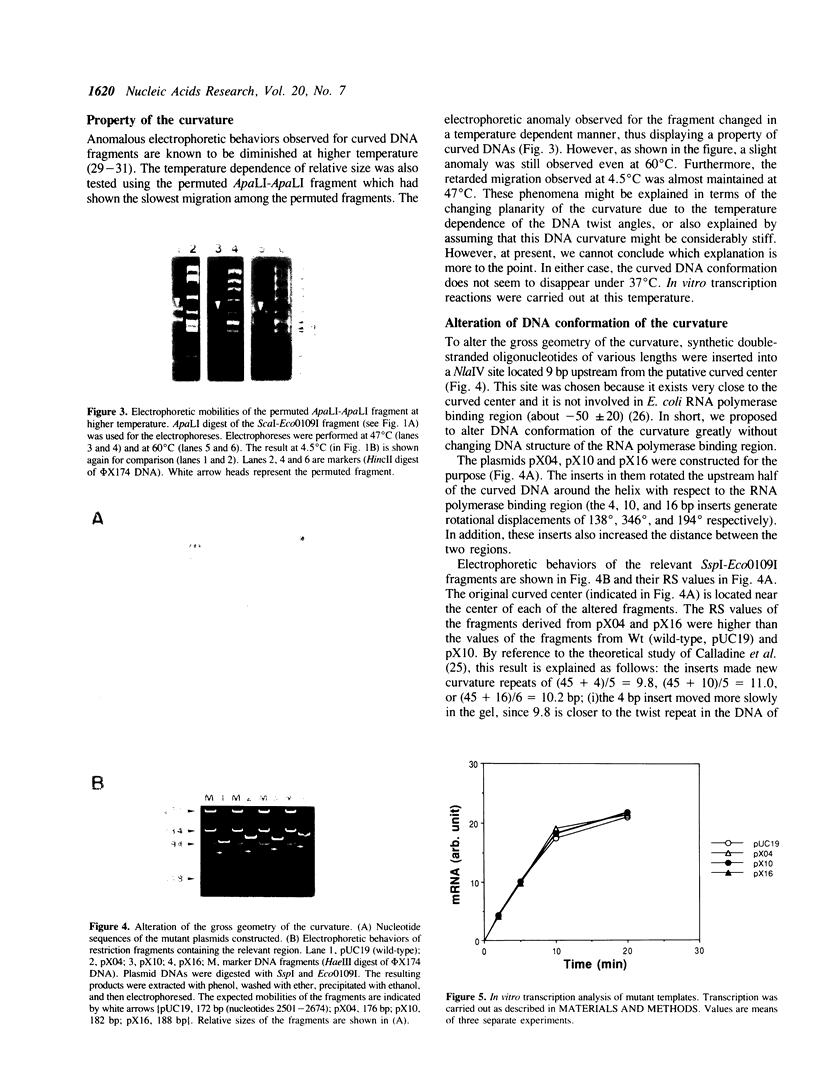
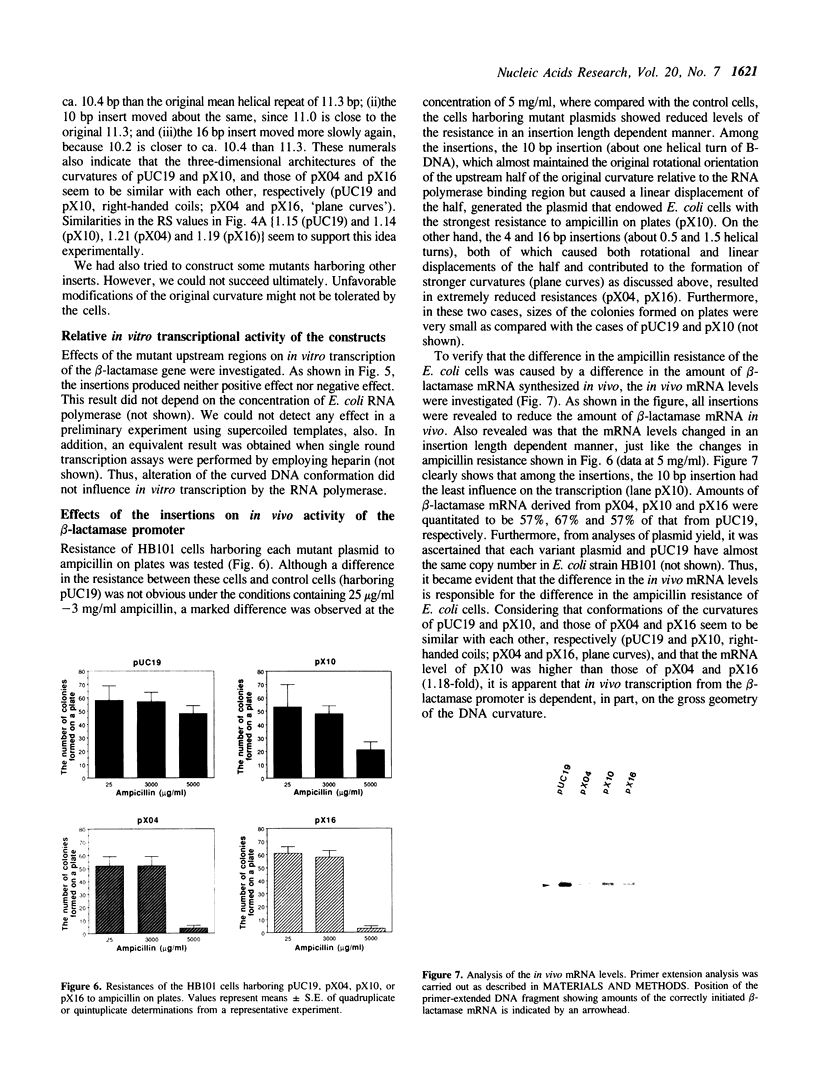
Abstract
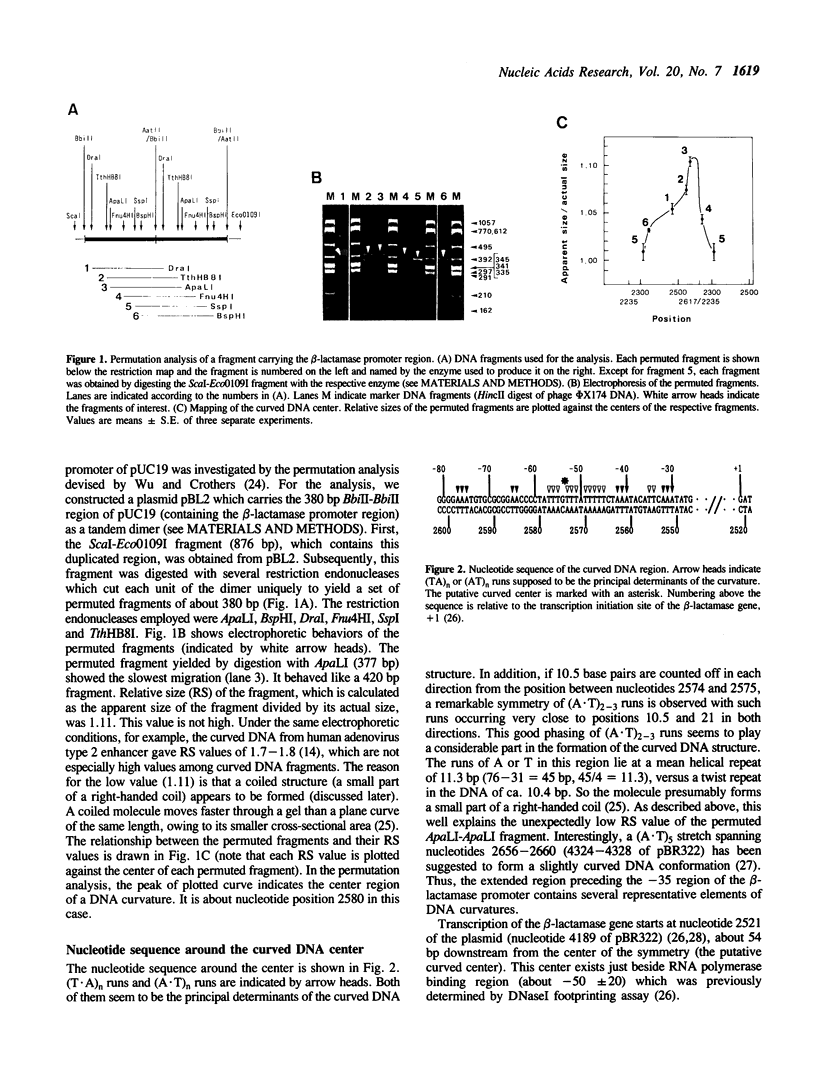
The region preceding the beta-lactamase promoter of Escherichia coli plasmid pUC19 has a curved DNA (bent DNA) structure. The center of the curvature was revealed to exist around nucleotide position 2580 of the plasmid, which is just beside RNA polymerase binding region. It was indicated that the identified region is curved even at 60 degrees C. The gross geometry of the curvature was altered by inserting synthetic double-stranded oligonucleotides between positions 2585 and 2586. Effect of the alteration on strength of the promoter was not detected in vitro. However, in vivo analyses showed that the promoter strength is apparently dependent, in part, on the gross geometry of the curvature. Insertions of 4 and 16 bp, both of which altered the gross geometry of the curvature greatly, caused considerable reductions of in vivo level of beta-lactamase mRNA. In vivo, overall three-dimensional structure of the region covering the promoter and the curvature seems to play some significant role in transcription of the gene.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amouyal M., Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987 Jun 20;195(4):795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Bracco L., Kotlarz D., Kolb A., Diekmann S., Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989 Dec 20;8(13):4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Cate R. L., Perlmutter A. P. Precise location of two promoters for the beta-lactamase gene of pBR322. S1 mapping of ribonucleic acid isolated from Escherichia coli or synthesized in vitro. J Biol Chem. 1982 Aug 10;257(15):9205–9210. [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R., McCall M. J. The intrinsic curvature of DNA in solution. J Mol Biol. 1988 May 5;201(1):127–137. doi: 10.1016/0022-2836(88)90444-5. [DOI] [PubMed] [Google Scholar]
- Cobbett C., Dickson B., Farmer L. The role of a static bend in the DNA of the aroF regulatory region of Escherichia coli. Gene. 1989 Jan 30;75(1):185–191. doi: 10.1016/0378-1119(89)90395-8. [DOI] [PubMed] [Google Scholar]
- Collis C. M., Molloy P. L., Both G. W., Drew H. R. Influence of the sequence-dependent flexure of DNA on transcription in E. coli. Nucleic Acids Res. 1989 Nov 25;17(22):9447–9468. doi: 10.1093/nar/17.22.9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res. 1987 Jan 12;15(1):247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S., Wang J. C. On the sequence determinants and flexibility of the kinetoplast DNA fragment with abnormal gel electrophoretic mobilities. J Mol Biol. 1985 Nov 5;186(1):1–11. doi: 10.1016/0022-2836(85)90251-7. [DOI] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. Synthetic DNA bending sequences increase the rate of in vitro transcription initiation at the Escherichia coli lac promoter. J Mol Biol. 1991 May 20;219(2):217–230. doi: 10.1016/0022-2836(91)90563-l. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., de Boer H. A., Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986 Jan 17;44(1):197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- Griffith J., Bleyman M., Rauch C. A., Kitchin P. A., Englund P. T. Visualization of the bent helix in kinetoplast DNA by electron microscopy. Cell. 1986 Aug 29;46(5):717–724. doi: 10.1016/0092-8674(86)90347-8. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Inokuchi K., Nakayama A., Hishinuma F. Sequence-directed bends of DNA helix axis at the upstream activation sites of alpha-cell-specific genes in yeast. Nucleic Acids Res. 1988 Jul 25;16(14B):6693–6711. doi: 10.1093/nar/16.14.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T., Makino K., Orita S., Nakata A., Kakunaga T. DNA bending and binding factors of the human beta-actin promoter. Nucleic Acids Res. 1989 Jan 25;17(2):523–537. doi: 10.1093/nar/17.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Lozinski T., Adrych-Rozek K., Markiewicz W. T., Wierzchowski K. Effect of DNA bending in various regions of a consensus-like Escherichia coli promoter on its strength in vivo and structure of the open complex in vitro. Nucleic Acids Res. 1991 Jun 11;19(11):2947–2953. doi: 10.1093/nar/19.11.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister C. F., Achberger E. C. Rotational orientation of upstream curved DNA affects promoter function in Bacillus subtilis. J Biol Chem. 1989 Jun 25;264(18):10451–10456. [PubMed] [Google Scholar]
- Mizuno T. Static bend of DNA helix at the activator recognition site of the ompF promoter in Escherichia coli. Gene. 1987;54(1):57–64. doi: 10.1016/0378-1119(87)90347-7. [DOI] [PubMed] [Google Scholar]
- Muzard G., Théveny B., Révet B. Electron microscopy mapping of pBR322 DNA curvature. Comparison with theoretical models. EMBO J. 1990 Apr;9(4):1289–1298. doi: 10.1002/j.1460-2075.1990.tb08238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T., Hashimoto S. Upstream half of adenovirus type 2 enhancer adopts a curved DNA conformation. Nucleic Acids Res. 1989 May 25;17(10):3845–3853. doi: 10.1093/nar/17.10.3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramstein J., Lavery R. Energetic coupling between DNA bending and base pair opening. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7231–7235. doi: 10.1073/pnas.85.19.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo F., Zaballos A., Salas M. Bend induced by the phage phi 29 transcriptional activator in the viral late promoter is required for activation. J Mol Biol. 1990 Feb 20;211(4):713–725. doi: 10.1016/0022-2836(90)90072-t. [DOI] [PubMed] [Google Scholar]
- Russell D. R., Bennett G. N. Characterization of the beta-lactamase promoter of pBR322. Nucleic Acids Res. 1981 Jun 11;9(11):2517–2533. doi: 10.1093/nar/9.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen N. C. Anomalous electrophoresis of deoxyribonucleic acid restriction fragments on polyacrylamide gels. Biochemistry. 1983 Dec 20;22(26):6186–6193. doi: 10.1021/bi00295a023. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Direct evidence for DNA bending at the lambda replication origin. Science. 1987 Apr 24;236(4800):416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. Catabolite activator protein-induced DNA bending in transcription initiation. J Mol Biol. 1991 May 20;219(2):201–215. doi: 10.1016/0022-2836(91)90562-k. [DOI] [PubMed] [Google Scholar]