Abstract
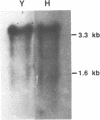
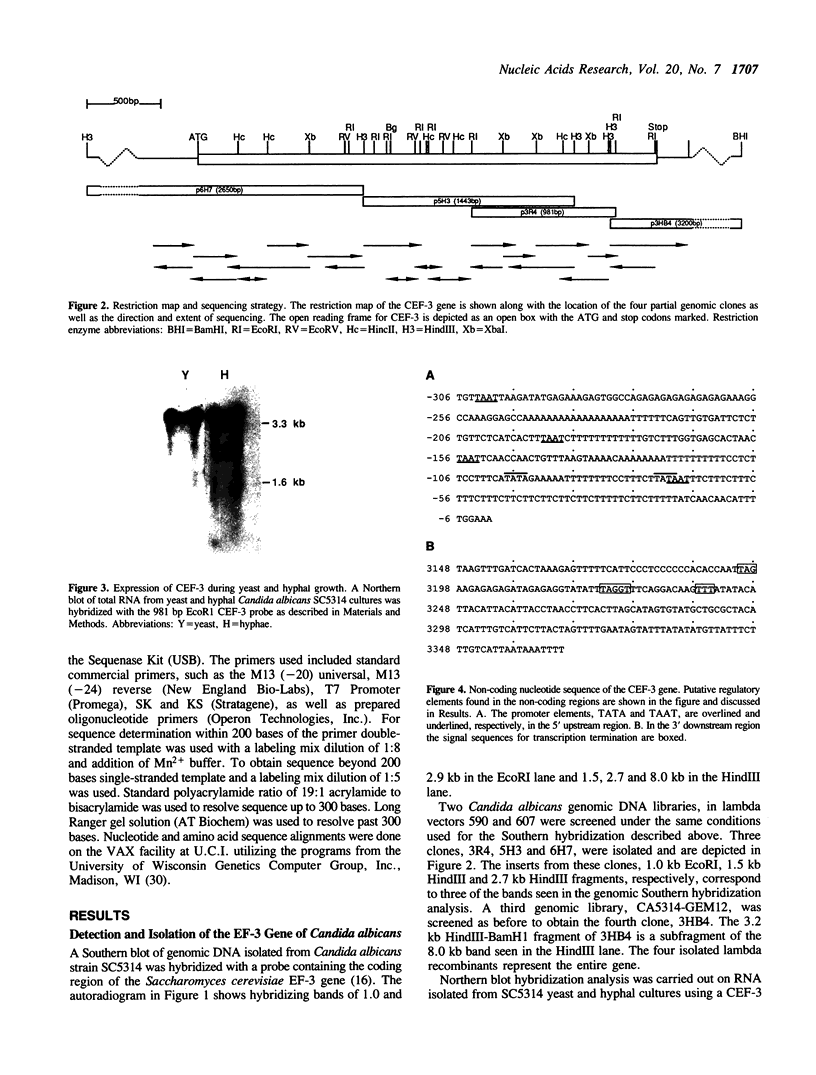
Elongation factor 3 (EF-3) is a unique and essential component of the translational system in fungi. The gene, CEF-3, encoding elongation factor 3 has been isolated from the dimorphic fungus Candida albicans. A heterologous gene probe containing the coding region of the EF-3 gene from Saccharomyces cerevisiae (YEF-3) was used to screen three Candida albicans genomic DNA libraries. The nucleotide sequences of four partial clones were determined and combined for a full-length of 3,671 base pairs (bp). A continuous open reading frame (ORF) of 3,147 bp encoding a predicted protein of 1,049 amino acids and Mr of 116,739 daltons has been identified. A transcript of 3,400 nucleotides is seen in Northern blot hybridization of Candida albicans total RNA using a CEF-3 gene probe. The single locus CEF-3 gene maps to chromosome 5 in the genome. Comparison of the deduced amino acid sequences of CEF-3 and YEF-3 shows 77.6%. identity. A higher degree of identity, 86.5%, is found when comparing the carboxy-terminal portions of the two proteins. At the nucleotide level, comparison of the coding regions of the two genes exhibit 79% identity while the upstream and downstream regions show 46% and 40% identity, respectively.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Chakraburtty K., Kamath A. Protein synthesis in yeast. Int J Biochem. 1988;20(6):581–590. doi: 10.1016/0020-711x(88)90096-1. [DOI] [PubMed] [Google Scholar]
- Colthurst D. R., Santos M., Grant C. M., Tuite M. F. Candida albicans and three other Candida species contain an elongation factor structurally and functionally analogous to elongation factor 3. FEMS Microbiol Lett. 1991 May 1;64(1):45–49. doi: 10.1016/0378-1097(91)90207-q. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Kurtz M. B., Kirsch D. R., Kelly R. The molecular genetics of Candida albicans. Microbiol Sci. 1988 Feb;5(2):58–63. [PubMed] [Google Scholar]
- Kurtz M. B., Kirsch D. R., Kelly R. The molecular genetics of Candida albicans. Microbiol Sci. 1988 Feb;5(2):58–63. [PubMed] [Google Scholar]
- Leer R. J., Van Raamsdonk-Duin M. M., Mager W. H., Planta R. J. Conserved sequences upstream of yeast ribosomal protein genes. Curr Genet. 1985;9(4):273–277. doi: 10.1007/BF00419955. [DOI] [PubMed] [Google Scholar]
- Magee B. B., Koltin Y., Gorman J. A., Magee P. T. Assignment of cloned genes to the seven electrophoretically separated Candida albicans chromosomes. Mol Cell Biol. 1988 Nov;8(11):4721–4726. doi: 10.1128/mcb.8.11.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M., Uritani M., Kagiyama H. Intrinsic ATPase activity of yeast peptide chain elongation factor 3(EF-3) and its direct interaction with various nucleotides. Nucleic Acids Symp Ser. 1986;(17):171–174. [PubMed] [Google Scholar]
- Qin S. L., Moldave K., McLaughlin C. S. Isolation of the yeast gene encoding elongation factor 3 for protein synthesis. J Biol Chem. 1987 Jun 5;262(16):7802–7807. [PubMed] [Google Scholar]
- Qin S. L., Xie A. G., Bonato M. C., McLaughlin C. S. Sequence analysis of the translational elongation factor 3 from Saccharomyces cerevisiae. J Biol Chem. 1990 Feb 5;265(4):1903–1912. [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Saporito S. M., Sypherd P. S. The isolation and characterization of a calmodulin-encoding gene (CMD1) from the dimorphic fungus Candida albicans. Gene. 1991 Sep 30;106(1):43–49. doi: 10.1016/0378-1119(91)90564-r. [DOI] [PubMed] [Google Scholar]
- Skogerson L. Separation and characterization of yeast elongation factors. Methods Enzymol. 1979;60:676–685. doi: 10.1016/s0076-6879(79)60063-0. [DOI] [PubMed] [Google Scholar]
- Skogerson L., Wakatama E. A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc Natl Acad Sci U S A. 1976 Jan;73(1):73–76. doi: 10.1073/pnas.73.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Sundstrom P., Smith D., Sypherd P. S. Sequence analysis and expression of the two genes for elongation factor 1 alpha from the dimorphic yeast Candida albicans. J Bacteriol. 1990 Apr;172(4):2036–2045. doi: 10.1128/jb.172.4.2036-2045.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teem J. L., Abovich N., Kaufer N. F., Schwindinger W. F., Warner J. R., Levy A., Woolford J., Leer R. J., van Raamsdonk-Duin M. M., Mager W. H. A comparison of yeast ribosomal protein gene DNA sequences. Nucleic Acids Res. 1984 Nov 26;12(22):8295–8312. doi: 10.1093/nar/12.22.8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uritani M., Miyazaki M. Characterization of the ATPase and GTPase activities of elongation factor 3 (EF-3) purified from yeasts. J Biochem. 1988 Mar;103(3):522–530. doi: 10.1093/oxfordjournals.jbchem.a122302. [DOI] [PubMed] [Google Scholar]
- Whelan W. L., Kirsch D. R., Kwon-Chung K. J., Wahl S. M., Smith P. D. Candida albicans in patients with the acquired immunodeficiency syndrome: absence of a novel of hypervirulent strain. J Infect Dis. 1990 Aug;162(2):513–518. doi: 10.1093/infdis/162.2.513. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]