Abstract
Transcriptional silencing in Saccharomyces cerevisiae is mediated by heterochromatin. There is a plethora of information regarding the roles of histone residues in transcriptional silencing, but exactly how histone residues contribute to heterochromatin structure is not resolved. We address this question by testing the effects of a series of histone H3 and H4 mutations involving residues in their aminoterminal tails, on the solvent-accessible and lateral surfaces of the nucleosome, and at the interface of the H3/H4 tetramer and H2A/H2B dimer on heterochromatin structure and transcriptional silencing. The general state, stability, and conformational heterogeneity of chromatin are examined with a DNA topology-based assay, and the primary chromatin structure is probed by micrococcal nuclease. We demonstrate that the histone mutations differentially affect heterochromatin. Mutations of lysine 16 of histone H4 (H4-K16) and residues in the LRS (loss of rDNA silencing) domain of nucleosome surface markedly alter heterochromatin structure, supporting the notion that H4-K16 and LRS play key roles in heterochromatin formation. Deletion of the aminoterminal tail of H3 moderately alters heterochromatin structure. Interestingly, a group of mutations in the globular domains of H3 and H4 that abrogate or greatly reduce transcriptional silencing increase the conformational heterogeneity and/or reduce the stability of heterochromatin without affecting its overall structure. Surprisingly, yet another series of mutations abolish or reduce silencing without significantly affecting the structure, stability, or conformational heterogeneity of heterochromatin. Therefore, histone residues may contribute to the structure, stability, conformational heterogeneity, or other yet-to-be-characterized features of heterochromatin important for transcriptional silencing.
DNA is packed via the formation of chromatin in eukaryotes. The basic unit of chromatin is the nucleosome consisting of 147 bp of DNA wrapping around a protein core made of two copies of each of core histones H2A, H2B, H3, and H4 (Luger et al. 1997). The primary chromatin structure composed of nucleosomes connected by linker DNAs can be folded further to form higher-order structures (Tremethick 2007). The degree of DNA compaction is not uniform across the genome, resulting in the existence of interspersed, highly condensed heterochromatin regions and decondensed euchromatin regions (Grewal and Moazed 2003; Dillon 2004). Formation of heterochromatin is mediated by silencing complexes consisting of repressor proteins and enzymes responsible for carrying out heterochromatin-specific histone modifications (Grewal and Moazed 2003). Nucleosomes in heterochromatin are arranged with a characteristically high regularity that is believed to be important for the folding of chromatin fiber into higher-order structures (Wallrath and Elgin 1995; Sun et al. 2001; Dillon 2004). Heterochromatin generally silences the expression of genes embedded in it, whereas euchromatin is permissive to gene expression. However, the exact mechanism of transcriptional silencing is not known.
In Saccharomyces cerevisiae, heterochromatin at the HML, HMR, and telomeric loci is formed via the association of the SIR complex consisting of Sir2p through Sir4p with nucleosomes (Rusche et al. 2003). Sir2p, which does not bind chromatin directly, is a histone deacetylase responsible for the hypoacetylation of histones in heterochromatin (Moazed 2001). Sir3p and Sir4p interact with the amino (N)-terminal tails of histones H3 and H4, and Sir3p also binds a region called LRS (loss of rDNA silencing) on the solvent-accessible surface of the nucleosome (Hecht et al. 1995; Park et al. 2002; Liou et al. 2005; Norris et al. 2008; Sampath et al. 2009). These interactions are believed to be key to the binding/propagation of SIR complex along chromatin.
As core histones are fundamental architectural components of chromatin, their mutations may affect the structure, dynamics, and positioning of nucleosomes, thereby affecting chromatin structure and function. In higher eukaryotes, each core histone is encoded by multiple genes at distinct loci (e.g., >10 genes are coding for histone H4 in humans) (Albig et al. 1997; Marzluff et al. 2002), making genetic analyses of histones very difficult. On the other hand, in S. cerevisiae, each core histone is encoded by only 2 genes (Hereford et al. 1979; Smith and Andresson 1983). This, combined with the high amenability of yeast, has made yeast an ideal system for genetic analyses of histone functions. Previous mutational studies have examined the roles of individual histone residues in cellular functions, including transcriptional silencing and DNA damage repair (Hyland et al. 2005; Dai et al. 2008). However, there is only limited information about how histone residues contribute to the structure of heterochromatin in vivo (E. Y. Xu et al. 2005; Yu et al. 2009).
We and others have previously developed a convenient assay to probe the state of chromatin structure at specific loci in the yeast genome by examining the topology of DNA at these loci (Bi and Broach 1997; Cheng et al. 1998). This is based on the fact that the supercoiling of eukaryotic DNA at a specific locus is mainly determined by the number of nucleosomes present there, as formation of each nucleosome constrains on average one negative supercoil on nucleosomal DNA (Simpson et al. 1985). Acetylation of histones reduces the negative supercoiling of DNA (Norton et al. 1989, 1990). Moreover, high-order chromatin structures that involve looping or coiling of the chromatin fiber may also contribute to DNA supercoiling. Therefore, the supercoiling of DNA can be used as an indicator of the overall state of chromatin. In the DNA topology-based assay for chromatin structure, the part of genome of interest is excised in vivo as a circular minichromosome via controlled site-specific recombination, and the DNA circle can then be isolated and analyzed by gel electrophoresis in the presence of a DNA intercalator to resolve the topoisomers of the circle (Figure 1B) (Bi and Broach 1997; Cheng et al. 1998).
Figure 1.—
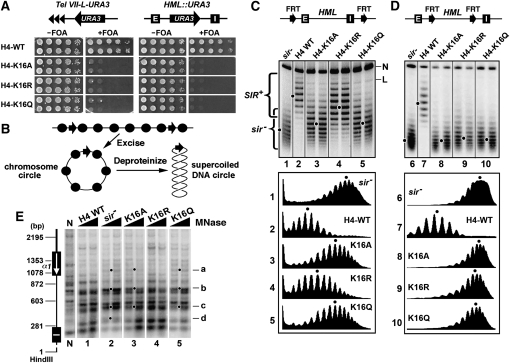
Effects of H4-K16 mutations on transcriptional silencing and heterochromatin structure. (A) Growth phenotypes of two independent clones of each of the strains UCC1111-WT, UCC1111-H4-K16A, -H4-K16R, and -H4-K16Q bearing URA3 integrated at Tel VII-L (left), as well as YQY736-WT, YQY736-H4-K16A, -H4-K16R, and -H4-K16Q bearing URA3 integrated at modified HML locus (right). Cells were grown to late log phase and serial 10-fold dilutions were spotted on synthetic complete medium with (+) or without (−) 1 mg/ml 5-fluoroorotic acid (FOA), and allowed to grow for 3 days. The silencing reporter constructs are illustrated at the top. Tel VII-L, left telomere of chromosome VII. E and I, the HML-E and -I silencers, respectively. Note that cells expressing URA3 are sensitive to FOA. (B) Strategy for examining the topology of chromosomal DNA. Two target sequences for a site-specific recombinase (arrows) are inserted to flank the chromosome region of interest. Recombination between these sequences excises the region as a minichromosome circle. After deproteinization, the topology of the DNA circle can be examined by electrophoresis in the presence of a DNA intercalator. Circles, nucleosomes. (C and D) Effects of H4-K16 mutations on the supercoiling of HML DNA. Two FRT (Flp1p recombination target) sites are inserted in direct orientation at HML to flank a region that includes (C, top) or excludes (D, top) the silencers. Middle: Cells of strains YQY695-WT, YQY695-H4-K16A, -H4-K16R, and -H4-K16Q (C), as well as YLO80-WT, YLO80-H4-K16A, -H4-K16R, and -H4-K16Q (D) were grown in YPR to late log phase, at which time galactose was added. After 2.5 hr of incubation, DNA was isolated and subjected to gel electrophoresis in the presence of 26 μg/ml chloroquine. Under this condition, more negatively supercoiled circles migrate more slowly. The positions of nicked (N) and linear (L) circles are indicated. Topoisomers of HML circle associated with silent or active chromatin were labeled as SIR+ or sir−, respectively. Densitometer scan of each lane was shown at the bottom. The distribution center of topoisomers in each sample is indicated by a dot. Note two independent clones of each H4-K16 mutant were examined. (E) Effects of H4-K16 mutations on HML heterochromatin as analyzed by MNase digestion. Cells of each of the strains YQY695-WT (H4-WT), YQY695-H4-K16A, -H4-K16R, and -H4-K16Q, as well as YQY695s (sir−), were first grown to log phase and then permeabilized and treated with MNase at 120 U/ml or 160 U/ml. DNA was isolated, digested with HindIII, and fractionated on an agarose gel. N, genomic DNA isolated from cells not treated with MNase and digested with HindIII. DNA fragments ending at the HindIII site were detected using a probe corresponding to a 200-bp sequence abutting this site. The positions of the HML-I silencer and the HMLα1 gene are shown on the left. Dots labeled a through d are sites that are uniquely sensitive in the sir− strain.
Using the DNA topology-based assay for chromatin structure, we have previously shown that DNA at the HML locus is more negatively supercoiled when the locus is transcriptionally silenced than when it is derepressed (Bi and Broach 1997), which is consistent with the higher regularity/density of nucleosomes and less acetylation of histones in heterochromatin vs. derepressed chromatin. Moreover, we showed that an increase in the negative supercoiling of HML DNA correlates with enhanced transcriptional silencing (Bi et al. 2004a), which further supports the notion that a higher degree of negative supercoiling reflects higher compaction of chromatin.
To gain insights into the mechanism by which histone residues contribute to heterochromatin structure and function, we used the DNA topology-based assay for chromatin structure together with chromatin mapping with micrococcal nuclease (MNase) to systematically examine the effects of a series of 106 histone H3 and H4 mutations on heterochromatin structure at the HML locus. These mutations represent a substantial portion of a recently described library of histone H3 and H4 mutations covering every residue in these proteins (Hyland et al. 2005; Dai et al. 2008). We identified histone mutations that alter the structure, stability, and/or conformational heterogeneity of heterochromatin, which is correlated with their effects on transcriptional silencing. On the other hand, interestingly, we also found some histone mutations to negatively affect transcriptional silencing without a detectable impact on the structure of heterochromatin.
MATERIALS AND METHODS
Plasmids and yeast strains:
Plasmids used in this study are listed in Table 1. The LEU2 gene in plasmids pYC318 through 348 was replaced by the TRP1 gene to make pXZ22 through pXZ25, respectively. Plasmid pXZ26 was similarly derived from pMS358. pH3-H4 is a CEN-TRP1-based plasmid carrying the HHT2-HHF2 cassette. Plasmid pH3-H4-K16Q is identical to pH3-H4 except carrying an hhf2 allele encoding H4-K16Q. The other plasmids carrying H3 or H4 single mutant alleles are similar named, but not listed in Table 1.
TABLE 1.
Plasmids
| Name | Description | Source/reference |
| pMS329 | CEN-URA3-HHT1-HHF1 | Morgan et al. (1991) |
| pRS412-HHT2-HHF2 | CEN-ADE2-HHT2-HHF2 | Kelly et al. (2000) |
| pH3-H4 | CEN-TRP1-HHT2-HHF2 | Hyland et al. (2005) |
| pH3-H4-K16Qa | CEN-TRP1-HHT2-hhf2-K16Q | Hyland et al. (2005) |
| pH3-H4-K8,16Qb | CEN-TRP1-HHT1-hhf1-K8,16Q | Zhang et al. (1998) |
| pH3-H4-K8,16Rc | CEN-TRP1-HHT1-hhf1-K8,16R | Zhang et al. (1998) |
| pYC318 | CEN-LEU2-hht1(Δ1–20)-HHF1 | C. Yu et al. (2006) |
| pYC328 | CEN-LEU2-hht1(Δ1–15)-HHF1 | C. Yu et al. (2006) |
| pYC338 | CEN-LEU2-hht1(Δ110)-HHF1 | C. Yu et al. (2006) |
| pYC348 | CEN-LEU2-hht1(Δ1–5)-HHF1 | C. Yu et al. (2006) |
| pMS358 | CEN-TRP1-hht1(Δ1–28)-HHF1 | Morgan et al. (1991) |
| pXZ22 | CEN-TRP1-hht1(Δ1–20)-HHF1 | This work |
| pXZ23 | CEN-TRP1-hht1(Δ1–15)-HHF1 | This work |
| pXZ24 | CEN-TRP1-hht1(Δ1–10)-HHF1 | This work |
| pXZ25 | CEN-TRP1-hht1(Δ1–5)-HHF1 | This work |
| pXZ26 | CEN-TRP1-hht1(Δ1–28)-HHF1 | This work |
Other CEN-TRP1 plasmids carrying mutant alleles of HHT2 or HHF2 are similarly named and are not listed here.
Also known as pXZ414-F47 (Zhang et al. 1998).
Also known as pXZ414-F49 (Zhang et al. 1998).
Yeast strains are listed in Table 2. Strains used to examine the effects of histone H3 and H4 mutations on the topology of HML DNA with silencers were derived from YXB94 (E. Y. Xu et al. 2005) as follows. The open reading frame (ORF) of HHT2 in YXB94 was replaced by natMX to make YQY671. The CEN-URA3-HHF1 plasmid in YQY671 was replaced by pH3-H4 (CEN-TRP1-HHT2-HHF2) to make strain YXB671a. YQY695-WT was derived from YQY671a by replacing its HHT1 with LoxP-kanMX-LoxP. YQY695s was derived from YXB695-WT by deleting its kanMX at hht1Δ::LoxP-kanMX-LoxP via Cre-mediated recombination, followed by replacement of SIR2 with kanMX. Plasmid pH 3-H4 in YQY695-WT was replaced by pMS329 (CEN-URA3-HHT1-HHF1) to make strain YQY699. Plasmids carrying mutant alleles of HHT1 or HHF1 (Table 4) derived from pH 3-H4 were used to individually replace pMS329 in YQY699 to make a series of histone H3 and H4 mutant strains for examining the topology of HML DNA-bearing silencers. For example, YQY695-H4-K16Q was made by replacing pMS329 in YQY699 with pH3-H4-K16Q (CEN-TRP1-HHT2-hhf2-K16Q).
TABLE 2.
Yeast strains
| Name | Genotype | Source/Reference |
| YXB94 | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 hhf1Δ::HIS3 hhf2Δ::LEU2 FRT-E-hml::β1-I-FRT [ciro] + p(CEN-URA3-HHF1) | E. Y. Xu et al. (2005) |
| YQY671 | YXB94, hht2Δ::natMX | This work |
| YQY671a | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 hhf1Δ::HIS3 hhf2Δ::LEU2 hht2Δ::natMX FRT-E-hml::β1-I-FRT [ciro] + pH3-H4 (CEN-TRP1-HHT2-HHF2) | This work |
| YQY695-WT | YQY671a, hht1Δ::LoxP-kanMX-LoxP | This work |
| YQY695s | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 hhf1Δ::HIS3 hhf2Δ::LEU2 hht1Δ::LoxP hht2Δ::natMX sir2Δ::kanMX FRT-E-hml::β1-I-FRT [ciro] + pH3-H4 (CEN-TRP1-HHT2-HHF2) | This work |
| YQY699 | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 hhf1Δ::HIS3 hhf2Δ::LEU2 hht1Δ::LoxP-kanMX-LoxP hht2Δ::natMX FRT-E-hml::β1-I-FRT [ciro] + pMS329 (CEN-URA3-HHT1-HHF1) | This work |
| YQY695 | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP | This work |
| -H4-K16Qa | hhf1Δ::HIS3 hhf2Δ::LEU2 hht1Δ::LoxP-kanMX-LoxP hht2Δ::natMX FRT- E-hml:: β1–I-FRT [ciro] + pH3-H4-K16Q (CEN-TRP1-HHT2-hhf2-K16Q) | |
| YXB102 | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 hhf1Δ::HIS3 hhf2Δ::LEU2 E-FRT-hml::β1-FRT-I [ciro] + p(CEN-URA3-HHF1) | Bi et al. (2004a) |
| YLO78 | YXB102, hht2Δ::natMX | |
| YLO79 | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 hhf1Δ::HIS3 hhf2Δ::LEU2 hht2Δ::natMX E-FRT-hml:: β1-FRT-I [ciro] + pH 3-H4 (CEN-TRP1-HHT2-HHF2) | This work |
| YLO80-WT | YLO79, hht1Δ::LoxP-kanMX-LoxP | This work |
| YLO80s | MATa ura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 hhf1Δ::HIS3 hhf2Δ::LEU2 hht1Δ::LoxP hht2Δ::natMX sir2Δ::kanMX E-FRT-hml::β1-FRT-I [ciro] + pH3-H4 (CEN-TRP1-HHT2-HHF2) | This work |
| YLO81 | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 hhf1Δ::HIS3 hhf2Δ::LEU2 hht1Δ::LoxP-kanMX-LoxP hht2Δ::natMX E-FRT-hml::β1-FRT-I [ciro] + pMS329 (CEN-URA3-HHT1-HHF1) | This work |
| YLO80- | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 | This work |
| H4-K16Qb | hhf1Δ::HIS3 hhf2Δ::LEU2 hht1Δ::LoxP-kanMX-LoxP hht2Δ::natMX E-FRT-hml:: β1-FRT-I [ciro] + pH3-H4-K16Q (CEN-TRP1-HHT2-hhf2-K16Q) | |
| UCC1111 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ ura3Δ Δhhf2-hht2::MET15 Δhhf1-hht1::LEU2 Adh4::URA3-TEL-VII-L + pRS412-HHT2-HHF2 (CEN-ADE2-HHT2-HHF2) | Kelly et al. (2000) |
| UCC1111- WT | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ ura3Δ Δhhf2-hht2::MET15 Δhhf1-hht1::LEU2 adh4::URA3-TEL-VII-L + pH3-H4 | This work |
| UCC1111 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 | This work |
| -H4-K16Qc | trp1Δ ura3Δ Δhhf2-hht2::MET15 Δhhf1-hht1::LEU2 adh4::URA3-TEL-VII-L + pH3-H4-K16Q | |
| YQY736 | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP | This work |
| -H4-K16 | hhf1Δ::HIS3 hhf2Δ::LEU2 hht1Δ::LoxP-kanMX-LoxP hht2Δ::natMX FRT- E-hml::β1-URA3-I [ciro] + pH3-H4-K16Q (CEN-TRP1-HHT2-hhf2-K16Q) | |
| YLO85 | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 hhf1Δ::HIS3 hhf2Δ::LEU2 hht1Δ::LoxP-kanMX-LoxP hht2Δ::natMX FRT- E-hml::β1-URA3-I [ciro] + pRS412-HHT2-HHF2 (CEN-ADE2-HHT2-HHF2) | This work |
| YQY736-WT | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 hhf1Δ::HIS3 hhf2Δ::LEU2 hht1Δ::LoxP-kanMX-LoxP hht2Δ::natMX FRT- E-hml::β1-URA3-I [ciro] + pH3-H4 | This work |
| YQY736 | MATaura3 his3 ade2 can1 trp1 his5 LEU2-GAL10-FLP1 | This work |
| -H4-K16Ad | hhf1Δ::HIS3 hhf2Δ::LEU2 hht1Δ::LoxP-kanMX-LoxP hht2Δ::natMX FRT- E-hml::β1-URA3-I [ciro] + pH3-H4-K16A |
YQY695-derived strains carrying other histone H3 or H4 mutant alleles are similarly named and are not listed here.
YLO80-derived strains carrying other histone H3 or H4 mutant alleles are similarly named and are not listed here.
UCC1111-derived strains carrying other histone H3 or H4 mutant alleles are similarly named and are not listed here.
YQY736-derived strains carrying other histone H3 or H4 mutant alleles are similarly named and are not listed here.
TABLE 4.
Effects of histone H3 and H4 mutations on transcriptional silencing and heterochromatin structure
| Histone H3 or H4 allele | Telomeric silencing | HML silencing | HML DNA supercoiling | HML chromatin stability |
| H3-WT | +++++a | +++++ | WTb | WT |
| H3-R2A | + | +++++ | WT | WT |
| H3-R2K | ++++ | +++++ | WT | WT |
| H3-T6A | ++++ | +++++ | WT | WT |
| H3-T6E | ++ | +++++ | WT | WT |
| H3-K9A | +++++ | +++++ | WT | WT |
| H3-K9R | +++++ | +++++ | WT | WT |
| H3-K9Q | +++++ | +++++ | WT | WT |
| H3-S10A | ++++ | +++++ | ||
| H3-S10E | ++++ | +++++ | ||
| H3-T11A | ++++ | +++++ | ||
| H3-T11E | ++++ | +++++ | ||
| H3-K14A | — | ++++ | ΔLkc = +1.5 | WT |
| H3-K14R | ++ | +++++ | WT | WT |
| H3-K14Q | + | ++++ | ΔLk = +1.5 | WT |
| H3-R17A | ++ | ++++ | ΔLk = +0.5 | WT |
| H3-R17K | +++++ | +++++ | ΔLk = −0.5 | WT |
| H3-K18A | ++++ | +++++ | WT | WT |
| H3-K18R | ++++ | +++++ | WT | WT |
| H3-K18Q | ++++ | +++++ | WT | WT |
| H3-S22F | +++++ | +++++ | WT | WT |
| H3-K23A | +++++ | +++++ | ||
| H3-K23R | +++++ | +++++ | ||
| H3-K23Q | +++++ | +++++ | ||
| H3-R26A | +++++ | +++++ | ||
| H3-R26K | +++++ | +++++ | ||
| H3-K27A | ++++ | +++++ | ||
| H3-K27R | +++ | +++++ | ||
| H3-K27Q | ++++ | +++++ | ||
| H3-S28A | +++++ | +++++ | ||
| H3-S28E | +++++ | +++++ | ||
| H3-K36A | +++ | +++++ | ΔLk = −1 | WT |
| H3-K36R | +++ | +++++ | ΔLk = −0.5 | WT |
| H3-K36Q | +++ | +++++ | ΔLk = −1 | WT |
| H3-R53A | +++++ | +++++ | ||
| H3-R53K | +++++ | +++++ | ||
| H3-R53Q | ++++ | +++++ | ||
| H3-K56A | ++++ | ++++ | ΔLk = −0.5 | Reduced |
| H3-K56R | + | +++ | ΔLk = −0.5 | WT |
| H3-K56Q | ++ | ++++ | WT | WT |
| H3-R72G | — | + | ΔLk = +1 | WT |
| H3-A75V | — | + | WT | WT |
| H3-F78L | +++++ | +++++ | ΔLk = −0.5 | WT |
| H3-K79A | — | — | ΔLk = +0.5 | Reduced |
| H3-K79R | +++ | +++++ | ΔLk = −0.5 | Reduced slightly |
| H3-K79Q | — | — | WT | Reduced |
| H3-K79E | — | — | ΔLk = +0.5/heterogeneousd | Reduced |
| H3-D81G | +++ | ++++ | WT | WT |
| H3-L82S | — | — | ΔLk = +2.5 | Reduced |
| H3-R83G | — | +++ | ΔLk = +1 | WT |
| H3-K115A | +++++ | +++++ | WT | WT |
| H3-K115R | +++++ | +++++ | WT | WT |
| H3-K115Q | +++++ | +++++ | WT | WT |
| H3-K122A | ++++ | ++++ | WT | WT |
| H3-K122R | +/− | +++ | WT | WT |
| H3-K122Q | ++++ | +++++ | WT | WT |
| H3Δ(1-5) | +/− | +++++ | WT | WT |
| H3Δ(1-10) | — | +++++ | WT | WT |
| H3Δ(1-15) | — | +++ | ΔLk = +1.5 | WT |
| H3Δ(1-20) | — | +++ | ΔLk = +1.5 | WT |
| H3Δ(1-28) | — | +++ | ΔLk = +1.5 | WT |
| H4-WT | +++++ | +++++ | WT | WT |
| H4-S1A | +++ | +++++ | ||
| H4-S1E | ++++ | +++++ | ||
| H4-R3A | +++++ | +++++ | ||
| H4-R3K | +++++ | +++++ | ||
| H4-K5A | +++++ | +++++ | ||
| H4-K5R | +++++ | +++++ | ||
| H4-K5Q | +++++ | +++++ | ||
| H4-K8A | +++++ | +++++ | ||
| H4-K8R | +++++ | +++++ | ||
| H4-K8Q | +++++ | +++++ | ||
| H4-K12A | +++ | ++++ | WT | WT |
| H4-K12R | ++++ | +++++ | WT | WT |
| H4-K12Q | +++ | +++++ | WT | WT |
| H4-K16A | — | — | ΔLk = +4.5 | Reduced |
| H4-K16R | — | — | ΔLk = +1.5 | Reduced |
| H4-K16Q | — | — | ΔLk = +4.5 | Reduced |
| H4-K8,16R | — | — | ΔLk = +3.5 | Reduced |
| H4-K8,16Q | — | — | ΔLk = +8 | N/A |
| H4-K20A | — | + | WT | WT |
| H4-K20R | +++++ | +++++ | WT | WT |
| H4-K20Q | — | — | WT | WT |
| H4-K31A | +++++ | +++++ | WT | WT |
| H4-K31R | +++++ | +++++ | WT | WT |
| H4-K31Q | +++++ | +++++ | WT | WT |
| H4-R39K | +++++ | +++++ | WT | WT |
| H4-S47A | +++ | +++++ | WT | WT |
| H4-S47E | — | +++ | ΔLk = +0.5 | WT |
| H4-K59A | — | — | WT/heterogeneous | WT |
| H4-K59R | — | +++ | WT | WT |
| H4-K59Q | — | — | WT/heterogeneous | Reduced |
| H4-V70F | — | — | WT | WT |
| H4-H75Y | ++++++ | ++++++ | WT | WT |
| H4-K77A | — | — | WT | WT |
| H4-K77R | +++++ | +++++ | WT | WT |
| H4-K77Q | ++ | +++++ | WT | WT |
| H4-K79A | — | + | WT | WT |
| H4-K79R | + | +++++ | ΔLk = −1 | WT |
| H4-K79Q | — | — | WT | WT |
| H4-K79M | — | — | WT | WT |
| H4-V81A | — | — | ΔLk = +1 | Reduced |
| H4-K91A | — | — | WT/heterogeneous | WT |
| H4-K91R | +++++ | +++++ | WT | WT |
| H4-K91Q | — | — | WT/heterogeneous | WT |
| H4-R92A | — | +++++ | WT | WT |
| H4-R92K | +++ | +++++ | WT | WT |
Transcriptional silencing in histone H3 and H4 wild-type (WT) strain is denoted +++++. Lack of silencing is denoted —.
WT indicates that the phenotype of a mutant strain is the same as, or similar to, that of the wild-type strain.
Linking number difference (ΔLk) relative to WT.
The topoisomers of the HML circle from the mutant are more heterogeneous than those in WT.
Strains used to analyze the effects of histone H3 and H4 mutations on the topology of HML DNA lacking silencers were derived from YXB102 (Bi et al. 2004a) as follows. HHT2 ORF in YXB102 was replaced by natMX to make YLO78. The CEN-URA3-HHF1 plasmid in YLO78 was replaced by pH3-H4 to make strain YLO79. YLO80-WT was derived from YLO79 by replacing its HHT1 with LoxP-kanMX-LoxP. YLO80s was derived from YLO80-WT by deleting its kanMX at hht1Δ::LoxP-kanMX-LoxP, followed by replacement of SIR2 with kanMX. Plasmid pH3-H4 in YLO80-WT was replaced by pMS329 to make strain YLO81. CEN-TRP1 plasmids carrying mutant alleles of HHT1 or HHF1 (Table 4) derived from pH3-H4 were used to individually replace pMS329 in YLO81 to make a series of histone H3 and H4 mutant strains for examining the topology of HML DNA lacking silencers. For example, YLO80-H4-K16Q was made by replacing pMS329 in YLO81 with pH 3-H4-K16Q.
Strains used to examine the effects of histone H3 and H4 mutations on telomeric silencing were derived from UCC1111 (Kelly et al. 2000). Plasmid pRS412-HHT2-HHF2 (CEN-ADE2-HHT2-HHF2) in UCC1111 was replaced by pH3-H4 to make UCC1111-WT. Plasmids carrying mutant alleles of HHT2 or HHF2 (Table 4) derived from pH3-H4 were used to individually replace pRS412-HHT2-HHF2 in UCC1111 to make a series of histone H3 and H4 mutant strains carrying URA3 integrated near the left telomere of chromosome VII. For example, UCC1111-H4-K16Q was made by replacing pRS412-HHT2-HHF2 in UCC1111 with pH3-H4-K16Q.
Strains used for examining the effects of histone H3 and H4 mutations on HML silencing were derived from YQY695-H4-K16Q. YQY736-H4-K16Q was made by transforming YQY695-H4-K16Q to Ura+ with pMB36 (Bi et al. 1999) digested with HindIII, effectively integrating URA3 at the PvuII site of the modified HML locus. YLO85 was made by replacing plasmid pH3-H4-K16 in YQY736-H4-K16Q with pRS412-HHT2-HHF2. YQY736-WT was made by replacing plasmid pRS412-HHT2-HHF2 with pH3-H4. Plasmids carrying mutant alleles of HHT2 or HHF2 (Table 4) were used to individually replace pRS412-HHT2-HHF2 in YLO85 to make a series of histone H3 and H4 mutant strains carrying HML::URA3. For example, YQY736-H4-K16A was made by replacing pRS412-HHT2-HHF2 in YLO85 with pH3-H4-K16A.
Analysis of DNA topology:
Cells were grown in YPR medium (1% yeast extract, 2% bacto-peptone, and 2% raffinose) to late log phase. Galactose (2%) was added to the culture that was further incubated for 2.5 hr to induce the expression of PGAL10-FLP1. Nucleic acids were isolated using the glass bead method, and fractionated on agarose gels supplemented with 26 μg/ml chloroquine. After Southern blotting, the DNA circles were detected by an HML-specific probe. The profiles of topoisomers from specific samples was obtained using the NIH image software.
Chromatin mapping with micrococcal nuclease and indirect end labeling:
MNase digestion of yeast chromatin and indirect end labeling were carried out as previously described (Bi et al. 2004b). About 1 × 109 log phase cells were turned into spheroplasts with zymolyase treatment. Spheroplasts were permeabilized with NP-40 as described (Kent et al. 1993). About 2 × 108 permeabilized spheroplasts were treated with MNase at 120 or 160 units/ml at 37° for 5 min. The reaction was stopped by 0.5% SDS and 25 mm EDTA, and DNA was isolated. An aliquot of DNA was determined to contain fragments suggesting the presence of nucleosome ladders by gel electrophoresis. Total genomic (naked) DNA was isolated from cells not treated with MNase and was digested with MNase at 7.5 units/ml. DNA from each sample was then digested with HindIII and run on a 1.0% agarose gel. DNA fragments ending the HindIII site near HML-I silencer were visualized by hybridization with a probe after Southern blotting.
RESULTS
We systematically examined the effects of a series of 106 histone H3 and H4 mutations on heterochromatin structure and transcriptional silencing in S. cerevisiae. The mutated residues include lysines, arginines, serines, and threonines that are known or potential targets for acetylation, methylation, or phosphorylation in yeast (Table 3), as well as residues within the LRS domain. Lysines were replaced with alanines or glutamines (to mimic the acetylated state) and arginines (to mimic an unmodified state). Arginines were changed to alanines and lysines. Serines or threonines were mutated to alanines and glutamic acids (the latter to mimic the phosphorylated form). Effects of the histone mutations on the silencing of the URA3 reporter integrated near the left telomere of chromosome VII (Tel VII-L) or at the HML locus were examined. We found that a mutation that reduces HML silencing always also reduces telomeric silencing to the same, or a greater, extent. A DNA topology-based assay was used to evaluate the overall state, as well as the stability and conformational heterogeneity of heterochromatin at the HML locus, and MNase was used to probe the primary structure of HML chromatin. Results from these experiments are summarized in Table 4, and those concerning histone mutants that markedly affect transcriptional silencing are elaborated as follows.
TABLE 3.
Histone H3 and H4 residues that are known or potential targets of acetylation, methylation, and phosphorylation in S. cerevisiae
| Histone residue | Modificationa | Exists in S. cerevisiaeb |
| H3-R2 | Methylation | |
| H3-K4 | Methylation | Yes |
| H3-K9 | Acetylation | Yes |
| H3-K9 | Methylation | |
| H3-S10 | Phosphorylation | |
| H3-14 | Acetylation | Yes |
| H3-R17 | Methylation | |
| H3-K18 | Acetylation | Yes |
| H3-K23 | Acetylation | Yes |
| H3-R26 | Methylation | |
| H3-K27 | Acetylation | Yes |
| H3-K27 | Methylation | |
| H3-S28 | Phosphorylation | |
| H3-K36 | Methylation | Yes |
| H3-R53 | Methylation | |
| H3-K56 | Acetylation | Yes |
| H3-K79 | Methylation | Yes |
| H3-K115 | Acetylation | |
| H3-K122 | Acetylation | |
| H4-S1 | Phosphorylation | Yes |
| H4-K5 | Acetylation | Yes |
| H4-K8 | Acetylation | Yes |
| H4-K12 | Acetylation | Yes |
| H4-K16 | Acetylation | Yes |
| H4-K20 | Acetylation | |
| H4-K20 | Methylation | |
| H4-K31 | Acetylation | |
| H4-S47 | Phosphorylation | |
| H4-K59 | Methylation | |
| H4-K77 | Acetylation | |
| H4-K79 | Acetylation | |
| H4-K79 | Methylation | |
| H4-K91 | Acetylation | Yes |
| H4-R92 | Methylation |
Identified in bovine (Zhang et al. 2003; Mersfelder and Parthun 2006). Some histone modifications are known to exist in S. cerevisiae.
Reference describing histone modifications in S. cerevisiae include Suka et al. (2001); Ng et al. (2002); van Leeuwen et al. (2002); Ye et al. (2005); F. Xu et al. (2005); Masumoto et al. (2005); Hyland et al. (2005); Cheung et al. (2005); Mersfelder and Parthun (2006); Zhang (2008).
Lysine 16 of histone H4 is essential for the structure of SIR-dependent heterochromatin:
H4-K16 is long known to play a critical role in transcriptional silencing (Park and Szostak 1990; Johnson et al. 1990). Consistently, we showed that changing H4-K16 to alanine (A), arginine (R), or glutamine (Q) abrogated the silencing of URA3 integrated near Tel VII-L or at the HML locus (Figure 1A). It is believed that a loss of transcriptional silencing is the result of a disruption of heterochromatin structure. However, it has not been experimentally tested whether histone mutations such as H4-K16A, -K16R, or -K16Q actually lead to a disruption of heterochromatin structure. Here we used the DNA topology-based assay for chromatin structure to address this question.
We made strain YQY699 that carries a CEN-URA3-HHT1-HHF1 plasmid and has the genomic HHT1-HHF1 and HHT2-HHF2 cassettes deleted. This strain allows convenient replacement of the CEN-URA3-HHT1-HHF1 plasmid with a set of CEN-TRP1-based plasmids bearing wild-type and mutant alleles of HHT2-HHF2. Moreover, in strain YQY699, the entire modified HML locus, including the E and I silencers, is bracketed by two tandem copies of Flp1p recombination target (FRT) for the site-specific recombinase Flp1p (Figure 1C, top), and the PGAL10-FLP1 (FLP1 under the control of the GAL10 promoter) construct was integrated at the LEU2 locus. Induction of PGAL10-FLP1 by galactose leads to the expression of Flp1p and subsequent excision of the HML locus as a circular minichromosome, capturing the chromatin structure at HML (Figure 1B). The HML DNA circle can then be isolated, and its supercoiling can be determined by resolving its topoisomers by agarose gel electrophoresis in the presence of the DNA intercalator chloroquine. Using this method, we have previously shown that HML DNA is far more negatively supercoiled when it is silenced (in a SIR+ strain) than when it is derepressed (in a sir− strain) (Figure 1C, compare lanes 2 and 1) (Bi and Broach 1997). In fact, silent HML DNA from the SIR+ strain has seven more negative supercoils than derepressed HML DNA from the sir− strain (Figure 1C, compare the center of distribution of topoisomers in 2 with that in 1). In other words, complete disruption of the silent state of HML chromatin causes a linking number change of 7 (ΔLk = 7) in HML DNA.
We found that the H4-K16A mutation sharply reduced the negative supercoiling of HML DNA by a ΔLk of 4.5 (Figure 1C, compare 3 and 2). Nevertheless, HML DNA in the H4-K16A mutant remained more negatively supercoiled (with a ΔLk of –2.5) than that in sir− mutant in which silent chromatin is completely dismantled (Figure 1C, compare 3 and 1). These results suggest that H4-K16A partially disrupts heterochromatin structure. H4-K16Q has a similar effect on the topology of HML DNA, and by inference, heterochromatin structure, as does H4-K16A (Figure 1C, compare 5 and 3). H4-K16R also reduced the negative supercoiling of HML DNA, but the change (ΔLk = 1.5) was significantly smaller than that caused by H4-K16A or -K16R (ΔLk = 4.5). Therefore, H4-K16R has a more moderate effect on heterochromatin structure relative to H4-K16A or -K16Q. Taken together, the above results demonstrate that although H4-K16A, -K16R, and -K16Q all abrogate HML silencing, they differentially compromise HML heterochromatin structure without completely dismantling it.
We next evaluated the stability of SIR-dependent chromatin in H4-K16 mutants. We have previously shown that an HML circle lacking silencers loses its silent state during cell-cycle progression, suggesting that heterochromatin is subject to disruption by cell-cycle-dependent processes upon disconnection from silencers (Bi and Broach 1997). The rate of loss of the silent state of silencer-free HML circles can be used as a proxy for the stability of HML chromatin.
We constructed histone H4-K16 wild-type and mutant strains in which FRTs bracket HML but excluding the silencers (Figure 1D, top). HML circles were excised from cells grown to late log phase. Under this condition (minimum cell growth), HML circle from wild-type strain maintained its high negative supercoiling relative to that from sir− strain (Figure 1D, compare 7 and 6). On the other hand, HML circle from H4-K16A, -K16R, or -K16Q mutant had lost its high negative supercoiling and adopted a topology similar to that of HML circle from sir− strain (referred to as sir− circle hereinafter) (Figure 1D, compare 8 through 10 with 6). Therefore, the SIR-dependent HML chromatin is less stable in the H4-K16 mutants than in wild-type cells.
Although DNA topology is a convenient indicator of the overall state of local chromatin, it does not reveal specific features of the primary chromatin structure. To complement our analyses of the effects of H4-K16 mutations on HML DNA topology, we also mapped HML chromatin directly using MNase digestion and indirect end labeling (Ryan et al. 1999; Zhang and Reese 2006). DNA from MNase-treated chromatin was isolated and subjected to digestion by HindIII that cleaves near the HML-I silencer (Figure 1E, diagram on the left) and electrophoresis and Southern blotting. DNA fragments from HML that ended at the HindIII site were detected by a probe abutting the HindIII site. Consistent with results form prior studies (Weiss and Simpson 1998; Q. Yu et al. 2006), we showed that the profile of MNase sensitive sites at HML was different between SIR+ and sir− strains (Figure 1E, compare 1 and 2). Specifically, sites a through d were more sensitive to MNase in the sir− vs. SIR+ strain (Figure 1E, compare 1 and 2). The profile of MNase sites in H4-K16A or H4-K16Q mutant was similar to that in sir− cells except that site d was cleaved less by MNase (Figure 1E, compare 3 and 5 with 2). On the other hand, MNase cleavage profile in H4-K16R mutant resembled that in SIR+ cells (Figure 1E, compare 4 and 1). These results are in line with our finding that the level of negative supercoiling of HML DNA in H4-K16A or -K16Q mutants was closer to that in sir− vs. SIR+ cells, and the opposite was the case for H4-K16R mutant (Figure 1C and 1D), further validating the use of DNA supercoiling as an indicator of chromatin structure.
A group of lysines in the globular domains of histones H3 and H4 are important for the stability and conformational heterogeneity of heterochromatin:
We found that individually changing a group of lysines within the globular domains of histones H3 and H4 including H3-K79, H4-K59, and H4-K91 to A, Q, or E (glutamic acid) increased the conformational heterogeneity and/or decreased the stability of HML chromatin, which was correlated with a loss of transcriptional silencing at HML.
H3-K79:
H3-K79 is located in the LRS domain of the solvent-exposed surface of the nucleosome (Luger et al. 1997; Park et al. 2002). Changing H3-K79 to A, Q, or E abolished silencing at the telomeric and HML loci, whereas the H3-K79R mutation moderately reduced silencing at Tel VII-L but had little or no effect on HML silencing (Figure 2A), consistent with results from earlier studies (Van Leeuwen et al. 2002; Hyland et al. 2005; Fry et al. 2006). Surprisingly, we found that none of the H3-K79 mutations significantly affected the negative supercoiling of HML DNA (Figure 2B). Note that in this work a significant change in DNA topology is defined as the absolute value of a ΔLk ≥ 1. Therefore, mutating H3-K79 apparently negatively affects transcriptional silencing without disrupting heterochromatin structure at HML.
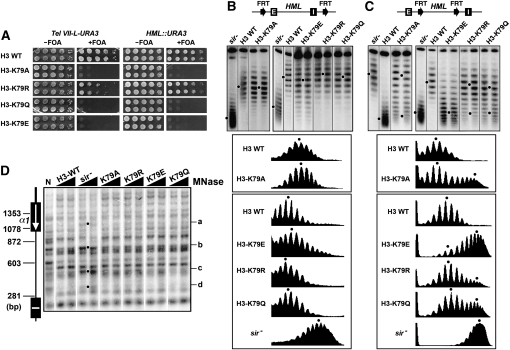
Figure 2.—
Effects of H3-K79 mutations on transcriptional silencing and heterochromatin structure. (A) Growth phenotypes of strains UCC1111-WT, UCC1111-H3-K79A, -H3-K79R, -H3-K79Q, and -H3-K79E (left), as well as YQY736-WT, YQY736-H3-K79A, -H3-K79R, -H3-K79Q, and -H3-K79E (right). Cells were grown to late log phase and serial 10-fold dilutions were spotted on synthetic complete medium with (+) or without (−) FOA. (B and C) Effects of H4-K79 mutations on the supercoiling of HML DNA. Top: Modified HML loci. Middle: HML circles from strains YQY695-WT, YQY695-H3-K79A, -H3-K79E, -H3-K79R, and -H3-K79Q (B), as well as YLO80-WT, YLO80-H3-K79A, -H3-K79E, -H3-K79R, and -H3-K79Q (C) were subjected to gel electrophoresis in the presence of chloroquine. Bottom: Densitometer scan of each lane. The distribution center of topoisomers in each sample is indicated by a dot. (D) Effects of H3-K79 mutations on HML heterochromatin. HML chromatin in each of the strains YQY695-WT, YQY695-H3-K79A, -H3-K79R, -H3-K79Q, and -H3-K79E as well as YQY695s was analyzed by MNase digestion and indirect end labeling as described in legend to Figure 1E.
To test whether H3-K79 is important for the maintenance/stability of heterochromatin we examined the effects of H3-K79 mutations on the topology of HML circle lacking silencers (Figure 2C). We found that the topoisomers of the HML circle in each mutant did not fit a single Gaussian distribution as those in H3-K79 wild-type strain did, but instead fit a profile consisting of two distinct and partially overlapping distributions, one of which was similar to that in H3 wild-type cells, and the other resembled the one in sir− cells (Figure 2C, note the existence of two distributions of topoisomers denoted by two dots in H3-K79A, -K79E, -K79R, or -K79Q). This result demonstrates that H3-K79 mutations increase the rate of disruption of heterochromatin on HML circle lacking silencers. In other words, these mutations decrease the stability of HML heterochromatin. H3-K79E had the biggest effect on the stability of HML chromatin, as the vast majority of silencer-free HML circles in H3-K79E cells lost their high negative supercoiling and adopted a topology similar to sir− circle over the period of less than one cell division (Figure 2C). On the other hand, silent HML chromatin in H3-K79R mutant was only slightly less stable compared to that in wild-type cells (Figure 2C, compare H3-K79R with H3-WT). Taken together, results in Figure 2C indicate an order of the degree of stability of HML chromatin in the H3-K79 mutants of H3-K79R > H3-K79A > H3-K79Q > H3-K79E.
We noted that the topoisomers of the silencer-bearing HML circle in the H3-K79E mutant were more heterogeneous (i.e., had a broader range of distribution) than those in wild-type cells (Figure 2B, compare H3-K79E and H3-WT). Given that each topoisomer/circle corresponds to HML chromatin in a single cell, a broader range of distribution of topoisomers reflects an increased conformational heterogeneity of HML chromatin (i.e., the range of distinct configurations HML chromatin adopts in a population of cells). Therefore, the H3-K79E mutation increases the conformational heterogeneity of HML heterochromatin, besides decreasing its stability.
We next examined whether the changes in the stability and/or conformational heterogeneity of HML chromatin caused by H3-K79 mutations were correlated with any alteration in the primary structure of chromatin. We mapped HML chromatin in H3-K79 mutants by MNase digestion and indirect end labeling. As shown in Figure 2D, the profile of MNase cleavage in each H3-K79 mutant was similar to that in wild-type cells. Therefore, the effects of mutating H3-K79 on the stability and conformational heterogeneity of HML locus are not caused by alterations in the primary structure of heterochromatin.
H4-K59:
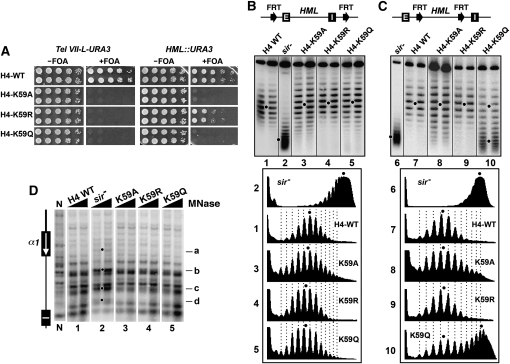
Like H3-K79, H4-K59 is also located on the solvent accessible surface of the nucleosome (Luger et al. 1997). H4-K59A and -K59Q abolished, while H4-K59R reduced, HML silencing (Figure 3A), which is consistent with, and extends, results from an earlier study (Zhang et al. 2003). None of the H4-K59 mutations altered the overall topology of HML circle as measured by the center of distribution of topoisomers (Figure 3B, compare 3 though 5 with 1), indicating a lack of significant change in HML chromatin structure. However, H4-K59A markedly increased the heterogeneity of topoisomers of HML circle (Figure 3B and 3C, compare 3 with 1, and 8 with 7). H4-K59Q modestly increased the heterogeneity of HML topoisomers, and in addition made a large portion of the silencer-free HML circles lose their high negative supercoiling (Figure 3B and 3C, compare 5 with 1, and 10 with 7). These results suggest that H4-K59A and, to a lesser extent, H4-K59Q increase the conformational heterogeneity of HML heterochromatin, and H4-K59Q also reduces the stability of HML chromatin. H4-K59R, on the other hand, did not seem to affect the overall structure, stability, or conformational heterogeneity of HML chromatin (Figure 3B and 3C, compare 4 with 1, and 9 with 7). The impact of H4-K59A or -K59Q on the conformational heterogeneity or stability of HML chromatin was not correlated with any significant change in the primary chromatin structure (Figure 3D, compare 3 and 5 with 1).
Figure 3.—
Effects of H4-K59 mutations on transcriptional silencing and heterochromatin structure. (A) Growth phenotypes of UCC1111-WT, UCC1111-H4-K59A, -H4-K59R, and -H4-K59Q (left), as well as YQY736-WT, YQY736-H4-K59A, -H4-K59R, and -H4-K59Q (right). (B and C) Effects of H4-K59 mutations on the supercoiling of HML DNA. Top: Modified HML loci. Middle: HML circles from the indicated strains were subjected to gel electrophoresis in the presence of chloroquine. Bottom: Densitometer scans of topoisomers from each sample. (D) Effects of H4-K59 mutations on HML heterochromatin. HML chromatin in the indicated strains was analyzed by MNase digestion and indirect end labeling.
H4-K91:
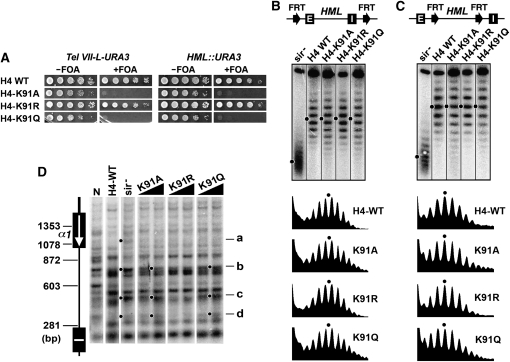
H4-K91 is located within the part of histone H4 that interacts with histone H2B and helps to stabilize H3/H4 tetramer-H2A/H2B dimer interaction involved in the formation of the histone octamer (Luger et al. 1997; Ye et al. 2005). Similar to H4-K59A and -K59Q mutations, H4-K91A and -K91Q also abolished telomeric and HML silencing (Figure 4A) and modestly increased the heterogeneity of the topoisomers of HML circle, hence the conformational heterogeneity of HML chromatin (Figure 4B). These mutations did not affect the stability of HML chromatin (Figure 4C). MNase cleavage profile of HML chromatin in H4-K91A or -K91Q mutant had certain characteristics that were similar to those in sir− strain (Figure 4D, bands b through d denoted by dots). Therefore, the increase in the conformational heterogeneity of HML chromatin induced by H4-K91A or -K91Q is correlated with partial disruption of the primary structure of heterochromatin. Unlike the H4-K91A and -K91Q mutations, H4-K91R did not affect telomeric or HML silencing (Figure 4A) and had no detectable effect on the structure, stability, or conformational heterogeneity of HML chromatin (Figure 4B through 4D).
Figure 4.—
Effects of H4-K91 mutations on transcriptional silencing and heterochromatin structure. (A) Growth phenotypes of UCC1111-WT, UCC1111-H4-K91A, -H4-K91R, and -H4-K91Q (left), as well as YQY736-WT, YQY736-H4-K91A, -H4-K91R, and -H4-K91Q (right). (B and C) Effects of H4-K91 mutations on HML DNA supercoiling. Top: Modified HML loci. Middle: Fractionation of HML circles from the indicated strains by gel electrophoresis in the presence of chloroquine. Bottom: Densitometer scans. (D) Effects of H4-K91 mutations on HML heterochromatin. HML chromatin in indicated strains was analyzed by MNase digestion and indirect end labeling.
Taken together, results described above suggest that H3-K79, H4-K59, and -K91 play roles in controlling the conformational heterogeneity of heterochromatin, and H3-K79 and H4-K59 (as well as H3-K56 discussed later) are also important for the maintenance of the stability of heterochromatin. An increase in the conformational heterogeneity or a decrease in the stability of heterochromatin seems to be sufficient to prevent transcriptional silencing.
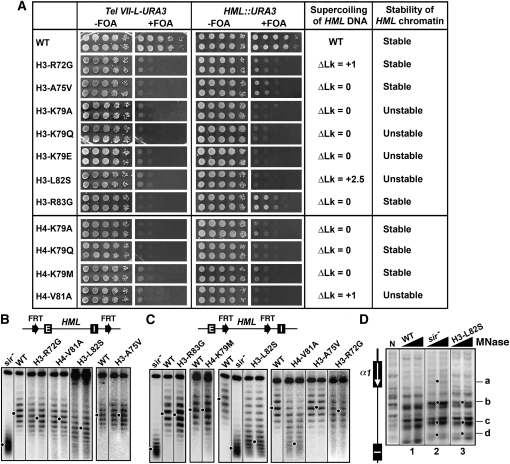
Roles of LRS residues in heterochromatin:
The LRS domain is composed of amino acids 72–83 of histone H3 and 78–81 of H4 (Park et al. 2002; Norris et al. 2008). We have just demonstrated above that LRS residue H3-K79 is involved in the stability and conformational heterogeneity of HML heterochromatin. We also examined the effects of a group of other LRS mutations on telomeric and HML silencing, as well as HML heterochromatin. These mutations eliminated or greatly reduced transcriptional silencing, but affected heterochromatin in distinct ways, as summarized in Figure 5A. The H3-L82S and H4-V81A mutations reduced the negative supercoiling of HML DNA and rendered HML chromatin unstable (Figure 5B and 5C). H3-R72G reduced the negative supercoiling of HML DNA, but had little or no effect on the stability of HML chromatin (Figure 5B and 5C). Yet the mutations H3-A75V and -R83G, as well as H4-K79A, -K79Q, and -K79M had no effect on the topology of HML DNA or HML chromatin stability (Figure 5B, 5C, 6B and 6C).
Figure 5.—
Effects of LRS mutations on transcriptional silencing and heterochromatin structure. (A) Growth phenotypes of LRS mutants bearing Tel VII-L-URA3 (left) or HML::URA3 (right). Note results for H3-K79A, -K79Q, and -K79E mutants have been shown in Figure 2 and were included here for comparison with results from other strains. (B and C) Effects of LRS mutations on HML DNA supercoiling. Top: modified HML loci. Bottom: Fractionation of HML circles from the indicated strains by gel electrophoresis in the presence of chloroquine. (D) Effect of H3-L82S on HML heterochromatin. HML chromatin was analyzed by MNase digestion and indirect end labeling.
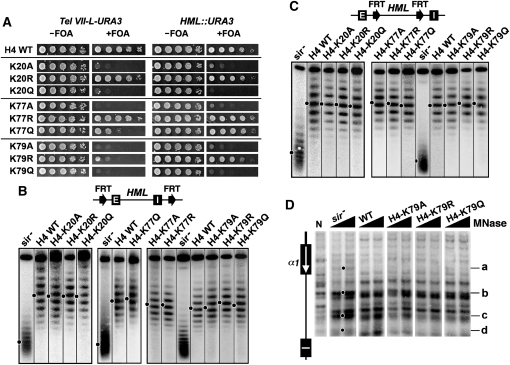
Figure 6.—
Effects of H4-K20, -K77, and -K79 mutations on transcriptional silencing and heterochromatin structure. (A) Growth phenotypes of H4-K20, -K77, and -K79 mutants bearing Tel VII-L-URA3 (left) or HML::URA3 (right). (B and C) Effects of H4-K20, -K77, and -K79 mutations on HML DNA supercoiling. Top: Modified HML loci. Bottom: Fractionation of HML circles from the indicated strains by gel electrophoresis in the presence of chloroquine. (D) Effects of H4-K79 mutations on HML heterochromatin. HML chromatin was analyzed by MNase digestion and indirect end labeling.
Of all the LRS mutations examined, H3-L82S had the greatest effect (ΔLk = 2.5) on the topology of HML DNA and the stability of HML chromatin (Figure 5B and 5C). It also induced significant changes in the profile of MNase digestion at HML (Figure 5D, compare 3 and 1). In fact, the profile of MNase sensitivity at HML in H3-L82S mutant had characteristics of that in the sir− strain (Figure 5D, compare 3 and 2), suggesting that H3-L82S disrupted the primary structure of HML heterochromatin. This is consistent with the fact that, of all the LRS mutations, only L82S has a significant effect on mating efficiency, consistent with a relatively severe silencing defect (Fry et al. 2006).
Some histone mutations greatly affect transcriptional silencing without significantly affecting heterochromatin structure:
A special heterochromatin structure is believed to be the basis of transcriptional silencing. However, surprisingly, we found that certain histone mutations abolished, or markedly reduced, transcriptional silencing without significantly affecting the structure, stability, or conformational heterogeneity of heterochromatin at HML. These mutations include LRS mutations H3-A75V, -R83G, H4-K77A, -K79A, -K79Q, and -K79M, as well as the H4-K20A, -K20Q, and V70F mutations (Figure 5, Figure 6, and Figure S1). As expected, H4-K79A and -K79Q also did not significantly affect the profile of MNase cleavage of HML chromatin (Figure 6D). This group of mutations may affect a yet-to-be-identified feature(s) of heterochromatin that escapes detection by the methods used here. Since a considerable body of evidence supports the idea that the LRS mutations reduce physical interactions with Sir3p (e.g., Norris et al. 2008), the above results suggest that avid Sir3p binding is not required for heterochromatin structure or stability.
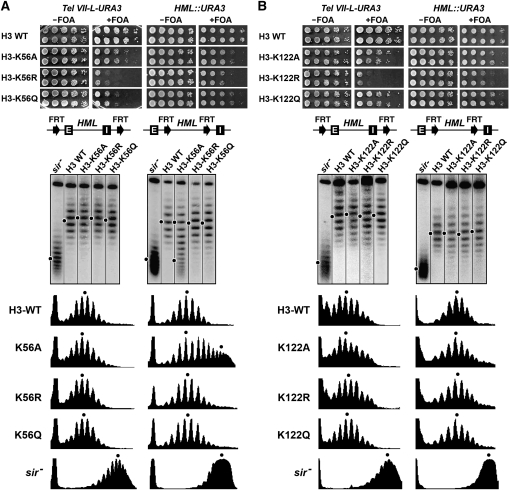
Unique roles of H3-K56 and H3-K122 in transcriptional silencing and heterochromatin structure:
Histones are rich in lysines, and the positive charges carried by them are generally important for chromatin assembly and structure. It is therefore not surprising that changing a lysine in histone H3 or H4 to R mimicking its unmodified state usually has less effect on silencing and heterochromatin structure than changing it to A or Q that does not carry positive charges (Table 4). However, we found this did not seem to apply to H3-K56 and -K122.
The H3-K56A, -K56R, and -K56Q mutations reduced telomeric and HML silencing to various extents, and it was H3-K56R that had the most severe effect (Figure 7A, top). Similarly, the H3-K122R mutation had a stronger effect on telomeric and HML silencing than H3-K122A and -122Q (Figure 7B, top). None of the H3-K56 and H3-K122 mutations affected HML DNA topology (Figure 7A and 7B, middle and bottom), and only H3-K56A destabilized HML chromatin (Figure 7A, middle right, compare K56A with sir− and H3-WT). H4-K56A or -K56R had no effect on the primary structure of HML chromatin as mapped by MNase digestion (Figure S2). H3-K56 is known to be acetylated in yeast and other eukaryotes, and H3-K122 is subject to acetylation in bovine and potentially also in yeast (Zhang et al. 2003). The above results suggest that preventing the acetylation of H3-K56 or -K122 (mimicked by the K → R mutation) has a negative impact on transcriptional silencing, despite the fact that histones, especially their N-terminal tails, are generally hypoacetylated in heterochromatin (Rusche et al. 2003).
Figure 7.—
Effects of H3-K56 and -K122 mutations on transcriptional silencing and heterochromatin structure. Results for (A) H3-K56 and (B) H3-K122 mutants are shown. Top: Growth phenotypes of H4-K56 (A) and -K122 (B) mutants bearing Tel VII-L-URA3 or HML::URA3. Middle: Fractionation of HML circles from the indicated strains by gel electrophoresis in the presence of chloroquine. Bottom: Densitometer scans.
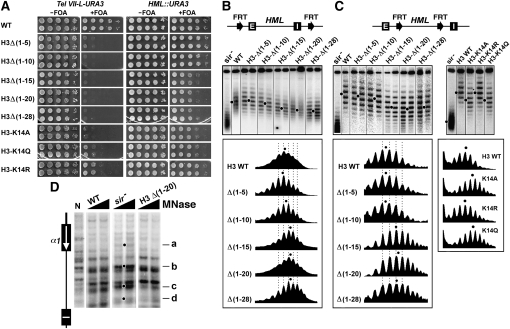
The amino terminus of histone H3 is important for the formation of fully silenced heterochromatin structure at HML:
The N terminus of histone H3 is required for telomeric silencing and is also important for efficient HML silencing (Mann and Grunstein 1992; Thompson et al. 1994). Here we reexamined the role of the N terminus of H3 in HML silencing using our HML::URA3 silencing reporter construct. We showed that deleting the first 10 residues of H3 (H3Δ(1–10)) did not affect, but H3Δ(1–15) significantly reduced the silencing of URA3 at HML (Figure 8A). Further deletion of up to 28 residues from the N terminus of H3 did not lead to more reduction in silencing (Figure 8A). These results suggest that residues 11–15 (T–G–G–K–A) of histone H3 play an important role in HML silencing. We found that mutating H3-K14 to A or Q had a similar effect on HML silencing as deletion of 15 or more residues from the H3 N terminus, whereas H3-K14R had no effect (Figure 8A).
Figure 8.—
Effects of mutations in histone H3 N-terminal domain on transcriptional silencing and heterochromatin structure. (A) Growth phenotypes of strains with H3 N-terminal truncations and H3-K14 mutations bearing Tel VII-L-URA3 (left) or HML::URA3 (right). (B and C) Effects of mutations in histone H3 N-terminal domain on the topology of HML DNA. Top: Modified HML loci. Middle: Fractionation of HML circles from indicated strains by gel electrophoresis in the presence chloroquine. Bottom: Densitometer scans. (D) Effects of H3Δ(1–20) truncation on HML heterochromatin. HML chromatin was analyzed by MNase digestion and indirect end labeling.
To test whether the N-terminal tail of histone H3 contributes to heterochromatin structure, we examined the effects of deleting increasingly larger portions of H3 N-terminal tail and mutating H3-K14 on the supercoiling of HML DNA. We found that H3Δ(1–5) and H3Δ(1–10) did not significantly affect the topology of HML DNA, but the H3Δ(1–15), H3Δ(1–20), and H3Δ(1–28) mutations all decreased the negative supercoiling of HML DNA by a ΔLk of ∼1.5 (Figure 8B and 8C). H3-K14A and -K14Q both also reduced HML DNA supercoiling by a ΔLk of ∼1.5, whereas H3-K14R had no effect (Figure 8C). These results suggest that residues 11–15 of the N-terminal tail of histone H3, especially H3-K14, are important for the formation of fully silenced HML chromatin, and that acetylation of H3-K14 (mimicked by H3-K14A or -K14Q) negatively affect silent chromatin. The H3 N-terminal truncations or H4-K14 mutations did not significantly affect the stability of HML chromatin (Figure 8C). We mapped HML chromatin in the H3Δ(1–20) mutant and showed that MNase cleavage profile in the mutant was generally similar to that in wild-type cells (Figure 8D), suggesting a lack of significant alteration in the primary structure of heterochromatin.
DISCUSSION
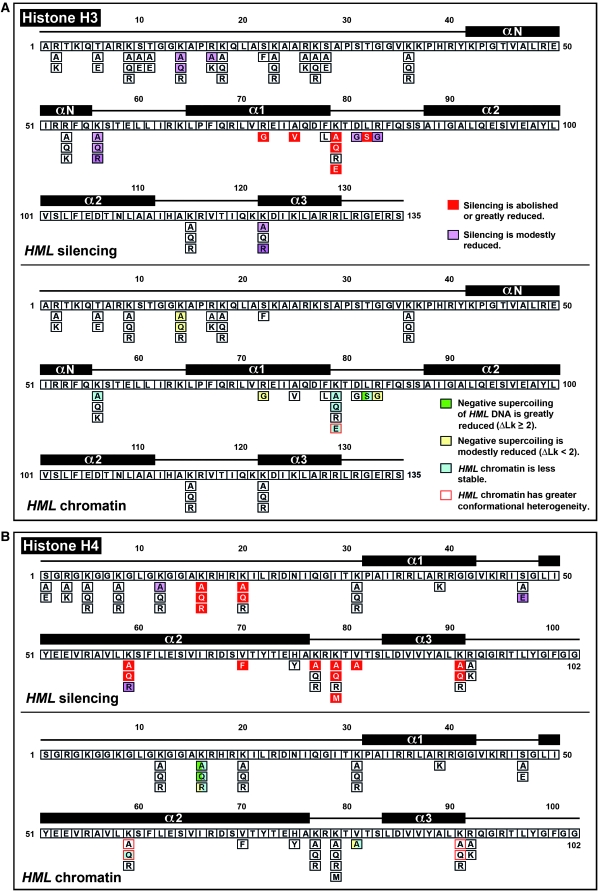
We find that histone H3 and H4 residues play various roles in transcriptional silencing and heterochromatin structure (summarized in Table 4 and Figure 9). As histones are components of chromatin, the effects of H3 and H4 mutations on heterochromatin are likely to be direct. However, it is formally possible that some mutations affect heterochromatin indirectly through, for example, modulating the expression of genes located elsewhere in the genome.
Figure 9.—
Summary of differential effects of histone H3 and H4 mutations on transcriptional silencing and heterochromatin structure at HML. (A) Effects of histone H3 mutations on HML silencing (top) and chromatin structure measured with a DNA supercoiling based assay (bottom). The histone fold domains of H3 are indicated above the sequence, and the substitutions of specific residues are indicated below the sequence. The phenotypes of H3 mutations are color coded. (B) Effects of histone H4 mutations on HML silencing (top) and chromatin structure (bottom). The phenotypes of H4 mutations are color coded similarly to those of H3 mutations.
H4-K16:
Of all the histone mutations examined, H4-K16A and -K16Q have the largest effects on HML heterochromatin, which is correlated with a loss of silencing. This is consistent with the finding that H4-K16 plays a critical role in the interaction between SIR complex and nucleosome (Hecht et al. 1995; Johnson et al. 2009). Histone acetylation is generally inhibitory to heterochromatin formation as Sir3p preferentially binds deacetylated chromatin in vitro (Johnson et al. 2009). H4-K16A disrupts Sir3p-chromatin interaction, suggesting a direct effect of H4-K16 acetylation on SIR–chromatin interaction. However, HML chromatin in H4-K16A or -K16Q mutant still bears some SIR-dependent characteristics. Therefore, H4-K16A or -K16Q alone may not completely block SIR–chromatin interaction in vivo. Consistent with this notion, we found that H4-K8,16Q double mutation completely disrupts HML heterochromatin, suggesting a synthetic effect of H4-K8Q and -K16Q on heterochromatin (Figure S3B).
H4-K16R has a smaller effect on heterochromatin than -K16A or -K16Q. This effect is likely the result of SIR complex being relocated from heterochromatin to ectopic euchromatin loci, as H4-K16R, mimicking global H4-K16 deacetylation, may promote promiscuous binding of SIR complex to euchromatin, thereby shrinking the pool of SIR complexes available for binding heterochromatin (Kimura et al. 2002; Suka et al. 2002). The H4-K8,16R double mutation has a greater effect on HML chromatin than H4-K16R (Figure 1C and Figure S3B), suggesting that H4-K8 and -K16 synergize to promote SIR–chromatin interaction,
LRS residues:
The LRS region of the solvent-accessible surface of the nucleosome is believed to interact with the BAH domain of Sir3p (Norris et al. 2008; Sampath et al. 2009). We show that a group of LRS mutations abrogate or reduce HML silencing and differentially affect the structure, stability, and conformational heterogeneity of HML heterochromatin (Figures 2 and 5). Therefore, LRS residues may contribute to transcriptional silencing by different mechanisms.
H3-K79 methylation is enriched in active chromatin and low at heterochromatin (van Leeuwen et al. 2002; Ng et al. 2003). H3-K79 methylation hinders the interaction between nucleosome and Sir3p-BAH in vitro (Onishi et al. 2007). In line with this, H3-K79R has little effect on silencing or heterochromatin (Figure 2). H3-K79A or -K79Q abolishes the positive charge, and -K79E introduces a negative charge at residue 79. The fact that these mutations all reduce the stability of HML chromatin with H3-K79E having the greatest effect suggests that the positive charge of K79 is important for heterochromatin stability. LRS mutations do not generally disrupt H3-K79 methylation or SIR complex association with HML in vivo (Fry et al. 2006). In fact, only H3-K79E caused a ∼30% decrease in SIR abundance at HML. Therefore, the reduction in silencing and changes in heterochromatin caused by LRS mutations are probably not a result of SIR dissociation from HML. As the solvent-accessible surface is involved in higher-order chromatin formation (Chodaparambil et al. 2007; Zhou et al. 2007), it is possible that LRS contributes to silencing by aiding the formation of high-order heterochromatin structure.
The N-terminal tail of histone H3:
Residues 4–20 of histone H3 were first found to be important for silencing by mutational analysis (Thompson et al. 1994). Residues 1–25 of H3 bind SIR proteins in vitro (Hecht et al. 1995). However, ChIP experiments indicate that, unlike H4 N-terminal tail, H3 tail is not required for SIR binding to chromatin, but instead contributes to the high compaction of HML heterochromatin (Sperling and Grunstein 2009). Consistently, H3 tail is involved in the folding and self-association of nucleosome arrays in vitro (Kan et al. 2007). We show here that H3 residues 11–15 are important for transcriptional silencing and high negative DNA supercoiling at HML. It is likely that the H3 tail-dependent high compaction of HML heterochromatin contributes to the high negative supercoiling of HML DNA.
The conformational heterogeneity of heterochromatin structure:
Elimination of silencing by H3-K79E, H4-K59A, -59Q, -K91A, or -K91Q is accompanied by an increase in the conformational heterogeneity of HML chromatin. H3-K79E and H4-K59A have no effect on the primary structure of HML chromatin, whereas H4-K91A and -K91Q mutations cause clear changes. Therefore, the conformational heterogeneity of heterochromatin may be determined by the primary structure or other yet-to-be-identified features of heterochromatin.
H3-K79 and H4-K59 are located on the solvent accessible surface of nucleosome and may affect high-order heterochromatin structure, thereby increasing its conformational heterogeneity. In addition, H3-K79E modestly reduces SIR association with chromatin (Fry et al. 2006). This may reflect a higher on- and off-rate of SIR associated with heterochromatin that possibly also contributes to the increased conformational heterogeneity of heterochromatin.
H4-K91 is located at the interface between H3/H4 tetramer and H2A/H2B dimer and is important for chromatin assembly (Ye et al. 2005). H4-K91A negatively regulates chromatin assembly presumably by weakening H3/H4 tetramer and H2A/H2B dimer interaction (Ye et al. 2005). Heterochromatin composed of “loosened” nucleosomes in H4-K91A or -K91Q mutant may be intrinsically more dynamic, leading to a higher conformational heterogeneity.
Residues on nucleosome lateral surface:
These residues follow the path of DNA around the histone octamer. Several bovine histone modifications involve H3 and H4 residues that either directly contact DNA or are positioned in close proximity to DNA and may interact with DNA indirectly (Zhang et al. 2003). These residues (H3-K56, -K115, -K122, H4-S47, -K77, and -K79) are conserved from yeast to mammals (Hyland et al. 2005). Mutating these residues has various effects on transcriptional silencing and heterochromatin structure at HML.
H3-K56 is near the DNA entry–exit points of nucleosome (Luger et al. 1997). H3-K56A, -K56R, and -K56Q all moderately reduce HML silencing, suggesting that the nature of the lysine side chain is important for silencing. As H3-K56 is acetylated only in S-phase (Masumoto et al. 2005), both cell-cycle-dependent acetylation and deacetylation of H3-K56 may be important for silencing; hence H3-K56A and -K56Q mimicking the persistently acetylated state and H3-K56R mimicking deacetylated state may both be inhibitory to silencing. None of the H3-K56 mutations affects heterochromatin structure, and only H3-K56A significantly reduces the stability of heterochromatin. The side chain of H3-K56 contacts DNA backbone though a H2O-bridged hydrogen bond (Luger et al. 1997; White et al. 2001). As the side chain of Q or R, but not A, can form hydrogen bonds, we reason that replacing K56 with A, but not Q or R, would prevent residue 56 of H3 to interact with DNA, thereby destabilizing heterochromatin.
Interactions between DNA and histones are at their highest at the nucleosome dyad (Luger and Richmond 1998; Hall et al. 2009). H3-K115 and -K122 lie below DNA at the dyad, and their side chain amines are poised for electrostatic interactions with the DNA backbone (Luger et al. 1997). Interestingly, acetylation of H3-K122, but not -K115, increases nucleosome mobility in vitro (Manohar et al. 2009). This may be why none of the K115A, -K115R, and -K115Q mutations affects HML chromatin or silencing. H3-K122A and -K122Q are predicted to increase nucleosome mobility and H3-K122R to reduce nucleosome mobility. Our finding that H3-K122R, but not -K122A or -K122Q, reduces HML silencing suggests that a certain degree of nucleosome mobility is important for efficient silencing. Similarly, having a certain amount of instability in Sir3p–LRS interaction was proposed to be critical for efficient silencing; either too strong or too weak a Sir3p–LRS interaction led to a loss of silencing (Norris and Boeke 2010). We think that maintaining an acetylatable (K) or acetylated (K → A or Q) state of K122 is necessary for the proper nucleosome mobility, while K → R makes nucleosomes too rigid and inhibits silencing. H4-S47 is also near the nucleosome dyad. H4-S47A mimicking unmodified S47 has no effect on HML silencing, whereas H4-S47E mimicking phosphorylated S47 reduces silencing (Figure S4), suggesting that H4-S47 phosphorylation is inhibitory to silencing.
H4-K77 and -K79 are near points of DNA-histone contacts around the circumference of nucleosome. It is interesting that H4-K77A abolishes HML silencing, whereas H4-K77Q and -K77R do not. Therefore, the nature of K77 side chain (possibly its polarity and/or the ability to form hydrogen bonds) is likely important for silencing. As for H4-K79, the K → A or Q mutation abolishes HML silencing, whereas K → R has little effect on silencing, suggesting that the unmodified state of this lysine is required for HML silencing.
Determinants of transcriptional silencing:
It is surprising that none of the aforementioned mutations on the lateral surface of nucleosome significantly affects HML chromatin despite that some of them abolish or significantly reduce HML silencing (Figure 9). Moreover, H3-A75V, H4-K20A, and -K20Q also abrogate or reduce silencing without affecting heterochromatin. What then determines transcriptional silencing? We posit that a conventionally defined heterochromatin structure with highly ordered nucleosomes and high negative DNA supercoiling is necessary but not sufficient for efficient silencing. Other properties of heterochromatin such as higher-order structures may also be required for silencing. The existence of a high-order, or more compact, structure at silent HML in vivo was supported by the finding that HML chromatin released from the genome migrates faster in sucrose gradient when it is silent vs. derepressed (Sperling and Grunstein 2009). Such a high-order structure likely involves the folding of the nucleosome array in heterochromatin. It will be interesting to investigate whether histone mutations that negatively affect silencing without affecting DNA topology and nucleosome positioning at HML alter the high-order structure or compactness of HML chromatin.
Acknowledgments
We thank Sharon Dent, Randall Morse, Mark Parthun, and M. Mitchell Smith for their gifts of plasmids and yeast strains. This work was supported by National Institutes of Health grants GM62484 to X.B. and U54 RR020839 to J.D.B.
LITERATURE CITED
- Albig W., Kioschis P., Poustka A., Meergans K., Doenecke D., 1997. Human histone gene organization: nonregular arrangement within a large cluster. Genomics 40: 314–322 [DOI] [PubMed] [Google Scholar]
- Bi X., Broach J. R., 1997. DNA in transcriptionally silent chromatin assumes a distinct topology that is sensitive to cell cycle progression. Mol. Cell. Biol. 17: 7077–7087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Braunstein M., Shei G. J., Broach J. R., 1999. The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc. Natl. Acad. Sci. USA 96: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Yu Q., Sandmeier J. J., Elizondo S., 2004a. Regulation of transcriptional silencing in yeast by growth temperature. J. Mol. Biol. 344: 893–905 [DOI] [PubMed] [Google Scholar]
- Bi X., Yu Q., Sandmeier J. J., Zou Y., 2004b. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol. Cell. Biol. 24: 2118–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T. H., Li Y. C., Gartenberg M. R., 1998. Persistence of an alternate chromatin structure at silenced loci in the absence of silencers. Proc. Natl. Acad. Sci. USA 95: 5521–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W. L., Turner F. B., Krishnamoorthy T., Wolner B., Ahn S. H., et al. , 2005. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15: 656–660 [DOI] [PubMed] [Google Scholar]
- Chodaparambil J. V., Barbera A. J., Lu X., Kaye K. M., Hansen J. C., et al. , 2007. A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat. Struct. Mol. Biol. 14: 1105–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Hyland E. M., Yuan D. S., Huang H., Bader J. S., et al. , 2008. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134: 1066–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon N., 2004. Heterochromatin structure and function. Biol. Cell 96: 631–637 [DOI] [PubMed] [Google Scholar]
- Fry C. J., Norris A., Cosgrove M., Boeke J. D., Peterson C. L., 2006. The LRS and SIN domains: two structurally equivalent but functionally distinct nucleosomal surfaces required for transcriptional silencing. Mol. Cell. Biol. 26: 9045–9059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S. I., Moazed D., 2003. Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- Hall M. A., Shundrovsky A., Bai L., Fulbright R. M., Lis J. T., et al. , 2009. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 16: 124–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S. M., Grunstein M., 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592 [DOI] [PubMed] [Google Scholar]
- Hereford L., Fahrner K., Woolford J., Jr., Rosbash M., Kaback D. B., 1979. Isolation of yeast histone genes H2A and H2B. Cell 18: 1261–1271 [DOI] [PubMed] [Google Scholar]
- Hyland E. M., Cosgrove M. S., Molina H., Wang D., Pandey A., et al. , 2005. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 25: 10060–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A., Li G., Sikorski T. W., Buratowski S., Woodcock C. L., et al. , 2009. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol. Cell 35: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. M., Kayne P. S., Kahn E. S., Grunstein M., 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87: 6286–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan P. Y., Lu X., Hansen J. C., Hayes J. J., 2007. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol. Cell. Biol. 27: 2084–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Qin S., Gottschling D. E., Parthun M. R., 2000. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol. Cell. Biol. 20: 7051–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent N. A., Bird L. E., Mellor J., 1993. Chromatin analysis in yeast using NP-40 permeabilised sphaeroplasts. Nucleic Acids Res. 21: 4653–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Umehara T., Horikoshi M., 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32: 370–377 [DOI] [PubMed] [Google Scholar]
- Liou G. G., Tanny J. C., Kruger R. G., Walz T., Moazed D., 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121: 515–527 [DOI] [PubMed] [Google Scholar]
- Luger K., Richmond T. J., 1998. DNA binding within the nucleosome core. Curr. Opin. Struct. Biol. 8: 33–40 [DOI] [PubMed] [Google Scholar]
- Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Mann R. K., Grunstein M., 1992. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 11: 3297–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar M., Mooney A. M., North J. A., Nakkula R. J., Picking J. W., et al. , 2009. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J. Biol. Chem. 284: 23312–23321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F., Gongidi P., Woods K. R., Jin J., Maltais L. J., 2002. The human and mouse replication-dependent histone genes. Genomics 80: 487–498 [PubMed] [Google Scholar]
- Masumoto H., Hawke D., Kobayashi R., Verreault A., 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436: 294–298 [DOI] [PubMed] [Google Scholar]
- Mersfelder E. L., Parthun M. R., 2006. The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 34: 2653–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., 2001. Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 13: 232–238 [DOI] [PubMed] [Google Scholar]
- Morgan B. A., Mittman B. A., Smith M. M., 1991. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol. Cell. Biol. 11: 4111–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Ciccone D. N., Morshead K. B., Oettinger M. A., Struhl K., 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 100: 1820–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., et al. , 2002. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16: 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A., Boeke J. D., 2010. Silent information regulator 3: the Goldilocks of the silencing complex. Genes Dev. 24: 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A., Bianchet M. A., Boeke J. D., 2008. Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction. PLoS Genet. 4: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton V. G., Imai B. S., Yau P., Bradbury E. M., 1989. Histone acetylation reduces nucleosome core particle linking number change. Cell 57: 449–457 [DOI] [PubMed] [Google Scholar]
- Norton V. G., Marvin K. W., Yau P., Bradbury E. M., 1990. Nucleosome linking number change controlled by acetylation of histones H3 and H4. J. Biol. Chem. 265: 19848–19852 [PubMed] [Google Scholar]
- Onishi M., Liou G. G., Buchberger J. R., Walz T., Moazed D., 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell 28: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Park E. C., Szostak J. W., 1990. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol. Cell. Biol. 10: 4932–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Cosgrove M. S., Youngman E., Wolberger C., Boeke J. D., 2002. A core nucleosome surface crucial for transcriptional silencing. Nat. Genet. 32: 273–279 [DOI] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Ryan M. P., Stafford G. A., Yu L., Cummings K. B., Morse R. H., 1999. Assays for nucleosome positioning in yeast. Methods Enzymol. 304: 376–399 [DOI] [PubMed] [Google Scholar]
- Sampath V., Yuan P., Wang I. X., Prugar E., van Leeuwen F., et al. , 2009. Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes. Mol. Cell. Biol. 29: 2532–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M., 1985. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell 42: 799–808 [DOI] [PubMed] [Google Scholar]
- Smith M. M., Andrésson O. S., 1983. DNA sequences of yeast H3 and H4 histone genes from two non-allelic gene sets encode identical H3 and H4 proteins. J. Mol. Biol. 169: 663–690 [DOI] [PubMed] [Google Scholar]
- Sperling A. S., Grunstein M., 2009. Histone H3 N-terminus regulates higher order structure of yeast heterochromatin. Proc. Natl. Acad. Sci. USA 106: 13153–13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N., Suka Y., Carmen A. A., Wu J., Grunstein M., 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8: 473–479 [DOI] [PubMed] [Google Scholar]
- Suka N., Luo K., Grunstein M., 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32: 378–383 [DOI] [PubMed] [Google Scholar]
- Sun F. L., Cuaycong M. H., Elgin S. C., 2001. Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol. Cell. Biol. 21: 2867–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. S., Ling X., Grunstein M., 1994. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369: 245–247 [DOI] [PubMed] [Google Scholar]
- Tremethick D. J., 2007. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell 128: 651–654 [DOI] [PubMed] [Google Scholar]
- van Leeuwen F., Gafken P. R., Gottschling D. E., 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- Wallrath L. L., Elgin S. C., 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9: 1263–1277 [DOI] [PubMed] [Google Scholar]
- Weiss K., Simpson R. T., 1998. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLalpha. Mol. Cell. Biol. 18: 5392–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. L., Suto R. K., Luger K., 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20: 5207–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu E. Y., Bi X., Holland M. J., Gottschling D. E., Broach J. R., 2005. Mutations in the nucleosome core enhance transcriptional silencing. Mol. Cell. Biol. 25: 1846–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Zhang K., Grunstein M., 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121: 375–385 [DOI] [PubMed] [Google Scholar]
- Ye J., Ai X., Eugeni E. E., Zhang L., Carpenter L. R., et al. , 2005. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol. Cell 18: 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Palumbo M. J., Lawrence C. E., Morse R. H., 2006. Contribution of the histone H3 and H4 amino termini to Gcn4p- and Gcn5p-mediated transcription in yeast. J. Biol. Chem. 281: 9755–9764 [DOI] [PubMed] [Google Scholar]
- Yu Q., Elizondo S., Bi X., 2006. Structural analyses of Sum1–1p-dependent Transcriptionally silent chromatin in Saccharomyces cerevisiae. J. Mol. Biol. 356: 1082–1092 [DOI] [PubMed] [Google Scholar]
- Yu Q., Kuzmiak H., Zou Y., Olsen L., Defossez P. A., et al. , 2009. Saccharomyces cerevisiae linker histone Hho1p functionally interacts with core histone H4 and negatively regulates the establishment of transcriptionally silent chromatin. J. Biol. Chem. 284: 740–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., 2008. Qualitative and quantitative analysis of lysine acetylation and methylation in yeast histone H3. Int. J. Mass Spectrom. 269: 101–111 [Google Scholar]
- Zhang L., Eugeni E. E., Parthun M. R., Freitas M. A., 2003. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma 112: 77–86 [DOI] [PubMed] [Google Scholar]
- Zhang W., Bone J. R., Edmondson D. G., Turner B. M., Roth S. Y., 1998. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17: 3155–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Reese J. C., 2006. Isolation of yeast nuclei and micrococcal nuclease mapping of nucleosome positioning. Methods Mol. Biol. 313: 245–255 [DOI] [PubMed] [Google Scholar]
- Zhou J., Fan J. Y., Rangasamy D., Tremethick D. J., 2007. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 14: 1070–1076 [DOI] [PubMed] [Google Scholar]