Abstract
CD4 binding on gp120 leads to the exposure of highly conserved regions recognized by the HIV co-receptor CCR5 and by CD4-induced (CD4i) antibodies. A covalent gp120-CD4 complex was shown to elicit CD4i antibody responses in monkeys, which was correlated with control of the HIV virus infection (DeVico, A., Fouts, T., Lewis, G. K., Gallo, R. C., Godfrey, K., Charurat, M., Harris, I., Galmin, L., and Pal, R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17477–17482). Because the inclusion of CD4 in a vaccine formulation should be avoided, due to potential autoimmune reactions, we engineered small sized CD4 mimetics (miniCD4s) that are poorly immunogenic and do not induce anti-CD4 antibodies. We made covalent complexes between such an engineered miniCD4 and gp120 or gp140, through a site-directed coupling reaction. These complexes were recognized by CD4i antibodies as well as by the HIV co-receptor CCR5. In addition, they elicit CD4i antibody responses in rabbits and therefore represent potential vaccine candidates that mimic an important HIV fusion intermediate, without autoimmune hazard.
Keywords: Antigen Presentation, Chemical Modification, Cysteine-mediated Cross-linking, HIV, Protein Cross-linking, AIDS Vaccine, Affinity Labeling, Molecular Mimicry
Introduction
Despite the development of effective drugs targeting the HIV virus, AIDS remains a major health problem worldwide. A preventive vaccine would be the best and cheapest defense against the epidemic, a crucial point for underprivileged countries where AIDS is most prevalent. The trimeric gp160 envelope protein (Env),3 which is composed of a noncovalent association between the gp41 membrane protein and the more accessible gp120 surface protein, is the main target of the humoral response. HIV resistance to antibody-mediated neutralization is mainly because of a number of amazing properties of the gp120 glycoprotein (1–4). Critical neutralizing epitopes are preserved among the different virus strains, but the virus has evolved ways to minimize the immunogenicity of its surface protein. The association of the envelope protein in a trimeric conformation shields potential epitopes (2, 4–7). The outer domain of gp120, heavily glycosylated with poorly or nonimmunogenic glycans (2, 4), prevents antibody access to the underlying peptide structure. This glycoprotein also exposes five variable loops (V1–V5), which are the result of the extraordinary ability of the virus to mutate at a very high rate (1–4, 8). These regions, in particular the V1/V2 domain, act as a global regulator of the sensitivity of primary HIV isolates to neutralization by antibodies (8, 9). Hypervariable loops therefore contribute to a rapid and constant selection of neutralizing antibody-escape variants during the course of infection. In contrast, conserved regions are less accessible (10) or hidden through conformational masking (11). Altogether, these factors explain why envelope-based immunogens tested to date have failed to elicit antibodies capable of neutralizing more than a minor fraction of primary isolates (4, 12). Consequently, phase III clinical trials have shown that gp120-based vaccines failed to protect or at best showed modest results against HIV infection (1, 4, 13, 14). A preventive HIV vaccine should elicit antibodies with reasonable affinity for the conserved regions of gp120, those particularly involved in virus attachment on targeted cells. HIV envelope glycoprotein binds to both CD4 receptor and some G-protein-coupled chemokine co-receptors, CCR5 or CXCR4 (15–18). However, induction of antibodies directed against these functional regions is impeded due to the inaccessibility of the CD4-binding site and the conformational masking of the co-receptor-binding site (11). To counteract the high entropy that prevents antibody access to the co-receptor-binding site, one approach is to stabilize the gp120 structure through interaction with CD4. Indeed, binding of gp120 to the host cell CD4 receptor causes the reorientation of the loop regions of gp120 and stabilizes the viral glycoprotein in an intermediate state that exposes the co-receptor-binding site (10, 11, 17–19). Complexes formed between gp120 and CD4 are recognized by a large panel of antibodies, called CD4-induced antibodies, whose epitopes overlap the co-receptor-binding site (20–23). Previous studies have shown that immunization with either a mixture of soluble CD4 and gp120 or a fusion protein between them elicited neutralizing antibodies effective against HIV-1 primary isolates (24–29). Moreover, vaccine trials have emphasized the benefit of a covalent linkage between CD4 and gp120 (25–27, 29–31). In addition, a stable linkage between CD4 and gp120 may be necessary to prevent dissociation that could be induced by the vaccine adjuvant and/or the high dilution of the complex in the blood.
Previously, a covalently linked gp120-CD4 complex was reported to elicit broadly neutralizing antibodies in macaques (27). More recently, a single chain construct formed by gp120BaL-rhesus macaque CD4D1D2, evaluated as an immunogen in rhesus macaques challenged with SHIVSF162P3, was found to elicit an antibody response that correlated with an accelerated clearance of plasma viremia and an absence of long term tissue viremia (26). Taken together, these data emphasize the usefulness of CD4-gp120 covalent complexes as a vaccine candidate. Nevertheless, the use of CD4 in a vaccine formulation should be avoided, because of potential autoimmune reactions. Therefore, the replacement of CD4 by appropriate synthetic mimetics possessing a high affinity for gp120 may solve this problem. Moreover, CD4 mimetics obtained by chemical synthesis can harbor specific moieties for HIV envelope cross-linking. Such CD4 mimetics were developed in our laboratory on the basis of structural similarities between the CDR2-like loop of CD4 and the β-hairpin region of a scorpion toxin (32). The last generation of these miniCD4s (27 residues) possesses a CD4-like affinity for gp120 and stabilizes the viral glycoprotein in a CD4-bound-like conformation, exposing both CD4i epitopes and the co-receptor-binding site (19, 33, 34).
This study describes the generation of a stable covalent complex between a specifically engineered miniCD4 and an Env-derived protein, with no antigenic properties altering the virus protein. This complex was shown to elicit a CD4i antibody response in rabbits, and the sera were able to neutralize a few tested virus strains.
EXPERIMENTAL PROCEDURES
HIV-1 Envelope Glycoprotein Expression
Stable CHO DG-44 cell lines were used to express gp120SF162 monomer or gp140ΔV2SF162 trimer (gp140 deleted of the V2 loop). SF162 envelope glycoproteins were purified as described previously (6, 7). Note that all the experiments carried out with the so-called gp140 in this study were in fact done with the V2 deleted version of gp140 (gp140ΔV2SF162).
CD4 Mimetic Peptide Synthesis
MiniCD4s were prepared on an Applied Biosystems synthesizer (model 433) by solid phase peptide synthesis using fluorenylmethyloxycarbonyl-protected amino acids and N-hydroxybenzotriazole/dicyclohexylcarbodiimide coupling strategy. Refolding was performed at 0.1 mg·ml−1 in 100 mm phosphate buffer, pH 7.8, in the presence of 5 mm reduced glutathione and 0.5 mm oxidized glutathione. Synthetic peptides were purified by reverse phase HPLC and checked by amino acid analysis and mass spectrometry. MiniCD4 sequences are shown below and in the supplemental material. For the disulfide bond exchange cross-linking strategy, an additional sulfhydryl group was introduced by derivatizing Lys4 on the M64U1 peptide. Fmoc-Lys(ivDde)-OH (where ivDde is (1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene)-3-methylbutyl and Fmoc is N-(9-fluorenyl)methoxycarbonyl) was used at position 4 during peptide synthesis. Selective deprotection of the ivDde moiety by 10 5-min treatments of the resin with 2% hydrazine was followed by N-succinimidyl S-acetylthiopropionate coupling. To allow determination of the cross-linking yield of miniCD4 to gp120, by fluorescence polarization or by chemiluminescence after SDS-PAGE, we introduced during the synthesis fluorescein or biotin moieties at a specific lysinyl residue (see below and supplemental material), after selective deprotection of an orthogonal methyltrityl protecting group and coupling of biotinamidohexanoic acid N-hydroxysuccinimidyl ester or 6-carboxyfluorescein N-hydroxysuccinimide ester.
MiniCD4 Peptides Used in This Study
A specific miniCD4 was designed for each cross-linking method (see also supplemental material). All the miniCD4s used in this study have a C-terminal carboxamide and share nanomolar affinities for gp120. The following sequences were used: M48U1, TpaNLHFCQLRCKSLGLLGRCAdPTU1CACV; M64U1-SH, TpaNL(K-Tpa)WCQKRCKSLGLLGRCAdPTU1CACV; and M64U1(Biot)-SH, TpaNL(K-Tpa)WCQKRC(K-Biot)SLGLLGRCAdPTU1CACV. Biot is biotin; Tpa is 3-mercaptopropanoyl (-COCH2CH2SH); dP is d-proline, and U1 is p-(cyclohexylmethyloxy)phenylalanine.
Cross-linking through Disulfide Bond Formation
MiniCD4 with an additional SH group introduced on the side chain of Lys4 (M64U1-SH) was incubated at a molar ratio of 3:1 with gp120 or 9:1 with the trimeric gp140SF162 for 1 h at room temperature in phosphate-buffered saline, pH 7.4 (PBS).
SDS-PAGE Analysis
Approximately 1–5 μg of cross-linked products, gp120 coupled to a biotinylated miniCD4, and their respective controls were separated by SDS-PAGE. After electrophoresis, the gel was transferred onto a PVDF membrane, and the biotinylated M64U1 was detected by subsequent treatment with streptavidin HRP and autoradiography using an enhanced chemiluminescence assay (ECL, GE Healthcare).
Cross-linking Yield Determination
40 μl of a 4 μm gp120-purified solution was diluted with 120 μl of 10 mm sodium phosphate buffer, pH 7.4, and reacted with various amounts of M64U1-SH for 30 min at room temperature. The complex was then treated with 3 μl of 1 mm 5-iodoacetamidofluorescein in ethanol for 45 min at room temperature and finally acidified with 30 μl of a 1.5 m citric acid solution to block all the reactions. The mixture was then injected on a C4 Vydac reverse phase column (150 × 4.6 mm; 1.5 ml·min−1; solvent A, H2O/TFA, 0.1%; solvent B, acetonitrile; gradient, 30–30% solvent B in 10 min and then 30–50% solvent B in 20 min, and 50–70% solvent B in 5 min). UV detection was carried out with a Waters 996 PDA detector. The outflow was then alkalinized with a triethylamine-saturated water solution, and fluorescence was detected at 522 nm (excitation, 500 nm). The area of the fluorescent peak was calibrated with fluorescein-labeled glutathione solutions of known concentration.
Labeled Residue Mapping
1.25 nmol of gp120SF162 was incubated with 3 eq of M64U1-SH in 260 μl of 1 m Tris citrate buffer, pH 8, for 1 h at room temperature. The complex was then reacted with 10 nmol of N-biotinoyl-N′-iodoacetylethylenediamine (1 mm stock solution in DMF) for 1 h at room temperature. The complex was reduced with DTT (90 mm) for 5 min and denatured in 6 m guanidinium chloride for 2 h at room temperature. 1.2 ml of Tris-HCl 1 m, pH 8 (to inhibit acidification through iodoacetamide reaction), was added to the mixture, and the proteins were reacted with a large excess of iodoacetamide (18 mg, 97 μmol) to block sulfhydryl groups. The mixture was acidified with TFA, and labeled gp120 was purified by HPLC on an analytical C4 reverse phase column (150 × 4.6 mm; 1 ml·min−1; solvent A, H2O/TFA 0.1%; solvent B, acetonitrile; gradient, 20–60% solvent B in 30 min; the expected protein came out at about 20.5 min). The protein was then lyophilized and solubilized in 50 μl of sodium phosphate buffer (20 mm, pH 7.8), Tergitol 0.75%, and deglycosylated with peptide:N-glycosidase F (Sigma) at 37 °C (three times the addition of 500 IUB milliunits in 30 h). The deglycosylated protein was then precipitated with 500 μl of cold ethanol. The precipitate was trypsinized in 20 μl of digestion buffer containing 100 mm NH4HCO3, pH 7.8, and trypsin protease (Promega) at 24 ng·μl−1. Trypsinization was performed at 50 °C and terminated after 2 h of incubation by adding 5 μl of concentrated TFA solution (50% in water). Digested samples were then desalted using C18 ZipTip from Millipore Waters and spotted on a MALDI plate using the dried droplet method with α-cyano-4-hydroxycinnamic acid matrix solution at 10 mg·ml−1 in 50% acetonitrile, water, 0.1% TFA.
MS and MS/MS spectra were registered using a 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Foster City, CA) in positive reflectron mode. Each MS spectrum was the result of 1000 shots, and calibration was applied by using trypsin autolysis fragments as internal standards. Analyses of the peptide mass fingerprints from base-line-corrected, noise-filtered de-isotoped spectra, and thus assignment of the tryptic fragments was performed using an in-house MASCOT search engine (Version 2.1.0, Matrix Science, London, UK). Search parameters against NCBInr data base updated in July, 2009, were as follows: enzymatic cleavage, trypsic; variable modifications, S-carboxamidomethylation (CAM) of cysteine residues; methionine oxidation, Asn and Gln deamidation, missed cleavages, 1; MS tolerance, 50 ppm; and MS/MS tolerance, 0.25 atomic mass units. MS/MS analyses were performed under post-source decay (PSD) mode. Each MS/MS spectrum was the result of 2000–4000 shots. The sequence of the tryptic fragment was identified by using MASCOT search engine as well as Data Explorer® processing software (version 4.9, Applied Biosystems) to assign the potential chemical modifications of the cysteine residues.
Surface Plasmon Resonance Biosensor Analysis
Experiments were conducted at 25 °C with 30 μl·min−1 flow rate in HBS (HEPES-buffered saline, 3 mm EDTA, 0.05% Biacore surfactant, pH 7.4) with a Biacore 3000 instrument (Biacore AB, Uppsala, Sweden). 48d mAb was immobilized at 3000 response units by the amine coupling kit (N-hydroxysuccinimide/N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide) provided by the manufacturer. Cross-linked complexes and their controls, at 10 nm, were tested for their binding activity toward 48d mAb. This monoclonal antibody was directed to CD4i epitopes, and it was also designated as a conformational antibody. All the sensorgrams were corrected by subtracting the signal from the reference flow cell.
Immunological Characterization Analysis
The binding of well characterized HIV-Env-specific mAbs was performed by a capture ELISA. Typically, the 96-well plates were coated overnight at 4 °C with lectin (concanavalin A, Sigma, 500 ng per well); wells were saturated with PBS, 5% BSA buffer, washed three times, and serial dilutions of both gp140-S-S-M64U1 complex and gp140 alone were added. The capture proteins were revealed by incubation with 50 ng of anti-Env mAbs (Centralized Facilities for AIDS reagents, National Institute for Biological Standards and Control) 1 h at room temperature. Several monoclonal antibodies were tested, directed against the carbohydrates moieties (2G12) to normalize the response, the gp41 domain (2F5), the V3 loop (447-52D and MN215), the CD4-induced epitopes (X5, 17b, and 48d), and the CD4-binding site (b12). Finally, plates were washed, and specifically bound antibodies were detected using peroxidase-conjugated antibodies as described earlier (34).
Cell Surface CCR5 Chemokine Receptor Binding Assays
Adherent CHO-K1 cells overexpressing CCR5 (35) were used to analyze envelope conformational change by flow cytometry FACS (BD Biosciences). Typically, 1 μm gp120s were preincubated or not with 3 eq of sCD4 (Progenic, Tarrytown, NY) or miniCD4 for 1 h at room temperature. This mixture was then diluted to 3 nm with PBS/BSA 0.5%, pH 7.4, and for competition experiments, 3 eq of X5 mAb were added. 100 μl of this solution were mixed to 2 × 105 CCR5+ cells. After 2 h of incubation at 4 °C, cells were washed with PBS/BSA 0.5%, pH 7.4, and incubated with D7324 mAb (Aalto Bio Reagents Ltd., Dublin, Ireland), which recognizes the C terminus of gp120. A phycoerythrin-tagged antibody directed against D7324 mAb further detected envelope binding to CCR5 co-receptor.
Rabbit Immunization
Samples were sterilized over 0.22-μm filters and diluted with an equal volume of adjuvant mixture (MF59TM) at the final concentration of 50 μg·ml−1. Each rabbit was immunized with 0.5 ml of adjuvanted protein complex per side intramuscular/Gluteus for a total of 1 ml per animal. Groups of five New Zealand White rabbits were immunized with either gp120, gp140, or M64U1-SH cross-linked to gp120 (gp120-S-S-M64U1) or M64U1-SH cross-linked to gp140 (gp140-S-S-M64U1). Four injections were administered at 0, 4, 12, and 24 weeks. Serum samples were collected before the first immunization and 2 weeks following the third and the fourth immunizations. CD4i antibody titers were determined by HIV-2 neutralization as described previously (41).
Virus Neutralization
Neutralization was assessed using molecularly cloned pseudoviruses and a luciferase reporter gene assay in TZM-bl cells as described previously (36, 37).
RESULTS
Cross-linking Strategies
One of the main objectives of this study was to couple a miniCD4 onto gp120 or gp140 without altering the antigenic properties of these glycoproteins, aiming ultimately to the stabilization of HIV envelope in the CD4-bound conformation, recognizing both CD4i Abs and the HIV co-receptor. Several first-tried standard cross-linking methods highlighted the difficulty of maintaining both cross-linking and antigenic properties of gp120 (see supplemental material). We thus had to develop a site-directed coupling reaction that allowed us to obtain a covalent complex with all the required properties.
Intermolecular Disulfide Bond Exchange Concept
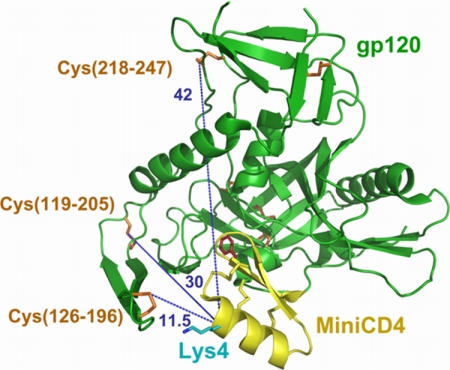
After the failure to conciliate cross-linking with antigenic properties using standard strategies, we developed an original coupling method between a miniCD4-SH and gp120 based on intermolecular disulfide bond exchange. Crystal structure of a core gp120-miniCD4 complex (38) shows that only the exposed Cys126–Cys196 disulfide bond on gp120 is well oriented and close enough to the miniCD4 molecule to permit a disulfide bond exchange with a sulfhydryl group properly positioned on the ligand (Fig. 1). Actually, the α-carbon of the fourth miniCD4 residue is 10.2 and 11.6 Å distant from the sulfur atom of Cys196 and Cys126, respectively. Therefore, modifying the ϵ-NH2 position of a lysine variant on position 4 of a miniCD4 with a thiopropyl moiety should bring the introduced thiol in an ideal position to react with this gp120 disulfide bond. The appropriate short length (11.5 Å) of this lysinyl-thiopropyl linker may not allow modification of the two other surface-accessible disulfide bonds on the core protein, i.e. Cys119–Cys205 and Cys218–Cys247, which are located on the other side of gp120 and are about 30 and 40 Å distant from Lys4 on the miniCD4, respectively. Such a miniCD4-SH was therefore developed and called M64U1-SH.
FIGURE 1.
Distance determination between accessible disulfide bonds on gp120 and the α-carbon on position 4 of the miniCD4. This picture was adapted from Stricher et al. (55), using PyMOL. MiniCD4 and gp120 are depicted in yellow and green, respectively. Disulfide bonds are shown in sticks (yellow for miniCD4 and orange for gp120). The three solvent-accessible cystines on gp120 are numbered. His4 was changed to lysine (cyan). The critical Phe23 residue of the miniCD4 is depicted as red sticks. Blue dotted lines show the distances in angstroms between the α-carbon of the reactive residue on the miniCD4 (Lys4) and the accessible disulfides on gp120.
Biochemical Characterization of the Disulfide Cross-linked Complexes
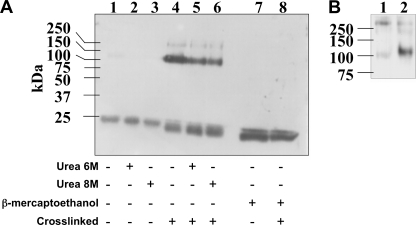
Complexes formed between M64U1-SH and gp120 were analyzed by nonreducing SDS-PAGE in the presence of urea. CD4 mimetics were visualized by chemiluminescence using biotin-tagged M64U1-SH and streptavidin HRP. Two bands could be observed, one below 20 kDa corresponding to free M64U1(Biot)-SH and one at ∼100 kDa corresponding to the miniCD4 covalently linked to gp120 (Fig. 2A). A faint band, at about 250 kDa, can also be seen and probably corresponds to a dimer of gp120, which is classically obtained by recombinant expression (39, 40). Covalent complexes formed by gp120 and M64U1(Biot)-SH remained detectable in 8 m urea (Fig. 2A, lane 6), although control complexes formed by gp120 and a miniCD4 without a free sulfhydryl group were only visible as a very weak band without urea (lane 1). Addition of 10% β-mercaptoethanol abolished the linkage in the cross-linked sample (Fig. 2A, lane 8), providing evidence of the existence of a disulfide bond between gp120 and the miniCD4.
FIGURE 2.
A, cross-linking analysis by SDS-PAGE. Uncross-linked or cross-linked complexes (lanes 1 and 4, respectively), preincubated in 6 m urea (lanes 2 and 5, respectively) or 8 m urea (lanes 3 and 6, respectively), were analyzed by SDS-PAGE in the presence (lanes 7 and 8) or not (lanes 1–6) of 10% β-mercaptoethanol. M64U1(Biot)-SH cross-linking was revealed by chemiluminescence through its biotin moiety via streptavidin HRP. B, protection experiment showing that M64U1(Biot)-SH is specifically covalently bound onto gp120. Separation on SDS-PAGE of gp120 preincubated or not with 100 eq of sCD4 (lanes 1 and 2, respectively) before mixing with 1 eq of biotinylated M64U1-SH. MiniCD4 binding was revealed through its biotin moiety as described above. Small quantities of labeled gp120 aggregates can be seen at about 250 kDa.
The coupling reaction specificity was further evaluated by a protection experiment, namely HIV-1 envelope glycoprotein gp120SF162 was preincubated with 100 eq of CD4 for 2 h and then mixed with an equimolar concentration of M64U1(Biot)-SH for 1 h. The resulting complexes were separated on SDS-PAGE, and miniCD4 binding was revealed through its biotin moiety as described above. Preincubation with sCD4 almost completely prevented binding of the miniCD4-SH (Fig. 2B, lane 1 compared with lane 2), thus demonstrating the specificity of the cross-linking.
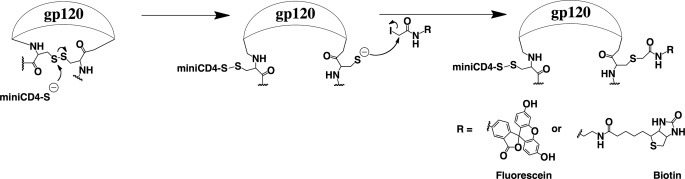
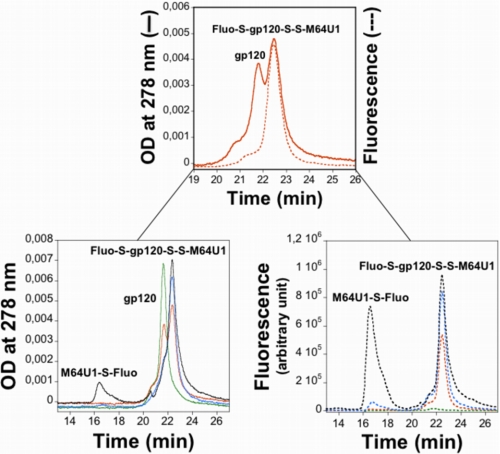
Formation of a disulfide bond between the envelope protein and M64U1-SH implies the release of a cysteinyl residue on the HIV surface protein, allowing an easy quantification of the cross-linking yield through modification of this unique sulfhydryl with 5-iodoacetamidofluorescein (Scheme 1). The fluorescein-labeled complex could be separated from the 5-iodoacetamidofluorescein in excess and even differentiated from the nonmodified gp120 by high performance reverse phase liquid chromatography (Fig. 3). Cross-linking was close to quantitative as observed on the UV profile, by the almost disappearance of the native gp120. A calibration with fluorescein labeled glutathione solutions of known concentrations allowed confirmation of this result by quantitation of the complex fluorescent peak area. As expected, no fluorescent labeling was observed on gp120, which was not cross-linked to M64U1-SH. These data also demonstrate the 1:1 covalent coupling ratio between gp120 and the miniCD4, as the chromatograms obtained for 1 and 3 eq of M64U1-SH are almost superimposable concerning gp120.
SCHEME 1.
Modification of the released cysteine on gp120. To demonstrate that M64U1-SH cross-linked gp120 through reaction on a disulfide bond and to quantify the cross-linking yield, we coupled 5-(iodoacetamido)fluorescein to the gp120-S-S-M64U1 complex. The released cysteine on gp120 was also coupled to N-biotinoyl-N′-iodoacetamidoethylenediamine for the labeling mapping analysis by mass spectrometry.
FIGURE 3.
HPLC profiles of fluorescently labeled cross-linked complexes. gp120 was reacted with various amounts of M64U1-SH (0 eq, green; 0.5 eq, orange; 1 eq, blue; 3 eq, black) for 30 min. The cross-linked complexes were then incubated with a large excess of 5-iodoacetamidofluorescein, for 45 min, and injected on an analytical Vydac C4 reverse phase column (5 μm, 4.6 × 150 mm; flow rate, 1.5 ml·min−1; solvent A, H2O, 0.1% trifluoroacetic acid; solvent B, acetonitrile; gradient, 30–30% solvent B in 10 min, then 30–50% solvent B in 20 min, and 50–70% solvent B in 5 min). Absorbance was measured at 278 nm (solid lines), and the fluorescence profile was obtained for a 500-nm excitation wavelength and a 522-nm emission wavelength (dotted lines). Note that chromatograms were focused on the area of interest.
The cross-linked residues on gp120 were then mapped by MALDI-TOF spectrometry. We labeled the released cysteine, during the cross-linking process, with an uncleavable sulfhydryl-specific biotin derivative (Scheme 1). After biotinylation of gp120, the reduced and carboxymethylated protein was deglycosylated and trypsinized. The biotinylated peptides were then detected by mass spectrometry.
All tryptic fragments of gp120 containing a cysteine residue in its different potential modified forms (CAM or Biot) were searched in the peptide mass fingerprint spectrum of trypsin-digested gp120 samples. The corresponding m/z of these fragments was then selected and isolated as precursor ion for PSD MS/MS sequence characterization. A few tryptic gp120 peptide fragments containing Cys residues were only detected in CAM form (data not shown), indicating that the corresponding cystines were not implied in the covalent association of gp120 to miniCD4. Only two gp120 peptides were detected with a biotinylated cysteine residue, namely a 15-mer peptide containing Cys196 and Cys205 (fragment 193–207) and a 14-mer peptide containing Cys126 and Cys131 (fragment 122–135). Interestingly, both peptides contain one cysteine residue of the Cys126–Cys196 disulfide bond that was expected to be modified.
PSD MS/MS fragmentation was performed on fragments 122–135 at m/z 1939.00 and fragment 193–207 at m/z 1988.96, and they showed that they contained one CAM-Cys and one Biot-Cys. The last peptide also had an asparagine to aspartate deamidation because of deglycosylation with peptide:N-glycosidase F. The obtained daughter ion spectra were then processed, and the peptide amino acid sequence was assigned to localize the Cys residues and identify their chemical modification. MS/MS spectrum of fragment at m/z 1939.00 showed that all daughter ions could be assigned if both sequences of Cys modification were considered (Fig. 4A). This result implies the presence of two peptides derived from the sequence 122–135 and corresponding to this mass, one having the combination CAM-Cys126 and Biot-Cys131 and the other Biot-Cys126 and CAM-Cys131. Therefore, Cys196 and Cys157, which are the disulfide bond partners of Cys126 and Cys131, respectively, are involved in cross-linking to miniCD4.
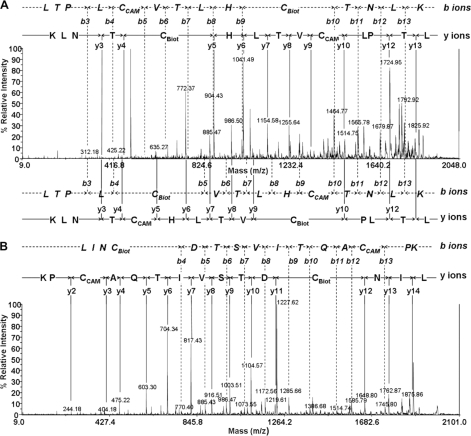
FIGURE 4.
Labeled residues mapping by mass spectrometry. A, identification of CAM- and Biot-Cys-modified fragment 122–135-residue sequence by PSD MS/MS analysis of precursor ion at m/z 1939.00. Daughter ions were assigned by b- and y-ions series. Two sequences were identified as follows: LTPLCBiotVTLHCCAMTNLK (top) and LTPLCCAMVTLHCBiotTNLK (bottom). B, identification of CAM- and Biot-Cys-modified fragment 193–207-residue sequence by PSD MS/MS analysis of precursor ion at m/z 1988.96. Daughter ions were assigned by b- and y-ions series. The unique sequence identified was LINCBiotDTSVITQACCAMPK.
The assignment of the daughter ion spectrum of MS/MS sequencing of fragments at m/z 1988.96 gave one unique b- and y-series ion corresponding to the sequence LINCBiotDTSVITQACCAMPK (Fig. 4B). This result unambiguously concludes that Cys126 had been cross-linked to the miniCD4.
We thus demonstrated that the miniCD4 reacted effectively with the expected disulfide bond but also cross-linked Cys157, which could not be anticipated as it belongs to the V1/V2 loop. This variable loop is in fact absent on all the gp120 structures to facilitate crystallization efforts. Nevertheless, the unexpected data indicate that, to be modified, this disulfide bridge must be located within 15 Å of the carbon α of Lys4 on M64U1. Similar results were obtained on the gp120ΔV2-labeled complex (data not shown).
Pharmacological Characterization of the Disulfide Cross-linked Complexes
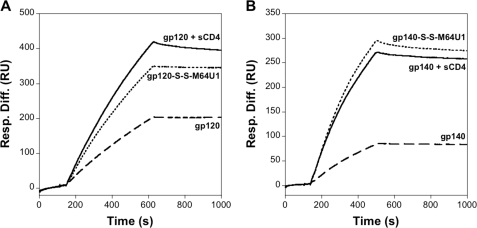
Cross-linked complexes formed between M64U1-SH and envelope proteins were analyzed for their ability to bind 48d CD4i mAb, by surface plasmon resonance (Fig. 5). The gp140-S-S-M64U1 complex presented a binding activity for the CD4i antibody similar to that of the uncross-linked one (gp140 + sCD4) and obviously much higher than gp140 alone (Fig. 5B). In the case of gp120 (Fig. 5A), the affinity for the CD4i antibody 48d is also enhanced for the covalent complex but to a lesser extent than for the ΔV2 version of gp140. This result might be correlated to the presence or not of the V2 loop, as seen later in a binding experiment on the CCR5 receptor.
FIGURE 5.
Binding analysis of the Env-S-S-M64U1 complexes to the 48d CD4i mAb. M64U1-SH or sCD4 (control) was incubated with gp120 or gp140, and CD4-induced epitope induction was evaluated by surface plasmon resonance. Experiments were conducted in HBS (HEPES-buffer saline, 3 mm EDTA, 0.05% Biacore surfactant, pH 7.4). Each sample was analyzed at 10 nm. Sample binding was tested on immobilized CD4i 48d mAb. Dashed lines, gp120 or gp140; dotted lines, M64U1-SH + (gp120 or gp140); solid lines, sCD4 + (gp120 or gp140). A, gp120 data; B, gp140 data.
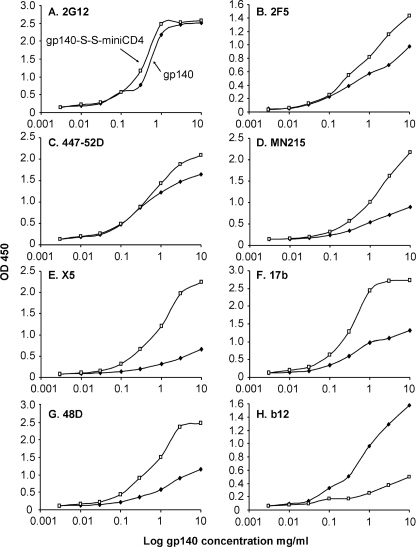
The antigenic integrity of the covalent complex gp140-S-S-M64U1 was analyzed by surface immunoprobing with a panel of mAbs of defined epitope specificities using capture ELISA (Fig. 6). We normalized the concentration of gp140 versus the gp140-S-S-M64U1 complex through ELISAs using 2G12 mAb (Fig. 6A), as this mAb binds carbohydrate moieties independently of the HIV envelope conformation. Then mAbs directed against gp41 (2F5) (Fig. 6B), V3 loop (447–52D, MN215) (Fig. 6, C and D, respectively), CD4i epitopes (X5, 17b and 48d; Fig. 6, E–G, respectively), and the CD4-binding site (b12; Fig. 6H) were tested for binding to gp140 alone and to gp140-S-S-M64U1. Binding of the cross-linked complex on 447–52D or MN215 was increased compared with that of gp140 alone (Fig. 6, C and D). This was somewhat expected as sCD4 enhances neutralization of HIV by V3-directed mAbs (41), notwithstanding a better accessibility of loop V3 to antibodies in the presence of CD4. This result highlights once again the mimicry of our miniCD4 with respect to CD4. A slight increase in binding was also observed for the 2F5 mAb on the complex (Fig. 6B), which might be related to the conformational change of gp120 after binding to the miniCD4, inducing movements in the extracellular part of gp41, which might better expose the 2F5 epitope. Corroborating the surface plasmon resonance analysis, binding of the cross-linked complex on CD4i Abs was strongly increased compared with that of gp140 alone (Fig. 6, E–G). In contrast, binding of the covalent complex to the anti-CD4-binding site antibody b12 is strongly reduced compared with gp140, providing additional support of the covalent coupling of M64U1-SH bound to the HIV envelope CD4-binding site (Fig. 6H).
FIGURE 6.
Immunochemical characterization of the gp140-S-S-M64U1 complex compared with gp140. Serial dilutions of gp140 (filled square) and gp140-S-S-M64U1 (open square) were preincubated on immobilized lectin (concanavalin A) and incubated with several mAbs as follows: 2G12 (A), 2F5 (B), 447–52D (C), MN215 (D), X5 (E), 17b (F), 48d (G), and b12 (H). Anti-gp120 mAbs binding was finally detected with peroxidase-labeled secondary antibodies.
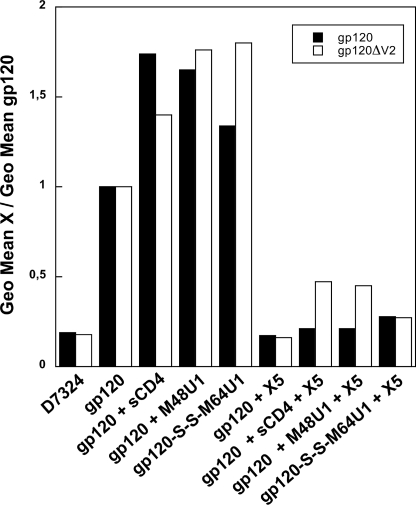
Finally, to better characterize the exposure of the co-receptor-binding site, we analyzed the binding ability of the complexes gp120-S-S-M64U1 and gp120ΔV2-S-S-M64U1, as gp140ΔV2-S-S-M64U1 surrogate, toward CCR5 chemokine receptor (Fig. 7). All the complexes, formed between gp120 and miniCD4 or CD4, were incubated at 3 nm with cells overexpressing the CCR5 receptor. We visualized binding of gp120 to CCR5 by flow cytometry (FACS), using mAb D7324, a gp120 C-terminal specific antibody. This antibody does not recognize gp140, the reason why we used gp120ΔV2-S-S-M64U1, as gp140ΔV2-S-S-M64U1 surrogate. mAb D7324 could be detected by a secondary phycoerythrin-labeled antibody. Consistent with the CD4i mAb binding, the association of gp120 and CCR5 is increased in the presence of sCD4 or miniCD4 (Fig. 7, 3rd to 5th lanes). gp120ΔV2 cross-linked complex clearly binds to CCR5 in an identical manner to noncovalent complexes formed with either a miniCD4 without an additional sulfhydryl moiety or sCD4 (Fig. 7, 5th lane compared with the 4th and 3rd lanes). This characteristic is slightly diminished in the case of full-length gp120. Actually, binding of the covalent complex to CCR5 cells was more efficient than that of gp120 but less efficient than that of noncovalently liganded gp120. To verify that the expected co-receptor-binding domain on gp120 mediated the binding to CCR5, the different complexes were tested for cell surface binding after treatment with 3 eq of X5, a CD4i antibody. We found that this mAb strongly inhibited both the binding of the covalent (Fig. 7, 9th lane) and noncovalent complexes (7th and 8th lanes) to CCR5.
FIGURE 7.
gp120-S-S-M64U1 binding to CCR5 chemokine receptor determined by flow cytometry. gp120 (2nd lane) and gp120 preincubated with either sCD4 (3rd and 7th lanes), M48U1 (4th and 8th lanes), or M64U1-SH (5th and 9th lanes) were incubated at 3 nm on cells overexpressing the CCR5 receptor. Each complex was also tested for cell surface binding after treatment with 3 eq of X5 CD4i antibody (6th to 9th lanes). gp120 binding to CCR5 was detected by using D7324 anti-gp120 mAb and a secondary antibody labeled with a phycoerythrin moiety. Background was evaluated by binding of D7324 on CCR5 in absence of gp120 (1st lane). Data are shown in full bars for gp120 full-length and in open bars for gp120ΔV2 (as a substitute of gp140ΔV2).
Taken together, these data demonstrate that our sulfhydryl-modified miniCD4 allows an efficient cross-linking of gp120 or gp140, and most importantly stabilizes the HIV surface protein in a conformation exposing both CD4i epitopes and the co-receptor-binding site. This last property seems to be the most evident on gp140 or gp120ΔV2, which are both V2 loop-deleted versions.
Rabbit Immunization
Following the encouraging in vitro results, we immunized rabbits with gp120-S-S-M64U1 or gp140-S-S-M64U1 complexes. Sera collected following the second, third, and fourth immunizations were analyzed for their capacity to neutralize the homologous SF162 virus. Except for sera obtained from rabbits immunized with the miniprotein alone (control), all other sera obtained at 2wp2 (2 weeks post-second immunization), 2wp3, and 2wp4 displayed neutralizing activity against the SF162 virus at the maximum concentration tested (dilution of 1:25) (data not shown). Interestingly, groups immunized with the complex elicited higher SF162 neutralizing titers than groups immunized with Env protein alone; however, although the neutralization titers induced with the CD4 miniprotein-Env complexes were higher, the means were not significantly different from those generated by Env alone (p > 0.05) (data not shown).
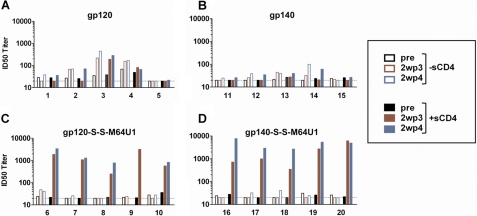
Importantly, covalent miniCD4-Env complexes elicited high levels of CD4i antibodies that neutralize HIV-2 in the presence of sCD4. Sera were analyzed for CD4i antibodies by testing their capacity to neutralize HIV-2 in the presence and absence of soluble CD4 (sCD4), as described previously (42). Individual rabbit sera were tested using the preimmunized 2wp3 and 2wp4 time points, and the ID50 neutralizing titers were determined (Fig. 8). Covalent complexes (gp120-S-S-M64U1 and gp140-S-S-M64U1) elicited high neutralizing titers for all immunized rabbits against the HIV-27312A/V434M pseudovirus in the presence of sCD4 and significantly higher CD4i neutralizing titers than the corresponding Env proteins alone (p = 0.0159) (Fig. 8). Interestingly, sera obtained with the gp140-S-S-M64U1 complex, a ΔV2 version, elicit on average almost three times more CD4i antibodies at 2wp4 than those elicited with gp120-S-S-M64U1, a full-length version. Although some neutralization was observed from a few of the sera from Env-only immunized rabbits, the titers obtained in the presence of sCD4 did not increase relative to those observed in the absence of sCD4. As expected, immunization with miniCD4 alone did not elicit CD4i neutralizing activity (supplemental Fig. S4). Stressing the importance of the covalent complexes, in subsequent studies, we included a group of rabbits immunized with a noncovalent mixture of gp120 and miniCD4, which was not effective in inducing antibodies against the CD4i epitopes.4
FIGURE 8.
CD4i antibody titer. Rabbits were immunized with gp120, gp140, or with M64U1-SH cross-linked to these two envelopes. Four protein immunizations were administered intramuscularly, in the gluteus, at weeks 0, 4, 12, and 24. Serum samples were collected before the first immunization (pre) and 2 weeks post third (2wp3) and fourth (2wp4) immunizations and titered for their CD4i antibodies. The 50% inhibition dose (ID50) for each serum corresponds to the CD4i antibody titer that neutralizes 50% of HIV-27312A/V434M in absence (open bars) or in presence (full bars) of sCD4. Note that rabbit number 9 expired 5 days before the fourth boost injection.
Neutralization of Subtype B Viruses
In addition to measuring neutralization activity against the homologous vaccine strain (SF162), we also tested for neutralization against a set of subtype B viruses using the 2wp4 sera. Results of a screen against several Tier 1 strains are shown in Table 1 and demonstrate that both gp120-S-S-M64U1 and gp140-S-S-M64U1 complexes were able to elicit virus neutralizing antibodies against some of these strains at high frequency. A follow-up study was performed in rabbits showing that both high frequency and high titer virus neutralization of Tier 1, as well as elevated titers against Tier 2 strains, can be achieved using the covalent miniCD4-Env complexes4 and that the response was predominantly directed to the CD4i epitope.
TABLE 1.
Percent of sera that neutralize subtype B viruses
Values indicate the percentage of rabbit sera (2wp4) from each group (five animals/group) that were able to neutralize the indicated pseudoviruses by ≥50% at a dilution of 1:25 in the TZM-bl assay.
| Immunization group | SF162 | IIIB | 89.6 | SS1196 | JRFL |
|---|---|---|---|---|---|
| gp120 | 100 | 100 | 20 | 20 | 0 |
| gp140 | 100 | 100 | 0 | 0 | 0 |
| gp120-S-S-M64U1 | 100 | 75 | 75 | 50 | 0 |
| gp140-S-S-M64U1 | 100 | 100 | 80 | 60 | 0 |
DISCUSSION
HIV-1 attachment and fusion require gp120 binding to both CD4 and chemokine co-receptors (15–18). gp120 binding to CD4 is known to stabilize the virus envelope in a conformation that presents an increased affinity for the chemokine receptors and CD4i antibodies (10, 11, 19–22). Therefore, isothermal titration calorimetry measurements indicated that, in complex with CD4, gp120 binds CD4i antibodies with little entropic change, at a typical level of entropy value for protein-antibody binding (11). In contrast, the prototypic CD4i antibodies faced an important entropic barrier to bind gp120 in its “unbound conformation” (11). These unusual negative entropies are indicative of large surface burial or protein folding and might influence efficiency with which CD4i antibodies are generated as well as the neutralization efficacy of these antibodies (19–22, 43). In addition, it has been shown that steric restrictions could prevent CD4i antibody binding to gp120 after HIV-1 attachment to CD4 receptor on the surface of targeted cells (44).
In contrast, several studies have emphasized the potential utility of CD4i Abs in vaccine development. Indeed, it has been shown that the chemokine co-receptor-binding site of HIV-1 from clades A–F and H and the circulating recombinant forms CRF01, CRF02, and CRF11 elicit high titers of CD4i antibody during natural human infection and that these antibodies bind and neutralize viruses as divergent as HIV-2 in the presence of soluble CD4 (sCD4) (42). The induction of CD4i Abs highlights the extraordinary degree of antigenic conservation linked to co-receptor binding exhibited by diverse HIV-1 and HIV-2 lineages and, at the same time, an ability of the human humoral immune system to exploit these constraints (42). A recent article argued that even if natural viral suppressors could show a low level of circulating Ab response against CD4i epitopes, CD4i-specific B lymphocyte memory cells were present at high frequencies in all natural viral suppressor subjects tested (45). Li et al. (46) also showed the contribution of uncharacterized CD4i mAbs to neutralization in broadly reactive HIV-1 patient sera. The induction of CD4i Abs in patients infected by HIV-1 indicates the exposure of the co-receptor-binding site on the virus surface, which may occur subsequently to the binding of gp120 to CD4 on the target cell despite the steric hindrance reported by Labrijn et al. (44) or because of CD4-independent variants exposing the co-receptor-binding site in most of the infected patients (47, 48). It may also be related to the presence of circulating soluble CD4 or of CD4-gp120 complexes at the surface of targeted cells, which could be due to the shedding of gp120 from gp41 and consequently from the virus particle following HIV binding to CD4. Taken together, these studies suggest that CD4i Abs play a role during the course of the HIV infection, in particular they may constrain the virus to CD4 dependence, with the binding to chemokine co-receptor being essential for virus entry (17, 18).
Although a post-infection induction of such antibodies is known to be without effect against the progression of the infection and may only select virus variants, a vaccine development based on the induction of CD4i Abs may however be interesting. Several studies, based on gp120-CD4 covalent complexes as vaccine candidates, aimed at inducing CD4i Abs have been undertaken (26, 27, 29, 31). It has been shown that gp120 cross-linked to CD4 D1D2 domains raised antibodies that neutralized primary viruses regardless of co-receptor usage (X4, R5 and dual-tropic X4/R5 isolates) and genetic subtype (subtypes A–C and E) in nonhuman primates (27). Recently, the benefit of CD4i antibodies has been reported in challenged nonhuman primates (26). Indeed, it has been shown that antibodies directed against CD4i epitopes were induced by a gp120BaL-rhesus macaque CD4D1D2 single chain. In a challenge with SHIVSF162P3 on rhesus macaques immunized with this fusion protein, this CD4i Ab response was correlated with the control of infection. This control did not seem to be due to anti-CD4 antibodies as no sera coming from macaques immunized with the gp120-CD4 fusion protein reacted with soluble CD4 in an enzyme-linked immunosorbent assay (26). Of course, such a correlation is not proof of the neutralizing efficiency of CD4i antibodies but shows that the presence of these mAbs is fully related to the CD4-bound state of gp120 in vivo. This conformation may present epitopes for neutralizing antibodies, which are still unknown to date. Actually, new families of neutralizing mAbs are still discovered in HIV-1-infected donors (49). Taken together, these studies indicate the importance of a vaccine strategy using a surface protein in its CD4-bound state.
Unfortunately, the development of an AIDS vaccine might be impeded by the use of CD4 with respect to autoimmune response. Therefore, we assessed the usefulness of replacing the CD4 by a CD4 mimetic. Although the miniCD4 presents high structural homology to CD4, the primary structures of these proteins are totally different. Moreover, the reduced size of a miniCD4 induces a low humoral response, which may be beneficial in a vaccine context. As the benefit of a covalent complex between gp120 and CD4 over a noncovalent one has been demonstrated previously (25, 27), we tested several standard cross-linking methods between miniCD4s and gp120. Only a site-directed affinity labeling of the envelope protein using a miniCD4 harboring a sulfhydryl moiety (M64U1-SH) allowed us to conciliate envelope modification and conformational change. An intermolecular disulfide bond could be formed through disulfide exchange with two gp120 cystines. We showed that Cys126, Cys196, and Cys157 could be labeled by M64U1-SH harboring a well positioned sulfhydryl moiety. These cysteine residues belong to cystine(126–196), which was expected to be modified, and cystine(131–157), which is located in the V1/V2 loop and could not be anticipated as this part of the protein is absent from all structures published to date. This approach led to specific cross-linking with an almost quantitative coupling yield. Above all, the cross-linked complex appeared strongly reactive toward both CD4i mAbs and the CCR5 chemokine receptor. A slight difference could be observed however between full-length gp120 and the envelope proteins gp120ΔV2 and gp140ΔV2, deleted of their V2 loop, in binding experiments of CD4i mAbs and the CCR5 receptor. This result might imply a specific role of V2 in the affinity-labeled virus protein, for which the fragment (131–157)-reduced disulfide is located close to this loop. In the absence of this loop (ΔV2), the covalently labeled envelope protein shares identical conformational properties to the noncovalently labeled one, exposing the overlapping CD4i and CCR5 receptor-binding sites. In the case of full-length gp120, the accessibility of these sites is enhanced too but not to the same extent. However, the precise reason of this phenomenon is unknown. Nevertheless, we provided evidence that this method may be applied to HIV envelope trimer as well as monomer.
Previous studies have shown that the absence of a disulfide bond between either residues 126 and 196 or residues 131 and 157 impaired neither gp160 folding nor cleavage between gp120 and gp41 nor CD4 binding to gp120 (50, 51). However, these mutants could not productively infect target cells and had difficulties in forming syncytia. The problem seems to depend on the assembly/budding of the virion (51). Here, we show that not only an envelope protein, lacking one of these two disulfide bonds, still binds a CD4 mimetic but also that the surface protein is stabilized in a conformation that exposes the co-receptor-binding site.
Immunization of rabbits with the gp120-S-S-M64U1 or the gp140-S-S-M64U1 cross-linked complexes and analysis of their sera showed the presence of high titers of CD4i Abs for all animals. Therefore, we demonstrated in this study that like the covalent gp120-CD4 complex or the gp120-CD4 fusion protein, our gp120-S-S-M64U1 or gp140-S-S-M64U1 covalent complexes elicit CD4i antibodies in immunized rabbits. This result also fully agrees with the fact that mutagenic conformational fixation of gp120 core proteins, in a similar manner to that achieved by CD4 binding, is associated with strikingly enhanced humoral immune responses against the co-receptor-binding site by epitope stabilization (52).
During the writing of this paper, a study on gp120-CD4 complex stabilization through targeted interchain disulfide exchange was published. Cerutti et al. (53) showed that a recombinant two-domain CD4 variant containing an S60C mutation could be covalently linked to gp120, probably through the Cys126–Cys196 disulfide bond at the stem of the V1/V2 variable loop, as this cystine is well positioned to react with Cys60. The authors demonstrated that this cross-linking increased the efficacy of CD4-mediated viral entry inhibition. Preliminary results showed that this does not seem to be the case for the miniCD4-SH (data not shown). However, it must be emphasized that M64U1-SH oxidizes relatively quickly in physiological medium, which could explain this result. Nevertheless, our goal in this study was not to find a new entry inhibitor of the virus, as our miniCD4s are already high affinity entry inhibitors (54), but to obtain a new potential vaccine candidate.
The encouraging results on virus neutralization described herein prompt us to test the gp140-S-S-M64U1 covalent complex as a preventive vaccine. We are therefore starting a pre-clinical vaccination study in nonhuman primates using this complex to determine whether antibodies elicited by the miniCD4-induced conformation of the envelope can protect against simian HIV challenge.
Supplementary Material
Acknowledgments
We acknowledge Dr. M. Parmentier for the generous gift of stable CHO-CCR5 cells and Drs. D. P. Burton, P. Parren, D. Katinger, A. Osterhaus, M. Schutten, and the National Institute for Biological Standards and Control Centre for AIDS Reagents (supported by the European Union Programme EVA (Contract QLKZ-CT-1999-00609) and the United Kingdom Medical Research Council. HIV-1 V3 mAb (obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health) was provided by Dr. Susan Zolla-Pazner.
This work was supported, in whole or in part, by National Institutes of Health HIV Research and Development Grant 5P01AI066287 from NIAID, Department of Health and Human Services (supervised by Novartis Vaccines and Diagnostics, Inc.). This work was also supported by the French National Agency for Research on AIDS and Viral Hepatitis Grant 447-52D.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
A. K. Dey, B. Burke, Y. Sun, K. Sirokman, K. Hartog, Y. Lian, A. R. Geonnotti, D. Montefiori, M. Franti, G. Martin, P. Kessler, A. Carfi, L. Martin, I. K. Srivastava, and S. Barnett, manuscript in preparation.
- Env
- envelope
- CAM
- S-carboxamidomethylation
- Biot
- biotinylated
- Ab
- antibody
- sCD4
- soluble CD4
- CD4i
- CD4-induced
- PSD
- post-source decay.
REFERENCES
- 1. Cohen J. (2003) Science 302, 1309–1310 [DOI] [PubMed] [Google Scholar]
- 2. Burton D. R., Desrosiers R. C., Doms R. W., Koff W. C., Kwong P. D., Moore J. P., Nabel G. J., Sodroski J., Wilson I. A., Wyatt R. T. (2004) Nat. Immunol. 5, 233–236 [DOI] [PubMed] [Google Scholar]
- 3. Gallo R. C. (2005) Lancet 366, 1894–1898 [DOI] [PubMed] [Google Scholar]
- 4. Girard M. P., Osmanov S. K., Kieny M. P. (2006) Vaccine 24, 4062–4081 [DOI] [PubMed] [Google Scholar]
- 5. Earl P. L., Sugiura W., Montefiori D. C., Broder C. C., Lee S. A., Wild C., Lifson J., Moss B. (2001) J. Virol. 75, 645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Srivastava I. K., Stamatatos L., Legg H., Kan E., Fong A., Coates S. R., Leung L., Wininger M., Donnelly J. J., Ulmer J. B., Barnett S. W. (2002) J. Virol. 76, 2835–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Srivastava I. K., Stamatatos L., Kan E., Vajdy M., Lian Y., Hilt S., Martin L., Vita C., Zhu P., Roux K. H., Vojtech L. C., Montefiori D., Donnelly J., Ulmer J. B., Barnett S. W. (2003) J. Virol. 77, 11244–11259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barnett S. W., Lu S., Srivastava I., Cherpelis S., Gettie A., Blanchard J., Wang S., Mboudjeka I., Leung L., Lian Y., Fong A., Buckner C., Ly A., Hilt S., Ulmer J., Wild C. T., Mascola J. R., Stamatatos L. (2001) J. Virol. 75, 5526–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pinter A., Honnen W. J., He Y., Gorny M. K., Zolla-Pazner S., Kayman S. C. (2004) J. Virol. 78, 5205–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwong P. D., Wyatt R., Robinson J., Sweet R. W., Sodroski J., Hendrickson W. A. (1998) Nature 393, 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kwong P. D., Doyle M. L., Casper D. J., Cicala C., Leavitt S. A., Majeed S., Steenbeke T. D., Venturi M., Chaiken I., Fung M., Katinger H., Parren P. W., Robinson J., Van Ryk D., Wang L., Burton D. R., Freire E., Wyatt R., Sodroski J., Hendrickson W. A., Arthos J. (2002) Nature 420, 678–682 [DOI] [PubMed] [Google Scholar]
- 12. Xu R., Srivastava I. K., Kuller L., Zarkikh I., Kraft Z., Fagrouch Z., Letvin N. L., Heeney J. L., Barnett S. W., Stamatatos L. (2006) Virology 349, 276–289 [DOI] [PubMed] [Google Scholar]
- 13. Mascola J. R., Snyder S. W., Weislow O. S., Belay S. M., Belshe R. B., Schwartz D. H., Clements M. L., Dolin R., Graham B. S., Gorse G. J., Keefer M. C., McElrath M. J., Walker M. C., Wagner K. F., McNeil J. G., McCutchan F. E., Burke D. S. (1996) J. Infect. Dis. 173, 340–348 [DOI] [PubMed] [Google Scholar]
- 14. Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., Premsri N., Namwat C., de Souza M., Adams E., Benenson M., Gurunathan S., Tartaglia J., McNeil J. G., Francis D. P., Stablein D., Birx D. L., Chunsuttiwat S., Khamboonruang C., Thongcharoen P., Robb M. L., Michael N. L., Kunasol P., Kim J. H. (2009) N. Engl. J. Med. 361, 2209–2220 [DOI] [PubMed] [Google Scholar]
- 15. Dalgleish A. G., Beverley P. C., Clapham P. R., Crawford D. H., Greaves M. F., Weiss R. A. (1984) Nature 312, 763–767 [DOI] [PubMed] [Google Scholar]
- 16. Klatzmann D., Champagne E., Chamaret S., Gruest J., Guetard D., Hercend T., Gluckman J. C., Montagnier L. (1984) Nature 312, 767–768 [DOI] [PubMed] [Google Scholar]
- 17. Moore J. P. (1997) Science 276, 51–52 [DOI] [PubMed] [Google Scholar]
- 18. Rizzuto C. D., Wyatt R., Hernández-Ramos N., Sun Y., Kwong P. D., Hendrickson W. A., Sodroski J. (1998) Science 280, 1949–1953 [DOI] [PubMed] [Google Scholar]
- 19. Zhang W., Canziani G., Plugariu C., Wyatt R., Sodroski J., Sweet R., Kwong P., Hendrickson W., Chaiken I. (1999) Biochemistry 38, 9405–9416 [DOI] [PubMed] [Google Scholar]
- 20. Xiang S. H., Doka N., Choudhary R. K., Sodroski J., Robinson J. E. (2002) AIDS Res. Hum. Retroviruses 18, 1207–1217 [DOI] [PubMed] [Google Scholar]
- 21. Xiang S. H., Wang L., Abreu M., Huang C. C., Kwong P. D., Rosenberg E., Robinson J. E., Sodroski J. (2003) Virology. 315, 124–134 [DOI] [PubMed] [Google Scholar]
- 22. Zhang M. Y., Shu Y., Sidorov I., Dimitrov D. S. (2004) Antiviral Res. 61, 161–164 [DOI] [PubMed] [Google Scholar]
- 23. Zhang M. Y., Shu Y., Rudolph D., Prabakaran P., Labrijn A. F., Zwick M. B., Lal R. B., Dimitrov D. S. (2004) J. Mol. Biol. 335, 209–219 [DOI] [PubMed] [Google Scholar]
- 24. Celada F., Cambiaggi C., Maccari J., Burastero S., Gregory T., Patzer E., Porter J., McDanal C., Matthews T. (1990) J. Exp. Med. 172, 1143–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Devico A., Silver A., Thronton A. M., Sarngadharan M. G., Pal R. (1996) Virology 218, 258–263 [DOI] [PubMed] [Google Scholar]
- 26. DeVico A., Fouts T., Lewis G. K., Gallo R. C., Godfrey K., Charurat M., Harris I., Galmin L., Pal R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17477–17482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fouts T., Godfrey K., Bobb K., Montefiori D., Hanson C. V., Kalyanaraman V. S., DeVico A., Pal R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 11842–11847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kang C. Y., Hariharan K., Nara P. L., Sodroski J., Moore J. P. (1994) J. Virol. 68, 5854–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Varadarajan R., Sharma D., Chakraborty K., Patel M., Citron M., Sinha P., Yadav R., Rashid U., Kennedy S., Eckert D., Geleziunas R., Bramhill D., Schleif W., Liang X., Shiver J. (2005) J. Virol. 79, 1713–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fouts T. R., Tuskan R., Godfrey K., Reitz M., Hone D., Lewis G. K., DeVico A. L. (2000) J. Virol. 74, 11427–11436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He Y., D'Agostino P., Pinter A. (2003) Vaccine 21, 4421–4429 [DOI] [PubMed] [Google Scholar]
- 32. Vita C., Drakopoulou E., Vizzavona J., Rochette S., Martin L., Ménez A., Roumestand C., Yang Y. S., Ylisastigui L., Benjouad A., Gluckman J. C. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13091–13096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martin L., Stricher F., Missé D., Sironi F., Pugnière M., Barthe P., Prado-Gotor R., Freulon I., Magne X., Roumestand C., Ménez A., Lusso P., Veas F., Vita C. (2003) Nat. Biotechnol. 21, 71–76 [DOI] [PubMed] [Google Scholar]
- 34. Stricher F., Martin L., Barthe P., Pogenberg V., Mechulam A., Menez A., Roumestand C., Veas F., Royer C., Vita C. (2005) Biochem. J. 390, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Samson M., Labbe O., Mollereau C., Vassart G., Parmentier M. (1996) Biochemistry 35, 3362–3367 [DOI] [PubMed] [Google Scholar]
- 36. Montefiori D. C. (2004) Current Protocols in Immunology (Coligan J. E., Kruisbeek A. M., Margulies D. H., Shevach E. M., Strober W., Coico R. eds) pp. 12.11.11–12.11.15, John Wiley & Sons, Inc., New York [Google Scholar]
- 37. Li M., Gao F., Mascola J. R., Stamatatos L., Polonis V. R., Koutsoukos M., Voss G., Goepfert P., Gilbert P., Greene K. M., Bilska M., Kothe D. L., Salazar-Gonzalez J. F., Wei X., Decker J. M., Hahn B. H., Montefiori D. C. (2005) J. Virol. 79, 10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang C. C., Stricher F., Martin L., Decker J. M., Majeed S., Barthe P., Hendrickson W. A., Robinson J., Roumestand C., Sodroski J., Wyatt R., Shaw G. M., Vita C., Kwong P. D. (2005) Structure 13, 755–768 [DOI] [PubMed] [Google Scholar]
- 39. Center R. J., Earl P. L., Lebowitz J., Schuck P., Moss B. (2000) J. Virol. 74, 4448–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Finzi A., Pacheco B., Zeng X., Kwon Y. D., Kwong P. D., Sodroski J. (2010) J. Virol. Methods 168, 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu X., Sambor A., Nason M. C., Yang Z. Y., Wu L., Zolla-Pazner S., Nabel G. J., Mascola J. R. (2008) Virology 380, 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Decker J. M., Bibollet-Ruche F., Wei X., Wang S., Levy D. N., Wang W., Delaporte E., Peeters M., Derdeyn C. A., Allen S., Hunter E., Saag M. S., Hoxie J. A., Hahn B. H., Kwong P. D., Robinson J. E., Shaw G. M. (2005) J. Exp. Med. 201, 1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Binley J. M., Wrin T., Korber B., Zwick M. B., Wang M., Chappey C., Stiegler G., Kunert R., Zolla-Pazner S., Katinger H., Petropoulos C. J., Burton D. R. (2004) J. Virol. 78, 13232–13252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Labrijn A. F., Poignard P., Raja A., Zwick M. B., Delgado K., Franti M., Binley J., Vivona V., Grundner C., Huang C. C., Venturi M., Petropoulos C. J., Wrin T., Dimitrov D. S., Robinson J., Kwong P. D., Wyatt R. T., Sodroski J., Burton D. R. (2003) J. Virol. 77, 10557–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guan Y., Sajadi M. M., Kamin-Lewis R., Fouts T. R., Dimitrov A., Zhang Z., Redfield R. R., DeVico A. L., Gallo R. C., Lewis G. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3952–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y., Svehla K., Louder M. K., Wycuff D., Phogat S., Tang M., Migueles S. A., Wu X., Phogat A., Shaw G. M., Connors M., Hoxie J., Mascola J. R., Wyatt R. (2009) J. Virol. 83, 1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wei X., Decker J. M., Wang S., Hui H., Kappes J. C., Wu X., Salazar-Gonzalez J. F., Salazar M. G., Kilby J. M., Saag M. S., Komarova N. L., Nowak M. A., Hahn B. H., Kwong P. D., Shaw G. M. (2003) Nature 422, 307–312 [DOI] [PubMed] [Google Scholar]
- 48. Zhang P. F., Bouma P., Park E. J., Margolick J. B., Robinson J. E., Zolla-Pazner S., Flora M. N., Quinnan G. V., Jr. (2002) J. Virol. 76, 644–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walker L. M., Phogat S. K., Chan-Hui P. Y., Wagner D., Phung P., Goss J. L., Wrin T., Simek M. D., Fling S., Mitcham J. L., Lehrman J. K., Priddy F. H., Olsen O. A., Frey S. M., Hammond P. W., Kaminsky S., Zamb T., Moyle M., Koff W. C., Poignard P., Burton D. R. (2009) Science 326, 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tschachler E., Buchow H., Gallo R. C., Reitz M. S., Jr. (1990) J. Virol. 64, 2250–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Anken E., Sanders R. W., Liscaljet I. M., Land A., Bontjer I., Tillemans S., Nabatov A. A., Paxton W. A., Berkhout B., Braakman I. (2008) Mol. Biol. Cell 19, 4298–4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dey B., Svehla K., Xu L., Wycuff D., Zhou T., Voss G., Phogat A., Chakrabarti B. K., Li Y., Shaw G., Kwong P. D., Nabel G. J., Mascola J. R., Wyatt R. T. (2009) PLoS Pathog. 5, e1000445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cerutti N., Mendelow B. V., Napier G. B., Papathanasopoulos M. A., Killick M., Khati M., Stevens W., Capovilla A. (2010) J. Biol. Chem. 285, 25743–25752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Van Herrewege Y., Morellato L., Descours A., Aerts L., Michiels J., Heyndrickx L., Martin L., Vanham G. (2008) J. Antimicrob. Chemother. 61, 818–826 [DOI] [PubMed] [Google Scholar]
- 55. Stricher F., Huang C. C., Descours A., Duquesnoy S., Combes O., Decker J. M., Kwon Y. D., Lusso P., Shaw G. M., Vita C., Kwong P. D., Martin L. (2008) J. Mol. Biol. 382, 510–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.