Abstract
An elevated level of homocysteine, a thiol amino acid, is associated with various complex disorders. The cellular effects of homocysteine and its precursors S-adenosylhomocysteine (AdoHcy) and S-adenosylmethionine (AdoMet) are, however, poorly understood. We used Saccharomyces cerevisiae as a model to understand the basis of pathogenicity induced by homocysteine and its precursors. Both homocysteine and AdoHcy but not AdoMet inhibited the growth of the str4Δ strain (which lacks the enzyme that converts homocysteine to cystathionine-mimicking vascular cells). Addition of AdoMet abrogated the inhibitory effect of AdoHcy but not that of homocysteine indicating that an increase in the AdoMet/AdoHcy ratio is sufficient to overcome the AdoHcy-mediated growth defect but not that of homocysteine. Also, the transcriptomic profile of AdoHcy and homocysteine showed gross dissimilarity based on gene enrichment analysis. Furthermore, compared with homocysteine, AdoHcy treatment caused a higher level of oxidative stress in the cells. However, unlike a previously reported response in wild type (Kumar, A., John, L., Alam, M. M., Gupta, A., Sharma, G., Pillai, B., and Sengupta, S. (2006) Biochem. J. 396, 61–69), the str4Δ strain did not exhibit an endoplasmic reticulum stress response. This suggests that homocysteine induces varied response depending on the flux of homocysteine metabolism. We also observed altered expression of mitochondrial genes, defective membrane potential, and fragmentation of the mitochondrial network together with the increased expression of fission genes indicating that the imbalance in homocysteine metabolism has a major effect on mitochondrial functions. Furthermore, treatment of cells with homocysteine or AdoHcy resulted in apoptosis as revealed by annexin V staining and TUNEL assay. Cumulatively, our results suggest that elevated levels of homocysteine lead to mitochondrial dysfunction, which could potentially initiate pro-apoptotic pathways, and this could be one of the mechanisms underlying homocysteine-induced pathogenicity.
Keywords: Apoptosis, Gene Expression, Homocysteine, Mitochondria, Reactive Oxygen Species (ROS), S-Adenosylmethionine (AdoMet), Yeast
Introduction
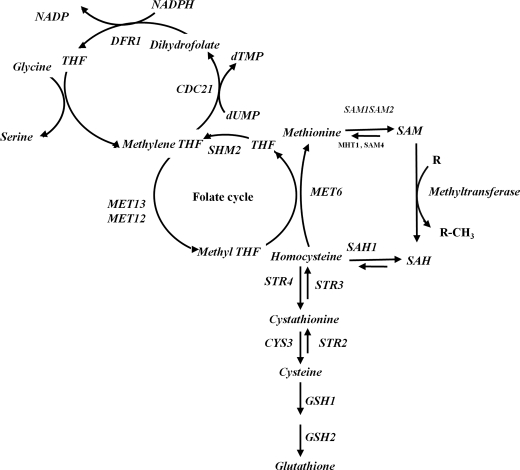
Homocysteine, a thiol-containing amino acid, occupies a pivotal position in the methionine metabolism. This cycle appears to be present in all normal mammalian cells and is necessary for the synthesis of several important metabolites, including S-adenosylmethionine (AdoMet),3 folic acid, polyamines, cysteine, and its derivatives. In human, methionine from dietary sources is first converted to AdoMet by the enzyme AdoMet synthase. AdoMet acts as a methyl donor for over 100 known transmethylation reactions, including methylation of macromolecules like DNA, RNA, and proteins. During these reactions AdoMet is converted by various methyltransferases to AdoHcy, which is then hydrolyzed to homocysteine and adenosine by S-adenosylhomocysteine hydrolase (Fig. 1). Homocysteine thus formed is immediately converted to methionine with the help of methionine synthase (remethylation process) or is metabolized into cystathionine and subsequently to cysteine and glutathione via the transulfuration process. In healthy well nourished individuals, the concentration of homocysteine is well regulated. However, inborn genetic defects, including single nucleotide polymorphism(s) in genes that are involved in homocysteine metabolism (1), deficiency of micronutrients like folate, B12, B6, or drugs that interfere with the methionine folate pathway, elevate the levels of homocysteine (2). An elevated level of homocysteine has been associated with various complex disorders like schizophrenia, Alzheimer disease, neural tube defect, diabetes, etc. (3–5) and has also been implicated as an independent risk factor for cardiovascular diseases (2, 6, 7).
FIGURE 1.
Homocysteine metabolism in yeast. The following abbreviations are used: STR4, cystathionine β-synthase homolog in yeast; SAH1, S-adenosyl-l-homocysteine hydrolase; MET6, methionine synthase; CDC21, thymidylate synthase; MET13, methylenetetrahydrofolate reductase; SHM2, serine hydroxymethyltransferase; dUMP, deoxyuridine 5-monophosphate; dTMP, deoxythymidine 5′-monophosphate; SAM1, SAM2, S-adenosylmethionine synthetase; MHT1, S-methylmethionine-homocysteine methyltransferase; THF, tetrahydrofolate; SAM, AdoMet; SAH, AdoHcy.
Although homocysteine has been associated with several diseases, the exact mechanism of homocysteine-induced pathogenesis is not yet clearly understood. Several hypotheses have been put forward to explain the deleterious effects of homocysteine. Prominent among these are the stress hypothesis (oxidative and endoplasmic reticulum stress) (8–10), molecular targeting hypothesis (ability of homocysteine to bind to free cysteine residues or break critical cysteine disulfide bonds thereby altering the structure and function of the protein) (11–14), and ability of homocysteine to alter the methylation status of DNA, RNA, proteins, and other metabolites (15). We believe that although these hypotheses have been proven beyond doubt under the conditions for which it has been tested, any one of these mechanisms cannot account for the toxicity of this thiol compound under all conditions. For instance, several studies have shown oxidative stress to be the single major cause of homocysteine-related pathogenicity (16–18), although others have shown little or no oxidative stress in the presence of homocysteine under their experimental conditions (19–21). We had also shown previously that addition of homocysteine and cysteine to the wild type yeast Saccharomyces cerevisiae results in growth defects that are probably mediated via endoplasmic reticulum stress and not oxidative stress (22). This may be due to the fact that although homocysteine is almost equally metabolized to methionine via the remethylation pathway and cystathionine via the trans-sulfuration process (23), under circumstances where the levels of homocysteine are elevated, the flux of homocysteine will be tilted more toward one of the pathways depending on why the homocysteine levels are elevated. For instance, deficiency of vitamin B12 or a variation in the methyl tetrahydrofolate reductase (MTHFR) gene (C677T) will reduce the conversion of homocysteine to methionine and hence would increase the flux of homocysteine toward the trans-sulfuration process, whereas mutations in cystathionine β-synthase (CBS) or in tissues that lack active CBS, like vascular tissues (24), the flux will be toward the remethylation process. Because the expression of the enzymes that are directly involved in the metabolism of homocysteine varies across tissues, it can be perceived that the physiological processes modulated by homocysteine will be tissue-dependent.
Apart from homocysteine, its metabolic precursor AdoHcy has been found to cause apoptosis in cultured pulmonary artery endothelial cells (25). Accumulation of AdoHcy has also been found to be cytotoxic in cultured T- and B-lymphocytes (26). Furthermore, Lin et al. (27) demonstrated that addition of AdoHcy in combination with homocysteine leads to synergistic DNA damage in BV2 cells. To better understand the effects of homocysteine and its precursors, we used the yeast S. cerevisiae as a model system, which is a good system for modeling human disease conditions and has unraveled complex mechanisms and pathways underlying many other human diseases like cancer, neurological diseases, etc. (28–31) and has proven to be a remarkably versatile model system for studies in molecular medicine. Most importantly, the yeast methionine cycle is similar to the human cycle with some minor variations, and deletion strains are readily available. In the post-genomics era, yeast has been used as a valuable tool in every aspect of high throughput biological research, from gene expression profiling to protein-protein interaction mapping (32–35). In this study using a strain (str4Δ) that lacks the expression of cystathionine β-synthase, we show that both homocysteine and AdoHcy inhibit the growth of yeast, but unlike in the wild type, endoplasmic reticulum stress may not play a role in the toxicity of homocysteine and/or its precursor AdoHcy.
EXPERIMENTAL PROCEDURES
Materials
dl-Homocysteine (Sigma H4628), S-(5′-adenosyl)-l-methionine chloride (Sigma A7007), S-(5′-adenosyl)-l-homocysteine (Sigma A9384), and mono-bromobimane were purchased from Sigma. The constituents of yeast media, including yeast extract, peptone, dextrose, and the amino acids, were purchased from HiMedia (India). All other chemicals used were of analytical grade.
Yeast Strain and Growth Conditions
The S. cerevisiae strains used in this study are as follows: BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and str4Δ (BY4742; Mat α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Str4::KanMX4); str4Δ Om45-GFP (BY4742; Mat α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 str4Δ::Hygro OM45-GFP::His3MX6); str4Δ Pcup1-GPX1 (BY4742; Mat α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 str4Δ::Hygro Pcup1-HA-GPX1::natNT2); str4Δ PGPD1-GPX1 (BY4742; Mat α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 str4Δ::Hygro PGPD1-HA-GPX1::natNT2); str4Δ Pcup1-TSA1 (BY4742; Mat α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 str4Δ::Hygro Pcup1-HA-TSA1::natNT2); str4Δ PGPD1-TSA1 (BY4742; Mat α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 str4Δ::Hygro PGPD1-HA-TSA1::natNT2). str4Δ and BY4742 strains were procured from the EUROSCARF (European S. cerevisiae archive for functional analysis) deletion collection (EUROSCARF, Institute of Microbiology, Johann Wolfgang Goethe-University Frankfurt, Frankfurt, Germany), and Om45 GFP or promoter strain were prepared as described elsewhere (36). Pre-cultures of wild type and str4Δ were prepared by growing a 5-ml yeast culture in YPD medium for 12–14 h. These cultures grown overnight were washed three times with sterile water and then resuspended in 10 ml of synthetic minimal media supplemented with amino acids (except methionine and serine) to an OD (600 nm) of 0.1. To study the effect of exogenously added homocysteine and its derivatives on yeast growth, homocysteine (5 mm), AdoHcy (600 μm), or AdoMet (600 μm), either singly or in combination, was added from freshly prepared stock solutions, and cells were grown on a rotary shaker at room temperature at 130 rpm. Yeast growth was monitored at 600 nm using a spectrophotometer (Eppendorf, BioPhotometer). 500 μm glutathione was added to satisfy the cysteine auxotrophy of the str4Δ strain. Intracellular homocysteine levels were determined by HPLC-FD method as described elsewhere (22).
RNA Isolation and Northern Blot Analysis
Total RNA from S. cerevisiae was isolated using glass bead lysis followed by the hot phenol method (37). For Northern blot analyses, the method of Ausubel et al. (37) was followed. Briefly, RNA was electrophoresed on 1.2% (w/v) denaturing formaldehyde gel, blotted on to nylon membrane (Hybond-N+; Amersham Biosciences), and immobilized by UV cross-linking. Gene-specific primers probes were designed and synthesized (Center for Genomic Applications, New Delhi, India). For gene expression analysis, specific probe primers were designed and synthesized by the Center for Genomic Applications. The amplified products were gel-purified using a Qiagen kit (Qiagen, Germany) according to the manufacturer's protocol. Radioactive [α-32P]dCTP was incorporated into a probe product by random primed labeling method using NEBLOT kit (New England Biolabs) according to the manufacturer's instructions. Prehybridization and hybridization were performed at 65 °C in rapid Hyb buffer (GE Healthcare). Signals were obtained using phosphorimager and quantification was done using ImageQuant software.
Microarray Experiment
Labeling, Hybridization, Data Analysis, and Bioinformatics
For microarray experiments, 10 μg of RNA was used to make double-stranded cDNA using a microarray cDNA synthesis kit (Roche Applied Science). The double-stranded cDNA was purified using a microarray target purification kit (Roche Applied Science) according to the manufacturer's protocol. Purified cDNA was concentrated using a SpeedVac and dissolved in 4 μl of 18 megohms of RNase-free water (Sigma). The purified cDNA was then used to prepare Cy-3/Cy-5 (Amersham Biosciences)-labeled cRNAs using microarray RNA target synthesis kit T7 (Roche Applied Science). Hybridization solution was prepared by mixing hybridization buffer (DIG Easy Hyb; Roche Applied Science), salmon testis DNA, and yeast tRNA (both from Sigma). The labeled cRNAs (control and treated) were pooled together, denatured at 65 °C, and applied onto cDNA microarray slides (yeast 6.4k) procured from the Microarray Center, Clinical Genomics Center, University Health Network (Toronto, Ontario, Canada). To exclude dye-specific bias, a dye-swap design was performed for each condition. The slides were scanned at a resolution of 10 μm using 70–100% laser power using a GenePix 4000A microarray scanner (Molecular Devices), and data were acquired using photomultiplier (PMT) settings in the range of 450–550. Image analysis for each array was done using the GenePix Pro 6.0 software (Molecular Devices), which produces fluorescence intensity pairs (cy3 and cy5) for each gene. After image acquisition, individual data spots on each microarray were visually inspected for size, signal-to-noise ratio, background level, and uniformity. Data for each spot were corrected for background, and the data from treated and reference samples were normalized to the total intensity. The genes showing 1.8-fold or more change in expression in dye-swap replicate microarrays were considered differentially expressed. Hierarchical clustering was performed using TIGR multiexperiment viewer software (MeV version 4.0). MIPS classification tool was used to understand the basic function and process of genes, and iPATH was used for analysis of metabolic pathways. DAVID (david.abcc.ncifcrf.gov) was used for identifying enriched biological processes in differentially expressed genes after Bonferroni correction for multiple hypotheses testing.
Intracellular Reactive Oxygen Species (ROS) Detection
The generation of intracellular reactive oxygen species in yeast in the presence of homocysteine and its derivatives was quantified by determining the incorporation of the fluorescent dye dihydroethidium (DHE, Invitrogen) using a flow cytometer (Guava EasyCyte, Millipore). Control and treated cells were harvested by centrifugation, washed, resuspended in milliQ water, and incubated in the dark with 10 μm DHE at room temperature for 1 h. This dye is normally cell-permeant and nonfluorescent, but upon oxidation it is converted to the fluorescent derivative ethidium and becomes impermeant (38, 39). Hydrogen peroxide, a known generator of ROS, was used as a positive control. Cells were treated with 1 mm hydrogen peroxide for 1 h and then incubated with DHE. The fluorescence intensity of >5000 cells was acquired for each sample by flow cytometry with an excitation at 488 nm and emission at 520 nm. Background fluorescence intensity was set based on a control sample, and higher fluorescence-containing cells were counted as DHE-positive cells. Fluorescence images of cells were also acquired using inverted epifluorescence microscope (TE200-U, Nikon).
Unfolded Protein Response Reporter Assay
To check for unfolded protein response, which is a hallmark of endoplasmic reticulum stress, a UPR reporter assay was performed. For this, the cells were transformed with pJC005 (kindly provided by Dr. Peter Walter), a 2-μm plasmid carrying the lacZ gene under the control of the UPRE from the KAR2 promoter (a chaperon that is overexpressed during endoplasmic reticulum stress). To compare lacZ expression in WT yeast versus str4 mutant yeast under quiescent or ER stress conditions, the β-galactosidase activity was measured from lysates using β-galactosidase assay kit (Promega). Alternatively, yeast cells treated with homocysteine and its derivatives were harvested and permeabilized by a drop of chloroform and 0.01% SDS. β-Galactosidase activity was measured from lysates using 2-nitrophenyl-β-d-galactopyranoside as a substrate and detected as an increase in absorbance at 420 nm.
Assessment of Mitochondrial Function and Integrity
The charged cationic green dye rhodamine 123 (Invitrogen) was used to assess mitochondrial membrane potential as described previously (40). Rh123 dye has been extensively used to detect the changes in ΔΨm in mammalian and yeast cells (40). Rh123 stains mitochondria directly, without passage through endocytotic vesicles and lysosomes, and distributes electrophoretically into the mitochondrial matrix depending upon change in mitochondrial membrane potential. Briefly, cells were harvested by centrifugation, and 1 ml of the cell suspension was loaded with 2 μm Rh123 (Molecular Probes) for 30 min. Fluorescence intensity was measured by flow cytometry, (Guava EasyCyte, Millipore) with excitation at 480 nm and emission at 530 nm. Cells displaying a defined threshold difference of green fluorescence intensity were considered having compromised mitochondrial or functional integrity. For visualization of the mitochondrial network, yeast cells were transformed with mitochondrially localized GFP marker plasmid Pvt100U (provided generously by Westermann and Neupert, 41) and visualized using inverted epifluorescence microscope (TE200-U, Nikon).
Apoptosis Assays
Detection of Chromatin Fragmentation
Yeast cells were collected and resuspended in 70% (v/v) ethanol for brief fixation and permeabilization. The cells were then incubated with DAPI (1 mg/ml; Sigma) for 15 min in the dark, washed twice with water, and examined under a fluorescence microscope. Cell images were recorded from an inverted epifluorescence microscope (TE200-U, Nikon) under 100× objectives.
Annexin V Staining
Phosphatidylserine exposure at the membrane surface was detected by a FITC-coupled annexin V assay kit using ApoAlert kit (Clontech) as described previously (42). Briefly, yeast cells were washed in sorbitol buffer (1.2 m sorbitol, 0.5 mm MgCl2, 35 mm potassium phosphate, pH 6.8), digested with 5.5% glusulase (PerkinElmer Life Sciences) and 15 units/ml lyticase (Sigma) for 2 h at 28 °C. Cells were then harvested, washed in binding buffer (10 mm Hepes/NaOH, pH 7.4, 140 mm NaCl, 2.5 mm CaCl2; Clontech) containing 1.2 m sorbitol buffer. Protoplasts were stained with annexin V-FITC and propidium iodide for 20 min at room temperature. The cells were harvested, suspended in binding buffer containing sorbitol, applied to a microscopic slide, and observed under a fluorescence microscope.
Terminal Deoxynucleotidyltransferase dUTP Nick End Labeling (TUNEL) Assay
TUNEL analysis was performed as described previously (42). DNA strand breaks were demonstrated by TUNEL using the in situ cell death detection kit (Roche Applied Science). In brief, yeast cells were fixed with 3.7% (v/v) formaldehyde for 30 min at room temperature and washed three times with phosphate-buffered saline (PBS), and cell walls were digested with 5.5% glusulase (PerkinElmer Life Sciences) and 15 units/ml lyticase (Sigma) for 2 h at 28 °C. Cells were incubated in permeabilization solution (0.1% (v/v) Triton X-100 and 0.1% (w/w) sodium citrate) for 2 min on ice and rinsed twice with PBS. Cells were subsequently incubated with 20 μl of TUNEL reaction mixture, containing terminal deoxynucleotidyltransferase and FITC dUTP, for 90 min at 37 °C. Finally cells were washed twice with PBS containing 1.2 m sorbitol buffer. Cells were then placed under a coverslip on microscopic slide and observed under a fluorescence microscope.
RESULTS
Effect of Homocysteine and Its Derivative on Yeast Growth
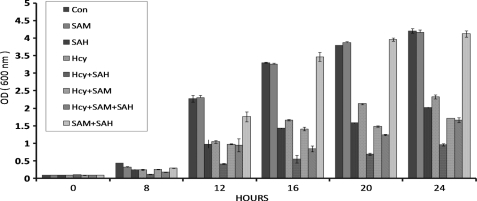
The objective of this study was to ascertain the cellular effects of homocysteine and its precursors on the yeast str4Δ deletion strain that lacks the enzyme necessary to catalyze the conversion of homocysteine to cystathionine in the trans-sulfuration process. For this, the str4Δ strain was grown in synthetic minimal media in the presence of homocysteine, AdoHcy, and AdoMet individually or in combination. Aliquots were withdrawn at indicated time points, and growth was monitored by measuring the absorbance at 600 nm. Addition of homocysteine resulted in a dose-dependent inhibition of yeast saturating around 5 mm homocysteine (data not shown). In a previous study, Christopher et al. (43) reported growth inhibition of str4Δ strain in the presence of 600 μm AdoHcy. Thus, we used these concentrations of homocysteine and AdoHcy for all further experiments. We found that addition of homocysteine and AdoHcy inhibited the growth of str4Δ strain, although addition of AdoMet had no effect on the growth (Fig. 2). We also found that when homocysteine and AdoHcy were added in combination, there was a synergistic effect on growth inhibition. Addition of AdoMet in combination with AdoHcy abrogated the inhibitory effect of AdoHcy, which is in agreement with the observation of Christopher et al. (43). However, AdoMet could not rescue the inhibitory effect of homocysteine. In fact, addition of AdoMet with homocysteine resulted in a slightly higher growth inhibition (at 20 and 24 h) as compared with homocysteine alone.
FIGURE 2.
Effect of AdoMet (SAM), AdoHcy (SAH), and homocysteine on yeast growth. Overnight grown cultures of the yeast strains str4Δ were diluted to an OD (600 nm) of 0.1 in synthetic minimal media supplemented with all amino acids except methionine and serine (see under “Experimental Procedures”). Yeast cells were grown in the presence or absence of 600 μm AdoHcy, 600 μm AdoMet, or 5 mm homocysteine. Aliquots of yeast cells were taken, and the growth was monitored at indicated time points at 600 nm. The data represent the means ± S.D. (n = 3).
To examine if the growth inhibition of str4Δ strain caused by homocysteine and AdoHcy was due to accumulation of homocysteine in the cells, the intracellular concentration of homocysteine was measured using HPLC after 16 h of incubation as the growth inhibition was maximal at this time point. We found that treating the cells with AdoHcy and AdoMet individually and/or in combination resulted in the increase in homocysteine levels as compared with controls to a similar extent (Table 1). Addition of AdoMet, AdoHcy, and AdoMet + AdoHcy increased the concentration of homocysteine from 1.46 pmol/107 cells in controls to 6.6, 6.4, and 7.1 pmol/107 cells, respectively. This suggests that AdoHcy-induced inhibition of str4Δ may not be due to an increase in intracellular homocysteine levels because its increase in the presence of AdoHcy is similar to that observed in the case of AdoMet or AdoMet + AdoHcy where growth inhibition was not observed. As expected, the intracellular homocysteine levels increased more than 18-fold (26.6 pmol/107 cells as compared with 1.46 pmol/107 cells in controls) after 16 h of homocysteine treatment (Table 1). Interestingly, addition of AdoMet and homocysteine in combination actually had a synergistic effect, and the level of intracellular homocysteine in this case was increased to about 39.7 pmol/107 cells.
TABLE 1.
Intracellular concentrations of homocysteine and methionine in str4Δ
Intracellular concentrations of homocysteine and methionine were measured at 16 h after exogenous addition of the homocysteine, AdoMet, AdoHcy, AdoMet + AdoHcy, Hcy + AdoMet, and methionine to yeast cells using HPLC-FD. The data represent the means of three experiments.
| Conditions | Homocysteine (pmol/107 cells) | Methionine (pmol/107 cells) |
|---|---|---|
| Control | 1.460 | 5.162 |
| AdoMet | 6.672 | 9.347 |
| AdoHcy | 6.470 | 10.173 |
| AdoMet + AdoHcy | 7.105 | 8.711 |
| Homocysteine | 26.672 | 7.3551 |
| Hcy + AdoMet | 39.739 | 16.279 |
| Methionine | 7.3402 | 40.220 |
Global Gene Expression Changes in str4Δ Cells in the Presence of Homocysteine and Its Derivatives
To understand the potential mechanism underlying yeast growth inhibition by homocysteine and its derivatives, we generated microarray gene expression profiles of yeast treated with these compounds alone or in combination. Because deletion of str4 might itself induce changes in the expression of certain genes, we also compared the gene expression profile of str4Δ with the wild type strain in an attempt to identify the genes and or the pathways that might be inherently affected due to deletion of str4. We found that a total of 386 genes were differentially expressed of which 191 were up-regulated and 195 down-regulated (supplemental Table 1). Functional annotation using Database for Annotation, Visualization, and Integrated Discovery (DAVID) revealed that the up-regulated genes were significantly over-represented in starch and sucrose metabolism (p < 9.07E-04) pathway, although amino acid metabolism, sulfur metabolism, and MAPK signaling pathway were significantly down-regulated.
Treatment of str4Δ strain with homocysteine and its derivatives singly or in combination resulted in the differential expression of 522, 824, 551, 427, and 519 genes in the presence of AdoMet, AdoHcy, AdoMet and AdoHcy, homocysteine, and homocysteine and AdoMet, respectively. The list of all the differentially transcribed genes is shown in supplemental Table 2.
Because both homocysteine and AdoHcy lead to growth inhibition of yeast, it is important to assess if the gene expression changes induced by homocysteine and AdoHcy are directly due to the addition of the metabolites or are an indirect result of the effects of cell growth. To address this, we looked at the cell cycle-regulated gene set from the Cycle database (44). This is a collection of cell cycle genes reported in literature. We compared this with the genes that were differentially expressed in the presence of homocysteine and AdoHcy in our study, and we found that the genes in the two sets were mostly dissimilar indicating that the majority (>90%) of genes that are differentially expressed in our study are independent of cell growth and therefore directly linked to homocysteine or AdoHcy stress (data not shown).
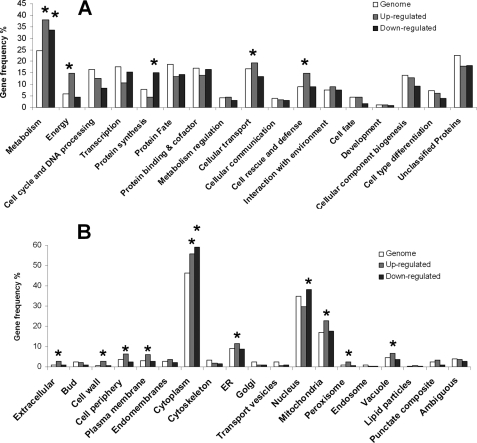
We then classified all the differentially expressed genes under all the conditions using MIPS classification. When compared with the yeast proteome, we found that the up-regulated genes were significantly over-represented in functional categories of metabolism (p < 3.07E-16), energy (p < 2.06E-19), cellular transport (p < 0.045), and cell rescue and defense (p < 1.88E-07), although the down-regulated genes were enriched in metabolism (p < 1.51E-10) and protein synthesis (p < 2.91E-15) (Fig. 3A). Using component-based classification, we found that the differentially expressed genes were enriched in various organelles like mitochondria, endoplasmic reticulum, nucleus, etc. (Fig. 3B). We also classified (functional and component-based) the differentially expressed genes obtained under each condition and found that the up-regulated genes were enriched in metabolism and energy under all conditions, whereas in the presence of AdoMet, AdoHcy, and AdoMet + AdoHcy, the down-regulated genes were also enriched in the functional category metabolism (supplemental Fig. 1). Interestingly, in the presence of AdoMet, AdoHcy, and AdoMet + AdoHcy, genes in the category of cell rescue and defense were up-regulated, although in the presence of homocysteine and Hcy +AdoMet genes in this category were down-regulated. This trend was also observed when the differentially expressed genes were segregated using hierarchical clustering (Fig. 4A). Some of the genes that were up-regulated in the presence of homocysteine were found to be down-regulated in the presence of AdoHcy and vice versa (Fig. 4A).
FIGURE 3.
Functional classification of differentially transcribed genes. Genes showing 2-fold or greater increase (up-regulated) or decrease (down-regulated) in response to homocysteine and its precursors were grouped in functional categories (A) and cellular compartments (B) according to the MIPS database. Categories and compartments that are significantly enriched (p value < 0.01) relative to the yeast proteome are marked with an asterisk.
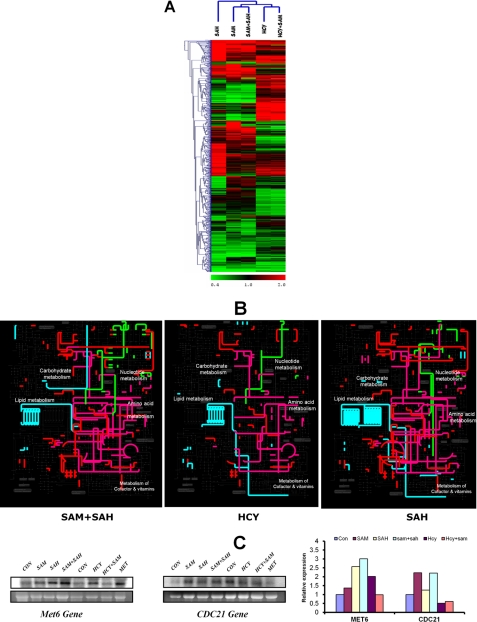
FIGURE 4.
Comparison of differentially expressed genes in str4Δ cells in presence of homocysteine and its precursors. A, heat map of the expression profiles of differentially expressed genes in the presence of AdoHcy (SAH), AdoMet (SAM), AdoMet + AdoHcy (SAM + SAH), homocysteine, Hcy + AdoMet (Hcy + SAM) were compared and analyzed by hierarchical clustering using TIGR Multiexperiment Viewer software (MeV version 4.0). The cluster of groups with similar expression profiles are shown at top and left. B, metabolic map of differentially expressed genes in the presence of AdoMet + AdoHcy, homocysteine, and AdoHcy. The differentially expressed genes were mapped on yeast metabolic network using the web-based tool iPATH. The subset of nucleotide metabolism gene is highlighted in green, lipid metabolism in light blue, amino acid metabolism in purple, and the rest in red. C, expression of Met6 and CDC21 genes was determined by Northern blotting. The cells were grown in the presence and absence of homocysteine and its precursors, and RNA was isolated from these cells. The lower panels in each blot represent gel loading of total RNA. Images were acquired using PhosphorImager and quantified by ImageQuant software.
We also mapped the differentially expressed genes on the yeast global metabolic network to visualize the global metabolic changes (Fig. 4B). A significantly different metabolic map was observed when the cells were grown in the presence of AdoMet + AdoHcy (where there was no inhibition) compared with cells grown in the presence of AdoHcy and homocysteine. For instance, lipid metabolism and nucleotide metabolism were different between AdoMet + AdoHcy and AdoHcy. The differential expression of genes like MET6 and CDC21 that are involved in nucleotide and one carbon metabolism were also checked using Northern experiments (Fig. 4C). We observed up-regulation of MET6 in the presence of AdoHcy and homocysteine. MET6 gene encodes for methionine synthase, an enzyme that converts homocysteine to methionine. Overexpression of this enzyme increases the conversion of homocysteine to methionine, and this might help cells to reduce its homocysteine levels. One of the key genes that regulate nucleotide synthesis is thymidylate synthase (CDC21), which is required for de novo biosynthesis of pyrimidine deoxyribonucleotides (Fig. 1). We found that thymidylate synthase expression is up-regulated only in the presence of AdoMet and AdoMet + AdoHcy, although it is down-regulated when the cells were treated with homocysteine and Hcy + AdoMet, where yeast growth inhibition was observed.
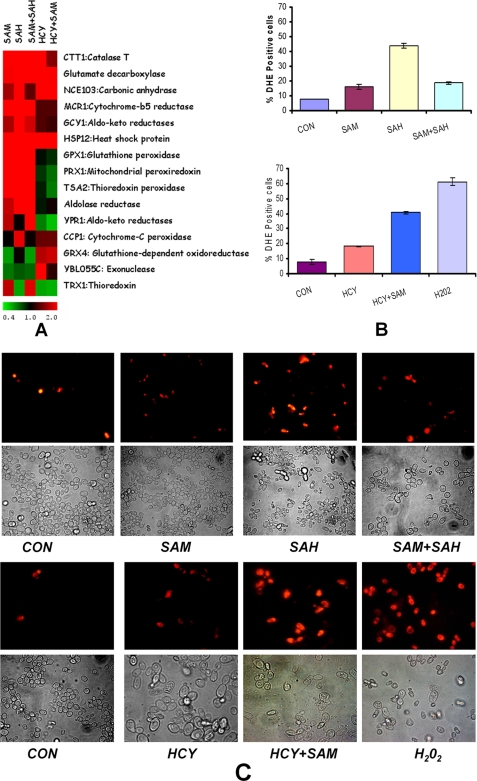
Downstream analysis also revealed enrichment for various biological processes in the differentially expressed genes under each of the five treatment conditions (supplemental Fig. 2). Of the various processes that were enriched, oxidation and reduction were found to be particularly striking because many processes in living organisms are driven by these reactions. A balance between pro- and anti-oxidant reagents that drive oxidation reduction reactions need to be maintained in living organisms. An excess of pro-oxidants, ROS, for example, causes oxidative stress that is considered to underlie development of several diseases, including cancer and cardiovascular and neurodegenerative disorders. The gene expression profiling thus provided a lead that ROS generation may underlie the pathogenicity of homocysteine and its derivatives in yeast. Furthermore, we used a web-based tool YEAS TRACT (45, 46) to ascertain if differentially expressed genes were enriched for particular transcription factors. This tool allows users to group a given gene list according to the transcription factors that are their potential regulators. We observed that the maximum numbers of differentially expressed genes were regulated under YAP1p (37.2%), a basic leucine zipper transcription factor required for oxidative stress tolerance. A hallmark of oxidative stress is the increased expression of antioxidant genes. From our transcriptomic data, we found that the expression of antioxidant genes like cytoplasmic glutathione peroxidase (GPX1), thioredoxin (TRX1), and catalase (CTT1) were up-regulated in presence of AdoMet, AdoHcy, and AdoMet + AdoHcy, although the expression of catalase was found to be up-regulated in the presence of homocysteine (Fig. 5A). However, most of the other antioxidant genes were not up-regulated in presence of homocysteine and Hcy + AdoMet. Cumulatively, the above results suggested that these metabolites might induce oxidative stress in yeast.
FIGURE 5.
Oxidative stress in str4Δ in the presence of homocysteine and its precursors. A, heat map of up-regulated oxidative stress response genes in the presence of homocysteine and its precursors was generated using Multiexperiment viewer software (MeV version 4.0b). B, flow cytometry measurement of ROS-positive cells. Yeast cells exposed to various conditions for 16 h or hydrogen peroxide for 1 h were harvested and incubated with DHE for 1 h and analyzed by Guava EasyCyte Flow cytometer. Error bars are representative of means ± S.D. (n = 3). C, fluorescence microscopy images were acquired using inverted Nikon epifluorescence microscope in similar experimental conditions. SAM, AdoMet; SAH, AdoHcy; CON, control.
Induction of Oxidative Stress
To ascertain if addition of AdoMet, AdoHcy, and homocysteine actually resulted in the generation of ROS, we determined intracellular reactive oxygen species using the fluorescent dye DHE. For this, cells were exposed to AdoMet, AdoHcy, homocysteine, AdoMet + AdoHcy, and Hcy +AdoMet up to 16 h, washed, and incubated with DHE for 1 h, and the fluorescence was measured using a flow cytometer. Cells treated with 1 μm hydrogen peroxide were considered as positive control. The percentage of DHE-positive cells, which is an indication of intracellular ROS, was found to be higher compared with control under all conditions. The highest percentage of DHE-positive cells (43.9%) was found in the presence of AdoHcy, which was considerably higher than the controls where 7–8% of the cells were found to be DHE-positive (Fig. 5B). Addition of AdoMet in combination with AdoHcy reduced the levels of DHE-positive cells to 18.6%, which was also similar to that obtained in presence of AdoMet alone. About 18% DHE-positive cells were found in the presence of homocysteine alone (Fig. 5B). However, addition of AdoMet in combination with homocysteine increased the percentage of DHE-positive cells (40%) thereby suggesting that AdoMet and homocysteine when added together lead to an increase in the generation of ROS. We also determined the levels of DHE-positive cells at different time points (12, 20, and 24 h) and observed similar results (data not shown). Similar results were obtained when the cells treated with DHE were visualized under a fluorescence microscope (Fig. 5C). We also checked if overexpression of antioxidant genes could rescue the growth defects of yeast. For this, we constructed the str4Δ strain with regulatable and constitutive overexpression of two important antioxidant genes, glutathione peroxidase (GPX1) and thiol-specific antioxidant (TSA1). The endogenous promoter of GPX1 and TSA1 genes were replaced by CUP1 and GPD promoters with HA epitope tag attached at the N terminus of genes (Fig. 6A) (47). The overexpression of enzyme was confirmed by Western blotting for HA epitope tag attached on endogenous proteins using anti-HA antibody. Both GPX1 and TSA1 genes were found to be overexpressed upon induction with copper sulfate within 4 h. The GPD promoter was also found to be strongly constitutively overexpressed (supplemental Fig. 3). However, when these overexpressed strains were grown in the presence of homocysteine and AdoHcy, they continued to show growth inhibition similar to that observed in str4Δ strains (supplemental Fig. 3). These results suggest that overexpression of at least these two antioxidant enzymes does not help to rescue yeast growth.
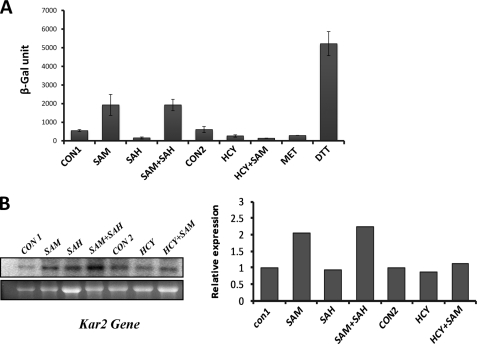
FIGURE 6.
ER stress response in str4Δ in the presence of homocysteine and its precursors. A, unfolded protein response in str4Δ cells. Cells were transformed with a plasmid (pJC005), a 2-μm plasmid carrying the lacZ gene under the control of the UPRE from the KAR2 promoter. Transformed cells were grown in respective conditions; cells were harvested at 16 h, and β-galactosidase activity was measured using 2-nitrophenyl-β-d-galactopyranoside as a substrate and detected as an increase in absorbance at 420 nm. B, expression of KAR2 gene determined by Northern blotting. The cells were grown in the presence and absence of homocysteine and its precursors for 16 h, and RNA was isolated from these cells. 20 μg of RNA was loaded per lane for Northern blot analysis. The lower panels in each blot represent gel loading of total RNA. Image was acquired using PhosphorImager and quantified by ImageQuant software. SAM, AdoMet; SAH, AdoHcy.
AdoHcy- and Homocysteine-mediated Growth Inhibition Is Not Due to ER Stress in str4Δ Cells
We had earlier shown using wild type yeast strain (BY4742) that homocysteine induced the expression of the KAR2 gene and cleaved hac1, which are characteristic hallmarks of ER stress (22). To ascertain if homocysteine or AdoHcy induced ER stress in str4Δ also, we assessed the induction of unfolded protein response, which is activated under ER stress. For this, we used a reporter plasmid PJC005 construct (a generous gift from the laboratory of Dr. Peter Walter) (48, 49) having lacZ gene fused to the UPRE. The UPRE element was originally identified from the promoter of KAR2, an ER chaperone protein that is up-regulated when Hac1 transcription factor binds to the UPRE. Thus, induction of lacZ was used as an index for the assessment of ER stress. Dithiothreitol (DTT), a known inducer of ER stress, was used as a positive control, and as expected, we observed significantly higher induction of lacZ in the presence of DTT (Fig. 6A). However, none of the three conditions where growth inhibition was observed (homocysteine, AdoHcy, or Hcy + AdoMet) resulted in the activation of lacZ (Fig. 6A). We also checked the expression of the ER stress-responsive gene KAR2 by Northern experiments. Consistent with the previous results, we did not find any induction of KAR2 gene expression in the presence of homocysteine, AdoHcy, or AdoMet + Hcy (Fig. 6B). These results indicate that ER stress might not be relevant under these conditions. Alternatively, it is also possible that deletion of STR4 gene by itself increases the cell's resistance to UPR activation, which could be a reason for our not detecting UPR in the presence of homocysteine or its derivatives. To check if the deletion of the str4 gene per se increases its resistance to UPR, we looked at the induction of lacZ in both wild type and str4Δ strain under quiescent and ER stress conditions (in the presence of DTT). We found that the induction of lacZ was similar in both the strains under quiescent or ER stress conditions (supplemental Fig. 4A). We also looked at the KAR2 gene expression using real time PCR in both wild type and str4Δ strains that were treated with DTT for 3 h. In both the strains, there was a similar induction of KAR2 gene expression (supplemental Fig. 4B). Furthermore, from the microarray analysis of wild type and the deletion strain, we compared the basal expression of the genes like IRE1, HAC1, and KAR2 that are involved in the ER stress pathway and found that the expression of these three genes in wild type were not significantly different from those in the str4Δ strains (supplemental Fig. 4C). These three facts clearly point to the fact that deletion of str4 does not have any impact on the ability of the cells to induce unfolded protein response.
AdoHcy and Homocysteine Leads to Mitochondrial Stress in str4Δ Cells
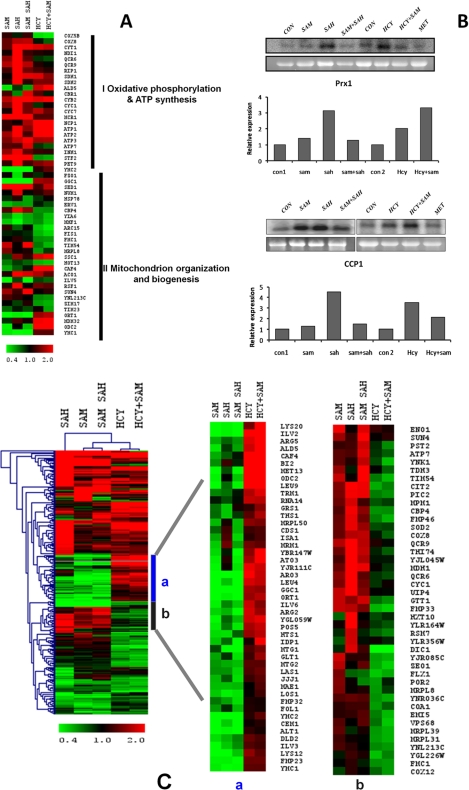
Our transcriptional profiling analysis showed that a significant proportion (p < 3.37E-05) of the total number of genes whose expression was modulated in the presence of the metabolites was mitochondrial genes. Out of 281 total mitochondrial genes, 22 genes representing oxidative phosphorylation and ATP synthesis-coupled electron transport were found to be affected (Fig. 7A). Most of these genes were found to be up-regulated in the presence of AdoHcy. Genes like ATP1, ATP2, and ATP3 were up-regulated both in the presence of AdoHcy and homocysteine, whereas NDI1, QCR6, QCR9, RIP1, CYB2, CYC1, CYC7, MCR1, INM1, and STF2 were up-regulated only in the presence of AdoHcy. Furthermore, we found 30 genes involved in mitochondrial organization and biogenesis to be affected among the total mitochondrial genes (Fig. 7A). Among these, we validated the up-regulation of two genes, mitochondrial cytochrome c peroxidase (CCP1) and peroxiredoxin (PRX1), by Northern blotting. Up-regulation of these genes in presence of AdoHcy and homocysteine suggests the induction of mitochondrial oxidative stress response under these conditions (Fig. 7B).
FIGURE 7.
Mitochondrial oxidative stress and transcriptomic response in the presence of AdoHcy and homocysteine. A, comparison of expression profiles of mitochondrial genes in str4Δ strain. Heat map of up-regulated mitochondrial genes in the presence of AdoMet (SAM), AdoHcy (SAH), AdoMet + AdoHcy, Hcy, and Hcy + AdoMet was generated using Multiexperiment viewer software (MeV version 4.0b). B, up-regulation of CCP1 and PRX1 genes in the presence of homocysteine and AdoHcy. The cells were grown in the presence and absence of these metabolites for 16 h, and RNA was isolated from these cells. 20 μg of RNA was loaded per lane for Northern blot analysis. The lower panels in each blot represent gel loading of ribosomal RNA. Image was acquired using PhosphorImager and quantified by ImageQuant software. C, effect of homocysteine and AdoHcy on mitochondrial gene expression. The expression profiles of up-regulated genes in the presence of AdoHcy, AdoMet, AdoMet + AdoHcy, Hcy, and Hcy +AdoMet were compared and analyzed by hierarchical clustering by using TIGR Multiexperiment Viewer software (MeV version 4.0). The portion of genes clustered with similar expression profiles are shown on the right. a, comparison of genes up-regulated in the presence of homocysteine and Hcy + AdoMet and with others. b, comparison of genes down-regulated in the presence of homocysteine and Hcy + AdoMet with others.
We also found that the gene expression profiles of the mitochondrial metabolism genes in the presence of AdoHcy and homocysteine were grossly different as many genes that were up-regulated in the presence of homocysteine were down-regulated in the presence of AdoHcy and vice versa (Fig. 7C). However, our gene expression profiling data suggested that homocysteine and AdoHcy might have a pronounced effect on mitochondrial genes. Thus, to explore the phenotypic effect of these changes on mitochondria, we checked mitochondrial membrane potential in the presence of homocysteine and AdoHcy. Changes in mitochondrial membrane potential play an important role in mitochondrial protein import, ATP generation, and lipid biogenesis. Mitochondrial transmembrane potential (ΔΨm) in str4Δ strain was evaluated using Rh123. Cells were analyzed using a flow cytometer. Sodium azide (NaN3; 20 mm), an electron transport inhibitor that decreases the mitochondrial membrane potential (ΔΨm) was used as a control. As expected, NaN3 treatment lead to hypopolarization of mitochondrial membrane potential (Fig. 8A). AdoHcy and homocysteine-treated cells also altered mitochondrial membrane potential as shown in Fig. 8B. However, no significant differences in mitochondrial membrane potential were observed in cells treated with AdoMet + AdoHcy (Fig. 8C). We observed a decrease in the fluorescence intensity in AdoHcy- and homocysteine-treated cells. We also checked the integrity of mitochondria in the presence of AdoHcy and homocysteine. For this, mitochondria were visualized by expressing mitochondrial matrix-localized fluorescent protein (subunit 9 of the F0-ATPase fused as mitochondrial presequence attached to the GFP protein) from plasmid Pvt100U (provided generously by the Westermann laboratory) (41). The str4Δ strain was transformed with plasmid Pvt100U, and cells were incubated in presence of AdoHcy and homocysteine for 12 h. Mitochondria were then visualized under a fluorescence microscope. In yeast, mitochondria are visualized as tubular networks as shown in the case of control cells (Fig. 8D). However, in the presence of both AdoHcy and homocysteine, we observed fragmentation of the mitochondrial network (Fig. 8D). In yeast, dynamic equilibrium of quality mitochondrial pool is maintained by the fusion and fission process to adjust its volume in accordance with cellular energy requirements, as mitochondrial autophagy (mitophagy) contributes both to selectively eliminate damaged mitochondria and reduce the amount of mitochondria.
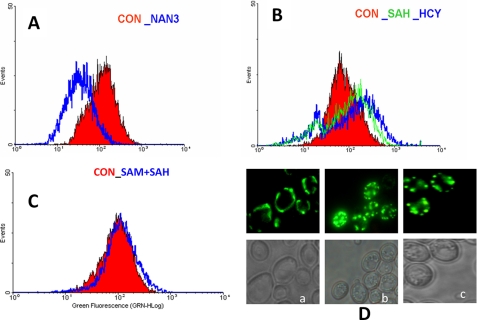
FIGURE 8.
AdoHcy and homocysteine induce mitochondrial membrane potential changes and mitochondrial fragmentation. Evaluation of mitochondrial membrane potential was in str4Δ cells. Yeast cells were grown for 16 h in minimal media containing AdoHcy (SAH) (600 μm), AdoMet (SAM) (600 μm), and homocysteine (5 mm) and after washing were incubated with Rh123 for 20 min and analyzed by flow cytometry. Overlay of fluorescence histograms was after staining with Rh123 dye. A, red histogram is for control, and blue is for sodium azide-treated yeast cells. B, red histogram is for control cells; green is for AdoHcy-treated cells, and blue is for homocysteine-treated cells. C, control cell is shown in red and AdoMet + AdoHcy is shown in blue. D, fragmentation of the mitochondrial network. Mitochondria were visualized using a mitochondria localized GFP protein. Mito-GFP protein-cloned plasmid (Pvt100U) constructs were transformed into str4Δ cells. Yeast cells expressing Mito-GFP were grown in the presence of AdoHcy and homocysteine for 12 h and observed under a fluorescence microscope. Panel a, control; panel b, AdoHcy; panel c, Homocysteine.
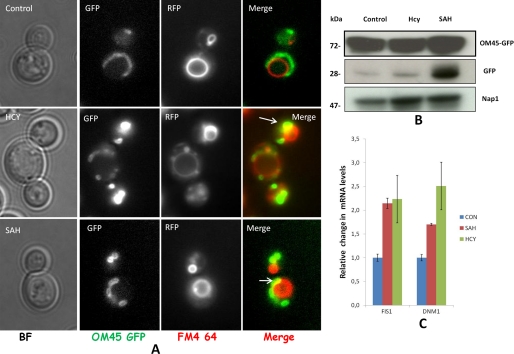
To see if treatment of homocysteine facilitates mitophagy, we used the Om45-GFP processing assay as described elsewhere (50) to monitor mitophagy. The Om45 protein localizes on the mitochondrial outer membrane, and following the uptake of mitochondria into the vacuole during mitophagy, Om45-GFP is degraded. However, the GFP is relatively stable within the vacuole and is often released as intact protein. The processed GFP then can be detected by immunoblotting as semiquantitative evidence for mitophagy. Similarly, the GFP fluorescence in vacuole can be observed in live cells using a fluorescence microscope. The OM45 protein was tagged with GFP as C-terminal tag in str4Δ strain and analyzed by Western blotting and fluorescence microscopy. In both AdoHcy- and homocysteine-treated cells, weak mitophagy response were observed as seen by some partial co-localization of FM4-64 vacuole staining with OM45, which was absent in WT type cells (Fig. 9A). Similarly, Western analysis shows the presence of free GFP that was found to be high after AdoHcy treatment as compared with homocysteine-treated cells (Fig. 9B). To check the mitochondrial dynamic equilibrium, we looked at the expression of mitochondrial fission genes. The mitochondrial fission process in yeast is regulated by two key proteins, Dynamin-related GTPase, Dnm1p, and mitochondrial FISsion protein 1, Fis1p. Dnm1p required for mitochondrial fission and morphology maintenance, assembles predominantly on the cytoplasmic face of mitochondrial tubules at sites at which division will occur, and associates with Fis1p. Yeast cells lacking DNM1 contain highly interconnected mitochondrial nets that are formed by ongoing fusion in the absence of fission activity although overexpression of DRP1 (mammalian homolog) in cells results in mitochondrial fragmentation (51, 52). We observed up-regulation of FIS1 and DNM1 in AdoHcy- and homocysteine-treated cells, which suggest that there is an increase in mitochondrial fission process in these cells (Fig. 9C). This also supports the earlier microscopic observation of more fragmented mitochondrial network in these conditions.
FIGURE 9.
Monitoring mitophagy response in str4Δ strain. A, co-localization of mtGFP and vacuoles. str4Δ strain containing OM45 protein tagged with GFP as the C-terminal tag was grown in the presence and absence of 5 mm homocysteine and 600 μm AdoHcy for 12 h. To stain the vacuole, FM4-64 (at a final concentration of 40 μm) was added for 2 h. Live cells were examined under a fluorescence microscope in their respective media to check proper vacuolar staining and Om45-GFP in live cells. Images were acquired with epifluorescence microscope and analyze by ImageJ software. B, detection of free GFP by immune blotting. Cells were collected at 12 h, lysed, and subjected to Western blot analysis with anti-GFP antibody or Nap1p antibody (loading control). The positions of full-length Om45-GFP and free GFP are indicated. C, mitochondrial fission gene expression. Cells were grown in the presence and absence of homocysteine and AdoHcy (SAH) for 16 h. RNA was isolated from these cells and subjected to real time PCR analysis using the gene-specific primers. IPP1 (inorganic pyrophosphatase) gene was used as the internal control (CON) gene for normalization. Error bars are representative of means ± S.D. (n = 2).
AdoHcy- and Homocysteine-induced Yeast Death Displays Characteristic Markers of Apoptosis
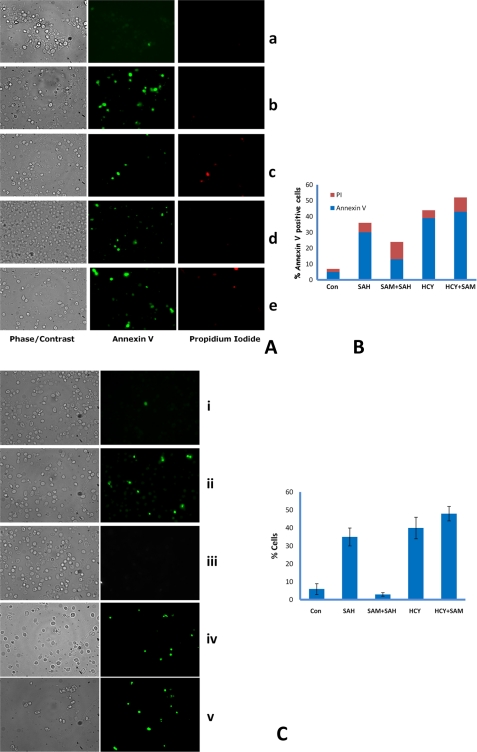
Mitochondria have been shown to play a key role in apoptosis, and mitochondrial fragmentation has been reported to activate the progression of apoptosis (53, 54). To check if addition of AdoHcy and homocysteine induce apoptosis in the str4Δ strain, we looked at morphological and biochemical features of apoptosis in str4Δ strain in the presence of homocysteine and its precursors. AdoHcy- and homocysteine-treated cells showed a fragmented nucleus when stained with DAPI in contrast to the untreated cells where the nuclei appeared as single round spots (data not shown). In mammalian and S. cerevisiae cells, exposure of phosphatidylserine at the outer leaflet of the plasma membrane is an early marker for apoptosis (42, 55). Hence, we examined the extent of phosphatidylserine externalization in str4Δ strain using the annexin-V-FITC assay. For this, yeast str4Δ cells were grown in the presence of AdoMet, AdoHcy, and homocysteine singly or in combination for 12 h to see early apoptotic cells, and spheroplasts of the cells were incubated with FITC-labeled annexin V antibody. In cells treated with homocysteine, AdoHcy, and Hcy + AdoMet, a significant proportion of the cells stained positive with annexin V indicating that homocysteine and AdoHcy can induce yeast apoptosis (Fig. 10, A and B). In contrast, few cells with FITC fluorescence were observed in untreated and cells treated with AdoMet + AdoHcy. To verify that binding of annexin V was not due to permeabilization of the plasma membrane during spheroplast preparation, cells were co stained with propidium iodide (PI).
FIGURE 10.
AdoHcy- and homocysteine-treated cells exhibit the typical markers of yeast apoptosis, DNA fragmentation, and annexin V staining. A, exposition of phosphatidylserine at the membrane surface. str4Δ cells were grown in minimal media containing AdoHcy (600 μm), AdoMet (600 μm), and homocysteine (5 mm), and cells were harvested after 12 h of incubation and stained with FITC-labeled annexin V for detection of exposed phosphatidylserine and propidium iodide for detection of damaged cells. Fluorescence and differential interference contrast micrographs showing normal annexin (−), PI (−) protoplasts, apoptotic cells as annexin (+) PI (−), and dead cells as annexin (+) PI (+). Panel a, control cells; panel b, AdoHcy; panel c, AdoMet + AdoHcy; panel d, homocysteine; panel e, Hcy + AdoMet. The results reported are for three independent experiments. B, quantitation of apoptotic cells; % of annexin V-positive cells were counted from >100 cells from different field views. C, DNA strand breakage visualized by TUNEL staining in str4Δ strain. str4Δ cells were grown in minimal media containing the following: AdoHcy (SAH) (600 μm), AdoMet (SAM) (600 μm), homocysteine (5 mm); cells were harvested after 12 h of incubation and stained for DNA strand breaks with TUNEL reaction containing fluorescently tagged dUTP. Quantitation of apoptotic cells; % positive cells were counted from >120 cells. Error bars are representative of means ± S.D. (n = 3). Left panel, phase contrast microscopy; right panel, fluorescence microscopy of the same cells. Panel i, control (Con) cells; panel ii, AdoHcy; panel iii, AdoMet + AdoHcy; panel iv, homocysteine; panel v, Hcy + AdoMet.
To further validate our results, we performed TUNEL assay for which cells were grown, fixed with formaldehyde, and subsequently digested with zymolyase to remove the cell wall. The cells were then stained with TUNEL reagent. Here, we also found that AdoHcy-, homocysteine-, and Hcy + AdoMet-treated cells were TUNEL-positive (Fig. 10C). In agreement with the annexin assay, even in TUNEL assay apoptotic cells were less (TUNEL-positive) after treatment with AdoMet + AdoHcy as compared with AdoHcy and homocysteine.
DISCUSSION
It is intriguing that a small amino acid like homocysteine is associated with a multitude of diseases. A single amino acid's association with so many disease conditions warrants its capacity to alter several basic cellular processes/pathways. Although several hypotheses have been proposed to explain the potentially harmful effects of homocysteine, a plausible mechanism that could explain how homocysteine could exert such a widespread effect remains unclear. We had earlier shown that growth inhibition of wild type yeast in the presence of homocysteine is not due to the generation of reactive oxygen species but is probably due to endoplasmic reticulum-associated stress (22).
Here, we studied the effects of homocysteine and its precursors on the yeast deletion strain str4Δ. We found that both homocysteine and AdoHcy inhibited the growth of str4Δ strain. In an earlier study, Christopher et al. (43) reported that addition of AdoHcy resulted in the inhibition of yeast growth, and AdoMet could abrogate the inhibitory effect of AdoHcy. They also proposed that the ratio of the concentrations of AdoMet to AdoHcy was important, and if the ratio exceeded a threshold value, there was no growth inhibitory effect. In agreement with their observations, we also found that the inhibitory effect of AdoHcy was abrogated in the presence of AdoMet. However, AdoMet failed to rescue the homocysteine-induced growth defect. In fact, addition of AdoMet marginally increased the growth inhibitory effect of homocysteine. Thus, although an increase in the AdoMet/AdoHcy ratio might be sufficient to overcome the inhibitory effect of AdoHcy, it is not sufficient to rescue the inhibitory effect of homocysteine. One of the potential reasons for this might be the ability of AdoMet to react directly with homocysteine forming methionine and AdoHcy. It has been shown by Vinci and Clarke (56) that the enzymes Sam4 and Mht1p are both capable of using (RS)-AdoMet as a methyl donor to convert homocysteine to methionine and AdoHcy. Thus, in effect, addition of AdoMet along with homocysteine might lead to an increase in the concentration of AdoHcy, reflecting a situation similar to addition of homocysteine, AdoMet, and AdoHcy in combination. Our growth curve studies also support this hypothesis. Furthermore, the concentration of methionine was found to be considerably higher in the presence of Hcy + AdoMet, indicating that addition of AdoMet with homocysteine results in the formation of methionine, in agreement with the observations of Vinci and Clarke (56). Furthermore, because the levels of homocysteine increased to a similar extent in the presence of AdoMet and AdoHcy, and AdoMet did not inhibit the growth of yeast, we believe that the inhibitory effect of AdoHcy may not be due to the accumulation of homocysteine. These results suggest that the mechanism by which AdoHcy and homocysteine mediate toxicity in yeast might be distinct. This observation was further supported by global gene expression profiling as transcriptional response of str4Δ strain showed differential transcriptomic response in the presence of homocysteine and its precursors.
Functional classification of the genes that were differentially expressed under each condition revealed that there were some key metabolic differences among the conditions studied. For instance, one of the key differences was observed in nucleotide metabolism genes. The gene thymidylate synthase (CDC21), which is required for de novo biosynthesis of pyrimidine deoxyribonucleotides, was up-regulated only in the presence of AdoMet and AdoMet + AdoHcy where growth inhibition were not present. However, this gene was down-regulated in the presence of homocysteine or Hcy +AdoMet. This gene catalyzes the conversion of deoxyuridine 5-monophosphate to deoxythymidine 5′-monophosphate and is essential for DNA synthesis. Down-regulation of TYMS genes could potentially lead to accumulation of UMP resulting in misincorporation of uracil in DNA, which may lead to replication defects. It has also been reported that disruption of methionine synthase in Schizosaccharomyces pombe leads to the accumulation of homocysteine, which is closely correlated with defective purine biosynthesis (57). Our results also points out that homocysteine and its derivative could disturb nucleotide metabolic processes in S. cerevisiae, which might play a role in their pathological mechanisms. We also observed changes in lipid metabolism, which could lead to imbalance of triglyceride and phospholipid synthesis and play an important role in disease conditions. A recent report by Malanovic et al. (58) also suggests that S-adenosyl-l-homocysteine hydrolase (Sah1) activity regulates AdoMet-dependent phosphatidylethanolamine methyltransferase reactions and thus has a major impact on cellular lipid homeostasis.
Our gene expression data revealed that several genes that were modulated in the presence of the metabolites are targets of YAP1p. Consistent with the gene expression profile, we found that there was accumulation of intracellular reactive oxygen species in cells under all conditions. However, there were prominent differences in the magnitude of ROS generated. AdoHcy was found to accumulate more reactive oxygen species compared with homocysteine-treated cells. In agreement with the growth studies, AdoMet reduced the burden of ROS in AdoHcy-treated cells. However, ROS accumulation in homocysteine-treated cells was found to increase in the presence of AdoMet, and this is probably due to the formation of AdoHcy in the presence of AdoMet and homocysteine as discussed above. However, overexpression of GPX1 or TSA1 did not help to rescue the growth defect. This suggests that probably the growth inhibition depends upon additional factors as oxidative stress alone could not account for cellular growth inhibition.
It has been suggested that altered mitochondrial membrane potential directly drives the formation of ROS (59–61). We observed altered mitochondrial membrane potential in AdoHcy- and homocysteine-treated cells as compared with control cells. Addition of AdoMet along with AdoHcy abrogated the impairment of mitochondrial membrane potential. Furthermore, we observed fragmentation of the mitochondrial network in the presence of AdoHcy and homocysteine, which indicates mitochondrial dysfunction under these conditions. Our gene expression profiling data suggest that a significant proportion of mitochondrial genes were differentially expressed in the presence of AdoHcy and homocysteine, but the gene expression data per se could not provide specific mitochondrial function or signaling pathways leading to mitochondrial dysfunction. However, using real time PCR, we observed up-regulation of FIS1 and DNM1 genes and weak mitophagy response in cells treated with AdoHcy and homocysteine, which suggest that the cells probably try to adjust the dynamic mitochondrial mass under these conditions. Earlier studies have shown that FIS1 overexpression promotes mitochondrial fragmentation, whereas FIS1 depletion produces interconnected mitochondrial nets (62, 63). Furthermore, overexpression of F1S1 has been reported to cause autophagy (64). Similarly DNM1 was identified in search of genes required for mitophagy as the dnm1Δ strain has been found to inhibit mitophagy (65, 66). Changes in the mitochondrial fission process and the presence of mitophagy in AdoHcy- and homocysteine-treated cells point to the fact that cells try to overcome mitochondrial stress possibly by adjusting mitochondrial mass and eliminating damaged mitochondria. However, further experiments are required to identify the events that trigger and control mitochondrial dynamics in the presence of AdoHcy and homocysteine.
Mitochondria play an important role in the apoptotic cascade (67). In fact, the fragmentation of the mitochondrial network is considered to be an early step in mammalian apoptosis, although its actual function, if any, remains unclear (68, 69). We indeed found the presence of apoptotic cells under the conditions of exogenous AdoHcy and homocysteine. Interestingly, AdoMet in combination with AdoHcy (AdoMet + AdoHcy) abrogated the AdoHcy-mediated apoptosis as evidenced by the dramatic reduction of apoptotic cells in the presence of AdoMet + AdoHcy. However, AdoMet along with homocysteine (Hcy + AdoMet) increased the presence of apoptotic cells. The increased apoptotic cells in the presence of Hcy + AdoMet support the presence of higher reactive oxygen species seen in earlier experiments.
Homocysteine has been shown to induce ER stress in various studies, including our previous work using wild type yeast strain (22, 70–72). ER stress induces adaptive cell signaling known as unfolded protein response, as a result of which ER-localized chaperones and proteins implicated in the ER-associated degradation pathway are induced. However, if the ER stress cannot be alleviated, it culminates into apoptosis (73, 74). In this study, we found that although both wild type and str4 deletion strains have similar basal unfolded protein responses in normal and under ER stress conditions, induction of unfolded protein response was not observed in the presence of AdoHcy and homocysteine in the deletion mutant, suggesting that apoptosis under these conditions might not be mediated through ER stress. Interestingly, we found unfolded protein response when the cells were treated with AdoMet and AdoMet + AdoHcy, the two conditions where there was no growth inhibition, again pointing to the fact that in the str4 mutant UPR may not be associated with yeast growth. However, the reason for induction of UPR in the presence of AdoMet is not clear. Because AdoMet is a universal methyl donor, the increase in the concentration of intracellular AdoMet might result in imbalances in methylation or post-translational modification of proteins in the ER. It has also been reported that AdoMet inhibits lipopolysaccharide-induced gene expression via modulation of histone methylation (75). Because AdoMet is involved in multiple pathways, it is possible that under these experimental conditions AdoMet induces ER stress via pathways that are independent of homocysteine. However, further experiments are needed to understand how AdoMet levels are linked to ER stress in the yeast str4 mutant strain.
In this study we have shown that addition of homocysteine and AdoHcy leads to growth inhibition in the yeast deletion strain that lack the enzyme str4. Although both homocysteine and AdoHcy inhibit the growth of yeast, addition of AdoMet abrogates the effects of AdoHcy but does not reverse the inhibitory effect of homocysteine. The results obtained with the str4Δ strain are also different from the wild type strain studied earlier wherein oxidative stress was not observed, although ER stress induction was observed in contrast to this study using the mutant strain where we did not find any induction of ER stress in the presence of homocysteine. These results indicate that homocysteine might exert different effects depending on the cell type, thereby affecting different stress signaling pathways. Our studies using wild type (22) and the str4Δ strain point to the fact that homocysteine could induce a varied response depending on the flux of homocysteine metabolism. Thus, the results obtained in this study may be valid only in tissues where the expression of str4, a homolog of human cystathionine β-synthase, is reduced or absent, e.g. vascular tissues.
Our study also indicates that the imbalance in homocysteine metabolism has a major effect on mitochondrial functions and results in the overexpression of the fission genes FIS1 and DNM1, which could lead to an increase in autophagy. Recently, in cell culture studies mitochondrial dysfunction has been shown to play a significant role in a complex disorder like neurological and cardiovascular diseases (76–80). The loss of mitochondrial functions leads to release of pro-apoptotic molecules that activate caspases and other cell death effectors (81). Although both AdoHcy and homocysteine were found to induce mitochondrial dysfunction, oxidative stress, and induction of apoptosis in cells, it was not clear how the AdoHcy-induced effect could be abrogated by addition of AdoMet. Further investigation is needed to understand how AdoMet, AdoHcy, and homocysteine regulate pro- and anti-apoptotic pathways in molecular terms. Also, studies using mammalian cells are necessary to confirm these findings.
Supplementary Material
Acknowledgments
We thank Farhan Mohammad for help with the microarray experiments, Anita Goel with flow cytometry experiments, and Dr. Beena Pillai for providing yeast strains. We are also thankful to Sumit kumar Nema and Faiz Ahmad, for their help in yeast growth assay and HPLC experiments. We sincerely thank Dr. Peter Walter and laboratory members and Dr. Benedikt Westermann and laboratory members for providing UPRE (PJC005) and mtGFP (Pvt100U) plasmids.
This work was supported in part by Grant CMM 0018/NWP 0034 from the Council of Scientific and Industrial Research, India.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and Tables 1 and 2.
- AdoMet
- S-(5′-adenosyl)-l-methionine chloride
- AdoHcy
- S-(5′-adenosyl)-l-homocysteine
- Hcy
- homocysteine
- ER
- endoplasmic reticulum
- ROS
- reactive oxygen species
- DHE
- dihydroethidium
- PI
- propidium iodide
- UPR
- unfolded protein response
- UPRE
- unfolded protein-response element.
REFERENCES
- 1. Kumar J., Garg G., Kumar A., Sundaramoorthy E., Sanapala K. R., Ghosh S., Karthikeyan G., Ramakrishnan L., Sengupta S. (2009) Circ. Cardiovasc. Genet. 2, 599–606 [DOI] [PubMed] [Google Scholar]
- 2. Verhoef P., Stampfer M. J., Buring J. E., Gaziano J. M., Allen R. H., Stabler S. P., Reynolds R. D., Kok F. J., Hennekens C. H., Willett W. C. (1996) Am. J. Epidemiol. 143, 845–859 [DOI] [PubMed] [Google Scholar]
- 3. Mills J. L., McPartlin J. M., Kirke P. N., Lee Y. J., Conley M. R., Weir D. G., Scott J. M. (1995) Lancet 345, 149–151 [DOI] [PubMed] [Google Scholar]
- 4. Applebaum J., Shimon H., Sela B. A., Belmaker R. H., Levine J. (2004) J. Psychiatr. Res. 38, 413–416 [DOI] [PubMed] [Google Scholar]
- 5. van Guldener C., Stehouwer C. D. (2003) Clin. Chem. Lab. Med. 41, 1412–1417 [DOI] [PubMed] [Google Scholar]
- 6. McCully K. S. (1996) Nat. Med. 2, 386–389 [DOI] [PubMed] [Google Scholar]
- 7. Clarke R., Daly L., Robinson K., Naughten E., Cahalane S., Fowler B., Graham I. (1991) N. Engl. J. Med. 324, 1149–1155 [DOI] [PubMed] [Google Scholar]
- 8. Starkebaum G., Harlan J. M. (1986) J. Clin. Invest. 77, 1370–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Austin R. C. (2009) Antioxid. Redox. Signal. 11, 2279–2287 [DOI] [PubMed] [Google Scholar]
- 10. Outinen P. A., Sood S. K., Liaw P. C., Sarge K. D., Maeda N., Hirsh J., Ribau J., Podor T. J., Weitz J. I., Austin R. C. (1998) Biochem. J. 332, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sengupta S., Wehbe C., Majors A. K., Ketterer M. E., DiBello P. M., Jacobsen D. W. (2001) J. Biol. Chem. 276, 46896–46904 [DOI] [PubMed] [Google Scholar]
- 12. Majors A. K., Sengupta S., Willard B., Kinter M. T., Pyeritz R. E., Jacobsen D. W. (2002) Arterioscler. Thromb. Vasc. Biol. 22, 1354–1359 [DOI] [PubMed] [Google Scholar]
- 13. Zinellu A., Sotgia S., Scanu B., Pintus G., Posadino A. M., Cossu A., Deiana L., Sengupta S., Carru C. (2009) Atherosclerosis 206, 40–46 [DOI] [PubMed] [Google Scholar]
- 14. Sundaramoorthy E., Maiti S., Brahmachari S. K., Sengupta S. (2008) Proteins 71, 1475–1483 [DOI] [PubMed] [Google Scholar]
- 15. Sharma P., Kumar J., Garg G., Kumar A., Patowary A., Karthikeyan G., Ramakrishnan L., Brahmachari V., Sengupta S. (2008) DNA Cell Biol. 27, 357–365 [DOI] [PubMed] [Google Scholar]
- 16. Eberhardt R. T., Forgione M. A., Cap A., Leopold J. A., Rudd M. A., Trolliet M., Heydrick S., Stark R., Klings E. S., Moldovan N. I., Yaghoubi M., Goldschmidt-Clermont P. J., Farber H. W., Cohen R., Loscalzo J. (2000) J. Clin. Invest. 106, 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kanani P. M., Sinkey C. A., Browning R. L., Allaman M., Knapp H. R., Haynes W. G. (1999) Circulation 100, 1161–1168 [DOI] [PubMed] [Google Scholar]
- 18. Tanriverdi H., Evrengul H., Enli Y., Kuru O., Seleci D., Tanriverdi S., Tuzun N., Kaftan H. A., Karabulut N. (2007) Cardiology 107, 313–320 [DOI] [PubMed] [Google Scholar]
- 19. Cavalca V., Cighetti G., Bamonti F., Loaldi A., Bortone L., Novembrino C., De Franceschi M., Belardinelli R., Guazzi M. D. (2001) Clin. Chem. 47, 887–892 [PubMed] [Google Scholar]
- 20. Weiss N., Heydrick S. J., Postea O., Keller C., Keaney J. F., Jr., Loscalzo J. (2003) Clin. Chem. Lab. Med. 41, 1455–1461 [DOI] [PubMed] [Google Scholar]
- 21. Weiss N., Zhang Y. Y., Heydrick S., Bierl C., Loscalzo J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12503–12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar A., John L., Alam M. M., Gupta A., Sharma G., Pillai B., Sengupta S. (2006) Biochem. J. 396, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stipanuk M. H. (2004) Annu. Rev. Nutr. 24, 539–577 [DOI] [PubMed] [Google Scholar]
- 24. Chen P., Poddar R., Tipa E. V., Dibello P. M., Moravec C. D., Robinson K., Green R., Kruger W. D., Garrow T. A., Jacobsen D. W. (1999) Adv. Enzyme Regul. 39, 93–109 [DOI] [PubMed] [Google Scholar]
- 25. Rounds S., Yee W. L., Dawicki D. D., Harrington E., Parks N., Cutaia M. V. (1998) Am. J. Physiol. 275, L379–L388 [DOI] [PubMed] [Google Scholar]
- 26. Palella T. D., Schatz R. A., Wilens T. E., Fox I. H. (1982) J. Lab. Clin. Med. 100, 269–278 [PubMed] [Google Scholar]
- 27. Lin H. C., Yang C. M., Liu C. L., Hu M. L. (2008) Biofactors 34, 81–95 [DOI] [PubMed] [Google Scholar]
- 28. Simon J. A., Bedalov A. (2004) Nat. Rev. Cancer 4, 481–492 [DOI] [PubMed] [Google Scholar]
- 29. Lindquist S., Krobitsch S., Li L., Sondheimer N. (2001) Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krobitsch S., Lindquist S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1589–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Priault M., Camougrand N., Kinnally K. W., Vallette F. M., Manon S. (2003) FEMS Yeast Res. 4, 15–27 [DOI] [PubMed] [Google Scholar]
- 32. Kumar A., Harrison P. M., Cheung K. H., Lan N., Echols N., Bertone P., Miller P., Gerstein M. B., Snyder M. (2002) Nat. Biotechnol. 20, 58–63 [DOI] [PubMed] [Google Scholar]
- 33. Kumar A., Snyder M. (2001) Nat. Rev. Genet. 2, 302–312 [DOI] [PubMed] [Google Scholar]
- 34. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 35. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 36. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 37. Ausubel F. M., Brent R., Kingston R., Moore D., Seidman J., Smith J. A., Struhl K. (eds) (1994) Current Protocols in Molecular Biology, pp. 13.12.1–13.12.5, 4.9.1–4.9.19, John Wiley & Sons, Inc., New York [Google Scholar]
- 38. Budd S. L., Castilho R. F., Nicholls D. G. (1997) FEBS Lett. 415, 21–24 [DOI] [PubMed] [Google Scholar]
- 39. Ludovico P., Sansonetty F., Côrte-Real M. (2001) Microbiology 147, 3335–3343 [DOI] [PubMed] [Google Scholar]
- 40. Johnson L. V., Walsh M. L., Bockus B. J., Chen L. B. (1981) J. Cell Biol. 88, 526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Westermann B., Neupert W. (2000) Yeast 16, 1421–1427 [DOI] [PubMed] [Google Scholar]
- 42. Madeo F., Fröhlich E., Fröhlich K. U. (1997) J. Cell Biol. 139, 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Christopher S. A., Melnyk S., James S. J., Kruger W. D. (2002) Mol. Genet. Metab. 75, 335–343 [DOI] [PubMed] [Google Scholar]
- 44. Gauthier N. P., Jensen L. J., Wernersson R., Brunak S., Jensen T. S. (2010) Nucleic Acids Res. 38, D699–D702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Monteiro P. T., Mendes N. D., Teixeira M. C., d'Orey S., Tenreiro S., Mira N. P., Pais H., Francisco A. P., Carvalho A. M., Lourenço A. B., Sá-Correia I., Oliveira A. L., Freitas A. T. (2008) Nucleic Acids Res. 36, D132–D136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Teixeira M. C., Monteiro P., Jain P., Tenreiro S., Fernandes A. R., Mira N. P., Alenquer M., Freitas A. T., Oliveira A. L., Sá-Correia I. (2006) Nucleic Acids Res. 34, D446–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gross A., Pilcher K., Blachly-Dyson E., Basso E., Jockel J., Bassik M. C., Korsmeyer S. J., Forte M. (2000) Mol. Cell. Biol. 20, 3125–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. (2000) Cell 101, 249–258 [DOI] [PubMed] [Google Scholar]
- 49. Chen Y., Feldman D. E., Deng C., Brown J. A., De Giacomo A. F., Gaw A. F., Shi G., Le Q. T., Brown J. M., Koong A. C. (2005) Mol. Cancer Res. 3, 669–677 [DOI] [PubMed] [Google Scholar]
- 50. Kanki T., Kang D., Klionsky D. J. (2009) Autophagy 5, 1186–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Otsuga D., Keegan B. R., Brisch E., Thatcher J. W., Hermann G. J., Bleazard W., Shaw J. M. (1998) J. Cell Biol. 143, 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smirnova E., Shurland D. L., Ryazantsev S. N., van der Bliek A. M. (1998) J. Cell Biol. 143, 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kissová I., Plamondon L. T., Brisson L., Priault M., Renouf V., Schaeffer J., Camougrand N., Manon S. (2006) J. Biol. Chem. 281, 36187–36197 [DOI] [PubMed] [Google Scholar]
- 54. Pozniakovsky A. I., Knorre D. A., Markova O. V., Hyman A. A., Skulachev V. P., Severin F. F. (2005) J. Cell Biol. 168, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rimon G., Bazenet C. E., Philpott K. L., Rubin L. L. (1997) J. Neurosci. Res. 48, 563–570 [DOI] [PubMed] [Google Scholar]
- 56. Vinci C. R., Clarke S. G. (2007) J. Biol. Chem. 282, 8604–8612 [DOI] [PubMed] [Google Scholar]
- 57. Fujita Y., Ukena E., Iefuji H., Giga-Hama Y., Takegawa K. (2006) Microbiology 152, 397–404 [DOI] [PubMed] [Google Scholar]
- 58. Malanovic N., Streith I., Wolinski H., Rechberger G., Kohlwein S. D., Tehlivets O. (2008) J. Biol. Chem. 283, 23989–23999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Skulachev V. P. (1996) Q. Rev. Biophys. 29, 169–202 [DOI] [PubMed] [Google Scholar]
- 60. Korshunov S. S., Skulachev V. P., Starkov A. A. (1997) FEBS Lett. 416, 15–18 [DOI] [PubMed] [Google Scholar]
- 61. Ricci J. E., Gottlieb R. A., Green D. R. (2003) J. Cell Biol. 160, 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yoon Y., Krueger E. W., Oswald B. J., McNiven M. A. (2003) Mol. Cell. Biol. 23, 5409–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. James D. I., Parone P. A., Mattenberger Y., Martinou J. C. (2003) J. Biol. Chem. 278, 36373–36379 [DOI] [PubMed] [Google Scholar]
- 64. Gomes L. C., Scorrano L. (2008) Biochim. Biophys. Acta 1777, 860–866 [DOI] [PubMed] [Google Scholar]
- 65. Kanki T., Wang K., Baba M., Bartholomew C. R., Lynch-Day M. A., Du Z., Geng J., Mao K., Yang Z., Yen W. L., Klionsky D. J. (2009) Mol. Biol. Cell 20, 4730–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nowikovsky K., Reipert S., Devenish R. J., Schweyen R. J. (2007) Cell Death Differ. 14, 1647–1656 [DOI] [PubMed] [Google Scholar]
- 67. Cheng W. C., Leach K. M., Hardwick J. M. (2008) Biochim. Biophys. Acta 1783, 1272–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Youle R. J., Karbowski M. (2005) Nat. Rev. Mol. Cell Biol. 6, 657–663 [DOI] [PubMed] [Google Scholar]
- 69. Pereira C., Silva R. D., Saraiva L., Johansson B., Sousa M. J., Côrte-Real M. (2008) Biochim. Biophys. Acta 1783, 1286–1302 [DOI] [PubMed] [Google Scholar]
- 70. Outinen P. A., Sood S. K., Pfeifer S. I., Pamidi S., Podor T. J., Li J., Weitz J. I., Austin R. C. (1999) Blood 94, 959–967 [PubMed] [Google Scholar]
- 71. Werstuck G. H., Lentz S. R., Dayal S., Hossain G. S., Sood S. K., Shi Y. Y., Zhou J., Maeda N., Krisans S. K., Malinow M. R., Austin R. C. (2001) J. Clin. Invest. 107, 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dickhout J. G., Sood S. K., Austin R. C. (2007) Antioxid. Redox. Signal. 9, 1863–1873 [DOI] [PubMed] [Google Scholar]
- 73. Rasheva V. I., Domingos P. M. (2009) Apoptosis 14, 996–1007 [DOI] [PubMed] [Google Scholar]
- 74. Reimertz C., Kögel D., Rami A., Chittenden T., Prehn J. H. (2003) J. Cell Biol. 162, 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ara A. I., Xia M., Ramani K., Mato J. M., Lu S. C. (2008) Hepatology 47, 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang X., Su B., Lee H. G., Li X., Perry G., Smith M. A., Zhu X. (2009) J. Neurosci. 29, 9090–9103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Szabadkai G., Duchen M. R. (2009) Apoptosis 14, 1405–1423 [DOI] [PubMed] [Google Scholar]
- 78. Su B., Wang X., Zheng L., Perry G., Smith M. A., Zhu X. (2010) Biochim. Biophys. Acta 1802, 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liesa M., Palacín M., Zorzano A. (2009) Physiol. Rev. 89, 799–845 [DOI] [PubMed] [Google Scholar]
- 80. Ballinger S. W., Patterson C., Knight-Lozano C. A., Burow D. L., Conklin C. A., Hu Z., Reuf J., Horaist C., Lebovitz R., Hunter G. C., McIntyre K., Runge M. S. (2002) Circulation 106, 544–549 [DOI] [PubMed] [Google Scholar]
- 81. Brüne B. (2005) Antioxid. Redox. Signal. 7, 497–507 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.