Abstract
Prior studies have shown that treatment with the peracetylated 4-fluorinated analog of glucosamine (4-F-GlcNAc) elicits anti-skin inflammatory activity by ablating N-acetyllactosamine (LacNAc), sialyl Lewis X (sLeX), and related lectin ligands on effector leukocytes. Based on anti-sLeX antibody and lectin probing experiments on 4-F-GlcNAc-treated leukocytes, it was hypothesized that 4-F-GlcNAc inhibited sLeX formation by incorporating into LacNAc and blocking the addition of galactose or fucose at the carbon 4-position of 4-F-GlcNAc. To test this hypothesis, we determined whether 4-F-GlcNAc is directly incorporated into N- and O-glycans released from 4-F-GlcNAc-treated human sLeX (+) T cells and leukemic KG1a cells. At concentrations that abrogated galectin-1 (Gal-1) ligand and E-selectin ligand expression and related LacNAc and sLeX structures, MALDI-TOF and MALDI-TOF/TOF mass spectrometry analyses showed that 4-F-GlcNAc 1) reduced content and structural diversity of tri- and tetra-antennary N-glycans and of O-glycans, 2) increased biantennary N-glycans, and 3) reduced LacNAc and sLeX on N-glycans and on core 2 O-glycans. Moreover, MALDI-TOF MS did not reveal any m/z ratios relating to the presence of fluorine atoms, indicating that 4-F-GlcNAc did not incorporate into glycans. Further analysis showed that 4-F-GlcNAc treatment had minimal effect on expression of 1200 glycome-related genes and did not alter the activity of LacNAc-synthesizing enzymes. However, 4-F-GlcNAc dramatically reduced intracellular levels of uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc), a key precursor of LacNAc synthesis. These data show that Gal-1 and E-selectin ligand reduction by 4-F-GlcNAc is not caused by direct 4-F-GlcNAc glycan incorporation and consequent chain termination but rather by interference with UDP-GlcNAc synthesis.
Keywords: Adhesion, Carbohydrate, Carbohydrate Structure, Glycoprotein Structure, Glycosylation Inhibitors, Lectin
Introduction
β-galactoside-binding galectins and sialyl Lewis X (sLeX)2-binding selectins (E-, P-, and L-selectin) are important in the development of effector/regulatory T cell subsets and for effector leukocyte recruitment into inflamed sites (1–6). Canonical galectin-binding determinants or N-acetyllactosamine (Galβ1,4GlcNAc or LacNAc) are converted to selectin-binding determinants upon α2,3-sialylation, α1,3-fucosylation, and, in some cases, sulfation. Hence, sharing of the same LacNAc core structure for ligand recognition coupled with their importance in controlling inflammation intensity provides the opportunity to target these moieties and help control inflammatory diseases, such as allergic dermatitis, psoriasis, graft versus host disease, multiple sclerosis, and lupus (1, 4, 5, 7–10).
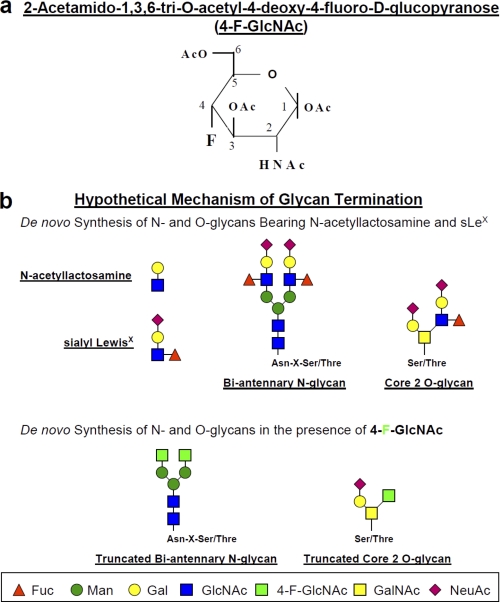
Our laboratory has studied the role of endothelial (E)-selectin ligands in the trafficking of T cells to sites of inflammation (7, 8, 11, 12) and, more recently, investigated the role of galectin-1 (Gal-1) ligands in expansion of immunosuppressive IL-10+ T cells and retraction of proinflammatory T helper (Th)1 and Th17 cell subsets (13). To highlight the importance and identity of E-selectin-binding determinants on effector T cells in inflammation, we routinely utilize the putative inhibitor of LacNAc formation, peracetylated 4-fluorinated glucosamine (4-F-GlcNAc), which is designed to passively enter cells, become deacetylated, and antagonize elongation of N- and O-glycan chains and sLeX formation in metabolically active cells (Fig. 1, a and b) (7, 8, 11, 12). Likewise, in related work, a peracetylated 4-fluorinated galactosamine (4-F-GalNAc) analog has been used to metabolically inhibit sLeX synthesis and related selectin ligand activities (14). Other glyco-metabolic inhibitors, such as disaccharide decoys, can be used by metabolically active cells, such as activated T cells and cancer cells, to divert endogenous glycosylation pathways from synthesizing key Lewis carbohydrate structures important for inflammation and metastasis (15–17).
FIGURE 1.
a, structure of 4-F-GlcNAc; b, putative mechanism of anti-glycosylation action.
In 4-F-GlcNAc efficacy studies, all evidence of glycan modulation has been generated using anti-sialyl Lewis antibodies or lectins, which help discriminate alterations in content and specific glycan structures on leukocyte surface glycoconjugates. Data using anti-sLeX monoclonal antibodies (mAbs) and Datura stramonium agglutinin (specificity: Galβ1,4GlcNAc; LacNAc) and Lycopersicon esculentum agglutinin (specificity: (LacNAc)n) lectins show that sLeX-like structures and LacNAc are markedly reduced following 4-F-GlcNAc treatment (7, 8, 11, 12). These findings support the notion that 4-F-GlcNAc is interfering with glycan synthesis by blocking the addition of galactose or fucose at the carbon 4-position upon incorporation into complex N-glycans and extended core 1 or core 2 O-glycans. Nonetheless, direct evidence on whether 4-F-GlcNAc actively incorporates into a glycan chain and terminates growing N-glycan antennae or core 2 O-glycans has not yet been found.
In this report, we applied matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) and MALDI-TOF/TOF mass spectrometry analyses to analyze the content, repertoire and complexity of N- and O-glycans released from 4-F-GlcNAc-treated human sLeX (+) T cells and leukemic KG1a cells. These leukocyte models contain high glyco-metabolic activities with concomitant elevated expression of sLeX and related selectin ligand activities (7, 11, 18–20) and, thus, are ideal for 4-F-GlcNAc mechanistic analysis. We found that 4-F-GlcNAc treatment on both leukocyte models 1) inhibited the synthesis of Gal-1 and E-selectin ligands and related carbohydrate-recognition determinants on glycoproteins, 2) reduced LacNAc and sLeX levels on N-glycans and on core 2 O-glycans, 3) reduced content and structural diversity of tri- and tetra-antennary N-glycans and of O-glycans while elevating biantennary N-glycan levels, and 4) did not result in higher levels of GlcNAc-terminated glycans. Importantly, MALDI-TOF MS analysis showed that masses (m/z ratios) of glycan structures did not correspond to theoretical structures containing a fluorine atom. Furthermore, expression profiling of glycome-related genes in 4-F-GlcNAc-treated cells revealed that alterations in the expression of nearly all 1200 glycome-related genes were negligible. We also found that LacNAc-dependent glycosyltransferase activities in 4-F-GlcNAc-treated cells were relatively unchanged from those assayed in untreated cells. 4-F-GlcNAc, however, reduced intracellular UDP-GlcNAc, a key precursor of LacNAc synthesis. These data show for the first time that 4-F-GlcNAc can effectively inhibit Gal-1 ligand and E-selectin ligand formation in glyco-metabolically active leukocytes without direct incorporation and truncation of glycan chains and that such inhibition may be due to reduction of UDP-GlcNAc formation.
EXPERIMENTAL PROCEDURES
Cells, Antibodies, Enzymes, and Chemicals
Human sLeX (+) T cells with E-selectin ligand activity were prepared from human peripheral blood mononuclear cells as described previously (7). Human leukemic KG1a cells, which also display high levels of sLeX and E-selectin ligand activity, were purchased from ATCC (Manassas, VA) and maintained as described previously (21).
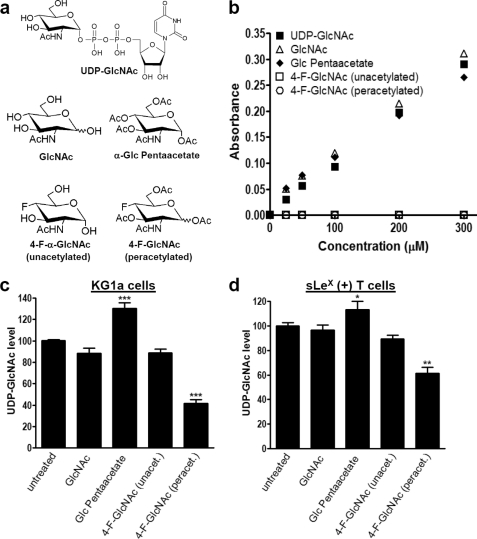
Anti-sLeX mAb (HECA-452) was purchased from BD PharMingen, Inc. (San Diego, CA). PerCP-mouse IgG1, PerCP-mouse IgG anti-human CD4 (clone SK3), PerCP-mouse IgG anti-human CD8 (SK1), allophycocyanin-goat anti-human Fc, mouse anti-PSGL-1 mAb KPL-1, and anti-β-actin mAb AC-15 were purchased from BD Biosciences. Mouse anti-CD44 mAb A3D8 was purchased from Sigma-Aldrich. Goat anti-mouse IgG, alkaline phosphatase (AP)-goat anti-rat IgM, and AP-goat anti-mouse IgG were purchased from Southern Biotechnology Associates, Inc. (Birmingham, AL). Mouse IgG anti-human CD3 and mouse IgG anti-human PSGL-1 monoclonal antibody (PL-2) was purchased from Beckman Coulter, Inc. (Brea, CA). Human IgG and bromelain were from Sigma. Recombinant mouse E-selectin-human IgG Fc1 (E-selectin-hFc) was purchased from R&D Systems (Minneapolis, MN). Mouse galectin-1-human IgG1e3-Fc1 (Gal-1hFc) was prepared in our laboratory as described (13). 2-Acetamido-1,3,6-tri-O-acetyl-4-deoxy-4-fluoro-d-glucopyranose (4-F-GlcNAc), which possesses a fluorine substitution at carbon 4 of the pyranose ring and is a putative inhibitor of LacNAc formation and relevant lectin-binding moieties (Fig. 1, a and b), was provided by the Chemical Resource Laboratory at Roswell Park Cancer Institute (7, 22–25). Unacetylated 4-F-α-GlcNAc 2-acetamido-2,4-dideoxy-4-(fluoro-α-d-glucopyranose) and fully acetylated α-GlcNAc (α-d-glucosamine pentaacetate) were purchased from Toronto Research Chemicals, Inc. (North York, Canada) (see Fig. 8). UDP-GlcNAc and GlcNAc were purchased from Sigma (see Fig. 8).
FIGURE 8.
Measurement of UDP-GlcNAc level in extracts of 4-F-GlcNAc-treated sLeX (+) KG1a and T cells. a, chair configurations of 4-F-GlcNAc (peracetylated) and structurally related control analogs, UDP-GlcNAc, GlcNAc, α-Glc pentaacetate, and 4-F-α-GlcNAc (unacetylated). b, assay specificity for detecting the sugars shown in a. c and d, level of UDP-GlcNAc measured in sLeX (+) KG1a or T cell extracts. KG1a cells were incubated for 48 h with 0.05 mm sugars. T cells were incubated for 48 h with 0.05 mm sugars or for 38 h with 0.01–0.025 mm 4-F-GlcNAc (peracetylated). Shown is mean UDP-GlcNAc level with S.E. indicated when compared with untreated control. Experiments were performed in triplicate on at least three separate occasions and donors. Statistically significant differences compared with untreated controls are shown as follows. *, p < 0.05; **, p < 0.01; ***, p < 0.001, one-way analysis of variance with Dunnett's post hoc test.
Cell Treatments
Logarithmically growing T cells and KG1a cell cultures were treated with 0.01–0.05 mm 4-F-GlcNAc or diluent controls for 36–48 h where indicated and then harvested for analysis in bioassays.
Western Blotting
Cell lysates were prepared, quantified by the Bradford method, and analyzed by SDS-PAGE/Western blotting as described previously (21, 26). Immunoblotting was performed using anti-sLeX mAb HECA-452 (1 μg/ml), Gal-1hFc (10 μg/ml), anti-PSGL-1 mAb KPL-1 (1 μg/ml), anti-β-actin mAb (1 μg/ml), mouse E-selectin-hFc (1 μg/ml), or respective isotype control antibody. Blots were then incubated with respective AP-goat anti-rat IgM, AP-goat anti-human IgG, or AP-rabbit anti-mouse IgG (Zymed Laboratories Inc., Inc., San Francisco, CA) and developed with Western Blue® AP-substrate (Promega, Madison, WI). Lanes pertaining to immunoblotted or lectin-blotted regions were scanned and analyzed by NIH ImageJ (National Institutes of Health, Bethesda, MD). Means ± S.D. of relative gray scale units were normalized to background staining and evaluated for statistical significance by comparing with untreated controls.
Flow Cytometry
Cells were harvested, washed, and suspended at 1 × 106 cells, 100 μl of cold PBS, 1% FBS. Primary mAbs (1 μg/test), Gal-1hFc (10 μg/ml), or isotype antibody control (2 μg/test) were incubated with cells for 60 min at 4 °C. After washing with cold PBS, 1% FBS, cells were incubated with fluorophore-conjugated secondary mAb for 30 min at 4 °C. After secondary antibody incubation, cells were pelleted, washed, and resuspended in 500 μl PBS, 1% FBS. Fluorescence measurements were collected within 1 h on a BD FACSCantoTM apparatus (BD Biosciences). Data were analyzed using FlowJo software (Tree Star, Inc., Ashland, OR). Cells were gated based on forward and side scatter.
Glycome-related Gene Expression Analysis
Cells were lysed in 1 ml of TRIzol® reagent (Invitrogen), RNA was prepared as previously described (27), and gene expression was analyzed on a GlycoV4 oligonucleotide array. The GlycoV4 oligonucleotide array is a custom Affymetrix GeneChip (Affymetrix, Santa Clara, CA) designed for the Consortium for Functional Glycomics. A complete description and annotation for the GlycoV4 array is available at the Functional Glycomics Web site. The GlycoV4 array includes probe sets for ∼1260 human and ∼1200 mouse transcripts encoding glyco-genes. Expression data normalization was performed using RMA Express 1.0 with quantile normalization, median polish, and background adjustment. The Limma package in the R software was used to find transcripts with differential expression. In Limma, S.E. values were estimated by fitting a linear model for each gene expression, and empirical Bayes smoothing was applied. Results were presented between two or more experimental conditions as a -fold change in expression level, the moderated t-statistic, the p value, and the adjusted p value. The adjusted p value is the p value adjusted for multiple testing using Benjamini and Hochberg's method to control the false discovery rate of 0.15 or less. The transcripts identified as differentially expressed were those with adjusted p value of <0.15 and -fold change of >1.4. Heat maps were generated with the dChip program (available on the World Wide Web). Red indicates increased and blue indicates decreased expression relative to the mean transcript expression value.
Glycan Profiling
Snap frozen KG1a and T cell pellets (25 million cells/pellet) prepared as above were treated as described previously (28, 29). Briefly, all samples were subjected to homogenization using a 130-watt Vibra-Cell ultrasonic processor (VC 130 PB, Sonics & Materials) within a sound-abating enclosure in extraction buffer (25 mm Tris, 150 mm NaCl, 5 mm EDTA, and 1% CHAPS at pH 7.4). The samples were then subjected to reduction in 4 m guanidine HCl (Pierce), carboxymethylation, and trypsin digestion, and the digested glycoproteins were purified by C18-Sep-Pak (Waters Corp., Hertfordshire, UK). N-Glycans were released by peptide:N-glycosidase F (EC 3.5.1.52, Roche Applied Science) digestion, whereas O-glycans were released by reductive elimination. N- and O-glycans were then permethylated using the sodium hydroxide procedure, and finally, the permethylated N- and O-glycans were purified by C18-Sep-Pak.
All permethylated samples were dissolved in 10 μl of methanol, and 1 μl of dissolved sample was premixed with 1 μl of matrix (20 mg/ml 2,5-dihydroxybenzoic acid in 70% (v/v) aqueous methanol), spotted onto a target plate (2 × 0.5 μl), and dried under vacuum. MS data were acquired using a Voyager-DE STR MALDI-TOF (Applied Biosystems, Darmstadt, Germany). MS/MS data were acquired using a 4800 MALDI-TOF/TOF (Applied Biosystems) mass spectrometer. The collision energy was set to 1 kV, and argon was used as collision gas. The 4700 calibration standard kit, Calmix (Applied Biosystems), was used as the external calibrant for the MS mode of both instruments, and [Glu1] fibrinopeptide B human (Sigma) was used as an external calibrant for the MS/MS mode of the MALDI-TOF/TOF instrument.
The MS and MS/MS data were processed using Data Explorer 4.9 Software (Applied Biosystems). The spectra were subjected to manual assignment and annotation with the aid of the glycobioinformatics tool, GlycoWorkBench (30). The proposed assignments for the selected peaks were based on 12C isotopic composition together with knowledge of the biosynthetic pathways. The proposed structures were then confirmed by data obtained from MS/MS experiments.
Protease and Glycosidase Digestions
Protease digestion was performed by treating cells for 1 h at 37 °C with 0.1% bromelain (Sigma), a protease with broad peptide specificity that cleaves all cell surface E-selectin glycoprotein ligands (11, 27, 31). Endo-β-galactoside (Escherichia freundii, EC 3.2.1.103, Glyco) digestion was carried out in 200 μl of sodium acetate buffer (37 °C, pH 5.8). 20 milliunits were added to the sample for 48 h with a fresh aliquot of the enzyme added after 24 h. Cleavage of all of the sialic acids (NeuAc) from KG1a and T cells was performed with a sialidase A (Arthrobacter ureafaciens, EC 3.2.1.18, Glyco) digestion. Samples were incubated in 200 μl of 50 mm sodium acetate buffer (37 °C, pH 5.5). 170 milliunits of sialidase were added for 24 h.
Gas Chromatography-Mass Spectrometry (GC-MS) Linkage Analysis
Partially methylated alditol acetates were prepared as described previously (28, 32). Linkage analysis of partially methylated alditol acetates was performed on a PerkinElmer Clarus 500 instrument fitted with a RTX-5 fused silica capillary column (30 meters × 0.32 mm inner diameter; Restek Corp.). All samples were dissolved in 50–250 μl of hexanes (Sigma) and injected manually (1–2 μl) to the injector set at 250 °C. A linear gradient oven temperature was set as follows. Initially, the oven was heated at 65 °C for 1 min and then heated to 290 °C at a rate of 8 °C/min, held at 290 °C for 5 min, and finally heated to 300 °C at a rate of 10 °C/min.
Glycosyltransferase Assays
Carbohydrate acceptors were from our previous study (33). 14C-Labeled UDP-galactose and UDP-N-acetyl-d-glucosamine (GlcNAc) were from PerkinElmer Life Sciences. Cells treated with 4-F-GlcNAc or vehicle control (PBS) as above were lysed in 100 mm Tris maleate buffer containing 2% Triton X-100. Galactosyltransferase (GalT) activity was measured in reaction mixture containing 100 mm HEPES (pH 7.0), 7 mm ATP, 20 mm manganese acetate, 1 mm UDP-Gal, 7.752 μm [14C]UDP-Gal (0.0024 μCi), 5 mm acceptor, and 0.3–1.5 mg/ml cell lysate. β1,3-N-acetylglucosaminyltransferase (GlcNAcT) activity was measured in a reaction mixture containing 47 mm HEPES buffer (pH 7.0), 4.7 mm GlcNAc 1,5-lactone (Toronto Research Chemicals), 9.33 mm manganese acetate, 66.67 mm GlcNAc, 2.316 μm UDP-GlcNAc (0.0025 μCi), 2 mm glycan acceptor, and 1.2–6.4 mg/ml cell lysate. All reactions proceeded for 20 h, at which point unreacted donor was separated from reaction product using silica gel 60 RP-18 reverse phase-thin layer chromatography (TLC) with water as the mobile phase. TLC plates were developed using a Super Resolution (SR, Medium) phosphor screen and the Cyclone Storage Phosphor System (PerkinElmer Life Sciences). Experiments were performed in triplicate, and acquired images were analyzed using NIH ImageJ. The absolute amount of product formed was determined in units of dpm by interpolation using 14C radioactive standards. Product formation was quantified in units of dpm/mg, based on the amount of protein in each reaction mixture. Reaction without acceptor was used as negative control. Percentage conversion quantifies the percentage of donor radioactivity transferred to acceptor, according to the equation, % conversion = dpm of product/(dpm of product + dpm of unreacted donor).
Measurement of UDP-GlcNAc
UDP-GlcNAc was measured spectrophotometrically by a modification of the Morgan and Elson color reaction of GlcNAc with p-dimethylaminobenzaldehyde (Ehrlich's reagent) as described previously (34). Briefly, carbohydrates were extracted by lysing cell pellets (35–50 × 106/tube) in 200 μl of chloroform/water (1:1) followed by centrifugation at 15,000 × g for 20 min at 4 °C. The top (aqueous) phase (100 μl) was added to a clean Eppendorf tube. To hydrolyze UDP-GlcNAc to GlcNAc, 10 μl of HCl (1 n) was added; the sample was heated for 20 min at 80 °C and neutralized with 10 μl KOH (1 n); and the resulting product, GlcNAc, was measured by the Morgan and Elson reaction as follows. 50 μl of 200 mm potassium tetraborate (Sigma) was added, and the sample was heated at 80 °C for 25 min and then cooled on ice for 5 min. 150 μl of Ehrlich's reagent (Sigma) (diluted 1:2 in acetic acid) was added to the sample, mixed, and incubated for 20 min at 37 °C. The sample was centrifuged at 15,000 × g for 20 min, 200 μl were added to a 96-well plate, and the absorbance was measured at 595 nm.
Statistical Analysis
To ascertain statistical significance, Student's t test was used. For comparisons between groups greater than 2, an analysis of variance with Dunnett's post hoc test was performed.
RESULTS
4-F-GlcNAc Reduced Gal-1- and E-selectin-binding Activities and Related Expression of Galβ1,4GlcNAc and sLeX on Glycometabolically Active Leukocytes
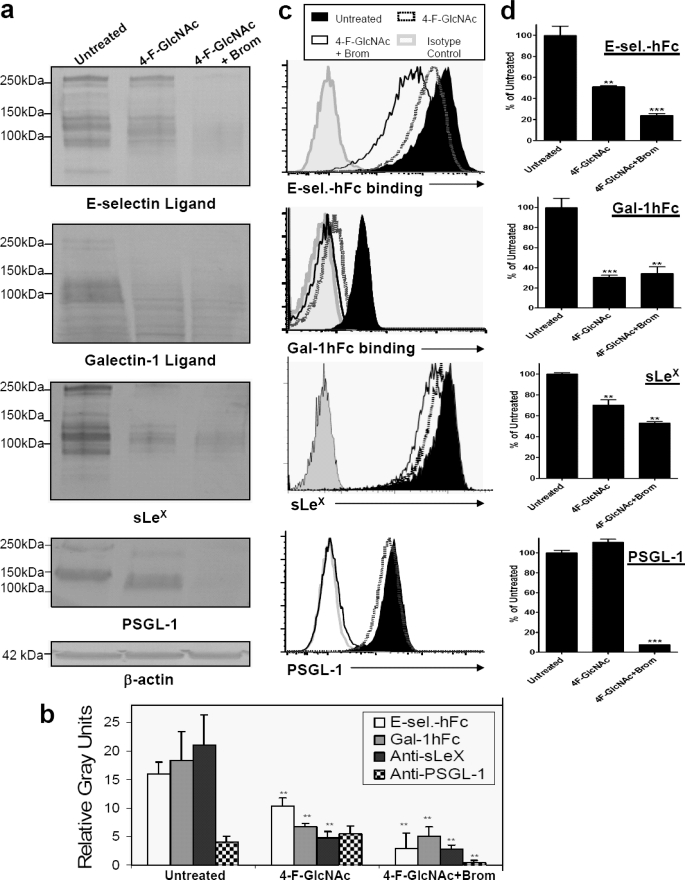
Early studies on 4-F-GlcNAc efficacy demonstrate that Gal-1 ligand expression on ovarian tumor cell surfaces is sensitive to 4-F-GlcNAc treatment (24, 25). More recently, we have shown that 4-F-GlcNAc treatment reduces E-selectin ligand and corresponding sLeX expression on glyco-metabolically active leukemic cells, sLeX (+) T cells, and murine effector Th cell subsets (7, 8, 11, 12). To authenticate the effect of 4-F-GlcNAc on E-selectin ligand/sLeX diminution and explore 4-F-GlcNAc inhibitory efficacy on leukocytic Gal-1 ligand expression, we performed Western blot and flow cytometric analysis with anti-sLeX mAb HECA-452, E-selectin-hFc, and Gal-1hFc using untreated or 0.05 mm 4-F-GlcNAc-treated human leukemic KG1a cells. At 0.05 mm, 4-F-GlcNAc selectively modulates glycosylation without a marked effect on protein synthesis (7, 8, 11, 12). In Western blots of KG1a cell lysates, we found that 4-F-GlcNAc significantly lowered E-selectin and Gal-1 ligands and related sLeX expression, whereas immunostaining intensity of PSGL-1 polypeptide was unchanged, validating 4-F-GlcNAc inhibitory efficacy on glycosylation and not protein synthesis (p < 0.01) (Fig. 2, a and b). In flow cytometry assays, PSGL-1 expression was unchanged, Gal-1 and E-selectin ligand expressions were markedly reduced, and, interestingly, sLeX was reduced by a lesser amount when compared with the level on Western blots (Fig. 2, c and d). To help clarify why disparate effects on sLeX expression were observed in Western blotting and flow cytometry studies, protease digestions with bromelain, which cleaves all E-selectin glycoprotein ligand and sLeX-bearing membrane protein (7, 11, 12, 19, 21, 26, 31, 35), were employed. When 4-F-GlcNAc-treated cells were treated with bromelain, Western blots showed that E-selectin ligand and sLeX expression on glycoproteins was completely removed (Fig. 2, a and b), whereas expression as determined by flow cytometry was not completely reduced (Fig. 2, c and d). As we have found previously, bromelain treatment can actually elevate sLeX expression, due to removal of membrane proteins and steric hindrance, which allows glycolipids heavily decorated with sLeX to bind mAb HECA-452 (11, 12, 21, 31).
FIGURE 2.
Analysis of 4-F-GlcNAc efficacy on E-selectin and Gal-1 ligand expression on sLeX KG1a cells. a, Western blots of KG1a cells treated with diluent control (Untreated), 0.05 mm 4-F-GlcNAc, or 4-F-GlcNAc and bromelain were stained with mouse E-selectin-human IgG Fc1 (E-sel.-hFc) (1 μg/ml), Gal-1hFc (10 μg/ml), anti-sLeX mAb (1 μg/ml), anti-PSGL-1 mAb (1 μg/ml), or anti-β-actin mAb (1 μg/ml). b, using NIH ImageJ, densitometric gray scale units of scanned lanes from 50 to 260 kDa of triplicate Western blots were measured and plotted as mean ± S.D. (error bars) in relative gray units. c, flow cytometry of KG1a cells treated with diluent control (Untreated), 0.05 mm 4-F-GlcNAc, or 4-F-GlcNAc and bromelain were stained with mouse E-selectin-human IgG Fc1 (1 μg/ml), Gal-1hFc (10 μg/ml), anti-sLeX mAb (1 μg/ml), anti-PSGL-1 mAb (1 μg/ml), or respective isotype control and respective fluorophore-conjugated secondary antibody. d, mean fluorescent intensities from triplicate flow cytometry experiments were compared with untreated control and presented as percentage of untreated control. Statistically significant differences when compared with untreated controls are shown as follows. **, p < 0.01; ***, p < 0.001.
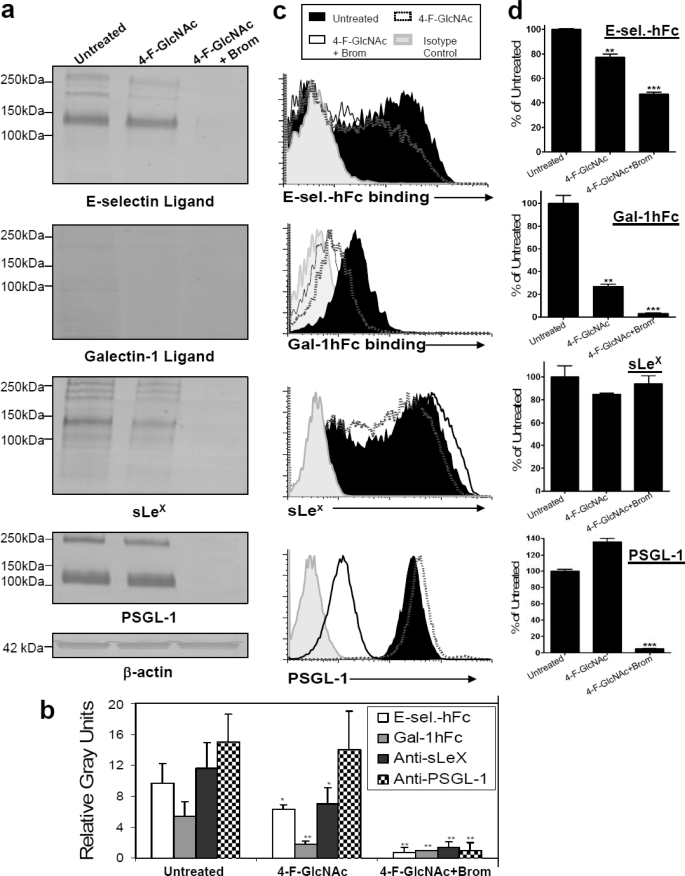
We subsequently performed analysis of 4-F-GlcNAc efficacy on ex vivo human sLeX (+) T cells, a model that bears sLeX and LacNAc moieties. Western blotting experiments indicated that E-selectin ligands and sLeX moieties were significantly reduced following 4-F-GlcNAc treatment compared with untreated controls (p < 0.05) (Fig. 3, a and b). Although Gal-1hFc blotting showed that sLeX (+) T cells expressed negligible Gal-1 ligand, differences in densitometric intensity between untreated and 4-F-GlcNAc-treated cell lysates (p < 0.01) were significant (Fig. 3, a and b). In flow cytometry experiments, although E-selectin-hFc and Gal-1hFc staining was significantly lowered (p < 0.01), sLeX staining was unchanged in 4-F-GlcNAc-treated cells compared with untreated controls (Fig. 3, c and d). When bromelain treatment was coupled with 4-F-GlcNAc treatment, flow cytometric analysis showed that sLeX expression was unchanged and E-selectin ligand was not completely cleaved, whereas Gal-1 ligands and PSGL-1 were absent (Fig. 3, c and d). When these dually treated cells were analyzed by Western blotting, complete removal of lectin/mAb-stained glycoproteins was noted (Fig. 3, a and b). Collectively, data from KG1a and T cell treatment analyses suggest that 4-F-GlcNAc effectively inhibited E-selectin ligand and sLeX formation on glycoproteins and that Gal-1 ligand activity was primarily found on 4-F-GlcNAc-sensitive glycoproteins.
FIGURE 3.
Analysis of 4-F-GlcNAc efficacy on E-selectin and Gal-1 ligand expression on sLeX (+) T cells. a, Western blots of T cells treated with diluent control (Untreated), 0.05 mm 4-F-GlcNAc, or 4-F-GlcNAc and bromelain were stained with mouse E-selectin-human IgG Fc1 (E-sel.-hFc) (1 μg/ml), Gal-1hFc (10 μg/ml), anti-sLeX mAb (1 μg/ml), anti-PSGL-1 mAb (1 μg/ml), or anti-β-actin mAb (1 μg/ml). b, using NIH ImageJ, densitometric gray scale units of scanned lanes from 50 to 260 kDa of triplicate Western blots were measured and plotted as mean ± S.D. (error bars) in relative gray units. c, flow cytometry of T cells treated with diluent control, 0.05 mm 4-F-GlcNAc, or 4-F-GlcNAc and bromelain were stained with mouse E-selectin-human IgG Fc1 (1 μg/ml), Gal-1hFc (10 μg/ml), anti-sLeX mAb (1 μg/ml), anti-PSGL-1 mAb (1 μg/ml), or the respective isotype control and respective fluorophore-conjugated secondary antibody. d, mean fluorescent intensities from triplicate flow cytometry experiments were compared with untreated control and presented as percentage of untreated control. Statistically significant differences when compared with untreated controls are shown as follows. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
4-F-GlcNAc Elicits Modest Effects on Glycome Gene Expression
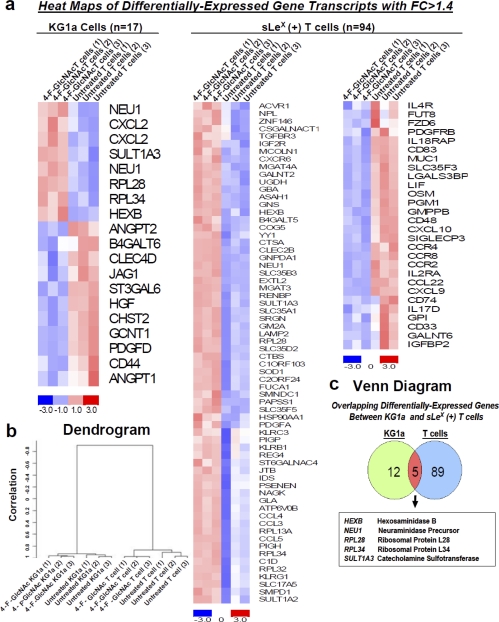
To examine whether 4-F-GlcNAc affected the transcription of glycosyltransferases/glycosidases capable of synthesizing LacNAc and sLeX moieties, we conducted expression analysis with a custom GlycoV4 oligonucleotide array from Affymetrix and analyzed data as described previously (36–39). RNA from untreated and 0.05 mm 4-F-GlcNAc-treated human leukemic KG1a and sLeX (+) T cells was prepared as described (31). Using Limma analysis with an adjusted p value of <0.15 and a -fold change (FC) of >1.4, we found that only 17 and 94 gene transcripts of ∼1200 genes were significantly differentially expressed from untreated versus treated KG1a and T cells, respectively. Heat maps of gene transcripts identified as differentially expressed showed the mean-scaled expression for these comparisons ranging from −3.0 (decreased) to 3.0 (increased) levels (Fig. 4a). A dendrogram of gene expression profiles from triplicate samples of untreated and 4-F-GlcNAc-treated KG1a and T cells showed clustering relative to the group differences, indicating high quality and consistent RNA preparations (Fig. 4b). A Venn diagram depicted the overlap of five gene transcripts that were increased in both untreated versus 4-F-GlcNAc-treated KG1a and T cell comparisons (Fig. 4c). These were 1) hexosaminidase B (HEXB) (KG1a comparison, FC = 1.4; T cell comparison, FC = 1.6); 2) sialidase 1 (NEU1) (from two distinct primer sets) (KG1a comparison, FC = 1.59; T cell comparison, FC = 1.62); 3) ribosomal protein L28 (RPL28) (KG1a comparison, FC = 1.41; T cell comparison, FC = 1.88); 4) ribosomal protein L34 (RPL34) (KG1a comparison, FC = 1.53; T cell comparison, FC = 2.35); and 5) sulfotransferase 3 (SULT1A3) (KG1a comparison, FC = 1.41; T cell comparison, FC = 2.01). Considering that RPL28 and -34 are housekeeping ribosomal protein genes (40); HEXB encodes the β-subunit of hexosaminidase A, which catalyzes the removal of terminal N-acetylhexosamines on GM2 gangliosides (41); NEU1 encodes a lysosomal N-acetylneuraminidase (42); and SULT1A3 encodes a sulfotransferase that catalyzes sulfate conjugation (43), the global effects of 4-F-GlcNAc on glycome gene expression appeared to be minor and did not selectively lower subsets of genes encoding α2,3-sialyltransferases, α1,3-fucosyltransferases, β1,4-galactosyltransferases, and β1,3-N-acetylglucosaminyltransferases that control the expression of LacNAc and sLeX.
FIGURE 4.
Gene expression analysis of 4-F-GlcNAc-treated sLeX (+) KG1a and T cells. a, heat maps of differentially expressed gene transcripts from untreated versus 4-F-GlcNAc-treated KG1a cells and untreated versus 4-F-GlcNAc-treated T cells depict increased (red) and decreased (blue) expression relative to the mean transcript expression values. At FC of 4-F-GlcNAc-treated/untreated ratios greater than 1.4, there were 17 and 94 differentially expressed gene transcripts from KG1a and T cells, respectively (adjusted p value of <0.1). b, gene expression clustering from untreated and 4-F-GlcNAc-treated KG1a and T cells of triplicate experiments was validated using centered correlation and average linkage and is represented in a dendrogram. c, a Venn diagram demonstrated that five genes were similarly modulated in KG1a and T cells (adjusted p value of <0.15).
4-F-GlcNAc Alters the Content and Complexity of N- and O-Glycans
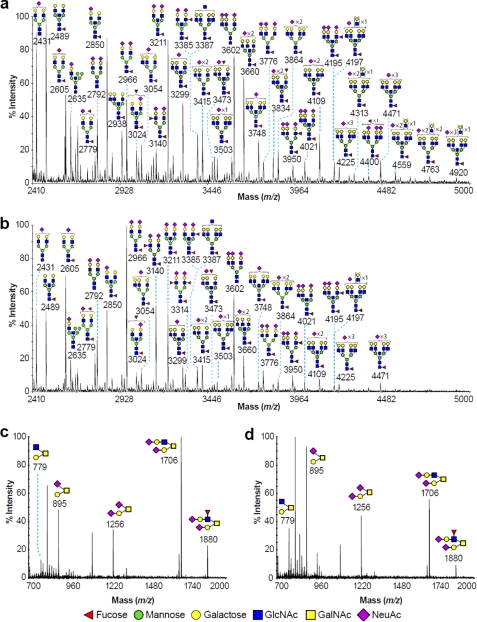
MALDI-TOF MS analysis of permethylated N-glycans from untreated KG1a cells is shown in Fig. 5a (for complete descriptions, see supplemental Table 1). The spectrum was mainly characterized by complex biantennary (m/z 2489, 2605, 2850, and 2966), triantennary (m/z 3054, 3299, and 3415), and tetra-antennary (m/z 3503, 3748, 3864, and 4109) N-glycan structures that were mainly core-fucosylated and either terminated with sialic acids (NeuAc) or contained undecorated LacNAc units. Several molecular ions had compositions consistent with the presence of bisected GlcNAc or unextended complex antennae. Of particular note were the ions at m/z 3140, 3385, 3950, and 4195 that corresponded to bi- and triantennary N-glycans terminated with sLeX epitopes. Larger tetra-antennary N-glycan structures with LacNAc extensions (m/z 4313, 4559, 4763, and 4920) were also observed.
FIGURE 5.
Partial MALDI-TOF spectra of permethylated N- and O-glycans derived from untreated and 4-F-GlcNAc-treated KG1a cells. Shown is the N-glycomic profile of untreated (a) and 4-F-GlcNAc-treated KG1a cells (b) and the O-glycomic profile of untreated (c) and 4-F-GlcNAc-treated KG1a cells (d). N- and O-glycomic profiles were obtained from the 50% MeCN fraction from a C18 Sep-Pak column (see “Experimental Procedures”). For clarity, major ions are shown. Schematic structures are according to the Consortium for Functional Glycomics guidelines. All molecular ions are [M + Na]+. Putative structures are based on composition, tandem MS, and biosynthetic knowledge. Structures that show sugars outside a bracket have not been unequivocally defined.
In contrast, the N-glycome of the 4-F-GlcNAc-treated KG1a cells (Fig. 5b; for complete descriptions, see supplemental Table 1) exhibited an increase in the abundance of biantennary N-glycans detected (e.g. at m/z 2966), accompanied, however, by a significant reduction in the relative abundance of tri- and tetra-antennary N-glycan structures (i.e. compare the ratio of m/z 2966:3776). These data indicated that 1) the sLeX-terminated biantennary N-glycans appeared to be present in higher relative abundance (m/z 3140 and 3314; Fig. 5b), whereas the sLeX-terminated triantennary N-glycans at m/z 3950 and 4195 were less abundant, and 2) the abundance of the larger tetra-antennary N-glycan structures with LacNAc extensions (m/z 4313, 4559, 4763, and 4920) was reduced.
The O-glycan profiles also exhibited differences in the relative abundances. The O-glycomes of untreated KG1a cells were composed mainly of mono- and disialylated core 1 (m/z 895 and 1256) and core 2 (m/z 779 and 1706) structures (Fig. 5c) as well as of sLeX core 2 structure (m/z 1880), whereas the O-glycomes of the 4-F-GlcNAc-treated KG1a cells were characterized from a significant decrease in core 2 and sLeX structures (Fig. 5d; m/z 1706 and 1880, respectively).
Similarly, human sLeX (+) T cells exhibited a reduction in the abundance of tetra-antennary N-glycans, and O-glycans showed a reduction in core 2 and sLeX structures (supplemental Fig. 1 and Table 2). Collectively, the data suggest that 4-F-GlcNAc altered N- and O-glycosylation of KG1a and T cells by reducing the abundance of tri- and tetra-antennary N-glycans, of LacNAc extensions, of mature core 2 O-glycans, and especially in KG1a cell sLeX-terminal epitopes.
Endo-β-galactosidase Digestion Revealed Changes in Polylactosamine-type Antennae
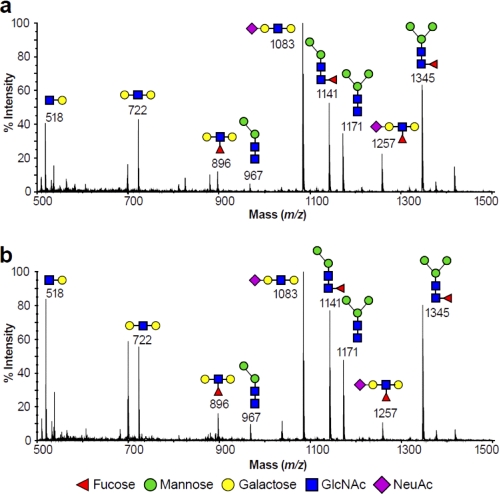
To further investigate changes in polylactosamine levels in N-glycans, we performed an endo-β-galactosidase digestion of the N-glycans of the untreated and 4-F-GlcNAc-treated KG1a and T cells (supplemental Fig. 2). The low mass permethylated digest products of the KG1a cells in both states (untreated and 4-F-GlcNAc-treated) are depicted in Fig. 6. Disaccharides released from within the polylactosamine chains gave a signal at m/z 518 corresponding to GlcNAc-Gal sequence (Fig. 6, a and b). The molecular ions at m/z 722, 896, 1083, and 1257 (Fig. 6, a and b) were consistent with the terminal sequences Gal-GlcNAc-Gal, Gal-(Fuc)GlcNAc-Gal, NeuAc-Gal-GlcNAc-Gal, and NeuAc-Gal-(Fuc)GlcNAc-Gal, respectively, which were predicted digestions of uncapped, fucosylated, sialylated, and sLeX polylactosamine chains, respectively. Evidence for the m/z 896 and 1257 components terminated by LewisX and sLeX epitopes was provided by MALDI-TOF-TOF MS/MS (data not shown). Molecular ions at m/z 967, 1141, 1171, and 1345 corresponded to N-glycan core structures Man2GlcNAc2, Man2GlcNAc-(Fuc)GlcNAc, Man3GlcNAc2, and Man3GlcNAc-(Fuc)GlcNAc, respectively, and they did not correspond to fragments released by the endo-β-galactosidase.
FIGURE 6.
Low mass MALDI-TOF mass spectra of permethylated N-glycans from endo-β-galactosidase digestion of KG1a cells. Profile of N-glycans of untreated (a) and 4-F-GlcNAc-treated KG1a cells (b). Profiles are from the 35% MeCN fraction from a C18 Sep-Pak column (see “Experimental Procedures”). All molecular ions are [M + Na]+. Putative structures based on composition, tandem MS, and the literature are shown. Schematic structures are according to the Consortium for Functional Glycomics guidelines.
Human sLeX (+) T cells upon treatment with endo-β-galactosidase digestion revealed a similar set of digestion product molecular ions in the low mass region (m/z 518, 722, 896, 1083, and 1257) (supplemental Fig. 3) as the KG1a cells.
Taken together, the data confirmed the presence of linear LacNAc units on both the untreated and 4-F-GlcNAc-treated KG1a and T cells. However, a comparison of the relative intensity of the digest product peaks with those of the N-glycan core structures, which would not be affected by the endo-β-galactosidase digestion, indicated a relative decrease in the sLeX-capped polylactosamine structures.
Supplemental Fig. 2 shows the spectra of the higher molecular weight region of the endo-β-galactosidase digestion of the N-glycans of the untreated and 4-F-GlcNAc-treated KG1a and T cells. The abundance of molecular ion signals, such as m/z 2734, 3095, 3456, 3544, 3632, and 3905, which all had compositions consistent with two unmodified GlcNAc residues and products of endo-β-galactosidase digestion, were all reduced in abundance in the 4-F-GlcNAc-treated KG1a cells. Additionally, the reduction in glycan complexity afforded by the endo-β-galactosidase digestion allowed for detection of additional high molecular weight glycans at m/z 4936 and 5007, both of which were consistent with tetra-antennary sLeX-capped glycans, neither of which was observed in the 4-F-GlcNAc-treated KG1a cells. Also, there was a general reduction in the amount of tetra-antennary structures (supplemental Fig. 2, a and b). Similar effects were also observed in the untreated and 4-F-GlcNAc-treated T cells (supplemental Fig. 2, c and d).
Linkage Analysis Revealed Differences in Polylactosamine-type Abundances
We performed GC-MS linkage analysis in order to compare the relative abundances of the 3-linked galactose and 4-linked GlcNAc residues (that both make up the LacNAc unit) between untreated and 4-F-GlcNAc-treated cells and to quantify respective levels of LacNAc units between the aforementioned cells. Because the 3-linked galactose residue could also potentially be produced from NeuAcα2–3Gal structures, linkage analysis was performed on desialylated N-glycans (supplemental Table 3). Results from linkage analysis of both untreated and treated KG1a and T cells showed that 1) the presence of 3-linked galactose (on desialylated N-glycans) and 4-linked GlcNAc residues was consistent with the presence of linear LacNAc units in both KG1a and T cells (untreated and 4-F-GlcNAc-treated); 2) when KG1a cells were treated with 4-F-GlcNAc, 3-linked galactose and 4-linked GlcNAc residue abundances were decreased by 79.7 and 88.5%, respectively, compared with the untreated KG1a cells, whereas for T cells, this decrease was 36.2 and 9.1% respectively; 3) the presence of 2,4-linked and 2,6-linked mannose was consistent with the MALDI-TOF data suggesting that KG1a and T cells expressed tri- and tetra-antennary N-glycans; 4) treatment of KG1a cells with 4-F-GlcNAc decreased 2,4-linked and 2,6-linked mannose by 84.0 and 74.5% respectively, whereas for the T cells, decreases were 50.1 and 26.8%, respectively, consistent with MALDI-TOF N-glycomic profiles showing reductions in tetra-antennary N-glycans of both cell types; and 5) 4-F-GlcNAc-treated KG1a cells exhibited a 85.3% decrease in the abundance of 3,4-linked GlcNAc residue, indicating an overall decrease in sLeX epitopes. These data corroborate endo-β-galactosidase results, which also showed decreases in sLeX structures, and demonstrated that, when compared with no treatment, both 4-F-GlcNAc-treated KG1a and T cells contained N-glycans with shorter LacNAc chains on fewer tri- and tetra-antennary structures. The presence of 3,4,6-linked mannose in all cell types provided compelling evidence for the presence of bisected N-glycans.
There Is No Evidence of Fluorine Incorporation into N- and O-Glycans
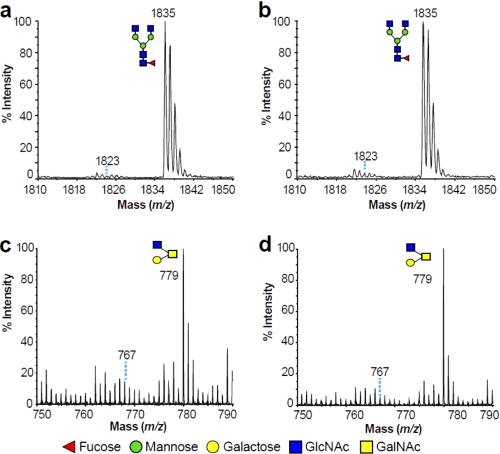
To determine whether 4-F-GlcNAc incorporated directly into glycans from 4-F-GlcNAc cells, we analyzed the spectrum of molecular masses for evidence of fluorine incorporation. Incorporation of 4-F-GlcNAc into a glycan would result in a molecular ion signal 12 units lower than the corresponding unmodified glycan. In Fig. 7, the spectrum of a GlcNAc-terminated biantennary N-glycan (m/z 1835) and a GlcNAc-terminated core 2 O-glycan (m/z 779) from the untreated and 4-F-GlcNAc-treated KG1a cells are depicted. Based on these spectral values, if a 4-F-GlcNAc was incorporated into these glycans, then the N- and O-glycomic profiles of 4-F-GlcNAc-treated KG1a cells would have had molecular ion signals at m/z 1823 and m/z 767, respectively (Fig. 7, b and d). However, the abundance of these ions was not altered in the treated cells. Moreover, MALDI-TOF-TOF MS/MS on the minor m/z 1823 and 767 ions observed in both sets of data verified no detectable incorporation of 4-F-GlcNAc for either the N- or the O-glycans (data not shown). Similarly, 4-F-GlcNAc-treated T cells showed no detectable incorporation of 4-F-GlcNAc (supplemental Fig. 4). Taken together, the data suggest that alteration of N- and O-glycosylation in KG1a and T cells was not due to incorporation of 4-F-GlcNAc.
FIGURE 7.
Partial MALDI-TOF spectra of permethylated N- and O-glycans derived from untreated and 4-F-GlcNAc-treated KG1a cells. Shown are enlarged scans of signals present at m/z 1835 (N-glycans) and 779 (O-glycans). N-glycomic profiles were obtained from the 35% MeCN fraction, whereas O-glycomic profiles were obtained from the 50% MeCN fraction (Fig. 5), both from a C18 Sep-Pak column (see “Experimental Procedures”). All molecular ions are [M + Na]+. Putative structures based on composition, tandem MS, and the literature are shown. Schematic structures are according to the Consortium for Functional Glycomics guidelines. Signals at m/z 1823 in a and b and at m/z 767 in c and d correspond to the theoretical incorporation of 4-F-GlcNAc.
4-F-GlcNAc Does Not Markedly Affect the Activity of Glycosyltransferases Involved in Poly-N-acetyllactosamine Formation
To further explore the mechanism of anti-glycan formation by 4-F-GlcNAc, we conducted glycosyltransferase assays using specific glycan acceptors and radiolabeled UDP-Gal or UDP-GlcNAc substrates to ascertain β1,3- and β1,4-galactosyltransferase (β1,3GalT or β1,4GalT) and β1,3-N-aceylglucosaminyltransferase (β1,3GlcNAcT) activities in 4-F-GlcNAc-treated lysates. Compared with lysates from diluent control treatments, lysates from sLeX (+) KG1a and T cells treated with 0.05 mm 4-F-GlcNAc (48 h) did not generate significant differences in β1,3- or β1,4GalT and β1,3GlcNAcT activities (Table 1). These data, in conjunction with unremarkable glycome expression analysis, indicate that the activities of β1,4GalT and β1,3GlcNAcT enzymes important for lactosamine synthesis were not significantly affected by glycan-modifying concentrations of 4-F-GlcNAc.
TABLE 1.
Activity of β1,3-galactosyltransferase, β1,4-galactosyltransferase, and β1,3-N-acetylglucosaminyltransferase in glycometabolically active KG1a and T cells treated with 4-F-GlcNAc
Data are presented as mean ± S.E. for three independent samples. Synthetic carbohydrate acceptors used are precursors of core 2 and LacNAc structures. ND, not detected.
| Cells | Treatment | β1,3GalT (GalNAc-para-aminobenzoic acid) | β1,4GalT (Galβ1,3(GlcNAcβ1,6) GalNAc-α-O-benzyl) | β1,3GlcNAcT (Galβ1,4GlcNAcβ1,3 Galβ1,4GlcNAc-β-O-benzyl) |
|---|---|---|---|---|
| Enzyme activity (dpm product formed/mg of protein in reaction mixture) | ||||
| KG1a | 5.5 ± 0.9 × 105 | 9.0 ± 2.5 × 104) | 10.2 ± 0.1 × 103 | |
| KG1a | 4-F-GlcNAc | 5.8 ± 1.6 × 105 | 13.7 ± 5.6 × 104) | 9.0 ± 0.5 × 103 |
| T cells | 6.4 ± 1.4 × 105 | ND | 4.5 ± 1.8 × 103 | |
| T cells | 4-F-GlcNAc | 5.5 ± 2.3 (×105) | ND | 4.7 ± 1.7 × 103 |
| Enzyme activity (percentage conversion of the radioactive donor into product) | ||||
| KG1a | 27.9 ± 4.1 | 5.2 ± 0.9 | 12.5 ± 1.0 | |
| KG1a | 4-F-GlcNAc | 29.0 ± 4.9 | 8.3 ± 2.8 | 12.2 ± 0.9 |
| T cells | 13.4 ± 1.6 | ND | 2.2 ± 0.7 | |
| T cells | 4-F-GlcNAc | 10.5 ± 1.4 | ND | 1.6 ± 0.1 |
4-F-GlcNAc Reduces UDP-GlcNAc
To determine whether 4-F-GlcNAc inhibits the formation of UDP-GlcNAc, the major precursor of LacNAc synthesis, we utilized a novel color-based assay (34). Because the assay reportedly detects GlcNAc that is either free or acid-hydrolyzed from UDP-GlcNAc, we performed preliminary experiments in the presence or absence of acid hydrolysis and found that UDP-GlcNAc was the major contributor of GlcNAc over free GlcNAc (data not shown). In other words, the assay selectively measured cellular UDP-GlcNAc level and not GlcNAc (34). As an additional control in this analysis, we next determined whether GlcNAc and/or 4-F-GlcNAc molecular variants were detected in the assay (Fig. 8a). GlcNAc and UDP-GlcNAc structures were detected in a dose-dependent manner (Fig. 8b). In addition, the assay also detected a fully acetylated variant of GlcNAc, α-d-glucosamine pentaacetate, indicating that acetyl groups lining the hexose ring did not interfere with detection (Fig. 8b). In contrast, neither 4-F-GlcNAc (unacetylated) nor 4-F-GlcNAc (peracetylated) was detectable, indicating that fluorine at carbon 4 blocked detection by this assay (Fig. 8b). However, 4-F-GlcNAc (peracetylated) did not interfere with the measurement of UDP-GlcNAc when both glycans were combined in the same tube (data not shown). In summary, these control experiments 1) validated the selectivity for measuring cellular UDP-GlcNAc and 2) confirmed that both peracetylated and unacetylated 4-F-GlcNAc, either in free form or acid-hydrolyzed from a possible cellular intermediate, UDP-4-F-GlcNAc, would not be detected and would not interfere with measurement of UDP-GlcNAc.
In cell-based experiments using KG1a and sLeX (+) T cells, we found that control GlcNAc treatment had minimal effect on intracellular UDP-GlcNAc level (Fig. 8, c and d). In contrast, the highly acetylated control form of GlcNAc, α-d-glucosamine pentaacetate, slightly elevated intracellular UDP-GlcNAc levels in KG1a and sLeX (+) T cells by 30 and 15%, respectively, which was probably due to greater cell uptake through acetylation of the hexosamine ring (Fig. 8, c and d). Although UDP-GlcNAc levels did not change significantly in response to 4-F-GlcNAc (unacetylated), 4-F-GlcNAc (peracetylated) decreased UDP-GlcNAc level significantly in KG1a cells by 55% and in sLeX (+) T cells by 35% relative to untreated control (Fig. 8, c and d). Moreover, the reduction in UDP-GlcNAc in KG1a cells treated with 4-F-GlcNAc (peracetylated) was 3-fold greater than cells grown in medium without glucose (data not shown). The reduced level of UDP-GlcNAc directly correlated with flow cytometric assays performed in parallel, wherein Gal-1 ligand expression was reduced significantly on KG1a and sLeX (+) T cells treated with 4-F-GlcNAc (peracetylated) but not treated with 4-F-GlcNAc (unacetylated), GlcNAc, or α-d-glucosamine pentaacetate (supplemental Fig. 5). Overall, these results indicated that 4F-GlcNAc (peracetylated) might reduce content and diversity of N- and O-glycans partly through depletion of UDP-GlcNAc, a key precursor of LacNAc biosynthesis, and that such depletion required both peracetylation and fluorination of the hexosamine ring.
DISCUSSION
The ability of peracetylated fluoro-glucosamine analogs to inhibit LacNAc and sLeX expression, which serve as critical carbohydrate determinants for Gal-1 and E-selectin binding, is well chronicled (7, 8, 11, 12, 14, 24, 25, 44). 4-F-GlcNAc, for example, has been hypothesized to elicit its anti-carbohydrate efficacy by directly incorporating into LacNAc structures, causing chain termination and truncated N- and/or O-glycans (7, 8, 11, 24, 25). Early studies proposed that isosteric substitution of the hydroxyl group for a fluorine at the carbon 4-position of GlcNAc would prevent glycosidic bonding at carbon 4 due to the inability of fluorine to function as a nucleophile or leaving group (22, 23, 45). Although most studies show that LacNAc or sLeX moieties related to Gal-1 or E-selectin ligand formation are down-regulated using anti-carbohydrate mAbs or lectins as probes, there is no evidence showing that 4-F-GlcNAc incorporates into LacNAc to block chain extension and/or the addition of fucose and/or N-acetylneuraminic acid. Interestingly, there is compelling evidence showing that 4-F-Glc[3H]NAc and fully acetylated [14C]4-F-GalNAc incorporate into the heavily glycosylated membrane protein, PSGL-1 (7, 14). In these experiments, however, it may be possible for the 3H-labeled N-acetyl side chain on 4-F-GlcNAc to be removed by amidases and/or de-N-acetylases and for the 14C-labeled O-acetyl side chains on 4-F-GalNAc to be removed by de-O-acetylases.3 Thus, radiolabeled N-/O-acetyl products could be further metabolized and potentially be used in de novo protein biosynthesis. What is still outstanding in defining the mechanism of 4-F-GlcNAc anti-carbohydrate action is whether it directly incorporates into intact glycan chains.
In this report, our intent was 1) to validate the glycan-modifying activity of 4-F-GlcNAc using Gal-1-LacNAc and E-selectin-sLeX binding interactions as well defined lectin-carbohydrate targets of 4-F-GlcNAc; 2) to provide firm structural evidence showing 4-F-GlcNAc incorporation (or lack thereof) into N-/O-glycans; 3) to determine whether 4-F-GlcNAc treatment alters the gene expression of glycosyltransferases involved in LacNAc or sLeX; 4) to ascertain whether 4-F-GlcNAc treatment interferes with the activities of glycosyltransferases involved in LacNAc synthesis; and 5) to determine whether 4-F-GlcNAc interferes with the metabolism of LacNAc biosynthesis. To attain these objectives, we utilized two leukocyte models, human leukemic KG1a cells and human ex vivo-generated sLeX (+) T cells, which express well defined glycosylation pathways requisite for biosynthesis of LacNAc and sLeX and corresponding Gal-1 and E-selectin ligand activities (7, 11, 12). In addition, we used identical concentrations of 4-F-GlcNAc previously shown to inhibit the synthesis of related Lewis antigen and LacNAc structures and diminish Gal-1 and E-selectin ligand activities (7, 11, 12, 24, 25).
As determined by Western blotting and flow cytometry, 4-F-GlcNAc significantly down-regulated LacNAc and sLeX moieties as well as Gal-1 and E-selectin-binding carbohydrates while eliciting little effect on the level of PSGL-1 expression in both KG1a and T cell models. Gal-1 ligand synthesis was more sensitive than E-selectin ligand to 4-F-GlcNAc treatment, indicating that LacNAc structures were more affected than sLeX, which can terminate either a single LacNAc moiety or an extended poly-LacNAc structure. Of note, employing protease bromelain digestion with 4-F-GlcNAc treatment largely eliminated Gal-1 ligand, suggesting that Gal-1 ligands are expressed on glycoprotein. These data suggested that 4-F-GlcNAc lowers the frequency of N-/O-glycans containing LacNAc terminated with sLeX and/or the abundance of LacNAc in N-/O-glycans.
To address whether 4-F-GlcNAc treatment down-regulated the expression of glycosyltransferases related to LacNAc and sLeX synthesis, we compared and contrasted the relative mRNA levels of glycome-related proteins, including glycosyltransferases, glycosidases, glycoproteins, and factors/receptors/signal proteins related to glycosylation, between untreated and 4-F-GlcNAc-treated cells. Using the GlycoV4 oligonucleotide array containing ∼1260 human probe IDs related to glyco-genes, data showed that 4-F-GlcNAc treatment caused minor differences (-fold change of >1.4) in a small subset of genes in KG1a (n = 17) and T cells (n = 94) that did not correspond to the repertoire of glycosyltransferases necessary for N- and O-glycan biosynthesis. In fact, only five genes were found to be differentially expressed in both cell models, none of which influence LacNAc formation and sLeX biosynthesis.
To further explore 4-F-GlcNAc effects on de novo biosynthetic pathways, we assayed the activities of LacNAc-synthesizing enzymes, β1,3GlcNAcT, β1,4GalT, and β1,3GalT. In both cell models, there was no appreciable change in β1,3GalT and β1,3GlcNAcT activity. In β1,4GalT assays, there was also no significant change between 4-F-GlcNAc-treated and untreated KG1a cells. Of note, LacNAc-core 2 glycan product in β1,4GalT assays was not detectable using T cell lysates, contrary to structural data showing the presence of sLeX-core 2 O-glycans on PSGL-1 and CD43 with specific mAbs (11). Because these ex vivo human T cells express sLeX on a single LacNAc backbone on a core 2 O-glycan recognized by mAb CHO-131 (11, 46), it is possible that minimal, experimentally undetectable β1,4GalT activity is needed for LacNAc synthesis or β1,4 transfer of Gal to GlcNAcβ1,6 on Galβ1,3(GlcNAcβ1,6)GalNAc-R. Collectively, these enzyme activity data parallel glycome expression analysis, in that LacNAc-synthesizing enzyme activities in 4-F-GlcNAc-treated cells are not significantly different from those in untreated cells.
To provide direct evidence of 4-F-GlcNAc-dependent glycan alterations, isolated glycans from untreated and 4-F-GlcNAc-treated cells were analyzed by MALDI-TOF MS and GC-MS linkage analyses. Data from these studies revealed that 1) 4-F-GlcNAc had no effect on the level of GlcNAc- terminated glycans, which indicated that 4-F-GlcNAc may not be incorporated and acting as a chain terminator; 2) 4-F-GlcNAc diminished LacNAc levels on N-glycans and on mature core 2 O-glycans and sLeX terminal epitopes especially on the KG1a cells, and 3) 4-F-GlcNAc reduced the content and structural diversity of tri- and tetra-antennary N-glycans and core 2 O-glycans, which resulted in higher levels of biantennary N-glycans and core 1 O-glycans, respectively. Structural determination of glycans from peracetylated 4-F-GlcNAc-treated leukocytes did not yield results consistent with incorporation of a fluoro-GlcNAc moiety in an oligosaccharide chain. The mechanism of 4-F-GlcNAc-dependent reductions in LacNAc and sLeX moieties and Gal-1 and E-selectin ligands activities is, thus, more complex than previously anticipated, in that 4-F-GlcNAc does not appear to function as an oligosaccharide chain terminator or to antagonize LacNAc-related glycosyltransferase level or activity. In fact, because 4-F-GlcNAc lowered UDP-GlcNAc levels, inhibition of LacNAc might occur further upstream of glycosyltransferase activity via inhibition of enzymes involved in UDP-GlcNAc biosynthesis. In that 4-F-GlcNAc reduced UDP-GlcNAc not only in KG1a and T cells but also in human prostate cancer PC-3 cells (data not shown), although at varying potencies, 4-F-GlcNAc is probably antagonizing UDP-GlcNAc formation via a common enzymatic step. One possible explanation, as illustrated in Fig. 9, is that 4-F-GlcNAc may passively diffuse into cells and inhibit one or more enzymes involved in UDP-GlcNAc synthesis, including UDP-N-acetylglucosamine pyrophosphorylase and/or upstream enzymes involved in GlcNAc 1-phosphate synthesis, an immediate precursor of UDP-GlcNAc (47). Also unclear, as illustrated in Fig. 9, is whether 4-F-GlcNAc is converted into a 4-F-GlcNAc 1-phosphate intermediate and converted to UDP-4-F-GlcNAc. However, given that fluorosugars were not detected de novo in glycans by the mass spectrometric data, it is unlikely that UDP-4-F-GlcNAc is generated by UDP-N-acetylglucosamine pyrophosphorylase. A recent study suggested that 4-F-GlcNAc could be interfering with 4-epimerization of UDP-GalNAc from UDP-GlcNAc, resulting in truncated glycans (48). This scenario could perhaps be the reason for observed reductions in core 2 O-glycans, which require UDP-GalNAc for initiating O-glycan synthesis but may not necessarily be the reason for the observed lowering in quantity and type of N-glycans. It has also been shown that 4-F-GlcNAc can deplete cellular UTP (22, 45). UTP is required to form nucleotide sugar donors, UDP-Gal and UDP-GlcNAc, which are used as substrates by β1,3/4GalT and β1,3GlcNAcT, respectively. In fact, our data here confirmed this assertion, wherein fully acetylated GlcNAc analogs are more effective than unacetylated GlcNAc analogs at raising cellular UDP-GlcNAc, due to their increased cell permeability and capacity to form UDP-GlcNAc (22). Following cellular entry and de-O-acetylation, 4-F-GlcNAc may be used as a decoy precursor to form UDP-4-F-GlcNAc. As a consequence, 4-F-GlcNAc may be lowering endogenous stores of UTP and diverting synthesis of UDP-Gal or UDP-GlcNAc pools, thereby lowering available substrates for LacNAc synthesis and N-glycan branching. Indeed, previous data on nucleotide sugar donor formation indicate that 4-F-GlcNAc treatment may alter the extent of radiolabeled galactose and glucosamine incorporation into newly synthesized glycoproteins (23, 25). In addition, a recent study suggested that the enzymes responsible for the tri- and tetra-antennary N-glycan branching are highly sensitive to the metabolic supply of UDP-GlcNAc (49). Another potential reason for diminished levels of tri-and tetra-antennary N-glycans may be that 4-F-GlcNAc is inhibiting one of several other GlcNAcT enzymes necessary for N-glycan branching not assayed here. A final potential explanation is that the 4-F-GlcNAc is influencing O-GlcNAc glycosylation. This novel form of intracellular glycosylation has been demonstrated to be functionally important for a diverse range of cellular regulatory proteins, such as transcription factors and signal transduction mediators, kinases, and phosphatases, all of which are involved in the complex regulation of glycosylation (50). These and other potential modes of 4-F-GlcNAc action may be the subject of future investigations.
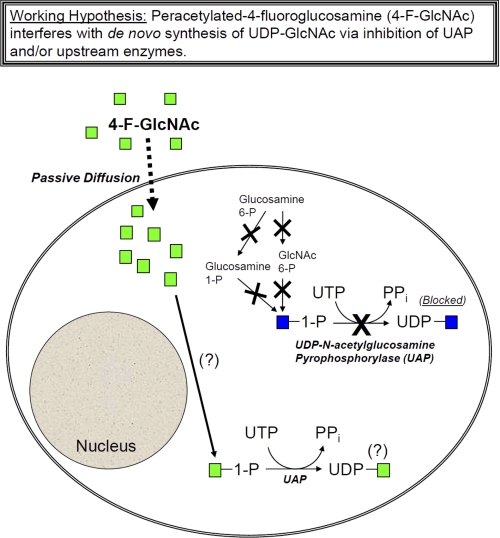
FIGURE 9.
Working hypothesis for 4-F-GlcNAc-mediated lowering of UDP-GlcNAc in cells. As shown in the illustration, UDP-GlcNAc is synthesized from upstream glycan precursors, including GlcNAc 1-phosphate (blue), glucosamine 6-phosphate (Glucosamine 6-P), glucosamine 1-phosphate (Glucosamine 1-P), and GlcNAc 6-phosphate. 4-F-GlcNAc (peracetylated) can passively diffuse into the cell aided by acetyl groups, wherein it subsequently inhibits UDP-GlcNAc formation via a mechanism that is still unknown. Shown are possible steps that might be inhibited by 4-F-GlcNAc that include UDP-N-acetylglucosamine pyrophosphorylase (UAP) along with upstream enzymatic steps involved in the generation of GlcNAc 1-phosphate. As emphasized with question marks, it is also unclear whether, upon entry into the cell, 4-F-GlcNAc is converted to 4-F-GlcNAc 1-phosphate and/or whether UAP can convert this potential intermediate into UDP-4-F-GlcNAc. Because 4-F-GlcNAc could not be detected by the cellular UDP-GlcNAc assay, formation of a UDP-4-F-GlcNAc intermediate is still unknown.
In summary, results presented in this report support a refined hypothesis whereby 4-F-GlcNAc may not function as an oligosaccharide chain terminator of glycans bearing LacNAc and sLeX structures but may work, at least in part, by reducing the pool of UDP-GlcNAc required for LacNAc synthesis. We are actively pursuing how 4-F-GlcNAc inhibits UDP-GlcNAc formation as a putative reason for lowering the content of LacNAc and sLeX on N- and O-glycans. Nevertheless, because sialylated Lewis structures on N- and O-glycans and highly branched N-glycans are associated with activated T cells and metastatic tumor cells (51), reducing Lewis antigens and diminishing N-glycan branching with 4-F-GlcNAc might blunt inflammation and cancer progression. Therefore, we continue to advance the notion that glycometabolically active cells, such as activated T cells and metastatic cancer cells, are selective targets for glycometabolic antagonism.
Supplementary Material
Acknowledgments
We thank Dr. Michael Ferguson (University of Dundee) for helpful discussions on cellular GlcNAc metabolism and GlcNAc side chain modulation. We also thank Dr. Sandra King for help in culturing and expanding sLeX (+) T cells.
This work was supported, in whole or in part, by National Institutes of Health (NIH), NCI, Grant RO1 CA118124 (to C. J. D.); NIH, NCCAM, Grant RO1 AT004268 (to C. J. D.); and NIH Analytical Glycotechnology Core (Core C) of the Consortium for Functional Glycomics Grant GM 62116. This work was also supported by Biotechnology and Biological Sciences Research Council Grants BBF0083091 and B19088 and the Wellcome Trust (to S. M. H. and A. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. 1–5.
M. Ferguson, personal communication.
- sLeX
- sialyl Lewis X
- AP
- alkaline phosphatase
- Fuc
- fucose
- GalT
- galactosyltransferase
- Gal-1
- galectin-1
- 4-F-GalNAc
- peracetylated 4-fluorinated galactosamine
- GlcNAcT
- β1,3-N-acetylglucosaminyltransferase
- LacNAc
- N-acetyllactosamine
- Man
- mannose
- NeuAc
- sialic acid
- 4-F-GlcNAc
- peracetylated 4-fluoro-glucosamine
- Th
- T helper
- FC
- -fold change
- Gal-1hFc
- galectin-1-human IgG1e3-Fc1
- GM2
- GalNAc-(NeuAc)-Gal-Glc-Cer
- E-selectin
- endothelial selectin.
REFERENCES
- 1. Liu F. T., Rabinovich G. A. (2010) Ann. N.Y. Acad. Sci. 1183, 158–182 [DOI] [PubMed] [Google Scholar]
- 2. Rabinovich G. A., Ilarregui J. M. (2009) Immunol. Rev. 230, 144–159 [DOI] [PubMed] [Google Scholar]
- 3. Rabinovich G. A., Toscano M. A. (2009) Nat. Rev. Immunol. 9, 338–352 [DOI] [PubMed] [Google Scholar]
- 4. Salatino M., Croci D. O., Bianco G. A., Ilarregui J. M., Toscano M. A., Rabinovich G. A. (2008) Expert Opin. Biol. Ther. 8, 45–57 [DOI] [PubMed] [Google Scholar]
- 5. Ley K., Kansas G. S. (2004) Nat. Rev. Immunol. 4, 325–335 [DOI] [PubMed] [Google Scholar]
- 6. Marth J. D., Grewal P. K. (2008) Nat. Rev. Immunol. 8, 874–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dimitroff C. J., Bernacki R. J., Sackstein R. (2003) Blood 101, 602–610 [DOI] [PubMed] [Google Scholar]
- 8. Dimitroff C. J., Kupper T. S., Sackstein R. (2003) J. Clin. Invest. 112, 1008–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dube D. H., Bertozzi C. R. (2005) Nat. Rev. Drug. Discov. 4, 477–488 [DOI] [PubMed] [Google Scholar]
- 10. Barthel S. R., Gavino J. D., Descheny L., Dimitroff C. J. (2007) Expert Opin. Ther. Targets 11, 1473–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Descheny L., Gainers M. E., Walcheck B., Dimitroff C. J. (2006) J. Invest. Dermatol. 126, 2065–2073 [DOI] [PubMed] [Google Scholar]
- 12. Gainers M. E., Descheny L., Barthel S. R., Liu L., Wurbel M. A., Dimitroff C. J. (2007) J. Immunol. 179, 8509–8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cedeno-Laurent F., Barthel S. R., Opperman M. J., Lee D. M., Clark R. A., Dimitroff C. J. (2010) J. Immunol. 185, 4659–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marathe D. D., Buffone A., Jr., Chandrasekaran E. V., Xue J., Locke R. D., Nasirikenari M., Lau J. T., Matta K. L., Neelamegham S. (2010) Blood 115, 1303–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown J. R., Fuster M. M., Whisenant T., Esko J. D. (2003) J. Biol. Chem. 278, 23352–23359 [DOI] [PubMed] [Google Scholar]
- 16. Brown J. R., Yang F., Sinha A., Ramakrishnan B., Tor Y., Qasba P. K., Esko J. D. (2009) J. Biol. Chem. 284, 4952–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuster M. M., Brown J. R., Wang L., Esko J. D. (2003) Cancer Res. 63, 2775–2781 [PubMed] [Google Scholar]
- 18. Dimitroff C. J., Lee J. Y., Fuhlbrigge R. C., Sackstein R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13841–13846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dimitroff C. J., Lee J. Y., Rafii S., Fuhlbrigge R. C., Sackstein R. (2001) J. Cell Biol. 153, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sackstein R., Dimitroff C. J. (2000) Blood 96, 2765–2774 [PubMed] [Google Scholar]
- 21. Dimitroff C. J., Descheny L., Trujillo N., Kim R., Nguyen V., Huang W., Pienta K. J., Kutok J. L., Rubin M. A. (2005) Cancer Res. 65, 5750–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bernacki R. J., Sharma M., Porter N. K., Rustum Y., Paul B., Korythyk W. (1977) J. Supramol. Struct. 7, 235–250 [DOI] [PubMed] [Google Scholar]
- 23. Sharma M., Bernacki R. J., Paul B., Korytnyk W. (1990) Carbohydr. Res. 198, 205–221 [DOI] [PubMed] [Google Scholar]
- 24. Woynarowska B., Dimitroff C. J., Sharma M., Matta K. L., Bernacki R. J. (1996) Glycoconj. J. 13, 663–674 [DOI] [PubMed] [Google Scholar]
- 25. Woynarowska B., Skrincosky D. M., Haag A., Sharma M., Matta K., Bernacki R. J. (1994) J. Biol. Chem. 269, 22797–22803 [PubMed] [Google Scholar]
- 26. Dimitroff C. J., Lechpammer M., Long-Woodward D., Kutok J. L. (2004) Cancer Res. 64, 5261–5269 [DOI] [PubMed] [Google Scholar]
- 27. Barthel S. R., Gavino J. D., Wiese G. K., Jaynes J. M., Siddiqui J., Dimitroff C. J. (2008) Glycobiology 18, 806–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sutton-Smith M., Delle A. (2006) Cell Biology: A Laboratory Handbook, pp. 415–425, Academic Press, Inc., San Diego, CA [Google Scholar]
- 29. Jang-Lee J., North S. J., Sutton-Smith M., Goldberg D., Panico M., Morris H., Haslam S., Dell A. (2006) Methods Enzymol. 415, 59–86 [DOI] [PubMed] [Google Scholar]
- 30. Ceroni A., Maass K., Geyer H., Geyer R., Dell A., Haslam S. M. (2008) J. Proteome Res. 7, 1650–1659 [DOI] [PubMed] [Google Scholar]
- 31. Barthel S. R., Wiese G. K., Cho J., Opperman M. J., Hays D. L., Siddiqui J., Pienta K. J., Furie B., Dimitroff C. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 19491–19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dell A., Reason A. J. (1993) Curr. Opin. Biotechnol. 4, 52–56 [DOI] [PubMed] [Google Scholar]
- 33. Marathe D. D., Chandrasekaran E. V., Lau J. T., Matta K. L., Neelamegham S. (2008) FASEB J. 22, 4154–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burghardt C., Kochan J. P. (November 21, 2007) European Patent Number EP1431397B1
- 35. Burdick M. M., Chu J. T., Godar S., Sackstein R. (2006) J. Biol. Chem. 281, 13899–13905 [DOI] [PubMed] [Google Scholar]
- 36. Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. (2001) Behav. Brain. Res. 125, 279–284 [DOI] [PubMed] [Google Scholar]
- 37. Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 38. Lockhart D. J., Dong H., Byrne M. C., Follettie M. T., Gallo M. V., Chee M. S., Mittmann M., Wang C., Kobayashi M., Horton H., Brown E. L. (1996) Nat. Biotechnol. 14, 1675–1680 [DOI] [PubMed] [Google Scholar]
- 39. Smyth G. K. (2004) Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 40. Kenmochi N., Kawaguchi T., Rozen S., Davis E., Goodman N., Hudson T. J., Tanaka T., Page D. C. (1998) Genome Res. 8, 509–523 [DOI] [PubMed] [Google Scholar]
- 41. Hou Y., Tse R., Mahuran D. J. (1996) Biochemistry 35, 3963–3969 [DOI] [PubMed] [Google Scholar]
- 42. Pshezhetsky A. V., Richard C., Michaud L., Igdoura S., Wang S., Elsliger M. A., Qu J., Leclerc D., Gravel R., Dallaire L., Potier M. (1997) Nat. Genet. 15, 316–320 [DOI] [PubMed] [Google Scholar]
- 43. Wood T. C., Aksoy I. A., Aksoy S., Weinshilboum R. M. (1994) Biochem. Biophys. Res. Commun. 198, 1119–1127 [DOI] [PubMed] [Google Scholar]
- 44. Dimitroff C. J., Pera P., Dall'Olio F., Matta K. L., Chandrasekaran E. V., Lau J. T., Bernacki R. J. (1999) Biochem. Biophys. Res. Commun. 256, 631–636 [DOI] [PubMed] [Google Scholar]
- 45. Bernacki R., Porter C., Korytnyk W., Mihich E. (1977) Adv. Enzyme Regul. 16, 217–237 [DOI] [PubMed] [Google Scholar]
- 46. Walcheck B., Leppanen A., Cummings R. D., Knibbs R. N., Stoolman L. M., Alexander S. R., Mattila P. E., McEver R. P. (2002) Blood 99, 4063–4069 [DOI] [PubMed] [Google Scholar]
- 47. Mio T., Yabe T., Arisawa M., Yamada-Okabe H. (1998) J. Biol. Chem. 273, 14392–14397 [DOI] [PubMed] [Google Scholar]
- 48. Nigro J., Wang A., Mukhopadhyay D., Lauer M., Midura R. J., Sackstein R., Hascall V. C. (2009) J. Biol. Chem. 284, 16832–16839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lau K. S., Partridge E. A., Grigorian A., Silvescu C. I., Reinhold V. N., Demetriou M., Dennis J. W. (2007) Cell 129, 123–134 [DOI] [PubMed] [Google Scholar]
- 50. Zeidan Q., Hart G. W. (2010) J. Cell Sci. 123, 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (2009) Essentials of Glycobiology, 2nd Ed., pp. 617–632, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.