Abstract
Traditional methods for detecting genes that affect complex diseases in humans or animal models, milk production in livestock, or other traits of interest, have asked whether variation in genotype produces a change in that trait’s average value. But focusing on differences in the mean ignores differences in variability about that mean. The robustness, or uniformity, of an individual’s character is not only of great practical importance in medical genetics and food production but is also of scientific and evolutionary interest (e.g., blood pressure in animal models of heart disease, litter size in pigs, flowering time in plants). We describe a method for detecting major genes controlling the phenotypic variance, referring to these as vQTL. Our method uses a double generalized linear model with linear predictors based on probabilities of line origin. We evaluate our method on simulated F2 and collaborative cross data, and on a real F2 intercross, demonstrating its accuracy and robustness to the presence of ordinary mean-controlling QTL. We also illustrate the connection between vQTL and QTL involved in epistasis, explaining how these concepts overlap. Our method can be applied to a wide range of commonly used experimental crosses and may be extended to genetic association more generally.
QUANTITATIVE trait locus (QTL) analysis has traditionally focused on detection of major genes controlling the expected mean of a phenotype. But there is substantial evidence that not only the mean but also the variance, that is, the stochastic variability of the phenotype about its average value, may itself be under genetic control. The identification of such variance-controlling loci, which we call vQTL, can be helpful in a variety of contexts, including selection of livestock for uniformity, evaluating predictability of response to medical treatment, identification of key biomolecular stabilizers, and assessment of population resilience in ecology and evolution.
One way of interpreting an increase in variability is as a decrease in stability. Waddington (1942) described the concept of canalization, whereby natural selection favors the relative constancy of some attributes, for example, well-formed organs and limbs, and thereby leads to the evolution of heritable architectures that buffer the impact of environmental or background genetic variation that would otherwise cause development to go astray. These architectures create virtual “canals” down which developmental programs flow. For a canalized phenotype, which modern usage expands to include nondevelopmental traits, the “zone of canalization” is the range of underlying liability over which potentially disruptive variation may be absorbed without serious consequence to the expressed trait value (Lynch and Walsh 1998). A well-studied example of a stabilizing architecture is that provided by heat-shock protein 90 (Hsp90), which buffers genetic and stochastic variation in the development of plants and flies (Rutherford and Lindquist 1998; Queitsch et al. 2002; Sangster et al. 2008).
But in absorbing variation, such stabilizing architectures also hide it from view, and a sensitizing change in the stabilizer that shifts liability outside the zone of canalization can have a dramatic effect on the phenotype. Such shifts release the combined effects of previously “cryptic” genetic variation: now decanalized, the phenotype is more sensitive to internal (including genetic) and external environment, and as a result varies more greatly between individuals (Dworkin 2005; Hornstein and Shomron 2006). In this vein, decanalization has been proposed to explain why the genetic architectures of some diseases in human populations seem more amenable than others to genetic dissection through genome-wide association (Gibson and Goldstein 2007). Specifically, whereas some disease phenotypes in homogeneous populations may be heavily canalized and thereby harder to dissect, others may have been decanalized by modern living conditions (e.g., inflammatory diseases) or modern admixture, while yet others are simply too recent in evolutionary history for buffering networks to have evolved (e.g., response to HIV).
Increased variability can also be adaptive. In natural populations disruptive selection favors diversity, with increased “capacitance” (Rice 2008) or “bet-hedging” (Beaumont et al. 2009) spreading risk over a variable fitness landscape. Feinberg and Irizarry (2010) recently proposed a heritable and selectable mechanism for this based on stochastic epigenetic variation. In controlled populations, variability can be increased through directional selection. For example, in a Drosophila selection experiment Clayton and Robertson (1957) reported increased bristle number variance, which is consistent with the idea that genotypes associated with higher environmental variance have a greater chance of being selected under directional selection (Hill and Zhang 2004). Moreover, genetic differences have been observed for phenotypic variability in body weight for chickens (Rowe et al. 2006) and snails (Ros et al. 2004) and litter size in rabbits (Ibanez-Escriche et al. 2008), sheep (Sancristobal-Gaudy et al. 1998), and pigs (Sorensen and Waagepetersen 2003).
In natural populations with stabilizing selection we should expect to find alleles minimizing variance for fitness traits (Lande 1980; Houle 1992), whereas directional selection during domestication will favor alleles that increase variance. One may therefore expect to find vQTL in experimental crosses between wild and domestic animals (see Andersson 2001). Nonetheless, genetic buffering that leads to phenotypic robustness need not require an evolutionary explanation to be observed, nor to be useful in medicine and agriculture. Plainly, detecting vQTL and inferring how they arose are separate questions; here we concentrate on the first.
Table 1 lists some sources of phenotypic variability in relation to the genetic groups studied. Unless otherwise qualified, we use “phenotypic variance” to describe the observed marginal variance of the phenotype in the population and distinguish “phenotypic variability” as the residual variance after controlling for main effects of QTL and other anticipated or manipulated environmental covariates. Phenotypic variability is thus the by-product of unmodeled interactions. Identifying major factors that influence variability requires defining groups between which variances would be contrasted (rows of Table 1). Our goal is to identify loci associated with differences in variance between such groups. For generality we concentrate on groups defined by the first row in Table 1, but note that the groupings defined in the remaining rows allow increasingly specific characterizations of vQTL effect. For instance, experimental crosses having multiple individuals within inbred lines will produce genetically identical individuals and the differences in phenotypic variability within each line are due to both environmental sensitivity and temporal fluctuation, but not epistasis.
TABLE 1.
Types of variance contributing to between-group differences in phenotypic variability
| Sources of phenotypic variability |
||||
| Variance groupa | Decanalization (epistasis) | Environmental sensitivity | Temporal fluctuation | Measurement error |
| Genetically distinct individuals with same allele at a vQTLb | • | • | • | • |
| Genetically identical individuals | • | • | • | |
| Same individual at different times | • | • | ||
| Same individual at the same time | • | |||
The group in which variance is assessed, and between which variance is compared.
The variance groups compared here.
Few studies have explicitly looked for vQTL. Among the more recent, Ordas et al. (2008) studied morphological traits and flowering time in maize. They detected vQTL by contrasting the residual variance between genotypes in replicates of recombinant inbred lines (RILs; see second row, Table 1). The effects were substantial, with alleles associated with a 30–40% increase over the average residual variance. Wittenburg et al. (2009) examined the sample variance of birth weight within pig litters as a gamma-distributed trait among 3914 sows, estimating a heritability of 0.1 for this trait using a generalized linear mixed model. Sangster et al. (2008) used Levene's test for detection of variance-controlling genes. In that test, the absolute values of the residuals are used as a response in an ANOVA (e.g., Faraway 2004). Mackay and Lyman (2005) studied Drosophila bristle number and found substantial differences in the coefficient of variation (CV) between inbred lines, comparing CV also using ANOVA. The methods used in these last two studies have the limitation of not being able to model confounding effects in the mean. Using residuals (as in Sangster et al. 2008; Wittenburg et al. 2009) can potentially incorporate covariates but involves conditioning on unknowns. There is thus considerable utility in a method that simultaneously estimates means and variances, flexibly accommodates covariates, applies to a wide range of experimental crosses, and is robust and fast enough for genome-wide analyses.
Regression-based models (Haley and Knott 1992; Martinez and Curnow 1992) have proven to be fast and powerful at detecting QTL controlling the mean of a complex trait in experimental crosses and flexible since they are straightforwardly extended to include epistatic effects and interactions (Carlborg and Haley 2004). Mott et al. (2000) developed the haplotype reconstruction method HAPPY and its associated regression model, which allows for a variable number of strains and may therefore be applied to vQTL mapping in, e.g., heterogeneous stocks (HS; Valdar et al. 2006,b) and multiparent advanced generation inbred cross resource populations (MAGIC lines; Cavanagh et al. 2008) such as the collaborative cross (CC; Churchill et al. 2004; Broman 2005; Valdar et al. 2006a; Chesler et al. 2008) and the Arabidopsis recombinant inbred lines of Kover et al. (2009).
Our aim is to develop a regression model for detection of major genes controlling phenotypic variance that can be applied genome wide. The estimation uses double generalized linear models (DGLMs; Smyth 1989) and its parameterization is based on the HAPPY formulation of inferred haplotypes. The method fits ordinary QTL and vQTL simultaneously in the same model. We apply it to simulated data from an F2 and the CC and real data from an F2 intercross of partially inbred lines.
MODELS AND METHODS
The standard regression model for interval mapping of a QTL uses the probability of line origin at a locus to describe its genetic state (Haley et al. 1994). In the simple case of mapping a single QTL in individuals arising from an F2 intercross of inbred founder lines A and B, the model is
| (1) |
where yi is the phenotype of individual i, xi is the ith row in a design matrix of suitable nongenetic covariates, qi represents the genetic state at the QTL, εi is the residual with variance σ2, and μ, β, and α are parameters estimated by the model. The QTL genotype gi is typically unknown but, thanks to information from linked markers M, its underlying haplotype pair is available indirectly as a probability distribution pi = (pi1, pi2, pi3), where pi1 = P(gi = AA|M), pi2 = P(gi ∈ {AB, BA}|M), and pi3 = P(gi = BB|M). The regression predictor qi in Equation 1 may therefore be formulated in terms of pi. For the additive genetic models considered here, qi = (qiA, qiB), where qiA = 2pi1 + pi2 and qiB = 2pi3 + pi2 correspond to the expected “doses” of haplotypes A and B respectively, and α corresponds to their estimated “dosage effects” on the phenotypic mean. In practice, to obviate the dependence induced by qiB = 2 – qiA, the regression model is fitted using qi = qiA, leading to a scalar effect α = α that estimates the dosage effect of A relative to B. The predictor qi may be alternatively formulated to accommodate more general effects, e.g., as qi = pi, or represent observed or imputed genotypes of known variants (e.g., Yalcin et al. 2005; Zheng et al. 2011). The QTL scan is usually summarized as a plot of the LOD score, F-statistic or, –log 10(P-value) (hereafter, log P), at each tested position along the genome. Chromosomal regions harboring QTL affecting the trait mean are located by the highest values of the test statistic above a suitable significance threshold (e.g., Broman and Sen 2009).
QTL regression model for detection of major loci controlling phenotypic variability:
We consider the regression
| (2) |
where zi contains nongenetic covariates affecting the residual variance of the model, γ is their corresponding effects vector, and θ is the dosage effect of each line on the residual variance, i.e., the additive vQTL effect. All other variables are defined as in Equation 1. The regression in Equation 2 is thus equivalent to Equation 1 but with εi ∼ N(0, σ2i) and , describing a model with separate effects for mean and variance.
Regression-based mapping of QTL (including vQTL) using Equation 2 assumes that
there are two founder lines,
the genetic state of the QTL is predicted accurately by marker data,
there is a single major QTL,
the QTL is fixed within each founder line,
the phenotype is Normally distributed, conditional on the QTL and covariate effects, and
the observed values yi are exchangeable, conditional on the QTL and covariate effects.
We present a fitting procedure for Equation 2 based on these assumptions and, thereafter, relax the assumptions one at a time to investigate the possibility of using Equation 2 for vQTL detection in empirical studies. In this article we assess Assumptions 1–4 using both simulations on F2 and the CC, and empirical results from a chicken F2 cross. We give theoretical solutions for how to relax Assumptions 5 and 6 and discuss these in the discussion. Our fitting procedure is based on DGLMs (see Appendix A) and uses the dglm package (Dunn and Smyth 2009) in R (R Development Core Team 2009).
Significance testing:
A general method for calculating P-values applicable to different trait distributions is available in dglm. The R code to extract the P-values from the dglm(.) function is given in the supporting information (File S1). We calculated 5% chromosome-wide significance thresholds by simulating chromosomes under a null model with no mean- and no variance-controlling QTL effects.
Estimation of line dosages:
We estimated line dosages at each putative QTL using probabilities estimated by the haplotype reconstruction program HAPPY (Mott et al. 2000). Given genotype data on individual i and its h founders, HAPPY uses a hidden Markov model (HMM) to infer probabilistically the haplotype pair underlying the genotypes at each marker. For every interval between adjacent pairs of markers, it then calculates the expected diplotype composition: that is, the average proportion of diplotype AA, AB, etc., that would be expected across the interval, given the interval’s length and its descent either side. The diplotype composition is reported as an h × h matrix Di for each individual i, and the expected line dosages are calculated as the h-vector qi = 1T(Di + DiT). Because qiT1 = 2 always, qi has h − 1 degrees of freedom and so we typically omit the hth element during model fitting.
Relaxing Assumption 1: More than two founder lines:
For experimental crosses with h > 2 founder lines, the predictors qi, α, and θ in Equation 2 expand to have h – 1 elements each.
Relaxing Assumption 2: Uncertain genotype states:
Uncertainty about the QTL genotype is most naturally modeled using a mixture distribution. When modeling QTL affecting the mean only, the marginal likelihood for observation i (omitting covariates) is the mixture , where pi = (pi1, pi2, pi3) is defined as above for two founder lines, N(.) is the normal density, and r = (–1, 0, 1). This is the likelihood used for maximum-likelihood (ML) estimation in interval mapping (Lander and Botstein 1989). The regression approach to interval mapping (Equation 1) treats pi as if it were an observed outcome, and as a result overestimates the residual variance for each observation by (Xu 1995; Xu 1998; Feenstra et al. 2006). When modeling mean- and variance-controlling QTL on the basis of Equation 2, the marginal likelihood is
| (3) |
The regression approach for this model overestimates residuals further, by (File S1). Although we could obtain ML estimates of the vQTL model using an EM-algorithm (Appendix B), as is done for interval mapping, we contend that the DGLM regression may be more useful. In particular, the EM-algorithm applied to this marginal likelihood is computationally slow, a marginal likelihood gives biased ML estimates for variances, and the regression approximation confers additional flexibility for modeling different distributions for the phenotype as well as different mean-variance relationships.
Relaxing Assumptions 1 and 2: Uncertain genotype states with more than two founders:
Uncertainty about line origin probability when there are multiple lines can produce multicollinearity among the genetic predictors in the regression framework. We overcome this technical problem using the dimension reduction approach described in Valdar et al. (2009), whereby the matrix of line dosage estimates from HAPPY is replaced by its informative eigenvectors.
Quantifying the effects of genotype uncertainty:
Regressing on expected dosages incorrectly models uncertainty in the predictors. At a mean-controlling QTL, individuals with less certain pi will be less well predicted by the mean part of the model, which could lead to inflated estimates of and, possibly, a false vQTL signal. We investigated this phenomenon empirically by monitoring the relationship between locus uncertainty and the proportion of false-positive vQTL in simulations of the CC. At each marker interval in each individual we quantified the uncertainty in line origin using the scaled selective information content (SIC), defined as follows. If P(gi = j) is the prior probability that individual i is in genetic state j given no marker information, and P(gi = j|M) is the posterior given marker data M, as estimated by the HAPPY HMM, then a measure of the information provided by M about the locus is the Kullback–Leibler divergence,
summed over all J possible states, with 0 log(0) ≡ 0. If we represent the states of the F2 cross as all possible phased founder diplotypes (i.e., AA, AB, BA, and BB for the cross of strains A and B) and of the CC as the set of homozygote diplotypes (i.e., AA, BB, … , HH; denoting founders by A–H), then P(gi = j) = J−1, ∀j. Rescaling as SIC = I(M, i)/log(J), the (scaled) selective information content at a locus for individual i ranges from 0, denoting equiprobable diplotypes and minimal certainty, to 1, denoting one diplotype with complete certainty.
Relaxing assumption 3: Multiple QTL:
Multiple QTL can be fitted by including additional predictors in Equation 2. Linked QTL may, however, affect the analysis if not included in the model. We therefore assessed, by means of simulations, the influence of additional independent and interacting (epistatic) QTL.
Relaxing assumption 4: QTL variation within lines:
Model 2 assumes that both the mean- and variance-controlling QTL have been fixed within the founder lines, which is reasonable for crosses from highly inbred lines but not necessarily for divergent outbred lines. It is possible that nonfixation of a mean-controlling QTL may be detected as a spurious vQTL. We illustrate this phenomenon for the Growth 2 QTL on chicken chromosome 1, which was found not to have been fixed in a divergent F2 cross (Kerje et al. 2003; Rönnegård et al. 2008).
SIMULATIONS AND DATA
We assessed the performance of our method by applying it to simulated F2 and CC populations, and to a real Red Jungle Fowl × White Leghorn F2 data set. Simulations were generated with software used for Valdar et al. (2006a) and Valdar et al. (2009) (see File S2).
Simulated F2 intercross:
Our simulated F2 population included 800 individuals each comprising a single 100-cM chromosome with 10 evenly spaced fully informative SNP markers. QTL, when simulated, were positioned midway between markers, at 45 cM. The simulated population size of 800 was chosen to reflect a typically sized F2 design such as the Red Jungle fowl × White Leghorn F2 cross described further below.
Simulated collaborative cross:
The CC is a panel of RILs descended from eight inbred founder strains: A/J, C57BL/6J, 129S1SvImJ, NOD/LtJ, NZO/H1LtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ (Churchill et al. 2004; Chesler et al. 2008). In keeping with previous simulations of this population (Valdar et al. 2006a), we simulated 1000 RIL individuals each generated from a separate breeding funnel. CC individuals comprised a single 100-cM chromosome with 1000 SNP markers drawn from the Mouse Diversity Genotyping Array (Yang et al. 2009; see http://cgd.jax.org/tools/diversityarray.shtml) and chosen to be roughly equally spaced and informative among the 8 founders. QTL, when simulated, were positioned at 28 and 68 cM, each midway between markers and in regions containing relatively informative markers.
Detection of QTL in F2 and CC with regression on known QTL genotypes:
We simulated QTL in the F2 and CC under four scenarios.
Scenario 1: No QTL effects
Scenario 2: Mean-controlling QTL effects
Scenario 3: Variance-controlling QTL effects
Scenario 4: Both mean- and variance-controlling QTL effects
Phenotypic values (Y) were generated as mean-controlling additive QTL effects (Q) plus a residual (E): Y = Q + E. For mean-controlling effects, Q = 1, 2, and 4 for QTL genotype aa, aA, and AA, respectively. These effects correspond to a QTL with a moderate effect explaining ∼2% of the phenotypic variance. For the scenarios with no vQTL, E was drawn from N(0, σ2) with σ2 = 100. For the scenarios with vQTL, σ2 = 100, 125, 156.25 for genotypes aa, aA, and AA, such that the the A allele had additive effects on the log scale and increase variance by 25% for each copy. These effects are moderate in size. Ordas et al. (2008) found variance-controlling QTL in maize where the allele effects resulted in an increase of the residual variance of 30–40%. For each scenario, we generated 10,000 replicates of the F2 simulation and 1000 replicates of the CC (positioning the QTL at 28 cM only). Model fitting was implemented using dglm function in R (R Development Core Team 2009), applying the regression in Equation 2 to the F2 populations and that in Equation 2 to the CC.
Detection of QTL for F2 and CC with regression on line dosage:
We repeated the simulations above at each marker interval fitting the DGLM to line dosages inferred by the HAPPY HMM. The simulations from scenario 1 (no effects) were used to obtain 5% chromosome-wise empirical significance levels.
Fitting a single QTL in the presence of linked QTL and epistasis:
To study a potential influence of linked QTL on estimated vQTL effects, we simulated three scenarios for the CC. DGLM regression was performed on line dosages as above, with 1,000 replicates simulated for each scenario.
Scenario E1: A single mean-controlling QTL at 28 cM
Scenario E2: Two linked mean-controlling QTL with additive effects at 28 cM and 68 cM
Scenario E3: Two linked mean-controlling QTL with epistatic effects at 28 cM and 68 cM
We generated phenotypes (Y) as Y = Q + E, with constant residual variance E ∼ N(0, 100) throughout. For Scenario E1, Q = 0, 2, or 4 for QTL genotype aa, aA, or AA. For Scenario E2, the additive QTL effects were calculated as Q = Q1 + Q2, where Qk = 0, 2, or 4 for aa, aA, or AA at QTL number k. For Scenario E3, QTL with interaction effects were simulated and Q was assigned values according to Table 2. For all three scenarios, we fitted a DGLM with line dosages as predictors for both mean and variance effects at the first QTL position only (28 cM).
TABLE 2.
Simulated epistatic effects for different QTL genotype combinations
| QTL 2 (68 cM) |
||||
| aa | aA | AA | ||
| QTL 1 (28 cM) | aa | 0 | 0 | 0 |
| aA | 0 | 2 | 0 | |
| AA | 0 | 0 | 4 | |
Red Jungle fowl × White Leghorn F2 cross: a worst-case scenario for assessing the effects of uncertainty in QTL genotype and nonfixation of QTL within founder lines:
In an F2 cross between the chicken lines Red Jungle fowl and White Leghorn, Kerje et al. (2003) had previously detected two QTL affecting body weight at 200 days of age on chromosome 1 around 100 cM (Growth 1) and 490 cM (Growth 2). This trait was chosen because the QTL Growth 1 and Growth 2 have been thoroughly studied previously (Kerje et al. 2003; Rönnegård et al. 2008) where Growth 1 has a very large effect and Growth 2 was not fixed within the founder lines. The cross was composed of four founder individuals (two from each line) and 756 F2 offspring. Although the QTL Growth 1 is easily shown to have a very strong effect on the mean (explaining 22% of the variance; Kerje et al. 2003), the analysis of Growth 2 is complicated by it being fixed within the Red Jungle fowl founders but variable within the White Leghorn line (Rönnegård et al. 2008), leading to additional uncertainty in the underlying QTL genotype. We use these data to explore the differences in estimates from the EM-algorithm (Appendix B), which explicitly takes account of this uncertainty, and the DGLM estimation, which does not, when there is a very strong mean-controlling QTL and moderate marker information. Furthermore, we study the effect of a QTL not being fixed within the founder lines. A sex effect was included as a covariate in both the mean and variance parts of the model. Line dosages were calculated as Haley–Knott probabilities (see File S2) and models were fit as above using the regression in Equation 2.
RESULTS
Detection of QTL for F2 and CC with regression on QTL genotypes:
For the F2 simulations the average of the estimated QTL effects was close to those simulated (Table 3). The false-positive rate was close to 0.05 (Table 4) for both F2 and CC, indicating that our DGLM approach produces the appropriate rate of false positives (type I error) when applied to known QTL genotypes.
TABLE 3.
Estimated QTL effects in simulated F2
| Regression on QTL genotypes |
Regression on line dosagesa |
|||
| Simulated effects | Ordinary QTL | vQTL | Ordinary QTL | vQTL |
| No QTL | 0.001 | −0.003 | 0.008 | 0.001 |
| Ordinary QTL | 1.997 | −0.003 | 2.063 | 0.023 |
| vQTL | −0.010 | 0.2175 | −0.004 | 0.238 |
| Ordinary and vQTL | 1.999 | 0.2173 | 1.913 | 0.247 |
Simulated value for mean-controlling QTL, 2.0; simulated value for variance-controlling QTL, 0.22.
Estimates from genome scan with regression on line dosages. Correction for shrunken line dosage estimates in HAPPY due to low marker information contents; corrected values = estimates from genome scan times half the range of line dosages.
TABLE 4.
Power to detect QTL at a 5% nominal level for regression on QTL genotypes
| F2 |
CC |
|||
| Simulated effects | Ordinary QTL | vQTL | Ordinary QTL | vQTL |
| No QTL | 0.052 | 0.052 | 0.054 | 0.053 |
| Ordinary QTL | 0.997 | 0.051 | 1.000 | 0.043 |
| vQTL | 0.051 | 0.966 | 0.057 | 0.998 |
| Ordinary and vQTL | 0.984 | 0.963 | 1.000 | 0.998 |
Detection of QTL for F2 and CC with regression on line dosage:
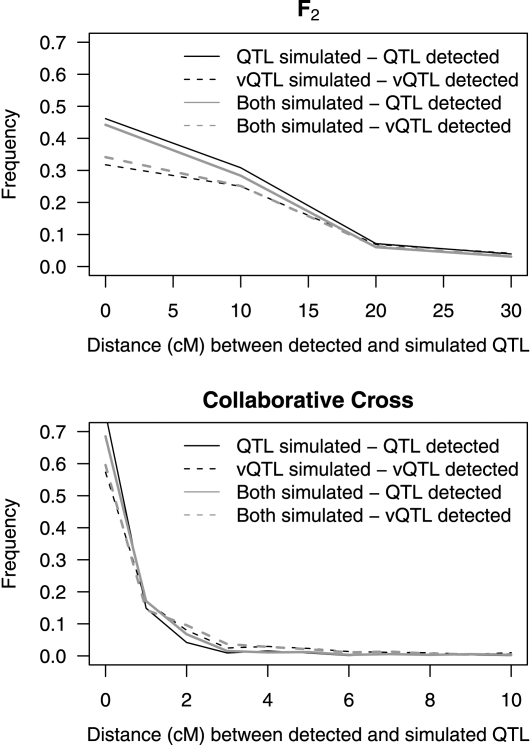
For the F2 simulations, the 5% chromosome-wide significance thresholds were log P = 2.02 and log P = 2.01 for the mean and variance parts of the model, respectively. Using these thresholds, the proportion of false positives was close to 0.05 (Table 5) both for Scenario 2 (mean-controlling QTL simulated) and Scenario 3 (vQTL simulated). The power to detect the mean-controlling QTL at a 5% genome-wide significance level was 91.7% (Table 5) and the power decreased slightly when a vQTL was added to the simulations due to the resulting increase in residual variance. The power to detect the vQTL was 77.0% (Table 5) and this was not substantially affected by including a mean-controlling QTL in the simulations (i.e., Scenario 4). For Scenario 4, with both ordinary QTL and vQTL being simulated, most QTL detected were positioned within, or close to, those simulated (Figure 1). The simulations gave similar results for Scenarios 2 and 3. The accuracy of the QTL position does not seem to be substantially affected if either a mean-controlling or a variance-controlling QTL is simulated vs. the Scenario 4 in which both effects are simulated (Table 6).
TABLE 5.
Power to detect QTL at a 5% chromosome-wide significance level for regression on line dosages
| F2 |
CC |
|||
| Simulated effects | Ordinary QTL | vQTL | Ordinary QTL | vQTL |
| No QTL | 0.050 | 0.050 | 0.050 | 0.050 |
| Ordinary QTL | 0.917 | 0.055 | 0.982 | 0.053 |
| vQTL | 0.059 | 0.770 | 0.046 | 0.808 |
| Ordinary and vQTL | 0.808 | 0.808 | 0.928 | 0.808 |
Figure 1.—
Distance (cM) between simulated and detected QTL for F2 (10,000 replicates) and the CC (1000 replicates).
TABLE 6.
Proportion of QTL that were detected at a 5% chromosome-wide significance level and whose chromosomal position was estimated accuratelya
| F2 |
CC |
|||
| Simulated effects | Ordinary QTL | vQTL | Ordinary QTL | vQTL |
| Ordinary QTL | 0.462 | — | 0.452 | — |
| vQTL | — | 0.318 | — | 0.262 |
| Ordinary and vQTL | 0.442 | 0.341 | 0.372 | 0.272 |
The chromosomal position was defined to be accurately estimated: (i) for the F2 cross if the QTL was detected within the correct marker interval, (ii) for the CC if the estimated position was within the correct ±0.3 cM.
Marker informativeness was small (SIC around 0.1) for the F2 simulations because markers were spaced 10 cM apart, where these intervals are considerably larger than in most QTL studies today (see, e.g., Kerje et al. 2003). For perfect information about QTL genotype the line dosage predictor qi is 0, 1, or 2, whereas for low information it has an attenuated range and is centered around 1.0. As a result, the regression on line dosage overestimated QTL effect (α and θ) for the F2 simulations. The extent of overestimation depends on the range of the line dosage (i.e., max(q) − min(q)), which is 2 with complete information but was 0.348 in the simulations. In Table 3 we therefore report estimates from the line dosage model after division by 2/0.348 = 5.75.
For the CC simulations the 5% chromosome-wide significance threshold for log P was 3.24 and 3.01 for the mean and variance parts of the model respectively. The power to detect the mean-controlling QTL at a 5% genome-wide significance level was 98.2% (Table 5) and, as above, this decreases slightly when a vQTL is added owing to the increased residual variance. The power to detect the vQTL was 80.8% (Table 5) and the power was not substantially changed when a mean-controlling QTL was included in the simulations. For the CC simulations, the proportion of QTL detected within the correct ±0.3 cM was highest for a vQTL when simulating both mean- and variance-controlling QTL (Table 6). The estimated QTL positions were well centered around the true simulated QTL position (Figure 1).
Fitting a single QTL in the presence of linked QTL and epistasis:
Fitting the DGLM model of Equation 2 to the simulated CC data with a single mean-controlling QTL (Scenario E1) resulted in 5.8% of replicates having P-values less than 0.05 in the variance part of the model. This is consistent with the P-values for the vQTL being robust to the presence of moderate-effect ordinary QTL. Under Scenario E2 (two linked QTL with additive effects) the proportion of replicates having P-values less than 0.05 was 4.0%, and the rate of false positives (type I error) did not seem to be substantially affected by an additional linked QTL acting additively. When there were two linked and interacting mean-controlling QTL (Scenario E3), 17.0% of the replicates had P-values for the variance submodel <0.05, indicating the proportion of false positives is substantially affected by linked QTL that act epistatically. By including interaction effects, between the two interacting loci, in the mean part of the model the false-positive rate was reduced to 5.6%.
By increasing the mean-controlling QTL effect, under Scenario E1, from 2.0 to 20, the empirical type I (false positive) error increased to 12.7%. Hence, vQTL detected close to mean-controlling QTL of large effect should be treated with caution and further analyzed using, e.g., an EM-algorithm where the effects of marker uncertainty are accounted for (see Appendix B).
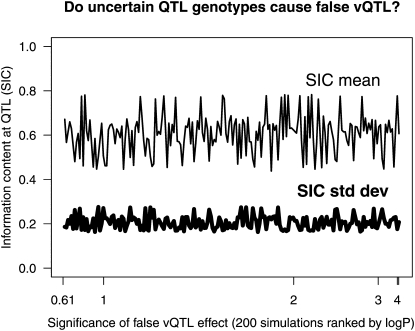
To investigate the effect of marker informativeness for the CC, we performed further simulations that repositioned the QTL at different locations in a 5-cM-spaced ladder between 25 and 70 cM. For these simulations (200 replicates per QTL location), the SIC varied considerably both within and between the QTL locations, but there was no clear relationship between SIC and the log P-values for false vQTL (Figure 2) when an ordinary QTL was simulated with additive effect of 2.0.
Figure 2.—
Relationship between log P-values for false vQTL and marker information content (SIC) when simulating a mean-controlling QTL in a CC population. For each of 200 simulations, ordered along the x-axis by their most significant vQTL peak, the plot shows the mean and standard deviation of SIC for 1000 mice. The SIC statistics are stationary, indicating no apparent tendency for marker uncertainty to produce false vQTL signals.
Red Jungle fowl × White Leghorn F2 cross: a worst-case scenario for assessing the effects of covariate uncertainty and nonfixation of QTL within founder lines:
In a preliminary model for chicken body weight without QTL effects, estimation of sex effects in a DGLM gave highly significant (P < 10−6) estimates of 410.0 and 0.509 for mean and variance predictors, respectively. The estimates being significant and having the same sign suggests a general mean-variance relationship. This was confirmed by applying the Box–Cox procedure (Box and Cox 1964), which suggested a square root transformation (, SE 0.11), and the extended quasi-likelihood (EQL) procedure of Nelder and Pregibon (1987), which indicated a linear mean-variance relationship (, SE 0.2) (see File S1).
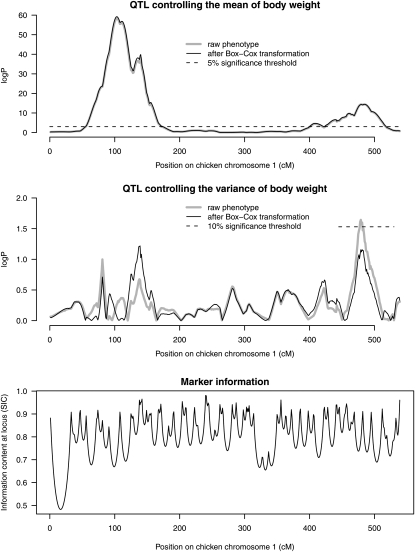
A chromosome scan using dglm revealed mean-controlling QTL (Figure 3) that were similar to those estimated by homoscedastic regression (Kerje et al. 2003). There were no large differences between the scans for ordinary QTL with Box–Cox transformed vs. original body weight as response. However, there were substantial differences between the chromosome scans for vQTL (Figure 3, middle graph), although no vQTL reached 5% significance.
Figure 3.—
Scan for QTL controlling the mean (top) and the variance (middle) of body weight at 200 days of age on chicken chromosome 1 in an F2 cross between Red Jungle Fowl and White Leghorn with 756 F2 offspring. (Bottom) Marker information contents (SIC). Genome-wide significance threshold calculated using 1000 permutations.
A moderate-sized peak was detected for the vQTL at approximately the same location as Growth 2 (nominal P-value = 0.02, chromosome-wise P-value = 0.10). This peak is likely due to the fact that the QTL alleles of Growth 2 were not fixed within the founder line of domestic White Leghorn hens (Rönnegård et al. 2008). The reason for this is that, if an F2 individual has an allele inherited from the domestic leghorn line, then the residual variance will be greater than if the allele was inherited from the Red Jungle Fowl line since Equation 2 assumes that the QTL alleles are fixed within founder lines. Consequently, the QTL around 490 cM is not a variance-controlling QTL but rather an effect of Growth 2 not being fixed within founder lines.
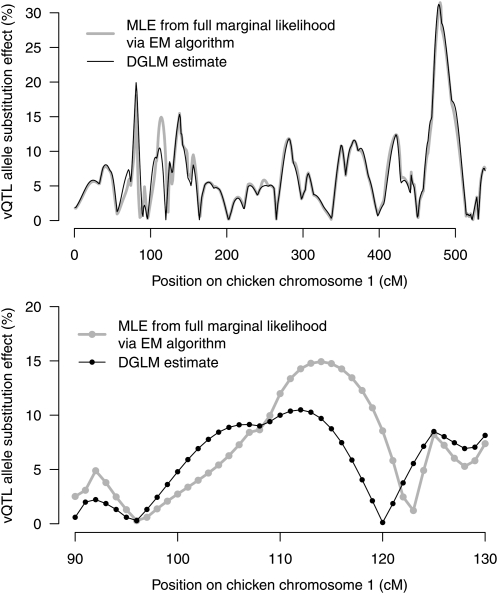
To assess spurious effects of uncertainty in genotype state, the additive effects for vQTL in Equation 2 were calculated using an EM-algorithm (Appendix B) and were subsequently compared with the DGLM estimates (Figure 4). The estimates are given as percentage change in residual variance for allele substitutions, i.e., . The estimates are almost identical except for the positions around 115 cM where there is a strong mean-controlling QTL and moderate marker information (log P = 57.3 and SIC = 0.7, Figure 3). Hence, no major improvement in the QTL detection was achieved by using the more theoretically correct EM-algorithm.
Figure 4.—
Estimates of vQTL effects given as percentage change in residual variance for allele substitutions for body weight at 200 days of age on chicken chromosome 1 in an F2 cross between Red Jungle Fowl and White Leghorn. Solid lines: Maximum-likelihood estimates from the full marginal likelihood. Shaded lines: DGLM estimates. Close up for 90–130 cM shown in bottom figure.
DISCUSSION
We have studied the potential of detecting variance-controlling QTL by fitting a double generalized linear model via a two-stage procedure to simulated phenotype and marker information. The model can detect QTL that affect both the mean and the variance or QTL that affect either the mean or the variance. Because we use line origin probabilities as predictors in the model, as calculated using the HAPPY HMM (Mott et al. 2000), our approach can be applied to a wide range of experimental crosses. There are, however, some important considerations to be emphasized.
Detecting vQTL can be more challenging in theory, practice, and subsequent interpretation than detecting QTL controlling the trait mean. This is unsurprising: at the most basic level, variances are harder to estimate than means, typically requiring five times as many observations to achieve comparable precision (cf. “Tukey's rule of 5” in Lee and Nelder 2006). Indeed, Visscher and Posthuma (2010) demonstrate analytically that detecting small effect vQTL among unrelated humans using traditional methods would require sample sizes in the 10,000s. More insidiously, inferences about differences in variances can be more sensitive to distributional assumptions than inferences about means. Specifically, it is common for raw phenotype measurements to exhibit a mean-variance relationship that naturally arises through the data generation process: for example, if body length is homoscedastic Gaussian then its square (e.g., body area) will not be. In the detection of vQTL, it is therefore especially important to recognize such relationships at an early stage and apply suitable normalizations or explore parametric alternatives to the normal distribution to avoid QTL affecting the mean appearing also to affect the variance. In File S1 we illustrate this problem along with an effective remedy via the Box–Cox transformation. Moreover, we set forth guidelines for how to approach a conservative vQTL analysis of data that are likely to be approximately normal but only after an unknown transformation, and of data that are more suitably modeled by other members of the exponential family, e.g., Poisson for count data, for which a known mean-variance relationship exists. Where it is felt such precautions may be inadequate or are impractical, we suggest resorting to the overconservative strategy of prioritizing “pure vQTL,” that is, strong vQTL with negligible mean effect, subject to all the usual requirements of deconfounding that apply to the detection of ordinary QTL.
Regression on genotype probabilities or expected line dosages can lead to inflated estimates of the residual variance (see models and methods). In File S1, we describe theoretically the effect of uncertain genotype states on the risk of detecting false vQTL and conclude that detected vQTL can be trusted as long as the marker informativeness is high or, if it is not, that the vQTL is not close to a mean-controlling QTL. Our simulations show that the power of detecting a vQTL in genome scans is largely unaffected by whether the same locus also affects the trait mean (Table 5), and we note that this includes our F2 simulation with a modest degree of informativeness from markers spaced 10 cM apart. In the analysis of chicken chromosome 1, we found that a mean-controlling QTL (Growth 1) having a very large effect (log P = 57.3) together with moderate marker information (SIC = 0.7) was needed to give any substantial bias in the vQTL estimation (Figure 4). We therefore conclude that the effect of inflated residual variance due to low marker information is not likely to be a major problem for our model.
We apply our model to populations of individuals who are genetically distinct and equally related, phenotyped in studies where genotype assignment is exchangeable across unmodeled environmental variation. A genetic locus whose genotype separates the population into groups with distinct variances is, by our definition, a putative vQTL. Table 1 illustrates how an effect detected in this way could arise from several different sources, each implying a potentially distinct paradigm of vQTL action. For example, it could represent canalizing epistasis, whereby some vQTL genotypes cushion the impact of genetic variation on the phenotype. Our simulations demonstrated that a mean-controlling QTL under epistatic control may indeed be detected as a vQTL with our proposed method, and this result is consistent with recent work by Paré et al. (2010) and Struchalin et al. (2010). Alternatively, it could represent a differential sensitivity to environmental variation. Specifically, increased variability under one genotype may manifest as temporally stable phenotypes varying between individuals, phenotypes being highly fluctuant within an individual but on average similar between, little actual variation but a tendency for increased error of measurement, or some combination of these. Dissecting the components of the induced variability could be accomplished by applying our DGLM framework to a more focused experimental design, e.g., incorporating replicates of the CC at different levels (e.g., guided by column 1 of Table 1). In some cases, further statistical exploration on the same data may also be helpful. For example, if the mechanism is epistatic, vQTL detection can be seen as a starting point for modeling the joint action of multiple interacting loci, and this is easily accommodated in our regression framework.
Levene’s test is a popular method for testing equality of variance. Paré et al. (2010) and Struchalin et al. (2010) used it to test for different variances between SNP genotypes. It is capable of modeling group effects and has the advantage of being quite insensitive to nonnormality. The DGLM approach, however, is far more flexible: it allows us to model continuous predictors and more complex general relationships of covariates on the variance, which Levene’s test cannot. Levene’s test also does not account for possible imbalance in the data since the estimated residuals mask varying uncertainty among groups. The DGLM approach, by contrast, allows modeling of nongenetic effects in the variance part of the model, which may be important if for instance the variances are different between batches or contemporary groups in a designed QTL experiment. Moreover, DGLMs can also be extended to include random effects, due to family or polygenic effects, through hierarchical generalized linear models (Lee and Nelder 1996; Lee et al. 2006; Rönnegård et al. 2010b) and double hierarchical generalized linear models (Lee and Nelder 2006; Rönnegård et al. 2010a). This modeling flexibility is necessary in QTL studies and gives a more general approach applicable to a wide range of experimental designs.
There are several possible extensions of our model. Dominance can be included similarly as in other QTL regression models (Haley et al. 1994), as can multiple loci (e.g., Valdar et al. 2009). Because DGLM allows for any response distribution from the exponential family (Smyth 2002), our model is straightforwardly extended to binomial, Poisson, or gamma-distributed traits. In particular, several studies have focused on QTL controlling the CV rather than the variance (e.g., Mackay and Lyman 2005; Ansel et al. 2008). Traits with a constant CV, given the explanatory variables, are naturally modeled using a gamma distribution (Mccullagh and Nelder 1989), and the DGLM method can be adapted to model such traits by setting the dispersion to CV2 (see File S1). If we do not know whether to search for variance-controlling or CV-controlling QTL (i.e., whether the trait should be normal or gamma distributed), we can use the EQL (Nelder and Pregibon 1987) to compare model fits (see File S1). Ideally, distributional assumptions should always be checked for a detected QTL using a QQ-plot for the deviance residuals (Mccullagh and Nelder 1989) from the mean part of the DGLM.
We anticipate identification of vQTL will be of interest in a wide range of genetics studies and applications. A clear application is in breeding systems, where vQTL detection could help selection for more robust livestock production (Mulder et al. 2007; Mulder et al. 2008). We believe there is also substantial scope for application in the study of animal models of human disease. For example, medical diagnosis of hypertension has traditionally focused on achieving reliable estimates of the underlying mean blood pressure. However, increasing evidence points to the dangers of temporal fluctuation about the mean (Parati et al. 2006; Brunelli et al. 2008; Rothwell et al. 2010). Our framework could be used to detect QTL affecting such variability in animal models and could be adapted to use in suitably controlled human studies.
In conclusion, we have developed a regression model for detection of major loci controlling phenotypic variance, which can be applied on a wide range of experimental crosses such as backcross, F2, and MAGIC lines such as the CC. We have studied the robustness of the model to varying marker information, misspecification of the response distribution, linked QTL, and epistasis, proposed recommendations for its use, and discussed the meaning of detected vQTL and how they might be further dissected. We expect detection of vQTL will have wide application in genetics.
APPENDIX A: DOUBLE GENERALIZED LINEAR MODEL THEORY
For known genotypes at the QTL, the ML estimates of the effect parameters β, α, γ, θ in Equation 2 can be estimated using Fisher scoring. Smyth (1989) showed this to be equivalent to predicting the mean effects using a linear model (Equation 1) and the squared residuals using a generalized linear model (GLM) with a gamma-distributed response and the log link.
ML gives biased variance component estimates and the estimates are also sensitive to influential observations with high leverages (Verbyla 1993). A restricted maximum likelihood (REML) approach, implemented in the dglm package, resolves these issues by incorporating leverage into the estimation (Smyth 2002; Lee et al. 2006). Specifically, the gamma GLM is used to predict , where the response is now the weighted deviance component di = /(1 – hii). Here, the hii is the leverage of observation i, equal to the ith diagonal element of the hat matrix where Xall is the design matrix for all fixed effects in the mean submodel and
Both the hat matrix and the associated leverages play an important role in ordinary regression theory (Hoaglin and Welsch 1978; Faraway 2004) and are used extensively in DGLM theory (Smyth 1989).
APPENDIX B: AN EM-ALGORITHM FOR ESTIMATING VQTL FOR NORMALLY DISTRIBUTED TRAITS IN F2 CROSSES
For an observation i, the full likelihood for vQTL with genotype uncertainty is
with variables other than fij defined as for Equation 3. Let li = log(Li), gij = log(fij), and ψ be the parameter vector, and assume observations are independent given the parameters such that Then,
| , |
where and
Here gij is the log-likelihood given the genotypes, so the gradient and Hessian of gij are those obtained from the logarithm of the normal density function fij. ML estimates of ψ are obtained by iterating the following EM steps until convergence:
Calculate the “weights” given the parameter estimates:
Calculate the parameter estimates given wij from: , which can be calculated using a Newton iterative method since we have the gradient and Hessian of gij.
Acknowledgments
We acknowledge Leif Andersson for providing data from the Red Jungle fowl × White Leghorn cross. We also acknowledge Gary Churchill for helpful discussions regarding use of the EM algorithm. L.R. recognizes financial support by the Swedish Research Council Formas. W.V. acknowledges partial support by National Institute of Mental Health/National Human Genome Research Institute grant P50 MH090338 and by a Career Development Award from the Medical Research Council, UK.
LITERATURE CITED
- Andersson L., 2001. Genetic dissection of phenotypic diversity in farm animals. Nat. Rev. Genet. 2: 1–11 [DOI] [PubMed] [Google Scholar]
- Ansel J., Bottin H., Rodriguez-Beltran C., Damon C., Nagaraj M., et al. , 2008. Cell-to-cell stochastic variation in gene expression is a complex trait. PLoS Genet. 4: e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont H. J., Gallie J., Kost C., Ferguson G. C., Rainey P. B., 2009. Experimental evolution of bet hedging. Nature 462: 90–93 [DOI] [PubMed] [Google Scholar]
- Box G. E. P., Cox D. R., 1964. An analysis of transformations (with discussion). J. R. Stat. Soc. B 26: 211–252 [Google Scholar]
- Broman K., 2005. The genomes of recombinant inbred lines. Genetics 169: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Sen S., 2009. A Guide to QTL Mapping with R/qtl. Springer, New York [Google Scholar]
- Brunelli S., Thadhani R. I., Lynch K. E., Ankers E. D., Joffe M. M., et al. 2008. Association between long-term blood pressure variability and mortality among incident hemodialysis patients. Am. J. Kidney Dis. 52: 716–726 [DOI] [PubMed] [Google Scholar]
- Carlborg O., Haley C. S., 2004. Epistasis: Too often neglected in complex trait studies? Nat. Rev. Genet. 5: 618–625 [DOI] [PubMed] [Google Scholar]
- Cavanagh C., Morell M., Mackay I., Powell W., 2008. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11: 215–221 [DOI] [PubMed] [Google Scholar]
- Chesler E. J., Miller D. R., Branstetter L. R., Galloway L. D., Jackson B. L., et al. , 2008. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm. Genome 19(6): 382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137 [DOI] [PubMed] [Google Scholar]
- Clayton G., Robertson A., 1957. An experimental check on quantitative genetical theory. II. The long-term effects of selection. J. Genet. 55: 152–170 [Google Scholar]
- Dunn P. K., Smyth G. K., 2009. dglm: double generalized linear models. R package version 1.6.1 [Google Scholar]
- Dworkin I., 2005. Canalization, cryptic variation, and developmental buffering: a critical examination and analytical perspective, pp. 131–158 Variation: A Central Concept in Biology, edited by Hallgrimsson B., Hall B. K. Elsevier Academic Press, New York [Google Scholar]
- Faraway J. J., 2004. Linear Models with R. Chapman & Hall/CRC, London [Google Scholar]
- Feenstra B., Skovgaard I. M., Broman K. W., 2006. Mapping quantitative trait loci by an extension of the Haley–Knott regression method using estimating equations. Genetics 173: 2269–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Irizarry R. A., 2010. Evolution in health and medicine Sackler colloquium: stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc. Natl. Acad. Sci. USA 107(Suppl): 1757–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G., Goldstein D. B., 2007. Human genetics: the hidden text of genome-wide associations. Curr. Biol. 17: 929–932 [DOI] [PubMed] [Google Scholar]
- Haley C. S., Knott S. A., 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324 [DOI] [PubMed] [Google Scholar]
- Haley C. S., Knott S. A., Elsen J. M., 1994. Mapping quantitative trait loci in crosses between outbred lines using least squares. Genetics 136: 1195–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Zhang X. S., 2004. Effects on phenotypic variability of directional selection arising through genetic differences in residual variability. Genet. Res. 83: 121–132 [DOI] [PubMed] [Google Scholar]
- Hoaglin D. C., Welsch R. E., 1978. The hat matrix in regression and ANOVA. Am. Stat. 32: 17–22 [Google Scholar]
- Hornstein E., Shomron N., 2006. Canalization of development by microRNAs. Nat. Genet. 38: S20–S24 [DOI] [PubMed] [Google Scholar]
- Houle D., 1992. Comparing evolvability and variability of quantitative traits. Genetics 130: 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Escriche N. D., Sorensen D., Waagepetersen R., Blasco A., 2008. Selection for environmental variation: a statistical analysis and power calculations to detect response. Genetics 180: 2209–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerje S., Carlborg O., Jacobsson L., Schutz K., Hartmann C., et al. , 2003. The twofold difference in adult size between the red jungle fowl and White Leghorn chickens is largely explained by a limited number of QTLs. Anim. Genet. 34: 264–274 [DOI] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., 1980. Genetic variation and phenotypic evolution during allopatric speciation. Am. Nat. 116: 463–479 [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Nelder J. A., 1996. Hierarchical generalized linear models (with Discussion). J. R. Stat. Soc. B 58: 619–678 [Google Scholar]
- Lee Y., Nelder J. A., 2006. Double hierarchical generalized linear models (with discussion). Appl. Stat. 55: 139–185 [Google Scholar]
- Lee Y., Nelder J. A., Pawitan Y., 2006. Generalized Linear Models With Random Effects: Unified Analysis via H-Likelihood. Chapman & Hall/CRC, London [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA [Google Scholar]
- Mackay T. F., Lyman R. F., 2005. Drosophila bristles and the nature of quantitative genetic variation. Phil. Trans. R. Soc. B. 360: 1513–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez O., Curnow R. N., 1992. Estimating the locations and the sizes of the effects of quantitative trait loci using flanking markers. Theor. Appl. Genet. 85: 480–488 [DOI] [PubMed] [Google Scholar]
- McCullagh P., Nelder J. A., 1989. Generalized Linear Models. Chapman & Hall/CRC, London [Google Scholar]
- Mott R., Talbot C. J., Turri M. G., Collins A. C., Flint J., 2000. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl Acad. Sci. USA 97: 12649–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder H. A., Bijma P., Hill W. G., 2007. Prediction of breeding values and selection response with genetic heterogeneity of environmental variance. Genetics 175: 1895–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder H. A., Bijma P., Hill W. G., 2008. Selection for uniformity in livestock production by exploiting genetic heterogeneity of environmental variance. Genet. Sel. Evol. 40: 37–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelder J. A., Pregibon D., 1987. An extended quasi-likelihood function. Biometrika 74: 221–232 [Google Scholar]
- Ordas B., Malvar R. A., Hill W. G., 2008. Genetic variation and quantitative trait loci associated with developmental stability and the environmental correlation between traits in maize. Genet. Res. 90: 385–395 [DOI] [PubMed] [Google Scholar]
- Parati G., Faini A., Valentini M., 2006. Blood pressure variability: its measurement and significance in hypertension. Curr. Hypertension Rep. 8: 199–204 [DOI] [PubMed] [Google Scholar]
- Paré G., Cook N. R., Ridker P. M., Chasman D. I., 2010. On the use of variance per genotype as a tool to identify quantitative trait interaction effects: a report from the women’s genome health study. PLoS Genet. 6: e1000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C., Sangster T., Lindquist S., 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624 [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2009. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Rice S. H., 2008. Theoretical approaches to the evolution of development and genetic architecture. Ann. NY Acad. Sci.s 1133: 67–86 [DOI] [PubMed] [Google Scholar]
- Rönnegård L., Besnier F., Carlborg O., 2008. An improved method for QTL detection and identification of within-line segregation in F2 intercross designs. Genetics 178: 2315–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnegård L., Felleki M., Fikse F., Mulder H. A., Strandberg E., 2010a. Genetic heterogeneity of residual variance estimation of variance components using double hierarchical generalized linear models. Genet. Sel. Evol. 42: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnegård L., Shen X., Alam M., 2010b. hglm: A package for fitting hierarchical generalized linear models. R J. 2(2): 20–28 [Google Scholar]
- Ros M., Sorensen D., Waagepetersen R., Dupont-Nivet M., SanCristobal M., et al. , 2004. Evidence for genetic control of adult weight plasticity in the snail helix aspersa. Genetics 168: 2089–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell P. M., Howard S. C., Dolan E., O’Brien E., Dobson J. E., et al. 2010. Prognostic significance of visit-to-visit variability, maximum systolic blood pressure, and episodic hypertension. Lancet 375: 895–905 [DOI] [PubMed] [Google Scholar]
- Rowe S. J., White I. M. S., Avendano S., Hill W. G., 2006. Genetic heterogeneity of residual variance in broiler chickens. Genet. Sel. Evol. 38: 617–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S., Lindquist S., 1998. Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342 [DOI] [PubMed] [Google Scholar]
- SanCristobal-Gaudy M., Elsen J. M., Bodin L., Chevalet C., 1998. Prediction of the response to a selection for canalisation of a continuous trait in animal breeding. Genet. Sel. Evol. 30: 423–451 [Google Scholar]
- Sangster T. A., Salathia N., Undurraga S., Milo R., Schellenberg K., et al. , 2008. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc. Natl. Acad. Sci. USA 105: 2963–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K., 1989. Generalized linear models with varying dispersion. J. R. Stat. Soc. B 51: 47–60 [Google Scholar]
- Smyth G. K., 2002. An efficient algorithm for REML in heteroscedastic regression. J. Comput. Graph. Stat. 11: 836–847 [Google Scholar]
- Sorensen D., Waagepetersen R., 2003. Normal linear models with genetically structured residual variance heterogeneity: a case study. Genet. Res. 82: 207–222 [DOI] [PubMed] [Google Scholar]
- Struchalin M. V., Dehghan A., Witteman J. C., Duijn C. V., Aulchenko Y. S., 2010. Variance heterogeneity analysis for detection of potentially interacting genetic loci: method and its limitations. BMC Genet. 11(1): 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W., Flint J., Mott R., 2006a. Simulating the Collaborative Cross: power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics 172: 1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W., Solberg L. C., Gauguier D., Burnett S., Klenerman P., et al. , 2006b. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nature Genetics 38: 879–887 [DOI] [PubMed] [Google Scholar]
- Valdar W., Holmes C. C., Mott R., Flint J., 2009. Mapping in structured populations by resample model averaging. Genetics 182: 1263–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbyla A. P., 1993. Modelling variance heterogeneity: residual maximum likelihood and diagnostics. J. R. Stat. Soc. B 55: 493–508 [Google Scholar]
- Visscher P. M., Posthuma D., 2010. Statistical power to detect genetic loci affecting environmental sensitivity. Behav. Genet. 40: 728–733 [DOI] [PubMed] [Google Scholar]
- Waddington C., 1942. Canalization of development and the inheritance of acquired characters. Nature 150: 563–565 [DOI] [PubMed] [Google Scholar]
- Wittenburg D., Guiard V., Teuscher F., Reinsch N., 2009. Comparison of statistical models to analyse the genetic effect on within-litter variance in pigs. Animal 2: 1559–1568 [DOI] [PubMed] [Google Scholar]
- Xu S., 1995. A comment on the simple regression method for interval mapping. Genetics 141: 1657–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., 1998. Further investigation on the regression method of mapping quantitative trait loci. Heredity 80: 364–373 [DOI] [PubMed] [Google Scholar]
- Yalcin B., Flint J., Mott R., 2005. Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics 171(2): 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Ding Y., Hutchins L., Szatkiewicz J., Bell T., et al. , 2009. A customized and versatile high-density genotyping array for the mouse. Nat. Meth. 6: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Li Y., Abecasis G. R., Scheet P., 2011. A comparison of approaches to account for uncertainty in analysis of imputed genotypes. Genet. Epidemiol. 35(2): 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]