Abstract
The Gram-negative bacterium Burkholderia pseudomallei is the etiological agent of melioidosis and is remarkably resistant to most classes of antibacterials. Even after months of treatment with antibacterials that are relatively effective in vitro, there is a high rate of treatment failure, indicating that this pathogen alters its patterns of antibacterial susceptibility in response to cues encountered in the host. The pathology of melioidosis indicates that B. pseudomallei encounters host microenvironments that limit aerobic respiration, including the lack of oxygen found in abscesses and in the presence of nitric oxide produced by macrophages. We investigated whether B. pseudomallei could survive in a nonreplicating, oxygen-deprived state and determined if this physiological state was tolerant of conventional antibacterials. B. pseudomallei survived initial anaerobiosis, especially under moderately acidic conditions similar to those found in abscesses. Microarray expression profiling indicated a major shift in the physiological state of hypoxic B. pseudomallei, including induction of a variety of typical anaerobic-environment-responsive genes and genes that appear specific to anaerobic B. pseudomallei. Interestingly, anaerobic B. pseudomallei was unaffected by antibacterials typically used in therapy. However, it was exquisitely sensitive to drugs used against anaerobic pathogens. After several weeks of anaerobic culture, a significant loss of viability was observed. However, a stable subpopulation that maintained complete viability for at least 1 year was established. Thus, during the course of human infection, if a minor subpopulation of bacteria inhabited an oxygen-restricted environment, it might be indifferent to traditional therapy but susceptible to antibiotics frequently used to treat anaerobic infections.
INTRODUCTION
Burkholderia pseudomallei is a resilient Gram-negative rod-shaped bacterium that is endemic to tropical areas of Southeast Asia and northern Australia (7). Infections with B. pseudomallei, through direct inoculation, inhalation, or ingestion, can result in a serious and often fatal disease called melioidosis (7). Typically, melioidosis is endemic to regions of Southeast Asia and northern Australia; recently, however, melioidosis has been increasingly recognized in Central and South America (1, 19). With worldwide reports of several sporadic cases of melioidosis, and with the increase in global travel by humans and global transport of animals, melioidosis is considered an emerging infectious disease.
The clinical presentations of melioidosis are diverse; it can manifest as an acute, chronic, or latent disease (58). B. pseudomallei can infect virtually all cell types and organs within the human body; however, the hallmark of melioidosis is the formation of multiple abscesses in a variety of organs, including the lungs, liver, and spleen (7). The most common clinical manifestation of melioidosis is pneumonia, which accounts for about half of all cases (7). Dissemination of B. pseudomallei into the bloodstream can result in acute fulminant septicemia, often with a fatal outcome. Chronic melioidosis usually manifests as a localized skin or soft tissue infection and accounts for 12% of cases (8). Additionally, reports have indicated that B. pseudomallei can cause latent infections, mostly asymptomatic for decades before reactivation (30, 36). This vast variety of clinical manifestations has earned B. pseudomallei the name “the great mimicker” and has often led to underdiagnosis of melioidosis or its misdiagnosis as tuberculosis (55).
Management of melioidosis remains a challenge despite the use of antibiotic therapy (7). B. pseudomallei is intrinsically resistant to most classes of antibiotics, which limits their therapeutic use during patient treatment (6). Antibiotic therapies using meropenem and imipenem (21), ceftazidime with or without trimethoprim-sulfamethoxazole, or chloramphenicol remain the regimens of choice for treating melioidosis (54). Yet successful eradication of B. pseudomallei infection requires a prolonged antibiotic regimen, up to 5 months. Despite this vigorous intervention, relapse occurs in 13 to 23% of the cases (7). In addition, reports have indicated the emergence of B. pseudomallei strains that are resistant to ceftazidime and trimethoprim (10, 17, 57). Therefore, the search for novel antibiotics and short treatment regimens effective at clearing B. pseudomallei infections remains a vital area of investigation. The high mortality rate associated with melioidosis, the difficulty of treatment, and the lack of a protective vaccine raise biosecurity concerns regarding the use of B. pseudomallei in biological weapons. These concerns led to the classification of B. pseudomallei as a category B select agent by the U.S. Centers for Disease Control and Prevention (40).
B. pseudomallei has evolved a free-living saprophytic lifestyle in wet soil and stagnant water as well as a parasitic lifestyle in humans and animals (7). One of the great challenges that face aerobic microorganisms is continued access to oxygen, which is required for growth. B. pseudomallei is an obligate respirer but is likely to encounter microaerobic and even anoxic microenvironments in its free-living and parasitic lifestyles. Free-living B. pseudomallei is often found deep in wet soil, a microenvironment that is known to have little to no oxygen (9, 26). Similarly, during the course of a human infection, B. pseudomallei is likely to encounter different oxygen tensions in different tissue types and organs within the human body (37). Abscesses, a common pathological finding in melioidosis, present the extreme end of anoxic conditions, since studies have shown that abscesses can become completely devoid of oxygen (37, 41, 51). An additional characteristic of abscesses is their often acidic pH (44), a condition that may exert additional stress on pathogens. B. pseudomallei can also form biofilms and microcolonies both in vivo and in vitro. Deeper layers of bacterial biofilms are known to be anaerobic (52, 56, 60). Therefore, the persistence of this pathogen under oxygen-limited conditions is likely an important factor for its survival in host microenvironments.
The anaerobic responses of several pathogens, such as Staphylococcus aureus, Pseudomonas aeruginosa, and Mycobacterium tuberculosis, are an area of intense investigation (15, 38, 49). The interest in the anaerobic responses of pathogenic bacteria stems from observations that anaerobic fitness of pathogens is often required for virulence and pathogenesis (14, 15, 38, 43). In addition, oxygen availability plays an important role during antibiotic treatment, since oxygen limitation often results in reduced antibacterial activity (32). Furthermore, the hypoxic response of M. tuberculosis has been intensely studied, because anaerobic bacilli enter a dormant state that is thought to occur during latent M. tuberculosis infections (50, 53).
To date, the ability of B. pseudomallei to survive anaerobic conditions has not been investigated. Here we demonstrate that B. pseudomallei can survive initial anaerobic conditions and that a subpopulation can survive in the absence of oxygen without replicating for at least a year. Using microarrays, we identified the early transcriptional response of B. pseudomallei to hypoxia. In addition, we demonstrate that conventional antibiotics become ineffective against anaerobic B. pseudomallei, while drugs that target anaerobes are highly efficient at killing this pathogen.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All experiments were performed using the clinical isolate Burkholderia pseudomallei K96243 (18). All manipulations of B. pseudomallei were carried out in a CDC select-agent certified biosafety level 3 facility at the University of Colorado—Denver. B. pseudomallei cultures were grown in Lennox broth (LB, comprising 10 g/liter pancreatic digest of casein [tryptone], 5 g/liter yeast extract, and 5 g/liter sodium chloride; Fisher Scientific, Hampton, NH) or in LB plus 0.75% glucose (LBG) at 37°C with shaking at 315 rpm. The liquid growth medium was sterilized by filtration through Nalgene Supor machV vacuum filtration devices. LB agar plates incubated at 37°C for 1 to 2 days were used to grow B. pseudomallei colonies.
Growth and survival under anaerobic conditions.
To study the effects of oxygen depletion on the growth and survival of B. pseudomallei, we adapted a model used to study the anaerobic dormancy of M. tuberculosis (53). The M. tuberculosis anaerobic model consists of a low-inoculum culture in an airtight sealed test tube with a culture-to-air headspace ratio of 2:1 (53). A small magnetic stir bar is used to stir the cultures slowly, thus allowing for the gradual depletion of oxygen by respiring bacteria while maintaining a homogenous culture. Anaerobic cultures of B. pseudomallei were grown in Hungate anaerobic glass tubes that were sealed with airtight Hungate rubber stoppers and screw caps (BellCo Glass, Vineland, NJ). Culture was performed as follows. Log-phase B. pseudomallei cultures grown in LBG were used to inoculate 14 ml of room-temperature LBG in tubes containing sterile magnetic bars to a final optical density at 600 nm (OD600) of 0.004 with a final culture-to-headspace ratio of 15:1. Tubes were sealed airtight using Hungate stoppers and screw caps and were incubated at 37°C with shaking at 275 rpm. Following the 24 h of incubation, tubes were transferred to a magnetic stirrer and were incubated with stirring at 37°C for as long as 1 year. Oxygen depletion was monitored by the decolorization of methyl blue injected into the tubes. Tubes were opened at the time points indicated in Fig. 1 in order to determine cell counts using serial dilution and plating onto LB agar for CFU determination. All experiments were repeated a total of three times.
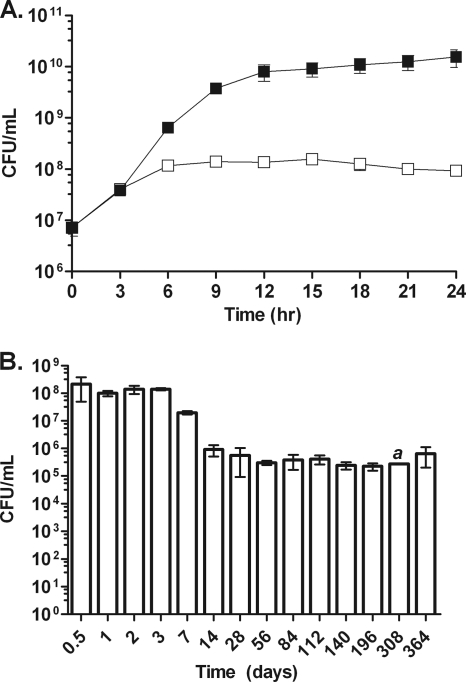
Fig. 1.
Growth and survival of B. pseudomallei in the anaerobic model. (A) Growth of B. pseudomallei in the shaking anaerobic tube model (open squares) compared to shaking aerated growth (filled squares) over the first 24 h. (B) Survival of B. pseudomallei in the anaerobic tube model over a period of 1 year. Error bars represent standard deviations for three independent cultures per time point. The letter a indicates a time point for which only one data point was available.
To examine the effect of pH on anaerobic survival, LBG was prepared at pH 6.0, pH 6.5, or pH 7.0 by addition of hydrogen chloride (HCl) or sodium hydroxide (NaOH) (both from Fisher). Tubes were opened at the time points indicated in Fig. 3 for CFU determination; supernatants were collected by centrifugation, sterilized by filtration through two 0.2-μm-pore-size filters, and stored at −80°C until all samples could be processed for both pH and ammonia content.
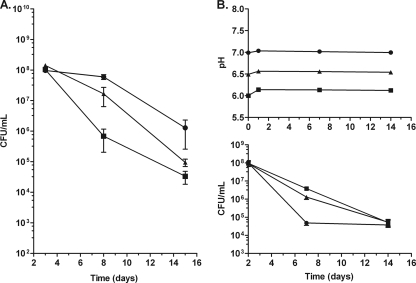
Fig. 3.
Anaerobic survival at an acidic pH. (A) Anaerobic survival in LBG medium adjusted to a pH of 6.0 (circles), 6.5 (triangles), or 7.0 (squares). (B) Change in pH and anaerobic survival in phosphoric acid-buffered LBG adjusted to a pH of 6.0, 6.5, or 7.0 (indicated as in panel A). Error bars represent standard deviations for three independent cultures per time point.
To examine the effect of buffering capacity on anaerobic survival, potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, St. Louis, MO) and sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich) were added to LBG to produce a final pH of 6.0, 6.5, or 7.0 with a total phosphoric acid concentration of 50 mM. Tubes were opened at the time points indicated in Fig. 3 for CFU determination; supernatants were collected by centrifugation, sterilized by filtration twice, and stored at −80°C until all samples could be processed for both pH and ammonia content.
To examine the effect of ammonia on anaerobic survival, 5 mM or 20 mM ammonium chloride (NH4Cl) (Sigma) was added to LBG. Tubes were opened at the indicated time points for CFU determination.
Ammonia quantification and pH determination.
Ammonia content was determined using an ammonia ion selective electrode (9512HPBNWP; Thermo Scientific, Waltham, MA). Supernatant samples were thawed, mixed, and diluted 1:40, 1:80, and 1:100; the ionic strength of each dilution was adjusted by the addition of NaOH to a final concentration of 0.1 M, and the conductivity of each dilution was determined by placing the electrode in the solution while the solution was stirred slowly. The ammonia content was calculated by back-calculation from a log-fit regression line fit to a series of standard solutions containing known quantities of ammonia.
pH was determined on a SevenEasy pH meter (Mettler-Toledo, Columbus, OH).
Nitrate reduction assays.
Two-day-old anaerobic B. pseudomallei cultures in Hungate tubes were injected with 200 μl of 3.5 M sterile sodium nitrate (Sigma-Aldrich) or sterile H2O and were incubated for an additional 48 h at 37°C with stirring. Tubes were then opened, and samples were removed to determine cell counts and nitrite production. Nitrite production was measured using the Griess reaction (16).
Antibiotic susceptibility testing.
All antibiotics used in this study were purchased from Sigma-Aldrich. Ceftazidime (CAZ) and trimethoprim-sulfamethoxazole (SXT) stock solutions were dissolved in 50% dimethyl sulfoxide (DMSO) and were used at final concentrations of 64 μg/ml and 256 μg/ml, respectively. Metronidazole (MTZ) and tinidazole (TNZ) stock solutions were dissolved in 100% DMSO and were used at final concentrations of 32 μg/ml and 12 μg/ml, respectively. Chloramphenicol (CHL) stock solutions were dissolved in ethanol and were used at a final concentration of 256 μg/ml. All antibiotic stock solutions were freshly prepared prior to use. The antibiotic susceptibility of aerated B. pseudomallei cultures was determined using log-phase cultures at 1 × 107 to 1.5 × 107 CFU/ml. Aerated cultures were grown in 14 ml LB in 125-ml vented plastic flasks at 37°C with shaking at 315 rpm. At time zero, samples were taken to determine initial bacterial counts, followed by the addition of 200 μl of the antibiotic stock solution or 200 μl of the respective solvent. Cultures were then incubated for 24 h at 37°C with shaking at 315 rpm, and samples were removed at 6 and 24 h for the determination of viable bacterial numbers. The antibiotic susceptibility of anaerobic B. pseudomallei cultures was determined using 7-day (1 × 107 to 1.5 × 107 CFU/ml) and 1-month (2 × 105 to 5 × 105 CFU/ml) anaerobic cultures. At time zero, a set of tubes was opened to determine the initial bacterial numbers. A separate set of tubes was injected with 200 μl of each antibiotic solution or their respective solvents. Cultures were then incubated for an additional 6 or 24 h at 37°C with stirring on a magnetic stirrer. Tubes were opened at 6 or 24 h, and viable bacterial numbers were determined by serial dilution and plating onto LB agar. The percentage of survival represents the number of viable cells after antibiotic treatment divided by the number of viable cells prior to antibiotic treatment, multiplied by 100. The results presented in this study represent averages for three independent cultures per treatment and time point.
For determination of the minimum bactericidal concentration (MBC), 500 μl of a 1.0 ×107-CFU/ml aerobic culture was added to each well of a 24-well plate, followed by the addition of the drug(s) at 10 μl per well. The 24-well plates were incubated at 37°C for 24 h before plating to determine the CFU count per milliliter. All assays included a drug vehicle control. The antibiotic susceptibility of anaerobic B. pseudomallei was determined using 7-day-old anaerobic cultures at 1.0 ×107 CFU/ml. Anaerobic testing was carried out in the same manner as aerobic testing, except that all work was done in a Bactron anaerobic environmental chamber (Shel Lab, Cornelius, OR).
Microarrays and data analysis.
Aerobic or anaerobic cultures used for RNA extraction were grown in 40 ml of LBG at 37°C to a final OD600 of 0.3. Aerobic cultures were grown in vented flasks with shaking at 315 rpm. Anaerobic cultures were grown in the same manner but were incubated for 4 h with shaking at 100 rpm in an anaerobic container in which oxygen was depleted using anaerobic BBL GasPak envelopes (Becton Dickinson, Franklin Lakes, NJ). RNA from bacterial cells was harvested, and cDNA was prepared, as described previously for M. tuberculosis and Burkholderia mallei (23, 50). Burkholderia microarrays were obtained through the NIAID-sponsored Pathogen Functional Genomics Resource Center. Following overnight hybridization, the microarrays were scanned using a Axon GenePix 4000B scanner as described previously (23, 50). Microarray images were obtained, and initial data analysis was performed using GenePix Pro software. Data were normalized as described previously (50). Microarray-determined ratios were calculated from 3 biological replicates and 2 microarrays for each biological replicate. Significance analysis of microarrays (SAM) (http://www-stat.stanford.edu/∼tibs/SAM/) was used to determine which genes were statistically significantly regulated during hypoxia (46). The genes included in Table 1 and in Tables S1 and S2 in the supplemental material conformed to these stringent selection criteria. They exhibited at least an average 2-fold induction or repression ratio and had a SAM false discovery q value of zero.
Table 1.
Main categories of hypoxia-induced genes
| Category and name | Gene | Hypoxic/aerobic expression ratio |
Gene producta | |
|---|---|---|---|---|
| Mean | SD | |||
| Arginine and pyruvate fermentation | ||||
| BPSS1711 | aceE | 4.1 | 1.3 | Pyruvate dehydrogenase E1 component |
| BPSS1944 | adhA | 20.8 | 6.7 | Alcohol dehydrogenase |
| BPSL1743 | arcA | 123.4 | 35.4 | Arginine deiminase |
| BPSL1744 | arcB | 73.8 | 23.8 | Ornithine carbamoyltransferase |
| BPSL1745 | arcC | 39.1 | 16.8 | Carbamate kinase |
| BPSL1742 | arcD | 20.9 | 3.9 | Arginine/ornithine antiporter |
| BPSS1956 | 6.2 | 3.8 | Acetate kinase | |
| ATP synthase | ||||
| BPSS1945 | atpG | 8.1 | 1.8 | Putative ATP synthase F1, gamma subunit |
| BPSS1946 | atpA | 11.4 | 2.6 | ATP synthase subunit A |
| BPSS1947 | 6.1 | 2.8 | ATP synthase B subunit | |
| BPSS1949 | 8.3 | 3.4 | FoF1 ATP synthase subunit A | |
| BPSS1951 | 3.2 | 1.4 | ATP synthesis-related protein | |
| BPSS1953 | atpD | 32.5 | 8.2 | ATP synthase subunit B |
| Electron transport | ||||
| BPSL0501 | cydB | 11.0 | 4.2 | Cytochrome d ubiquinol oxidase subunit II |
| BPSL0502 | cydA | 7.2 | 0.7 | Cytochrome d ubiquinol oxidase subunit I |
| BPSL0637 | ubiE | 2.9 | 0.8 | Quinone biosynthesis methyltransferase |
| BPSL0640 | ubiB | 2.4 | 1.1 | Putative ubiquinone biosynthesis protein |
| BPSL1256 | 108.9 | 46.0 | Putative cytochrome c precursor | |
| BPSL1257 | 19.8 | 4.1 | Putative cytochrome c precursor | |
| BPSL1259 | 13.4 | 2.4 | Putative cytochrome c oxidase-related protein | |
| BPSL1260 | 10.4 | 1.8 | Cytochrome c oxidase subunit 1 | |
| BPSL1261 | 4.8 | 3.0 | Putative cytochrome c-related protein | |
| BPSL1600 | 7.0 | 3.8 | Putative cytochrome c-related lipoprotein | |
| BPSL2861 | ubiA | 4.4 | 2.1 | 4-Hydroxybenzoate octaprenyltransferase |
| BPSL3076 | 2.8 | 0.5 | 3-Octaprenyl-4-hydroxybenzoate carboxy-lyase | |
| BPSL3121 | petC | 2.3 | 0.2 | Ubiquinol-cytochrome c reductase, cytochrome c1 |
| BPSL3122 | petB | 8.8 | 2.4 | Cytochrome b |
| BPSL3123 | petA | 3.7 | 1.4 | Ubiquinol-cytochrome c reductase subunit |
| BPSL3179 | 2.3 | 0.9 | Putative cytochrome c biogenesis protein | |
| BPSL3181 | 3.2 | 0.9 | Cytochrome c | |
| BPSS1147 | 11.3 | 6.2 | NADH dehydrogenase subunit | |
| BPSS1159 | 2.4 | 0.8 | Nitrate reductase, alpha subunit | |
| BPSS1729 | 23.9 | 7.0 | Cytochrome c | |
| BPSS1896 | cyoB | 4.2 | 1.3 | Ubiquinol oxidase, subunit I |
| BPSS1897 | cyoA | 5.1 | 1.7 | Ubiquinol oxidase, subunit II |
| Motility and chemotaxis | ||||
| BPSL0225 | 2.1 | 0.5 | Putative flagellar hook length control protein | |
| BPSL0226 | fliJ | 2.1 | 0.8 | Flagellar export protein |
| BPSL0227 | fliI | 3.6 | 2.1 | Flagellum-specific ATP synthase |
| BPSL0230 | fliF | 3.8 | 2.3 | Flagellar MS ring protein |
| BPSL0267 | 3.5 | 0.4 | Putative flagellar synthesis protein | |
| BPSL0268 | 4.1 | 2.0 | Putative negative regulator of flagellin synthesis | |
| BPSL0269 | flgA | 2.3 | 0.4 | Flagellar basal body P ring biosynthesis protein |
| BPSL0271 | flgC | 2.6 | 1.1 | Flagellar basal body rod protein |
| BPSL0272 | flgD | 3.4 | 1.3 | Putative basal-body rod modification protein |
| BPSL0280 | flgK | 3.1 | 0.3 | Flagellar hook-associated protein |
| BPSL0281 | flgL | 3.6 | 0.5 | Flagellar hook-associated protein |
| BPSL0706 | 9.2 | 5.6 | Putative aerotaxis sensor receptor | |
| BPSL1611 | 3.7 | 1.1 | Putative chemotaxis transmembrane protein | |
| BPSL3293 | flhF | 3.0 | 1.5 | Flagellar biosynthesis regulator |
| BPSL3300 | cheY | 2.2 | 0.3 | Chemotaxis protein |
| BPSL3301 | cheB | 4.4 | 1.9 | Chemotaxis-specific methylesterase |
| BPSL3302 | 2.6 | 1.0 | Chemoreceptor glutamine deamidase CheD | |
| BPSL3303 | cheR | 3.5 | 1.9 | Chemotaxis protein methyltransferase |
| BPSL3304 | tsr | 2.8 | 1.0 | Methyl-accepting chemotaxis protein I |
| BPSL3306 | cheA | 2.7 | 0.8 | Chemotaxis two-component sensor kinase |
| BPSL3307 | cheY1 | 2.6 | 0.6 | Chemotaxis two-component response regulator |
| BPSL3309 | motA | 2.7 | 0.5 | Flagellar motor protein |
| BPSL3311 | flhD | 3.9 | 1.6 | Transcriptional activator |
| BPSL3320 | fliD | 2.2 | 0.4 | Flagellar hook-associated protein |
| BPSL3338 | 6.8 | 3.1 | Putative methyl-accepting chemotaxis protein | |
| BPSS0204 | tsr | 3.9 | 1.7 | Methyl-accepting chemotaxis protein I |
| BPSS0276 | aer | 26.6 | 13.0 | Aerotaxis receptor |
| BPSS0733 | 3.0 | 0.8 | Methyl-accepting chemotaxis protein II | |
| BPSS0814 | 3.3 | 0.6 | Chemotaxis protein | |
| BPSS0933 | 4.4 | 0.7 | Methyl-accepting chemotaxis protein | |
| Stress related | ||||
| BPSL0899 | clpA | 5.5 | 2.6 | ATP-dependent Clp protease |
| BPSL1323 | hsp | 19.8 | 10.7 | Putative heat shock protein |
| BPSL1484 | clpB | 11.6 | 4.6 | ClpB heat shock protein |
| BPSL2267 | 3.5 | 1.4 | Putative Hsp33 chaperonin | |
| BPSL2863 | 20.2 | 6.4 | Ferritin DPS family DNA binding protein | |
| BPSS0032 | 2.6 | 0.8 | Universal stress-related protein | |
| BPSS0836 | 13.3 | 2.0 | Universal stress protein | |
| BPSS0838 | 4.0 | 0.5 | Universal stress protein | |
| BPSS1140 | 3.3 | 0.7 | Universal stress family protein | |
| BPSS1934 | 2.5 | 0.9 | Universal stress protein | |
| BPSS2288 | 87.5 | 35.6 | HSP20/alpha crystalline family protein | |
| Adherence | ||||
| BPSL1008 | 2.5 | 0.7 | Putative exported fimbria-related chaperone | |
| BPSL1892 | 4.2 | 1.5 | Putative fimbria-related outer membrane protein | |
| BPSL1896 | 4.3 | 1.9 | Putative membrane fimbrial assembly protein | |
| BPSL1899 | 6.8 | 1.1 | Putative fimbrial assembly-related protein | |
| BPSS1602 | pilT | 2.1 | 0.7 | Twitching motility protein |
| Polyhydroxybutyrate synthesis | ||||
| BPSS1954 | phbC | 89.5 | 25.9 | Poly-β-hydroxybutyrate polymerase |
| BPSS0017 | 2.4 | 0.8 | 3-Hydroxybutyrate dehydrogenase | |
| BPSS0354 | 2.3 | 0.2 | 3-Hydroxybutyrate dehydrogenase | |
| BPSL2298 | phaP | 5.9 | 1.2 | Phasin-like protein |
Gene symbols and gene product function are based on locus annotation found at http://www.burkholderia.com/ and in the Burkholderia pseudomallei K96243 genome sequence (18).
RESULTS
Anaerobic model for B. pseudomallei.
When we used the anaerobic model described by Wayne and Hayes (53) for use with M. tuberculosis, B. pseudomallei formed a pellicle at the air-liquid interface regardless of the stirring speed or the addition of detergents to the growth medium. To prevent pellicle formation, tubes containing low-inoculum cultures with a culture-to-air volume ratio of 15:1 were sealed in airtight glass tubes and were vigorously shaken at 275 rpm for 24 h. After 24 h, the cultures were stirred on magnetic stir plates to maintain homogenous culture conditions and to prevent culture aggregation. Under these conditions, no aggregation or pellicle formation was visible.
Growth and survival in the anaerobic model.
The growth rate of B. pseudomallei in the sealed tubes resembled that of aerated cultures during the first 3 h of growth (Fig. 1 A). Initial growth was supported by the dissolved oxygen in the culture medium. As the density of the culture increased, the bacterial growth rate slowed, and growth eventually stopped after 6 to 7 generations, between the first 9 to 12 h (Fig. 1A). Growth arrest coincided with oxygen depletion by respiring bacteria, as determined by the decolorization of methylene blue (data not shown). These results demonstrate that B. pseudomallei was unable to grow in rich LB medium supplemented with glucose when oxygen was absent, although the ability of B. pseudomallei to proliferate under oxygen-limiting conditions in other growth media has yet to be determined. The survival of anaerobic B. pseudomallei was monitored over a 1-year period. Anaerobic cultures were opened at various time points, and the numbers of bacteria capable of forming a colony on LB agar under ambient air were determined. The viability of B. pseudomallei showed a gradual decline from 1 to 4 weeks after the establishment of anaerobic conditions (Fig. 1B). The survival of B. pseudomallei dropped by about 1 log10 unit with each passing week, and by 1 month, roughly 2 × 105 CFU/ml remained (Fig. 1B). Similar survival kinetics were obtained from experiments using either sealed glass Hungate tubes or anaerobic gas packs, using LB medium without added glucose (data not shown). After the first month under anaerobic conditions, no further decrease in viability was observed to at least 1 year. Taken together, these results demonstrate that B. pseudomallei is unable to grow under anaerobic conditions and that, while the majority of the culture survived for a few days to a week, a subset was able to survive indefinitely in an apparently nonreplicating, persistent anaerobic state for as long as a year.
B. pseudomallei strain K96243 can grow under anoxic conditions via nitrate respiration.
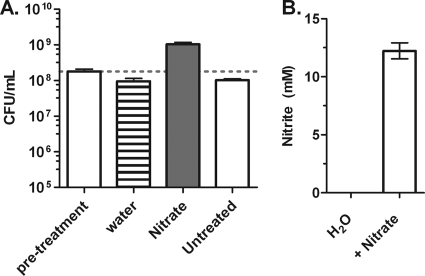
Several aerobic bacteria, including B. pseudomallei, can grow in the absence of oxygen by using nitrate as a terminal electron acceptor (59). To confirm that the clinical isolate B. pseudomallei strain K96243 could grow via anaerobic respiration, 2-day-old anaerobic cultures were injected with either 25 mM sodium nitrate or water, and growth was determined after 2 days. The addition of nitrate to the anoxic cultures led to bacterial growth resulting in a 5-fold increase in viable bacterial counts (Fig. 2 A). In contrast to nitrate supplementation, the addition of water resulted in no growth, and culture viability declined to a level similar to that of untreated controls. These results indicate that the amount of dissolved oxygen introduced during the injection process did not sustain significant growth and that the growth observed was a direct result of nitrate addition (Fig. 2A). To confirm that the anoxic growth of B. pseudomallei upon the addition of nitrate was a result of nitrate reduction, the production of nitrite was measured. Only nitrate-treated cultures were positive for nitrite production, converting half of the nitrate added into nitrite within 2 days (Fig. 2B). Taken together, these results confirm that B. pseudomallei strain K96243 can grow in the absence of oxygen by reducing nitrate.
Fig. 2.
Anoxic growth of B. pseudomallei via nitrate respiration. (A) Anoxic growth of B. pseudomallei cultures, as assessed by CFU determination, upon the addition of 25 mM sodium nitrate after 2 days of anaerobic conditions. (B) Reduction of nitrate to nitrite by anaerobic B. pseudomallei. Anaerobic cultures either were injected with water or nitrate or were left untreated and were assayed for nitrite production by using the Griess reaction. Error bars represent standard deviations for three independent cultures per treatment.
Effect of pH on anaerobic survival.
Anaerobic survival studies were conducted at acidic pHs to determine if a more physiological pH range found in abscesses could affect the survival of nonrespiring B. pseudomallei. Growth media prepared at a slightly acidic pH—either 6.5 or 6.0—were more hospitable for anaerobic survival than a neutral medium (Fig. 3 A). Near-100% viability was maintained for 1 week under anaerobic conditions for B. pseudomallei at a pH of 6.0, while a >100-fold drop in viability was observed for bacteria at a neutral pH. To determine if anaerobic cultures were able to change the pH of their growth culture media, the final pH of spent medium was determined using a pH meter at 1 week following anaerobiosis. The pH of the cultures starting at 6.0 increased to 6.5 after 1 week. The pH of cultures starting at 6.5 increased to 6.8 after 1 week. The pH of cultures starting at 7.0 increased to 7.1 after 1 week and remained at 7.1 for at least 1 month (data not shown). Media buffered with 50 mM phosphoric acid exhibited only minor increases in pH (Fig. 3B), but pH stabilization did not increase survival, indicating that an increase in pH was not ultimately responsible for the death of the culture (Fig. 3C). The growth rates of aerobic B. pseudomallei grown in LBG at pH 6.0, 6.5, or 7.0 or in LBG buffered with phosphoric acid at pH 6.0, 6.5, or 7.0 were identical (data not shown).
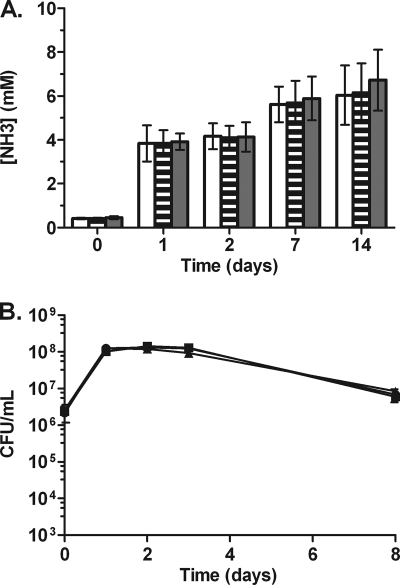
Effect of ammonia on anaerobic survival.
The increase in pH observed in the anaerobic cultures could have been the result of ammonia excretion from the metabolism of amino acids. To determine if amino acids were metabolized under anaerobic conditions, we measured ammonia levels. Ammonia accumulated significantly during the more metabolically active aerobic and hypoxic phases in the first 24 h of the model and continued to accumulate for the entire 2 weeks under anaerobic conditions, albeit at a slower pace (Fig. 4 A). At 2 weeks, ammonia levels reached a maximum concentration of 7 mM. These findings indicate that amino acids were metabolized anaerobically, resulting in ammonia accumulation in the spent medium. To determine if ammonia is toxic to B. pseudomallei under anaerobic conditions, LBG medium was supplemented with either 5 mM or 20 mM ammonia, and B. pseudomallei survival in the anaerobic model was monitored. No change in viability was observed under anaerobic conditions with ammonia added at more than 2-fold the levels that accumulated by amino acid metabolism in the absence of exogenous ammonia (Fig. 4B). No change in the growth rate was observed for B. pseudomallei grown aerobically in LBG plus ammonia (5 mM or 20 mM) (data not shown).
Fig. 4.
Ammonia accumulation and toxicity in anaerobic B. pseudomallei cultures. (A) Ammonia accumulation in LBG at pH 6.0 (open bars), 6.5 (striped bars), or 7.0 (filled bars). Error bars represent standard deviations for three independent cultures per time point. (B) Anaerobic survival of B. pseudomallei over the course of 1 week in LBG with no added ammonia (circles), 5 mM ammonia (squares), or 20 mM ammonia (triangles).
Transcriptional response of B. pseudomallei to hypoxia.
To determine the initial transcriptional response of B. pseudomallei to hypoxic conditions, whole-genome expression profiling was performed using Burkholderia oligonucleotide microarrays. RNA was isolated from B. pseudomallei cultures incubated for 4 h in anaerobic chambers and was compared with RNA from control aerated log-phase cultures as described in Materials and Methods. The expression ratios of genes induced and repressed in response to hypoxia are presented in Tables S1 and S2 in the supplemental material, respectively. Many of the genes induced by hypoxia encode proteins involved in 7 general areas, including (i) arginine and pyruvate fermentation, (ii) ATP synthase, (iii) electron transport, (iv) motility and chemotaxis, (v) stress-related functions, (vi) adherence functions, and (vii) polyhydroxybutyrate synthesis (Table 1). Increased motility of hypoxic and anaerobic B. pseudomallei cultures was confirmed by microscopic observation of wet-mount slides (data not shown).
As for genes involved in hypoxic and anaerobic metabolism, the arcABCD operon, involved in arginine import and fermentation, was highly induced in response to hypoxia. In addition, genes encoding pyruvate dehydrogenase, acetate kinase, and alcohol dehydrogenase, which are involved in the anaerobic metabolism of pyruvate, were also induced. Surprisingly, a second ATP synthase operon located on the small chromosome of B. pseudomallei was also induced under hypoxic conditions. This ATP synthase operon is part of a genomic region spanning the genes BPSS1944 to BPSS1958 and includes genes encoding a putative alcohol dehydrogenase, a polymerase, an acetate kinase, a 6-phosphofructokinase, an acetyltransferase, and hypothetical proteins, all of which were induced by hypoxia. Moreover, the expression of the high-oxygen-affinity cytochrome oxidase, cytochrome bd oxidase, as well as that of several cytochrome c biogenesis genes, was also induced. Taken together, these results suggest that B. pseudomallei possesses a robust transcriptional network that allows it to respond to oxygen depletion.
Hypoxic conditions lead to the repression of large number of genes, including those involved in protein synthesis, indicating a hypoxia-induced stringent response (see Table S2 in the supplemental material). Another set of genes repressed in response to hypoxia comprised those located on the large chromosome that encode the apparent aerobic ATP synthase.
Anaerobic B. pseudomallei is tolerant to conventional antibiotics but highly susceptible to the nitroimidazole antibiotics.
To evaluate the effects of anoxia on antibiotic efficiency, the susceptibilities of aerobic and anaerobic B. pseudomallei cultures to antibiotics were determined. Two classes of antibiotics were evaluated for their anti-B. pseudomallei activities. One class consisted of antibiotics used in melioidosis treatment, including ceftazidime, which targets cell wall synthesis; trimethoprim-sulfamethoxazole, which targets DNA synthesis; and chloramphenicol, which targets protein synthesis. The other class of antibiotics consisted of two nitroimidazole derivatives, metronidazole and tinidazole, that are commonly used to treat anaerobic bacterial and amebic infections. Nitroimidazole antibiotics are inactive prodrugs that become activated inside anaerobic cells through the reduction of the nitro group into nitro radical anions. These radical anions damage the biomolecules of anaerobic cells, leading to cell death (28, 33). It has been shown recently that nitroimidazoles kill M. tuberculosis via the production of nitric oxide (45) and that B. mallei is acutely sensitive to nitric oxide (23). We have observed that anaerobic B. pseudomallei is sensitive to nitric oxide (unpublished observation). Antibiotic concentrations were used based on their in vitro anti-B. pseudomallei activities from previous studies and their abilities to maintain antimicrobial activity after 24 h of incubation under our experimental conditions (48).
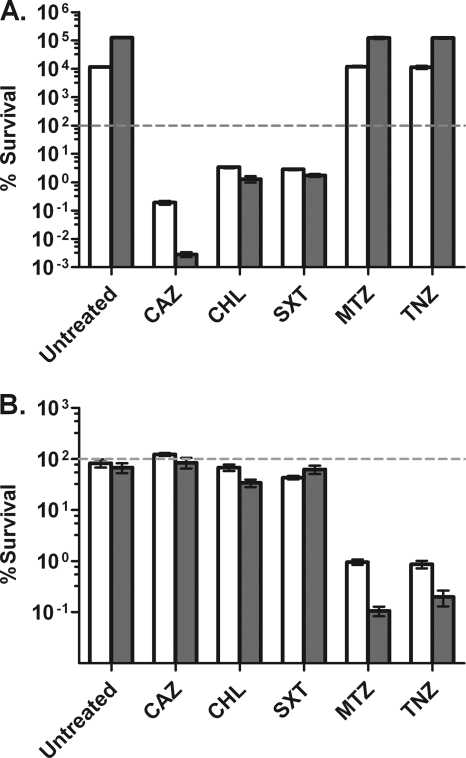
Aerated or 1-week-old anaerobic cultures were treated with each antibiotic for 6 and 24 h, after which survival was determined by plating for CFU. Treatment of aerated cultures with trimethoprim-sulfamethoxazole or chloramphenicol resulted in a >95% reduction in viability after 6 h and had no additional killing effect after an extended incubation of 24 h (Fig. 5 A). Ceftazidime treatment of aerated cultures resulted in 2 log10 and 5 log10 reductions in viability after 6 and 24 h, respectively. Neither metronidazole nor tinidazole had any effect on the growth rate or viability of aerated B. pseudomallei cultures (Fig. 5A). Treatment of anaerobic B. pseudomallei cultures with trimethoprim-sulfamethoxazole, chloramphenicol, or ceftazidime at the same concentrations used against aerated cultures, and at the same bacterial cell density, had no significant killing activity, and cell counts remained equal to those for untreated or mock-treated cultures (Fig. 5B). However, when anaerobic cultures were treated with metronidazole or tinidazole, 2 log10 and 3 log10 reductions in the number of viable bacteria were observed after 6 h and 24 h, respectively. MBCs were determined for each drug under standard aerobic and anaerobic conditions (Table 2). With chloramphenicol, 99.9% killing could not be achieved at any concentration, so an MBC at which 99% killing was observed (MBC99) was reported. The same trends observed with the other drug sensitivity experiments were observed again when the MBCs for the drug panel were determined. Bactericidal concentrations of ceftazidime rose from <32 μg/ml for aerated cultures to >256 μg/ml for anaerobic cultures. Bactericidal concentrations of trimethoprim-sulfamethoxazole also rose from <128 μg/ml for aerated cultures to >256 μg/ml for anaerobic cultures. Meanwhile, bactericidal concentrations of metronidazole and tinidazole fell from more than 256 μg/ml for aerated cultures to less than 128 μg/ml and 64 μg/ml for anaerobic cultures, respectively (Table 2). These results demonstrate that under anaerobic conditions, nonreplicating B. pseudomallei becomes highly tolerant to antibiotics used to treat melioidosis but exquisitely sensitive to nitroimidazole antibiotics.
Fig. 5.
Antibiotic susceptibilities of aerated and anaerobic B. pseudomallei cultures. Cultures (14 ml) were treated with antibiotics for 6 h (open bars) or 24 h (shaded bars) at final concentrations of 64 μg/ml ceftazidime (CAZ), 256 μg/ml chloramphenicol (CHL), 256 μg/ml trimethoprim-sulfamethoxazole (SXT), 32 μg/ml metronidazole (MTZ), and 12 μg/ml tinidazole (TNZ). (A) Susceptibility of aerated cultures grown in vented shaking flasks to antibiotic challenge. (B) Susceptibility of 1-week anaerobic cultures grown in sealed tubes to antibiotic challenge. The percentage of survival was calculated by dividing cell counts at 6 or 24 h by cell counts prior to treatment, multiplied by 100. Error bars represent standard deviations for three independent cultures for each treatment per time point presented.
Table 2.
MBCs for aerobic and anaerobic cultures
| Antibiotic | MBC (μg/ml) for: |
|
|---|---|---|
| Aerobic cultures | Anaerobic cultures | |
| Metronidazole | >256 | ≤128 |
| Tinidazole | >256 | ≤64 |
| Ceftazidime | ≤32 | >256 |
| Trimethoprim-sulfamethoxazole | ≤128 | >256 |
| Chloramphenicol | ≤256 (MBC99)a | >256 (MBC99)a |
An MBC with 99.9% killing could not be achieved for chloramphenicol under the conditions assayed; therefore, an MBC99 is reported.
Drug tolerance of anaerobic persisters.
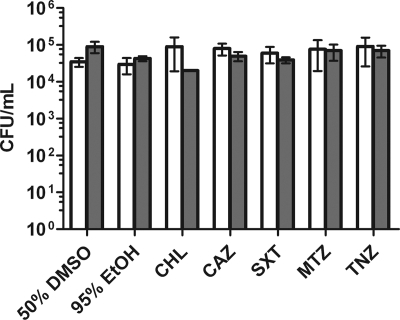
Approximately 0.1% of the anaerobic culture remained viable for at least 1 year, and likely much longer (Fig. 1B). To determine the drug susceptibility of this subpopulation, we treated cultures that had been anaerobic for 1 month with antibiotics (Fig. 6). Interestingly, this subpopulation was entirely tolerant not only to the antibiotics effective during aerobic conditions but also to the drugs effective during the early stages of anaerobiosis. These data indicated that the subpopulation of bacteria that survived indefinitely under anaerobic conditions was also a pure population of drug-tolerant persisters. We confirmed that the drug tolerance of long-term anaerobic bacteria was not genetic by recovering long-term anaerobic drug-treated bacteria under aerated conditions and then testing them for antibiotic susceptibility under aerobic and anaerobic conditions. These bacteria remained susceptible to antibiotics, and therefore it is unlikely that genetic mutations were responsible for the drug resistance observed under long-term anaerobic conditions (data not shown).
Fig. 6.
Antibiotic susceptibilities of 1-month anaerobic B. pseudomallei cultures. One-month anaerobic cultures were treated for 6 h (open bars) or 24 h (shaded bars) at final concentrations of 64 μg/ml ceftazidime (CAZ), 256 μg/ml chloramphenicol (CHL), 256 μg/ml trimethoprim-sulfamethoxazole (SXT), 32 μg/ml metronidazole (MTZ), and 12 μg/ml tinidazole (TNZ). EtOH, ethyl alcohol. Error bars represent standard deviations for three independent cultures for each treatment per time point presented.
DISCUSSION
The oxygen tension in the human host differs in different tissues and locations within abscesses and lesions; thus, the ability of pathogenic bacteria to sense and respond metabolically to low oxygen levels is crucial for their fitness and survival as they encounter respiration-limited microenvironments. To date, a substantial body of evidence has been accumulated for the anaerobic responses of bacterial pathogens to medically and clinically relevant processes, including pathogenesis, virulence, persistence, antibiotic tolerance, and latency (4, 13, 14, 49, 52). In this report, we describe the response of B. pseudomallei to oxygen-limited conditions and the role of oxygen restriction in antibiotic tolerance.
Burkholderia species, including B. pseudomallei, are known to grow anaerobically using nitrate as a terminal electron acceptor (59). In this work, we confirmed that the clinical isolate B. pseudomallei strain K96243 could grow in the absence of oxygen when nitrate was provided as a terminal electron acceptor. The presence of some nitrite and nitrate in lesions and abscesses is expected from the oxidation of nitric oxide produced by inflammatory cells. The amounts of nitrite produced under anaerobic growth were roughly half the amount of nitrate added to the cultures. This is most likely due to the further reduction of nitrite to nitrogen gas that has been shown to occur in B. pseudomallei (34). The ability of B. pseudomallei to grow under anoxic conditions in the presence of an alternative respiratory electron acceptor demonstrates that it is an obligate respirer.
To gain insight into the adaptation that occurs during the shift of B. pseudomallei from aerobic to anaerobic conditions, we used microarray analysis to determine the transcriptional changes that occur during this shift. Hypoxia led to the repression of genes involved in ribosomal biogenesis, suggesting an overall reduction in protein synthesis during oxygen depletion, corresponding to a reduced growth rate and eventual growth cessation. These data indicate that hypoxia initiated a stringent response in B. pseudomallei. The reduction of protein synthesis in anaerobic B. pseudomallei was not unexpected, since in the absence of an electron acceptor, the ability to regenerate ATP and the reducing power via oxidative phosphorylation were limited, leading to eventual growth arrest. Thus, under these metabolic conditions, decreasing the rate of protein synthesis is likely beneficial for energy conservation.
Genes involved in the arginine deamination pathway were among those most highly induced in response to hypoxia. The deamination of arginine to ornithine can be used to generate ATP via substrate-level phosphorylation under anaerobic conditions (61). In P. aeruginosa, the arginine deaminase pathway has been well characterized and is required for bacterial viability and growth under anaerobic conditions (47). Thus, it is very likely that B. pseudomallei utilizes the same pathway for energy generation during respiratory stress. Additionally, three genes involved in pyruvate fermentation in P. aeruginosa were induced in B. pseudomallei in response to hypoxia. In P. aeruginosa, pyruvate fermentation into acetate and lactate can sustain anaerobic survival but not anaerobic growth (12). As with P. aeruginosa, the addition of pyruvate to anaerobic B. pseudomallei cultures enhanced their long-term survival; however, the addition of arginine decreased survival (unpublished observations). Thus, as in P. aeruginosa, nitrate respiration and pyruvate fermentation are viable metabolic pathways for energy production in B. pseudomallei under anaerobic conditions, while the role of arginine deamination is less clear.
We found that under acid anaerobic conditions that more closely resemble those found in bacterial abscesses, the survival of B. pseudomallei was enhanced but eventually the majority of the culture died. Bacterial survival was extended to 1 week under slightly acid conditions. One explanation for the eventual loss of viability may be the lack in the growth medium of specific components normally found in the host that are required for anaerobic survival. The accumulation of ammonia demonstrated that amino acids were metabolized during anaerobiosis, and it is possible that preferred amino acids were consumed within the first week under anaerobic conditions and that the lack of these amino acids resulted in the eventual death of the culture over the ensuing weeks. In some anaerobic survival paradigms, glucose fermentation supports the anaerobic survival of bacteria; however, it does not appear that glucose fermentation alone is capable of supporting the survival of B. pseudomallei, since a vast excess of glucose was present in the medium and significant death was still observed. The ability of pyruvate to extend survival indicates that carbohydrates other than glucose or amino acids that feed into pyruvate may be beneficial for anaerobic B. pseudomallei. Another possible explanation for the death of B. pseudomallei under anaerobic conditions is that the bacteria are incapable of extended survival without respiration and that in the absence of oxygen or other terminal electron acceptors, only a very small subpopulation of dormant B. pseudomallei persists.
An interesting observation was the reciprocal expression of the two ATP synthases from the large and small chromosomes of B. pseudomallei in response to hypoxia. It appears that B. pseudomallei utilizes two separate and nonoverlapping F-type ATP synthases in the presence or absence of oxygen. The exact function and significance of the second ATP synthase, expressed from the small chromosome during hypoxia, are unknown. In the absence of a terminal electron acceptor, it is possible that the ATP synthase functions in reverse by hydrolyzing ATP to maintain a proton motive force and membrane potential that are required for various processes, such as solute transport and motility. Perhaps the second ATP synthase is more efficient in catalysis of the reverse reaction of ATP synthesis than the ATP synthase expressed under aerobic conditions. Other possibilities are that the hypoxic ATP synthase functions specifically in an anaerobic electron transport system or that it has another role under conditions in which respiration and proton translocation would be severely limited. More studies are required to address the significance of the second ATP synthase and its possible role in the adaptation of B. pseudomallei to hypoxia and anaerobiosis.
Several hallmark genes of the M. tuberculosis DosR regulon, which responds to conditions that inhibit respiration (50), were also induced by B. pseudomallei during hypoxia. One of the most highly hypoxia induced genes in both M. tuberculosis and B. pseudomallei encodes a member of the alpha crystalline (Acr) protein family, which is induced 87.5-fold in B. pseudomallei (Table 1). Also, six genes that encode universal stress proteins (Usp) are part of the DosR regulon, and five are induced by hypoxia in B. pseudomallei (Table 1). Acr is a small heat shock protein thought to function as a protein chaperone. Acr expression is induced in various bacteria in response to several stress conditions, including hypoxia (22). The specific function of alpha crystalline in M. tuberculosis or B. pseudomallei is unknown; however, studies of this protein in other bacteria suggest that alpha crystallines act as chaperones for other proteins to prevent aggregation (39). The universal stress proteins are a family of bacterial proteins, as their name suggests, and are induced in response to several stress conditions, such as starvation, hypoxia, heat, oxidative stress, and acid stress (24, 35). The mechanisms by which they protect bacteria from stress are not currently known; however, in P. aeruginosa, a Usp is essential for long-term anaerobic survival, suggesting an important role in anaerobic adaptation (42).
Another adaptive feature of the hypoxic transcriptome of B. pseudomallei is the induction of stress-related proteins. The ferritin-like protein, Dps, was highly induced by hypoxia in B. pseudomallei. Dps is a nonspecific DNA-binding protein that functions to protect DNA and/or sequester iron (2). In B. pseudomallei, Dps is induced under oxidative stress conditions, and this protein protects DNA from damage caused by acid and oxidative stress (27). Additionally, under hypoxic conditions, B. pseudomallei induced the expression of clpAB. ClpAB protease is a well-studied protease that plays a major role in protein quality control and the stress responses of various bacteria (5, 11). Under anaerobic conditions, the expressed ClpAB protease could be used to degrade proteins expressed during aerobic growth that are no longer needed during an anaerobic nonreplicating state, thus in effect changing the aerobic proteome into an anaerobic proteome. The induction of dps, acr, usp, and clpAB suggests that B. pseudomallei has evolved a robust response to adapt to stressful anaerobic conditions. Under anaerobic conditions, the growth of B. pseudomallei is inhibited, and overall metabolism is most likely minimal. Thus, the expression of these stress proteins during entry into an anaerobic state might provide protection to nonreplicating B. pseudomallei by protecting cellular proteins and DNA.
Under anaerobic conditions, B. pseudomallei became highly tolerant to the antibiotics used for the treatment of melioidosis. These results are in line with the idea that nonreplicating bacterial cells with low metabolic activity are often indifferent to antibiotics that target active cellular processes such as cell wall, DNA, and, protein synthesis (3, 13). Surprisingly, nonreplicating anaerobic B. pseudomallei cultures were exceptionally sensitive to the nitroimidazole antibiotics metronidazole and tinidazole. Our findings regarding the sensitivity of anaerobic B. pseudomallei to metronidazole and tinidazole may have important implications for the treatment of melioidosis. During the course of a human infection, B. pseudomallei is likely to encounter hypoxic and anoxic conditions such as those found in abscesses or biofilms. Under these conditions, treatment with antibiotics such as ceftazidime, trimethoprim-sulfamethoxazole, or chloramphenicol would likely be ineffective at eradicating B. pseudomallei. However, in these anoxic microenvironments, the nitroimidazole antibiotics should be highly efficacious at killing B. pseudomallei when the conventional antibiotics fail. Abscesses and microcolonies are common pathological findings in human melioidosis, and both often contain little to no oxygen (31, 37, 56). Both metronidazole and tinidazole are considered safe for human use and are widely used to treat amebic and anaerobic bacterial infections (20, 29). The low penetration of antibiotics into abscesses is an additional confounding factor for the treatment of pathogens in these microenvironments (51). More-comprehensive studies using animal models are warranted to evaluate if metronidazole or tinidazole can offer therapeutic value against melioidosis, such as shortening the treatment time or preventing relapse.
Our data demonstrate that a small proportion (0.1%) of the anaerobic population is capable of long-term persistence. This persister population is tolerant not only to the drugs effective against aerobic B. pseudomallei but also to drugs that target anaerobic bacteria. Interestingly, drugs that are effective against anaerobic B. pseudomallei are capable of killing 99.9% of the population when administered before long-term anaerobiosis has eliminated all but the persisters. It appears that the anaerobic drugs are capable of killing the population that is still metabolically active and will eventually die under anaerobiosis; however, the drugs are not effective against the apparently dormant persisters. The anaerobic persisters appear to be a true persister population that is likely analogous to the persisters described for other bacteria (25). Significant work has shown that persisters become much more prevalent during stationary phase than during early-log phase. It appears that the stress of anaerobiosis may initiate a conversion similar to that induced by the stress of stationary phase, in which a small portion of the population shifts to a dormant state. Persisters from stationary phase are difficult to study, since they are at maximum 1% of the population, and the other 99% of viable bacteria remain drug sensitive. Traditionally, in order to enrich and isolate a persister population in a bacterial culture, antibiotics are added to eliminate nonpersister cells. In the anaerobic model described here, only persisters survived long-term anaerobiosis; thus, the culture does not require antibiotic treatment to eliminate the nonpersisters. Therefore, this model allows for the direct examination of a persister population without antibiotic elimination of the nonpersister population. The extreme stability of the persister population and the ability to observe a pure persister population provide a new model for the study of persisters and should allow for further insight into the mechanisms of persistence.
In conclusion, our work aims at understanding the hypoxic and anaerobic microenvironments that B. pseudomallei is likely to encounter during melioidosis. The current report demonstrates that a subpopulation of B. pseudomallei is capable of stable long-term anaerobic survival. Additionally, this work shows that anaerobiosis induces two stages of antibiotic tolerance. First, under initial oxygen-limiting conditions, the entire population becomes tolerant to traditional melioidosis antibiotics. The second population is uncovered during long-term anaerobiosis, in which the surviving bacteria are tolerant to all antibiotics tested. These data suggest that respiratory stress may be a key factor behind the need for lengthy treatment regimens and the high frequency of relapse after treatment.
Supplementary Material
ACKNOWLEDGMENTS
This research project was supported by Rocky Mountain Regional Center of Excellence grant U54 AI-065357. Burkholderia microarrays were obtained through the Pathogen Functional Genomics Resource Center, managed and funded by the Division of Microbiology and Infectious Diseases, NIAID, NIH, DHHS, and operated by the J. Craig Venter Institute.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 2 May 2011.
REFERENCES
- 1. Aardema H., et al. 2005. Changing epidemiology of melioidosis? A case of acute pulmonary melioidosis with fatal outcome imported from Brazil. Epidemiol. Infect. 133:871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Almirón M., Link A. J., Furlong D., Kolter R. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6:2646–2654 [DOI] [PubMed] [Google Scholar]
- 3. Anderson G. G., O'Toole G. A. 2008. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol. 322:85–105 [DOI] [PubMed] [Google Scholar]
- 4. Boulette M. L., Payne S. M. 2007. Anaerobic regulation of Shigella flexneri virulence: ArcA regulates fur and iron acquisition genes. J. Bacteriol. 189:6957–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Capestany C. A., Tribble G. D., Maeda K., Demuth D. R., Lamont R. J. 2008. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J. Bacteriol. 190:1436–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chaowagul W. 2000. Recent advances in the treatment of severe melioidosis. Acta Trop. 74:133–137 [DOI] [PubMed] [Google Scholar]
- 7. Cheng A. C., Currie B. J. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Currie B. J., Fisher D. A., Anstey N. M., Jacups S. P. 2000. Melioidosis: acute and chronic disease, relapse and re-activation. Trans. R. Soc. Trop. Med. Hyg. 94:301–304 [DOI] [PubMed] [Google Scholar]
- 9. Dance D. A. 2000. Ecology of Burkholderia pseudomallei and the interactions between environmental Burkholderia spp. and human-animal hosts. Acta Trop. 74:159–168 [DOI] [PubMed] [Google Scholar]
- 10. Dance D. A., Wuthiekanun V., Chaowagul W., White N. J. 1989. The antimicrobial susceptibility of Pseudomonas pseudomallei. Emergence of resistance in vitro and during treatment. J. Antimicrob. Chemother. 24:295–309 [DOI] [PubMed] [Google Scholar]
- 11. Ekaza E., Teyssier J., Ouahrani-Bettache S., Liautard J. P., Köhler S. 2001. Characterization of Brucella suis clpB and clpAB mutants and participation of the genes in stress responses. J. Bacteriol. 183:2677–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eschbach M., et al. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 186:4596–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Field T. R., White A., Elborn J. S., Tunney M. M. 2005. Effect of oxygen limitation on the in vitro antimicrobial susceptibility of clinical isolates of Pseudomonas aeruginosa grown planktonically and as biofilms. Eur. J. Clin. Microbiol. Infect. Dis. 24:677–687 [DOI] [PubMed] [Google Scholar]
- 14. Fink R. C., et al. 2007. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J. Bacteriol. 189:2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fuchs S., Pané-Farré J., Kohler C., Hecker M., Engelmann S. 2007. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 189:4275–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Green L. C., et al. 1982. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal. Biochem. 126:131–138 [DOI] [PubMed] [Google Scholar]
- 17. Häußler S., Rohde M., Steinmetz I. 1999. Highly resistant Burkholderia pseudomallei small colony variants isolated in vitro and in experimental melioidosis. Med. Microbiol. Immunol. 188:91–97 [DOI] [PubMed] [Google Scholar]
- 18. Holden M. T. G., et al. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U. S. A. 101:14240–14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inglis T. J. J., Rolim D. B., Sousa A. D. Q. 2006. Melioidosis in the Americas. Am. J. Trop. Med. Hyg. 75:947–954 [PubMed] [Google Scholar]
- 20. Inoue K., Sugano K. 2002. Metronidazol (MNZ). Nippon Rinsho 60(Suppl. 2):671–675(In Japanese.) [PubMed] [Google Scholar]
- 21. Jenney A. W., Lum G., Fisher D. A., Currie B. J. 2001. Antibiotic susceptibility of Burkholderia pseudomallei from tropical northern Australia and implications for therapy of melioidosis. Int. J. Antimicrob. Agents 17:109–113 [DOI] [PubMed] [Google Scholar]
- 22. Jobin M. P., Delmas F., Garmyn D., Diviès C., Guzzo J. 1997. Molecular characterization of the gene encoding an 18-kilodalton small heat shock protein associated with the membrane of Leuconostoc oenos. Appl. Environ. Microbiol. 63:609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones-Carson J., et al. 2008. Inactivation of [Fe-S] metalloproteins mediates nitric oxide-dependent killing of Burkholderia mallei. PloS One 3:e1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kvint K., Nachin L., Diez A., Nyström T. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6:140–145 [DOI] [PubMed] [Google Scholar]
- 25. Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56 [DOI] [PubMed] [Google Scholar]
- 26. Liesack W., Schnell S., Revsbech N. P. 2000. Microbiology of flooded rice paddies. FEMS Microbiol. Rev. 24:625–645 [DOI] [PubMed] [Google Scholar]
- 27. Loprasert S., Whangsuk W., Sallabhan R., Mongkolsuk S. 2004. DpsA protects the human pathogen Burkholderia pseudomallei against organic hydroperoxide. Arch. Microbiol. 182:96–101 [DOI] [PubMed] [Google Scholar]
- 28. Ludlum D. B., Colinas R. J., Kirk M. C., Mehta J. R. 1988. Reaction of reduced metronidazole with guanosine to form an unstable adduct. Carcinogenesis 9:593–596 [DOI] [PubMed] [Google Scholar]
- 29. Manes G., Balzano A. 2004. Tinidazole: from protozoa to Helicobacter pylori—the past, present and future of a nitroimidazole with peculiarities. Expert Rev. Anti Infect. Ther. 2:695–705 [DOI] [PubMed] [Google Scholar]
- 30. Mays E. E., Ricketts E. A. 1975. Melioidosis: recrudescence associated with bronchogenic carcinoma twenty-six years following initial geographic exposure. Chest 68:261–263 [DOI] [PubMed] [Google Scholar]
- 31. Mikamo H., Ninomiya M., Tamaya T. 2003. Anaerobic abscess. Nippon Rinsho 61(Suppl. 2):481–484(In Japanese.) [PubMed] [Google Scholar]
- 32. Morrissey I., Smith J. T. 1994. The importance of oxygen in the killing of bacteria by ofloxacin and ciprofloxacin. Microbios 79:43–53 [PubMed] [Google Scholar]
- 33. Müller M. 1983. Mode of action of metronidazole on anaerobic bacteria and protozoa. Surgery 93:165–171 [PubMed] [Google Scholar]
- 34. Murray P. R., Baron E. J., Jorgensen J. H., Pfaller M. A., Yolken R. H. (ed.). 2003. Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC [Google Scholar]
- 35. Nachin L., Nannmark U., Nyström T. 2005. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187:6265–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ngauy V., Lemeshev Y., Sadkowski L., Crawford G. 2005. Cutaneous melioidosis in a man who was taken as a prisoner of war by the Japanese during World War II. J. Clin. Microbiol. 43:970–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park M. K., Myers R. A., Marzella L. 1992. Oxygen tensions and infections: modulation of microbial growth, activity of antimicrobial agents, and immunologic responses. Clin. Infect. Dis. 14:720–740 [DOI] [PubMed] [Google Scholar]
- 38. Platt M. D., et al. 2008. Proteomic, microarray, and signature-tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late-stage cystic fibrosis airway conditions. J. Bacteriol. 190:2739–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reddy G. B., Kumar P. A., Kumar M. S. 2006. Chaperone-like activity and hydrophobicity of alpha-crystallin. IUBMB Life 58:632–641 [DOI] [PubMed] [Google Scholar]
- 40. Rotz L. D., Khan A. S., Lillibridge S. R., Ostroff S. M., Hughes J. M. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rubinstein E., Dreznik Z., Mark Z. 1982. Gentamicin and cefsulodin efficacy in a rat abscess model. Surg. Gynecol. Obstet. 155:363–368 [PubMed] [Google Scholar]
- 42. Schreiber K., et al. 2006. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J. Bacteriol. 188:659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sengupta N., Paul K., Chowdhury R. 2003. The global regulator ArcA modulates expression of virulence factors in Vibrio cholerae. Infect. Immun. 71:5583–5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simmen H.-P., Blaser J. 1993. Analysis of pH and pO2 in abscesses, peritoneal fluid, and drainage fluid in the presence or absence of bacterial infection during and after abdominal surgery. Am. J. Surg. 166:24–27 [DOI] [PubMed] [Google Scholar]
- 45. Singh R., et al. 2008. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science (New York, N.Y.) 322:1392–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tusher V. G., Tibshirani R., Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vander Wauven C., Piérard A., Kley-Raymann M., Haas D. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vorachit M., Chongtrakool P., Arkomsean S., Boonsong S. 2000. Antimicrobial resistance in Burkholderia pseudomallei. Acta Trop. 74:139–144 [DOI] [PubMed] [Google Scholar]
- 49. Voskuil M. I. 2004. Mycobacterium tuberculosis gene expression during environmental conditions associated with latency. Tuberculosis (Edinb.) 84:138–143 [DOI] [PubMed] [Google Scholar]
- 50. Voskuil M. I., et al. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wagner C., Sauermann R., Joukhadar C. 2006. Principles of antibiotic penetration into abscess fluid. Pharmacology 78:1–10 [DOI] [PubMed] [Google Scholar]
- 52. Walters M. C., Roe F., Bugnicourt A., Franklin M. J., Stewart P. S. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wayne L. G., Hayes L. G. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. White N. J., et al. 1989. Halving of mortality of severe melioidosis by ceftazidime. Lancet ii:697–701 [DOI] [PubMed] [Google Scholar]
- 55. Wiersinga W. J., van der Poll T., White N. J., Day N. P., Peacock S. J. 2006. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 4:272–282 [DOI] [PubMed] [Google Scholar]
- 56. Wong K. T., Puthucheary S. D., Vadivelu J. 1995. The histopathology of human melioidosis. Histopathology 26:51–55 [DOI] [PubMed] [Google Scholar]
- 57. Wuthiekanun V., et al. 2005. Trimethoprim/sulfamethoxazole resistance in clinical isolates of Burkholderia pseudomallei. J. Antimicrob. Chemother. 55:1029–1031 [DOI] [PubMed] [Google Scholar]
- 58. Wuthiekanun V., Peacock S. J. 2006. Management of melioidosis. Expert Rev. Anti Infect. Ther. 4:445–455 [DOI] [PubMed] [Google Scholar]
- 59. Yabuuchi E., et al. 1992. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36:1251–1275 [DOI] [PubMed] [Google Scholar]
- 60. Yoon S. S., et al. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593–603 [DOI] [PubMed] [Google Scholar]
- 61. Zúñiga M., Pérez G., González-Candelas F. 2002. Evolution of arginine deiminase (ADI) pathway genes. Mol. Phylogenet. Evol. 25:429–444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.