Abstract
Early steps in the biogenesis of Photosystem II (PSII) in the cyanobacterium Synechocystis sp. PCC 6803 are thought to occur in a specialized membrane fraction that is characterized by the specific accumulation of the PSII assembly factor PratA and its interaction partner pD1, the precursor of the D1 protein of PSII. Here, we report the molecular characterization of this membrane fraction, called the PratA-defined membrane (PDM), with regard to its lipid and pigment composition and its association with PSII assembly factors, including YCF48, Slr1471, Sll0933, and Pitt. We demonstrate that YCF48 and Slr1471 are present and that the chlorophyll precursor chlorophyllide a accumulates in the PDM. Analysis of PDMs from various mutant lines suggests a central role for PratA in the spatial organization of PSII biogenesis. Moreover, quantitative immunoblot analyses revealed a network of interdependences between several PSII assembly factors and chlorophyll synthesis. In addition, formation of complexes containing both YCF48 and Sll0933 was substantiated by co-immunoprecipitation experiments. The findings are integrated into a refined model for PSII biogenesis in Synechocystis 6803.
Keywords: Membrane Biogenesis, Photosynthesis, Photosynthetic Pigments, Protein Assembly, Protein Targeting, Photosystem II, Synechocystis, Chlorophyllide a
Introduction
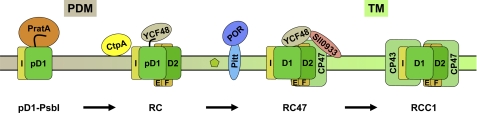
Cyanobacteria are Gram-negative bacteria that perform oxygenic photosynthesis. They contain three types of membranes: the outer membrane, the plasma membrane (PM),2 and the thylakoid membrane (TM) system. The outer membrane and PM form the cell envelope and delimit the periplasmic space, whereas TMs are localized within the cell and house the large pigment-protein complexes of the photosynthetic electron transfer chain, i.e. Photosystem (PS) II, PSI, the cytochrome b6f complex, and the ATP synthase (1). It is not clear whether direct connections exist between the different membrane systems and, in particular, where synthesis of TMs is initiated (2–4). In the cyanobacterium Synechocystis sp. PCC 6803, an intermediate membrane subfraction (PDM) was recently identified, which is defined by the presence of a membrane-bound form of the PSII assembly factor PratA (5). PratA is a periplasmic tetratricopeptide repeat protein that has been shown to interact with the C-terminal segment of pD1, the precursor of the PSII reaction center core protein D1 and to affect its maturation (5, 6). In Synechocystis 6803, pD1 contains a C-terminal extension of 16 amino acids that has to be processed by the endoprotease CtpA to allow proper assembly of the oxygen-evolving complex on the lumenal side of PSII (7–10). Because substantial amounts of pD1 accumulate in PDMs, these were hypothesized to represent a specialized membrane region close to the PM where early steps in the de novo assembly of PSII take place (4, 5). Several intermediates in this early assembly process have been detected, including the so-called reaction center (RC) complex containing pD1 and D2, as well as subunits PsbI, PsbE, and PsbF (11). Successive attachment of the inner antennal proteins CP47 and CP43 yields complexes RC47 and RCC1, respectively. RCC1 represents the monomeric PSII core complex, which is capable of oxygen evolution (11).
In addition to PratA, several other D1-associated factors have been suggested to function during the early steps of PSII biogenesis not only in cyanobacteria but also in higher plants (for a recent review, see Ref. 11). Thus, the cyanobacterial YidC/Oxa1/Alb3 homolog Slr1471 has been shown to interact directly with the D1 protein during its integration into the membrane (12). Similarly, a direct interaction has been revealed between pD1 and YCF48, the Synechocystis homolog of HCF136 from Arabidopsis thaliana, which seems to be involved in facilitating formation of PSII RC intermediates (13, 14). The A. thaliana protein PAM68 and its Synechocystis homolog Sll0933 have also been shown to be required for proper assembly of PSII, and a direct interaction of PAM68 with D1 was suggested in yeast two-hybrid analyses (15).
As the assembly of photosynthetic protein complexes has to be coordinated with the integration of pigments to form functional photosystems, it appears likely that chlorophyll synthesis is synchronized with the expression of chlorophyll-binding proteins. This would not only ensure the availability of sufficient pigments to build up the photosynthetic apparatus but would also prevent the accumulation of free and potentially harmful chlorophyll and/or chlorophyll precursor molecules (16, 17). Such coordination may be favored by co-localization of the machineries required for protein and pigment synthesis, as suggested by studies of the Pitt protein, which interacts with the light-dependent protochlorophyllide oxidoreductase (POR), an enzyme involved in the conversion of protochlorophyllide a into chlorophyllide a (4, 11, 18, 19), a precursor of chlorophyll a. Intriguingly, both Pitt and POR are present in PDM fractions of wild-type cells, suggesting a role for PDMs not only in protein synthesis and assembly but also in pigment synthesis and insertion (4, 18).
Here, we present a comprehensive characterization of the distribution of membrane-associated D1 assembly factors in the PDM subfraction from Synechocystis 6803. Additionally, we looked at qualitative differences in the lipid and pigment composition of PDMs and TMs to uncover correlations between pigment and protein synthesis and assembly. The results reveal a PDM-localized network for pigment-protein complex assembly and underline the importance of the PratA factor for the spatial organization of the assembly process. Moreover, further evidence for a direct link between YCF48 and Sll0933 was obtained. Together, they appear to mediate crucial steps at the PDM/TM interface.
EXPERIMENTAL PROCEDURES
Growth Conditions and Construction of Cyanobacterial Strains
Glucose-tolerant wild-type Synechocystis 6803 and relevant mutant strains were grown under continuous irradiation with 30 μmol of photons m−2 s−1 at 30 °C in BG11 medium containing 5 mm glucose (20). For construction of a ctpA− mutant strain, the coding region was amplified by PCR using the primer pair THctpAa (GAATTCATGGGTAAACGGACAAGGCGGTTT) and THctpAb (CTCGAGTTAGTTAGTTGGGCTTGTGAGCCG). The ctpA gene was disrupted by insertion of a kanamycin resistance cassette into the single MunI restriction site, and wild-type cells were transformed with this construct as described (21). Complete segregation of the mutated gene was confirmed by PCR analysis (data not shown). Mutant strains pratA−, pitt−, ins0933, ycf48−, and psbB− and psbA deletion strain TD41 were constructed as described previously (6, 7, 14, 15, 18, 22).
Antibody Production
For generation of specific antibodies, the sequence encoding amino acid residues 275–342 of YCF48 was amplified using primers 2034-5 (AAGGATCCGAAGAAGTATGGGTAGCGGG) and 2034-3 (AACTCGAGCTAGGGAACCATTGCCACCT). The subcloned PCR fragment was inserted into the BamHI and XhoI sites of expression vector pGEX-4T-1. Overexpression of the GST fusion proteins in Escherichia coli BL21(DE3) cells and subsequent purification using glutathione-Sepharose 4B (GE Healthcare) were carried out according to the manufacturer's instructions. Polyclonal antisera were raised in rabbits (BioGenes GmbH, Berlin, Germany). Generation of antibodies against Synechocystis 6803 PratA, Pitt, and Sll0933 has been described previously (6, 15, 18). Antibodies specific for CP47 (global), A. thaliana CP43, Chlamydomonas reinhardtii PsaA, and RbcL (global) were obtained from Agrisera (Vännäs, Sweden).
Protein Extraction, Membrane Fractionation, and Immunoblot Analysis
Protein extraction, two-step membrane fractionation on sucrose gradients, and subsequent immunoblot analysis were carried out as described previously (5, 18). For quantification, proteins of different strains were isolated, separated by SDS-PAGE, blotted, and probed with various antibodies. After densitometric scanning, quantification of signals was performed using AIDA software (version 3.52.046), and the levels of mutant strains were compared with those of the wild-type strain (set to 100%). Means ± S.D. were calculated from three independently inoculated cultures.
Two-dimensional Blue Native/SDS-PAGE
Synechocystis membranes were isolated as described (23), and two-dimensional Blue native/SDS-PAGE was performed as described previously (5).
Co-immunoprecipitation
For co-immunoprecipitation experiments, 50 ml of wild-type cells were harvested by centrifugation (4000 × g, 10 min, 4 °C), resuspended in 750 μl of thylakoid buffer (50 mm HEPES/NaOH (pH 7.0), 5 mm MgCl2, 25 mm CaCl2, and 10% glycerol), and broken with glass beads (0.4–0.6 mm) using a Mini-BeadBeater (Glen Mills). Unbroken cells were removed by a short centrifugation step (20,000 × g, 45 s) before membranes were sedimented (20,000 × g, 30 min, 4 °C) and washed twice with thylakoid buffer. The sediment was subsequently resuspended in 50 μl of Tris/NaCl buffer (50 mm Tris-HCl (pH 8) and 150 mm NaCl) and solubilized for 30 min on ice with 1.3% β-dodecyl maltoside. After another centrifugation step (20,000 × g, 30 min, 4 °C), the supernatant was diluted 1:10 with Tris/NaCl and incubated overnight at 4 °C in the presence of 10 μl of anti-YCF48 antiserum or preimmune serum. Protein A-agarose was added (5 mg; Roche Applied Science), and incubation was continued for another hour at room temperature. The agarose beads were sedimented by centrifugation and washed five times, and bound proteins were eluted by incubation with 30 μl of 2× Roti®-Load 1 (Roth, Karlsruhe, Germany) for 5 min at 95 °C.
Pigment and Lipid Analyses
Pigments were extracted from concentrated membrane subfractions (5) by vortexing in the presence of added chloroform (2 ml) and methanol (1 ml). After the addition of 3 ml of chloroform and 3 ml of water, tubes were inverted, and the two phases were separated by centrifugation (3000 × g, 5 min, 4 °C). The organic phase was dried, and pigments/lipids were redissolved in 100 μl of chloroform and subjected to TLC on silica gel plates (SIL G-25 UV254, Macherey-Nagel, Düren, Germany) in a mixture of acetone, toluene, and water (91:30:8). For visualization of lipids, the plates were sprayed with a staining solution (36 mm FeSO4, 5.7 mm KMnO4, and 3% (v/v) H2SO4) and incubated for 10 min at 120 °C.
Prior to analysis by HPLC, pigments were extracted from concentrated membrane subfractions with acetone. The analysis was performed on a Flux instrument equipped with a C18 column (Grom Sil 120 ODS-5, 3-μm particle size, 150-mm length, 2-mm inner diameter). Elution was carried out for 3 min with a 60:40 acetone/water mixture (pH 3.5), followed by a linear gradient to 100% acetone applied over a period of 20 min. The column was then rinsed for 20 min with 100% acetone. Absorption spectra were measured with a Thermo diode array detector in the 350–750-nm range. Pigments were identified based on their retention times and absorption spectra.
RESULTS
Pigments in PDMs and TMs
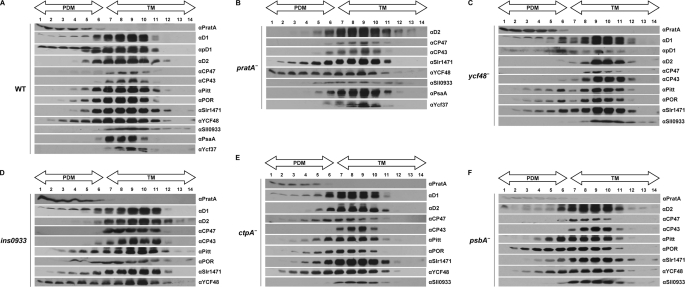
PDMs have recently been suggested to represent a special membrane subcompartment where early steps in TM biogenesis take place (4, 5). As a first step toward a comprehensive characterization of PDMs, we examined the main pigments and lipids in PDMs and TMs. Membranes of wild-type Synechocystis 6803 cells were separated into PDM and TM fractions by two rounds of sucrose density gradient centrifugation (first on a step and then on a linear gradient), and the distribution of marker proteins was analyzed using antibodies (5). The inner antennal proteins of PSII, i.e. CP47 and CP43, serve as markers for TMs (10, 24) and were almost exclusively detected in fractions 7–11, hereafter referred to as TM fractions (see Fig. 2A) (5). In contrast, PDM fractions 1–6 were defined by the accumulation of PratA and pD1 (see Fig. 2A) (5). The majority of the PSII RC core protein D2 was located in TMs; however, a minor portion was detected in PDM fractions 4–6 (see Fig. 2A). Although it cannot be excluded that also minor amounts of CP43 and CP47 were present in the PDM fractions, it is obvious that this dual localization was more pronounced for D2 (see Fig. 2A). Hence, CP47 and CP43 can be regarded as suitable TM markers with the membrane fractionation method used. Subsequent TLC of membrane material from isolated fractions revealed no obvious qualitative differences in the main lipids between the two subfractions (supplemental Fig. 1).
FIGURE 2.
Membrane sublocalization of different PSII assembly factors. Synechocystis cell extracts of the wild-type strain (A) and pratA− (B), ycf48− (C), ins0933 (D), ctpA− (E), and psbA− (F) mutant strains were separated by two consecutive rounds of sucrose density gradient centrifugation (5). The second linear gradient from 20 to 60% sucrose was apportioned into 14 fractions, which were analyzed by immunoblotting using the indicated antibodies. Fractions 1–6 represent PDMs, and fractions 7–14 represent TMs. To facilitate comparison between gradients, sample volumes were normalized to the volume of fraction 7 that contained 40 μg of protein. Due to the sharp fall in the levels of pD1-containing RC complexes in ins0933, no pD1 signal could be detected in this mutant (15). Because no mature D1 can accumulate in the ctpA− mutant, pD1 was detected using anti-D1 antiserum. For the pD1 signal of pratA−, see Ref. 5.
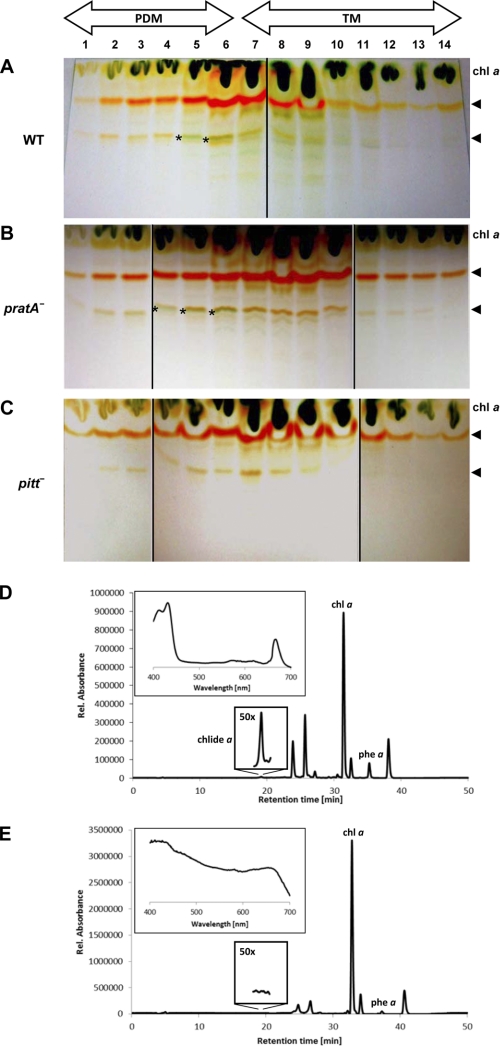
Similarly, most of the lipophilic pigments, i.e. carotenoids and chlorophyll derivatives, were detectable in both PDMs (Fig. 1A, fractions 1–6) and TMs, although relatively more chlorophyll a was found in TMs (fractions 7–11). The only exception was a minor green band that was identified only in PDM fractions 5 and 6 of the wild-type material (Fig. 1A). HPLC-based analyses comparing the pigment compositions of PDM fraction 5 (Fig. 1D) and TM fraction 9 (Fig. 1E) revealed that the only pigment of green color that could exclusively be detected in PDM fraction 5 represents chlorophyllide a, a late intermediate in chlorophyll biosynthesis (Fig. 1D). Therefore, the PDM-specific band visible on thin-layer plates seems to be caused by chlorophyllide a accumulation (Fig. 1A, asterisks). This result is in agreement with earlier reports demonstrating that the highest concentrations of chlorophyllide a and its precursor, protochlorophyllide a, were detected in a membrane subfraction hypothesized to resemble contact sites between PMs and TMs called “thylakoid centers” (25). The PDM-specific accumulation of chlorophyllide a argues that PDMs represent a subcompartment in which not only protein complex assembly but also later steps in chlorophyll synthesis and probably insertion into polypeptides take place. This specific membrane localization of chlorophyllide a is independent of the presence of the PDM-specific PratA protein because it was not altered in a pratA− mutant (Fig. 1B).
FIGURE 1.
Pigment analysis of PDM and TM fractions. Synechocystis PDMs and TMs of wild-type (A), pratA− (B), and pitt− (C) cells were obtained by two rounds of sucrose density gradient centrifugation. Equal volumes of hydrophobic pigments extracted from each fraction were then subjected to TLC. Asterisks mark a pigment accumulation in PDM fractions that seems to represent chlorophyllide a. Arrows indicate the positions of carotenoids. D and E, HPLC-based pigment analysis of PDM fraction 5 and TM fraction 9, respectively, of wild-type cells. The PDM-specific peak (shown also magnified ×50) was identified as chlorophyllide a (chlide a) based on its retention time and absorption spectrum (inset). At the comparable retention time in TM fraction 9, no characteristic absorption spectrum for chlorophyll derivatives was detectable (inset). Due to their retention times and absorption spectra, other peaks were identified as chlorophyll a (chl a), pheophytin a (phe a), and carotenoids (all unlabeled peaks). Carotenoids were not further specified. Rel., relative.
It was recently shown that lack of the POR-interacting protein Pitt causes a reduction in light-dependent chlorophyll synthesis (18). Therefore, we examined whether the accumulation and/or membrane localization of chlorophyllide a is affected in the pitt− mutant. Fig. 1C shows that the distribution of chlorophyll a and carotenoids in pitt− cells resembled that in the wild-type cells, whereas the putative chlorophyllide a band was completely absent. Therefore, loss of Pitt seems to affect synthesis of chlorophyllide a and, in particular, its accumulation in PDMs.
Localization of PSII Assembly Factors in PDMs
The accumulation of PratA and the pD1 precursor protein in PDMs implies that other factors involved in early steps in de novo PSII assembly might be located in the same membrane subfraction. To test this prediction, the accumulation of various assembly-related factors in membrane subfractions was monitored by immunoblotting. As described previously (18), the chlorophyll synthesis factors Pitt and POR were localized mainly in TMs, with only minor portions present in PDMs (Fig. 2A). The early PSII assembly factors Slr1471 and YCF48 accumulated predominantly in TMs, but substantial amounts of these proteins, especially YCF48, were also detected in PDM fractions, underlining a role of PDMs during early PSII assembly (Fig. 2A) (12, 14). Interestingly, Sll0933, a factor suggested to be involved in integration of CP47 into PSII subcomplexes, showed a different membrane localization (15). This protein was found solely in TM fractions (Fig. 2A), supporting the idea that the assembly of the inner PSII antenna takes place in TMs (10).
To determine whether PDMs are also involved in PSI assembly, the subcellular distribution of the PSI core protein PsaA and the PSI assembly factor YCF37 was investigated (23). Both proteins showed the same pattern as CP47/CP43 and therefore appear to be restricted to TMs (Fig. 1A). Thus, PDMs seem to represent a membrane subcompartment harboring factors involved in the early steps of PSII assembly only.
Effects of Inactivation of Individual PSII-related Assembly Factors on Localization of Other PSII-associated Proteins
We have previously shown that inactivation of PratA affects the distribution of pD1, the POR enzyme, and its interaction partner Pitt. In the absence of PratA, all three proteins are shifted toward lighter fractions of the sucrose gradient, i.e. they show a more pronounced accumulation in PDMs (5, 18). Hence, we tested whether the membrane localization of these and other photosynthesis-related proteins is affected in various mutant backgrounds.
In the pratA− mutant, the PSII-associated proteins Slr1471 and YCF48 accumulated to higher levels in PDM fractions compared with wild-type cells (Fig. 2, A and B). In addition, the localization of Sll0933 was moderately affected upon inactivation of pratA. In wild-type cells, this protein was found in TMs only, whereas in a pratA− mutant background, it was also detected in PDM fractions 5 and 6 (Fig. 2, A and B). In contrast, the distribution of D2 remained unaltered (Fig. 2B). Similarly, the PSI-related proteins PsaA and Ycf37 continued to co-localize with TM markers CP43 and CP47 (Fig. 2B). Thus, the localization of the PSII assembly factors tested, but not of PSI-associated proteins, depends on the presence of PratA. For this reason, the distribution of PsaA and YCF37 was not analyzed in other PSII mutants described below.
Because YCF48 had been postulated to play a role in the transfer of PSII precomplexes to TMs (14), we also investigated the distribution of PSII-associated proteins in PDM and TM fractions from ycf48− mutant cells. The data revealed a moderate shift of Pitt, POR, and Slr1471 toward PDM fractions, whereas the distribution of all other proteins tested was not affected (Fig. 2, A and C, compare fractions 2–4, respectively). Hence, although YCF48 might be involved in the membrane organization of selected PSII assembly factors, it is apparently not strictly required for directing PSII intermediates from PDMs to TMs.
Recently, the list of PSII assembly factors was further extended by the identification of PAM68 in A. thaliana (15). Its homolog in Synechocystis, Sll0933, was shown to affect accumulation of the RC complex (15). Here, a minor alteration in membrane organization was observed in the sll0933− mutant ins0933, as suggested by a slight shift of Slr1471 and YCF48 toward the top of the gradient. The distribution of other proteins tested did not change (Fig. 2D). This indicates some involvement of Sll0933 in the spatial organization of early PSII assembly steps in the PDM.
Previous membrane fractionation studies on a pitt− mutant had revealed increased accumulation of POR and the pD1 precursor protein in PDMs (18). Therefore, it was postulated that Pitt plays a role in the spatial organization of early PSII assembly, in addition to its involvement in chlorophyll synthesis (18). However, it did not alter the membrane distribution of other PSII proteins or assembly factors additionally tested in this study (supplemental Fig. 2A).
In summary, various factors are required to coordinate the membrane localization of PSII intermediates and their associated proteins during the assembly of PSII. Inactivation of PratA leads to the most obvious changes in localization. Because PratA was initially found to affect the C-terminal maturation of pD1 (8, 26), we next tested whether inhibiting C-terminal processing by inactivation of CtpA, the endoprotease that removes the D1 extension, compromises PDM/TM membrane organization. Surprisingly, in this mutant, the localization of all proteins, including immature pD1, was similar to that seen in wild-type cells (Fig. 2, A and E) with one striking exception: the inner antennal protein CP47 was detected in fractions 4–11 in the ctpA− mutant but in fractions 7–11 in the wild-type cells (Fig. 2, A and E). In contrast, CP43 was not affected by the lack of mature D1 protein (Fig. 2, A and E). Hence, a defect in pD1 processing results in the accumulation of CP47 in PDMs, but cleavage of the C-terminal extension of D1 is not essential for the transport of PSII subcomplexes to TMs.
When D1 itself is lacking due to the inactivation of all three psbA gene copies in the psbA− mutant strain TD41 (7), only POR showed a more pronounced abundance in PDM fractions; no effects on the distribution of other tested factors were noted (Fig. 2F). Finally, we analyzed factor localization in a psbB− mutant lacking CP47 (22). One major difference was observed relative to the wild-type cells, i.e. the absence of substantial amounts of pD1 in PDM fractions (supplemental Fig. 2B). This suggests that, in the absence of ongoing PSII assembly, PDM-localized pD1 is rapidly degraded or is quickly transferred to TMs.
Accumulation of PSII Assembly Factors in Different PSII Assembly Mutants
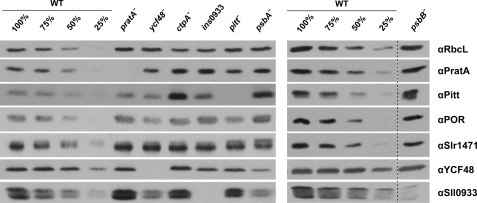
The membrane distribution of PSII assembly factors described above suggests at least a degree of interdependence between some of them. To determine whether their accumulation is affected in various PSII assembly mutants, whole cell protein extracts were prepared from the wild-type cells and mutants of interest and quantified by immunoblot analyses. No effects were observed in the case of the Oxa homolog Slr1471, which accumulated to wild-type levels in all mutants tested (Fig. 3 and Table 1). Intriguingly, the accumulation of PSII assembly factors was not altered in the pratA− mutant either, in contrast to the strong effects on their membrane localization observed upon inactivation of PratA (Figs. 2, A and B, and 3 and Table 1). The amounts of PratA itself increased to ∼150% of wild-type levels in the ctpA− mutant (in which pD1 accumulates) and to ∼140% in the D1-deficient strain psbA−. This suggests some feedback control of pratA gene expression in the absence of functional D1.
FIGURE 3.
Accumulation of PSII assembly factors in various mutants. Whole cell extracts (30 μg of protein) from wild-type cells and pratA−, ycf48−, ctpA−, ins0933, pitt−, psbA−, and psbB− mutants were fractionated by SDS-PAGE, blotted onto nitrocellulose membrane, and probed with the indicated antibodies. Representative results of three independent experiments are shown.
TABLE 1.
Summary of the effects of the indicated PSII mutants on membrane localization and relative levels of different PSII assembly factors
The entries for proteins that differ in their membrane localization in certain mutants compared with wild-type cells are in italic (compare Fig. 2 and supplemental Fig. 2). The numbers refer to the levels of membrane-localized PSII assembly factors in the indicated mutants relative to those in wild-type cells (arbitrarily set to 100). Values are means ± S.D. of three independent experiments. The clearest differences from wild-type levels are highlighted in boldface; examples of the underlying experiments are shown in Fig. 3.
| Protein | Mutant |
||||||
|---|---|---|---|---|---|---|---|
| pratA− (5) | pitt− (18) | ycf48− | ins0933 | ctpA− | psbB− | psbA− | |
| PratA | 113 ± 20 | 108 ± 33 | 105 ± 22 | 149 ± 17 | 110 ± 55 | 138 ± 4 | |
| Slr1471 | 103 ± 25 | 83 ± 28 | 106 ± 21 | 105 ± 10 | 109 ± 16 | 110 ± 38 | 89 ± 14 |
| YCF48 | 99 ± 35 | 88 ± 16 | 70 ± 20 | 104 ± 2 | 101 ± 17 | 49 ± 23 | |
| Sll0933 | 108 ± 31 | 82 ± 11 | 51 ± 8 | 77 ± 17 | 15 ± 9 | 48 ± 13 | |
| Pitt | 101 ± 32 | 103 ± 21 | 178 ± 32 | 295 ± 122 | 206 ± 62 | 244 ± 108 | |
| POR | 101 ± 15 | 52 ± 16 | 52 ± 16 | 104 ± 20 | 67 ± 25 | 107 ± 28 | 96 ± 35 |
Inactivation of ctpA has an even more pronounced effect on the POR interaction partner Pitt than on PratA. Steady-state levels of Pitt were 3-fold higher in the ctpA− mutant than in the wild-type cells, whereas a decrease in the amount of POR to ∼70% and a slighter decrease in the amount of Sll0933 to ∼80% were observed (Fig. 3 and Table 1). A similar increase in Pitt accumulation was noted in the psbA− mutant and, to a lesser extent, in the psbB− and ins0933 mutants also (Fig. 3 and Table 1).
On the other hand, lack of the Pitt protein resulted in a minor decrease in Sll0933 to ∼80% of wild-type amounts, in addition to the previously reported reduction in the level of POR (18). The level of POR was also reduced to ∼50% in the ycf48− mutant. This suggests that Sll0933 and YCF48 represent additional candidates that might play a role in coordinating protein and cofactor synthesis during PSII assembly (Fig. 3 and Table 1).
Furthermore, the amounts of Sll0933 and YCF48 were found to be reduced by 50% relative to wild-type levels in the psbA− mutant, and the amount of Sll0933 was also reduced by half in the ycf48− mutant (Fig. 3 and Table 1). Conversely, lack of Sll0933 resulted in a 30% drop in the steady-state levels of YCF48, suggesting an interrelationship between YCF48 and Sll0933. However, one striking exception to the consistent coregulation of YCF48 and Sll0933 was the dramatic decrease in Sll0933 seen in the psbB− mutant, in which YCF48 levels were not affected. This argues for a close relationship between Sll0933 and CP47 (Fig. 3 and Table 1).
Sll0933 and YCF48 Form Part of a Complex
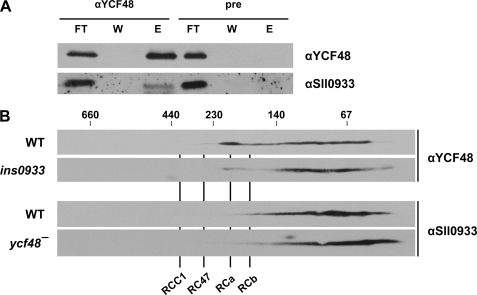
The interdependence between the levels of YCF48 and Sll0933 is compatible with a direct interaction between them. Indeed, this has already been postulated based on yeast two-hybrid studies of their respective A. thaliana homologs HCF136 and PAM68 (15). To test this hypothesis further, solubilized membrane proteins of wild-type cells were subjected to co-immunoprecipitation analyses. The anti-YCF48 antiserum quantitatively precipitated the YCF48 factor, together with lesser but significant amounts of Sll0933 (Fig. 4A). Neither protein could be precipitated by the preimmune serum. Due to non-quantitative precipitation, no conclusive results were obtained when, in the reciprocal experiment, an anti-Sll0933 antiserum was used for co-immunoprecipitation. Nevertheless, the data suggest that YCF48 and Sll0933 are, at least transiently, components of the same complex. This is further supported by the fact that the two proteins co-migrated, at least to some extent, in the range between 150 kDa and the free proteins on two-dimensional Blue native/SDS gels (Fig. 4B). In the ycf48− mutant, a subtle effect on Sll0933 migration was observed: relatively more Sll0933 was associated with the smaller complexes than was the case in the wild-type cells (Fig. 4B). The more pronounced but still moderate accumulation of YCF48 in RC complexes was reduced in the ins0933 mutant, with a concomitant increase in accumulation in the lower molecular mass range (Fig. 4B). We therefore conclude that subpopulations of YCF48 and Sll0933 molecules form a transient complex. Overall, the analysis of PSII-related assembly factors in the different mutant backgrounds reveals a complex regulatory network that links factors involved in chlorophyll synthesis and protein complex assembly.
FIGURE 4.
Interaction of YCF48 and Sll0933. A, membrane proteins from wild-type cells were solubilized with 1.3% β-dodecyl maltoside and co-immunoprecipitated using anti-YCF48 antiserum or preimmune serum (pre). FT, of the flow-through containing unbound proteins; W, of the supernatant from the last washing step; E, one-half of the eluted fraction containing co-immunoprecipitated proteins. B, two-dimensional Blue native/SDS-PAGE of wild-type, ycf48−, and ins0933 membranes (each fraction is equivalent to 20 μg of chlorophyll) was performed as described previously (5). RCa and RCb are RC complexes a and b (these complexes are not defined in text), RC47 is the PSII core complex lacking CP43, and RCC1 is the monomeric PSII core complex. The positions of size markers are indicated at the top. The indicated proteins were detected using the respective antibodies.
DISCUSSION
Assignment of Stages in PSII Biogenesis to Specific Membrane Fractions
Previous membrane fractionation studies have identified a PDM fraction in Synechocystis 6803 in which early steps of PSII assembly were suggested to occur (4, 5). Here, we have demonstrated that PratA itself plays an essential role in the spatial organization of PDMs as a PSII biogenesis platform because its inactivation (in contrast to that of other factors) severely affects the sublocalization of all other PSII assembly factors tested, i.e. Slr1471, YCF48, and Sll0933 (Fig. 2B). Additional support for a role of PDMs during the initial steps of PSII assembly is provided by the preferential accumulation of pD1 in this membrane subcompartment (5). In contrast, the inner antennal proteins CP47 and CP43, which are attached to PSII RC complexes during later steps in PSII biogenesis, are localized to TMs (Fig. 2A). The immediate assembly partner of D1, the D2 subunit of PSII, was found in both PDMs and TMs, suggesting that assembly proceeds from lighter to more dense membrane fractions in the order D1 + D2 + CP47/CP43. This is consistent with current models of PSII biogenesis based on the analysis of PSII assembly intermediates by two-dimensional Blue native/SDS-PAGE (11) and is further supported by the relative accumulation of the early PSII assembly factors Slr1471 and YCF48 in PDMs (Fig. 2A). Intriguingly, D2-containing PDM fractions 4–6, which probably mark the positions of RC complexes, represent the only membrane fractions in which the chlorophyll a precursor chlorophyllide a accumulates to detectable levels. However, the POR enzyme is localized mainly in TMs and only to smaller proportions in fractions 5 and 6 (Fig. 2A). Possibly, the chlorophyllide a synthesized in TMs is converted faster into chlorophyll a. In chloroplasts, the respective enzyme, the chlorophyll synthase, has been localized exclusively in TMs, whereas activity measurements in Synechocystis revealed its presence in both TMs and perhaps the PDMs resembling thylakoid centers (19, 25). As discussed previously (4), it also remains to be established whether the pool of chlorophyllide a in PDMs is synthesized by the light-dependent POR enzyme or the light-independent POR system in cyanobacteria (27). To answer this question, the ChlL subunit of light-independent POR was used for antiserum production, but due to unspecific binding of the antibody, no conclusions could be drawn (data not shown). Nevertheless, the strong reduction of chlorophyllide a accumulation in PDMs in the pitt− mutant suggests a direct involvement of POR in the synthesis of PDM-localized chlorophyll and, concomitantly, a more efficient conversion of residual childe a to chlorophyll a in the pitt− mutant. Thus, the data suggest that chlorophyll a synthesis/integration is correlated with RC complex formation. This is further evidenced by the finding that inactivation of the RC assembly factor YCF48 (14) clearly affects both the localization and accumulation of POR (Figs. 2C and 3 and Table 1). Therefore, the chlorophyll synthesis enzyme POR and its interaction partner Pitt appear to be integrated into the assembly network. Moreover, Pitt levels are clearly increased in the ins0933, ctpA−, psbB−, and psbA− strains, suggesting that, in these mutants, regulatory pathways that are dedicated to the compensation of photosynthetic deficiencies are activated. Overall, the results obtained therefore substantiate a tight connection between membrane protein assembly and pigment insertion.
A Network of Assembly Factors Mediates PSII Assembly
The patterns of co-localization among assembly factors and their response to genetic perturbation point to certain interdependences between them. This is most striking in the case of YCF48 and Sll0933. (i) Sll0933 inactivation shifts the distribution of YCF48 toward PDM fractions. (ii) Accumulation of each is decreased in the absence of the other. (iii) The levels of both proteins decline in the absence of D1. (iv) The two factors co-migrate, at least partially, in two-dimensional Blue native/SDS gels, and the absence of Sll0933 affects the migration behavior of YCF48. (v) Immunoprecipitation of YCF48 with a cognate antiserum also brings down Sll0933, demonstrating that the two factors form part of the same complex. Moreover, yeast two-hybrid assays have shown that the Sll0933 homolog from A. thaliana, PAM68, interacts directly with several PSII subunits and assembly factors, including the plant YCF48 homolog HCF136 (15). In addition, YCF48 has previously been shown to recognize the pD1 C-terminal extension, whereas Sll0933 has been implicated in the accumulation of PSII RC complexes that lack the inner antennal protein CP47 but contain YCF48 and pD1/D1 (14, 15). In the mutant ins0933, no stable accumulation of RC complexes and no pD1 precursor protein could be detected, suggesting rapid conversion of RC complexes into monomeric PSII core complexes (RCC) (15). Considering that the two factors localize to different membrane fractions and that Sll0933 is strongly dependent on the presence of CP47 (Figs. 2A and 3 and Table 1), the following scenario for the early steps of de novo PSII assembly can be envisaged (Fig. 5). In PDMs, the pD1 precursor protein first associates with the periplasmic PratA protein. Following the addition of D2 to form the RC complex, YCF48 is then attached via the C terminus of pD1 (14). The resulting complex then moves to TMs, where Sll0933, possibly together with CP47, binds to YCF48, thereby facilitating the correct integration of CP47 into the RC to yield the RC47 complex. This process appears to be accelerated in the ins0933 mutant strain because no RC complexes and pD1 accumulate in ins0933 cells despite the presence of wild-type levels of functional PSII complexes (15). This suggests rapid conversion of pD1 into D1 and formation of RC47 complexes.
FIGURE 5.
Working model for PSII biogenesis in Synechocystis 6803. PSII biogenesis begins in PDMs close to the PM with the interaction of PratA-bound pD1 protein with PsbI. After the addition of D2 and PsbE/F subunits, RC complexes are formed. Insertion of chlorophyll a (small green pentagon) is probably mediated by the Pitt-POR and YCF48-Sll0933 complexes, with the concomitant insertion of CP47. The resulting RC47 complexes are transferred to the TM, where CP43 is attached to form RCC1 monomers. For further explanation, see “Discussion.”
The question arises as to the function of the Sll0933-mediated delay of PSII assembly. One possibility is that a slow down in PSII assembly might allow for more efficient integration of other factors, such as chlorophylls, into RC complexes. This idea is especially appealing in light of the relationship between YCF48 and the POR enzyme discussed above, as well as the accumulation of chlorophyllide a in fractions likely to contain RC complexes. Further evidence suggesting a connection between chlorophyll integration and YCF48/Sll0933 (and thus RC complex formation) is provided by the finding that Sll0933 levels fall upon Pitt inactivation (Fig. 3 and Table 1). In addition, inhibition of maturation of RC-localized pD1 in the ctpA− mutant is accompanied by a reduction in the levels of both POR and Sll0933 (Fig. 3 and Table 1) and aberrant accumulation of CP47 in PDM fractions.
Alternatively or in addition, it is conceivable that Sll0933 participates in a quality-control step during movement of RCs into TMs. Obviously, more detailed studies are required to elucidate the precise functions and interrelationships of assembly factors like PratA, Pitt, and YCF48/Sll0933. All of them seem to interact at the interface between PDMs and TMs to coordinate early steps in the PSII maturation pathway.
One aspect that has not been addressed in this work concerns the reassembly of PSII during repair after photodamage to the D1 protein. It is still unclear whether D1 repair and de novo PSII assembly are spatially separated. In chloroplasts of C. reinhardtii, repair and de novo synthesis indeed take place in different TM subfractions, raising the possibility that the same holds for cyanobacteria (28). The factor Psb27 has recently been shown to be involved in facilitating the assembly of the manganese cluster of PSII during the repair process (29, 30). However, when we analyzed the membrane distribution of PSII assembly factors in a psb27− mutant, no alterations were observed (data not shown). Hence, the question remains open whether PSII reassembly in general (and Psb27 in particular) is integrated into the network responsible for the de novo assembly of PSII.
Conclusions and Future Perspectives
The emerging picture of TM biogenesis in cyanobacteria is dominated by a strict spatiotemporal organization, especially of the early steps in PSII assembly. Initially, the pD1 precursor protein is integrated into regions that contact the PM. Progressive maturation of PSII, with sequential formation of the intermediate complexes RC, RC47, and RCC1, then occurs in a subcompartment that is linked to the TMs. This physical and functional link is represented by the PDM. The maturation process also includes the insertion of chlorophyll a and probably other cofactors like carotenoids and the non-heme iron atom of PSII. Apparently, YCF48 and Sll0933 play a crucial role in this process at the interface between PDMs and TMs. It is tempting to speculate that the connecting PDMs are identical to previously described thylakoid centers that are located at the ends of converging TMs close to the PM (3, 4). Future work will have to determine the precise location of PDMs within the cell using high resolution imaging of live cells and electron microscopy.
Supplementary Material
Acknowledgments
We thank A. Wilde, J. Soll, N. Murata, and Y. Nishiyama for providing anti-Ycf37, anti-Slr1471, anti-POR, and anti-pD1 antibodies and J. Komenda and P. Nixon for providing the psbB− and psbA− mutants, respectively.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB TR1 TPC10 (to J. N.) and a Bayerische Gleichstellungsförderung (BGF) scholarship from the Ludwig-Maximilians-Universität München (to A. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- PM
- plasma membrane
- TM
- thylakoid membrane
- PS
- Photosystem
- PDM
- PratA-defined membrane
- RC
- reaction center
- POR
- protochlorophyllide oxidoreductase.
REFERENCES
- 1. Vermaas W. F. J. (2001) in Encyclopedia of Life Sciences, pp. 245–251, Nature Publishing Group, London [Google Scholar]
- 2. Liberton M., Howard Berg R., Heuser J., Roth R., Pakrasi H. B. (2006) Protoplasma 227, 129–138 [DOI] [PubMed] [Google Scholar]
- 3. van de Meene A. M., Hohmann-Marriott M. F., Vermaas W. F., Roberson R. W. (2006) Arch. Microbiol. 184, 259–270 [DOI] [PubMed] [Google Scholar]
- 4. Nickelsen J., Rengstl B., Stengel A., Schottkowski M., Soll J., Ankele E. (2011) FEMS Microbiol. Lett. 315, 1–5 [DOI] [PubMed] [Google Scholar]
- 5. Schottkowski M., Gkalympoudis S., Tzekova N., Stelljes C., Schünemann D., Ankele E., Nickelsen J. (2009) J. Biol. Chem. 284, 1813–1819 [DOI] [PubMed] [Google Scholar]
- 6. Klinkert B., Ossenbühl F., Sikorski M., Berry S., Eichacker L., Nickelsen J. (2004) J. Biol. Chem. 279, 44639–44644 [DOI] [PubMed] [Google Scholar]
- 7. Nixon P. J., Trost J. T., Diner B. A. (1992) Biochemistry 31, 10859–10871 [DOI] [PubMed] [Google Scholar]
- 8. Anbudurai P. R., Mor T. S., Ohad I., Shestakov S. V., Pakrasi H. B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8082–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roose J. L., Pakrasi H. B. (2004) J. Biol. Chem. 279, 45417–45422 [DOI] [PubMed] [Google Scholar]
- 10. Zak E., Norling B., Maitra R., Huang F., Andersson B., Pakrasi H. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13443–13448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nixon P. J., Michoux F., Yu J., Boehm M., Komenda J. (2010) Ann. Bot. 106, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ossenbühl F., Inaba-Sulpice M., Meurer J., Soll J., Eichacker L. A. (2006) Plant Cell 18, 2236–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Plücken H., Müller B., Grohmann D., Westhoff P., Eichacker L. A. (2002) FEBS Lett. 532, 85–90 [DOI] [PubMed] [Google Scholar]
- 14. Komenda J., Nickelsen J., Tichý M., Prásil O., Eichacker L. A., Nixon P. J. (2008) J. Biol. Chem. 283, 22390–22399 [DOI] [PubMed] [Google Scholar]
- 15. Armbruster U., Zühlke J., Rengstl B., Kreller R., Makarenko E., Rühle T., Schünemann D., Jahns P., Weisshaar B., Nickelsen J., Leister D. (2010) Plant Cell 22, 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yaronskaya E., Ziemann V., Walter G., Averina N., Börner T., Grimm B. (2003) Plant J. 35, 512–522 [DOI] [PubMed] [Google Scholar]
- 17. He Q., Vermaas W. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5830–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schottkowski M., Ratke J., Oster U., Nowaczyk M., Nickelsen J. (2009) Mol. Plant 2, 1289–1297 [DOI] [PubMed] [Google Scholar]
- 19. Eckhardt U., Grimm B., Hörtensteiner S. (2004) Plant Mol. Biol. 56, 1–14 [DOI] [PubMed] [Google Scholar]
- 20. Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. (1979) J. Gen. Microbiol. 111, 1–61 [Google Scholar]
- 21. Eaton-Rye J. J. (2004) Methods Mol. Biol. 274, 309–324 [DOI] [PubMed] [Google Scholar]
- 22. Eaton-Rye J. J., Vermaas W. F. (1991) Plant Mol. Biol. 17, 1165–1177 [DOI] [PubMed] [Google Scholar]
- 23. Dühring U., Irrgang K. D., Lünser K., Kehr J., Wilde A. (2006) Biochim. Biophys. Acta 1757, 3–11 [DOI] [PubMed] [Google Scholar]
- 24. Bergantino E., Brunetta A., Touloupakis E., Segalla A., Szabò I., Giacometti G. M. (2003) J. Biol. Chem. 278, 41820–41829 [DOI] [PubMed] [Google Scholar]
- 25. Hinterstoisser B., Chichna M., Kuntner O., Peschek G. A. (1993) J. Plant Physiol. 142, 407–413 [Google Scholar]
- 26. Shestakov S. V., Anbudurai P. R., Stanbekova G. E., Gadzhiev A., Lind L. K., Pakrasi H. B. (1994) J. Biol. Chem. 269, 19354–19359 [PubMed] [Google Scholar]
- 27. Armstrong G. A. (1998) J. Photochem. Photobiol. B 43, 87–100 [Google Scholar]
- 28. Uniacke J., Zerges W. (2007) Plant Cell 19, 3640–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nowaczyk M. M., Hebeler R., Schlodder E., Meyer H. E., Warscheid B., Rögner M. (2006) Plant Cell 18, 3121–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roose J. L., Pakrasi H. B. (2008) J. Biol. Chem. 283, 4044–4050 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.