microcentrifuge tubes. The hydrolysis tubes were sealed and incubated at 100 °C for 2 h. A set of typical monosaccharide standards, 5 nmol each of Ara, Man, Glc, Gal, Xyl, Rib, rhamnose, GlcNAc, Fuc, O-methylated-derived monosaccharide standards, 3-O-MeGlc (Sigma), 6-O-MeGal (City Chemical, West Haven, CT), and 6-O-MeMan, or a blank sample were treated similarly. After cooling to room temperature, each reaction mixture was evaporated to dryness in a vacuum centrifuge. The residues were each dissolved in 200 μl of 2-propanol (analytical grade, Sigma) followed by evaporation to dryness to remove residual trifluoroacetic acid.
2AB Derivatization
Glycans released by the action of PNGase F, Endo H, and N-glycosidase A, monosaccharides from the hydrolyzed glycans, and monosaccharide standards were derivatized with 2AB according to the method described by Bigge et al. (37). All the derivatives were analyzed by HPLC. In addition, the PNGase F, N-glycosidase A, and Endo H-released glycans derivatives were also analyzed by mass spectrometry. When needed, the identification of the derivatized monosaccharides was also verified using mass-spectrometry.
NP-HPLC Analysis of 2AB-derivatized Samples
Glycans and monosaccharide derivatives were analyzed by NP-HPLC as described previously (38). The glycans were separated on a TSK amide-80 column (4.6 × 250 mm, 5-μm pore size; Tosoh Bioscience, Montgomeryville, PA) at 30 °C using a Waters 2695 separations module and a Waters 2475 fluorescence detector. The following solvents were used: A, 50 mm ammonium formate solution (prepared from formic acid (analytical grade, Merck) and ammonium hydroxide solution (Fluka, Buchs, Switzerland)), pH 4.4; B, 100% acetonitrile, HPLC far-UV grade (Sigma). The linear gradient conditions used were: 0–152 min, 20–58% A at 0.4 ml/min; 152–155 min, 58% A at 0.4 ml/min; 155–157 min, 100% A at 0.4 ml/min; 157–163 min, 100% A at 1 ml/min; 178.5–180 min, 20% A at 0.4 ml/min. The total run time was 180 min, and samples were injected in 95 μl of a 20% DDW, 80% acetonitrile mixture. Glucose unit (GU) values were determined by standardizing each run against a ladder of glucose oligomers obtained from a partial hydrolysate of dextran. The amount of each derived oligosaccharide present in the samples was calculated by measuring individual peak areas with Waters Empower software and then comparing those values to values obtained from a standard 2AB curve. The monosaccharides from the hydrolyzed glycans were identified by HPLC by comparative monosaccharide analysis using a standard series of fluorescently labeled monosaccharides. An aqueous sample was used as a blank and was treated identically to the sugar samples. The blank NP-HPLC chromatogram base line was straight, indicating the absence of sugars.
Weak Anion-exchange (WAX)-HPLC Analysis of 2AB-derivatized Samples
To investigate the charge carried by the 2AB-labeled N-glycans, their WAX-HPLC chromatogram was compared with that of labeled bovine serum fetuin N-glycans that are known to carry from one to four negative charges. WAX chromatography was performed with a 0.75 × 5-cm DEAE column (301 VHP 575P, Vydac, Columbia, Maryland) at 30 °C, with the same HPLC noted above. The gradient used was that described by Guile et al. (39). The solvents were: A, 50 mm ammonium formate, pH 9; B, DDW. Initial conditions were 0% A and 100% B at a flow rate of 1 ml/min followed by a linear gradient reaching 5% A and 95% B over 12 min, an increase to 21% A and 79% B over 13 min, and a further increase to 80% A and 20% B over 25 min and then holding for 5 min. The column was washed with 100% A for 5 min at a flow rate of 1 ml/min before being re-equilibrated in 0% A for the next sample. The dried samples were injected in 100 μl of DDW and the labeled N-glycan standards.
Synthesis of 6-O-MeMan
6-O-Acetyl-2,3,4-tri-O-benzyl-d-mannopyranose (40, 41) was deacetylated under Zemplen conditions (catalytic (0.1 eq) sodium methoxide in methanol for 16 h at room temperature) to afford 2,3,4-tri-O-benzyl-d-mannopyranose. This compound was methylated by treatment with NaH (3 eq) in dimethylformamide for 15 min followed by methyl iodide MeI (2.5 eq) for 16 h at room temperature. Acetolysis of the resulting 2,3,4-tri-O-benzyl-1,6-di-O-methyl-d-mannopyranose was performed by refluxing with glacial acetic acid and 2 m H2SO4 (5:1 mixture by volume) for 3 h to gave the lactol 2,3,4-tri-O-benzyl-6-O-methyl-d-mannopyranose. Finally, hydrogenolysis in methanol with a paladium/carbon catalyst gave the target 6-O-MeMan.
RP-HPLC of 2AB-derivatized Monosaccharide Samples
2AB-labeled monosaccharides were separated on a Vydac (Columbia, MD) 218TP54 RP C18 HPLC column (4.6 × 250 mm, 5-μm pore size). The solvents used were: A, 0.2% N,N,N′,N′-tetramethylethylenediamine (Pierce) adjusted to pH 4.5 with H3PO4 followed by filtering with a Stericup, 0.22-μm GP Express PLUS membrane (Millipore, Billerica, MA); B, 100% acetonitrile. Initial conditions were 95% A and 5% acetonitrile at a flow rate of 0.4 ml/min followed by a linear gradient of 95–93% A over 30 min. The column was washed with 100% A for 5 min followed by acetonitrile at a flow rate of 1 ml/min before re-equilibration in the initial solvent system. The samples were injected in 100 of μl A and were identified by comparative monosaccharide analysis using a standard series of fluorescently labeled monosaccharides.
Exoglycosidase Digestions
Arrays of exoglycosidases were used in combination with NP-HPLC to investigate the antenna structure. The reaction mixtures consisted of a 10-μl solution containing 2AB-labeled glycans, exoglycosidases, and the appropriate reaction buffer as recommended by the manufacturers. Briefly, a 5-pmol sample of labeled glycans was incubated overnight at 37 °C with each of the following enzymes separately as follows: 1 μl of Arthrobacter ureafaciens sialidase (EC 3.2.1.18, 1 units/ml), 2 μl of bovine kidney α-fucosidase (EC 3.2.1.51, 1 units/ml), 2 μl of bovine testes β-galactosidase (EC 3.2.1.23, 2 units/ml), 2 μl of Streptococcus pneumoniae β-hexosaminidase (EC 3.2.1.52, 120 milliunits/ml), 2 μl of Xanthomonas β 1,2-xylosidase (EC 3.2.1.37, 10 units/ml). Each sample was also incubated with 3.5 μl of jack bean α-mannosidase (EC 3.2.1.24, 100 units/ml) for 24 h. After 24 h, an additional aliquot of enzyme (3.5 μl) was added to the sample mixture and incubated for another 24 h. Each sample was also incubated with 1 μl of Aspergillus saitoi 1,2-α-d-mannosidase (EC 3.2.1.113, 200 milliunits/ml) for 24 h. A second aliquot of enzyme (1 μl) was then added, and the digestion was continued for a further 24 h. After enzyme digestion, the samples were separated from the exoglycosidases by allowing the digested sample to disperse onto a protein binding filter (Millipore, Watford, UK) and inserted into a microcentrifuge for subsequent HPLC analysis. All the enzymes were purchased from Prozyme, San Leandro, CA, with the exception of the xylosidase, which was purchased from Calbiochem.
Positive Matrix-assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF) and MS/MS
The 2AB-labeled glycans (about 500 pmol each) and monosaccharides obtained by hydrolysis were collected manually after NP-HPLC separation. To remove the HPLC solvents, the contents of each vial were washed with 500 μl of DDW and dried by vacuum centrifugation (×3). Then, the pellet was suspended in 1 μl of DDW before MALDI-MS analysis. In addition, total pooled 2AB-labeled N-glycans (about 500 pmol) were dried and resuspended in 1 μl DDW, washed through a protein-binding filter (EZ filter), dried, and resuspended in 1 μl of DDW for subsequent MALDI-MS analysis.
Aliquots (1 μl) of samples were dialyzed for 30 min on a piece of Nafion 117 membrane (Aldrich) floating on DDW (42). The MALDI matrix was prepared as follows: dihydroxybenzoic acid (Sigma), 0.5 mg, was dissolved by vortexing in 100 μl of DDW:acetonitrile (1:1, v/v) and sonicated for 5 min to give a homogeneous saturated solution. This solution was centrifuged for 1 min at 13,000 rpm. Aliquots of the supernatant (1 μl) were applied to the MALDI target and were mixed with the dialyzed samples and allowed to dry under ambient conditions. Separated 2AB-labeled N-glycans were prepared similarly before being analyzed by positive MS/MS.
Positive-ion MALDI-TOF MS and MS/MS were performed on a Bruker-Daltonics Reflex IV mass spectrometer. A nitrogen laser of 337 nm was used. Spectra were run in a positive mode in a linear flight path with an extraction voltage of 20 kV. Spectra were taken from at least 400 random shots across the sample. The instrument was calibrated with standard I (Bruker-Daltonics) containing compounds with molecular masses ranging from 3 to 17 kDa. The X-Mass software (Bruker-Daltonics) was used for the analysis. For collision-induced dissociation spectra (positive MS/MS), the precursor ion was selected by the timed ion selector, and the spectra were acquired at a 30-kV accelerating voltage, whereas the reflector voltage was decreased step by step, allowing the detection of various fragments. The collision cell was filled with argon until the pressure in the source chamber reached ∼ 5 × 10−6 bars.
Negative-ion Electrospray Ionization (ESI) and MS/MS
Mass spectrometry was performed with a Waters quadrupole-time-of-flight (Q-Tof) Ultima Global instrument (Waters MS-Technologies, Manchester, UK) in negative ion mode. Samples in 1:1 (v:v) methanol:water were infused through Proxeon nanospray capillaries (Proxeon Biosystems, Odense, Denmark). The ion source conditions were: temperature, 120 °C; nitrogen flow, 50 liters/h; infusion needle potential, 1.2 kV; cone voltage, 100 V; RF-1, voltage 150 V. Spectra (2-s scans) were acquired with a digitization rate of 4 GHz and accumulated until a satisfactory signal-to-noise ratio was obtained. For MS/MS data acquisition collision-induced dissociation, the parent ion was selected at low resolution (about 3 m/z mass window) to allow transmission of isotope peaks and fragmented with argon at a pressure (recorded on the instrument's pressure gauge) of 0.5 millibar. The voltage on the collision cell was adjusted in terms of mass and charge to give an even distribution of fragment ions across the mass scale. Typical values were 80–120 V. Other voltages were as recommended by the manufacturer. Instrument control, data acquisition, and processing were performed with MassLynx software Version 4.0.
RESULTS
NP-HPLC of N-Glycans after Release with Endoglycosidases
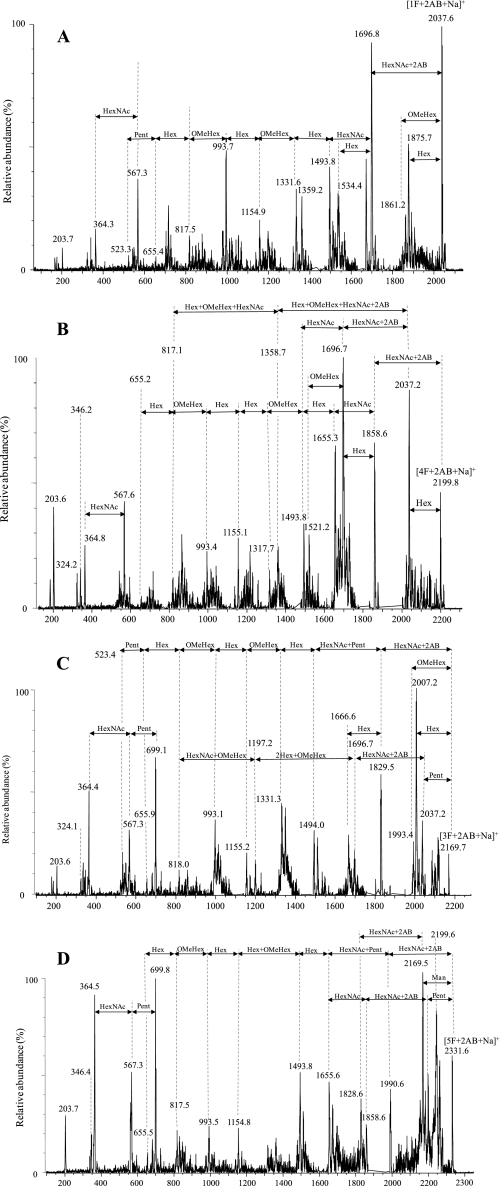
NP-HPLC of the derivatized N-glycans, which had been released after digestion of the 66-kDa glycoprotein by PNGase F, revealed four main peaks (labeled 1F+2AB, 3F+2AB, 4F+2AB, 5F+2AB in Fig. 1A), indicating a minimum of four different N-glycan fractions (1F, 3F, 4F, 5F) in the sugar moieties of the 66-kDa glycoprotein. The NP chromatogram of the N-glycosidase A-released glycans was the same as that of the PNGase F-released glycans, indicating the absence of a core α 1,3-linked Fuc (data not shown).
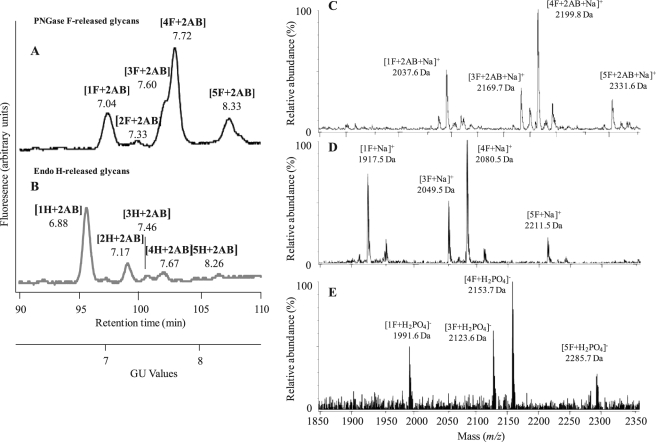
FIGURE 1.
A and B, NP-HPLC profiles of 2AB-labeled N-glycans released from the sugar moieties of the 66-kDa glycoprotein after PNGase F (A) and Endo H digestion (B) are shown. Retention times were converted to a GU scale relative to a dextran ladder. GU values are shown beneath the designation of each. C–E, mass determination of PNGase F-released glycans from the 66-kDa glycoprotein is shown. Positive ion MALDI-TOF MS of the total 2AB-labeled N-glycans (C) and unlabeled N-glycans (D) released from the 66-kDa glycoprotein by PNGase F is shown. E, negative ion ESI mass spectra of the total N-glycans released from the 66-kDa glycoprotein by PNGase F are shown. Interpretation of peak masses in terms of sugar composition and the calculated masses of the ions are summarized in Table 1.
The N-glycan chromatogram obtained after Endo H digestion exhibited one main peak (designated 1H+2AB in Fig. 1B) and four minor peaks (designated 2H+2AB, 3H+2AB, 4H+2AB, and 5H+2AB in Fig. 1B), indicating the existence of oligomannose and/or hybrid type N-glycans.
WAX-HPLC Analysis of N-Glycans
The possible presence of negatively charged glycans existing in the N-glycan pool was examined by running the derivatized samples on WAX-HPLC. All the glycans were found to be neutral.
Exoglycosidase Array Digestion
NP-HPLC analysis of 2AB-linked glycans released by PNGase F and Endo H and then digested with the exoglycosidases listed under “Experimental Procedures” showed that these enzymes did not have any effect on the glycans. This finding indicates that the mixture of N-linked glycans obtained from the 66-kDa glycoprotein of Porphyridium sp. differs considerably from N-glycans from other species that are normally substrates for these enzymes.
Identification of the N-Glycan-derived Monosaccharides
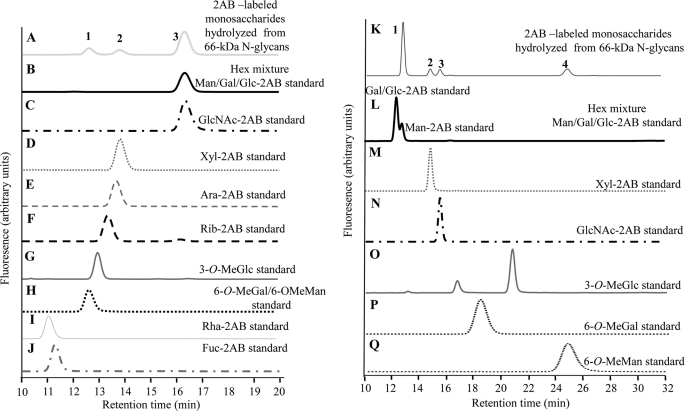
The identity of the constituent monosaccharides of the N-glycans was determined by a combination of NP-HPLC and RP-HPLC analysis. The NP-HPLC chromatogram of the monosaccharides derived from the hydrolyzed glycans showed only three peaks (peaks 1–3, Fig. 2A), whereas the RP-HPLC chromatogram showed four peaks (peaks 1–4, Fig. 2K). The chromatograms thus suggest a minimum of four different monosaccharides in the hydrolyzed glycan samples.
FIGURE 2.
A–J, NP-HPLC separation of 2AB-labeled sugars is shown. Shown are monosaccharides derived from hydrolyzed N-glycans (A), a hexose mixture (Gal, Glc, Man) (B), GlcNAc (C), Xyl (D), Ara (E), Rib (F), 3-O-MeGlc (G), 6-O-MeGal and 6-O-MeMan mixture (H), rhamnose (I), Fuc (J). K–Q, RP-HPLC separation of 2AB-labeled sugars is shown. Shown are monosaccharides were derived from hydrolyzed N-glycans (K), a hexose mixture (Gal, Glc, Man) (L), Xyl (M), GlcNAc (N), 3-O-MeGlc (O), 6-O-MeGal (P), and 6-O-MeMan (Q).
The most prominent peaks detected in the NP- and RP-HPLC chromatograms were Peak 3 (Fig. 2A) and Peak 1 (Fig. 2K), respectively. The retention time of Peak 3 in the NP-HPLC chromatogram matched that of the hexose residues, Glc, Gal, or Man and GlcNAc (Fig. 2, B and C). To reveal the specific identity of Peak 3 (Fig. 2A), the compound was collected and run on a RP-HPLC. The RP-HPLC chromatogram of the isolated Peak 3 showed two peaks. The retention time of the first (Fig. 2K, Peak 1) and second (Fig. 2K, Peak 3) peaks matched those of Man and GlcNAc standards, respectively (Fig. 2, L and N), which were run under the same conditions. To further confirm that Peak 1 in RP-HPLC is a Man residue, we collected the separated peak and mixed it with each one of the hexose standards (Glc, Gal, Man) and separated the mixture again on RP-HPLC. Only the Man mixture chromatogram exhibited one sharp peak, thus identifying the hexose as Man.
Peak 2 in the NP-HPLC chromatogram (Fig. 2A) was collected and further subjected to RP-HPLC separation. Results indicate that this peak in NP-HPLC correlates to Peak 2 found in the RP-HPLC chromatogram (Fig. 2K). Comparing this peak to standards showed that the retention time matches that of either Xyl or Ara (Fig. 2, D and E). To determine whether this pentose was Xyl or Ara, the compound was isolated and mixed with each of the pentose standards. The mixtures were analyzed on a NP-HPLC column and gave a sharp peak with Xyl and a wide peak with Ara (data not shown), strongly suggesting that Peak 2 is Xyl. Comparison of the GU value of Xyl standard with the GU value of Ara in the RP-HPLC chromatograms yielded the same GU value, indicating that these pentose residues cannot be separated by each other by using RP-HPLC columns conditions as described above.
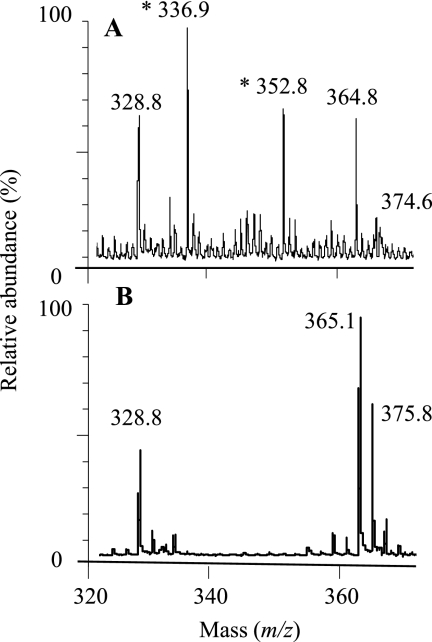
Both the NP- and RP-HPLC chromatograms exhibited a peak (Peak 1 in Fig. 2A and Peak 4 in Fig. 2K, respectively) that did not match any of the typical glycan standards. To identify the unknown monosaccharide, a separated fraction of Peak 1 from the NP-HPLC chromatogram and its corresponding region of the hydrolyzed blank (11.8–13 min) were each collected and subjected to analysis by MALDI-TOF MS. Comparison of the two MS spectra (hydrolyzed monosaccharide sample and the corresponding blank sample) showed that the two prominent peaks that appear in the spectrum of the hydrolyzed monosaccharide but not in the blank spectrum had masses of 336.9 and 352.8 Da (Fig. 3). These ions correlated with [O-MeHex+2AB+Na]+ (calculated m/z 337.1) and [O-MeHex+2AB+K]+ (calculated m/z 353.1) ions, respectively, indicating the hydrolyzed monosaccharide to be O-MeHex. The other peaks observed in the MALDI-TOF MS spectra may probably be attributed to noise derived from the hydrolysis method or the measurement.
FIGURE 3.
Mass determination of peak 1 from Fig. 2 (unknown monosaccharide derived from N-glycans). A, shown is a positive-ion MALDI-TOF MS spectrum of Peak 1. B, shown is an MS analysis of the blank of Peak 1 (DDW that underwent the same procedure as the N-glycans). m/z values indicate the molecular mass of the monosaccharide including additional [Na]+ or [K]+ ions and the 2AB residue. The monosaccharide mass values are indicated with asterisks.
To elucidate the position of the methyl group and the identity of the hexose of the unknown monosaccharide (Peak 1, Fig. 2A; Peak 4, Fig. 2K), we ran three available methylated hexose standards, 3-O-MeGlc (Fig. 2G/Fig. 2O), 6-O-MeGal (Fig. 2H/Fig. 2P), and 6-O-MeMan (Fig. 2H/Fig. 2Q), and compared their retention times with the retention time of the unknown methylhexose. The retention time of the O-MeHex, derived from the hydrolyzed N-glycans, in the NP-HPLC chromatogram (Peak 1, Fig. 2A) matched the retention time of both the 6-O-MeGal and 6-O-MeMan (Fig. 2H) but did not match 3-O-MeGlc (Fig. 2G). To reveal the specific hexose identity of the derived O-MeHex, the compound was isolated and mixed with each one of the 6-O-MeHex standards, and each mixture was run on NP-HPLC. The mixture containing the 6-O-MeMan gave one sharp peak, whereas the mixture with the 6-O-MeGal gave a rather broad peak, suggesting that our O-MeHex is 6-O-MeMan. To confirm this finding we also analyzed the 6-O-MeHex by separation on RP-HPLC. The retention time, derived from hydrolyzed N-glycans, in the RP-HPLC chromatogram (Peak 4, Fig. 2K) matched only the retention time of 6-O-MeMan (Fig. 2Q). Similarly, to reveal the specific identity of the derived O-MeHex, the compound was isolated and mixed with each one of the O-MeHex standards and was run again on RP-HPLC. Whereas injecting a mixture of derivatized 6-O-MeMan and the unknown O-MeHex resulted in appearance of one sharp peak, co-injection experiments indicated that the unknown did not co-elute with either 6-O-MeGal or 3-O-MeGlc. These data suggest that the O-MeHex is indeed 6-O-MeMan, a conclusion consistent with the results from the negative ion MS/MS experiment (see below).
Mass Spectrometry of N-Glycans by Positive-ion MALDI-TOF MS and Negative-ion ESI-MS/MS
The molecular weights of the 2AB-labeled and unlabeled PNGase F-released glycans from 66-kDa glycoprotein were measured by MALDI-TOF MS (Fig. 1, C and D), and their deduced compositions, in terms of the monosaccharides identified above, are listed in Table 1. In addition, HPLC-separated 2AB-labeled glycans were also collected manually, and their masses were determined. These masses showed a direct correlation with the GU values. For example, the glycan with the smallest GU value on the NP-HPLC (peak 1F+2AB, Fig. 1A) had the lowest mass of 2037.6 Da by MALDI-TOF MS (peak [1F+2AB+Na]+, Fig. 1C).
TABLE 1.
Composition of the observed 66-kDa N-glycans according to the positive-ion MALDI-TOF MS and negative-ion ESI-MS spectra
| Glycan | Positive ion [M+Na]+ |
Positive ion [M+2AB+Na]+ |
Negative ion [M+H2PO4]− |
Composition |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Found | Calculated | Found | Calculated | Found | Calculated | Hex | O-Me Hex | Pentose | HexNAc | |
| m/z | ||||||||||
| 1F | 1917.5 | 1917.70 | 2037.6 | 2037.77 | 1991.6 | 1991.65 | 5 | 3 | 1 | 2 |
| 3F | 2049.5 | 2049.74 | 2169.7 | 2169.81 | 2123.6 | 2123.69 | 5 | 3 | 2 | 2 |
| 4F | 2080.5 | 2079.75 | 2199.8 | 2200.82 | 2153.7 | 2153.70 | 6 | 3 | 1 | 2 |
| 5F | 2211.5 | 2211.79 | 2331.6 | 2331.86 | 2285.7 | 2285.74 | 6 | 3 | 2 | 2 |
The unlabeled glycans were also analyzed by negative ESI-MS (Fig. 1E). All MS spectra, both negative and positive (Fig. 1, C–E), indicated the presence of at least four N-glycans. Their masses did not match those of any N-glycans previously found in other organisms. The masses of oligosaccharides, assigned as 1F and 3F, differed by 132 Da, suggesting a mass difference of a pentose. The difference between oligosaccharides assigned as 1F and 4F was 162 Da, suggesting a mass difference of a hexose. The mass values measured in the MS profiles of the negative ESI-MS were found to be even masses, indicating the presence of an even number of nitrogens in the oligosaccharide molecules.
Negative MS/MS Spectra of N-Glycans
To obtain a more detailed analysis, underivatized oligosaccharides from PNGase F release were subjected to negative mode MS/MS (Fig. 4). The negative ion MS/MS spectra were typical of neutral glycans run as phosphate adducts (phosphate was the anion used to ionize the compounds). The spectra, which were interpreted according to published data (43–46), all contained a major ion of 259 mass units less than that of the molecular ion consistent with a 2,4A fragmentation (Domon and Costello nomenclature (47)) of the core GlcNAc (loss of 161 mass units and the phosphate adduct) after abstraction of the 3-proton by the phosphate. This mass loss showed no substitution of the core GlcNAc. A second ion, 60 mass units below this ion, was also present in the spectra of all compounds and corresponds to a BR cleavage (the subscript R is used here to refer to “reducing terminus”) consistent with a β(1→4)-linkage.
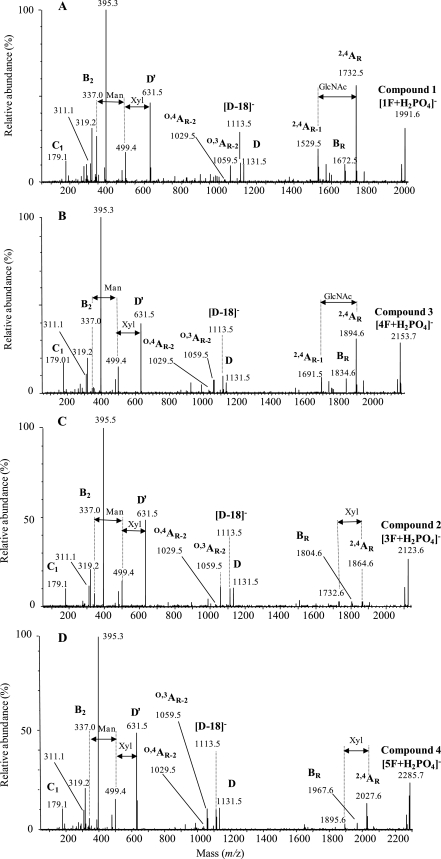
FIGURE 4.
Negative ion ESI-collision-induced dissociation (phosphate adducts) of N-linked glycans released from 66-kDa glycoprotein by PNGase F. Shown are compound 1 at m/z 1991 ([1F+H2PO4]− ion) (A), compound 3 at m/z 2153 ([4F+H2PO4]− ion) (B), compound 2 at m/z 2123 ([3F+H2PO4]− ion) (C), and compound 4 at m/z 2286 ([1F+H2PO4]− ion) (D). Ion nomenclature follows the method of Domon and Costello (47).
The spectra of Compounds 1 [1F+H2PO4]− and 3 [4F+H2PO4]− contained an additional ion 203 mass units below that of the 2,4AR ion, corresponding to a similar cleavage of the penultimate GlcNAc residue (Fig. 4, A and B, 2,3AR-1 ion). The spectra of Compounds 2 [3F+H2PO4]− (Fig. 4C) and 4 [5F+H2PO4]− (Fig. 4D), however, which had an extra Xyl residue, did not contain this ion, suggesting that the Xyl was attached to the 3-oxygen of the penultimate GlcNAc, blocking the abstraction of a proton at this site and accounting for the absence of this 2,4AR-1 ion. These two spectra contained an ion representing loss of a pentose from the 2,4AR ion (m/z 1732.6 and 1895.6), showing that it was not attached to the reducing-terminal GlcNAc.
Normally, in plants and insects Xyl is found attached to the 2-position of the branching Man. However, the negative ion MS/MS spectrum of [Man]3[GlcNAc]2[Xyl]1[Fuc]1 from horseradish peroxidase, which contains such a 2-linked Xyl, contained a very abundant ion corresponding to the 2,4AR-1 fragment (m/z 677), consistent with the 3-proton being available for abstraction.4 Thus, it appears that the compounds with two Xyl residues have one Xyl attached to the 3-position of the penultimate GlcNAc residue.
In all other respects the negative ion MS/MS spectra of all four compounds were virtually identical (Fig. 4, A–D). The group of ions at m/z 1131, 1113, 1059, and 1029 (weak) are similar to those from high mannose glycans and correspond to D, [D-18]−, O,3AR-2, and O,4AR-2, respectively (the D ion is formed by loss of the 3-antenna and chitobiose core and, thus, contains the intact 6-antenna and branching core Man). The similarity of these ions with those in the high mannose glycans again suggests that there is no Xyl substitution on the core Man because fragmentation around and across this residue is responsible for these four fragments. The mass of the D ion (m/z 1131), which contains the 6-antenna, indicated a composition of [Man]4[O-MeHex]2[Xy]l1, leaving after subtraction of the core GlcNAc residues, a composition of [Man]1[O-MeHex]1 for the 3-antenna in Compounds 1 and 2 and [Man]2[O-MeHex]1 for the 3-antenna in Compounds 3 and 4. Because all four glycans have the same structure for the 6-antenna and core, the extra Man in the glycans of mass m/z 2153 and 2286 must be at the non-reducing end of the 3-antenna.
At the low mass end of the spectrum, the major C1 ion is at m/z 179, corresponding to non-reducing terminal Man. Low intensity ions at m/z 337 and 355 correspond to B2 and C2 ions with compositions of [Man]1[O-MeMan]1. Therefore, the antenna(e) appears to terminate with Man-O-MeMan. The ion at m/z 311 corresponds in mass to a C2 ion consisting of Xyl1Man1 (179 + 132), indicating attachment of Xyl onto a non-reducing terminal Man and not to O-MeMan.
The similarity of the spectra with those of high mannose glycans suggests a similar topology, and therefore, the two branches of the 6-antenna exhibit Man-[O-MeMan] and Xyl-Man-[O-MeMan] compositions. The composition of the ion at m/z 631 appears to be [Man]2[O-MeMan]1[Xyl]1, which is consistent with that of a D′ ion (the 3,6-disubstitution pattern of the Man α 1,6-linked to the core mannose is the same as that of the core Man itself).
The ion at m/z 499 is the result of a further loss of Xyl from the ion at m/z 631 (the 6-branch of the 6-antenna). Thus, the Xyl appears to be attached to this portion of the molecule. This conclusion is supported by the presence of the major ion at m/z 395 that appears to have a composition of Xyl-Hex plus 101 mass units. The equivalent ion without Xyl (m/z 263) is seen in the spectra of high mannose glycans that have two Man residues in the 6-branch of the 6-antenna. The ion at m/z 319 has a composition of [Man]1[O-MeHex]1-H2O. Corresponding ions are not seen in the spectra of high mannose glycans. The most probable source of this ion is loss of Xyl-Hex from m/z 631. There is a very weak ion at m/z 149, consistent with a non-reducing terminal Xyl. One antenna, therefore, appears to have the composition Xyl-Man-[O-MeHex].
The ion at m/z 395, mentioned above, has the composition Xyl-Man+101. Ions of similar composition are found in the spectra of complex carbohydrates, where the 101 mass units contain carbon atoms 1, 2, 3, and 4 of the attached Man residue. Assuming that m/z 395 has a similar structure, the 101 mass units would be from an O-MeHex that had lost carbons 5 and 6 together with the methyl group. Therefore, the methoxy group must be at C6, giving 6-O-MeHex. Based on these observed MS/MS data and the derived monosaccharide determination (based on the HPLC chromatogram), we conclude that the 6-O-MeHex is most likely a 6-O-MeMan residue.
The central region of the spectrum of the compound producing peak 1 was more complex than the others and contained prominent ions at m/z 969, 951, and 897 corresponding to the D, [D-18]− and O,3AR-1 ions from an analog of the major 1F glycan (see Fig. 7B) containing one less Man residue in the 6-antenna (see Fig. 7C). This Man residue must, therefore, be attached to the 3-antenna, indicating the presence of a fifth compound.
FIGURE 7.
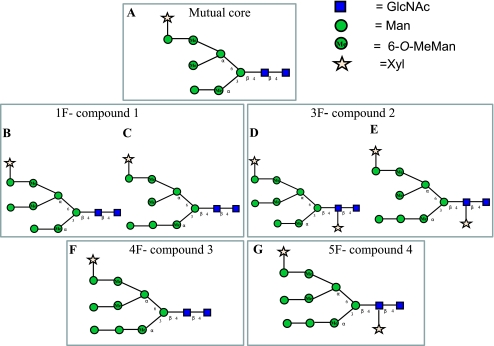
Suggested structures of the 66-kDa N-linked glycan fractions. A, a mutual core exists in 1F, 3F, 4F, 5F. Shown are prominent (B) and minor (C) N-glycan in 1F fraction, prominent (D) and minor (E) N-glycan in 3F fraction, 4F N-glycan (F), and 5F N-glycan (G).
Positive MS/MS Spectra of N-Glycans
To collect more information on the glycan structures, each of the 2AB-labeled PNGase-F-released glycans was isolated by NP-HPLC and examined by positive ion MS/MS (Fig. 5). The interpretations of the positive MS/MS spectra were in good agreement with those of the negative MS/MS spectra. The positive ion MS/MS spectra of all four derivatized N-glycan fractions were very similar (Fig. 5, A–D) in that they all contained the following ions: m/z 203, 364, 567, 655, 993, 817, 1155, and 1494. The structural interpretations of these compounds are shown in supplemental Tables ST1. The above ions indicate the same core structure with composition [O-MeMan]2[Man]4[Xyl]1[GlcNAc]2 in each one of the N-glycans (the suggested structure is shown as the glycans core; see Fig. 7A). All four spectra show the presence of fragments resulting from the loss of one or both core GlcNAc residues; the intensity of [GlcNAc]1-2AB and [GlcNAc]2-2AB fragments is also relatively high.
FIGURE 5.
Positive ion MS/MS spectra of the 2AB-labeled N-glycans released from the 66-kDa glycoprotein by PNGase F. Shown are glycan at m/z 2038 ([1F+2AB+Na]+ ion) (A), at m/z 2200 ([4F+2AB+Na]+ ion) (B), positive ion MS/MS spectra of the glycan at m/z 2170 ([3F+2AB+Na]+ ion) (C), and at m/z 2332 ([5F+2AB+Na]+ ion) (D). The arrows on the spectra indicate mass differences corresponding to glycan residues and do not necessarily imply a fragmentation sequence.
The presence of the ion at m/z 699 in the positive ion MS/MS spectra of both compounds that contained an additional Xyl (Fig. 5, C and D) and the absence of this ion in the spectra of the other two compounds ([1F+2AB+Na]+, Fig. 5A; [4F+2AB+Na]+, Fig. 5B) indicate that the m/z 699 fragment has the probable composition of [Xyl]1[GlcNAc]2-2AB. The ions at m/z 699, 567, and 364 may indicate on a series; Abstracting the Pent mass residue (132 Da) from m/z 699 yields the m/z 567 ion ([GlcNAc]2-2AB). Another loss of a further HexNAc residue (203 Da) from m/z 567 ion yields the m/z 364 ion ([GlcNAc]1-2AB).
Because the negative MS/MS data showed that the reducing terminal GlcNAc does not have an additional Xyl, it is evident that the Xyl of these two compounds is attached to the penultimate GlcNAc. In addition, the spectra of the compounds [3F+2AB+Na]+ and [5F+2AB+Na]+ contained ions at m/z 1494 and 1656, which indicated the presence of an additional Xyl attached at the non-reducing terminus.
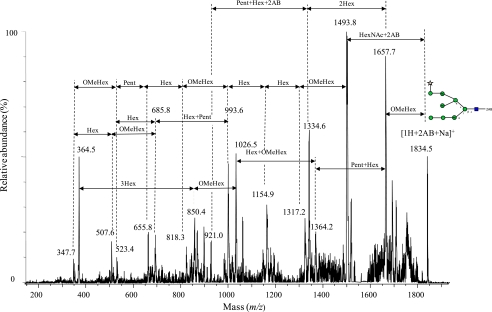
The most abundant 2AB-labeled glycan released by Endo H was also separated (Peak [1H+2AB], Fig. 1B) by NP-HPLC and subjected to positive MS/MS analysis. As expected, the observed mass of this glycan was m/z 1834, i.e. 203 Da (which is compatible with the mass of GlcNAc) less than the mass of glycan 1F that was released after PNGase F digestion ([1H+2AB+Na]+, Fig. 6). The MS/MS spectrum (Fig. 6) and the MS/MS interpretations of the Endo H-released glycan (1H, supplemental Tables ST2) appears to be different from the other MS/MS spectrum. The spectrum of [1H+2AB+Na]+ (Fig. 6) is the only one that contains a prominent ion (m/z 1334.6), which indicates the loss of Hex2. The ion at m/z 1657.7 shown in the [1H+2AB+Na]+ spectrum (Fig. 6) is also prominent, indicating a prominent loss of O-MeMan (176 Da less than the parent ion [1H+2AB+Na]+). The same phenomenon is evident in the other spectra (m/z 1861.2, Fig. 5A; m/z 1993.4, Fig. 5C); however, the ions derived from the loss of non-reducing terminal O-MeHex in these spectra are minor in comparison to the ion m/z 1657.7 that is shown in the positive MS/MS [1H+2AB+Na]+ spectrum (Fig. 6). In the positive ion MS/MS spectra of the PNGase F-released glycans, a prominent loss of O-MeMan appears only after Man has been lost. In addition, the negative ion MS/MS spectrum of Compound 1 also indicates the presence of a non-reducing O-MeHex (m/z 193, Fig. 4A). It is evident that the non-reducing O-MeHex in the glycans without the additional Man (1F, 3F) is located in the 3-antenna, as it cannot be located in the 6-antenna, whose terminal consists of Xyl and Man.
FIGURE 6.
Positive ion MS/MS spectra of the 2AB-labeled N-glycans released after Endo H digestion at m/z 1834. 5 ([1H+2AB+Na]+ ion). The arrows on the spectra indicate mass differences corresponding to glycan residues and do not necessarily imply a fragmentation sequence.
Proposed N-Glycan Structure
Because the only hexose residues that were found in the N-glycans are Man residues, and because the negative MS/MS fragments are typical of the oligomannose topology, the glycan structure is most certainly of the oligomannose type. Based on the negative and positive ion spectra of the N-glycans (assuming high mannose topology), the proposed structures of the four glycan fractions are presented in Fig. 7. Most of the fragment ions derived from the 1F fraction indicate a specific glycan structure (shown in Fig. 7B). They also indicate the presence of another glycan structure but in low abundance (shown in Fig. 7C). It can be concluded that glycan 1F contains a mixture of two isomers (Fig. 7, B and C). However, according to the positive ion MS/MS spectrum of the 1H glycan, it can be suggested that most of the compound released after Endo H digestion contains the minor isomer (with non-reducing O-MeMan on the 3-branch of the 6-antenna) derived from the 1F glycan (shown in Fig. 6). This can be explained by differential release by Endo H. Thus, it can be suggested that the minor isomeric glycan in the fraction containing glycan 1F is released more rapidly than the major compound. A similar situation should exist for 3F, compound 2 (Fig. 7, D and E). Presumably, the presence of the Xyl on the core GlcNAc inhibits the action of Endo H, explaining the low yield of compound 2 released after Endo H digestion. Assuming that the same core structure is present in all compounds, the suggested structures of 4F and 5F are shown in Fig. 7, F and G, respectively.
The glycan profile can be explained, assuming that the hexoses are mannose, by postulating that a specific enzyme is able to remove only the terminal Man on the 3-antenna of the mannose-9 analog (Compounds 4F and 5F). After this, Man is removed, to give compounds 1F and 3F, yielding the Man 8A isomer, and further processing stops. It can be suggested that this enzyme may be a Golgi endomannosidase (48, 49) that specifically cleaves the α1–2 linkage between the glucose-substituted mannose residue and the more internal portion of its polymannose branch, leading to the formation of [Man]8[GlcNAc]2 (Man 8A) isomer (50). By searching for red algae endomannosidase genome sequence and domain similarity (based on the data of the National Center for biotechnology Information of the National Institutes of Health), no homology was found, although limited red algae genome data are known. As a result, the existence of endomannosidase cannot be ruled in red algae.
DISCUSSION
This study constitutes the first investigation of the N-glycan structures of a glycoprotein from within the cell wall polysaccharide of the red microalga Porphyridium sp. Glycan analysis showed four prominent N-linked glycans, based on the oligomannose topology, that were different from structures found so far in other organisms. These novel structures contain the same four monosaccharides: GlcNAc, Man, 6-O-MeMan, and Xyl. All have the same core structure with the characteristic structure [Man]4[O-MeMan]3[Xyl]1[GlcNAc]2, containing methylated Man residues (Fig. 7A).
Several studies have reported methylated sugars as constituents of glycoproteins in other organisms, including nematodes, yeasts, snails, algae, and planarians (51–62). For example, in the study by Hall et al. (52), 3-O-MeGal was found in hemocyanin from Helix pomatia (Roman snail), and both 3-O-MeGal and 3-O-MeMan were identified in hemocyanin from Lymnaea stagnalis (a freshwater snail). In mollusks, 3-O-MeMan and/or 3-O-MeGal were found in some hemocyanins (53–56), and in nematodes, O-linked glycans were shown to contain 2-O-methylated fucose (57, 58). However, only a few studies have investigated the overall glycan structure (composition and monosaccharide sequence) of molecules that contain methylated sugars. For example, the structures of N-linked glycans from a lectin of the giant clam Hippopus hippopus were found to be primarily of the oligomannose type, but in addition, some contained a 6-O-methylated group on the terminal Man residue of the chain (59). Another study investigating the primary structures of two biantennary N-glycans of the glycoprotein from Rapana venosa (marine snail) hemocyanin showed that the glycans contain a 3-O-methyl group on the terminal Gal and/or GlcNAc (58). A terminal 3-O-MeMan was also found in N-glycans from the gastropod Arion lusitanicus (61) and from the planarian Dugesia japonica (62).
Although the organisms described above are able to methylate terminal monosaccharides (Man and Gal) in glycans, the red microalga Porphyridium sp. is the first organism to be described that is able to methylate internal Man residues, i.e. residues other than those directly at the non-reducing terminus. The presence of three O-MeMan groups and a Xyl on the antennae in the glycans is probably also the reason that the terminal Man residues were not removed by mannosidase digestion.
Xyl residues are found in N-glycans from plants (63), insects (64), molluscs (53), and in rare examples of parasitic helminths (65) but not normally in mammals. In addition, the position and linkage of Xyl (attached to the 2-position of the core branching Man) is the same in all the organisms mentioned above. In this study we found for the first time a Xyl residue attached to the Man of the 6-antenna and linked 1 → 3- to the penultimate GlcNAc of the core). These findings indicate the existence of special glycosyltransferases and glycosylation pathways, unique to the red microalgae. It is not known whether the Xyl residues reported here have a similar allergenic nature to the Xyl residues found in other glycans (26, 29). The glycosylation pattern might be important when using the red microalgae as cell factories for biopharmaceuticals.
The overall structures of the N-glycans in Porphyridium sp. are novel, but the glycans appear to exhibit a core oligomannose topology that is common in plant and yeast N-glycans. Thus, the N-glycans of the Porphyridium sp. glycoprotein combine structural features from plants and those from the giant clam H. hippopus (which also contains 6-O-Man).
Supplementary Material
Acknowledgments
We thank Prof. Pauline Rudd of the National Institute for Bioprocessing, Research, and Training, Professor of Glycan Biology at University College Dublin for encouragement during this study. We also thank the Wellcome Trust for an equipment grant to purchase the Q-TOF Ultima Global mass spectrometer.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables ST1 and ST2.
O. Levy-Ontman, S. M. Arad, D. J. Harvey, T. B. Parsons, A. Fairbanks, and Y. Tekoah, unpublished observation.
- PNGase F
- N-glycosidase F
- 2AB
- 2-aminobenzamide
- DDW
- double-distilled water
- Endo H
- endoglycosidase H
- ESI
- electrospray ionization
- GU
- glucose unit
- Hex
- hexose
- m/z
- mass/charge ratio
- NP
- normal phase
- WAX
- weak anion-exchange
- Ara
- arabinose
- Xyl
- xylose
- Fuc
- fucose.
REFERENCES
- 1. Varki A. (1993) Glycobiology 3, 97–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dwek R. A. (1996) Chem. Rev. 96, 683–720 [DOI] [PubMed] [Google Scholar]
- 3. Varki A., Cummings R., Esko J., Freeze H., Hart G., Marth J. (1999) Essentials of Glycobiology, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 4. Wormald M. R., Dwek R. A. (1999) Structure 7, R155–R160 [DOI] [PubMed] [Google Scholar]
- 5. Taylor M. E., Drickamer K. (2003) Introduction to Glycobiology, 2nd Ed., Oxford University Press, Oxford [Google Scholar]
- 6. Weerapana E., Imperiali B. (2006) Glycobiology 16, 91R-101R [DOI] [PubMed] [Google Scholar]
- 7. Grunow A., Becker B., Melkonian M. (1993) Eur. J. Cell Biol. 61, 10–20 [PubMed] [Google Scholar]
- 8. Becker B., Perasso L., Kammann A., Salzburg M., Melkonian M. (1996) Planta 199, 503–510 [Google Scholar]
- 9. Becker B., Dreschers S., Melkonian M. (1995) Eur. J. Phycol. 30, 307–312 [Google Scholar]
- 10. Becker D., Melkonian M. (1992) Eur. J. Cell Biol. 57, 109–116 [PubMed] [Google Scholar]
- 11. Gödel S., Becker B., Melkonian M. (2000) Protist 151, 147–159 [DOI] [PubMed] [Google Scholar]
- 12. Balshüsemann D., Jaenicke L. (1990) Eur. J. Biochem. 192, 231–237 [DOI] [PubMed] [Google Scholar]
- 13. Lapidot M., Shrestha R. P., Weinstein Y., Arad (Malis) S. (2010) Cellular Origin, Life in Extreme Habitats, and Astrobiology: Red Algae in Genomic Age (Seckbach J., Chapman D. J. eds) Vol. 13, Part 3, pp. 205–225, Springer, Dordrecht, The Netherlands [Google Scholar]
- 14. Arad (Malis) S., Weinstein Y. (2003) Biomedic (Israel) 1, 32–37 [Google Scholar]
- 15. Arad S. M., Levy-Ontman O. (2010) Curr. Opin. Biotech. 21, 358–364 [DOI] [PubMed] [Google Scholar]
- 16. Arad (Malis) S. (1999) Chemicals from Microalgae (Cohen Z. ed) pp. 282–287, Taylor and Francis, New York [Google Scholar]
- 17. Arad (Malis) S., Richmond A. (2004) Handbook of Microalgal Culture: Biotechnology and Applied Phycology (Richmond A. ed) pp. 289–297, Blackwell Publishing Ltd; Oxford [Google Scholar]
- 18. Arad (Malis) S. (1988) Algal Biotechnology (Stadler T., Mollion J., Verdus M. C., Karamanos Y., Morvan H., Christiaen D. eds) pp. 65–87, Elsevier Applied Science, London [Google Scholar]
- 19. Geresh S., Arad (Malis) S. (1991) Bioresour. Technol. 38, 195–201 [Google Scholar]
- 20. Heaney-Kieras J., Chapman D. J. (1976) Carbohydr. Res. 52, 169–177 [DOI] [PubMed] [Google Scholar]
- 21. Lupescu N., Arad (Malis) S., Geresh S., Bernstein M., Glazer R. (1991) Carbohydr. Res. 210, 349–352 [Google Scholar]
- 22. Geresh S., Lupescu N., Arad (Malis) S. (1992) Phytochemistry 31, 4181–4186 [Google Scholar]
- 23. Geresh S., Dubinsky O., Arad S. M., Christiaen D., Glaser R. (1990) Carbohydr. Res. 208, 301–305 [DOI] [PubMed] [Google Scholar]
- 24. Geresh S., Arad S. M., Levy-Ontman O., Zhang W., Tekoah Y., Glaser R. (2009) Carbohydr. Res. 344, 343–349 [DOI] [PubMed] [Google Scholar]
- 25. Gloaguen V., Ruiz G., Morvan H., Mouradi-Givernaud A., Maes E, Krausz P., Strecker G. (2004) Carbohydr. Res. 339, 97–103 [DOI] [PubMed] [Google Scholar]
- 26. van Ree R., Cabanes-Macheteau M., Akkerdaas J., Milazzo J. P., Loutelier-Bourhis C., Rayon C., Villalba M., Koppelman S., Aalberse R., Rodriguez R., Faye L., Lerouge P. (2000) J. Biol. Chem. 275, 11451–11458 [DOI] [PubMed] [Google Scholar]
- 27. Shrestha R. P., Weinstein Y., Bar-Zvi D., Arad S. M. (2004) J. Phycol. 40, 568–580 [Google Scholar]
- 28. Simon-Bercovitch B., Bar-Zvi D., Arad S. M. (1999) J. Phycol. 35, 78–83 [Google Scholar]
- 29. Garcia-Casado G., Sanchez-Monge R., Chrispeels M. J., Armentia A., Salcedo G., Gomez L. (1996) Glycobiology 6, 471–477 [DOI] [PubMed] [Google Scholar]
- 30. Ucko M., Shrestha R. P., Mesika P., Bar-Zvi D., Arad (Malis) S. (1999) J. Phycol. 35, 1276–1281 [Google Scholar]
- 31. Küster B., Wheeler S. F., Hunter A. P., Dwek R. A., Harvey D. J. (1997) Anal. Biochem. 250, 82–101 [DOI] [PubMed] [Google Scholar]
- 32. Tarentino A. L., Plummer T. H., Jr., Maley F. (1974) J. Biol. Chem. 249, 818–824 [PubMed] [Google Scholar]
- 33. Trimble R. B., Tarentino A. L., Plummer T. H., Jr., Maley F. (1978) J. Biol. Chem. 253, 4508–4511 [PubMed] [Google Scholar]
- 34. Altmann F., Schwihla H., Staudacher E., Glossl J., Marz L. (1995) J. Biol. Chem. 270, 17344–17349 [DOI] [PubMed] [Google Scholar]
- 35. Altmann F., Paschinger K., Dalik T., Vorauer K. (1998) Eur. J. Biochem. 252, 118–123 [DOI] [PubMed] [Google Scholar]
- 36. Navazio L., Miuzzo M., Royle L., Baldan B., Varotto S., Merry A. H., Harvey D. J., Dwek R. A., Rudd P. M., Mariani P. (2002) Biochemistry 41, 14141–14149 [DOI] [PubMed] [Google Scholar]
- 37. Bigge J. C., Patel T. P., Bruce J. A., Goulding P. N., Charles S. M., Parekh R. B. (1995) Anal. Biochem. 230, 229–238 [DOI] [PubMed] [Google Scholar]
- 38. Guile G. R., Rudd P. M., Wing D. R., Prime S. B., Dwek R. A. (1996) Anal. Biochem. 240, 210–226 [DOI] [PubMed] [Google Scholar]
- 39. Guile G. R., Wong S. Y., Dwek R. A. (1994) Anal. Biochem. 222, 231–235 [DOI] [PubMed] [Google Scholar]
- 40. Tennant-Eyles R. J., Davis B. G., Fairbanks A. J. (2000) Tetrahedron Asymmetry 11, 231–243 [Google Scholar]
- 41. Murakata C., Ogawa T. (1992) Carbohydr. Res. 235, 95–114 [DOI] [PubMed] [Google Scholar]
- 42. Börnsen K. O., Mohr M. D., Widmer H. M. (1995) Rapid Commun. Mass Spectrom. 9, 1031–1034 [DOI] [PubMed] [Google Scholar]
- 43. Harvey D. J. (2005) J. Am. Soc. Mass Spectrom. 16, 622–630 [DOI] [PubMed] [Google Scholar]
- 44. Harvey D. J. (2005) J. Am. Soc. Mass Spectrom. 16, 631–646 [DOI] [PubMed] [Google Scholar]
- 45. Harvey D. J. (2005) J. Am. Soc. Mass Spectrom. 16, 647–659 [DOI] [PubMed] [Google Scholar]
- 46. Harvey D. J., Royle L., Radcliffe C. M., Rudd P. M., Dwek R. A. (2008) Anal. Biochem. 376, 44–60 [DOI] [PubMed] [Google Scholar]
- 47. Domon B., Costello C. E. (1988) Glycoconj. J. 5, 397–409 [Google Scholar]
- 48. Lubas W. A., Spiro R. G. (1987) J. Biol. Chem. 262, 3775–3781 [PubMed] [Google Scholar]
- 49. Lubas W. A., Spiro R. G. (1988) J. Biol. Chem. 263, 3990–3998 [PubMed] [Google Scholar]
- 50. Moore S. E., Spiro R. G. (1990) J. Biol. Chem. 265, 13104–13112 [PubMed] [Google Scholar]
- 51. Vliegenthart J. F. G., Montreuil J. (1995) Glycoproteins (Montreuil J., Schachter H., Vliegenthart J. F. G. eds) pp. 13–28, Elsevier Science B.V., Amsterdam [Google Scholar]
- 52. Hall R. L, Wood E. J., Kamberling J. P., Gerwig G. J., Vliegenthart F. G. (1977) Biochem. J. 165, 173–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kamerling J. P., Vliegenthart J. F. G. (1997) Glycoproteins II (Montreuil J., Vliegenthart J. F. G., Schachter H. eds) pp. 123–161, Elsevier Science B.V. Amsterdam [Google Scholar]
- 54. Lommerse J. P., Thomas-Oates J. E., Gielens C., Préaux G., Kamerling J. P., Vliegenthart J. F. G. (1997) Eur. J. Biochem. 249, 195–222 [DOI] [PubMed] [Google Scholar]
- 55. Stoeva S., Rachev R., Severov S., Voelter W., Genov N. (1995) Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 110, 761–765 [DOI] [PubMed] [Google Scholar]
- 56. Stoeva S., Schütz J., Gebauer W., Hundsdörfer T., Manz C., Markl J., Voelter W. (1999) Biochim. Biophys. Acta 1435, 94–109 [DOI] [PubMed] [Google Scholar]
- 57. Khoo K. H., Maizels R. M., Page A. P., Taylor G. W., Rendell N. B., Dell A. (1991) Glycobiology 1, 163–171 [DOI] [PubMed] [Google Scholar]
- 58. Guérardel Y., Balanzino L., Maes E., Leroy Y., Coddeville B., Oriol R., Strecker G. (2001) Biochem. J. 357, 167–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Puanglarp N., Oxley D., Currie G. J., Bacic A., Craik D. J., Yellowlees D. (1995) Eur. J. Biochem. 232, 873–880 [PubMed] [Google Scholar]
- 60. Dolashka-Angelova P., Beck A., Dolashki A., Beltramini M., Stevanovic S., Salvato B., Voelter W. (2003) Biochem. J. 374, 185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gutternigg M., Ahrer K., Grabher-Meier H., Bürgmayr S., Staudacher E. (2004) Eur. J. Biochem. 271, 1348–1356 [DOI] [PubMed] [Google Scholar]
- 62. Natsuka S., Hirohata Y., Nakakita S., Sumiyoshi W., Hase S. (2011) FEBS J. 278, 452–460 [DOI] [PubMed] [Google Scholar]
- 63. Lerouge P., Cabanes-Macheteau M., Rayon C., Fischette-Lainé A. C., Gomord V., Faye L. (1998) Plant Mol. Biol. 38, 31–48 [PubMed] [Google Scholar]
- 64. Altmann F., Staudacher E., Wilson I. B., März L. (1999) Glycoconj. J. 16, 109–123 [DOI] [PubMed] [Google Scholar]
- 65. Khoo K. H., Chatterjee D., Caulfield J. P., Morris H. R., Dell A. (1997) Glycobiology 7, 663–677 [DOI] [PubMed] [Google Scholar]
- 66. Shrestha R. P. (1999) A Non-covalenty Bound Cell-wall Glycoprotein of the Red Microalga Porphyridium sp.: Characterization and Functions. Ph.D. thesis, Ben-Gurion University of the Negev, Beer-Sheva, Israel [Google Scholar]
- 67. Simon-Bercovitch B. (1997) Cell-wall Formation in the Red Microalga Porphyridium sp. Ph.D. thesis, Ben-Gurion University of the Negev, Eilat, Israel [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.