Abstract
Sumoylation regulates a wide range of cellular processes. However, little is known about the regulation of the SUMO machinery. In this study, we demonstrate that two lysine residues (Lys-153 and Lys-157) in the C-terminal region of the yeast E2-conjugating enzyme Ubc9 are the major and minor autosumoylation sites, respectively. Surprisingly, mutation of Lys-157 (ubc9K157R) significantly stimulates the level of Ubc9 autosumoylation at Lys-153. The functional role of Ubc9 autosumoylation is exemplified in our findings that cell cycle-dependent sumoylation of cytoskeletal septin proteins is inversely correlated with the Ubc9 autosumoylation level and that mutation of the Ubc9 autosumoylation sites results in aberrant cell morphology. Our study elucidates a regulatory mechanism that utilizes automodification of the E2 enzyme of the sumoylation machinery to control substrate sumoylation.
Keywords: Cell Cycle, Enzymes, Eukaryote, Ubiquitin, Ubiquitination, Autosumoylation, SMT3, Septin, Sumoylation, Ubc9
Introduction
Signal transduction relies on post-translational modifications, such as acetylation, methylation, phosphorylation, carboxylation, glycosylation, ubiquitination, and sumoylation, to transmit information within and across cells (1). Post-translational modification of proteins by small ubiquitin-related modifier (SUMO)3 plays a major role in regulating diverse cellular processes, including cell cycle regulation, transcriptional regulation, cellular localization, maintenance of genome integrity, chromosome segregation, and protein stability (2–5). For example, sumoylation causes changes in the subcellular localization of RanGAP1 by targeting it to the nuclear pore complex (6, 7). Sumoylation counteracts ubiquitination and subsequently blocks the degradation of IκB, thereby negatively regulating NF-κB activation (8). Sumoylation stimulates the transcriptional activities of transcription factors such as p53 and HSF2 (9–11). By contrast, sumoylation represses the transcriptional activities of transcription factors such as LEF (12) and c-Myb (13). In budding yeast, sumoylation of bud neck-associated septin proteins (Cdc3, Cdc11, and Shs1) may play a role in the disassembly of the septin ring (14). Thus, SUMO conjugation is a key regulatory mechanism in gene expression and growth control pathways in both yeast and higher eukaryotes.
Sumoylation is a highly evolutionarily conserved pathway from yeast to humans. Mammals contain four SUMO family members, SUMO-1 to SUMO-4, whereas Saccharomyces cerevisiae has a single SUMO gene, SMT3. In the sumoylation pathway, SUMO is covalently attached to lysine residues of substrate proteins by a series of enzymatic reactions similar to ubiquitination (7, 15, 16). In yeast, Smt3 is first synthesized as a precursor and subsequently processed by Ulp1 (17), one of the specific ubiquitin-like proteases, to its mature form with a Gly-Gly motif at its C terminus. In an ATP- and Mg2+-dependent reaction, the C terminus of mature Smt3 forms a thioester bond with the cysteine residue at the active site of the E1-activating enzyme Aos1/Uba2. Next, Smt3 is transferred to the unique E2-conjugating enzyme Ubc9 and forms a thioester bond with the active cysteine residue by a transesterification reaction. At the final step, the C terminus of mature Smt3 forms a stable isopeptide bond with the ϵ-amino group of the target lysine residue on the substrate by the combined activity of Ubc9 and SUMO E3 ligases such as Siz1, Siz2, Mms21, and the meiotic E3 Zip3, which enhance SUMO transfer from Ubc9 to the target proteins (18). The target lysine residue is usually located within the consensus sequence ΨKXE (where Ψ denotes a large hydrophobic amino acid and X is any amino acid) (19). SUMO modification is reversible, and Smt3 can be removed from the substrate by a family of ubiquitin-like proteases including Ulp1 and Ulp2. Most of the genes (SMT3, AOS1, UBA2, UBC9, MMS21, and ULP1) involved in the SUMO conjugation pathway are essential for cell viability with notable exceptions of SIZ1, SIZ2, and ULP2.
For ubiquitination, cells express dozens of E2 and hundreds of E3 enzymes to achieve high substrate specificity (20). Compared with the ubiquitination machinery, the sumoylation machinery is much simpler. For sumoylation, the cell only carries a single E2 (Ubc9) and four E3 ligases (4, 18, 21, 22). Although a number of E3 ligases for specific sumoylation have now been identified, they are not essential for the transfer of SUMO to the target in vitro (23). By contrast, Ubc9 is required to connect all SUMO machinery components including the E1, SUMO, and the E3s at different sumoylation stages. It also plays a role in target recognition by binding to the consensus ΨKXE sequence often present in substrates (19). Therefore, Ubc9 appears to function as the major determinant in the sumoylation pathway.
Modification of mammalian Ubc9 with SUMO has been observed in in vitro sumoylation assays (24–26). In addition, sumoylation of Ubc9 was observed in yeast cells in an attempt to identify novel sumoylation targets (27–30). Previous studies by Knipscheer et al. (26) have uncovered that mammalian Ubc9 is sumoylated at Lys-14 and that this autosumoylation regulates target discrimination. The authors also identified Lys-153 as the sumoylation site of yeast Ubc9 (26). However, the function of yeast Ubc9 sumoylation remains unclear. Because the sumoylation sites in yeast and mammals are located near the C and N terminus of Ubc9, respectively, it is possible that other lysine residues in Ubc9 besides Lys-14 and Lys-153 may be subjected to sumoylation. It is also conceivable that autosumoylation of Ubc9 at separate sites may represent different regulatory functions.
In this study, by performing a detailed mutagenesis analysis, we found that yeast Ubc9 forms isopeptide bonds with Smt3 at Lys-153 (major site) and Lys-157 (minor site) both in vivo and in vitro. Mutation of both Lys-153 and Lys-157 to arginine (ubc9K153R,K157R) abolishes Ubc9 autosumoylation but contrastingly increases sumoylation of the target proteins, as demonstrated with bud neck-associated septin proteins.
EXPERIMENTAL PROCEDURES
Yeast Strains
All of the Saccharomyces cerevisiae strains used in this study were derived from BY4705 (MATα, ade2::hisG, his3 Δ200, leu2 Δ0, lys2 Δ0, met15 Δ, trp Δ63, ura3 Δ0) (31) and are listed in supplemental Table S1. To create gene deletion mutants, the cells were transformed with PCR products bearing a selection marker (KanMX or HphMX) flanked by sequences complementary to the genes to be deleted. To generate ubc9 mutant strains, the LEU2 plasmids encoding mutated Ubc9 were transformed to the UBC9 shuffle strain whose genomic UBC9 gene was replaced by the HphMX marker through PCR-mediated gene replacement (32, 33) and complemented by a centromeric URA3 plasmid encoding wild-type Ubc9. The URA3 plasmid encoding wild-type Ubc9 was then evicted from the cells by selecting the transformed cells on plates containing 5-fluoroorotic acid (34). To generate strains carrying C-terminally FLAG3-His6-tagged CDC3 and CDC11, the PCR-generated DNA fragment containing the FLAG3-His6 sequence and KanMX marker was integrated immediately downstream of the individual ORF at the original chromosomal locus by one-step gene targeting. Gene deletion and epitope tagging were confirmed by colony PCR and Western analysis.
Construction of Plasmids
To express Ubc9 and Smt3 recombinant proteins, the ORFs of UBC9 and SMT3 were amplified by PCR from S. cerevisiae genomic DNA using primers carrying a BamHI restriction site at the 5′-end and a HindIII restriction site at the 3′-end. Fragments were subsequently cloned into the Escherichia coli expression vector pQE30 (Qiagen) linearized with restriction endonucleases BamHI and HindIII. The rFc-hTOP1(110–125) fusion protein was prepared as described previously (25). pRS416-UBC9 and pRS415-UBC9 plasmids were gifts from Mary-Ann Bjornsti (35). The individual Ubc9 mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol and confirmed by DNA sequencing.
Synchronization of Yeast Cultures
Hydroxyurea (10 mg/ml) and nocodazole (15 μg/ml) were added to logarithmically growing cultures for 2 h to arrest cell growth at the S and G2/M phases, respectively.
Purification of Recombinant Proteins
Overexpression of His6-tagged yeast E1 in E. coli was done as described previously (36). For the preparation of His-tagged Ubc9 and Smt3, individual pQE vector-based plasmids containing His-tagged Ubc9 and Smt3, respectively, were transformed into Escherichia coli strain TOP10 (Invitrogen). The resulting transformants were cultured at 37 °C to mid-log phase in LB medium containing ampicillin (50 μg/ml). Each recombinant protein was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside to a final concentration of 1 mm, and the cells were cultured for an additional 3 h at 37 °C. The cells were harvested by centrifugation at 4,000 × g for 20 min and then resuspended in 1× SUMO binding buffer (20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 5 mm immidazole, 1 mm phenylmethylsulfonyl fluoride) for cell lysis by sonication. After sonication, the extracts were centrifuged at 13,000 × g for 30 min at 4 °C, and supernatants were loaded onto nickel-nitrilotriacetic acid columns (Qiagen). The columns were washed with binding buffer containing 20 mm imidazole and eluted with binding buffer containing 100 mm imidazole and 10% glycerol. The purity of the eluted protein was determined by 15% SDS-PAGE, followed by staining with Coomassie Blue. Purified proteins were stored at −70 °C until use. The recombinant rabbit Fc fusion protein, rFc-hTOP1(110–125) was expressed and purified as described (25).
Purification of Sumoylated Ubc9
Sumoylated Ubc9 was prepared by incubating 5 mg of Aos1-Uba2, 15 mg of Ubc9, 15 mg of Smt3, and 2 mm ATP in 300 ml of buffer containing 20 mm Hepes (pH 7.5), 5 mm MgCl2 for 4–6 h at 37 °C. Ubc9*SUMO and Ubc9 in the resulting reaction mixture were separated by ion exchange chromatography on Mono Q 5/50 column (GE Healthcare). Purified proteins were concentrated and stored at −80 °C.
Purification of Cdc3-FLAG3-His6
Yeast total extract from yeast strains containing Cdc3 with a C-terminal FLAG3 His6 tag was used for immunoprecipitation of Cdc3. Yeast total extract (1 mg) was mixed with 20 μl of anti-FLAG M2 affinity gel (Sigma) in 1× SUMO binding buffer (see above) and incubated for 2 h at 4 °C. The anti-FLAG gel beads with bound Cdc3 were washed three times with 1 ml of 1× SUMO binding buffer. Cdc3 was eluted with 500 μl of elution buffer containing 1 mg/ml triple-FLAG peptide (Sigma).
In Vitro Sumoylation Assay
Reaction mixtures of 1 μg of His6-Ubc9, 1 μg of His6-Smt3, 400 ng of His6-Aos1-Uba2-His6, and 1 μg of substrate protein were incubated at 37 °C for indicated time periods in 20 μl of buffer containing 20 mm Hepes (pH 7.5), 5 mm MgCl2, and 2 mm ATP. The reaction was terminated by adding 4× LDS sample buffer (Invitrogen) containing 100 mm DTT to assay the isopeptide-linked SUMO-Ubc9. All of the samples were heated at 65 °C for 10 min, separated on 4–12% Bis-Tris gels (Invitrogen), and transferred to 0.45 mm PVDF, followed by immunoblotting with appropriate antibodies. All of the experiments were repeated at least twice.
Preparation of Yeast Cell Extracts and Immunoblotting Analysis
Yeast total extracts were prepared according to a TCA precipitation method (32). Protein separation by SDS-PAGE and Western blotting were performed according to standard procedures. Antibodies used in this study were mouse anti-FLAG monoclonal antibody (1/5000; Sigma), goat anti-Ubc9 polyclonal antibody (1/2000; yC-19; Santa Cruz Biotechnology), mouse anti-Ubc9 polyclonal antibody (1/2000; homemade), rabbit anit-Ubc9 polyclonal antibody (1/5000; homemade), mouse anti-Smt3 polyclonal antibody (1/2000; homemade), and rabbit anti-Smt3 polyclonal antibody (1/5000; homemade). Primary antibodies were detected using corresponding horseradish peroxidase-conjugated secondary antibodies (1/5000; Santa Cruz Biotechnology). Final detection was performed using the ECL plus and ECL advanced Western blotting detection system (Millipore). The percentage of the substrate conjugated by Smt3 was determined by quantitative analysis.
Immunoprecipitation
Cells carrying indicated V5-tagged Ubc9 variants were collected by centrifugation, washed once, and resuspended with immunoprecipitation buffer (100 mm NaCl, 0.02% Triton X-100, 50 mm Tris-HCl, pH 7.5, plus protease inhibitors as described above). The cells were lysed by grinding in liquid nitrogen, and lysates were clarified by centrifugation (35,000 × g for 60 min at 4 °C). The clarified extracts were transferred to fresh tubes, and centrifugation was repeated. Aliquots (200 μl, containing ∼3 mg of total protein) of the second supernatants were used for immunoprecipitation. The cell extracts were then incubated with 20 μl of anti-V5-agarose affinity gel (Sigma). After incubation for 2 h at 4 °C with rocking, the beads with bound proteins were collected by low speed centrifugation and washed with 1 ml of immunoprecipitation buffer. Washing was repeated four more times. The beads with bound Ubc9 variants were resuspended with SDS-PAGE sample buffer, boiled, and analyzed by SDS-PAGE and immunoblotting with antibodies against Ubc9 and Smt3.
Examination of Cell Morphology
To observe the morphology of the cells, mid-log phase cells cultured for 5 h were washed once with PBS and viewed under a Zeiss Axioimager Z1 upright fluorescence microscope. Images of random fields of cells were acquired for each sample, visualized, and processed with Adobe Photoshop.
RESULTS
Modification of Yeast Ubc9 by Smt3
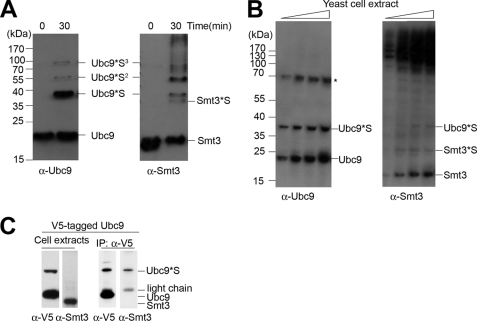
Utilizing an in vitro sumoylation assay, we observed in our previous work that human Ubc9 (hUbc9) is sumoylated (25). In addition, Knipscheer et al. (26) demonstrated that both human and yeast Ubc9 are sumoylated, and sumoylated human Ubc9 exhibits altered sumoylation activity toward different substrates. To further investigate the role of yeast Ubc9 sumoylation in vivo and in vitro, we established an in vitro sumoylation assay using purified yeast E1 (His6-Aso1-Uba2-His6), E2 (His6-Ubc9), and SUMO (His6-Smt3) proteins (supplemental Fig. S1). The appearance of gel bands in electrophoresis corresponding to the Ubc9-Smt3 complex provides evidence for isopeptide bond formation (Fig. 1A). Additionally, we observed in vivo Ubc9 autosumoylation in yeast cells under logarithmic growth (Fig. 1B) and confirmed the existence of sumoylated Ubc9 through immunoprecipitation of V5 epitope-tagged Ubc9 from yeast (Fig. 1C).
FIGURE 1.
Modification of yeast Ubc9 by Smt3. A, yeast Ubc9 autosumoylation in vitro. In vitro sumoylation assay was performed in the presence of 400 ng of Aos1-Uba2, 1 μg of Ubc9, 1 μg of Smt3, and 5 mm ATP at 37 °C for 0 or 30 min. The reaction mixture was subjected to SDS-PAGE, followed by immunoblotting with anti-Ubc9 antibody (left panel) and anti-Smt3 antibody (right panel). Ubc9*S, Ubc9*S2, and Ubc9*S3 denote Ubc9 conjugated to monomeric, dimeric, and trimeric forms of Smt3. B, yeast Ubc9 autosumoylation in vivo. Increasing amounts of extracts from log phase cell culture (A600 nm = 0.04, 0.08, 0.16, and 0.32) of yeast strain BY4705 were separated by SDS-PAGE, followed by immunoblotting with anti-Ubc9 antibody (left panel) and anti-Smt3 antibody (right panel). The asterisk denotes a nonspecific band. Smt3*S indicates the Smt3 dimer. C, immunoprecipitation (IP) of Ubc9 and SUMO-conjugated Ubc9. Whole cell lysates from cells expressing V5-tagged UBC9 were immunoprecipitated with anti-V5-agarose affinity gel and subjected to immunoblotting using anti-V5 and anti-Smt3 antibodies.
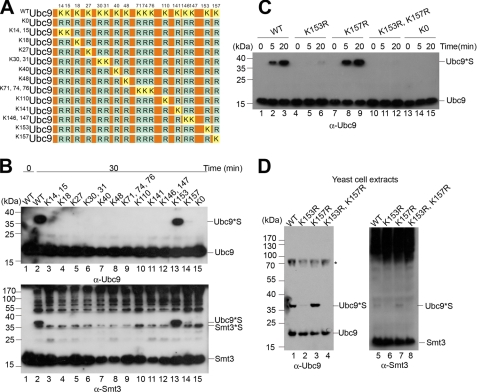
Ubc9 Is Sumoylated Only at Lys-153 and Lys-157
Using mass spectrometry analysis, it has been shown previously that yeast Ubc9 is sumoylated at Lys-153 (26). We then mutated Lys-153 of yeast Ubc9 to arginine and confirmed that Lys-153 is the major in vitro SUMO conjugation site. Interestingly, we found that K153RUbc9 still maintained a certain level of Ubc9 sumoylation (see below; Fig. 2C, lane 6), suggesting that Ubc9 contains at least two lysine residues for isopeptide bond formation with Smt3. In an effort to identify the sumoylation site(s), we performed a systematic mutational analysis of Ubc9. We mutated all of the lysine residues of Ubc9 to arginines and examined the autosumoylation and conjugating activity of this non-lysine variant (designated K0Ubc9; Fig. 2A and supplemental Fig. S2A). We found that no autosumoylated K0Ubc9 was detectable (Fig. 2B, lane 15, and supplemental Fig. S2B). To examine whether autosumoylation of K0Ubc9 is prevented because of a loss of E2 conjugating activity of this variant, we performed in vitro sumoylation assays. We used a synthetic polypeptide containing a short sequence of human topoisomerase 1 (amino acids 110–125) fused to the rabbit immunoglobulin Fc fragment [rFc-TOP(110–125)] as the substrate for the SUMO conjugation assay. This polypeptide sequence (FSSPPQIKDEPEDDGY) contains a lysine residue (underlined) within the consensus sequence for SUMO conjugation (25). SUMO-conjugated products of rFc-TOP1(110–125) can be detected by Western blotting analysis using an antibody against rabbit immunoglobulin. As shown in supplemental Fig. S2B, the rate of Smt3 conjugation to the synthetic substrate was significantly increased with K0Ubc9 as compared with the wild-type Ubc9, suggesting that nonsumoylated Ubc9 has a higher E2 conjugating activity.
FIGURE 2.
Ubc9 is sumoylated only at Lys-153 and Lys-157. A, yeast Ubc9 wild type (WTUbc9), non-lysine variant (K0Ubc9), and lysine reversion variants (K14,15Ubc9 to K157Ubc9). The positions of lysine residues, highlighted in yellow boxes, are shown on the top. The non-lysine Ubc9 variant has all endogenous lysine residues replaced by arginine, highlighted in blue boxes. B, SUMO conjugation of Ubc9 lysine reversion variants. In vitro sumoylation assay was performed in the presence of WT (lane 2) or indicated Ubc9 revertants (K14,15 to K0) (lanes 3–15). The Smt3-Ubc9 conjugate (Ubc9*S) was revealed by probing with anti-Ubc9 (upper panel) and anti-Smt3 antibodies (lower panel) in the immunoblotting analysis. Smt3*S, poly-Smt3 conjugate. C, SUMO conjugation of Ubc9 variants. In vitro sumoylation assay was conducted for WT or indicated Ubc9 variants for 0, 5, or 20 min, followed by immunoblotting with anti-yUbc9 antibody to reveal the SUMO-Ubc9 conjugate (Ubc9*S). Ubc9 variants containing single or double lysine-to-arginine mutations at Lys-153 and Lys-157 were studied in the sumoylation analysis. The K0Ubc9 variant was used as a nonsumoylation control. D, Ubc9 autosumoylation of Ubc9 mutants in vivo. Whole cell extracts from yeast with wild-type Ubc9 or Ubc9 variants were analyzed by SDS-PAGE, followed by immunoblotting with anti-Ubc9 antibody (left panel) and anti-Smt3 antibody (right panel). The asterisk denotes a nonspecific band.
We subsequently reintroduced single or multiple lysine residues on the non-lysine (K0Ubc9) background to generate Ubc9 variants containing between one and three lysine residues (designated as K14, 15Ubc9 to K157Ubc9; Fig. 2A). In vitro sumoylation reactions carried out with the purified Ubc9 revertants (supplemental Fig. S2A) showed a strong modification of Ubc9 for the variant K153Ubc9 containing a single lysine at Lys-153 (Fig. 2B, lane 13). In addition, a weak modification signal was detected for K157Ubc9, the single-lysine Ubc9 variant containing Lys-157 (lane 14). These data suggest that both Lys-153 and Lys-157 are the sumoylation sites of Ubc9, of which the former is the major site.
To further confirm that Lys-153 and Lys-157 are the only two sumoylation sites on Ubc9, we performed in vitro sumoylation assays with single-point mutation variants K153RUbc9, K157RUbc9, and the double-mutation variant K153R,K157RUbc9. As shown in Fig. 2C, no sumoylated Ubc9 was detected for K153R,K157RUbc9 (lanes 11 and 12). Furthermore, when the major sumoylation site Lys-153 was mutated to arginine, the level of autosumoylation was significantly reduced, although not fully abolished (lane 6). Interestingly, SUMO conjugation at Lys-153 is stimulated when Lys-157 is mutated (lanes 2 and 3 versus lanes 8 and 9).
To further investigate the sumoylation sites of Ubc9 in yeast cells, we constructed a UBC9 plasmid shuffle strain that contains the chromosomal UBC9 gene replaced by a drug resistance cassette (ubc9::HphMX) and a CEN URA3 plasmid with UBC9 under its endogenous promoter (pRS416-UBC9) to compensate for the chromosomal deletion. The UBC9 shuffle strain was then transformed with a CEN LEU2 plasmid carrying the coding sequence of either wild-type UBC9 or ubc9 mutants with lysine-to-arginine mutations at positions 153 and/or 157. The resulting yeast transformants were spotted onto selective medium supplemented with glucose. Each individual transformant was then replica-plated onto a plate containing 5-fluoroorotic acid to select for cells that have lost the URA3-marked plasmid (pRS416-UBC9). All of the yeast Ubc9 mutant strains were viable. Total cell extracts were prepared from individual transformants containing ubc9 mutants and subjected to immunoblotting analysis. In agreement with our in vitro studies, we found that a small fraction of mutant ubc9K153R was sumoylated, whereas no sumoylated Ubc9 was detected in the ubc9K153R,K157R extracts (Fig. 2D). Moreover, the in vivo sumoylation level for ubc9K157R was slightly increased compared with that of wild-type extracts (Fig. 2D; WT versus K157R). We further applied immunoprecipitation assay to isolate Ubc9 from yeast extracts and quantified the SUMO conjugates of Ubc9 variants (supplemental Fig. S3). We observed an increased amount of SUMO-conjugated Ubc9 for the K157R variant compared with the wild-type Ubc9 (supplemental Fig. S3B). Taken together, we conclude that mutation of the minor sumoylation site (Lys-157) to arginine increases Ubc9 sumoylation at Lys-153.
Ubc9 Autosumoylation Negatively Regulates Sumoylation in Vitro
To determine whether sumoylation at Lys-153 and Lys-157 residues of Ubc9 affects substrate sumoylation, we performed in vitro sumoylation assays using WTUbc9 and Ubc9 variants (K153RUbc9, K157RUbc9, and K153R,K157RUbc9) and a synthetic polypeptide rFc-TOP1(110–125) as the substrate for the SUMO conjugation assay. To measure the rate of SUMO conjugation on the substrate, we utilized a time course sumoylation assay and quantified the amount of accumulation of SUMO-modified substrates. As shown in supplemental Fig. S4, variants K153RUbc9 and K153R,K157RUbc9, both of which lack the major Ubc9 autosumoylation lysine site (Lys-153), significantly increased the rate of rFc-TOP1(110–125) sumoylation. This result indicates that Ubc9 autosumoylation may negatively regulate substrate sumoylation caused by either the reduced conjugating activity of the sumoylated Ubc9 or the competition for SUMO conjugation between Ubc9 and the substrate.
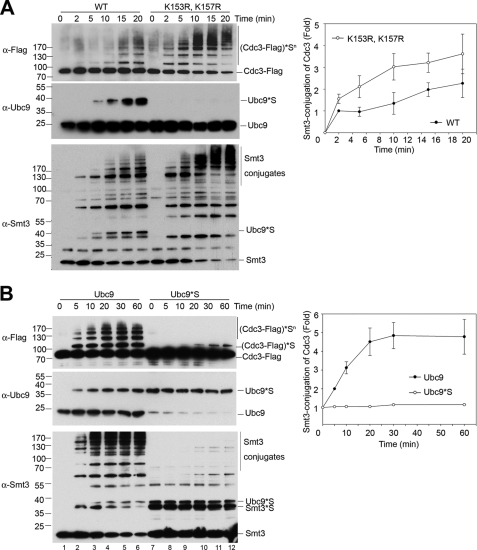
Ubc9 Autosumoylation Negatively Regulates Sumoylation of Septins in Vitro and in Vivo
Although the sumoylated human Ubc9 exhibits different sumoylation activity toward substrates in vitro (26), the physiological function of sumoylated Ubc9 remains unclear. Previous studies in budding yeast have shown that the evolutionarily conserved septin components (for example, Cdc3 and Cdc11) are the prominent sumoylation substrates, and SUMO conjugation of septins is required for septin ring formation during cytokinesis (14, 37). We therefore examined whether Ubc9 autosumoylation affects sumoylation of septins. The C-terminally FLAG-tagged Cdc3 was isolated from yeast cellular extracts by affinity purification with anti-FLAG antibody-conjugated agarose beads and subsequently incubated with wild-type Ubc9 or autosumoylation-deficient variant K153R,K157RUbc9. The rate of Cdc3 sumoylation by the K153R,K157RUbc9 variant was significantly increased compared with that by wild-type Ubc9 (Fig. 3A).
FIGURE 3.
Ubc9 autosumoylation negatively regulates septin sumoylation of septins in vitro. A, SUMO conjugating activity of the wild-type Ubc9 and the K153R,K157RUbc9 variant. In vitro sumoylation was performed in the presence of partially purified Cdc3-FLAG3-His6 as the substrate and either wild-type Ubc9 (WT; lanes 1–6) or K153R,K157RUbc9 variant (K153R,K157R; lanes 7–12) for the indicated times. Smt3-Cdc3, Smt3-Ubc9, and total Smt3 conjugates were analyzed by immunoblotting with anti-FLAG antibody (top panel), anti-Ubc9 antibody (middle panel), and anti-Smt3 antibody (bottom panel), respectively. (Cdc3-FLAG)*S, SUMO-conjugated Cdc3; Ubc9*S, SUMO-conjugated Ubc9. The results shown represent the means ± S.D. of four independent experiments. Smt3 conjugation of Cdc3 in wild-type Ubc9 was set to 1. B, SUMO conjugating activity of Ubc9 and Smt3-conjugated Ubc9. In vitro sumoylation was performed using either purified unmodified Ubc9 (lanes 1–6) or Smt3-conjugated Ubc9 (Ubc9*S; lanes 7–12) and partially purified Cdc3-FLAG3-His6 as substrate. Reaction mixtures were subjected to SDS-PAGE, followed by immunoblotting with anti-FLAG antibody (top panel), anti-Ubc9 antibody (middle panel), and anti-Smt3 antibody (bottom panel), respectively. The results shown are the means ± S.D. of three independent experiments. Smt3 conjugation of Cdc3 in wild-type Ubc9 was set to 1.
To further confirm the down-regulation effect of Ubc9 autosumoylation on sumoylation of septins, we performed in vitro sumoylation assays with purified Smt3-conjugated Ubc9. Large scale in vitro Ubc9 sumoylation was performed to generate the sumoylated Ubc9, which was then purified from the nonsumoylated Ubc9 by ion exchange chromatography (supplemental Fig. S5). Utilization of the sumoylated Ubc9 in SUMO conjugation of Cdc3 in vitro demonstrated that the sumoylated Ubc9 exhibited significantly lower activity (Fig. 3B).
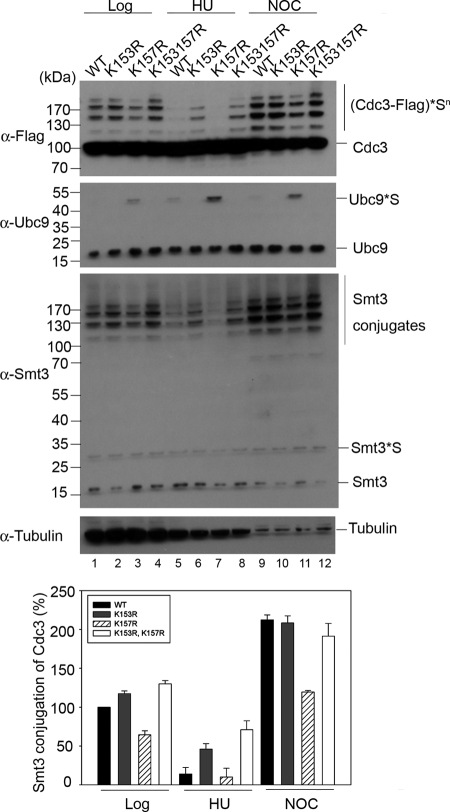
Next, we analyzed sumoylation levels of Cdc3 during progression of the cell cycle in Ubc9 lysine mutation strains by immunoblotting with anti-FLAG antibodies (Fig. 4, α-Flag). In log phase cells, the level of Cdc3 sumoylation is slightly increased in ubc9K153R (17%) and ubc9K153R,K157R (30%) strains, as compared with that in the UBC9WT strain (Fig. 4, α-Flag, lanes 1–4). By contrast, a 35% reduction in Cdc3 sumoylation was observed in ubc9K157R cells (Fig. 4, α-Flag, lane 3). In addition, the total cellular sumoylation pattern revealed by immunoblotting using anti-Smt3 antibody is similar to that of Cdc3 sumoylation, likely because of the predominant contribution of SUMO conjugates from septin components in yeast cells (Fig. 4, α-Smt3). Interestingly, the sumoylation levels of Cdc3 and total cellular sumoylation are inversely correlated with the observed level of Ubc9 autosumoylation, because the level of Ubc9 autosumoylation in the ubc9K157R mutant strain was elevated, and those of the ubc9K153R and ubc9K153R,K157R mutant strains were reduced or no longer detectable (Figs. 2D and 4, α-Ubc9).
FIGURE 4.
Ubc9 autosumoylation negatively regulates septin sumoylation of septins in vivo. Cdc3 sumoylation in vivo is shown. Cdc3 sumoylation was analyzed in cells grown to log phase (lanes 1–4) or arrested in S or G2/M phase with HU (lanes 5–8) and nocodazole (NOC) (lanes 9–12), respectively. Whole cell extracts were analyzed by SDS-PAGE, followed by immunoblotting. All of the strains contain a C-terminally FLAG-tagged CDC3. Anti-FLAG antibody was used to probe SUMO-Cdc3 conjugates (upper panel; (Cdc3-FLAG)*Sn), anti-Ubc9 antibody was applied to probe Ubc9 and SUMO-Ubc9 (Ubc9*S), anti-Smt3 antibody was applied to probe overall Smt3 conjugates, and anti-tubulin antibody was applied to provide the loading control (bottom panel). Bottom histogram, relative sumoylation level normalized to the level for the WT Ubc9 strain at the log phase. The results shown are the means ± S.D. of three independent experiments. Smt3 conjugation of Cdc3 in wild-type Ubc9 cells at log phase was set to 100%.
We further examined Cdc3 sumoylation in the S phase by treatment with hydroxyurea (HU). Under HU treatment, Cdc3 sumoylation was decreased in the wild-type and ubc9K157R mutant strains (Fig. 4, α-Flag, log versus HU), consistent with previous findings of reduced SUMO conjugation of Cdc3 in the S phase of the cell cycle (14, 37). By contrast, the sumoylation levels of the ubc9K153R and ubc9K153R,K157R mutant strains showed only mild reduction (Fig. 4, α-Flag, lanes 6 and 8), which could indicate that these mutant cells might confer improper cell cycle control. We next examined Cdc3 sumoylation in cells arrested at the G2/M phase with nocodazole treatment. In accordance with previous observations (14, 37), all of the wild-type and ubc9 mutant strains showed increased Cdc3 sumoylation compared with the levels of the log phase and HU-treated cells. However, we found that the level of Cdc3 sumoylation in the ubc9K157R mutant strain is relatively lower. Furthermore, the relative levels of Cdc3 sumoylation in the four yeast strains under HU and nocodazole treatments correlate inversely with the amount of Ubc9 SUMO conjugates (Fig. 4, α-Flag versus α-Ubc9).
In addition to Cdc3, we also examined the sumoylation levels of another septin component, Cdc11 (supplemental Fig. S6). Similar to the pattern observed for Cdc3 sumoylation, we also observed an inverse correlation between the level of Cdc11 sumoylation and Ubc9 autosumoylation (supplemental Fig. S6). Taken together, our in vivo and in vitro results strongly support the notion that Ubc9 autosumoylation negatively regulates sumoylation of septins.
Autosumoylation of Ubc9 Is Important for Maintaining Cell Morphology
To investigate the functional importance of Ubc9 sumoylation in regulating sumoylation of septins and cell cycle progression in yeast cells, the phenotypes of strains carrying different Ubc9 variants (UBC9WT, ubc9K153R, ubc9K157R, and ubc9K153R,K157R) were analyzed. Microscopic examination (Fig. 5A) of cells after 5 h of growth in liquid culture at the permissive temperature (30 °C) reveals that mutant cells exhibit aberrant cell morphology similar to the cytoskeletal protein septin mutants (39). The percentage of cells with aberrant morphology was calculated (≥400 cells were counted for each analysis). The results showed that UBC9WT cells exhibit only 0.2% aberrant morphology, whereas ubc9K153R and ubc9K153R,K157R mutant cells have ∼5.1% aberrant morphology, and ubc9K157R cells have ∼3.5% aberrant morphology (Fig. 5B).
FIGURE 5.
Autosumoylation of Ubc9 is important for maintaining cell morphology. A, micrographs showing the cell morphology of cells with Ubc9 variants (UBC9WT, ubc9K153R, ubc9K157R and ubc9K153R,K157R). The arrows indicate cells with aberrant morphology. B, bar graph showing calculated percentages of cells with morphology defects (≥400 cells were counted for each analysis). UBC9WT cells exhibit only 0.2% aberrant morphology, ubc9K153R and ubc9K153R,K157R mutant cells have ∼5.1% aberrant morphology, and ubc9K157R cells have ∼3.5% aberrant morphology. The results shown are the means ± S.D. of two independent experiments.
DISCUSSION
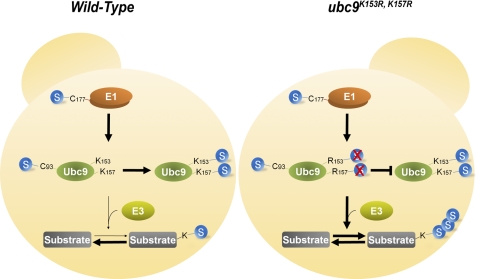
Sumoylation is an important post-translational modification that regulates the function of proteins in a number of cellular pathways. However, how sumoylation itself is regulated is not well understood. In this study, we utilized a mutagenesis approach to identify Ubc9 sumoylation sites and investigated the effect of Ubc9 autosumoylation on septin sumoylation in S. cerevisiae. We observed that septin sumoylation is negatively regulated by autosumoylation of Ubc9 (Fig. 6).
FIGURE 6.
Model for the regulatory role of Ubc9 autosumoylation in SUMO conjugation. SUMO conjugation in cells with or without Ubc9 autosumoylation is depicted. SUMO conjugation is achieved through successive thioester bond formation to the catalytic cysteine of E1 (Aos1/Uba2) and to the catalytic cysteine of E2 (Ubc9) and subsequent isopeptide bond linkage between SUMO and target lysines. Autosumoylation of Ubc9 diverts cellular Ubc9 to form the SUMO-Ubc9 isopeptide bond conjugate inactive in further substrate sumoylation. When the level of Ubc9 autosumoylation is elevated under conditions such as hydroxyurea treatment, cellular SUMO conjugation is down-regulated (Wild-type; left panel). When Ubc9 autosumoylation is reduced or abolished, cellular sumoylation level is elevated because of increased SUMO conjugation activity by the nonsumoylated Ubc9 (ubc9K153, R157R; right panel).
SUMO modification typically occurs at the lysine residue within the consensus sequence ΨKXE (see above) that is directly recognized by Ubc9. However, Ubc9 may also interact with its substrates at multiple positions and recognize alternative sites to modify nonconsensus sequences. A number of nonconsensus SUMO conjugations were reported including, for instance, E2–25K (40), Cdc3 (14), proliferating cell nuclear antigen (41), human PML (42), and human Ubc9 (26, 43). As revealed in our study, yeast Ubc9 autosumoylation at the C-terminal amino acid residues Lys-153 and Lys-157 appears to follow the pattern of nonconsensus SUMO conjugation, and the resulting modification regulates the function of Ubc9. Indeed, as we have demonstrated in our in vivo and in vitro analysis, Ubc9 autosumoylation at Lys-153 and Lys-157 negatively affects SUMO conjugation to septins. The fact that we obtained a similar result with the artificial substrate rFc-TOP1(110–125) suggests that Ubc9 autosumoylation likely negatively affects SUMO conjugation to other substrates as well (supplemental Fig. S4).
Interactions between Ubc9 and Smt3 include the formation of a covalent bond, either through lysine or through the catalytic cysteine and noncovalent Smt3 binding to the SUMO-binding motif of Ubc9. Covalently linked Smt3 could also bind noncovalently to another Ubc9 molecule. Previous studies have shown that the noncovalent interaction between Ubc9 and Smt3 enhances the conjugation activity of Ubc9. In the noncovalent Ubc9-Smt3 complex, the C-terminal glycine of Smt3 is located near the C-terminal Lys-153 and Lys-157 of Ubc9 (44–46). Therefore, one possible explanation for the negative effect of Ubc9 autosumoylation is that Smt3 conjugation at Lys-153 or at Lys-157 hinders the noncovalent association of Ubc9 and SUMO at the SUMO-binding motif necessary for SUMO conjugation. In addition, the E1-activating enzyme was found to compete directly with Smt3 for Ubc9 binding because E1 binds to Ubc9 near the SUMO-binding motif (24). SUMO conjugation at Lys-153 or Lys-157 could therefore also compromise the interaction between Ubc9 and E1. Thus, we propose that Ubc9 autosumoylation at Lys-153 and Lys-157 physically obstructs its interactions with Smt3- and E1-activating enzyme, resulting in its reduced SUMO conjugation activity. This hypothesis is further supported by results obtained with Ubc9-Smt3 fusion proteins. Fusion of Smt3 to the C terminus of Ubc9 severely reduced the conjugating activity of Ubc9 in vitro, whereas fusion of Smt3 to the N terminus of Ubc9 did not exhibit any obvious effect (data not shown).
Although lysines 153 and 157 of Ubc9 are located close to each other, SUMO modifications at these two positions are likely not redundant but appear to generate different functional consequences. As demonstrated in our mutational study, the substitution of Lys-157 by arginine to deactivate SUMO conjugation at Lys-157 increased Ubc9 autosumoylation at Lys-153 compared with wild-type Ubc9 and decreased the SUMO conjugating activity of Ubc9. On the contrary, deactivation of SUMO conjugation at Lys-153 by arginine mutation resulted in decreased Ubc9 autosumoylation and elevated Ubc9 SUMO conjugating activity. Lys-153 appears to be the major SUMO conjugation site, and SUMO conjugation to Lys-157 inhibits SUMO conjugation to Lys-153. Interestingly, a similar self-inhibitory effect was observed for the three human Ubc9 autosumoylation lysine residues, namely Lys-14, Lys-49, and Lys-153 (43), and mutation of Lys-153 also increased human Ubc9 autosumoylation (26). Despite the different lysine residues involved in Ubc9 autosumoylation, this self-modification process is conserved from yeast to humans and functions to regulate both Ubc9 autosumoylation and SUMO conjugating activities.
Protein sumoylation is highly dynamic and tightly regulated. Conjugation of SUMO requires the concerted coordination of E1-activating (Aos1/Uba2), E2-conjugating (Ubc9), and E3-ligating (Siz1 and Siz2) enzymes, and deconjugation depends on the action of SUMO isopeptidases (Ulp1 and Ulp2). SUMO-conjugating and -deconjugating enzymes must collaboratively maintain homeostasis of cellular sumoylation. Although SUMO isopeptidases reduce the level of sumoylation through deconjugation of SUMO linkages, other mechanisms also exist that modulate SUMO conjugation. Sumoylation has been shown to vary during the cell cycle as evidenced by the findings that specific sets of SUMO conjugates accumulate during mitosis and that the sumoylation level endures a dramatic decrease during the G1 and S phases (37). In this study, we found that the Ubc9 autosumoylation level is increased during hydroxyurea treatment and that sumoylation of septins is regulated by Ubc9 autosumoylation at different stages of cell cycle progression. We propose that during S phase, in addition to the deconjugation activity of isopeptidases, Ubc9 autosumoylation is raised to inhibit the SUMO conjugating activity of Ubc9, thus resulting in attenuation of septin sumoylation. During the G2/M phase, SUMO conjugating activity is contrastingly elevated because of a combined decrease in SUMO deconjugation and Ubc9 autosumoylation, resulting in elevated sumoylation of septins. The importance of Ubc9 autosumoylation in mediating SUMO conjugation to septins is evidenced by the finding that a mutation at Lys-153 or Lys-157 of Ubc9 causes a cell morphological phenotype.
Automodification is commonly present in post-translational modification pathways and plays an important role in the regulation of cell function (47, 48). For example, Cdc34, an ubiquitin E2 enzyme, is autoubiquitinated in the presence of SCF (Skp1, Cdc53/Cull, and F-box protein), inhibiting its ubiquitin conjugating activity to transfer ubiquitin to separate Cdc34 molecules (49). Knipscheer et al. (26) recently reported that mammalian Ubc9 autosumoylation regulates its target discrimination. In addition, other sumoylation components such as E1-activating enzyme (Aos1/Uba2), E3-ligating enzyme (Siz1/Siz2), and isopeptidases (SENPs) have been reported to be modified by SUMO (38). However, the functional consequence of SUMO modification of these enzymes is currently not understood.
In this study, we found that sumoylated yeast Ubc9 negatively regulates its activity to transfer SUMO to substrates. The effect of Ubc9 autosumoylation is summarized and diagrammatically shown in Fig. 6. Our work reveals an important self-inhibitory mechanism for controlling the SUMO conjugating activity of Ubc9, which greatly expands our understanding of the functional role of automodification of the key regulatory components of the post-translational modification systems.
Supplementary Material
Acknowledgments
We thank Drs. Leroy F. Liu, Chung Wang, and Rey-Huei Chen for stimulating discussions; Drs. Marry-Ann Bjornsti and Ting-Fang Wang for providing plasmids; Drs. Leroy F. Liu, Juu-Chin Lu, and Heiko Kuhn for manuscript editing; and Chiao-En Chen, Ruei-Yuan Tsai, and Hsiao-Ling Chiang for preparation of antibodies.
This work was supported by National Science Council in Taiwan Grant NSC98-2311-B-001-008-MY3 and by the Institute of Molecular Biology, Academia Sinica.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S6.
- SUMO
- small ubiquitin-related modifier
- TOP
- topoisomerase
- HU
- hydroxyurea
- NOC
- nocodazole
- DTT
- dithiothreitol
- SENPs
- sentrin-specific proteases
- PML
- promyelotic leukaemia.
REFERENCES
- 1. Bossis G., Melchior F. (2006) Mol. Cell 21, 349–357 [DOI] [PubMed] [Google Scholar]
- 2. Pichler A., Melchior F. (2002) Traffic 3, 381–387 [DOI] [PubMed] [Google Scholar]
- 3. Gill G. (2004) Genes Dev. 18, 2046–2059 [DOI] [PubMed] [Google Scholar]
- 4. Johnson E. S. (2004) Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 5. Hay R. T. (2005) Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 6. Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. (1997) Cell 88, 97–107 [DOI] [PubMed] [Google Scholar]
- 7. Matunis M. J., Coutavas E., Blobel G. (1996) J. Cell Biol. 135, 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Desterro J. M., Rodriguez M. S., Hay R. T. (1998) Mol. Cell 2, 233–239 [DOI] [PubMed] [Google Scholar]
- 9. Rodriguez M. S., Desterro J. M., Lain S., Midgley C. A., Lane D. P., Hay R. T. (1999) EMBO J. 18, 6455–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gostissa M., Hengstermann A., Fogal V., Sandy P., Schwarz S. E., Scheffner M., Del Sal G. (1999) EMBO J. 18, 6462–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goodson M. L., Hong Y., Rogers R., Matunis M. J., Park-Sarge O. K., Sarge K. D. (2001) J. Biol. Chem. 276, 18513–18518 [DOI] [PubMed] [Google Scholar]
- 12. Sachdev S., Bruhn L., Sieber H., Pichler A., Melchior F., Grosschedl R. (2001) Genes Dev. 15, 3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bies J., Markus J., Wolff L. (2002) J. Biol. Chem. 277, 8999–9009 [DOI] [PubMed] [Google Scholar]
- 14. Johnson E. S., Blobel G. (1999) J. Cell Biol. 147, 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saitoh H., Sparrow D. B., Shiomi T., Pu R. T., Nishimoto T., Mohun T. J., Dasso M. (1998) Curr. Biol. 8, 121–124 [DOI] [PubMed] [Google Scholar]
- 16. Müller S., Ledl A., Schmidt D. (2004) Oncogene 23, 1998–2008 [DOI] [PubMed] [Google Scholar]
- 17. Li S. J., Hochstrasser M. (1999) Nature 398, 246–251 [DOI] [PubMed] [Google Scholar]
- 18. Schwartz D. C., Hochstrasser M. (2003) Trends Biochem. Sci. 28, 321–328 [DOI] [PubMed] [Google Scholar]
- 19. Sampson D. A., Wang M., Matunis M. J. (2001) J. Biol. Chem. 276, 21664–21669 [DOI] [PubMed] [Google Scholar]
- 20. Hershko A., Ciechanover A., Varshavsky A. (2000) Nat. Med. 6, 1073–1081 [DOI] [PubMed] [Google Scholar]
- 21. Melchior F. (2000) Annu. Rev. Cell Dev. Biol. 16, 591–626 [DOI] [PubMed] [Google Scholar]
- 22. Kerscher O., Felberbaum R., Hochstrasser M. (2006) Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 23. Wilkinson K. A., Henley J. M. (2010) Biochem. J. 428, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bencsath K. P., Podgorski M. S., Pagala V. R., Slaughter C. A., Schulman B. A. (2002) J. Biol. Chem. 277, 47938–47945 [DOI] [PubMed] [Google Scholar]
- 25. Yang M., Hsu C. T., Ting C. Y., Liu L. F., Hwang J. (2006) J. Biol. Chem. 281, 8264–8274 [DOI] [PubMed] [Google Scholar]
- 26. Knipscheer P., Flotho A., Klug H., Olsen J. V., van Dijk W. J., Fish A., Johnson E. S., Mann M., Sixma T. K., Pichler A. (2008) Mol. Cell 31, 371–382 [DOI] [PubMed] [Google Scholar]
- 27. Wohlschlegel J. A., Johnson E. S., Reed S. I., Yates J. R., 3rd (2004) J. Biol. Chem. 279, 45662–45668 [DOI] [PubMed] [Google Scholar]
- 28. Zhou W., Ryan J. J., Zhou H. (2004) J. Biol. Chem. 279, 32262–32268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Denison C., Rudner A. D., Gerber S. A., Bakalarski C. E., Moazed D., Gygi S. P. (2005) Mol. Cell Proteomics 4, 246–254 [DOI] [PubMed] [Google Scholar]
- 30. Hannich J. T., Lewis A., Kroetz M. B., Li S. J., Heide H., Emili A., Hochstrasser M. (2005) J. Biol. Chem. 280, 4102–4110 [DOI] [PubMed] [Google Scholar]
- 31. Brachmann R. K., Yu K., Eby Y., Pavletich N. P., Boeke J. D. (1998) EMBO J. 17, 1847–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Knop A. E., Arndt A. J., Raponi M., Boyd M. P., Ely J. A., Symonds G. (1999) Gene Ther. 6, 373–384 [DOI] [PubMed] [Google Scholar]
- 33. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 34. Boeke J. D., LaCroute F., Fink G. R. (1984) Mol. Gen. Genet. 197, 345–346 [DOI] [PubMed] [Google Scholar]
- 35. Jacquiau H. R., van Waardenburg R. C., Reid R. J., Woo M. H., Guo H., Johnson E. S., Bjornsti M. A. (2005) J. Biol. Chem. 280, 23566–23575 [DOI] [PubMed] [Google Scholar]
- 36. Cheng C. H., Lo Y. H., Liang S. S., Ti S. C., Lin F. M., Yeh C. H., Huang H. Y., Wang T. F. (2006) Genes Dev. 20, 2067–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takahashi Y., Iwase M., Konishi M., Tanaka M., Toh-e A, Kikuchi Y. (1999) Biochem. Biophys. Res. Commun. 259, 582–587 [DOI] [PubMed] [Google Scholar]
- 38. Bailey D., O'Hare P. (2004) J. Biol. Chem. 279, 692–703 [DOI] [PubMed] [Google Scholar]
- 39. Gladfelter A. S., Kozubowski L., Zyla T. R., Lew D. J. (2005) J. Cell Sci. 118, 1617–1628 [DOI] [PubMed] [Google Scholar]
- 40. Pichler A., Knipscheer P., Oberhofer E., van Dijk W. J., Körner R., Olsen J. V., Jentsch S., Melchior F., Sixma T. K. (2005) Nat. Struct. Mol. Biol. 12, 264–269 [DOI] [PubMed] [Google Scholar]
- 41. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 42. Kamitani T., Kito K., Nguyen H. P., Wada H., Fukuda-Kamitani T., Yeh E. T. (1998) J. Biol. Chem. 273, 26675–26682 [DOI] [PubMed] [Google Scholar]
- 43. Subramaniam S., Mealer R. G., Sixt K. M., Barrow R. K., Usiello A., Snyder S. H. (2010) J. Biol. Chem. 285, 28428–28432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Knipscheer P., van Dijk W. J., Olsen J. V., Mann M., Sixma T. K. (2007) EMBO J. 26, 2797–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Capili A. D., Lima C. D. (2007) J. Mol. Biol. 369, 608–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Duda D. M., van Waardenburg R. C., Borg L. A., McGarity S., Nourse A., Waddell M. B., Bjornsti M. A., Schulman B. A. (2007) J. Mol. Biol. 369, 619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arnold J. E., Gevers W. (1990) Biochem. J. 267, 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Banerjee A., Gregori L., Xu Y., Chau V. (1993) J. Biol. Chem. 268, 5668–5675 [PubMed] [Google Scholar]
- 49. Scaglione K. M., Bansal P. K., Deffenbaugh A. E., Kiss A., Moore J. M., Korolev S., Cocklin R., Goebl M., Kitagawa K., Skowyra D. (2007) Mol. Cell. Biol. 27, 5860–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.