Abstract
We have screened 47 locked nucleic acid (LNA) antisense oligonucleotides (ASOs) targeting conserved (>95% homology) sequences in the hepatitis C virus (HCV) genome using the subgenomic HCV replicon assay and generated both antiviral (50% effective concentration [EC50]) and cytotoxic (50% cytotoxic concentration [CC50]) dose-response curves to allow measurement of the selectivity index (SI). This comprehensive approach has identified an LNA ASO with potent antiviral activity (EC50 = 4 nM) and low cytotoxicity (CC50 >880 nM) targeting the 25- to 40-nucleotide region (nt) of the HCV internal ribosome entry site (IRES) containing the distal and proximal miR-122 binding sites. LNA ASOs targeting previously known accessible regions of the IRES, namely, loop III and the initiation codon in loop IV, had poor SI values. We optimized the LNA ASO sequence by performing a 1-nucleotide walk through the 25- to 40-nt region and show that the boundaries for antiviral efficacy are extremely precise. Furthermore, we have optimized the format for the LNA ASO using different gapmer and mixomer patterns and show that RNase H is required for antiviral activity. We demonstrate that RNase H-refractory ASOs targeting the 25- to 40-nt region have no antiviral effect, revealing important regulatory features of the 25- to 40-nt region and suggesting that RNase H-refractory LNA ASOs can act as potential surrogates for proviral functions of miR-122. We confirm the antisense mechanism of action using mismatched LNA ASOs. Finally, we have performed pharmacokinetic experiments to demonstrate that the LNA ASOs have a very long half-life (>5 days) and attain hepatic maximum concentrations >100 times the concentration required for in vitro antiviral activity.
INTRODUCTION
Hepatitis C virus (HCV) infection is a major cause of liver disease, with >170 million infected people worldwide being at risk from liver failure and hepatocarcinoma. Current standard of care (SOC) using pegylated alpha 2a interferon (IFN-α2a)-ribavirin is failing 40 to 50% of treated HCV patients, so new therapies are urgently required. New directly acting antiviral therapies for hepatitis C are becoming available, but these will eventually fail many HCV patients due to emerging drug-resistant mutations in HCV strains (6, 7). HCV genomic RNA is an attractive antiviral target because it holds genetic information for viral proteins and contains regions of highly conserved sequence required for HCV replication/translation. Many groups have identified antisense oligonucleotides (ASOs) capable of inhibiting HCV RNA replication and viral polyprotein synthesis in vitro. Clinical trials on chronically HCV-infected patients show that modified ASOs targeting HCV can result in >2-log-unit decreases in viral loads, although the mechanism driving clinical antiviral activity has yet to be fully validated as antisense (14, 28, 33). Moreover, locked nucleic acid (LNA) represents a new generation of ASOs with improved affinity of binding to RNA targets, increased sequence specificity, greater biostability against nucleases, and reportedly lower toxicity (3, 15, 22, 37, 40).
HCV is a positive-strand RNA virus with a linear genome of ∼9,500 nucleotides (nt). Different isolates show considerable nucleotide variability, leading to the division of HCV genomes into many different genotypes and subtypes, including the most clinically relevant HCV genotypes, 1a and 1b. In all genotypes, the viral RNA contains a large open reading frame that encodes a polyprotein precursor of ∼3,020 amino acids, giving rise to core (C), envelope (E1, E2), and nonstructural (NS2 to NS5b) proteins. The coding sequence is preceded by a 5′ noncoding region of ∼340 nt (highly conserved among all HCV genotypes and subtypes) and folds into a stable secondary structure, serving as an internal ribosomal entry site (IRES), essential for efficient translation of the viral proteins (36). Studies using a cell-based HCV replication system (replicon model) have revealed that a host microRNA (miR-122) plays a positive role in HCV replication in vitro and is recruited directly to a 25- to 40-nt region in the IRES via two adjacent binding sites at 23-CACUCC-28 and 38-CACUCC-43, respectively (19, 23). LNA-based oligomers have been used to target miR-122 in primates, and these studies demonstrated that effective doses of LNA-based ASOs can be delivered to the liver for 12 weeks with no adverse effects (12, 25).
We have generated a set of 47 LNA ASOs directed against highly conserved HCV sequences and developed a medium-throughput 96-well microplate-based, reverse transfection protocol using the well-characterized HCV subgenomic replicon system (9B replicon cells described in reference 13) to generate full dose-response curves for both antiviral and cytotoxic activities. To alleviate the logistical hurdles for this extensive dual primary screen, multiple vials of 9B replicon cells were stored frozen and individual vials were thawed immediately prior to transfection. The antiviral activities of the LNA ASOs were measured via a luciferase reporter that correlates with replicon replication. Each LNA ASO was titrated from ∼1 μM to 0.3 nM, and ASO concentrations required to produce a 50% inhibition in replicon activity (50% effective concentration [EC50]) and the concentrations required to produce a 50% reduction in cell viability (50% cytotoxic concentration [CC50]) were calculated. The ratio of CC50/EC50 gives the in vitro selectivity index (SI), providing a metric for LNA ASO-specific anti-HCV activity.
Any hits from the primary screen using frozen/thawed 9B replicon cells were analyzed using freshly passaged 9B cells to confirm antiviral activity. Mismatch ASOs were used to validate the antisense mechanism of action (MoA) before extensive optimization of the initial hit ASO, including a 1-nucleotide walk through the target region and comprehensive evaluation of alternative LNA ASO formats (gapmer, mixomer, different lengths). These different formats were assayed for both antiviral activity and pharmacokinetic (PK) properties (tissue loading and half-life [t1/2]). In summary, this is the first report showing a comprehensive walkthrough of the HCV IRES using LNA-based ASOs.
MATERIALS AND METHODS
Cells.
The Huh-7 cell clone 9B replicon containing I389/NS3-3′/LucUbiNeo-ET has been described previously (13). Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 22 U penicillin G/ml, 220 μg/ml streptomycin, 1 mM sodium pyruvate, 2 mM glutamine, and 0.5 mg/ml G418. Frozen 107-cell aliquots of these cells were prepared by GE Healthcare and stored in liquid nitrogen prior to use.
Generation of LNA ASOs.
All LNA ASOs were synthesized by Exiqon A/S (Denmark) and dissolved in 10 mM Tris, pH 7.5–1 mM EDTA.
Generation of antiviral EC50 and CC50 data.
EC50 potencies were generated using a lipofection reverse transfection protocol using transfection reagent (Dharmafect3; Dharmacon, United Kingdom) on 9B replicon cells cultured in 96-well plates in the presence of 750 nM stuffer DNA (5′-GACCACTTGCCACCCATC-3′). A 100 μM stock solution of test ASO LNA was diluted in a 1/3 dilution series in buffer (3 μM stuffer DNA in OptiMEM [Gibco]) and incubated with an equal volume of 1% transfection reagent-OptiMEM for 20 min at room temperature. An equal volume of 9B replicon cell culture (density, 3 × 105 cells/ml in 2× medium) was added, and the mixture was incubated for 40 h at 37°C in a humidified atmosphere with 5% CO2. An equal volume of luciferase assay reagent (BritelitePlus; Perkin Elmer) was added, and the chemiluminescence was quantified using an Envision 2104 multilabel plate reader (Perkin Elmer). The negative control for inhibition activity was measured in the absence of added LNA ASO. The percent inhibition of replicon activity by each ASO relative to that of 10 IU IFN-α2a (100% positive control for antiviral activity) was then calculated after background correction. For reverse transcription-PCR (RT-PCR) experiments, total RNA was extracted (RNAeasy; Qiagen) and first-strand cDNA was synthesized using reverse transcriptase (cDNA Archive; Applied Biosystems). Quantitative PCR was performed using TaqMan universal PCR master mix (Applied Biosystems), HCV replicon forward primer (5′-TCCCGGGAGAGCCATAGTG) and reverse primer (5′-GGCATTGAGCGGGTTGATC), and a 6-carboxyfluorescein-labeled probe (5′-CCGGAATTGCCAGGACGACCG). Parallel reactions using the actin probe set (4326315E; Applied Biosystems) were also set up. Reactions were performed using an Applied Biosystems 7900 HT apparatus.
CC50 cytotoxicity measurements of LNA ASO transfections were generated using the same transfection protocol described above. After 40 h incubation, 10 μl WST-1 reagent (catalog no. 11644807; Roche Applied Science) was added to each well and the plate was incubated for 1 h. The plate was read at 430 nm in a Perkin Elmer plate reader. The LNA ASO cytotoxic activity was calculated as a percentage of total maximum toxicity, measured using a final assay concentration 100 μM cycloheximide as a positive control. The negative control for the cytotoxicity assay was measured in the absence of LNA ASO. The percent cytotoxic effect of LNA ASO transfection was then calculated as a percentage of that for the positive 100 μM cycloheximide control after background correction.
Pharmacokinetic analysis.
CD-1 mice were dosed by intravenous (i.v.) bolus injection with oligonucleotide prepared in sterile filtered phosphate-buffered saline (pH 7.4) at a concentration of 0.5 mg/ml (dosing volume, 10 ml/kg of body weight). Mice were euthanized at 1, 7, 24, 72, 120, and 168 h. Liver and kidneys were harvested from each animal at necropsy and immediately frozen on dry ice. Samples were stored at −80°C prior to analysis. All experiments with animals were carried out in compliance with United Kingdom legislation and were subjected to local ethical review.
Tissue samples were analyzed for LNA ASOs using a modified high-performance liquid chromatography (HPLC) and tandem mass spectrometry (MS/MS) approach developed within our facilities for oligonucleotide quantitation (submitted for publication). In brief, tissue samples were diluted 1:4 (wt/vol) in sterile water and homogenized in a Precelly 24-cell disrupter (Bertin Technologies). Aliquots (100 μl) were used for analysis alongside calibration standards (0.05 to 100 μg/g) and quality control (QC) samples (0.1, 5, and 50 μg/g) prepared in blank tissue homogenate, and a 16-mer internal standard was added from the LNA ASO series. Samples were diluted with 500 μl 5% ammonium hydroxide, 10 μl 1 M dithiothreitol, and 100 μl phenol-chloroform solution (2:1, vol/vol) containing 1 mM −4-(4-diethylaminophenylazo)-1-(4-nitrobenzyl)-pyridinium bromide and then mixed thoroughly. Following centrifugation, the aqueous fraction was removed and further washed in 200 μl dichloroethane, before being isolated again and dried under nitrogen at 50°C. The residue was reconstituted in 100 μl 8 mM triethylamine (TEA) prior to injection of 20-μl aliquots.
The HPLC separation was performed using a C18 monolithic column (2 by 50 mm; Onyx; Phenomenex) at 60°C and operating with a flow rate of 0.25 ml/min. The aqueous mobile phase was 8.6 mm TEA and 200 mM heptafluoroisopropanol, which acted as the ion pair reagents, and the analytes were eluted using methanol applied as a gradient from 10 to 35% over 6 min. The column was washed in 95% methanol for 1 min and then reequilibrated for 3 min. The analytes were monitored with an AB Sciex API 4000 Qtrap mass spectrometer operated in negative ion mode with an electrospray ionization interface. Transition ions representing the [M-7H]7−, [M-8H]8−, and [M-9H]9− charge states of the parent LNA ASO nominal mass, all fragmented to the singular m/z 95 phosphorothioate (PS) daughter ion, were monitored and summed together during the peak integration process. The collision energy was −125 eV, and the ion source was set at 450°C. Under these conditions, each assay had a lower limit of quantitation of approximately 50 ng/g tissue for the LNA ASO analyzed. The level of interassay precision (coefficient of variation) demonstrated with QC replicates in liver and kidney (n = 4) ranged from 1 to 14%, whereas the QC accuracy ranged from 93 to 109%. Data were analyzed using the WinNonLin (version 5.2) program (Pharsight) and noncompartmental analysis to derive basic pharmacokinetic parameters from tissue data.
RESULTS
Screening of an LNA ASO library targeting HCV sequences to generate SI values.
We have generated LNA ASOs targeting all IRES sequences (from 1 to 350 nt using an 8-nucleotide overlap) and, additionally, all highly conserved (>95% homology) HCV sequences outside the IRES. The format of the individual LNA ASOs and the corresponding HCV nucleotide regions targeted are given in Table S1 in the supplemental material. The majority of ASOs were 16-mers using a 4-8-4 gapmer format (two terminal regions of 4 LNA-modified nucleotides flanking a central region of 8 DNA nucleotides), although we included all-LNA ASOs for the highly conserved HCV regions of <11 nucleotides in length and alternative gapmer formats to target regions of between 12 and 16 nucleotides. Each ASO was screened at 650 nM to 0.3 nM for both antiviral activity and general cytotoxicity using a modified reverse transfection plate-based procedure. To ensure uniform transfection conditions across all different ASO concentrations, we maintained a fairly constant (<2-fold change) oligonucleotide concentration across all points of the dose-response curve by adding a fixed amount of stuffer oligonucleotide. The 9B subgenomic HCV replicon cell line was used in all assays (13).
To validate the specificity of our screening approach, we included several negative and positive-control LNA ASOs in our library. A nonspecific negative-control ASO was inactive in our replicon assay, having neither antiviral activity nor cytotoxic activity using either the 4-6-4 or 4-8-4 format (sequence codes [seqCODEs] CONT-14 and CONT-16, rows 1 and 2, respectively; Table 1). Similarly, two positive-control ASOs targeting the firefly luciferase sequences of the 9B replicon, also using either the 4-6-4 or 4-8-4 format, were added to the library and were identified to be nontoxic active hits (seqCODEs LUC-14 and LUC-16, rows 3 and 4, respectively; Table 1). We observe a slight 3- to 4-fold increase in potency using the 4-8-4 format. It has been reported that the efficacy of target RNA cleavage improves with increasing gapmer size, giving ∼100% cleavage with a gap of 8 nt (24).
Table 1.
Antiviral EC50 data and cytotoxic CC50 data for the ASOs used in the primary screen of 9B subgenomic replicon cellsa
| Row | seqCODE | EC50 (nM) | CC50 (nM) | SI |
|---|---|---|---|---|
| 1 | CONT-14 | >833 (n = 9) | >833 (n = 8) | ∼0 |
| 2 | CONT-16 | >520 (n = 9) | >455 (356–582, n = 10) | ∼0 |
| 3 | LUC-14 | 188 (0.567–62,100, n = 2) | >833 (n = 2) | >4.44 |
| 4 | LUC-16 | 56.7 (0.502–6,400, n = 3) | >833 (n = 2) | >14.7 |
| 5 | seq134 | 51.8 (5.66–475, n = 4) | 147 (43.2–500, n = 4) | 2.83 |
| 6 | seq132 | 4.07 (2.89–5.74, n = 24) | >74.5 (40.7–136, n = 25) | >18.3 |
| 7 | seq131 | >32.7 (2.91–367, n = 4) | >35.8 (1.62–792, n = 4) | ∼0 |
| 8 | seq129 | >456 (151–1,380, n = 6) | >833 (n = 4) | ∼0 |
| 9 | seq60-90c | >833 (n = 2) | >833 (n = 3) | ∼0 |
| 10 | seq60-90a | >833 (n = 3) | >614 (166–2,280, n = 3) | ∼0 |
| 11 | seq124 | >351 (90.0–1,370, n = 4) | 149 (7.50–2,950, n = 3) | 0.423 |
| 12 | seq123 | >102 (0.749–14,000, n = 3) | 84.1 (16.5–428, n = 4) | 0.822 |
| 13 | seq110-140a | >55.7 (0.402–7,730, n = 3) | 82.1 (14.2–476, n = 4) | 1.47 |
| 14 | seq110-140c | 39.4 (0.0147–105,000, n = 2) | 32.1 (4.17–247, n = 4) | 0.814 |
| 15 | seq118 | 58.6 (8.90–386, n = 3) | 142 (34.7–577, n = 3) | 2.42 |
| 16 | seq116 | 42.4 (0.825–2,180, n = 3) | 27.3 (0.206–3,610, n = 2) | 0.643 |
| 17 | seq115 | 66.6 (18.7–237, n = 4) | 88.0 (7.38–1,050, n = 3) | 1.32 |
| 18 | seq166 | 3.37 (0.175–64.9, n = 4) | 8.19 (1.43–46.9, n = 4) | 2.43 |
| 19 | seq170-200c | >833 (n = 3) | >833 (n = 3) | ∼0 |
| 20 | seq170-200b | 5.25 (0.131–210, n = 3) | 14.0 (4.54–43.4, n = 3) | 2.67 |
| 21 | seq170-200a | 400 (144–1,110, n = 3) | 385 (94.5–1,570, n = 3) | 0.962 |
| 22 | seq111 | >489 (49.3–4,840, n = 3) | 177 (5.47–5,730, n = 3) | 0.362 |
| 23 | seq207-250c | >833 (n = 3) | >815 (744–894, n = 3) | ∼0 |
| 24 | seq207-250b | 145 (6.88–3,070, n = 3) | >704 (83.8–5,920, n = 2) | >4.84 |
| 25 | seq207-250a | 12.9 (4.19–39.8, n = 5) | 187 (42.4–822, n = 5) | 14.5 |
| 26 | seq103 | 20.8 (6.84–63.4, n = 3) | 77.4 (3.22–1,860, n = 3) | 3.71 |
| 27 | seq101 | 233 (0.219–248,000, n = 2) | 264 (0.661–105,000, n = 2) | 1.13 |
| 28 | seqT6 | 49.4 (0.00459–532,000, n = 2) | 24.1 (0.0379–15,400, n = 2) | 0.488 |
| 29 | seq100 | 154 (21.3–1,120, n = 3) | 170 (0.649–44,700, n = 2) | 1.1 |
| 30 | seq99 | 417 (203–856, n = 3) | >738 (158–3,440, n = 2) | >1.77 |
| 31 | seq98 | 79.7 (8.27–768, n = 3) | 171 (64.9–450, n = 3) | 2.14 |
| 32 | seq310-330a | >31.3 (0.521–1,880, n = 4) | >161 (12.4–2,090, n = 5) | ∼0 |
| 33 | seq310-330c | 20.1 (3.29–123, n = 4) | 23.6 (0.478–1,160, n = 3) | 1.17 |
| 34 | seq94 | 1.57 (0.0795–31.0, n = 4) | 3.10 (0.320–30.0, n = 3) | 1.97 |
| 35 | seq92 | 164 (1.55–17,400, n = 3) | 69.6 (1.37–3,540, n = 3) | 0.424 |
| 36 | seq151 | >16.7 (0.310–901, n = 4) | 39.6 (19.9–79.0, n = 4) | 2.37 |
| 37 | seq149 | >311 (4.50–21,500, n = 3) | >833 (n = 2) | ∼0 |
| 38 | seqT19 | >520 (n = 2) | >520 (n = 2) | ∼0 |
| 39 | seq163 | >833 (n = 2) | >833 (n = 3) | ∼0 |
| 40 | seq155 | >833 (n = 2) | 71.9 (0.972–5,320, n = 2) | 0.0863 |
| 41 | seq138 | >833 (n = 2) | >833 (n = 2) | ∼0 |
| 42 | seq165 | >833 (n = 2) | >833 (n = 3) | ∼0 |
| 43 | seq601 | 34.8 (0.644–1,880, n = 2) | >520 (n = 2) | >15.0 |
| 44 | seq602 | 4.41 (2.10–9.25, n = 2) | 45.4 (2.88–716, n = 2) | 10.3 |
| 45 | seq139 | 114 (30.9–418, n = 5) | >255 (56.0–1,160, n = 5) | >2.24 |
| 46 | seq600 | 21.7 (0.920–510, n = 2) | >405 (16.9–9,720 n = 2) | >18.7 |
| 47 | seqT22 | 10.8 (10.8–10.9, n = 2) | 135 (0.00639–2.84E6, n = 2) | 12.4 |
| 48 | seq153 | >833 (n = 2) | >475 (0.384–588,000, n = 2) | ∼0 |
| 49 | seq154 | >833 (n = 2) | >833 (n = 2) | ∼0 |
| 50 | seq146 | 33.9 (5.66–203, n = 6) | >412 (160–1,060, n = 4) | >12.2 |
| 51 | seq158 | >163 (7.73–3,450, n = 4) | 60.7 (8.48–434, n = 3) | 0.372 |
| 52 | seq132a | 6.47 (4.30–9.74 n = 2) | 241 | 37.3 |
| 53 | seq132b | >19.8 (1.45–272, n = 5) | >67.5 (6.24–730, n = 5) | ∼0 |
| 54 | seq132c | >77.9 (2.63E9–2.32E12, n = 2) | >139 (8.88–2,180, n = 2) | ∼0 |
| 55 | seq132d | >520 (n = 2) | >520 (n = 2) | ∼0 |
| 56 | seq132e | >520 (n = 2) | >520 (n = 2) | ∼0 |
| 57 | seq132f | 196 (65.2–591, n = 2) | No summarized data | |
| 58 | seq132g | 4.68 (0.00879–2,500, n = 2) | >520 (n = 2) | >111 |
| 59 | seq132h | 7.26 (0.00136–38,800, n = 2) | >520 (n = 2) | >71.7 |
| 60 | seq132i | >520 (n = 2) | >330 (1.01–107,000, n = 2) | ∼0 |
| 61 | seq132j | >72.4 (0.0354–148,000, n = 3) | >202 (9.74–4,180, n = 4) | ∼0 |
| 62 | seq401 | >159 (33.6–754, n = 4) | 199 (90.6–439, n = 4) | 1.25 |
| 63 | seq400 | >405 (137–1,190, n = 3) | >520 (n = 3) | ∼0 |
| 64 | seq500 | 123 (0.124–122,000, n = 2) | >278 (0.0991–781,000, n = 2) | >2.27 |
The seqCODEs correspond to the identity of individual ASOs described in the text. SI is the ratio of CC50/EC50. An SI value of ∼0 indicates that ASOs lack both antiviral activity and cytotoxic activity. The 95% confidence limits are given in parentheses; n refers to the number of separate repeats for each experiment to generate confidence limits. These values were generated by the in-house Pfizer database SIGHTS.
The LNA ASO library was constructed using a PS backbone to maintain the RNase H compatibility of the ASO, take advantage of the improved serum protein binding properties over those of a phosphodiester (PO) backbone (these properties reduce renal excretion and increase coverage of liver tissue), and improve nuclease stability (3). Where the conserved sequences were <16 nt, the ASOs were all-LNA to assess whether translational arrest could be a possible mechanism for antiviral activity. The ASO gapmers were generated with a melting temperature (Tm) of >60°C with an upper limit of <90°C (to reduce nonspecificity). The self-hybridization score was generally <40°C and at least 30°C below the Tm. The difference between the score for hybridization to the target and self-hybridization was at least 40°C.
Forty-seven ASOs were screened in the 9B replicon cell line at n = 2 (Table 1, rows 5 to 53), and seven hits with SIs of >10 were identified: five hits targeting NS5a (seq600, seq601, seq602, seqT22, and seq146, rows 46, 43, 44, 47, and 50, respectively, in Table 1) and two hits targeting IRES (seq132 and seq207-250a, rows 6 and 25, respectively, in Table 1).
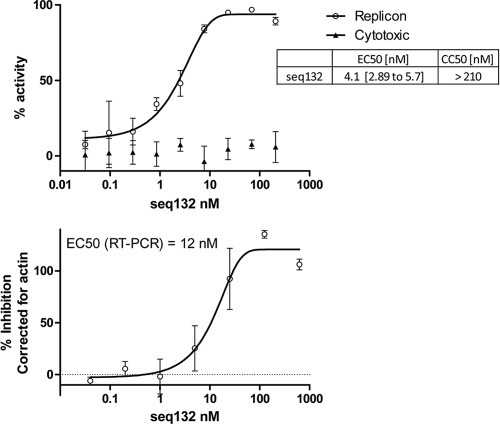
Follow-up titrations using freshly passaged replicon cells revealed that most of the hits exhibit dose-response curves with maxima of <100% (see Fig. S1 in the supplemental material). Only the seq132 ASO exhibited reproducible, potent antiviral (4 nM) activity with no cytotoxicity (>880 nM) and a >95% maximal asymptote for the dose-response curve in the replicon assay (Fig. 1). In addition, we also used an RT-PCR assay readout using replicon-directed probe-primer pairs to demonstrate that seq132 transfection resulted in the dose-dependent loss of replicon RNA at a similar potency with an EC50 of ∼12 nM (Fig. 1).
Fig. 1.
(Top panel) 9B subgenomic replicon cells (freshly passaged) were transfected with increasing concentrations of seq132 ASO as described in Materials and Methods. After 48 h, the luciferase activity (empty circles) and cytotoxicity (filled triangles) were measured. Antiviral activity was expressed as the percentage of HCV replicon inhibition relative to that achieved with 10 IU/ml IFN-α2a and was plotted against the seq132 concentration. Nonlinear regression analysis was used to calculate each EC50 using GraphPad Prism software (version 5.01). Error bars represent the range of duplicate data points. Results of a representative experiment using freshly passaged replicon cells are shown. This experiment was performed >10 times, with similar results each time. The EC50 and CC50 values shown in the inset are from 25 experiments, including experiments in both freshly passaged and frozen/thawed cell experimental formats, and include 95% confidence limits. (Bottom panel) 9B replicon cells transfected as described for the top panel were lysed for RT-PCR analysis using a replicon-specific probe/primer pair (empty circles). The data were corrected for actin expression, and percent inhibition relative to that achieved with 10 IU/ml IFN-α2a treatment was calculated and plotted against seq132 concentration. This experiment was independently performed three times, with similar seq132 potencies each time. Nonlinear regression analysis was used to calculate each EC50 using GraphPad Prism (version 5.01). Error bars represent the range of duplicate data points.
We note that the SI for seq132 obtained when seq132 is transfected into freshly passaged replicon cells is improved over that when it is transfected into the frozen/thawed cells used in the primary screen (SI > 100 in Fig. 1 compared to SI > 18 in row 6, Table 1). This improved SI results from the reduced cytotoxicity observed when freshly passaged replicon cells are used in the follow-up assay (CC50 > 210 nM) and frozen/thawed replicon cells are used in the primary screen (CC50 > 50 to 100 nM). Our results showed that the logistical advantage of using frozen/thawed cells for initial screening was partially offset by slightly increased susceptibility (∼3-fold) to cytotoxicity.
Validation of antisense mechanism of action.
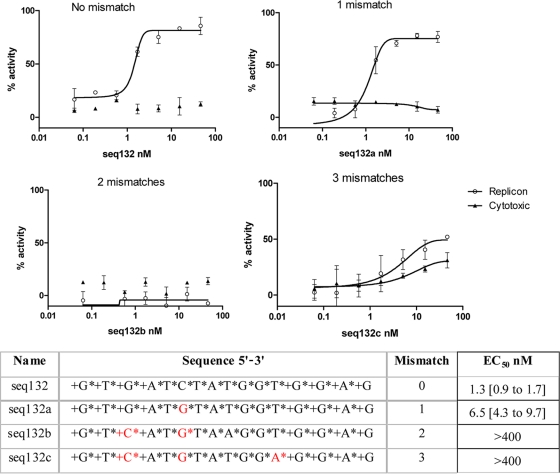
To confirm the antisense mechanism of action for LNA ASO seq132, we synthesized analogues of seq132 containing 1, 2, or 3 mismatches to the 25- to 40-nt HCV target sequence. Figure 2 shows that efficacy is reduced after 1 mismatch and is completely inactive after 2 mismatches. Similar results were obtained using the frozen/thawed cells in the primary screen (rows 52 to 55, Table 1). The apparent increase in activity at 3 mismatches is driven by the nonspecific toxicity emerging as the LNA ASO sequence loses specificity for the 25- to 40-nt target region.
Fig. 2.
9B subgenomic replicon cells (freshly passaged) were transfected with increasing concentrations of various analogues of ASO seq132 as described in Materials and Methods. The sequences of the mismatch ASOs are shown at the bottom, where + indicates LNA and * indicates phosphorothioate linkage. After 48 h, the luciferase activity (empty circles) and cytotoxicity (filled triangles) were measured, and percent inhibition relative to that achieved with 10 IU/ml IFN-α2a was calculated and plotted against seq132 concentration. Nonlinear regression analysis was used to calculate each EC50 using GraphPad Prism (version 5.01). Error bars represent the range of duplicate data points. These experiments were performed three times, with similar results each time. The EC50s in the table are shown with 95% confidence limits.
Optimization of target sequence and LNA ASO format.
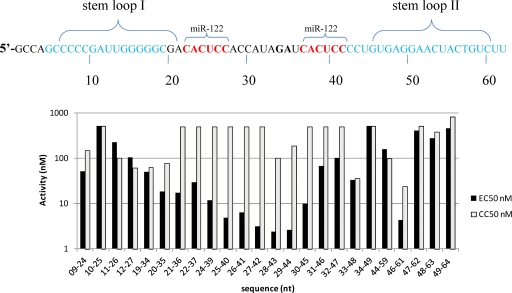
We optimized the LNA ASO sequence using a 1-nucleotide walk through the sequences surrounding the 25- to 40-nt binding site for seq132 (Fig. 3). Twenty-one LNA ASOs were synthesized and screened in the replicon assay to measure both CC50s and EC50s. This approach identified two LNA ASOs with 2- to 3-fold increased potency relative to that of the original seq132 hit (Fig. 3). The boundaries of the target sequences were remarkably precise, with log-unit-fold drops in antiviral activity caused by a 2-nt shift (for example, the EC50 for seq29 to seq44 is 5 nM, whereas that for seq31 to seq46 is >50 nM; Fig. 3, bottom panel).
Fig. 3.
(Top panel) The nucleotide region of the 5′ end of HCV IRES (subtype gt1b) is shown, with stem-loops I and II shown along with binding sites for miR-122. The dinucleotide G35A36 referred to in the text is shown in boldface. (Bottom panel) 9B subgenomic replicon cells (frozen/thawed) were transfected with increasing concentrations of the ASOs as described in Materials and Methods. All ASOs are 16-mers in 4-8-4 gapmer format. After 48 h, the luciferase activity dose-response curves (two independent experiments, n = 2 for each experiment) and cytotoxicity dose-response curves (two independent experiment, n = 2 for each) were plotted using GraphPad Prism (version 5.01). Nonlinear regression analysis was used to calculate each EC50 and CC50 value from dose-response curves, and EC50s and CC50s are plotted against the ASO used for the transfection. The seqCODE labels (09-24, 10-25, etc.) on the x axis correspond to nt sequences in the 9B IRES region.
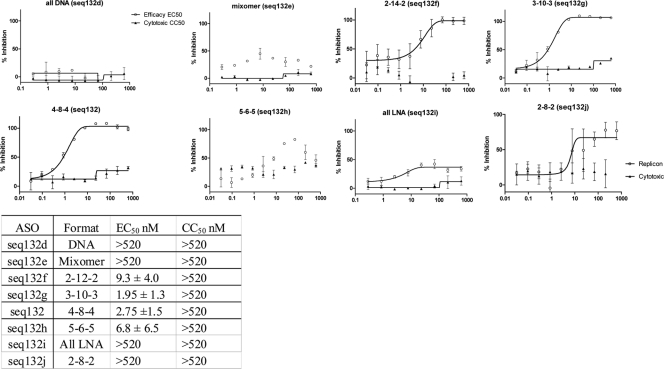
We then studied the effect of changing the format of seq132. The gapmer format was altered by increasing the terminal LNA regions from 2 nucleotides to 5 nucleotides, with the corresponding central LNA-free core being reduced from 12 to 6 nucleotides to generate an expanded family of ASOs, including all DNA (seq132d), a mixomer (seq132e), 2-12-2 (seq132f), 3-10-3 (seq132g), 5-6-5 (seq132h), all LNA (seq132i), and a 12-mer 2-8-2 (seq132j). The mixomer contains 50% LNA bases randomly across the ASO. These analogues were transfected into freshly passaged 9B replicon cells, and the all-LNA and mixomer formats for seq132 were shown to have lost antiviral potency (seq132e and seq132i in Fig. 4). Both these formats of ASO are refractory to RNase H activity (3, 5, 15, 24). The 3-10-3 format showed potent antiviral activity (seq132g in Fig. 4) that was on a par with that of the original 4-8-4 format for seq132, confirming the requirement for an internal non-LNA–RNA heteroduplex to act as a substrate for RNase H and subsequent RNA degradation. These ASOs were also screened using the 9B primary-screen frozen/thawed transfection protocol, with essentially identical results (rows 55 to 61, Table 1).
Fig. 4.
9B subgenomic replicon cells (freshly passaged) were transfected with increasing concentrations of seq132 ASO analogs as described in the text. After 48 h, the luciferase activity (empty circles) and cytotoxicity (filled triangles) were measured. Antiviral activity was expressed as the percentage of HCV replicon inhibition relative to that achieved with 10 IU/ml IFN-α2a and plotted against seq132 concentration. Nonlinear regression analysis was used to calculate each EC50 using GraphPad Prism (version 5.01). Error bars represent the range of duplicate data points. This experiment was performed >3 times, with similar results each time. The table at the bottom shows the calculated EC50 and CC50 values ± standard deviations.
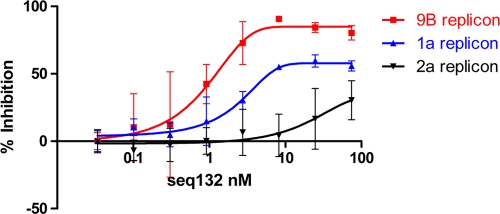
We note that the 25- to 40-nt regions of clinically relevant HCV subtypes gt1a and gt1b differ at nucleotide positions 35 and 36, which read A35G36 in gt1a and G35A36 in gt1b (Fig. 3). Further sequence analyses reveal that HCV subtype gt2a has an additional change in this region (namely, A29 to G29), making a total of 3 mismatches between gt1b and gt2a. We obtained both HCV gt1a and gt2a subgenomic replicons (4, 21) to assay the antiviral activity of ASO seq132. Figure 5 shows that seq132 has reduced potency and efficacy against HCV sequences containing mismatches in the 25- to 40-nt region. In an attempt to generate a single LNA ASO capable of antisense activity in both gt1a and gt1b HCV, we generated analogues of seq132 in 4-8-4 gapmer format containing either abasic residues or promiscuous bases (inosine) at the ASO position corresponding to the G35A36 in the target region. Table 1 shows that the abasic analogue (seq400) was completely inactive, whereas the inosine analogue (seq500) had much reduced antiviral activity compared to that of seq132 (EC50 = 100 nM; rows 63 and 64, respectively, Table 1).
Fig. 5.
9B, 1a, and 2a subgenomic replicon cells were transfected with increasing concentrations of seq132 ASO as described in Materials and Methods. After 48 h, the luciferase activities were measured and percent inhibition relative to that achieved with 10 IU/ml IFN-α2a was calculated. Each replicon line was tested in duplicate, and each experiment was independently repeated at least 2 times. Nonlinear regression analysis was used to calculate the EC50s using GraphPad Prism (version 5.01). Error bars represent the range of duplicate data points. Results of a representative experiment are shown.
Effect of different gapmer/mixomer formats on pharmacokinetic properties of seq132.
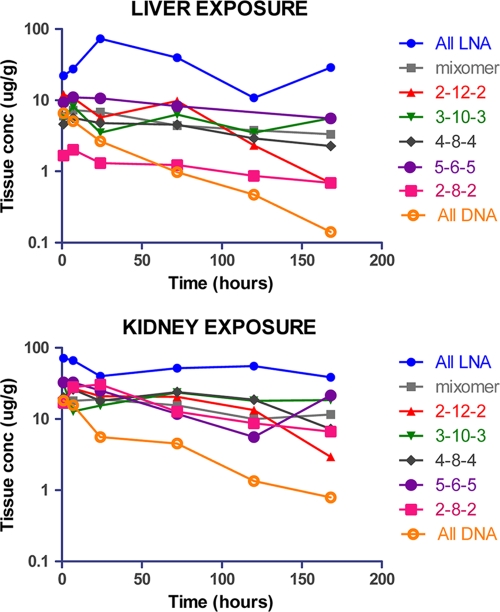
The seq132 LNA ASO analogues used for the experiment whose results are shown in Fig. 4 were also used to dose mice with a single i.v. bolus at 5 mg/kg. LNA ASO liver and kidney levels were measured over a time course of up to 7 days by LC-MS/MS (chromatograms of the extracted ASO are shown in Fig. S2 in the supplemental material). Figure 6 shows that the all-LNA format (seq132i) is absorbed better than all other formats, both in liver and in kidney, with maximum concentrations (Cmaxs) being equal to 71 and 73 μg/g, respectively (Table 2). The liver Cmaxs for all other remaining ASO formats range from 2 to 8 μg/g liver, with the Cmax for the 12-mer version (seq132j) being at the lower end of the range (Fig. 6; Table 2). The kidney Cmaxs for all seq132 ASO formats are in the range of 17 to 33 μg/g and are therefore uniformly greater than liver Cmaxs, especially with regard to the 12-mer version (seq132j), with a kidney Cmax 15-fold higher than that observed for liver. This could be driven by the fact that shorter oligonucleotides are known to have lower plasma protein binding capability, and therefore, a larger proportion of the dose will pass through the glomerulus and into the proximal tubules, where the majority of oligonucleotide is reabsorbed. Once in the liver and/or kidney, most LNA-containing seq132 analogs have roughly the same half-life (>5 days), with the exception of the 2-12-2 analog (seq132f), which is cleared at a higher rate, similar to that for the all-DNA format (seq132d) (Table 2).
Fig. 6.
Liver and kidney exposure from mice dosed at 5 mg/kg with seq132 analogues (seq132d to seq132i), as detailed in the legend to Fig. 4. Profiles shown are exposure composites, with each point being a singular necropsy point.
Table 2.
Pharmacokinetic properties of LNA ASOs in different formats
| ASO | Formatb | Livera |
Kidneya |
||
|---|---|---|---|---|---|
| t1/2 (h) | Cmax (μg/g) | t1/2 (h) | Cmax (μg/g) | ||
| seq132d | DNA | 35 | 6.56 | 47 | 18.2 |
| seq132e | mixomer | 143 | 7.27 | 174 | 18.9 |
| seq132f | 2-12-2 | 43 | 12 | 53 | 26.9 |
| seq132g | 3-10-3 | 151 | 8.19 | 185 | 31 |
| seq132 | 4-8-4 | 125 | 5.65 | 92 | 26.1 |
| seq132h | 5-6-5 | 155 | 11 | 65 | 32.6 |
| seq132i | All LNA | 155 | 73.2 | 246 | 71.5 |
| seq132j | 2-8-2 | 147 | 2.01 | 67 | 30.4 |
n = 1 for each mouse.
For the 16-mer gapmer format, the flanking numbers refer to the number of LNA residues at each terminus and the internal number refers to a central core of unmodified DNA. The mixomer format has LNA residues distributed throughout the 16-mer.
DISCUSSION
This is the first report showing a comprehensive walk through of the conserved HCV genome using LNA-based ASOs in the subgenomic replicon system. We highlight three important points: (i) the unveiling of a new IRES binding site accessible to antisense oligomers; (ii) the stringent requirement for RNase H activity for the antiviral mechanism, providing insight into the function of miR-122 binding to the IRES and into the HCV life cycle; and (iii) the advantages and disadvantages of an anti-HCV therapeutic modality based on LNA ASOs, including the favorable pharmacokinetic properties of seq132. We also present our data in the context of recent publications showing the antiviral activity of an LNA-based oligonucleotide (SPC3649) targeting miR-122 (25).
Our results have identified a new target site for LNA-based HCV therapeutics. Of the 47 LNA ASOs used in this primary screen, over a third showed significant cytotoxicity (defined as a CC50 of <100 nM). Of the remaining 30 noncytotoxic ASOs, 14 showed some degree of antiviral activity (potency, <400 nM), of which 7 ASOs had an SI of >10, including 5 targeting NS5b and 2 targeting the IRES. The low number of hits suggests that the stem-loop structures of the IRES are refractory to invasion by LNA ASO, despite the superior Tm values of LNA-based chemistries. Our data might indicate that the kinetics and the thermodynamics of ASO hybridization might be severely attenuated when a high energetic cost is required to unfold secondary structure of the target RNA sequences: the increase in enthalpy from LNA-RNA heteroduplex formation may not compensate for the cost of unwinding RNA-RNA stem-loop structures (27). Alternatively, taking into account the biochemical functions of the IRES, any antisense sequence invading the structure would likely be facing quarternary ribonucleoprotein structures, imparting further thermodynamic barriers on efficacy.
Follow-up titrations using freshly passaged replicon cells revealed that the seq132 ASO exhibited reproducibly potent antiviral activity (4.1 nM; 95% confidence limits, 2.89 to 5.74 nM at n = 24) with low cytotoxicity (>880 nM in Fig. 1, 2, and 4) and a >95% maximal asymptote for the dose-response curve in the replicon assay. In addition, we used an RT-PCR readout using replicon-directed probe-primer pairs to demonstrate that seq132 transfection resulted in the dose-dependent loss of replicon RNA. Finally, we demonstrated the sequence specificity for the seq132 mechanism of action by showing that two mismatches result in complete negation of antiviral activity, as expected for antisense modalities.
Previous studies aiming to select ASOs targeting conserved IRES sequences have not identified the 25- to 40-nt region as a priority target region but instead have pointed to other IRES regions, namely, the region around stem-loop III and the region around the initiation ATG codon (1, 2, 16, 27, 41). In some of these studies, the authors have omitted the 25- to 40-nt region from the experimental system (1, 2, 16). The remaining studies had used in vitro translation systems, and both sets of studies concluded that the IRES sequences flanking the initiation ATG were optimal (27, 41). It was surprising that we did not select ASOs targeting loop IIId (250 to 280 nt) or the initiation codon (342 to 344 nt). Our library contained three ASOs targeting loop IIId (seq103, seq101, and seqT6) and two ASOs targeting the initiation codon region (seq92 and seq94). Table 1 shows potent antiviral activity among these ASOs but considerable cytotoxic activity, giving poor SI values of <5. These data excluded these sequences from our selection criterion of an SI of >10. We note that none of the studies mentioned above performed cytotoxicity assessments on the ASOs targeting the initiation codon. Interestingly, the non-LNA second-generation ASO ISIS 14803 targeting the initiation codon region has been used in phase 1 and phase 2 trials but showed limited efficacy and some self-resolving toxicity (14, 28, 33).
Our results have added insights into HCV biology. miR-122 has been suggested to be required for HCV replication in vitro and is recruited directly to the 25- to 40-nt region via the two adjacent binding sites at 23-CACUCC-28 and 38-CACUCC-43, respectively (19, 23). This binding is responsible for enhanced viral RNA synthesis and a positive stimulation effect on viral translation (17, 18, 26). Other groups have reported additional indirect miR-122-dependent mechanisms for enhanced HCV replication (29, 38). Finally, it is known that miR-122 modulates the levels of many different host mRNAs, some of which encode enzymes involved in lipid metabolism, possibly enhancing HCV replication in cell culture models and in vivo through these pathways (20, 42).
Figure 3 shows the ASOs used in the 1-nucleotide walk-through study and the positions of both distal and proximal miR-122 binding sites at 23-CACUCC-28 and 38-CACUCC-43. The antiviral activity of the ASOs corresponds to the hybridization footprint covering either distal or proximal miR-122 binding sites within the 25- to 40-nt region and decreases when the ASO footprint encroaches on the secondary structure of stem-loop I (on the 5′ side of the ASO footprint) or stem-loop II (on the 3′ side of the ASO footprint). The loss of antiviral activity upon encroachment on stem-loop II is remarkably stark, with a log-unit drop in antiviral activity after only a 2-nucleotide shift.
Several antiviral mechanisms are possible due to seq132 binding to the 25- to 40-nt region: (i) antagonism of miR-122 binding, (ii) blockage of the availability of the 25- to 40-nt region to partake in regulatory processes, (iii) loss of HCV sequences due to RNase H activity, and (iv) local destabilization of IRES secondary structure.
Is the seq132 antiviral MoA due to antagonism of miR-122 binding or due to loss of HCV sequences from induced RNase H activity? The RNase H-independent analogues of seq132 (mixomer and all LNA) were not antiviral in this assay, whereas the RNase H-dependent gapmer format was antivirally active. Clearly, the MoA has a requirement for RNase H, an observation compatible with the fact that replicon RNA is decreased after transfection with seq132. It is likely, therefore, that the antiviral mechanism of action is driven by RNase H-mediated HCV genome loss and is not necessarily due to blockage of miR-122 binding. Going one step further, our observation that both the all-LNA and mixomer analogues of seq132 are not antiviral in the replicon assay suggests strongly that the antiviral mechanism cannot be due to blockage of miR-122 binding because both of these ASOs can target the 25- to 40-nt region with increased affinity (higher Tm) compared with that of seq132, with the 4-8-4 gapmer format. It is interesting that the all-LNA and mixomer seq132 formats do not result in overt antiviral activity and is somewhat surprising, given the important role of miR-122 in replicon biology. One possible explanation is that occupation of both distal and proximal miR-122 sites with any incoming hybridizing nucleic acid ligand (miR or seq132 analogues) would be sufficient to maintain replicon activity (in the absence of RNase H activity). In this respect, it is interesting that occupation of this region by all-LNA or mixomer analogues of seq132 would be expected to disrupt the long-range annealing (LRA) interaction between the 25- to 40-nt region and other internal complementary sequences in the replicon RNA (11, 31). It has recently been reported that the 25- to 40-nt region engages in LRA interactions to result in a closed IRES conformation refractory to translation. The binding of miR-122 (or oligonucleotides targeting miR-122 sites) in vitro is able to block these inhibitory regulatory LRAs, resulting in an open IRES available for translation (11). In this manner, the RNase H-resistant mixomer and all-LNA analogues of seq132 could act as surrogates for the miR-122 role in vivo, explaining the lack of antiviral activity, despite binding to the 25- to 40-nt region.
As mentioned above, the binding of miR-122 to the IRES has been extensively studied in vitro, and there is general agreement concerning the importance for miR-122 binding sites for HCV function. However, in vivo studies highlight two somewhat counterintuitive observations: (i) the levels of miR-122 do not correlate with intrahepatic HCV copy number, and (ii) low levels of miR-122 are predictive of high levels of HCV in nonresponding IFN-treated patients (32). One question that has not been explored is whether complete ablation of endogenous miR-122 expression blocks HCV replication, and if this was achievable, it would be interesting to check whether the all-LNA version of seq132 could be used as an experimental tool to rescue HCV.
We note that seq132 attained 95 to 98% antiviral activity, and we rarely saw 100% inhibition (defined by treatment with 10 IU/ml IFN-α2a in our system). This is of obvious concern because one could argue that 2 to 5% of the replicons are still active and thus offer a quick route to establishing potentially resistant quasispecies within the patient. This is in contrast to other antiviral modalities, such as the modality of interferon, which gives a 100% dose-response curve using the same assay. The lack of total inhibition may be caused by a number of situations, including the possibility that (i) some cells receive a suboptimal dose of LNA, (ii) 2 to 5% of HCV replicons contain escape mutants in the target sequence (i.e., primary resistance), (iii) some cells have local defects in their antisense machinery, or (iv) physiological/functional agonism results from titrating out unknown regulatory host cell factors as levels of incoming LNA ASOs approach saturation point. Clearly, further in vivo testing in HCV disease efficacy models is required to fully understand the clinical efficacy of this modality.
Sequence analysis reveals that 94% of the clinically relevant HCV gt1b strains have a perfect match to the target region for seq132 and are therefore susceptible to therapeutic intervention. However, only 9.4% of gt1a HCV strains have a perfect match, with the majority containing the G35A36 dinucleotide inverted (A35G36 in gt1a sequences). On this basis, seq132 is unlikely to be 100% efficacious against this population. In attempting to design a pan-HCV version of seq132, we introduced promiscuous inositol bases (or abasic linkages) at the corresponding positions for the dinucleotide. This approach resulted in much decreased efficacy. Several strategies could be envisaged to generate pan-HCV cover using seq132, including treating patients with chronic HCV infection with both analogues of seq132 such that gt1a and gt1b HCV strains are targeted simultaneously.
We observe Cmaxs equal to 5 to 10 μg/g mouse liver for seq132 (or ∼1 to 2 μM), representing a concentration 500 times higher than the EC50 calculated from our in vitro experiments using the replicon cell line 9B. We note that the average HCV copy number in the HCV subgenomic replicon is 5,000 to 10,000, whereas it has been estimated that the number of HCV genomes in an infected hepatocyte in vivo is <10 copies per hepatocyte (9, 30, 34, 39). Thus, the molarity of HCV target sites in our experimental system is much higher than that which might be encountered in vivo. We also observe a plasma half-life of >5 days, suggesting that exposure for LNA ASO modalities might indeed be on a par with that for current once-weekly dosing regimes of pegylated IFN. Assuming a 1:1 molar stoichiometry in the antisense mechanism of action, one would theoretically expect antisense modalities to be efficacious in the femtomolar range. Our data, however, reinforce observations by others that collectively point to a rate-limiting step(s), including cellular uptake, intracellular transport, and differential cellular compartmentalization, of HCV and oligonucleotide therapeutics (3, 8, 10, 25).
Finally, we note that an LNA-based oligonucleotide (SPC3649) directed against miR-122 is capable of delivering antiviral activity (12, 25). Lanford et al. and other investigators have shown that weekly i.v. dosing of HCV-infected chimpanzees at 5 mg/kg SPC3649 results in a virological response (VR) of a >2-log-unit drop in plasma HCV RNA levels and decreased expression of cellular mRNA carrying the miR-122 seed region (12, 25). Partly due to these multiple effects, the Lanford group (25) had difficulty in defining whether the VR mechanism was due to a direct antagonistic effect by blockage of miR-122 binding to the 25- to 40-nt HCV region or an indirect effect due to a change in host mRNA repertoires. Of particular note, the published data for SPC3649 show a very slowly evolving VR over a period of 3 to 4 weeks. In contrast, a directly acting, specifically targeted antiviral therapy for HCV (STAT-C) induces a VR within a few days. In addition, the antagonism of miR-122 as an anti-HCV strategy is not clear-cut because it has been reported that low levels of miR-122 in liver biopsy specimens of treated patients are predictive of increased viral load and IFN treatment failure, whereas low levels of miR-122 (if it is a critical factor for HCV persistence) would be expected to increase the frequency of a VR (32). Despite the lack of clarity surrounding the VR mechanism, the PK properties of SPC3649 (Cmax = 25 μg/ml and plasma t1/2 = ∼20 days) are very favorable and are similar to our own PK data.
There has been some controversy regarding the safety of LNA-modified ASOs, with some studies reporting toxicity issues (35) and others showing little or no toxicity (12, 25). The precise mechanism by which toxicity may arise is presently unclear. As a consensus emerges on the impact of the trafficking bottleneck on oligonucleotide therapeutic efficacy, one cannot fail to consider its contribution to safety. The question remains whether in vivo cytotoxicity (if it is present) is a consequence of off-target (i.e., sequence-independent) effects. To observe on-target efficacy, an antisense LNA routinely needs to achieve liver concentrations >1,000-fold higher than the target RNA copy number. There are significant gains to be made, therefore, in resolving the cell-trafficking bottleneck in terms of both safety and dose requirement, which are of material importance to the costs of manufacturing such therapeutic modalities. Notwithstanding the requirement for further exploration of the safety, intracellular trafficking, and optimal mechanism of action for this class of therapeutics, it is important to highlight the significant practical advantages associated with their potential use as therapeutics: (i) reduced manufacturing costs compared to those of 2nd-generation ASO and biologics, (ii) favorable pharmacokinetics compared to those of small interfering RNA, and (iii) substantial stability in lyophilized format, potentially eliminating cold storage requirements.
We have taken a comprehensive screening approach to identify the 25- to 40-nt region in the HCV replicon as a specific target for LNA ASO therapeutics, which have excellent in vivo hepatic exposure and a >5-day half-life. Our studies also unearth some interesting insight into the regulatory role that the 25- to 40-bp region can exert on replicon biology.
Supplementary Material
ACKNOWLEDGMENTS
We thank Thomas Lehmann for providing the milligram quantities of LNA oligonucleotides for the PK study and Oona Adams for organizing delivery of the RT-PCR data.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 18 April 2011.
REFERENCES
- 1. Alt M., et al. 1999. Comparative inhibitory potential of differently modified antisense oligodeoxynucleotides on hepatitis C virus translation. Eur. J. Clin. Invest. 29:868–876 [DOI] [PubMed] [Google Scholar]
- 2. Alt M., Renz R., Hofschneider P. H., Paumgartner G., Caselmann W. H. 1995. Specific inhibition of hepatitis C viral gene expression by antisense phosphorothioate oligodeoxynucleotides. Hepatology 22:707–717 [PubMed] [Google Scholar]
- 3. Bennett C. F., Swayze E. E. 2010. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50:259–293 [DOI] [PubMed] [Google Scholar]
- 4. Blight K. J., McKeating J. A., Marcotrigiano J., Rice C. M. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braasch D. A., Liu Y., Corey D. R. 2002. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res. 30:5160–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chary A., Holodniy M. 2010. Recent advances in hepatitis C virus treatment: review of HCV protease inhibitor clinical trials. Rev. Recent Clin. Trials 5:158–173 [DOI] [PubMed] [Google Scholar]
- 7. Cooper C. 2010. Viral response to specifically targeted antiviral therapy for hepatitis C and the implications for treatment success. Can. J. Gastroenterol. 24:385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crooke S. T.(ed.). 2007. Antisense drug technology: principles, strategies and applications, p. 537–558 CRC Press, Boca Raton, FL [Google Scholar]
- 9. Dahari H., Ribeiro R. M., Rice C. M., Perelson A. S. 2007. Mathematical modeling of subgenomic hepatitis C virus replication in Huh-7 cells. J. Virol. 81:750–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Detzer A., Overhoff M., Mescalchin A., Rompf M., Sczakiel G. 2008. Phosphorothioate-stimulated cellular uptake of siRNA: a cell culture model for mechanistic studies. Curr. Pharm. Des. 14:3666–3673 [DOI] [PubMed] [Google Scholar]
- 11. Diaz-Toledano R., Ariza-Mateos A., Birk A., Martinez-Garcia B., Gomez J. 2009. In vitro characterization of a miR-122-sensitive double-helical switch element in the 5′ region of hepatitis C virus RNA. Nucleic Acids Res. 37:5498–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elmen J., et al. 2008. LNA-mediated microRNA silencing in non-human primates. Nature 452:896–899 [DOI] [PubMed] [Google Scholar]
- 13. Frese M., et al. 2003. Hepatitis C virus RNA replication is resistant to tumour necrosis factor-alpha. J. Gen. Virol. 84:1253–1259 [DOI] [PubMed] [Google Scholar]
- 14. Gordon S. C., et al. 2003. Treatment of chronic hepatitis C with ISIS 14803, an antisense inhibitor, given for 12 weeks. Hepatology 38:306A [Google Scholar]
- 15. Grunweller A., Hartmann R. K. 2007. Locked nucleic acid oligonucleotides: the next generation of antisense agents? BioDrugs 21:235–243 [DOI] [PubMed] [Google Scholar]
- 16. Hanecak R., et al. 1996. Antisense oligonucleotide inhibition of hepatitis C virus gene expression in transformed hepatocytes. J. Virol. 70:5203–5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henke J. I., et al. 2008. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27:3300–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jangra R. K., Yi M., Lemon S. M. 2010. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J. Virol. 84:6615–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jopling C. L., Yi M., Lancaster A. M., Lemon S. M., Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 20. Kapadia S. B., Chisari F. V. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. U. S. A. 102:2561–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kato T., et al. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817 [DOI] [PubMed] [Google Scholar]
- 22. Kaur H., Babu B. R., Maiti S. 2007. Perspectives on chemistry and therapeutic applications of locked nucleic acid (LNA). Chem. Rev. 107:4672–4697 [DOI] [PubMed] [Google Scholar]
- 23. Krutzfeldt J., et al. 2005. Silencing of microRNAs in vivo with ‘antagomirs. ’ Nature 438:685–689 [DOI] [PubMed] [Google Scholar]
- 24. Kurreck J., Wyszko E., Gillen C., Erdmann V. A. 2002. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 30:1911–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lanford R. E., et al. 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327:198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lewis A. P., Jopling C. L. 2010. Regulation and biological function of the liver-specific miR-122. Biochem. Soc Trans. 38:1553–1557 [DOI] [PubMed] [Google Scholar]
- 27. Lima W. F., Brown-Driver V., Fox M., Hanecak R., Bruice T. W. 1997. Combinatorial screening and rational optimization for hybridization to folded hepatitis C virus RNA of oligonucleotides with biological antisense activity. J. Biol. Chem. 272:626–638 [PubMed] [Google Scholar]
- 28. McHutchison J. G., et al. 2006. A phase I trial of an antisense inhibitor of hepatitis C virus (ISIS 14803), administered to chronic hepatitis C patients. J. Hepatol. 44:88–96 [DOI] [PubMed] [Google Scholar]
- 29. Norman K. L., Sarnow P. 2010. Modulation of hepatitis C virus RNA abundance and the isoprenoid biosynthesis pathway by microRNA miR-122 involves distinct mechanisms. J. Virol. 84:666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Randall G., Grakoui A., Rice C. M. 2003. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. U. S. A. 100:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romero-Lopez C., Berzal-Herranz A. 2009. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA 15:1740–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sarasin-Filipowicz M., Krol J., Markiewicz I., Heim M. H., Filipowicz W. 2009. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat. Med. 15:31–33 [DOI] [PubMed] [Google Scholar]
- 33. Soler M., McHutchison J. G., Kwoh T. J., Dorr F. A., Pawlotsky J. M. 2004. Virological effects of ISIS 14803, an antisense oligonucleotide inhibitor of hepatitis C virus (HCV) internal ribosome entry site (IRES), on HCV IRES in chronic hepatitis C patients and examination of the potential role of primary and secondary HCV resistance in the outcome of treatment. Antivir. Ther. 9:953–968 [PubMed] [Google Scholar]
- 34. Stiffler J. D., et al. 2009. Focal distribution of hepatitis C virus RNA in infected livers. PLoS One 4:e6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Swayze E. E., et al. 2007. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 35:687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsukiyama-Kohara K., Iizuka N., Kohara M., Nomoto A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Veedu R. N., Wengel J. 2009. Locked nucleic acid as a novel class of therapeutic agents. RNA Biol. 6:321–323 [DOI] [PubMed] [Google Scholar]
- 38. Villanueva R. A., et al. 2010. miR-122 does not modulate the elongation phase of hepatitis C virus RNA synthesis in isolated replicase complexes. Antiviral Res. 88:119–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vona G., et al. 2004. Intrahepatic hepatitis C virus RNA quantification in microdissected hepatocytes. J. Hepatol. 40:682–688 [DOI] [PubMed] [Google Scholar]
- 40. Wahlestedt C., et al. 2000. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. U. S. A. 97:5633–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wakita T., Wands J. R. 1994. Specific inhibition of hepatitis C virus expression by antisense oligodeoxynucleotides. In vitro model for selection of target sequence. J. Biol. Chem. 269:14205–14210 [PubMed] [Google Scholar]
- 42. Ye J. 2007. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 3:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.