Abstract
Summary: Riboflavin [7,8-dimethyl-10-(1′-d-ribityl)isoalloxazine, vitamin B2] is an obligatory component of human and animal diets, as it serves as the precursor of flavin coenzymes, flavin mononucleotide, and flavin adenine dinucleotide, which are involved in oxidative metabolism and other processes. Commercially produced riboflavin is used in agriculture, medicine, and the food industry. Riboflavin synthesis starts from GTP and ribulose-5-phosphate and proceeds through pyrimidine and pteridine intermediates. Flavin nucleotides are synthesized in two consecutive reactions from riboflavin. Some microorganisms and all animal cells are capable of riboflavin uptake, whereas many microorganisms have distinct systems for riboflavin excretion to the medium. Regulation of riboflavin synthesis in bacteria occurs by repression at the transcriptional level by flavin mononucleotide, which binds to nascent noncoding mRNA and blocks further transcription (named the riboswitch). In flavinogenic molds, riboflavin overproduction starts at the stationary phase and is accompanied by derepression of enzymes involved in riboflavin synthesis, sporulation, and mycelial lysis. In flavinogenic yeasts, transcriptional repression of riboflavin synthesis is exerted by iron ions and not by flavins. The putative transcription factor encoded by SEF1 is somehow involved in this regulation. Most commercial riboflavin is currently produced or was produced earlier by microbial synthesis using special selected strains of Bacillus subtilis, Ashbya gossypii, and Candida famata. Whereas earlier RF overproducers were isolated by classical selection, current producers of riboflavin and flavin nucleotides have been developed using modern approaches of metabolic engineering that involve overexpression of structural and regulatory genes of the RF biosynthetic pathway as well as genes involved in the overproduction of the purine precursor of riboflavin, GTP.
INTRODUCTION
Riboflavin [7,8-dimethyl-10-(1′-d-ribityl)isoalloxazine, vitamin B2] (RF) is an obligatory component of human and animal diets, as it serves as a precursor of the flavin coenzymes flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD), which are involved in oxidative metabolism and other processes. Commercially produced RF is used for animal feed, as a dietary supplement, and as an additive by the food industry.
The biosynthetic pathway of RF synthesis in microorganisms and plants has been elucidated. It starts from GTP and ribulose-5-phosphate and proceeds through pyrimidine and pteridine intermediates. Flavin nucleotides are synthesized in two consecutive reactions from riboflavin in prokaryotes and eukaryotes. The pathway for the synthesis of the natural RF analog 5-deazariboflavin shows some similarities to that for RF. The antibiotic roseoflavin, which is a natural RF analog [7-methyl-8-dimethylamino-10(1′-d-ribityl)isoalloxazine], is probably synthesized from RF. Some microorganisms and all animal cells are capable of RF uptake, and many microorganisms, especially RF overproducers, have distinct systems for RF excretion (efflux) to the medium.
Regulation of RF synthesis occurs at the level of enzyme activity and synthesis. In yeasts, the first enzyme of RF synthesis, GTP cyclohydrolase II, is regulated by allosteric inhibition exerted by FAD and other nucleotides containing an adenylic moiety. The physiological role of this regulation is not known. Regulation of RF synthesis at the gene level differs in bacteria, yeasts, and fungi. In bacteria, production of RF is repressed at the transcriptional level by FMN, which binds to nascent noncoding mRNA and blocks further transcription (the so-called riboswitch). In flavinogenic molds, RF overproduction starts at the stationary phase due to derepression of enzymes involved in RF synthesis and is accompanied by sporulation and mycelial lysis. In flavinogenic yeasts, transcriptional repression of RF synthesis is caused by iron ions. The putative transcription factor encoded by SEF1 is somehow involved in this regulation.
Most commercial RF is currently produced by microbial synthesis. For this, special selected strains of the bacterium Bacillus subtilis, the mold Ashbya gossypii, and the yeast Candida famata (Candida flareri) are used. Whereas earlier RF overproducers were isolated by classical selection, current producers of RF and flavin nucleotides have been developed using modern approaches of metabolic engineering that involve overexpression of structural and regulatory genes of the RF biosynthetic pathway as well as genes involved in the overproduction of the purine precursor of riboflavin, GTP.
There are numerous reviews and one monograph on the topic of the synthesis of riboflavin and other flavins. Most of them appeared several years ago and are cited in appropriate places in the current review. Readers are also referred to the two most recent reviews (115, 178), which cover biochemical pathways and the biotechnology of RF synthesis, respectively. Here we have tried to provide balanced narratives on different aspects of the biosynthesis and transport of RF, flavin nucleotides, and some other natural flavins, including the biochemistry, enzymology, and metabolic and genetic regulation of these processes, construction of flavin overproducers using classic selection and modern approaches of metabolic engineering, and data on industrial production of RF. This review is apparently the first in English to summarize the data on iron-dependent regulation of RF synthesis in flavinogenic yeasts and construction of yeast overproducers of RF and flavin nucleotides.
RIBOFLAVIN AND FLAVIN NUCLEOTIDES (FMN AND FAD) AND THEIR ROLE IN METABOLISM
Discovery and Occurrence
Riboflavin (RF) (vitamin B2) was discovered in 1879 as a yellow pigment from milk and called lactoflavin. Its chemical structure was deciphered by Paul Karrer (Zurich, Switzerland) and Richard Kuhn (Heidelberg, Germany) in the 1930s (210, 244). They were awarded the Nobel Prize for this and other achievements in 1937 and 1938, respectively. Kuhn first proved that RF is an essential growth factor, viz., vitamin B2.
The main sources of RF in diets are milk, dairy, and meat products. In the United Kingdom, for example, milk and dairy products contribute 51% of RF intake in preschool children, 35% in schoolchildren, 27% in adults, and 36% in the elderly (356). Cereals, meats, and fatty fish are also good sources of RF, and certain fruits and vegetables, especially dark green vegetables, contain reasonably high RF concentrations. The recommended daily intake of RF is 1.3 mg/day for men and 1.1 mg/day for women (122). RF deficiency is endemic in populations that lack dairy and meat products. RF deficiency may contribute to increased concentrations of plasma homocysteine, with an associated increased risk of cardiovascular disease. Deficiency may also cause impairment in iron metabolism and night blindness (356).
Chemical Structure and Properties
RF [7,8-dimethyl-10-(1′-d-ribityl)isoalloxazine] is a heterocyclic compound produced by all plants and most microorganisms. Animals and rare prokaryotic and eukaryotic microorganisms (e.g., Corynebacterium pyogenes, Streptococcus pyogenes, Listeria monocytogenes, some lactic acid bacteria, mycoplasmas, spirochetes, rickettsiae, and protists) cannot synthesize RF and need to obtain it from their diets (or from the medium), so RF must be considered a vitamin for them (85, 132, 208, 241, 356, 455, 494, 504). The growth response of Lactobacillus casei to RF has been used for developing a microbiological assay for this vitamin (456). Proof of RF auxotrophy differs between organisms, e.g., based on the absence or presence of RF biosynthetic genes or direct determination of growth dependence on exogenous RF.
In animals, RF deficiency results in retarded growth, failure to thrive, and eventual death (76). Experimental RF deficiency results in growth failure, weakness, ataxia, and inability to stand. Animals collapse, become comatose, and die. Deficiency leads to dermatitis, hair loss, corneal opacity, cataracts, hemorrhagic adrenals, fatty degeneration of the kidney and liver, and inflammation of the mucous membrane of the gastrointestinal tract (180). RF deficiency also leads to developmental abnormalities (321, 523). Postmortem studies of animals fed an RF-deficient diet showed that they had only a third of the normal amount of RF in the liver (129), the main storage organ for RF. RF deficiency is rarely observed in developed countries, though groups with a risk of low intake of RF are common (pregnant and lactating women, children, athletes, and some categories of patients on certain medicines (125, 356).
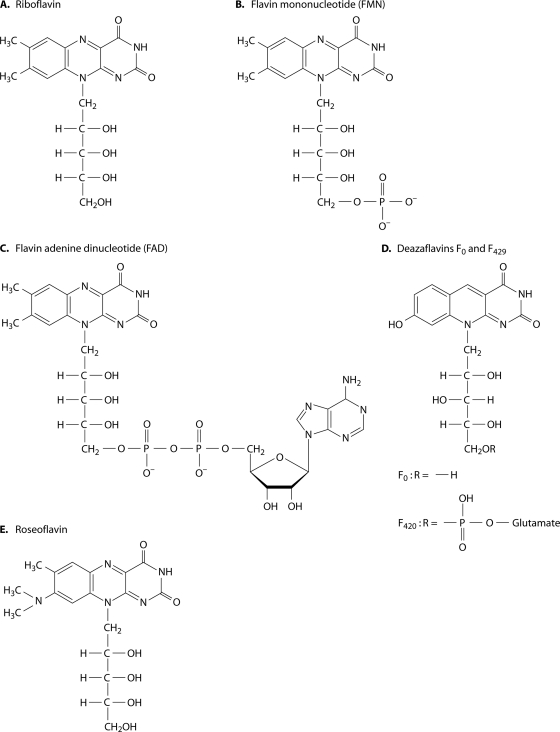
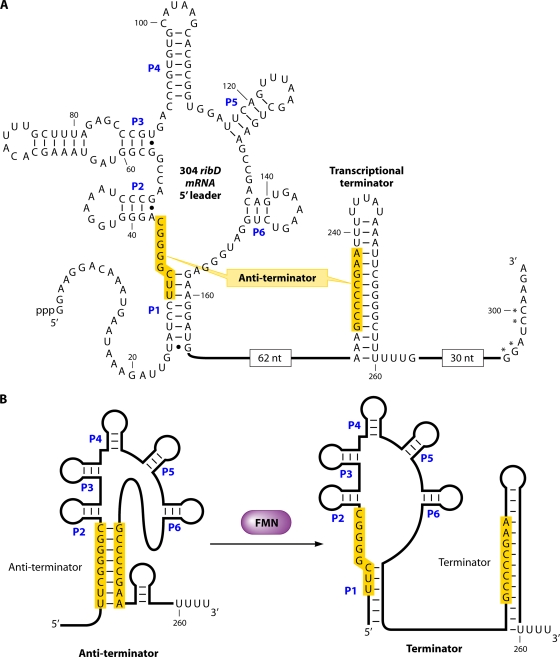
The currently used name RF recognizes the presence of the sugar alcohol ribitol in the molecule of this vitamin and the yellow color of the substance. RF (Fig. 1A) usually does not have direct metabolic functions in the living cell but serves as a precursor for the synthesis of derivatives known as flavin nucleotides or flavin coenzymes, i.e., riboflavin-5′-phosphate (flavin mononucleotide [FMN]) (Fig. 1B) and flavin adenine dinucleotide (FAD) (Fig. 1C). RF, FMN, and FAD are the main representatives of the group of substances known as “flavins.” In general, flavins are designated as derivatives of the dimethylisoalloxazine {7,8-dimethylbenzo[g] pteridine-2,4(3H,10H)-dione} skeleton, with a substitution in the 10 position. The properties of natural flavins have been well studied (30, 33, 525a). RF and most other flavins are yellow compounds with a characteristic yellow-green fluorescence in UV light. Peak absorbances by aqueous solutions of RF occur at 223, 266, 373, and 445 nm. The maximum fluorescence emission of neutral aqueous RF solution is at 535 nm. Light absorbance and intense fluorescence are used in the analytical determination of flavins. Different forms can be easily separated by liquid chromatography (503). RF is slightly soluble in water and ethanol; its solubility is 100 to 130 mg/liter and 45 mg/liter at room temperature in water and absolute ethanol, respectively (30). It is poorly soluble in allyl and benzyl alcohols, amyl acetate, and phenol and practically insoluble in ether, chloroform, acetone, and benzene. RF is soluble in alkaline solution but becomes unstable under these conditions. FMN is much more soluble in water (30 to 50 g/liter) (30, 279).
Fig. 1.
Chemical structures of flavins.
In an acid medium, flavin nucleotides are hydrolyzed to free RF. Flavin molecules possess amphoteric properties. They fluoresce only in the oxidized state. Optimal fluorescence occurs at pH 3 to 8. The molecular coefficients of RF and FMN fluorescence are very close, whereas that of FAD is significantly lower (∼20% of the molar fluorescence of RF and FMN) due to fluorescence quenching by the adenylic moiety (204). Irradiation of flavins leads to their decomposition, producing derivatives with totally or partially degraded ribityl chains; lumiflavin (7,8,10-trimethylisoalloxazine) is accumulated in alkaline solutions, whereas lumichrom (7,8-dimethylisoalloxazine) is produced at neutral and acid pHs. RF and other flavins produced water-insoluble complexes with salts of metal ions, e.g., Mg2+, Hg2+, Cu2+, Fe2+, Co2+, and Ni2+ (33, 374).
Analytical Methods
As mentioned above, flavins intensely absorb light and fluoresce, making these properties important in most assays (30, 503, 549). More complicated is assaying individual flavins within mixtures. Thus, different approaches have been proposed, e.g., paper (30, 56), ion-exchange (536), and thin-layer (147) chromatography, paper (533) and capillary (186) electrophoresis, specific RF extraction by 2-phenylethanol (478), or separation on silica gel and other resins (93). Currently, flavins are separated by high-performance liquid chromatography (379, 503, 549) with subsequent fluorescence detection, approaches that are approved by AOAC International (240).
Other Natural Flavins
In addition to RF, FMN, FAD, and products of their photolysis (lumiflavin and lumichrome), there are other natural flavins. Most well known are 5-deazaflavins, which are derivatives of flavins in which nitrogen in the 5 position of the isoalloxazine heterocyclic structure is replaced by carbon. One deazaflavin, 7,8-didemethyl-8-hydroxy-5-deazaflavin, has a ribitylated derivative known as coenzyme F0, and an oligoglutamylated derivative of F0 (coenzyme F420) (Fig. 1D) is involved in hydride transfer during reductive transformation of carbon dioxide and acetate into methane in methanogenic archaea (103, 151, 258, 299, 514). Coenzyme F420 has also been found in certain streptomycetes, in which it serves as a cofactor in the biosynthesis of tetracycline and lincomycin (74, 189, 347, 369). Mycobacterium and Nocardia spp. use coenzyme F420 as a cofactor of glucose-6-phosphate dehydrogenase (359, 360). Cofactor F420 is required for the activation of experimental antituberculosis drugs by Mycobacterium tuberculosis and Mycobacterium bovis strain BCG (43, 476). Although F420 contains a 5-deazariboflavin moiety, its biochemistry is more similar to that of NAD(P) than to that of FMN/FAD (42). In its deprotonated 8-hydroxy form, coenzyme F0 is known to act as a cofactor of DNA photolyases from the cyanobacteria Synechocystis spp. as well as the eukaryotes Scenedesmus spp., Ostreococcus tauri, and Drosophila melanogaster (104, 105, 146, 385).
Some natural flavins have a reddish-orange color, e.g., the antibiotic roseoflavin [7-methyl-8-dimethylamino-(1′-d-ribityl)isoalloxazine] (Fig. 1E), which is produced by Streptomyces davawensis and is active against Gram-positive bacteria (333, 334). The basidiomycete Schizophillum commune produces two RF derivatives, known as schizoflavins: 7,8-dimethyl-l0-(2,3,4-trihydroxy-4-carboxybutyl)isoalloxazine (RF acid or riboflavinoic acid) and 7,8-dimethyl-l0-(2,3,4-trihydroxy-4-formylbutyl)isoalloxazine (RF-aldehyde or riboflavinal) (488). Their exact metabolic functions are unknown. Other closely related compounds are molybdopterins (279), which consist of a pyranopterin, a complex heterocycle featuring a pyran fused to a pterin ring. In addition, the pyran ring has two thiolates that serve as ligands in molybdo- and tungstoenzymes (198). Natural flavins found as prosthetic groups of several enzymes in the strict anaerobe bacterium Peptostreptococcus elsdenii are 6-hydroxy-7,8-dimethyl-isoalloxazine and 7-methyl-8-hydroxy-isoalloxazine (142). Nekoflavin, identified as 8α-hydroxyriboflavin, was isolated from the choroid of cat eyes (297). This flavin, together with another hydroxyl derivative, 7α-hydroxyriboflavin, was also found in human urine (326). Glycoside derivatives of RF and other isoalloxazines are quite common, i.e., RF glucosides, RF galactosides, and RF oligosaccharides, produced by some species of bacteria, yeasts, and mycelial fungi (485). Lampteroflavin, the riboflavinyl α-ribofuranoside, proved to be a light emitter in the mushroom Lampteromyces japonicus (499). Plants frequently secrete RF and its derivatives RF-5′-sulfate and RF-3′-sulfate under conditions of iron starvation (484).
Chemical syntheses resulted in a large collection of analogs of RF (30, 33, 249), and their biological activities have been studied in bacteria and animal models (152, 250, 279, 515). Some of them possessed significant antibacterial or antiprotist activities. Strong antibacterial activity also was found for 8-N-alkyl analogs of roseoflavin (212, 213).
Biological Role of Flavins
The chemical entity responsible for the diverse biological activity of flavin is the isoalloxazine moiety. It exists in three redox states: (i) the oxidized or quinone state, (ii) the one-electron reduced or semiquinone (radical) state, and (iii) the two-electron reduced (fully reduced) or hydroquinone state. Flavin is an amphoteric molecule existing as neutral, anionic, and cationic species in all three redox states (290). The redox potential for the two-electron reduction of the flavin is about −200 mV. However, this value can greatly vary in flavoproteins due to the crucial role of the protein environment in the properties of flavins, spanning a range from approximately −400 mV to +60 mV. In general, the proximity of a positive charge is believed to increase the redox potential, and a negative charge or a hydrophobic environment is expected to lower it (130).
Flavins are essential to the nutrition of all prokaryotic and eukaryotic cells. Their significant biological role in most cases is connected with the coenzyme functions of FMN and FAD. These nucleotides bind to proteins, producing flavoproteins (flavoenzymes). There is one exception: free RF is the active redox cofactor for the Na+-pumping NADH:quinine oxidoreductase in Vibrio cholerae (200). Hundreds of flavoproteins are currently known (290), and new ones are being reported every year. It is currently estimated that on average of 1 to 3% of the genes in bacterial and eukaryotic genomes encode flavin-binding proteins (81). Most flavoproteins contain noncovalently bound FAD and, more rarely, FMN. Most flavin-protein interactions involve the N-10 side chain, i.e., the ribityl side chain of FMN or FAD. Relatively few flavoproteins contain covalently bound coenzymes (FAD). Covalent binding of coenzymes increases the oxidative power of the enzyme (131). Covalent attachment occurs between the 8 position of the flavin ring system and a histidine and/or between the 6 position and a thiol group of a cysteine residue (527). This covalent linkage is a result of autoxidation (80).
Redox reactions.
Flavins fulfill their biological functions through an ability to transfer one and two electrons from hydrogen atoms and hydride ions. Therefore, they can participate in redox reactions as either a one- or a two-electron mediator, making the flavoenzymes very versatile in terms of substrate and type of reactions, which is a major reason for the ubiquity of flavin-dependent enzymes in biological systems. In contrast, the other redox cofactors usually catalyze exclusively either one- or two-electron processes (100). The reactions catalyzed by a flavoenzyme always involve two separate half-reactions, i.e., reductive and oxidative half-reactions, both of which are necessary for the turnover of the enzyme. The International Union of Biochemistry adopted a classification for flavoenzymes based on their reaction substrates, recognizing five classes of flavoenzymes that catalyze reactions with net redox change: (i) transhydrogenase, where two-electron equivalents are transferred, along with the appropriate hydrogen ions, from one organic substrate to another; (ii) dehydrogenase-oxidase, where two-electron equivalents are transferred to the flavin from an organic substrate, where molecular oxygen is the oxidizing substrate, being reduced to H2O2; (iii) dehydrogenase-monooxygenase, where the flavin is reduced generally by a reduced pyridine nucleotide and where on oxidation with O2 in the presence of a cosubstrate, one atom of oxygen is inserted into the cosubstrate while the other is reduced to H2O; (iv) dehydrogenase-electron transferase, where the flavin is reduced by two-electron transfer from a reduced substrate and then reoxidized in sequential single-electron transfers to acceptors, such as cytochromes and iron-sulfur proteins; and (v) electron transferase, where the flavin is reduced and reoxidized in one-electron steps (171, 192, 298, 311).
It has to be pointed out that isoalloxazine chromophore is involved in redox reactions, whereas the side chain serves for binding to apoflavoproteins. Flavocoenzymes can form very complex catalytic sites involving more than one flavin coenzyme (both FMN and FAD), modified flavins, and/or additional cofactors, such as iron-sulfur clusters (69, 271, 332, 375).
Reactions with no net redox change.
Although most flavoproteins carry out reactions with net redox changes, there are a number of unusual flavoproteins that catalyze reactions with no net redox change. These fall into 4 groups: (i) those that utilize two-electron flavin chemistry (N-methylglutamate synthase and 5-hydroxyvaleryl-coenzyme A [CoA] dehydratase), (ii) others that involve free radical flavin chemistry [chorismate synthase, DNA photolyase, (6–4)photolyase, and 4-hydroxybutyryl-CoA dehydratase], (iii) a number in which the role of flavin remain unclear [(R)-2-hydroxyacyl-CoA dehydratases, isopentenyl diphosphate isomerase, and UDPgalactopyranose mutase], and (iv) those that apparently do not involve the flavin directly in catalysis (acetohydroxyacid synthases and hydroxynitrile lyase) (reviewed in reference 42).
Light emission and other processes.
Flavins can also be involved in nonrelated processes, one of them being phototropism. FMN is the cofactor in phototropins of plants (53), which are the blue light-sensitive photoreceptors responsible for phototropism (bending responses of plants toward or away from light sources) (188), chloroplast movement (166), and many other functions. FMN is noncovalently bound in phototropin. The cofactors of cryptochromes are FAD and methenyl tetrahydrofolate (269). FAD is also the redox- and light-sensitive noncovalently bound chromophore in BLUF proteins (148). They are also involved in a variety of nonredox processes, such as blue-light sensing in plants (70, 262, 386). FMN-containing fluorescent proteins have been engineered from the blue-light photoreceptors of Bacillus subtilis and Pseudomonas putida; after codon optimization, they have been heterologously expressed in bacteria and yeasts (95, 489, 496). In contrast to the green fluorescent protein (GFP), the FMN-containing fluorescent proteins fluoresce in both the presence and absence of oxygen, which is important for studying proteins under anaerobic conditions.
Flavins are also involved in circadian rhythm (135, 191, 384). RF induces disease resistance in plants by activating a signal transduction pathway (5, 94, 550) and is involved as the apoptosis-inducing factor in a mitochondrial flavoprotein (306).
Lumazine proteins.
Some fluorescent bacteria of the genera Photobacterium and Vibrio produce fluorescent proteins that are also known as lumazine proteins. These proteins use 6,7-dimethyl-8-ribityllumazine, an RF immediate biosynthetic precursor, as the noncovalently bound prosthetic group (236, 328, 349). In addition to 6,7-dimethyl-8-ribityllumazine, RF, FMN, and 6-methyl-7-oxo-8-ribitylllumazine (the product of 6,7-dimethyl-8-ribityllumazine oxidation) can be used as prosthetic groups. Lumazine proteins act as optical transponders in the above-mentioned fluorescent bacteria.
FMN as the precursor of coenzyme B12.
FMN (but not FAD) also possesses an important biological function in being the biosynthetic precursor of the dimethylbenzimidazol part of coenzyme B12 (63, 366, 367). Conversion of FMN to dimethylbenzimidazol involves a unique transformation reaction with no precedent in chemistry, which includes retro-aldol condensation sandwiched between two 2-electron oxidations (154, 519). The corresponding oxidoreductase BluB from Sinorhizobium meliloti triggers the unprecedented fragmentation and contraction of the bound FMNH2 and cleavage of the ribityl tail to form dimethylbenzimidazol and d-erythrose 4-phosphate (488a).
Free flavins.
Some biological functions can be fulfilled by free flavins, including RF, as a rule when secreted from the cells. In Helicobacter pylori, the excreted RF is thought to have a role in Fe3+ reduction and hence in iron acquisition (529), and a similar role in Campylobacter jejuni has been suggested (78). Secreted FMN and RF mediate extracellular electron transfer involved in Fe3+ and other cation reduction in chemotrophic bacteria of the genus Shewanella (77, 287, 509). Thus, free flavins participate as electron shuttles in the so-called Mtr respiration pathway (important for a chemotrophic mode of nutrition), in insoluble ion solubilization, and usually in some geochemical cycles. Secretion of RF by microorganisms, the physiological role of this process, and its regulation are described below. Avian eggs contain RF, FMN, FAD, and RF-binding protein (RBP), which are required for the active transport of RF into the egg and storage of the RF needed later in development (523, 535). Archaea also contain RF-binding proteins (dodecins), also known as lumichrome-binding proteins, that are involved in the regulation of flavin homeostasis (157, 158).
BIOCHEMICAL PATHWAYS OF RIBOFLAVIN SYNTHESIS IN BACTERIA, FUNGI, AND PLANTS
Animals and a few prokaryotes (e.g., some lactic acid bacteria) cannot synthesize RF de novo. All plants and fungi and most bacteria are capable of RF production and are a source of vitamin B2 for animals, including humans. At the same time, all organisms, including animals, convert RF to the flavin coenzymes FMN and FAD.
The biochemical pathway of RF synthesis was mostly established before 2000, based on research conducted in the United States, Japan, Ukraine, Russia, and Germany. The crucial breakthroughs in deciphering the pathway of RF synthesis were made by Adelbert Bacher and his colleagues in Munich, Germany. Biochemical reactions leading to synthesis of flavin coenzymes FMN and FAD were established many years ago (216, 218, 230, 403). At about the same time, the RF synthase reaction leading to synthesis of RF from two molecules of its immediate precursor 6,7-dimethyl-8-ribityllumazine was described (165, 355). The role of purine compounds as precursors of RF was known from the works of Goodwin in the middle of the 20th century (150). The carbon atom of the purine precursor and all carbon atoms of the pyrimidine ring were incorporated into the RF molecule (7). Later it was established that some guanylic compound at the nucleoside or nucleotide level acts as precursor of RF and that this ribose moiety of the purine precursor is transferred to the ribityl side chain of RF (18, 27, 272, 282). Finally, the first reaction of the pathway leading to RF, that with GTP cyclohydrolase II, was described (123, 416). After this finding, it became clear that RF synthesis starts from GTP, which in the GTP cyclohydrolase reaction is converted to a phosphorylated pyrimidine derivative. Such phosphorylated ribosylated pyrimidine has to be converted in some way to nonphosphorylated ribitylated pteridine (the above-mentioned 6,7-dimethyl-8-ribityllumazine) and then again to the nonphosphorylated ribitylated modified isoalloxazine (RF). Further work on the pathway was hampered by the instability of phosphorylated ribosylated pyrimidine intermediates of RF synthesis. The most intricate was the identification of reactions involved in the conversion of the pyrimidine precursor of RF to the pteridine precursor. In these reactions an intermediate of the pentose phosphate pathway, ribose-5-phosphate or its derivative (273, 275a, 276), later identified as ribulose-5-phosphate (506, 507), is involved.
Various approaches have been used to decipher the RF biosynthesis pathway. To identify the nature of what were thought to be the purine precursors of RF, cell feeding with radioactively labeled purines, inhibitors of purine interconversion, and mutants defective in specific steps of purine metabolism were used (reviewed in references 10, 14, and 414). Identification of the 4-carbon compound involved in converting the pyrimidine precursor of RF to the pteridine precursor was based on synthesis of the pteridine precursor (6,7-dimethyl-8-ribityllumazine) by cell extracts of wild-type cells and RF-defective mutants after addition of the putative source of the 4-carbon compound (275a, 276, 506). The intermediates in RF synthesis were investigated after their accumulation in the culture media of RF-deficient mutants of the yeasts Saccharomyces cerevisiae and Pichia (Candida) guilliermondii and of Bacillus subtilis; they were subsequently identified by physicochemical methods (16, 17, 272, 331, 420). The tentative pathway was confirmed after isolation of the corresponding enzymes, followed by cloning and mutation of the structural genes involved in the pathway (14, 115, 414). Additional model organisms used for studying RF biosynthesis are the flavinogenic yeast Candida famata (Candida flareri) and the molds Ashbya gossypii and Eremothecium ashbyii.
The biochemical pathways of RF synthesis appeared to be similar, but not identical, in bacteria, fungi, and plants (see below). Surprisingly, the pathways of RF synthesis are identical in eubacteria and plants but different in fungi and archaea (112–115). One important step in RF synthesis remains unknown, namely, the conversion of the phosphorylated pyrimidine derivative of RF to its nonphosphorylated derivative (the dephosphorylation step). The biochemical pathways leading to production of other natural flavins, such as deazaflavins and roseoflavin, remain to be elucidated and await future study.
Comprehensive reviews of the biochemistry of RF synthesis have been published by Bacher and his colleagues (13, 14, 112–115). The following brief overview of the RF biosynthesis pathway is based on these reviews and recent publications.
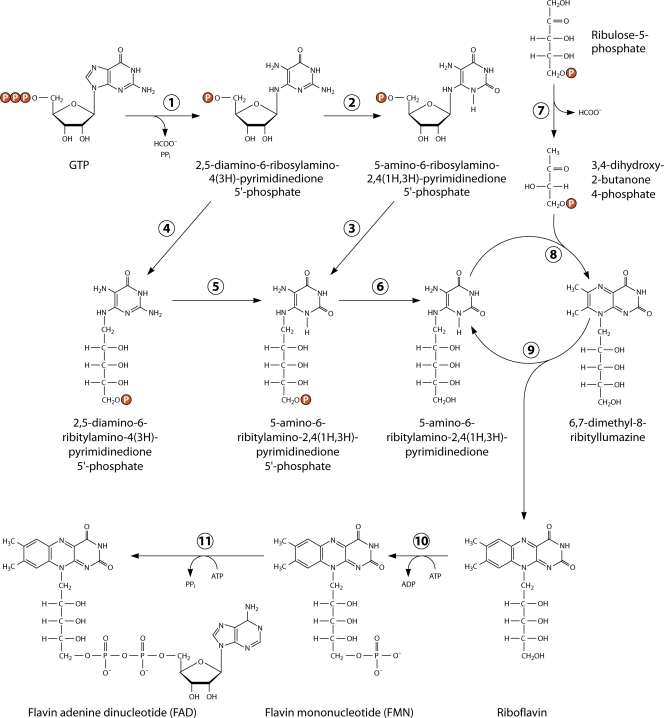
The pathway of RF synthesis (Fig. 2) starts from two precursors, GTP (one molecule) and ribulose-5-phosphate (two molecules).
Fig. 2.
Biosynthesis of riboflavin and flavocoenzymes. (Reproduced from reference 112 with permission of the Royal Society of Chemistry.)
GTP Cyclohydrolases II and III
The first reaction of RF biosynthesis is catalyzed by GTP cyclohydrolase II (EC 3.5.4.25); this term is used to distinguish it from GTP cyclohydrolase I (EC 3.5.4.16), which is involved in biosynthesis of folic acid and biopterin. GTP cyclohydrolase II removes C-8 from GTP, producing formate; the enzyme also removes pyrophosphate. It was first isolated from cell extracts of Escherichia coli (123); however, the role of the enzyme in RF biosynthesis was first established using RF auxotrophs of the flavinogenic yeast P. guilliermondii (416, 425). To some extent, the mechanisms of the GTP cyclohydrolase I and II reactions are similar, although the final products of the reactions are different (46, 112). The product of the GTP cyclohydrolase II reaction is 2,5-diamino-6-ribosylamino-4(3H)- pyrimidinedione phosphate (123, 124). Alternatively, GMP is formed as the reaction product with a rate of ∼10% of that for the major pyrimidine product (373). GTP cyclohydrolase II from E. coli is the homodimer and contains Zn2+ ions as a cofactor per subunit, being activated by Mg2+ (206, 257, 371). The enzyme properties were studied in the bacteria E. coli (35, 123, 206), B. subtilis (185), Helicobacter pylori (32), and Streptomyces coelicolor (461), the flavinogenic yeast P. guilliermondii (412, 418, 425), and the model plant Arabidopsis thaliana (174). Yeasts and some bacteria (e.g., E. coli) contain separate genes coding for GTP cyclohydrolase II (in yeasts designated RIB1) (266, 330, 331, 420, 546), whereas plants and other bacteria (e.g., B. subtilis) contain the fused gene coding for a protein with two domains, one with GTP cyclohydrolase II activity and another with the activity of 3,4-dihydroxy-2-butanone 4-phosphate synthase (see below) (174, 185, 308). The three-dimensional structure of GTP cyclohydrolase II is known (365). The active center is formed by 3 cysteine residues, Cys54, Cys65, and Cys67, which bind Zn2+ as well as Arg128 and Tyr105 (206, 365). The reaction starts by pyrophosphate release, which appeared to be the rate-limiting step of the overall reaction, after which imidazole ring opening and elimination of formate occur (206, 373).
In H. pylori, GTP cyclohydrolase II is involved in determining its hemolytic phenotype; heterologous expression of the corresponding gene ribAB in E. coli induces hemolytic activity of the recipient strain (32, 108).
Archaea and some eubacteria also contain another GTP cyclohydrolase, GTP cyclohydrolase III, which catalyzes the conversion of GTP to 2-amino-5-formylamino-6-ribosylamino-4(3H)-pyrimidinone 5-phosphate, i.e., the formylated derivative of the product of GTP cyclohydrolase II (151, 181, 461). In other words, GTP cyclohydrolase III, in contrast to GTP cyclohydrolase II, hydrolyzes the imidazole ring of GTP but does not remove the resulting formyl group from the formamide. It is noteworthy that the product of GTP cyclohydrolase III is the intermediate of the GTP cyclohydrolase II reaction (151, 402). In the archaeon Methanocaldococcus jannaschii, GTP cyclohydrolase III is apparently involved in biosynthesis of RF (and deazaflavin), as the product of this reaction undergoes formate cleavage by the specific formamide hydrolase (160). Thus, the 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinedione phosphate, the product of GTP cyclohydrolase II, is produced in archaea by the consecutive action of GTP cyclohydrolase III and formamide hydrolase. It is also interesting that the gene coding for GTP cyclohydrolase III from archaea has no homology with genes coding for GTP cyclohydrolase II, whereas the genes display high homology in S. coelicolor (461).
Reductase and Deaminase
In the next two reactions, deamination of the amino group at position 2 and reduction of the ribosyl side chain to ribityl take place (Fig. 2). The sequence of deamination and reduction is distinct in fungi and archaea on the one hand and bacteria and plants on the other (14, 112, 113). In yeasts and fungi, the enzyme catalyzing the second reaction of RF biosynthesis, 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5-phosphate reductase [another name is 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5-phosphate synthase] (EC 1.1.1.193), uses NADPH for reduction of the product of the GTP cyclohydrolase II reaction (21, 179, 305, 330, 377, 417). In S. cerevisiae the corresponding gene is RIB7 (330), whereas in P. guilliermondii it is RIB2 (420). In archaea and rare eubacteria, 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5-phosphate reductase, like enzymes from fungi and yeasts, also acts as the distinct enzyme in the second step of RF biosynthesis (67, 153, 377). In archaea, it uses both NADPH and NADH as reductants. The genes coding for reductases from the pathogenic yeast Candida glabrata, the extremophilic eubacterium Aquifex aeolicus, and the archaeon M. jannaschii were cloned and expressed in E. coli, and the corresponding proteins from these organisms were isolated in a purified state. All three enzymes catalyze an identical reaction (67, 377). The 3-dimensional structure of the enzyme from M. jannaschii has been determined by X-ray crystallography (67).
In the third step, hydrolytic deamination of the 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5′-phosphate to 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate occurs in A. gossypii and S. cerevisiae (21, 179, 323). The corresponding enzyme (EC 3.5.4.26) is encoded by genes RIB2 in S. cerevisiae and RIB3 in P. guilliermondii (331, 420). The enzymes have been partially purified; however, the properties of the fungal deaminase involved in RF biosynthesis have not been studied in detail. No enzyme classification numbers have been assigned for reductases and deaminases from fungi and archaea. In archaea, the deaminase gene has not yet been identified.
In eubacteria, e.g., E. coli and B. subtilis, the hydrolytic deamination of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5-phosphate occurs before reduction of the ribosyl side chain (14, 60, 119, 372). These bacteria possess bifunctional (2-domain-containing) deaminase-reductase enzymes encoded by one gene, called ribD in E. coli and ribG in B. subtilis (372). The deamination domain, diaminohydroxyphosphoribosylaminopyrimidine deaminase (EC 3.5.4.26), is located in the C-terminal part of the protein, whereas the reductase domain (EC 1.1.1.193) is located in the N-terminal region. Expression of truncated genes encoding only deaminase or reductase domains gave active enzymes, but these were unstable. Bifunctional deaminases-reductases, the products of the E. coli ribD and B subtilis ribG genes, were isolated in their homogenous state, and X-ray crystallography was used for studying their structure (68, 473). The rate of the E. coli deaminase reaction exceeds that of the reductase reaction, suggesting that there is no channeling between the two active sites (281).
Apparently plants, like eubacteria, contain a gene coding for deaminase and reductase. The gene from A. thaliana has been cloned and has homology with the ribG gene of B. subtilis, coding for the bifunctional deaminase-reductase. A synthetic gene with optimized codon sequences encoding the N-terminal part of the A. thaliana gene product was expressed in E. coli. The resulting protein was obtained in a homogenous state and catalyzed the diaminohydroxyphosphoribosylaminopyrimidine deaminase reaction of the RF biosynthetic pathway (119). The mechanism of the reductase reaction in plants has not been studied to date. The corresponding plant enzyme could not be identified by genome comparison, apparently having a very specific amino acid sequence (113).
Dephosphorylation of 5-Amino-6-Ribitylamino-2,4(1H,3H)-Pyrimidinedione 5′-Phosphate
5-Amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate, which is produced after the first three reactions of RF biosynthesis, undergoes further dephosphorylation giving 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione, and only this last compound is used in the lumazine synthase reaction, the next step in RF synthesis (112, 113, 115). However, the mechanism of dephosphorylation is not known. The corresponding mutants have not been isolated after screening of hundreds of RF auxotrophs of B. subtilis, E. coli, S. cerevisiae, P. guilliermondii, and C. famata (23, 331, 420, 511). The involvement of nonspecific phosphatases in the biosynthesis of RF is unlikely, as this enzyme would not discriminate between the products of GTP cyclohydrolase II, reductase and deaminase, which are all phosphorylated. One may assume that an unknown phosphatase is involved simultaneously in the biosynthesis of RF and some other compound; therefore, the corresponding mutants cannot be detected among RF monoauxotrophs. The activity of this hypothetic phosphatase has to be very high, as industrial B. subtilis recombinant RF producers were isolated by overexpression of the RF operon, which apparently does not contain the phosphatase gene (185, 343).
3,4-Dihydroxy-2-Butanone 4-Phosphate Synthase
The pyrimidine precursor of RF [5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione] is then converted to the pteridine compound, 6,7-dimethyl-8-ribityllumazine. Conversion of one ring of the pyrimidine compound to two condensed-ring pteridines requires the joining of a 4-carbon compound. The origin of these 4 carbons was the subject of long-lasting discussions. It has been suggested that the donor could be diacetyl, acetoin, intermediates of pentose phosphate pathway, hexoses, trioses, and the ribityl residue of 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione (20, 112, 414). The structure of the 4-carbon precursor of RF was elucidated using a cell-free system of RF synthesis in the flavinogenic yeast P. guilliermondii (276). The pyrimidine precursor of RF, 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione, was converted to RF in cell extracts of the wild-type strain and to 6,7-dimethyl-8-ribityllumazin in extracts of rib7 mutants defective in RF synthase. Production of 6,7-dimethyl-8-ribityllumazin was stimulated by addition of ribose-5-phosphate but did not occur in the P. guilliermondii rib6 RF auxotroph (273). Later it was shown that the 4-carbon RF precursor was 3,4-dihydroxy-2-butanone 4-phosphate, which is produced in a single enzymatic step from ribulose-5-phosphate. The corresponding enzyme was isolated from P. guilliermondii and then characterized (506–508). The enzyme (EC 4.1.99.12) catalyzes the elimination of carbon 4 of the substrate as formate. Work with the purified enzyme confirmed the skeletal rearrangement postulated on the basis of in vivo studies. This reaction is characterized by extraordinary complexity. The 3,4-dihydroxy-2-butanone 4-phosphate synthase has now been isolated from many organisms (112). The enzyme from E. coli is a homodimer with a molecular mass of 47 kDa (220, 370). Similarly, in B. subtilis and plants 3,4-dihydroxy-2-butanone 4-phosphate synthase is the part of the fused protein also containing GTP cyclohydrolase II (113, 174). GTP cyclohydrolase is located at the C-terminal end of the fused protein, whereas 3,4-dihydroxy-2-butanone 4-phosphate synthase occupies the N-terminal part of the protein. The structure of this synthase was studied by X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy (99, 220, 263, 264, 465, 466). In addition to its known function in RF synthesis, the synthase also functions somehow in the regulation of mitochondrial respiration, as the corresponding S. cerevisiae rib3 knockout mutant grew with RF in a glucose medium but not in a glycerol or ethanol medium (197).
The active site of 3,4-dihydroxy-2-butanone 4-phosphate synthase was localized by crystallographic analysis of the enzymes from the archaeon M. jannaschii and the pathogenic yeast Candida albicans in a complex with ribulose-5-phosphate (99, 465, 466). A highly conserved loop comprised of several acidic amino acid residues is essential for catalysis, as shown by studies with a variety of mutant proteins (120).
Lumazine Synthase
6,7-Dimethyl-8-ribityllumazine synthase, or lumazine synthase (EC 2.5.1.B6), catalyzes the condensation of 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione with 3,4-dihydroxy-2-butanone 4-phosphate. The enzyme was first isolated from B. subtilis as a complex with RF synthase, known as “heavy RF synthase” (12). This designation led to some confusion. Studying lumazine synthase became possible after identification of the aliphatic 4-carbon precursor of RF, 3,4-dihydroxy-2-butanone 4-phosphate (506, 507). Currently, lumazine synthases have been purified from many prokaryotic and eukaryotic organisms, and the corresponding genes have been cloned and sequenced (47, 137, 226, 248, 315, 316, 318, 346).
The lumazine synthase proteins can be divided into 2 groups; enzymes from fungi, yeasts, and certain eubacteria form c5-symmetric homopentamers, whereas those from plants, archaea, and many eubacteria are represented by capsids of 60 identical subunits that are characterized by icosahedral 532 symmetry and a mass of ∼1 MDa. The icosahedral lumazine synthases can be best described as dodecamers of pentamers. The subunit folds are all very similar. The topologically equivalent active sites (5 in the case of the pentameric enzymes and 60 in the case of the icosahedral enzymes) are all located at the interfaces between adjacent subunits in the pentamer motif. The structural complexity of some of these proteins is in surprising contrast with the absence of any amino acid residues that can individually be of major importance for the enzyme-catalyzed reaction (112, 118). In Bacillaceae, lumazine synthase and RF synthase form a complex comprising an icosahedral capsid of 60 lumazine synthase subunits and a core of 3 RF synthase subunits (see below for more details) (12, 19). In spite of the difference in protein organization, alignment of amino acid sequences shows high homology among all lumazine synthases for which the crystal structure is known (139). The pentameric enzymes of S. cerevisiae and Brucella abortus contain inserts of 4 amino acids between helices a4 and a5, which is hypothesized as being responsible for their inability to form an icosahedral capsid as a consequence of steric hindrance (116, 231). Pathogenic bacteria belonging to the genus Brucella contain 2 types of lumazine synthases; one type has very low enzymatic activity and an unusual decametric structure that appeared to be an immunodominant antigen (556).
Lumazine synthase from the fission yeast Schizosaccharomyces pombe is characterized by a bright yellow color, unlike the case for all other lumazine synthases, which are colorless. The yellow color is due to noncovalent binding of RF together with small amounts of 6,7-dimethyl-8-ribityllumazine (116). The molecular structure of lumazine synthases has been analyzed in considerable detail by X-ray structure and electron microscopy analyses. This was done for the pentameric enzymes of S. cerevisiae and S. pombe and the icosahedral enzyme of the hyperthermophilic eubacterium Aquifex aeolicus, in complex with various structural analogs of substrate and product and of putative intermediates (139, 231, 303, 551, 552).
Remarkably, the lumazine synthase reaction can proceed without enzyme catalysis in dilute neutral-pH solutions at room temperature, and thus the acceleration caused by the enzyme is rather limited. Moreover, the activation energy of the noncatalyzed reaction is lower than that of the enzyme-catalyzed reaction, at least in the case of the enzymes from eubacteria and spinach (118, 225).
Riboflavin Synthase
The final step in RF biosynthesis catalyzed by RF synthase (EC 2.5.1.9) involves dismutation of 2 molecules of 6,7-dimethyl-8-ribityllumazine, in which an exchange of a 4-carbon unit occurs, thus transforming one of them into RF and the other one into 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione, the substrate of the lumazine synthase reaction (Fig. 2). This enzyme was identified many years ago as the first enzyme of the pathway of RF biosynthesis in flavinogenic fungi (246, 284, 291). Later, RF synthases from the flavinogenic fungus A. gossypii and baker's yeast (S. cerevisiae) were isolated in a purified form, and the mechanism of the reaction was studied in detail by Plaut and his colleagues (165, 352, 353, 355, 513). Currently, genes coding for RF synthases have been cloned from S. cerevisiae and other organisms, including the yeast S. pombe, the eubacteria B. subtilis and E. coli, and the plant A. thaliana. The properties of the corresponding enzymes have also been studied in detail (117, 140, 265, 389). All the enzymes mentioned are homotrimers. However, RF synthases from archaea are homopentamers, with unrelated amino acid sequences (121, 362).
Whereas the RF synthases of the archaea M. jannaschii and Methanobacterium thermoautotrophicum share no similarity with those of eubacteria and eukaryotes, they have significant sequence similarity with 6,7-dimethyl-8-ribityllumazine synthases (121), thus suggesting that RF synthases of archaea are paralogs of lumazine synthase.
The reaction catalyzed by RF synthase can be formally described as a dismutation involving the transfer of a 4-carbon moiety between 2 identical substrate molecules of 6,7-dimethyl-8-ribityllumazine. One molecule serves as the donor of the 4-carbon moiety used for conversion of the pteridine ring of the substrate to the isoalloxazine ring of RF (340, 354). The second product of that dismutation, 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione, serves as the substrate for the preceding step of RF biosynthesis catalyzed by lumazine synthase and is recycled by this enzyme. By their joint action, lumazine synthase and RF synthase generate 1 mol of RF from 1 mol of GTP and 2 mol of ribulose 5-phosphate.
It is interesting that a very complex RF synthase reaction, similar to the lumazine synthase reaction, goes spontaneously without a catalyst. Appropriate conditions for the nonenzymatic conversion of 2 molecules of 6,7-dimethyl-8-ribityllumazine to RF and 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione are boiling water solutions at acidic or neutral pH (28, 29, 380).
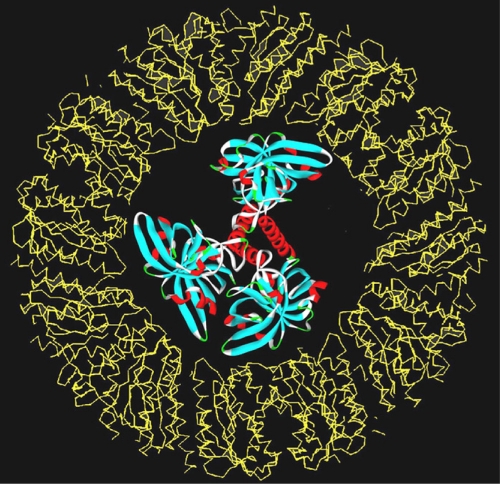
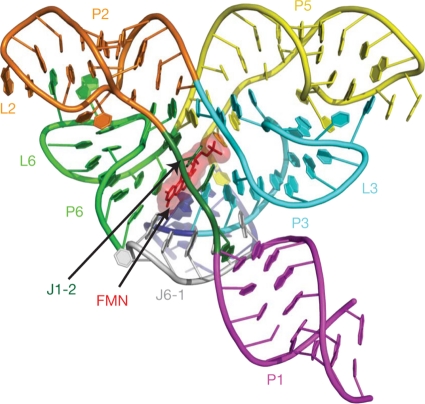
A lumazine synthase/RF synthase complex with an unusual quaternary structure has been found in Bacillaceae and has been studied in more detail in B. subtilis (15, 247, 248). The complex, referred to before as “heavy RF synthase,” consists of an RF synthase homotrimer enclosed in the central core space of the icosahedral lumazine synthase capsid (11, 114) (Fig. 3). The structural details of this complex remain unknown. The enzyme complex catalyzes the formation of one equivalent of RF from 1 mol of 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione and 2 mol of 3,4-dihydroxy-2-butanone 4-phosphate. Kinetic analysis under steady-state conditions showed that RF is formed more rapidly from the above-mentioned lumazine synthase substrates than from 6,7-dimethyl-8-ribityllumazine. This anomalous kinetic behavior has been attributed to substrate channeling due to the confinement of the RF synthase module inside the icosahedral capsids (114, 224). It is noteworthy that the cavity of recombinant icosahedral lumazine synthase capsids can be used in vitro for the containment of nanocrystalline iron oxide (428).
Fig. 3.
Computer-generated model of a heavy riboflavin synthase complex. The capsid was generated using the coordinates from the lumazine synthase 60-mer of B. subtilis (Protein Data Bank [PDB] entry 1RVV) and from the riboflavin synthase trimer of E. coli (PDB entry 1I8D). (Reproduced from reference 114 with permission of Elsevier.)
The 3-dimensional structures of RF synthases from E. coli, S. pombe, and plants have been determined by X-ray crystallography, and their intramolecular sequence structures are very similar. Structural studies show a folding pattern of 2 domains with close topologic similarity (140, 265, 401). This 2-domain architecture has important implications for the dismutation mechanism. Recently, RF synthases without apparent sequence similarity to the enzymes from eubacteria, fungi, and plants have been cloned and characterized from the archaea Methanobacterium thermoautotrophicum and M. jannaschii (98, 121, 190). Sequence comparison indicates that the pentameric archaeal RF synthases must have separated from the lumazine synthase branch at a very early time in evolution. In archaea, there is a fundamental difference in the stereochemistry of the pentacyclic intermediate involved in the dismutation of 6,7-dimethyl-8-ribityllumazine into RF and 5- amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione in the last step of RF biosynthesis (190). Notably, the active sites of the RF synthase from archaea have the same basic topology as that of lumazine synthase (362).
Fluorescent lumazine proteins, as already mentioned, possess sequence homology to RF synthase, although they are devoid of enzymatic activity and are monomeric, in contrast to trimeric RF synthases (236, 302, 349). The fluorescent proteins may play a role in light emission from the respective host organism. More specifically, energy is presumably transmitted from the excited state of bacterial luciferase to a luminescent protein, and thus the fluorescent proteins can serve as optical transponders that modulate both the wavelength distribution and the quantum efficiency in this process (254, 255). The crystal structure of the lumazine protein from Photobacterium kishitanii has been studied (393).
In general, the catalytic efficiencies and reaction rates of the enzymes involved in RF biosynthesis are very low, frequently around 1 catalytic cycle per min per enzyme subunit; only reductases from yeasts and eubacteria possess much higher catalytic rates (112). Low reaction rates of lumazine and RF synthases are especially surprising, as these reactions occur spontaneously without a catalyst. The above-mentioned low efficiency of the catalysis of RF biosynthesis enzymes apparently indicates the cell demand for a very small amount of RF and flavin coenzymes and can be the limiting factor in the construction of more efficient industrial RF overproducers.
Riboflavin Synthesis in Different Organisms
Unexpectedly, the order of the reduction/deamination reactions of the RF pyrimidine precursor is identical in fungi and archaea (reduction followed by deamination) and different in eubacteria and plants (112). Both eubacteria and plants possess bifunctional GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase, whereas fungi have separate enzymes for each reaction (174). In bifunctional enzymes, the GTP cyclohydrolase II domain is located at the C-terminal end of the fusion protein. Presumably, there was either a single evolutionary event in the gene or an unknown selection pressure that led to the fusion of the corresponding coding sequences (174). Interestingly, the N-terminal sequences of plant RF biosynthesis enzymes contain putative signals for chloroplast targeting (199). Thus, RF biosynthesis apparently occurs inside chloroplasts, which may have evolved from ancient cyanobacteria.
Also unexpectedly, archaeal RF biosynthesis is similar to that in fungi but unlike that in eubacteria. Archaea and fungi first reduce and subsequently deaminate the pyrimidine precursor of RF. The archaeal RF biosynthesis pathway has several unique features in that it uses GTP cyclohydrolase III for catalysis of the first reaction and an additional specific hydrolase (see above). The RF synthases of archaea are dissimilar to those from other organisms, although they have high homology to lumazine synthases (153).
BIOSYNTHESIS AND DEGRADATION OF FLAVIN NUCLEOTIDES FMN AND FAD
Organisms generally do not need free RF, which almost always serves only as a precursor of flavin nucleotides. In contrast to RF biosynthesis, which occurs only in plants, fungi, and most prokaryotes, all organisms, including animals, can synthesize the flavin nucleotides FMN and FAD (for further details, see a recent review on microbial synthesis of flavin nucleotides [540]).
Riboflavin Kinase
FMN (RF-5′-phosphate) is produced by specific phosphorylation of RF at the 5′ position of the ribityl chain in a reaction catalyzed by RF kinase (EC 2.7.1.26). The corresponding activity in plants had been described many years ago (145, 310); however, a detailed study of the enzyme properties was made possible after the corresponding gene had been cloned and overexpressed. Two groups of RF kinases are recognized. One group is represented in fungi, plants, animals, archaea, and (rarely) eubacteria by monofunctional RF kinase proteins (10, 26, 73, 211, 214, 289, 387, 390, 458, 537). The structures of S. pombe and human RF kinases showed a novel family of phosphoryl-transferring enzymes (26, 211). The role of monofunctional B. subtilis RF kinase in FMN synthesis in vivo has yet to be clarified (177, 457, 458). In addition to monofunctional RF kinases, bifunctional RF kinase/FAD synthetase is the main enzyme involved in eubacterial flavin nucleotide biosynthesis (101, 280, 286, 320). Regarding the bifunctional enzymes, the N-terminal and C-terminal domains are related to nucleotidyltransferases and RF kinases, respectively (133). In plants, another type of bifunctional RF kinase has been discovered, which contains an FMN hydrolase domain (387). Located in the N-terminal domain of the bifunctional protein, the FMN hydrolase belongs to the haloacid dehalogenase superfamily of enzymes. Eukaryotic RF kinases share sequence similarity to the RF kinase parts of bifunctional enzymes from eubacteria.
RF kinases use oxidized RF and ATP as substrates, although there is a B. subtilis RF kinase that uses reduced RF (219), and the enzyme from Brevibacterium ammoniagenes uses metaphosphate as phosphate donor (320). The RF kinase reaction is irreversible. In the case of bifunctional RF kinase/FAD synthetase, phosphorylation of RF to FMN is essentially irreversible, while adenylylation of FMN to FAD is readily reversible (101). In addition to providing cells with FMN, RF kinase can fulfill other functions. For example, in Streptococcus agalactiae, a gene (mreA) coding for RF kinase is responsible for resistance to macrolide antibiotics (73).
The archaebacterium M. jannaschii contains different RF kinases with no homology to RF kinases from other organisms (4, 289). Archaeal enzymes represent a unique class of kinases that use CTP instead of ATP as the phosphate donor. Another EC number (EC 2.7.1.161) has been assigned to this RF kinase.
Localization of RF kinases has been studied in several eukaryotes. For example, in S. cerevisiae, RF kinase is found in both microsomes and inner mitochondrial membranes (22, 390). A mitochondrial location of RF kinases has been reported in rat liver (24) and plants (143). However, a bifunctional RF kinase/FMN hydrolase from A. thaliana is apparently cytosolic (387). In S. cerevisiae, the RF kinase is a vital enzyme, since deleting FMN1 is lethal (390). Interestingly, RF kinase directly interacts with receptors of the tumor necrosis factor (TNF), activates NADPH oxidase, and consequently stimulates production of reactive oxygen species in mouse and human cells. Exogenous FMN and FAD could substitute for RF kinase in this stimulation (543). RF kinase is rate limiting in the synthesis of FAD, an essential prosthetic group of NADPH oxidase.
FAD Synthetase
The enzyme of FAD synthesis, FAD synthetase or FMN adenylyltransferase (EC 2.7.7.2), catalyzes transfer of adenylyl moieties from ATP to FMN. In eukaryotic organisms, only monofunctional FAD synthetases are known, whereas in bacteria, FAD synthetases that act as part of bifunctional RF kinase/FAD synthetase were found (133, 134). Genome sequence analysis of nearly 800 prokaryotes revealed a bifunctional RF kinase/FAD synthetase with conservation of several consensus regions and highly conserved residues. The N-terminal and C-terminal domains of the products of analyzed genes are related to nucleotidyltransferases and RF kinases, respectively. The structure of the bifunctional enzyme in the thermophilic bacterium Thermotoga maritima has been reported. This structure shows that the enzyme is folded in 2 domains and comprises one ATP-binding site in each of the domains, with a single flavin-binding site (517, 518). A structural model of the bifunctional enzyme from Corynebacterium ammoniagenes has been proposed (133, 173). In B. subtilis, a bifunctional RF kinase/FAD synthetase, specific for reduced flavins, has been reported (219).
Amino acid sequencing shows that bacterial and eukaryotic FAD synthetases belong to 2 different protein superfamilies, which apparently utilize different sets of active-site residues to accomplish the same reaction (184). Monofunctional eukaryotic FAD synthetases have been cloned and overexpressed from yeasts (403, 531), mammals (54, 300, 327), and plants (388). There is only one FAD synthetase in yeasts, whereas human and plant cells contain 2 isoforms and corresponding genes (54, 388, 531). There are reports of cytosolic (531) and mitochondrial (22) localization of FAD synthetase in S. cerevisiae, whereas mitochondrially located FAD synthetases have been reported in plant cells (144, 388). In human cells, one isoform is cytosolic whereas the second one is mitochondrial (497). FAD synthetase, similarly to RF kinase, is an essential gene, with knockout of FAD1 in S. cerevisiae being lethal (531). Apparently, exogenous FMN and FAD cannot penetrate yeast cells to recover growth of yeast mutants defective in RF kinase and FAD synthetase. The crystal structure of yeast FAD synthetases has been studied (184, 259).
Degradation of Flavin Nucleotides
FMN and FAD can be degraded to RF and FMN, respectively. FMN degradation is catalyzed by the nonspecific phosphohydrolase FMN hydrolase. As discussed above, plants contain bifunctional enzymes that have RF kinase and FMN hydrolase domains (387), but the physiological function of the hydrolase domain is unknown. No specific FMN hydrolase has been described so far, and the corresponding enzymes appear to be nonspecific toward many monophosphoric esters (2, 24, 437, 443, 479). No EC number has been assigned to enzymes hydrolyzing FMN. FAD can be hydrolyzed to FMN and AMP by FAD pyrophosphatases (EC 3.6.1.18), which are also nonspecific enzymes since they also hydrolyze NAD, NADH, and CoA (24, 222, 256, 363, 408). The physiological roles of enzymes hydrolyzing FMN and FAD in maintaining intracellular and intraorganellar levels of free RF, FMN, and FAD have yet to be elucidated.
BIOSYNTHESIS OF NATURAL FLAVIN ANALOGS
As discussed above (“Other Natural Flavins”), there are numerous natural flavins. However, their metabolic functions and biosynthetic pathways remain unknown. In this section, we briefly summarize the available data on the biosynthesis of 2 important natural flavin analogs, 5-deazariboflavins (flavins with coenzyme functions) and the antibiotic roseoflavin.
5-Deazaflavins
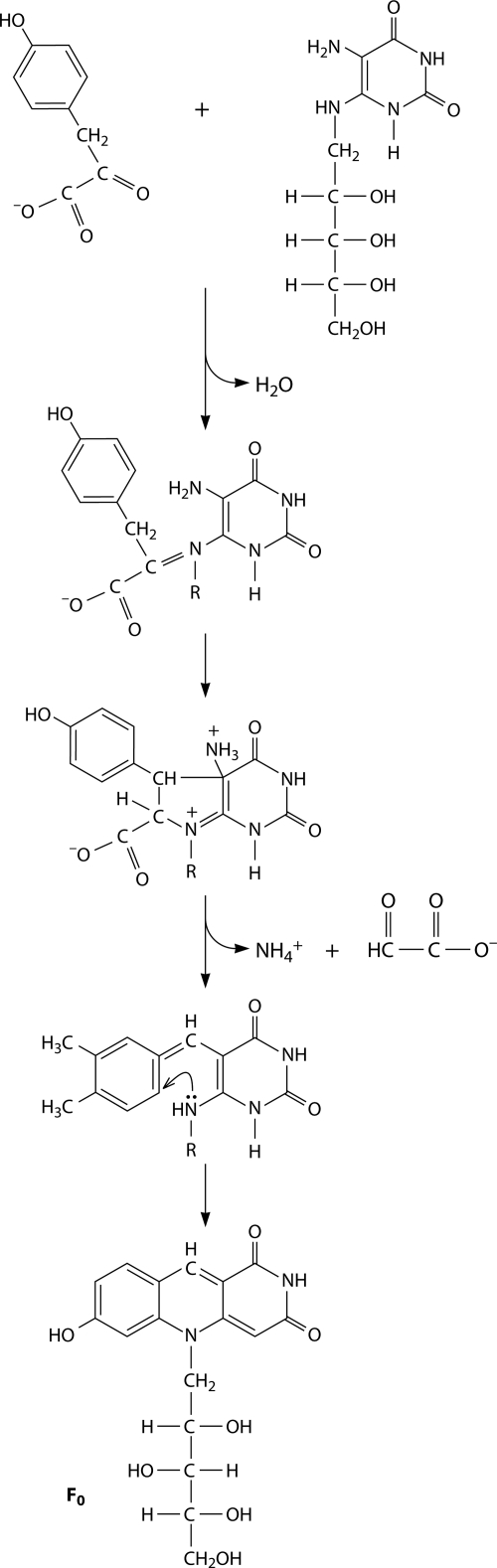
Biosynthetic pathways for coenzymes F0 and F420 have been studied in methanogens. 7,8-Didemethyl-8-hydroxy-5-deazaflavin (and its N-10-ribitylated derivative known as coenzyme F0) is produced by the interaction of the RF precursor 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione with 4-hydroxyphenylpyruvate (151, 368). Thus, 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione is the final common intermediate in the synthesis of RF and deazaflavines. Specific, detailed steps in deazaflavin synthesis are not known. Oxalate is somehow involved in F0 biosynthesis in Methanobacterium thermoautotrophicum (524). In M. jannaschii and Methanosarcina mazei, the synthesized 7,8-didemethyl-8-hydroxy-5-deazaflavin (and coenzyme F0) are further converted to coenzyme F420 by the coupling of 2-phospho-l-lactate (126, 159). The enzyme of M. jannaschii MJ0768 catalyzes the GTP-dependent addition of 2 glutamates to the l-lactyl phosphodiester of 7,8-didemethyl-8-hydroxy-5-deazariboflavin (F0) to form F420-glutamyl-glutamate (261). This enzyme has no sequence similarity to any previously characterized protein. A hypothetical pathway of F0 cofactor synthesis is shown in Fig. 4. Nothing is known about deazaflavin synthesis in eukaryotic cells, which contain F0 as a cofactor of DNA photolyase (104, 105, 146). There are homologs of archaeal enzymes involved in F0 synthesis from 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione and 4-hydroxyphenylpyruvate in eukaryotic green algae (146). For other eukaryotes that use deazaflavins as cofactors of DNA photolyase, these compounds can be considered vitamins provided by some prokaryotes.
Fig. 4.
Biosynthesis of the F0 cofactor. (Reproduced from reference 151 with permission of the Royal Society of Chemistry.)
Roseoflavin
The antibiotic roseoflavin [7-methyl-8-dimethylamino-10-(1′-d-ribityl)-isoalloxazine], produced by Streptomyces davawensis, is apparently synthesized from RF. Data on radioactive guanine incorporation into roseoflavin and radioactivity dilution in roseoflavin by nonradioactive RF support this hypothesis (296). Possible intermediates in the pathway from RF to roseoflavin are 8-amino- and 8-methylamino derivatives of RF (201). The mechanism of substitution of a methyl group at position 7 of RF with an amino group remains unknown, but it may include some additional intermediate(s). The corresponding enzymes involved in the conversion of RF to roseoflavin are also unknown. Intracellular roseoflavin is converted by RF kinase to roseoflavin-5′-phosphate (an analog of FMN) and further by FAD synthetase to roseoflavin adenine dinucleotide (515). Roseoflavin derivatives of FMN and FAD are inactive as coenzymes, apparently due to loss of their oxidizing ability following an intramolecular charge transfer from the 8-dimethylamino group to the pteridine moiety (434). Roseoflavin producers normally convert roseoflavin to analogs of FMN and FAD, so presumably defects in these conversions cannot explain S. davawensis resistance to roseoflavin (155). In roseoflavin-sensitive B. subtilis strains, roseoflavin binds to the FMN riboswitch and thus inhibits transcription of the RF operon (253, 335). Riboswitch mutations in roseoflavin-resistant mutants disrupt ligand binding to FMN-binding aptamers. Perhaps the reason for the insensitivity of S. davawensis to roseoflavin is the inability of the antibiotic produced to bind with the FMN riboswitch. RF biosynthesis genes have been cloned from S. davawensis and expressed in E. coli (156). However, genes involved in RF conversion to roseoflavin remain unidentified.
RIBOFLAVIN TRANSPORT INTO AND OUT OF THE CELL
RF must be capable of readily penetrating cells of animals and lactic acid bacteria that are RF auxotrophs, since their growth depends on the uptake of exogenous RF (356). The ability of RF prototrophic cells to take up exogenous RF is not obligatory and also is not that obvious. As discussed above, the flavin antibiotic roseoflavin is active only against Gram-positive bacteria. Many Gram-negative bacteria are resistant to roseoflavin, probably due to the inability of this RF structural analog to enter cells (23, 156, 333, 334). Many microorganisms are capable of RF oversynthesis and accumulation in a medium (see below), suggesting that these organisms can efficiently secrete RF. Lactating mammary gland cells actively secrete RF into milk (187, 501). In eukaryotic organisms, there probably exist specific systems which provide transport of RF and/or flavin coenzymes to organelles (mitochondria, vacuoles, etc.). The data suggest the existence of specific cellular transport systems for RF in many micro- and macroorganisms.
Below, the peculiarities of RF transport in different groups of organisms are considered. No direct data on RF transport in archaea and plants appear to be available in the literature.
Bacteria
RF transport has been studied experimentally in B. subtilis and Lactococcus lactis. E. coli neither transports exogenous RF nor possesses genes homologous to RF transport genes from other bacteria (505). Hence, RF auxotrophic mutants of E. coli need very high concentrations of RF to grow (23, 422). Analysis of RF genes coding for RF transporters (which are subject to RF feedback regulation with the so-called RFN element [see below]) predicts the existence of 3 homolog classes: (i) homologs of ypaA (ribU) of B. subtilis, (ii) homologs of ribM of L. lactis, and (iii) homologs of impX of Fusobacterium nucleatum (504).
The last class of genes has not yet been functionally characterized in bacteria. According to other classifications, ypaA from B. subtilis belongs to the bile/arsenite/RF transporter (BART) superfamily, which involves the family of RF transporters (285). The RF transporter family comprises 58 proteins from eubacteria and archaea; all those with 5 transmembrane domains have their N termini outside and their C termini inside the cells. Members of this family can be subdivided into 4 clusters, with the ypaA protein being a member of the third cluster (285).
Studies with B. subtilis have shown that RF transport is a specific carrier-mediated process with an extremely high affinity (apparent Km between 5 and 20 nM). RF transport was low in RF prototrophic cells and was greatly stimulated in RF auxotrophs, apparently due to transporter protein repression by RF (65). RF transport occurred against a concentration gradient (50), and RF accumulated inside the cells in its coenzyme forms, FMN and FAD (65). Membrane vesicles isolated from B. subtilis cells bound RF with high affinity, and the solubilized RF-binding activity was characterized, like RF transport, by high substrate specificity and affinity. Inhibitors of energy metabolism blocked RF transport (65, 505). However, it was not determined whether inhibitors suppress an RF transport process per se or subsequent ATP-dependent RF conversion to flavin nucleotides. The gene ypaA proved to code for the RF transport protein in B. subtilis (243). Its knockout abolished RF transport and drastically increased demand of RF auxotrophs in exogenous RF, whereas overexpression activated RF transport (243, 505). FMN and FAD inhibited uptake of radioactive RF, but it is unclear if these nucleotides act in their intact forms or after conversion to RF as inhibitors of RF transport. The corresponding gene codes for a protein with 5 transmembrane domains with a cytoplasmic C terminus (505). Exogenous RF repressed synthesis of ypaA protein (504).
RF transport has also been studied in detail in the lactic acid bacterium Lactobacillus lactis (57). The gene ribU is responsible for RF transport in this organism. Exogenous roseoflavin and FMN inhibited RF transport, and deletions in ribU gene abolished the process. Exogenous RF and FMN repressed synthesis of a transport protein. RF transport occurred in the absence of an energy source; it is probably driven by equilibration of internal and external RF pools via an exchange (counterflow) mechanism. This possibility is supported by the fact that accumulated radiolabeled RF in energized cells could be chased out of the cell with excess unlabeled exogenous RF (57). The ribU gene was overexpressed and the corresponding membrane protein solubilized from membranes before purification. RF, FMN, and roseoflavin bound this RibU protein with extremely high affinity (Kd for RF, 0.6 nM), and Trp68 was involved in binding RF (97). In L. lactis, the RF transporter is classified as an energy-coupling factor (ECF) transporter (376), but the reported absence of energy dependency for RF transport (57) confuses the overall picture of the energy needs for this ECF transport system. A similar situation was found in Corynebacterium glutamicum. The corresponding structural gene, pnuX (otherwise designated ribM), was isolated and expressed in E. coli, resulting in transformants that acquired the ability to take up RF from the medium (57). This process showed saturation kinetics with an apparent Km of 11 μM. Only roseoflavin inhibited RF uptake; FMN was ineffective. Active transport inhibitors had no effect on RF uptake. RF auxotrophs of E. coli that expressed the heterologous ribM-encoded transporter may be involved in the synthesis of flavoproteins with modified cofactors, e.g., with roseoflavin derivatives (293). A putative RF transporter-encoding gene (ribM) has also been identified in the roseoflavin producer, S. davawensis. Expression of this gene in E. coli resulted in RF-transporting transformants that were highly sensitive to roseoflavin (156).
Recently, many bacterial RF transport proteins have been classified as members of a novel class of modular transporters that mostly involve vitamin transporters (376). According to their proposed modular structure, vitamin transporters consist of substrate-specific integral membrane proteins and are responsible for substrate recognition and translocation, together with additional energy-coupling modules (167, 172, 376): module A, containing ATPase (the same as in ABC superfamily transporters [79]); module T, a characteristic transmembrane protein with unknown function; and module S, which is specific to its own substrate. The A and T modules can be shared by different transport systems. This new group of modular transporters are known as ECF (energy-coupling factor) transporters (376). The RF transporter from B. subtilis belongs to an ECF transport system, since deletion of the ecfT gene, which encodes module T, completely abolished RF transport, similarly to mutants defective in the ypaA gene. In contrast to ABC transporters, ECF transporters never use soluble periplasmic substrate-binding proteins and share A and T modules between many quite different S components (79, 376). Although the L. lactis ribU gene has also been included in this classification (376), it apparently acts as facilitator, so the precise role of A and T modules in RF transport remains unexplained.
Thus, numerous RF transport genes have been identified in prokaryotes. They form separate families of membrane proteins of the bile/arsenite/RF transporter (BART) superfamily and apparently consist of several modules. The affinities of investigated bacterial RF transporters for their substrates are very high. Synthesis of RF transporters is repressed by RF. As substrate specificity of RF transporters from pathogenic bacteria can differ from that of RF transporters from animals, identification of specific inhibitors of bacterial RF transport will be of interest for development of novel antimicrobial agents. Studies of bacterial RF transport deal only with RF uptake from the medium, and while some bacteria (e.g., B. subtilis) overproduce and excrete RF as an industrial process, little is known about the mechanisms of RF excretion in bacteria. A simple exchange (counterflow) of intra- and extracellular RF has been postulated for L. lactis (57), but this fails to account for unidirectional excretion of RF during its oversynthesis. RF cellular transport mechanisms await further elucidation.
Yeasts and Filamentous Fungi
The characteristics of RF transport (both uptake and excretion) have been studied in some detail in baker's yeast (S. cerevisiae), the flavinogenic yeast P. guilliermondii, and to a lesser extent the flavinogenic fungus A. gossypii.
Saccharomyces cerevisiae.
RF auxotrophs of S. cerevisiae require significant amounts of RF for growth (1 to 10 μg/ml), which significantly exceed the levels for the growth of RF auxotrophs of B. subtilis (50, 331, 364). RF prototrophic cells of S. cerevisiae were unable to transport RF from the medium. However, anaerobically grown RF auxotrophic mutants can take up radioactive RF (345). RF transport has saturation kinetics (Km, 15 μM; pH optimum, 7.5), and RF analogs inhibit uptake. Uptake by S. cerevisiae RF auxotrophs could not be reproduced in other studies (364). Nevertheless, the gene coding for the RF transporter has been cloned by selection for multicopy suppressors that allow RF auxotrophs to grow in greatly reduced concentrations of exogenous RF. This transporter gene showed strong homology to genes coding for monocarboxylate transporters and was designated MCH5 (364). The corresponding protein was localized to the plasma membrane. Uptake of radioactive RF was measurable only in RF auxotrophic Δrib4 and Δrib5 transformants overexpressing Mcf5p. Exogenous RF repressed synthesis of Mch5 protein in S. cerevisiae, but the mechanism is unclear. Heterologous expression of MCH5 in S. pombe produced transformants with an ability to take up RF from the medium, seemingly by facilitated diffusion, as inhibitors of energy metabolism did not affect the process. The data raise the question of the physiological roles of the transport system, since S. cerevisiae and other yeast species are invariably RF prototrophs, implying that the transport systems are inoperative in wild-type cells.
RF is taken up by cells of RF-deficient mutants and released from S. cerevisiae cells during incubation in a vitamin-free medium (345). This efflux, similar to uptake, shows saturation kinetics (Km, 48 μM) and a pH optimum of 5.0. Exogenous sugars stimulated RF excretion but did not influence RF uptake. Although RF excretion occurs in RF prototrophs, RF uptake was observed only in anaerobically grown RF auxotrophs, which suggests that RF uptake and efflux are catalyzed by separate transporters (345). Nevertheless, other work has discussed the role that the RF transporter Mcf5p plays in facilitating RF flux into and out of the cell (364). Activation of the transcription factor encoded by PUT3 is involved in adapting S. cerevisiae RF auxotrophs to low concentrations of exogenous RF (460).
Flavins have to be transported inside the cells to different organelles. In S. cerevisiae, RF transport from the cytosol to mitochondria is catalyzed by at least two separate transport systems (22). FAD produced in mitochondria is exported into the cytosol using a specific transport system distinct from that for RF mitochondrial uptake. The FLX1 gene, coding for the mitochondrial FAD exporter, has been isolated and functionally characterized. Flx1p is somehow involved in the regulation of synthesis or processing of mitochondrial flavoproteins. The mitochondrial FAD exporter also seems to exist in plants (144). Some indirect data on a specific S. cerevisiae FAD transporter into the lumen of the cytoplasmic reticulum have been reported (358).
Pichia guilliermondii.
P. guilliermondii is the only yeast species that possesses a system for active RF transport (involving an RF permease). This enzyme is cryptic in wild-type strains, but it can be manifested in RF auxotrophs adapted to growth in a medium with very low concentrations of RF.
(i) RF permease I.
The wild-type cells of the flavinogenic yeast P. guilliermondii, like those of S. cerevisiae, cannot take up RF from the medium, and RF auxotrophs of P. guilliermondii grow only at very high concentrations of exogenous RF, much higher than those for S. cerevisiae. Optimal growth of P. guilliermondii RF auxotrophs occurred only at very high RF concentrations of ∼200 μg/ml (420, 449). These initial RF auxotrophs were used for the isolation of auxotrophs with much lower exogenous RF requirements of ∼2 μg/ml. The resultant strains still did not take up exogenous RF from the medium but were characterized by multiple sensitivity to structurally and functionally unrelated antibiotics and antimetabolites that normally do not inhibit yeast growth (actinomycin D, structural analogs of RF, etc.) (447). One of these RF auxotrophs (rib2 mutant) with decreased requirements for exogenous RF was used to isolate RF auxotrophs growing at RF concentrations 1 order of magnitude lower (0.1 to 0.3 μg/ml). One of the strains, MS1-3, has been studied in detail. It formed bright yellow colonies in a medium with a high RF concentration (200 μg/ml). Washed cells grown in a sucrose medium with a low RF concentration (0.5 μg/ml) and incubated in carbon-free or glycerol-containing media with a high RF concentration (100 to 200 μg/ml) took up RF against a concentration gradient, accumulating it in vacuoles as crystals (427, 449, 450). The RF concentration in cells could reach 20 mg/g (dry weight), characterized by saturation kinetics (Km of 0.17 mM, i.e., 1 order of magnitude higher than that in S. cerevisiae) (345), at a pH optimum of 5.8. The corresponding transport system was designated an RF permease (449, 450). Its activity was strongly inhibited by energy poisons and showed strict substrate specificity. It is noteworthy that RF permease was totally devoid of the ability to transport FMN and FAD (449). The ability to transport RF in strains displaying RF permease unexpectedly appeared to be a recessive trait. RF prototrophs isolated from initial auxotrophic RF-transporting strains took up this vitamin with the same velocity; thus, RF permease was not regulated by RF. RF permease was strongly and competitively inhibited by glucose (Ki of 5.7 mM). However, RF permease did not transport glucose, since it inhibited RF permease with unchanged efficiency in a mutant that was totally defective in glucose utilization and transport (442). In addition to glucose, RF permease was inhibited by sucrose, maltose, trehalose, and α-methylglucoside and slightly by 2-deoxyglucose, whereas fructose and mannose had no effect. Glucose also strongly inhibited uptake of the RF analog 8-piperidyl-10-(1′-d-ribityl)isoalloxazine, which competed with RF for uptake into the cell; however, in contrast to RF, it could not have been excreted by the cells into the medium. Thus, glucose inhibition of RF transport cannot be explained by activation of RF efflux.
RF permease was synthesized only in media with α-glucosides, including sucrose, maltose, α-methylglucoside, and melizitose, whereas cells grown on other substrates (glucose, fructose, etc.) did not take up RF from the medium at all, and an RF auxotroph in a glucose medium required high concentrations of exogenous RF (100 μg/ml) for growth (452). Coordinated induction of RF permease and α-glucosidase has been found. Genetic control of RF permease synthesis showed two regulatory genes of negative action, designated RPF80 and RPF81, and one gene of positive action, RPF82 (440). Recessive mutations rfp80 and rfp81 and dominant mutation RFP82c led to the constitutive synthesis of RF permease (and α-glucosidase) in a glucose medium without α-glucosides, whereas recessive mutation rfp82 rendered strains unable to synthesize RF permease in media containing α-glucosides. However, the molecular nature of the these regulatory genes, as well as that of the structural gene of RF permease, was not identified, as at that time the molecular genetics of P. guilliermondii had not been developed.
Selection for growth of the RF auxotroph at very low concentrations of RF in the presence of the structural nonmetabolizable analog 8-piperidyl-10-(1′-d-ribityl)isoalloxazine (which competes with RF for transport) made it possible to isolate P. guilliermondii mutants with increased RF permease affinity to RF (Km of 31 μM, which is close to the value for the RF transport system of S. cerevisiae). Additional, unidentified mutations introduced in this strain allowed constitutive synthesis of RF permease in glucose media and resistance to glucose inhibition of RF uptake. The corresponding RF auxotroph, strain 9i, grew in media containing 0.005 μg RF/ml and appeared to be very effective for microbiological RF assay, as well as for removal of RF from solutions and its overaccumulation inside cells (A. A. Sibirny, unpublished data).
(ii) RF permease II.
The isolation of independent strains of P. guilliermondii capable of RF uptake was attempted. To do so, another RF auxotroph, an rib3 mutant, was selected for its ability to grow with very low RF concentrations. The mutant RA68-2 was selected, which produced yellow colonies on plates with RF. Washed cells of this strain took up RF from the medium against a concentration gradient and accumulated it in vacuoles as crystals. However, the properties of the RF transport systems in strains MS1-3 and RA68-2 appeared to be quite different. The transport velocity of RA68-2 cells was 1 order of magnitude lower than that of strain MS1-3; it was identical in cells grown in sucrose or glucose media, and these sugars did not inhibit RF uptake. Due to such differences, the RF transport system in strain MS1-3 and its derivatives was designated RF permease I, and that in strain RA68-2 was designated RF permease II (426). Activity of RF permease II, like that of RF permease I, did not depend on RF auxotrophy. The genes coding for RF permeases I and II seem not to be linked.
Wild-type P. guilliermondii seems to contain a cryptic gene for RF permease that is not expressed for several reasons, one of which could be the absence of an efficient promoter. During positive selection for RF auxotrophs growing at very low concentrations of exogenous RF, this gene is transferred under the control of an efficient promoter or fused to a functional gene that is being expressed. Differences in the properties of RF permeases I and II are related to sites of insertion of this cryptic gene. Gene transfer could hypothetically take place using mobile genetic elements or some other recombination event. Cloning of the gene(s) coding RF permease(s) is a prerequisite for understanding the cryptic nature of this transport system in wild-type strains and its visualization during selection for RF auxotrophs growing with very low RF concentrations. It could also shed light on the nature of the vacuolar RF transport responsible for accumulation of RF crystals in vacuoles. The development of P. guilliermondii molecular genetics (see below) opened up good prospects for these studies, which are now in progress. Some data show that the cells of the distinct wild-type strain of P. guilliermondii can take up exogenous RF and accumulate it (267, 477). This finding could not be reproduced with other strains of this species; however, those authors suggest that there does exist in many cases a cryptic RF permease that expresses its activity under certain conditions.
(iii) RF excretase.
P. guilliermondii cells of the RF auxotroph MS1-3 that were loaded with RF excreted this RF on incubation in a vitamin-free medium. Sugars strongly activated this process (by 10 to 13 times), whereas inhibitors of energy metabolism severely suppressed it (427, 451). The RF excretion system was characterized by substrate specificity that differed from that of RF permease; e.g., the RF structural analog 8- piperidyl-10-(1′-d-ribityl)isoalloxazine efficiently competed with RF for transport into the cells and completely abolished the ability to leave the cell. RF excretion was a temperature-sensitive process, not occurring at 0°C, and it had a pH optimum of 7.0. Data on the different effects of carbon substrates on RF uptake and excretion and on the different substrate specificities of the systems for RF uptake and excretion suggest the existence of a distinct transport system for efflux of RF from the cells, an RF excretase. This system exists in wild-type strains, which excrete and accumulate considerable amounts of RF from the culture medium, especially under conditions of iron deficiency (415). The RF prototrophic strains of P. guilliermondii displaying RF permease I activity reabsorb a considerable proportion of synthesized and excreted RF in an iron-deficient medium. Mutants of strains with RF permease I activity that overproduce RF in an iron-rich medium have been isolated. They grew as yellow colonies due to reabsorption by the cells of overproduced RF (Sibirny, unpublished data). From them, the next generation of mutants has been isolated and was totally devoid of the ability to excrete RF into the medium, apparently due to a genetic defect in RF excretase (445). Cloning of the RF excretase gene in P. guilliermondii is under way. Its overexpression can speed up RF excretion from the cells and consequently lead to an increase in RF synthesis and its accumulation in the culture medium.
Ashbya gossypii (Eremothecium gossypii).
Some peculiarities of RF transport have been studied in the flavinogenic fungus A. gossypii (127), which is used for the industrial production of RF. RF prototrophs are unable to take up exogenous RF, although a rib5 auxotroph displayed RF transport from the medium. The uptake system was characterized by high affinity to RF (Km of 40 μM) but very low activity. The RF uptake system was characterized by substrate specificity and sensitivity to the energy inhibitor 2,4-dinitrophenol. RF can be excreted from mycelia; however, the authors did not study the excretion process per se due to the inability of the rib5 auxotroph to accumulate sufficient RF. Therefore, an RF overproducer resistant to itaconic acid (400) has been used to study RF efflux (127). During the experiments, the mold synthesized and excreted RF, and the authors examined the total accumulation process. The alkylating agent N-ethylmaleimide inhibited, whereas FMN activated, RF accumulation in the medium. The mechanisms of these phenomena are not known. Like the strains of P. guilliermondii that express RF permease, A. gossypii accumulates part of the synthesized RF in vacuoles. Although the transport system responsible for RF accumulation in vacuoles was not identified, the process is apparently energized by vacuolar ATPase. Disruption of the corresponding gene, VMA1, totally blocked RF accumulation in vacuoles and increased overall RF production (128). Thus, further study of the RF transport is of biotechnological importance.
There has been a report of the ability of another fungus, a Phycomyces sp., to transport exogenous RF (82). dar mutants resistant to RF analogs were unable to transport RF. The gene dar probably codes for the RF transporter.
Animals
For animals, RF is an essential growth factor (vitamin), and hence the lives of entire organisms and particular cells depend on RF uptake. Milk is a rich source of RF for human babies and mammal cubs, and thus efflux of RF from cells of the mammary gland into milk is also of vital importance. RF transport has been studied in many animal tissues, cells, and membrane vesicles, especially in the small intestine and colon (382, 545). Renal transport is also very important (202, 459). Biochemical data suggest the existence of two membrane RF transport mechanisms, one being mediated by an energy-dependent transport with saturation kinetics and the other possibly being the passive diffusion process observed under conditions of nonphysiological high RF concentrations (125, 555). FMN and FAD strongly inhibited RF transport and apparently shared the same transporter (182). The carrier-mediated pathway is apparently regulated by the Ca2+/calmodulin pathway. A role of receptor-mediated endocytosis in RF transport has also been suggested. Specific soluble RF-binding proteins have been found in bird eggs and pregnant mammals (523). They are used for storage of RF and its delivery to developing embryos. Apparently, delivery to cells and tissues of RF that is bound to its binding protein relies on endocytosis, whereas penetration of free soluble RF takes place via membrane carriers (transporters) (125).
Molecular mechanisms of RF membrane transport in mammals were unknown until recently, when two putative RF transporters, RFT1 and RFT2, were identified (538, 544). The genes shared homology with each other and with the bacterial RF transporter gene impX from Halobacterium nucleatum (504, 538). Human and rat RFT1- and RFT2-encoding genes were isolated from kidney cDNA library clones. The RFT1 protein consists of 10, and the RFT2 protein of 11, putative transmembrane domains, and both proteins localized to cytoplasmic membranes. Overexpression of human and rat RFT1 and RFT2 increases the rate of RF transport, and small interfering RNA (siRNA) targeting RFT1 significantly decreased this process (538, 544). Lumiflavin, FMN, and FAD strongly inhibited RFT2-mediated transport. RF was, however, the preferred substrate of the RFT2 transporter (538).
The protein responsible for RF secretion from the mammary gland into milk has long been known as the multidrug transporter called breast cancer resistance protein (BCRP). Its synthesis is strongly induced during pregnancy and lactation, and it belongs to the ABC family of multidrug transporters. In Bcrp1−/− mice, RF secretion into milk was reduced by >60-fold and that of FMN by 6-fold compared to that in wild-type mice (501). Apparently, RF secretion into milk is the main function of the identified transporter. A similar transporter was found in humans. There is also an alternative flavin exporter responsible for FAD efflux into milk; however, the corresponding gene has not yet been identified. It has been suggested that the assay of RF excretion could be used as a novel marker of multidrug resistance in malignant cells (187).
Summarizing this section on RF transport, although RF transporters involved in RF uptake have been isolated in several organisms, it is noteworthy that the mechanisms of RF efflux in microorganisms remain elusive. Regulation of RF transport in fungi and the mechanism of the cryptic status of RF permeases in wild-type strains also need to be elucidated.
REGULATION OF RIBOFLAVIN SYNTHESIS
Regulation of RF synthesis occurs mostly at the level of the synthesis of its biosynthetic enzymes. Effectors of this regulation are flavins and, unexpectedly, iron ions and, to a lesser extent, other metals (cobalt, chromium, zinc in flavinogenic yeasts, and magnesium in some in fungi) in many organisms.
Feedback Inhibition of GTP Cyclohydrolase II
Very little is known about the regulation of RF synthesis at the level of modulation of enzyme activity. Apparently, GTP cyclohydrolase II is the rate-limiting reaction in RF synthesis, at least in B. subtilis (185). As a rule, activities of key enzymes catalyzing the first limiting step of the anabolic pathways are regulated by allosteric feedback inhibition exerted by the product of the reaction or entire pathway. However, very little is known about the possible feedback control of RF biosynthesis at the level of GTP cyclohydrolase II, although feedback control is well documented for GTP cyclohydrolase I, which is involved in the biosynthesis of folates and tetrahydrobiopterin (44, 237, 283). While the structures of bacterial GTP cyclohydrolases from E. coli and B. subtilis have been studied in detail (206, 257, 365, 373), no information on feedback inhibition of this enzyme by RF, flavin nucleotides, or other metabolites is available. GTP cyclohydrolase II from the flavinogenic yeast P. guilliermondii was allosterically inhibited by FAD (412, 425). This inhibition was retained in P. guilliermondii GTP cyclohydrolase II overexpressed in E. coli, whereas the native bacterial enzyme showed no such inhibition (546). RF and FMN had no effect on enzyme activity. The physiological role of this inhibition remains unclear, since in addition to FAD, other nucleotides containing an adenylic group (5′-AMP, 3′,5′-AMP, ADP, ATP, NAD, and NADP) inhibited enzyme activity to a similar extent (412, 425). No data are available on the effect of FAD and other adenylic nucleotides on the activity of GTP cyclohydrolases from other yeasts, including S. cerevisiae. No P. guilliermondii mutants have proved to be defective in this kind of inhibition.
There is apparently another possible feedback regulation of RF biosynthetic enzymes, namely, through 6,7-dimethyl-8-ribityllumazine synthase from P. guilliermondii, which is inhibited by RF but not FAD, whereas neither RF nor FAD influenced the activities of 3,4-dihydroxy-2-butanone 4-phosphate synthase and RF synthase (274, 275). There are no data on the physiological significance of 6,7-dimethyl-8-ribityllumazine synthase inhibition obtained in experiments on the isolated enzyme.
Transcriptional Regulation in Bacteria Using the Riboswitch Mechanism
The discovery of the riboswitch mechanism involved in regulation of the RF operon in B. subtilis has been among the major achievements in modern molecular genetics.
Riboswitch (RFN element) in Bacillus.
The regulation of RF synthesis has been studied in detail in B. subtilis. This organism was initially chosen for investigation, and the first principal results on the RF operon were obtained by S. E. Bresler, D. A. Perumov, and colleagues in Russia. Later, investigators from Germany, the United States, and China were involved in these studies. In 1969, Bresler and his colleagues described a mutant of B. subtilis that overproduced RF (52). Its isolation, as well as that of RF auxotrophs, initiated a series of experiments on the biochemical and genetic aspects of RF synthesis in B. subtilis. All known mutations leading to RF auxotrophy are linked and are localized close to genes involved in lysine biosynthesis (49, 51).
The RF operon (rib operon) consists of 5 genes (ribGBAHT) that encode the catalytic enzymes for RF synthesis from GTP and ribulose-5-phosphate. The operon is transcribed as one polycistronic RNA of nearly 4,300 bp (308). The untranslated leader region, designated ribO, of ∼300 bp (originally thought to be the operator region), has been identified upstream of the first gene in the operon (309). Although mutations in ribO led to RF overproduction (221), searches for the gene coding for the repressor protein were unsuccessful. Mutations in two trans-acting genes, ribC and ribR, which caused RF oversynthesis have been identified (163, 280, 458); however, they coded for bifunctional RF kinase/FAD synthetase and monofunctional RF kinase, respectively. Genes ribC and ribR are not linked to the RF operon. The available data suggest that RibC and RibR proteins do not interact with the ribO region. At the same time, mutants defective in RF kinases still proved to be sensitive to repression of RF synthesis after addition of exogenous FMN (48, 280). Inspection of the leader region of the rib operon revealed a sequence that could fold into a characteristic and evolutionarily well-conserved RNA structure of ∼140 bp called the RFN element. Analysis of 23 different RFN elements showed that 10 of 20 bp of the inverted repeats were completely conserved and that of the 47 nucleotides from the single-strand RNA filament, 24 were conserved (138, 221, 504). The RFN element directly binds with FMN, and the formation of this complex is important in the regulation of the rib operon (307, 528). Direct binding of the small molecule FMN to nascent mRNA of the RFN element leads to a conformational change resulting in terminator hairpin formation, which leads to attenuation of transcription. In the absence of FMN, the conformation of the RFN element forms an antiterminator structure that allows normal transcription of the entire operon (307). The RFN element serves as the receptor for a metabolite-dependent riboswitch that directly binds FMN in the absence of proteins (528). The RFN element is a natural FMN-binding aptamer, the allosteric character of which is used for tight control in the expression of the RF operon (Fig. 5). RFN elements have been found upstream of RF biosynthesis and RF transporter genes in many bacteria (504).
Fig. 5.
FMN causes transcription termination of the ribD RNA in vitro. (A) Sequence and secondary structure of the leader sequence ribD RNA. Shaded regions identify nucleotides that are complementary and might serve as an antiterminator structure. Nucleotides denoted with an asterisk have been altered from the wild-type sequence to generate a restriction site. The nucleotide sequence comprising the 62- and 30-nucleotide regions (not shown) is presented in references 16 and 17. (B) Model for the FMN-dependent riboswitch. Shaded regions identify the putative antiterminator structure that is disrupted after binding of FMN and formation of the P1 structure. (Reproduced from reference 528 with permission of the publisher. Copyright 2002 National Academy of Sciences, U.S.A.)
Similar metabolite-binding riboswitches are involved in the regulation of other operons and separate genes in bacteria. Riboswitches are found in many classes of eubacteria but only a few archaea and eukaryotes (25). Riboswitches involved in vitamin and coenzyme synthesis constitute the most abundant class of untranslated mRNA regions capable of direct sensing of cellular metabolites (75, 325, 378, 405). Riboswitches are the nontranslated mRNAs that bind effectors (low-molecular-weight metabolites) using selective binding domains (otherwise called ligand-binding pockets or aptamers) without a requirement for any proteins. Regulation of gene expression by the riboswitch mechanism without participation of regulatory proteins is probably a molecular relic of the ancestral RNA world.
Riboswitches typically contain disordered regions in their conserved aptamer cores that become structured upon metabolite binding. These changes may trigger rearrangements in additional expression platform structures located outside the aptamer, giving two alternative conformations. Riboswitches are able to interconvert between these ligand-bound and ligand-free structures in the context of the full-length RNA (25, 405, 406). The crystal structure of the FMN riboswitch controlling the putative RF transporter encoded by impX in Fusobacterium nucleatum bound with FMN, RF, and roseoflavin has been determined (404). Binding of the riboswitch to FMN depends on Mg2+ ions and is further enhanced by K+ ions. FAD, RF, and lumiflavin have much lower affinity for the riboswitch than FMN. The roseoflavin-bound structure adopts a conformation similar to that of the RF-bound structure, with some additional conformational adjustments. The data explain the effect of regulatory mutations in the FMN riboswitch on gene expression and can be used for developing strategies to search for specific inhibitors of RF synthesis in pathogenic bacteria by acting on the FMN riboswitch. The structure of the FMN-bound riboswitch is shown in Fig. 6.
Fig. 6.
Overall riboswitch structure in a ribbon representation. P1 to P6, domains; L1 to L6, loops. The J1-2 segment participates in antiterminator formation in the absence of FMN, whereas in the FMN-bound state, it is locked up in the junction. (Reproduced from reference 404 with permission of Macmillan Publishers Ltd., copyright 2009.)
The carboxy terminus of the B. subtilis ribR monofunctional RF kinase binds to the FMN riboswitch, but this binding is partially lost in cells bearing mutations in the RFN element (177). The data suggest that riboswitch regulation using FMN as an effector can be supplemented with the action of monofunctional RF kinase, thus acting as a regulatory protein.
Regulation in E. coli.
It is noteworthy that in many groups of bacteria, e.g., in Gammaproteobacteria, including Enterobacteriaceae, the FMN riboswitch attenuates translation, or both transcription and translation, mechanisms (25, 504). It uses FMN-induced structure changes of the riboswitch structure to block the initiation of translation. Unlike riboswitches with transcription control mechanisms that require very specific terminator structures in their expression platforms, the RNA structures that prevent translation initiation can be more varied.
In physiological terms, RF synthesis in E. coli occurs constitutively (422, 526). Genes of RF synthesis are scattered on the E. coli chromosome (23, 495). 5′ noncoding regions of the RF structural genes contain RFN elements apparently acting at the translation level (25, 504); however, the physiological role of these riboswitches in the regulation of RF synthesis in E. coli remains unknown. The E. coli ribA gene, coding for GTP cyclohydrolase II, belongs to the soxRS regulon, which is induced by superoxide-generating agents. The localization of the superoxide-responsive element in the promoter has been determined (232, 233). The exact role of this regulation is unknown; however, it may indicate that GTP cyclohydrolase II or the final product of the pathway, RF or flavin nucleotides, is somehow involved in defense against oxidative stress. Reactive oxygen forms convert GTP to 8-hydroxy-dGTP, which acts as a mutagen. GTP cyclohydrolase II fulfills a supplementary role in the detoxification of 8-hydroxy-dGTP to mutT protein (229).
Other bacteria.
The ribBA gene of H. pylori, coding for bifunctional GTP cyclohydrolase II//3,4-dihydroxybutanone phosphate synthase, is derepressed under conditions of iron deficiency, which leads to increased flavin synthesis (529). Heterologous expression of this gene in E. coli doubled RF synthesis and ferrireductase activity.
As mentioned above, flavins participate in bacterial luminescence. In Photobacterium phosphoreum and Photobacterium leiognathi, RF biosynthesis genes are linked to the luciferase (lux) operon and are transcribed as one polycistronic mRNA (251, 252, 270). In other luminescent species, e.g., Vibrio fischeri, an unlinked ribB gene homologous to the E. coli gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase forms a joint regulon with the lux operon (61).
It is well known that Clostridium acetobutylicum is capable of overproducing RF under conditions of iron deficiency (175), but the mechanism of regulation remains obscure.
Transcriptional Regulation in Mycelial Fungi
Of the mycelial fungi that naturally overproduce RF, two (the phytopathogenic [cotton pathogens] fungi Eremothecium ashbyii [162] and Eremothecium [Ashbya] gossypii [525]) belong to the group of the most flavinogenic natural organisms known (83). RF accumulates in the mycelia, giving them a bright yellow color (128). The ability to overproduce RF is an unstable feature in E. ashbyii, and highly flavinogenic clones easily lose their potential during lyophilization or storage at room temperature (313). Therefore, most work has been conducted on E. gossypii (its generally used name remains A. gossypii, which is used throughout this review). However, E. ashbyii overproduces not only RF but also FAD, whereas A. gossypii does not overproduce flavin nucleotides (337, 534). A. gossypii is characterized by a high level of gene order conservation (synteny) between its genome and that of the yeast S. cerevisiae and thus became a popular model for the biology of fungal development (350, 522). Methods of molecular genetics for A. gossypii, including insertional mutagenesis, have been developed (391, 467, 468, 530). The genome of A. gossypii has been sequenced and has strong similarities to that of S. cerevisiae. With a size of only 9.2 Mb, carrying 4,718 protein-coding genes, the A. gossypii genome is the smallest of those of free-living eukaryotes yet characterized (45, 86).
RF oversynthesis by both of the above-mentioned fungi occurs after vegetative growth has ceased, during mycelium lysis and spore formation (209, 463). As a rule, similar kinetics of production have been seen during microbial synthesis of secondary metabolites, e.g., antibiotics, but not primary metabolites. Therefore, RF overproduction by the mycelial fungi E. ashbyii and A. gossypii has been called “pseudosecondary biosynthesis” (66). The physiological reasons for RF overproduction by E. ashbyii and A. gossypii during initiation of sporulation are unknown, although the processes are closely linked. A mutant of A. gossypii that has lost the ability to sporulate is characterized by reduced RF synthesis (66), and cyclic AMP (cAMP) inhibited both sporulation and RF oversynthesis (463). RF protects spores of A. gossypii against UV light (463). That fungi can produce pigments to attract insects for spore spreading is well known (3), but one cannot exclude the possibility that RF overproduced during sporulation by E. ashbyii and A. gossypii attracts insects spreading their spores to new plants (438).
Flavinogenic activity of A. gossypii depends on the cultivation temperature, dropping significantly at 38°C. It has been suggested that at elevated temperatures the specific repressor of RF biosynthesis is activated, although no direct evidence has been presented (83). In E. ashbyii the shift from growth to the production phase was accompanied by derepression of GTP cyclohydrolase II and FAD synthetase, whereas the activity of RF synthase was only slightly changed (239). In A. gossypii, the RF production phase was characterized by a strong increase in transcription of several RF biosynthesis genes (RIB3, RIB4, and RIB5 but not RIB2 and RIB7), as well as many other genes involved in transcription, translation, membrane transport, cell architecture, and metabolism (209, 292, 396, 397). The mechanisms involved in the shift from growth to production phase and in the induction of RF synthesis and the nature of the messenger(s) that induces RF oversynthesis remain unknown. It was hypothesized that some kind of stress factors that accumulated during cessation of growth promote transcription activation and RF production (397).
Transcriptional Regulation in Flavinogenic Yeasts by Iron
Flavinogenic yeasts include the group of strains that overproduce RF under iron-restrictive conditions (492). This group includes P. guilliermondii (asporogenic strains of this species are designated Candida guilliermondii), C. flareri (teleomorph, Debaryomyces subglobosus), C. famata, Debaryomyces hansenii, Schwanniomyces occidentalis, the pathogenic yeast C. albicans, and some others (227, 322, 415, 436, 437, 441, 516). The reasons for this and the physiological role of this overproduction are unknown. Some have suggested that the stimulation of RF production induced by iron limitation spreads among different organisms. In addition to these yeasts, the list includes bacteria, i.e., C. acetobutylicum (175), H. pylori (529), C. jejuni (78), and Shewanella (77), as well as plants such as tobacco, sunflower, and others (176, 502, 512, 521). Several reasons for the stimulation of RF production by iron deficiency have been discussed, including a direct role of RF as an electron donor either for iron reduction or as a cofactor for the activity of intra- and extracellular enzymes. This is well documented for some bacterial species, but no experimental data exist for flavinogenic yeasts. In our own experiments, a mutant of the flavinogenic yeast C. flareri (C. famata) which is defective in RF oversynthesis in iron-deficient medium due to mutation in the putative transcription factor gene SEF1 (91) grows more slowly in an iron-deficient medium than the wild-type strain, which overproduced RF under these conditions (K. Dmytruk, O. Lyzak, and A. Sibirny, unpublished observation). Similar data on growth defects in an iron-deficient medium were obtained with the P. guilliermondii rib83 mutant, which is unable to overproduce RF (471). The rib83 mutation was recently found to be allelic to sef1 (D. Fedorovych, V. Boretsky, and A. Sibirny, unpublished observation). It can be hypothesized that in flavinogenic yeasts, RF is involved in iron (Fe3+) assimilation, e.g., by nonenzymatic reduction of practically insoluble Fe3+ to much more soluble Fe2+. However, the vast majority of yeast species do not overproduce RF under conditions of iron limitation, which suggests different mechanisms of iron acquisition by flavinogenic and nonflavinogenic yeasts. In contrast to the case in B. subtilis, RF and flavin nucleotides do not repress their own synthesis in yeasts (415).
Among flavinogenic yeasts, P. guilliermondii is the model organism, whereas C. flareri (C. famata) represents organisms with the highest flavinogenic potential. Studies on the iron-dependent regulation of RF synthesis in yeasts have been hampered by the absence of methods for classical and molecular genetic analyses of this group of organisms. Finally, methods of classical genetics for analyzing dominance-recessiveness and meiotic segregation have been developed for P. guilliermondii, while methods of molecular genetics have been developed for both organisms (Table 1) (1, 40, 41, 436, 437, 441, 448, 454, 511). Structural genes of RF biosynthesis have been cloned from these organisms (90, 266, 277, 510, 546). A promoter assay system has been developed for C. famata, and several strong promoters have been cloned (194). The genome of C. guilliermondii, the anamorph of P. guilliermondii, is publicly available (http://www.broad.mit.edu, C. guilliermondii sequencing project). It should be mentioned that strain C. famata VKM Y-9 used in these experiments has recently been reclassified as a C. flareri strain (322), but since all previous publications and patents have used C. famata, only that name is used throughout this review. The genome of D. hansenii CBS 767, also a flavinogenic strain (its anamorph is C. famata, but not the recently reclassified C. flareri VKM Y-9), has been sequenced in the framework of the French National Program Genolevures (http://cbi.labri.fr/Genolevures/elt/DEHA)).
Table 1.
Comparison of developed methods of genetic analysis for the flavinogenic yeasts P. guilliermondii and C. famata (C. flareri)
| Method or characteristic | P. guilliermondii | C. famata |
|---|---|---|
| Hybridization and meiotic segregation | + | − |
| Selective markers | ||
| Recessive | URA3 | LEU2, ADE1 |
| Dominant | ble, IMH3, ARO4 | |
| Maximum transformation frequency (transformant/μg DNA) | 104 | 105 |
| Gene library | + | + |
| ARS elements | + | + |
| Strong promoters | TEF1 | TEF1 |
| Multicopy integration events | Up to 3 copies | Up to 3 copies |
| Insertion mutagenesis | + | + |
| Gene deletion technique | + | − |
| Cloning of RF structural genes | RIB1, RIB7 | RIB1, RIB2, RIB5, RIB6, RIB7 |
| Cloning of RF regulatory genes | SEF1, YAP1, TUP1 | SEF1, MET2 |
Iron deficiency leads to derepression of all of the P. guilliermondii enzymes involved in RF synthesis, with the exception of the reductase, an enzyme in the second step of the pathway that is constitutively synthesized (417). Iron content did not influence synthesis of enzymes involved in RF conversion to flavin coenzymes (408, 424). Derepression occurred at the transcriptional level, as iron starvation caused increased RIB1 (GTP cyclohydrolase II) and RIB7 (RF synthase) mRNA accumulation (38). Regulation by iron at the transcriptional level was also seen for the RIB1 and RIB3 (3,4-dihydroxy-2- butanone-4-phosphate synthase) genes of a strain of another flavinogenic species, Candida membranifaciens subsp. flavinogenie W14-3 (516). The activity of the RIB2 product, reductase, is so high that when it dropped below the detection level in the leaky rib2 mutant of P. guilliermondii, this did not cause RF auxotrophy (417).
Numerous mutants of P. guilliermondii that are defective in the regulation of RF synthesis by iron have been isolated by several innovative selection procedures. One of them seems to be straightforward, since it has been based on the isolation of mutants resistant to RF analogs. The assumption was that exogenous RF must alleviate the inhibitory action of RF analogs on yeast growth, and thus RF overproducers must be more resistant to these compounds. However, wild-type strains of P. guilliermondii were resistant to very high concentrations of RF analogs, apparently due to their inability to penetrate the yeast cell. This obstacle was overcome either by using mutants with multiple sensitivity to antibiotics and antimetabolites (447) or by addition of high concentrations of phosphates or sulfates that make wild-type cells susceptible to the inhibitory action of RF analogs (444). In the mutants resistant to RF analogs that have been isolated, overproduction of RF in the medium was seen, as expected, when a sufficient amount of iron was present (421, 407). The second method for the selection of mutants overproducing RF in an iron-rich medium was with leaky rib2 mutants partially defective in activity of reductase (the second enzyme in biosynthesis). Activity was so low that the leaky strains required exogenous RF in the medium and a high iron content, although they grew without RF in an iron-deficient medium. The leaky rib2 suppressor mutants that acquired the ability to grow in a medium with a high iron content without RF were characterized by derepression of GTP cyclohydrolase II, and after substitution of the leaky rib2 with the wild-type RIB2 allele, RF was overproduced in an iron-rich medium (411, 419). Another method was similar in using a mutant with a thermosensitive GTP cyclohydrolase II that grew like the parental strain without exogenous RF only in an iron-deficient medium. Mutants with impaired regulation of RF synthesis were isolated as revertants growing at the minimal restrictive temperature in an iron-sufficient medium without exogenous RF. In meiotic segregants that contained the wild-type allele of the RIB1 gene, mutants overproduced RF in an iron-rich medium (9). An additional approach was based on isolation of rib1 mutants with decreased requirements for exogenous RF (472) or on the leaky rib1 mutant (with a missense mutation leading to substitution G620A) growing without exogenous RF in an iron-rich medium (38, 469, 470). Mutants with derepressed synthesis of RF in an iron-rich medium were also isolated from the yeast C. famata by selection of mutants resistant to the RF structural analog 7-methyl-8-trifluoromethyl-10(1′-d-ribityl)isoalloxazine in a medium with a high sulfate concentration (439; Sibirny, unpublished data).
In P. guilliermondii, all isolated mutations were analyzed using hybridization and meiotic segregation analyses. All led to RF overproduction and derepression of RF biosynthetic enzymes in iron-rich media and were recessive. They were divided into 9 complementation classes: rib80, rib81, hit1, and red1 to red6 (407, 411, 470, 472). Mutations rib81 and red6 showed constitutive derepression of RIB1 gene transcription in an iron-rich medium (38). Most RF-overproducing mutants also had derepressed iron transport and an elevated content of nonhemin iron (109, 110, 409, 410, 415). These data suggest a coordinated regulation of RF synthesis and iron acquisition in P. guilliermondii. RF overproduction in P. guilliermondii causes Co2+ ion stress (106) and oxidative stress (357). Later, it was shown that Co2+ ions, iron deficiency, and the mutations rib80, rib81, and hit1 all lead to oxidative stress (39). These observations can be partially explained by hypothesizing that RF acts as an antioxidant.
Elucidation of the identified gene action required cloning of the genes as the first step. However, mutants overexpressing RF do not differ significantly from wild-type cells in their growth characteristics, which hampered transformant selection with the restored wild-type phenotype. One additional RF-overproducing mutant of P. guilliermondii was obtained using the molecular approach of knockout of the yeast frataxin homolog gene YFH1, encoding a mitochondrial protein involved in iron trafficking and storage (8, 64, 392). The resulting Δyfh1 mutant, like mutants isolated by conventional mutagenesis, overproduced RF in an iron-rich medium, possessed an elevated intracellular iron content, and was hypersensitive to oxidative stress. However, the Δyfh1 mutation was nonallelic to mutations rib80, rib81, hit1, and red6 (361). It seems that mutations in numerous genes involved in iron transport, assimilation, and storage lead to RF overproduction in P. guilliermondii. Knowledge of the precise role of the identified genes in regulation of RF synthesis awaits further studies. Recently, insertion mutagenesis was used in the isolation of P. guilliermondii mutants with derepressed RF synthesis in iron-sufficient medium. Among them, the gene VMA1, coding for vacuolar ATPase, was tagged. Insertion and knockout vma1 mutants overproduced RF in an iron-rich medium (41a). The role of vacuolar ATPase in the regulation of RF synthesis remains obscure.
Mutants with the opposite phenotype, i.e., an inability to overproduce RF in iron-deficient media, were also isolated from flavinogenic yeasts. In P. guilliermondii, such mutants were isolated as mutants lacking the ability to excrete RF in iron-deficient medium. The corresponding recessive mutation was designated rib83 (413). Mutation rib83 is epistatic over mutations leading to RF oversynthesis, i.e., rib80, rib81, and hit1 (471; D. Fedorovych, unpublished data). The iron content in rib83 cells was similar to that in the wild-type strain (39). In C. famata, insertion mutagenesis was used to isolate RF-nonoverproducing mutants and tag the corresponding genes. Three mutants unable to overproduce RF contained an insertional cassette in the promoter regions of the structural gene coding for GTP cyclohydrolase II (RIB1) and in homologs of the gene MET2, involved in methionine biosynthesis, and the gene SEF1, coding for a putative transcription factor (91). Intact orthologs of these genes from D. hansenii restored normal regulation of RF synthesis. Data on Sef1p functions in other yeasts are poor. Kluyveromyces lactis SEF1 and its S. cerevisiae homolog can suppress a mutation in Rpm2p, which is a protein subunit of yeast mitochondrial RNase P, an enzyme responsible for the 5′ maturation of mitochondrial tRNAs. DNA sequence analysis of the K. lactis SEF1 gene showed that it contained the Zn(2)-Cys(6) binuclear cluster motif found in a growing number of yeast transcription factors (161). Disruption of the SEF1 homolog in P. guilliermondii led to isolation of mutants unable to overproduce RF. Complementation analysis showed that Δsef1 P. guilliermondii is allelic to rib83, which was isolated earlier by classical selection (D. Fedorovych, V. Boretsky, and A. Sibirny, unpublished data).
Expression of C. famata SEF1 is derepressed under conditions of iron starvation, which suggests autoregulation of gene expression. In the nonflavinogenic yeasts S. cerevisiae and Pichia stipitis, expression of this gene is not regulated by iron. The industrial strain C. famata dep8 possesses constitutively derepressed SEF1 which does not depend on iron content (K. Dmytruk and A. Sibirny, unpublished data). Introduction of the D. hansenii SEF1 gene into Δsef1 C. famata restored the ability to overproduce RF, whereas introduction of SEF1 from the nonflavinogenic species P. stipitis in the same Δsef1 C. famata strain did not. The coding sequences of SEF1 are very similar in flavinogenic and nonflavinogenic yeast species; we therefore suggest that the inability of P. stipitis SEF1 to restore RF overproduction in Δsef1 C. famata depends on its low constitutive expression. SEF1 may play a central role in regulating RF synthesis in flavinogenic yeasts, but the mechanism of its action remains to be elucidated.
THE RIBOFLAVIN BIOSYNTHESIS PATHWAY AS A TARGET FOR ANTIMICROBIAL DRUGS
Most microorganisms, including pathogens, synthesize RF de novo, and many of them (especially Gram-negative bacteria and pathogenic fungi) cannot take up RF from the growth medium. Therefore, for microorganisms that cannot transport RF, specific inhibitors of RF synthesis enzymes should be potent antimicrobial drugs where they can penetrate the cell. Mammals do not possess RF biosynthetic enzymes and thus have no target for potential antimicrobial drugs; they are capable of very efficient RF transport. In the light of the rapid development of resistance to virtually all current antibiotics, exploration of new targets and development of novel anti-infective drugs are urgent medical needs (37, 112, 278, 365).
Inhibitors of metabolic reactions have rarely been used as antimicrobial agents, although sulfonamides, the first antibacterial agents discovered before antibiotics, act against a broad spectrum of bacterial pathogens by competing with the natural substrate of dihydropteroate synthase, p-aminobenzoate, thereby inhibiting this enzyme, which is involved in folic acid biosynthesis (301). In this case, sulfonamides, but not folic acid, can be transported by pathogens, whereas human cells effectively absorb folic acid and do not have an intrinsic pathway for biosynthesis of folic acid. Thus, the search for inhibitors of the enzymes involved in RF synthesis as potential antimicrobial agents is based on the premise that they will have the same mode of action on folic acid biosynthesis as sulfonamides.
All RF biosynthetic enzymes are potential targets for inhibition. However, GTP cyclohydrolase II, reductase, deaminase, and 3,4-dihydroxy-2-butanone 4-phosphate synthase act on phosphorylated substrates and products; most probably, therefore, structural inhibitors of these enzymes would not penetrate the microbial cells in their native form. Lumazine synthase and RF synthase are more promising targets for inhibition because they act with nonphosphorylated compounds as substrates or products [5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione and 6,7-dimethyl-8-ribityllumazine, respectively]. It would probably be difficult for the latter compound (being a pteridine) to penetrate the cell envelope. Rybitiled pyrimidine derivatives are more suitable as inhibitors of lumazine and RF synthases, although many pyrimidine analogs could have an influence on other processes, such as nucleotide metabolism and nucleic acid biosynthesis.
Throughput screening methods that are based on the competitive binding of tested inhibitors with lumazine synthase or RF synthase were developed (68, 205). Almost 100,000 compounds have been tested for inhibition of RF biosynthetic enzymes (279, 314, 316, 491, 553). Some compounds were very efficient inhibitors in vitro, and a few of them displayed antibiotic activity against M. tuberculosis. Therefore, this work is promising.
Alternative use of the RF biosynthetic pathway as a target for novel antimicrobial drug can be explored for pathogenic Gram-positive bacteria that can effectively transport RF. As described above, the natural RF analog roseoflavin binds with the FMN riboswitch, thereby repressing RF operon transcription (253, 335, 404). If roseoflavin binding to the FMN riboswitch is the major target for its antimicrobial action, the search for structural analogs of this antibiotic with more potent or specific binding to the riboswitch could be another use of the RF biosynthetic pathway for the identification of new antimicrobial drugs (36, 278). Specificity of the action of a selective corepressor of RF biosynthesis could be achieved due to the absence of this pathway in the host organism (humans or animals). These RF or roseoflavin analogs, however, should not be metabolized to analogs of flavin nucleotides, which might be toxic for host organisms.
FLAVIN SYNTHESIS BY FLAVINOGENIC MICROORGANISMS
Flavins are present in each cell of living organisms. Cellular flavins are present preferentially in nucleotide forms, with FAD being the most abundant. The content of free intracellular RF does not exceed 7% of the total flavins (438). The pool of free flavins (not bound to proteins) in microorganisms varies between 20 and 200 μg/g biomass. Some microorganisms have much larger intracellular pools of free flavins, e.g., the flavinogenic fungi E. ashbyii and A. gossypii (66). Many microorganisms excrete some portion of synthesized flavins to the medium, preferentially as free RF (83). This feature allows RF biosynthesis to be considered a poorly controlled process. For example, exponentially growing wild-type cells of E. coli, B. subtilis, and Pseudomonas fluorescens secrete into the medium several times more flavins than they contain inside the cells in coenzyme forms (526). The role of the excreted RF remains unknown, but it could be hypothesized that it participates in the mobilization of practically insoluble Fe3+ by its reduction to the more soluble Fe2+ form (287).
According to Demain (83), microorganisms that accumulate >10 mg RF/liter are overproducers. They can be divided into 3 groups: weak (producing around 10 mg RF/liter), moderate (producing up to 600 mg RF/liter), and highly active or flavinogenic (producing >10 g RF/liter). The overproducers include the bacteria C. acetobutylicum, Mycobacterium smegmatis, and Corynebacterium diphtheriae, the yeasts Candida (Pichia) guilliermondii, Candida ghoshii, Candida parapsilosis, C. flareri, C. famata, D. subglobosus, D. hansenii, S. occidentalis, and Torulopsis candida, and the mycelial fungi Aspergillus niger, A. gossypii, E. ashbyii, and others. The first observation of RF overproduction by bacteria was by Japanese authors who described the accumulation of RF in C. acetobutylicum (539). RF overproduction in yeasts was reported at approximately the same time (59, 348). The maximal amount of RF produced by yeast under conditions of iron deficiency was 560 mg RF/liter in C. flareri (260). In addition to the concentration of iron ions, some other factors act on RF synthesis. Strong stimulation of RF synthesis in flavinogenic yeasts was caused by Co2+ (87, 106), and some stimulation was seen with Cr6+, Mn2+, and Zn2+ (111, 228, 487). In the fungus A. niger, RF synthesis was stimulated by a deficiency of Mg2+ ions (319, 435). Some species of flavinogenic yeasts overproduce RF in iron-sufficient media containing n-alkanes as the sole carbon source (88, 329, 423), but the mechanisms of these stimulatory effects remain unknown.
INDUSTRIAL PRODUCTION OF RIBOFLAVIN AND FLAVIN NUCLEOTIDES AND THEIR PRACTICAL APPLICATIONS
The vitamin market was $2.3 billion in 2003, with most vitamins being produced microbiologically. In 2003, the annual production of RF was 4,600 tons, with a total value of $134 million (84). In 2008, the total world riboflavin production capacity was around 10,000 tons, whereas annual demand is around 6,000 tons estimated to be worth between $150 million and $500 million depending on the price per kilogram (http://www.articlepros.com/business/Chinese-Marketing/article-660667.html and http://www.articlesbase.com/management-articles/riboflavin-market-development-situation-and-future-trends -2077296.html). Most RF is currently produced biotechnologically using advanced microbial producers constructed from B. subtilis and A. gossypii (178). Production with the yeast C. famata was shut down several years ago by ADM Co. due to the instability (at that time) of the available producer, dep8. The largest producer of RF currently is China (Hubei Guangji Pharmaceuticals, Shanghai Desano Vitamins Co.), although this vitamin is also produced in other countries in Europe and Asia (Aventis, BASF, Daicel, DSM, Kyowa, Mitsui, Roche, and Takeda).
Over 80% of RF is used in agriculture as an additive to premixes for animal feeding (poultry, pigs, etc.) (462). RF also is used in the food industry as the yellow colorant E-101 for beverages, as well as in medicine as a component of vitamin mixtures and for treatment of cataracts, migraine, malaria, methemoglobinemia, some organic acidurias, and other diseases of the skin, eyes, and nervous system (31, 483). RF in the presence of long-wavelength UV irradiation effectively inactivates different viruses, which can be important for vaccine development (62).
Flavin nucleotides are used by the pharmaceutical and food industries. FMN is 200 times more soluble in water, so it is often offered by pharmaceutical companies. However, it should be pointed out that FMN offered as medicine is synthesized chemically and contains >30% of impurities of a flavin nature (324), which can act as antivitamins and/or RF antagonists. Reduced FMN has an interesting potential application in the detoxification of chlorinated xenobiotics (72). FAD is used in medicine and is produced biotechnologically (430). It was recently found that FAD could be an efficient treatment of some inheritable diseases, e.g., chronic granulomatous diseases caused by mutations in leukocyte NADPH oxidase genes (183) and Friedreich ataxia caused by the lack of the mitochondrial protein frataxin (149).
CONSTRUCTION OF INDUSTRIAL RIBOFLAVIN PRODUCERS BY CLASSIC MUTAGENESIS AND SELECTION AND BY METHODS OF METABOLIC ENGINEERING
The history of industrial RF production has been described in detail in previous reviews (83, 178, 438, 462). After the first period of microbial production of RF based on natural microbial isolates and a subsequent period of chemical production, most RF is currently produced using genetically engineered microorganisms. Both methods of classic mutagenesis and selection and modern methods of metabolic engineering have been used. Naturally, these approaches differed depending on the organism used. In the case of B. subtilis, most effort was directed at impairment of regulation of the RF operon and amplification of the copy number of the structural gene. In A. gossypii, most attention has been paid to increasing the RF pathway supply with purine precursors and activation of the glyoxylic acid cycle, which improves catabolism of oil, the preferred carbon substrate for RF synthesis. In C. famata, improvement of RF productivity has been achieved by introducing several unidentified mutations leading to resistance against different toxic agents and, recently, by increasing the copy number of transcription factor SEF1 along with the genes involved in purine nucleotide interconversion and the RF biosynthetic pathway. As some plants overproduce RF under conditions of iron deficiency, they can be used for selection of RF-enriched plants as the source of vitamin B2 in food supplements (176).
Below, a short description of the main approaches used in the isolation of efficient RF producers from bacteria, mycelial fungi, and yeasts is given.
B. subtilis
Wild-type strains of B. subtilis do not overproduce RF. RF-overproducing mutants are easy to isolate but produce very small amounts (52). Industrial RF producers of B. subtilis have been isolated using several selection steps for resistance to different antimetabolites and subsequently introducing regulatory mutations alleviating repression by FMN and increasing the copy numbers of the RF operon and of the ribA gene, coding for the first enzyme of RF biosynthesis (178, 341–344). Additionally, RF production in B. subtilis can be increased by derepression of the ZWF1 gene, encoding glucose-6-phosphate dehydrogenase (96), and by protein engineering of GTP cyclohydrolase II to improve its kinetic characteristics (257).
Work on B. subtilis RF industrial producers started after isolation of mutants showing, successively, resistance to 8-azaguanine, decoyinine, and methionine sulfoxide (343). Mutants resistant to the purine analog 8-azaguanine often arose due to derepression of purine nucleotide synthesis (395). Decoyinine inhibits GMP synthetase, and decoyinine-resistant mutants can possess derepressed levels of this enzyme (295). Mutants resistant to methionine sulfoxide seem to possess elevated activity of IMP dehydrogenase (294). An isolated triple mutant (producing small amounts of RF [∼7 mg/liter]) was used to select roseoflavin-resistant mutants. About 1 to 2% of them grew as yellow colonies and possessed mutations in the ribC gene, coding for bifunctional RF kinase/FAD synthetase (163, 280). The yellow roseoflavin-resistant mutant RB50 accumulated elevated titers of RF (up to 40 mg/liter) due to derepression of RF operon transcription (343).
Several copies of the DNA fragment containing the RF operon were introduced into the genome of the RB50 strain, which led to a 10-fold increase in RF accumulation by the transformants (up to 700 mg/liter). Subsequently, their own RF operon promoters were replaced with the P15 promoter of B. subtilis phage SPO1. After optimization of the cultivation conditions in the fermentor for the constructed strain, 14 g RF/liter was obtained in 48 h and 15 g/liter was accumulated in 56 h (344). Finally, introduction in this strain of one additional copy of the ribA gene under the control of the P15 promoter, coding for bifunctional GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase, led to strain VB2XL1, which was characterized by a further 25% increase in RF accumulation (185). Overexpression of other individual genes of the RF operon did not increase RF synthesis (178). A similar protocol for the isolation of RF-overproducing B. subtilis strains has been used by Chinese authors (429).
However, the achieved RF yield in B. subtilis is quite low compared to the theoretical maximal yield (394). Recent work shows that B. subtilis possesses the potential for further increases in RF synthesis. A series of mutated GTP cyclohydrolases has been obtained using error-prone mutagenesis; several of them have shown improved kinetic characteristics (specific activity and Vmax). Apparently the introduction of this kinetically improved enzyme (which is known in B. subtilis to catalyze the rate-limiting reaction of the overall pathway) into industrial RF producers can further increase RF yield and productivity (257). RF synthesis in the RF-overproducing strain has been improved by a genetic defect in transketolase (141) or by redirecting carbon flow through phosphoenolpyruvate carboxinase (547). Overexpression of the ZWF1 gene, coding for glucose-6-phosphate dehydrogenase, in the RF-overproducing strain also increased the RF yield by 25% (96). Reduction of maintenance metabolism by knockout of the major cytochrome oxidase that catalyzes the less efficient branch of the respiratory chain significantly increased the yield in an RF-overproducing strain (548). Transposon mutagenesis of B. subtilis showed novel and unexpected genes involved in the regulation of RF synthesis: disruption of the fliP, yjaU, and yloN genes led to a 25 to 35% increase in RF yield, whereas transposon insertion in numerous genes (nearly 30) significantly decreased RF synthesis (493). The effect of many such insertions cannot yet be rationally explained. Overexpression of one of the identified genes, gapB (coding for gluconeogenic glyceraldehyde-3-phosphate dehydrogenase), due to knockout of its regulator ccpN, improved yield in the industrial RF producer. These data clearly show that insertion mutagenesis is a promising approach for the identification of novel genes involved in RF synthesis and their subsequent manipulation (493). Transcriptome analysis of a B. subtilis wild-type strain and the RF-overproducing mutant RH33 suggested that the potential bottleneck in RF overproduction is the low pool of phosphoribosyl pyrophosphate. Simultaneous overexpression of two genes, prs and ywlF, involved in the biosynthetic pathway of phosphoribosyl pyrophosphate from ribulose-5-phosphate in the RF overproducer RH33 increased RF production by 25% (429). Synthesis of RF by some of the constructed strains has been optimized using model-predictive control, based on artificial neural networks (242) and statistical design (532). Flux response studies of RF-overproducing B. subtilis suggested a role of the transhydrogenase-like mechanism in maintaining the balance of reducing equivalents during RF oversynthesis (381).
Genetic manipulation of central carbon metabolism could also have a positive effect on RF production in B. subtilis. For example, disruption of the pta gene (encoding phosphotransacetylase) and simultaneous overexpression of the als gene (encoding acetolactate synthase) increased RF production (554). The authors explained this phenomenon as due to an elevated intracellular level of ATP.
Other Bacteria
Corynebacterium ammoniagenes.
Corynebacterium ammoniagenes is used for the industrial production of purine and pyrimidine nucleotides (136, 317) and therefore was chosen for developing an alternative bacterial RF producer. In the first step, the structural genes of RF, located in a cluster, were cloned (234). To isolate the RF overproducer, a strong promoter of C. ammoniagenes was cloned and substituted for the natural promoter that controls the expression of the RF biosynthesis genes in the wild-type strain. This led to a 3-fold increase in RF yield (235). Additionally, the ribosome-binding sequence of the bifunctional gene coding for GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase was optimized, which strongly increased RF synthesis. Under optimized conditions, the engineered strain accumulated 15.3 g RF/liter in 72 h, which is comparable to the yield from B. subtilis. No selection for resistance to RF analogs was used in this work.
Lactic acid bacteria and propionibacteria.
Selection of roseoflavin-resistant mutants seemed an efficient approach for the isolation of RF-overproducing strains of several species of lactic acid bacteria (L. lactis, Lactobacillus plantarum, and Leuconostoc mesenteroides) as well as Propionibacterium freudenreichii (58, 486). All the analyzed mutations were localized in the riboswitch regions of RF gene clusters and not in the ribC gene, coding for RF kinase/FAD synthetase. The resulting mutants showed rather modest RF production, several thousandfold lower than that of industrial RF producers. For example, roseoflavin-resistant mutants of L. lactis accumulated to only near 1.2 mg RF/liter (486), mutants of L. mesenteroides and L. plantarum produced maximally 0.5 to 0.6 mg RF/liter, and P. freudenreichii accumulated up to 3 mg RF/liter (58). However, yogurts obtained with RF-overproducing mutants of lactic acid bacteria and propionibacteria as starter cultures could be used to increase the RF content in milk products.
A. gossypii
With A. gossypii, much attention was paid to optimization of fermentation conditions favoring RF oversynthesis (83, 268). RF overproduction occurs on different residues, including oily waste-activated bleaching earth (490). Selection for resistance to RF analogs was not reported, apparently due to the resistance of A. gossypii to these compounds (462). No information is available on A. gossypii mutants resistant to analogs of purines, although exogenous purines and the purine precursor glycine activate RF production (268). As oil stimulates RF production in A. ashbyii, activation of the glyoxylic acid cycle should be important for strain improvement. Itaconate inhibits the key enzyme of the glyoxylate shunt, isocitrate lyase, and mutants resistant to itaconate produced enhanced levels of RF (399, 400). Improvement of RF synthesis was also obtained in oxalate- and itaconate-resistant mutants (339, 481). The exact nature of these mutations was not elucidated.
The flavinogenic potential of A. gossypii was substantially improved using modern approaches of metabolic engineering. These were directed at activation of the glyoxylate cycle and supply with purine precursors of RF. At the same time, no information is available on improvement of RF production by overexpression of RF structural genes. Introduction of an additional copy of the ICL1 gene, encoding isocitrate lyase, enhanced RF production in a medium containing soybean oil (203). Overexpression of the second enzyme of the pathway, malate synthase, also improved oil consumption and RF production (480). Disruption of VMA1, encoding vacuolar ATPase, which energized active RF transport from the cytoplasm to the vacuole, resulted in complete excretion of synthesized RF into the medium and increased the total production of RF (128). However, disruption of VMA1 in the flavinogenic yeast P. guilliermondii also resulted in RF oversynthesis, although normally this organism does not accumulate the RF produced in vacuoles (41a).
More attention has been paid to activation of RF synthesis in A. gossypii with the purine precursor. Overexpression of GLY1, encoding the glycine biosynthetic enzyme threonine aldolase, strongly enhanced RF production, apparently due to an improved supply of glycine, the purine precursor (312). An increase in RF production was also achieved by disruption of the SHM2 gene, encoding one of two isoenzymes of serine hydroxymethyltransferase, due to a defect in the conversion of glycine to serine, which again improved the glycine supply (398). Conversion of glyoxylate into glycine was improved by heterologous expression of the alanine:glyoxylate aminotransferase gene from S. cerevisiae (217). To enhance the metabolic flux through the purine nucleotide biosynthesis pathway, the key regulatory gene ADE4, encoding phosphoribosyl pyrophosphate amidotransferase, was cloned and overexpressed under the control of the strong constitutive promoter of glyceraldehyde phosphate dehydrogenase to abolish repression caused by exogenous adenine (195). To alleviate feedback inhibition of this enzyme by ATP and GTP, site-specific mutagenesis of the ADE4 gene by replacement of three amino acid residues was carried out. Constitutive overexpression of this engineered enzyme resulted in a 10-fold increase in RF synthesis (195). A. gossypii contains two phosphoribosyl pyrophosphate synthetases, encoded by genes PRS2,4 and PRS3. Overexpression of each of them resulted in an increase of RF production. The activity of these enzymes is feedback inhibited by GDP. Two conserved amino acid residues in the products of the PRS2,4 and PRS3 genes were changed to release this inhibition. Overexpression of the engineered PRS2,4 and PRS3 genes produced strains with further elevated RF production, almost twice that of the wild-type strain (196). A protocol for insertion mutagenesis of A. gossypii has been developed (391), and a disruption mutant defective in gene BAS1 (encoding a member of the Myb family of transcription factors) has been tagged. Corresponding disruption and knockout strains overexpressed RF (292). BAS1 proved to be involved in adenine-mediated repression of the de novo biosynthesis of purine nucleotides. Furthermore, it participates in the regulation of the glycine pathway, spore germination, and the duration of the trophic phase. Deletion of the C-terminal part of BAS1 resulted in constitutive activation of the adenine and glycine pathways, thereby enhancing RF overproduction (292).
Data indicating that A. gossypii industrial RF producers accumulate >15 g RF/liter (462) did not take into account more recent publications, and so current producers could accumulate even higher titers of RF.
E. ashbyii
E. ashbyii, which is closely related to A. gossypii (304, 336), had been used for industrial RF production in the United States (462) and in Pinsk (Belarus) in the former Soviet Union. However, these businesses were shut down, mainly due to the relatively low productivity of E. ashbyii (well below 4 g RF/liter) (207) and its high genetic instability (475). However, E. ashbyii, in contrast to A. gossypii, overproduces, in addition to RF, small amounts of FAD (291, 534). As FAD has pharmaceutical and neutraceutical applications and is produced commercially (430), E. ashbyii still possesses its own biotechnological potential.
Much attention has been paid to optimization of media and cultivation conditions for E. ashbyii (34, 207, 238, 336, 337). Mutants resistant to 8-azaguanine with RF production enhanced by 2 to 4 times have been isolated (474). No tools for molecular research on E. ashbyii have been reported.
C. famata (C. flareri)
The yeast industrial RF producer C. famata dep8 (ATCC 20849) was isolated by conventional mutagenesis from strain C. famata VKM Y-9 without application of molecular tools. It needs to be pointed out that strain VKM Y-9 has been recently reidentified as C. flareri (322). However, as scientific and patent literature designates the yeast RF producer C. famata, this name is used throughout this review.
Selection consisted of 10 steps of mutagenesis and one step of protoplast fusion (168–170). At the first stage of selection, a mutant with deregulated purine synthesis growing as a pink colony was isolated. It accumulated 3-fold more RF than the wild-type strain. Independently, the wild-type strain was mutagenized and a mutant overproducing RF in the presence of 5′-AMP was picked up with a 6-fold increased productivity. Both these mutants were used for protoplast fusion, of which one hybrid colony accumulated RF crystals inside the cells, produced 14-fold more RF, and was picked up for further selection. After mutagenesis and plating on a poor medium, one mutant growing as a yellow colony was selected. Its RF productivity had increased 27-fold relative to the wild-type strain. Further, mutants resistant to 2-deoxyglucose were isolated, as it had been suggested that resistant strains should possess enhanced glucose conversion to RF. The most flavinogenic of them was used at the next step, where the adenine analog 4-aminopyrazolo-(3,4-d)pyrimidine (a known inhibitor of purine biosynthesis) (351) was used as the selection agent. The best selected strain accumulated up to 7.5 g RF/liter after 6 days of cultivation in shake flasks and 1.1 g RF/liter in test tubes. This strain was mutagenized and selected for resistance to 2-deoxyglucose. The most flavinogenic mutant, designated strain D, accumulated 1.58 mg RF/liter in test tubes. Mutagenized cells of this strain D were plated onto a medium with a high concentration of FeCl3, and yellow colonies were picked up. The most flavinogenic strain was mutagenized and plated onto an exhausted medium obtained by filtering the culture medium after prolonged cultivation of this strain supplemented with glycine and sucrose. Several yellow colonies were picked up, of which the most flavinogenic mutant accumulated 2.5 g RF/liter and was designated strain A (also dep8, deposited as ATCC 20849). Only this strain was used for industrial production of RF.
Several attempts were made to isolate even more productive strains. For this, strain dep8 was mutagenized and plated onto a diluted exhausted medium obtained after its cultivation for 10 days with supplements of sucrose and glycine. Among the mutants growing in this medium, strain F accumulated 3.4 g RF/liter of medium. At the next stage, cells of strain F were mutagenized and plated onto a medium with the antibiotic adenosine analog tubercidin (7-deaza-adenosine). The most flavinogenic strains, B and G, accumulated up to 3.8 mg RF/liter. As their high flavinogenic activity was an unstable feature, strain B was used for isolation of a UV-sensitive mutant. The corresponding strain, designated H, accumulated 3.1 g RF/liter (168, 170). The molecular nature of the introduced mutations is unknown. Instability remains the most serious drawback in the industrial strain dep8.
Cultivation conditions in fermentors that favored maximal RF accumulation were developed for strain A (dep8). In a 14-liter fermentor, 16 g RF/liter was accumulated after 240 h of cultivation, whereas in a 450-liter fermentor, the concentration reached 21 g/liter after 200 h (169, 170).
Strain C. famata dep8 appeared to be rather unstable, reverting to totally nonflavinogenic variants, although these differed from the wild-type strains of C. famata because iron deficiency did not stimulate RF production by revertants (K. Dmytruk, O. Lyzak, and A. Sibirny, unpublished data). It was also observed that strain dep8 failed to use ethanol as the sole carbon source, whereas the wild-type strain and nonflavinogenic revertants normally grow on ethanol. The reasons for the defects in ethanol utilization, and correlations between the inability to utilize ethanol and RF overproduction in strain dep8, remain unknown. Reversions occurred invariably as a result of frameshifts and creation of nonsense codons in the earlier-identified gene SEF1, coding for a putative transcription factor (91; Dmytruk et al., unpublished data). Introduction of an additional copy of the SEF1 gene ortholog isolated from the sequenced strain D. hansenii CBS 767 into strain dep8 greatly improved the stability of the resultant transformants; moreover, it resulted in enhancement of RF production (Dmytruk et al., unpublished data).
An independent C. famata RF overproducer has recently been isolated using a combination of random mutagenesis and rational approaches of metabolic engineering. First, the RF-overproducing strain AF-4 was isolated from the wild-type strain C. famata VKM Y-9 in six consecutive steps of conventional mutagenesis (439). The selection scheme is given in Table 2. In the first step, mutants resistant to the RF structural analog 7-methyl-8-trifluoromethyl-10-(1′-d-ribityl)isoalloxazine were selected. Isolation of the resistant mutants was carried out on plates with the RF analog (200 mg/liter) and 0.6 M K2SO4. The most flavinogenic strain was used for isolation of mutants resistant to 8-azaguanine (200 mg/liter). The most flavinogenic strain was used in the third step of selection with 6-azauracil (200 mg/liter). 6-Azauracil inhibits yeast IMP dehydrogenase (107), and exogenous guanine alleviates the inhibitory action of 6-azauracil on C. famata growth. The most flavinogenic strain was selected for resistance to 6-diazo-5-oxo-l-norleucine (5 mg/liter), which inhibits purine biosynthesis (71), and the inhibition of C. famata growth by 6-diazo-5-oxo-l-norleucine was restored with exogenous guanine. It was unexpectedly found that C. famata growth was inhibited by the natural nucleoside guanosine (100 mg/liter), and this inhibition could not be restored by guanine. It was decided to isolate guanosine-resistant mutants and check them for RF oversynthesis. The best RF producer obtained in the previous step was mutagenized and plated with guanosine (300 mg/liter). In the last step of the conventional selection procedure, the original observation that cultivation of C. famata RF-overproducing mutants on plates with a buffered medium at pH 6.8 to 7.0 strongly inhibited their RF production but did not suppress growth and the colonies appear white, was used. After mutagenesis and plating, the best RF producer was isolated in medium with guanosine on plates containing 0.1 M phosphate buffer (pH 6.8), and following an analysis of ∼3,000 colonies, brightly yellow ones were picked up. By testing them in liquid medium, the most flavinogenic mutant, designated AF-4, was selected, which accumulated about 680 mg RF/liter, whereas the wild-type strain under these conditions accumulated only 2 to 3 mg RF/liter (92, 439) (Table 2).
Table 2.
Steps for selection of C. famata RF-overproducing strain AF-4
| Step | Strain | Selective factor | Riboflavin, mg/litera |
|---|---|---|---|
| I | AA | 7-Methyl-8-trifluoromethyl-10- (1′-d-ribityl)isoalloxazine | 35 |
| II | AB | 8-Azaguanine | 78 |
| III | AC | 6-Azauracil | 176 |
| IV | AD | 2-Diazo-5-oxo-l-norleucine | 380 |
| V | AE | Guanosine | 450 |
| VI | AF-4 | Search for yellow colonies at high pH | 688 |
Riboflavin synthesis was analyzed on a semisynthetic medium supplemented with yeast extract on the fifth day of incubation on a shaker (200 rpm).
This strain, AF-4, was relatively stable and, in contrast to industrial strain C. famata dep8, did not revert to RF-nonoverproducing revertants during several (∼10) years of storage on agar slants and cultivation. Its flavinogenic activity was, however, ∼30% lower than that of the industrial RF producer C. famata dep8 (Fig. 7). Molecular events that evolved during each step of selection of strain AF-4 have not been studied.
Fig. 7.
Evaluation of the improved C. famata riboflavin overproducers isolated by metabolic engineering. RF synthesis was assayed after 5 days of cultivation in yeast extract-peptone-dextrose (YPD) medium. (Reproduced from reference 92 with permission of Elsevier.)
Metabolic engineering approaches for the selection of the advanced RF producer involved introduction of additional copies of transcription factor gene SEF1 and IMH3 (encoding IMP dehydrogenase) orthologs from D. hansenii and the homologous genes RIB1 (encoding GTP cyclohydrolase II) and RIB7 (encoding RF synthase), which encoded the first and last enzymes of the RF biosynthesis pathway. Overexpression of all these genes in the RF overproducer AF-4, obtained by classical selection, resulted in a 4.1-fold increase of RF production in shake flask experiments, although separate introduction of an additional one or two copies of SEF1and overexpression of IMH3 also had considerable positive effects on RF production (92) (Fig. 7). The constructed strain is also much more productive than the industrial strain C. famata dep8. These authors believe that the constructed strain might well be employed in biotechnological production of vitamin B2 because it is very productive and nonreverting.
P. guilliermondii
The flavinogenic potential of P. guilliermondii is weaker than that of C. famata (C. flareri), and thus the known P. guilliermondii RF overproducers cannot compete with those isolated from other organisms (245). Nevertheless, some P. guilliermondii mutants, especially those capable of RF uptake and accumulation, can also be applied in biotechnology. RF permease entirely lacks the ability to transport FMN, though it efficiently takes up and accumulates RF in cells (449). It was proposed that this feature could be used to concentrate RF inside cells from diluted solutions of this vitamin to thereby enrich yeast with RF (453) and to separate RF from FMN, which is important for assaying RF kinase (215). RF-overproducing mutants with genetic defects in RF excretase accumulate all synthesized RF inside the cells. The RF content of these cells exceeds 500 to 1,000 times the normal RF content in yeast cells (445). Dried RF-enriched biomass can be directly used as an inexpensive source of RF for animal feeding. RF prototrophic strains of P. guilliermondii overproducing the RF biosynthetic precursor 6,7-dimethyl-8-ribityllumazine have been isolated (446).
Methylotrophic Yeasts
Methylotrophic yeasts grown on methanol synthesize huge amounts of the FAD-containing enzyme alcohol oxidase, which catalyzes the first reaction of methanol catabolism (338). Under certain cultivation conditions, alcohol oxidase can comprise nearly 30% of soluble protein in the cells. Correspondingly, during methylotrophic growth, yeast cells contain an elevated content of FAD (up to 1.5 mg FAD/g dry biomass), with most of it being bound to flavoproteins (6, 89, 500). Methanol induces synthesis of alcohol oxidase as well as RF synthase, RF kinase, and FAD synthetase in methylotrophic yeasts (55, 102, 431, 432). The mechanisms of this induction are not known. A mutant of Hansenula polymorpha unable to grow on methanol was characterized by a 10-fold decrease in both the amount of alcohol oxidase protein and the level of total intracellular FAD (498). The content of free intracellular FAD, however, was identical in methanol-induced cells of the wild-type strain and also in the mutant of the glucose-grown cells of both strains. The content of FAD in another FAD-containing enzyme, d-amino acid oxidase, remained unchanged in the mutant cells. Apparently, the absence of the apoprotein in some way prevents induction of FAD-synthesizing enzymes. It is noteworthy that single mutations in the H. polymorpha gene GCR1, coding for a glucose transporter homolog, allow constitutive synthesis of FAD-containing alcohol oxidase in a glucose medium without methanol (464). The available data suggest an involvement of alcohol oxidase apoprotein in the regulation of FAD synthesis. High-cell-density cultures of methylotrophic yeasts grown on methanol accumulate flavins, including free RF (482). However, the amounts of accumulated flavins in culture liquids of wild-type cells of methylotrophic yeasts are very small.
Overexpression of the first gene of RF synthesis (P. pastoris RIB1 or a heterologous gene) coding for GTP cyclohydrolase II, under the control of a strong constitutive promoter of glyceraldehyde-3-phosphate dehydrogenase substantially increased RF production in Pichia pastoris. Overexpression of all 6 structural genes of this pathway led to accumulation of 20 mg RF/liter in a glucose medium during shake flask experiments and of 175 mg RF/liter during high-density cultivation in a fermentor (288). The data show that RF producers can be engineered in nonflavinogenic yeasts; however, the RF yield achieved is very low relative to those found in industrial producers. Apparently the RF pathway in the constructed strain could not be sufficiently supplied with the purine precursor GTP. It could be interesting to incubate cells of the constructed strain with methanol, which, being the natural inducer of flavin synthesis, could provide cells with abundant amounts of some signals or substrates which limit RF oversynthesis and are produced in insufficient amounts during glucose cultivation.
CONSTRUCTION OF PRODUCERS OF FLAVIN NUCLEOTIDES
Readers are referred to a recent review on construction of flavin nucleotide producers (540). Flavin nucleotides are used in the pharmaceutical and food industries because they are more efficient than RF in treating some diseases and have significantly higher solubility in water. FMN (RF-5′-phosphate) is currently produced by chemical phosphorylation of RF; however, even the most purified preparations of chemically synthesized FMN contain ∼25% of impurities of a flavin nature, i.e., isomeric RF phosphates, RF cyclophosphates, and RF bisphosphates (33, 324). These compounds can act as antimetabolites and thus be toxic. FAD is produced biotechnologically, with an annual production of 10 tons (430); however, this compound is much more expensive than RF. FAD (95% purity) is ∼1,000 times more expensive than RF, and synthetic FMN containing 30% impurities is even more expensive (see http://www.sigmaaldrich.com/united-states.html for prices).
FAD could be isolated from the mycelium of E. ashbyii (291, 534) or produced by biotransformation of exogenous FMN or RF and ATP using bacterial cells. The first process in FAD production used a mutant of Sarcina lutea that is defective in adenosine deaminase in a medium supplemented with FMN. d-Cycloserine stimulated this process, and the maximal yield of FAD reached 0.7 g/liter after 5 days of cultivation (383, 520). Enzymatic conversion of FMN and ATP to FAD using FAD synthetase from Arthrobacter globiformis is known but is very expensive (433). More recently, efficient transformation of exogenous FMN and ATP to FAD was achieved using cells of a recombinant strain of Brevibacterium ammoniagenes with a 20-fold derepression of bifunctional RF kinase/FAD synthetase (164). Under optimal conditions, 1.2 g FAD/liter was produced from exogenously added FMN and ATP. When metaphosphate was used as the phosphate donor instead of ATP, the recombinant strain accumulated FMN from RF and ATP without the production of FAD. FMN production reached 10 g/liter (320). Heterologous overexpression of bifunctional RF kinase/FAD synthetase in E. coli, Enterobacter, and Pseudomonas resulted in increased maximal concentrations of FAD in a medium with FMN and ATP of up to 18 g/liter. A strong disadvantage of these processes is that expensive additives (FMN and especially ATP) are used in high concentrations. Apparently there is an intrinsic drawback in the development of bacterial organisms for production of flavin nucleotides because FMN is a corepressor of RFN riboswitches, which prevents accumulation of large amounts of FMN and FAD during de novo biosynthesis.
Flavinogenic yeasts possess a quite different regulatory system where normally iron, and not flavin nucleotides, represses the enzymes involved in RF biosynthesis. Therefore, it is possible to construct yeast strains that are capable of FMN and FAD oversynthesis de novo. To construct an effective producer of FMN, FMN1, encoding RF kinase, has been cloned from the sequenced flavinogenic strain D. hansenii CBS 767 and overexpressed in C. famata under the control of its own strong regulatory promoter RIB1 and its own strong constitutive promoter TEF1. C. famata transformants with several (6 to 8) integrated copies of the native D. hansenii FMN1 gene under the control of the TEF1 promoter had a 200-fold-enhanced activity of RF kinase compared to the wild-type strain and accumulated much larger amounts of FMN in an iron-deficient medium (in which no FMN accumulation occurred) (193). To construct FMN producers that can overproduce this nucleotide in an iron-rich medium, the D. hansenii FMN1 gene was integrated in several (5 or 6) copies in the C. famata RF-overproducing strain AF-4. The resulting transformants had high RF kinase activity and accumulated 130 mg FMN/liter in the culture medium after 24 h of cultivation (542). Optimization of the medium content and cultivation conditions raised this to nearly 300 mg FMN/liter in 30 h (Fig. 8) (541).
Fig. 8.
RF kinase activity (A) and FMN accumulation in the culture medium (B) by C. famata strains overexpressing the FMN1 gene. wt, wild type. (Reproduced from reference 542 with permission of Elsevier.)
To construct an FAD overproducer, the gene FAD1 of D. hansenii, encoding FAD synthetase under the control of TEF1 promoter, was integrated in 2 copies into a strain capable of FMN overproduction. The resulting transformants had a 15-fold-increased activity of FAD synthetase compared to the parental strain, and in optimized medium they accumulated 450 mg FAD/liter after 40 h of cultivation (V. Yatsyshyn, D. Fedorovych, and A. Sibirny, unpublished data). Constructed producers of FMN and FAD still accumulate a large portion of RF in the culture medium, which could be explained by the high activity of FMN- and FAD-degrading enzymes in these cases. To construct more efficient overproducers of FMN and FAD, multicopy integration of FMN1 and FAD1 genes has to be achieved in the more efficient RF producers that are available (92), and genes responsible for hydrolysis of FMN and FAD have to be inactivated.
UNRESOLVED ISSUES AND FUTURE PROSPECTS
There are unresolved questions in both basic and applied research on flavin synthesis. In the biochemistry of RF synthesis, one stage—namely, dephosphorylation of 5-amino-6-ribitylamino-2,4(1H,3H)-pyrimidinedione 5′-phosphate—remains to be deciphered. In the physiology of RF synthesis, the significance of the very frequent secretion of part of the synthesized RF into the medium remains a mystery. Such secretory activity could be the result of imprecise regulation of this minor biosynthetic pathway, which was suggested many years ago (83). It cannot be excluded, however, that secreted RF plays some role, such as being involved in iron mobilization or oxidative stress defense. Especially important are investigations into the reasons for and mechanisms of RF overproduction in some bacteria, flavinogenic yeasts, and plants under conditions of iron deficiency. There are apparently strong reasons for the wide distribution of iron-dependent repression of RF synthesis among prokaryotes, fungi, and plants.
In applied studies of flavinogenesis, the search for potent and specific inhibitors of RF synthesis as effective anti-infectives remains a promising and important task. For Gram-negative bacteria and pathogenic fungi, which are incapable of RF transport into the cell, investigations need to be concentrated on the search for specific and effective inhibitors of the intermediate reactions of RF synthesis. For Gram-positive bacteria, which efficiently transport exogenous RF, the search for RF and roseoflavin analogs that are not converted to analogs of flavin nucleotides, while they potently block RF biosynthesis through riboswitch binding, will continue. Much needs to be done to improve current RF biotechnological producers since they are not all free of serious drawbacks. Low productivity is apparently the most important problem, as usually not more than 4% of the carbon source is converted to RF in the available strains. For yeast and bacterial industrial producers, genetic stability is an additional concern. For many purposes, RF has to be replaced by flavin coenzymes, especially in medicine and the food industry. However, current biotechnological producers of FMN and FAD give low productivities and yields. These problems nevertheless provide a good perspective for successful developments in the nearest future.
ACKNOWLEDGMENTS
A.A.S. is grateful to colleagues from the Institute of Cell Biology, NAS of Ukraine, Lviv, namely, Y. R. Boretsky, K. V. Dmytruk, D. V. Fedorovych, V. M. Ubiyvovk, and V. Y. Yatsyshyn, for critical reading of the manuscript and stimulating discussions. We are grateful to Denys N. Wheatley for helpful comments on the manuscript and to BioMedES Limited (Aberdeen, United Kingdom) for help in presenting this review in the best possible English.
Biographies

Charles A. Abbas holds a B.S. in microbiology from the University of Minnesota, an M.S. in microbiology from the University of Montana, and a Ph.D. in microbiology and cell science from the University of Florida. Currently he is the Director of Yeast and Renewable Research at Archer Daniels Midland Company. Areas of research include bioprocessing of commodity crops to high-value-added products and development of yeast strains for the large-scale microbial fermentation production of ethanol, polymers, amino acids, enzymes, vitamins, carotenoids, and organic acids. Dr. Abbas was the recipient of the Alltech Medal of Excellence for Alcohol Production in 2001, was Chair of ASM Division O, has given numerous talks at scientific meetings, and has chaired sessions on topics including yeast biotechnological applications, emerging biorefineries, food security, and system biology. He is the author of more than 30 research articles, review articles, and book chapters, more than 100 abstracts, and over a dozen patents and patent applications.

Andriy A. Sibirny graduated from Lviv National University (Ukraine); his Ph.D. and Dr.Sc. studies on yeast biochemistry and genetics were performed at Lviv Division of Institute of Biochemistry, NAS of Ukraine. He had sabbaticals in Russia, Switzerland, the United States, and Italy. Currently, he works as Director and Head of the Department of Molecular Genetics and Biotechnology, Institute of Cell Biology, NAS of Ukraine (Lviv). He is also the Head of the Department of Biotechnology and Microbiology, University of Rzeszow (Poland). His scientific interests include yeast cell biology, namely, mechanisms of autophagic peroxisome degradation, catabolite repression, and stress response, as well as yeast biotechnology, which involve regulation of riboflavin synthesis, alcoholic fermentation, and production of heterologous proteins by nonconventional yeasts. He is the Chair of the International Commission on Yeasts, the Corresponding Member of the NAS of Ukraine, and the President of the Ukrainian Society for Cell Biology.
Footnotes
This review is dedicated to the memory Prof. Georgiy M. Shavlo-vsky, one of the pioneers in studies of riboflavin biosynthesis by yeasts, the mentor of one of the authors (A.A.S.).
REFERENCES
- 1. Abbas C., et al. March 2007. Transformation systems for flavinogenic yeast. U.S. patent 7,009,045
- 2. Akiyama T., Selhub J., Rosenberg I. H. 1982. FMN phosphatase and FAD pyrophosphatase in rat intestinal brush borders: role in intestinal absorption of dietary riboflavin. J. Nutr. 112:263–268 [DOI] [PubMed] [Google Scholar]
- 3. Alexopoulos C. J., Mims C. W., Blackwell M. 1996. Introductory mycology, 4th ed. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 4. Ammelburg M., et al. 2007. A CTP-dependent archaeal riboflavin kinase forms a bridge in the evolution of cradle-loop barrels. Structure 15:1577–1590 [DOI] [PubMed] [Google Scholar]
- 5. Asai S., Mase K., Yoshioka H. 2010. A key enzyme for flavin synthesis is required for nitric oxide and reactive oxygen species production in disease resistance. Plant J. 62:911–924 [DOI] [PubMed] [Google Scholar]
- 6. Ashin V. V., Trotsenko Y. A. 1998. Formation and distribution of modified FAD between isozymes of alcohol oxidase in the methylotrophic yeast Pichia methanolica. Biochemistry (Moscow) 63:1407–1413 [PubMed] [Google Scholar]
- 7. Audley B. G., Goodwin T. W. 1962. Studies on the biosynthesis of riboflavin. 7. The incorporation of adenine and guanine into riboflavin and into nucleic acid purines in Eremothecium ashbyii and Candida flareri. Biochem. J. 84:587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Babcock M., et al. 1997. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276:1709–1712 [DOI] [PubMed] [Google Scholar]
- 9. Babyak L. Y., Sibirnyi A. A., Shavlovskii G. M. 1993. Selection and some properties of the mutants rib81 with impaired regulation of riboflavin biosynthesis. Tsitol. Genet. 27:28–32 (In Russian.) [Google Scholar]
- 10. Bacher A. 1991. Biosynthesis of flavins, p. 215–249 In Müller F. (ed.), Chemistry and biochemistry of flavoenzymes, vol. 1 CRC Press, Boca Raton, FL [Google Scholar]
- 11. Bacher A., Bauer R., Eggers U., Harders H., Schnepple H. 1976. Riboflavin synthases of Bacillus subtilis, p. 729–732 In Singer T. P. (ed.), Flavins and flavoproteins. Elsevier, Amsterdam, Netherlands [Google Scholar]
- 12. Bacher A., et al. 1980. Riboflavin synthases of Bacillus subtilis. Purification and properties. J. Biol. Chem. 255:632–637 [PubMed] [Google Scholar]
- 13. Bacher A., et al. 2001. Biosynthesis of riboflavin. Vitam. Horm. 61:1–49 [DOI] [PubMed] [Google Scholar]
- 14. Bacher A., Eberhardt D., Fischer M., Kis K., Richter G. 2000. Biosynthesis of vitamin B2 (riboflavin). Annu. Rev. Nutr. 20:153–167 [DOI] [PubMed] [Google Scholar]
- 15. Bacher A., et al. 1997. Biosynthesis of riboflavin: lumazine synthase and riboflavin synthase. Methods Enzymol. 280:389–399 [DOI] [PubMed] [Google Scholar]
- 16. Bacher A., Lingens F. 1970. Biosynthesis of riboflavin. Formation of 2,5-diamino-6-hydroxy-4(1′-d-ribitylamino)pyrimidine in riboflavin auxotroph. J. Biol. Chem. 245:4647–4652 [PubMed] [Google Scholar]
- 17. Bacher A., Lingens F. 1971. Biosynthesis of riboflavin. Formation of 6-hydroxy-2,4,5-triaminopyrimidine in RIB7 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 246:7018–7022 [PubMed] [Google Scholar]
- 18. Bacher A., Mailander B. 1973. Biosynthesis of riboflavin. The structure of the purine precursor. J. Biol. Chem. 248:6227–6231 [PubMed] [Google Scholar]
- 19. Bacher A., Mailander B. 1978. Biosynthesis of riboflavin in Bacillus subtilis: function and genetic control of the riboflavin synthase complex. J. Bacteriol. 134:476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bacher A., et al. 1993. Biosynthesis of flavins, p. 147–192 In Dugas H., Schmidtchen F. P. (ed.), Bioorganic chemistry frontiers. Springer Verlag, Berlin, Germany [Google Scholar]
- 21. Bacher A., et al. 1997. Biosynthesis of riboflavin: GTP cyclohydrolase II, deaminase, and reductase. Methods Enzymol. 280:382–389 [DOI] [PubMed] [Google Scholar]
- 22. Bafunno V., et al. 2004. Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J. Biol. Chem. 279:95–102 [DOI] [PubMed] [Google Scholar]
- 23. Bandrin S. V., Beburov M. I., Rabinovich P. M., Stepanov A. I. 1979. Riboflavin auxotrophs of Escherichia coli. Genetika 15:2063–2065 (In Russian.) [PubMed] [Google Scholar]
- 24. Barile M., et al. 1997. Flavin adenine dinucleotide and flavin mononucleotide metabolism in rat liver—the occurrence of FAD pyrophosphatase and FMN phosphohydrolase in isolated mitochondria. Eur. J. Biochem. 249:777–785 [DOI] [PubMed] [Google Scholar]
- 25. Barrick J. E., Breaker R. R. 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 8:R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bauer S., et al. 2003. Crystal structure of Schizosaccharomyces pombe riboflavin kinase reveals a novel ATP and riboflavin-binding fold. J. Mol. Biol. 326:1463–1473 [DOI] [PubMed] [Google Scholar]
- 27. Baugh C. M., Krumdieck C. L. 1969. Biosynthesis of riboflavine in Corynebacterium species: the purine precursor. J. Bacteriol. 98:1114–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beach R., Plaut G. W. 1969. The formation of riboflavin from 6,7-dimethyl-8-ribityllumazine in acid media. Tetrahedron Lett. 40:3489–3492 [DOI] [PubMed] [Google Scholar]
- 29. Beach R. L., Plaut G. W. 1970. Stereospecificity of the enzymatic synthesis of the o-xylene ring of riboflavin. J. Am. Chem. Soc. 92:2913–2916 [DOI] [PubMed] [Google Scholar]
- 30. Beinert H. 1960. Flavin coenzymes, p. 339–416 In Boyer P., Lardy H., Myrback K. (ed.), The enzymes, vol. 2 Academic Pres, New York, NY [Google Scholar]
- 31. Bender D. A. 2003. Nutritional biochemistry of the vitamins, 2nd ed., p. 172–199 Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 32. Bereswill S., et al. 1998. Hemolytic properties and riboflavin synthesis of Helicobacter pylori: cloning and functional characterization of the ribA gene encoding GTP-cyclohydrolase II that confers hemolytic activity to Escherichia coli. Med. Microbiol. Immunol. 186:177–187 [DOI] [PubMed] [Google Scholar]
- 33. Berezovskii V. M. 1973. Chemistry of vitamins. Food Industry Publishing House, Moscow, USSR [Google Scholar]
- 34. Bigelis R. 1989. Industrial products of biotechnology: application of gene technology, p. 243 In Rehm H. J., Reed G. (ed.), Biotechnology, vol. 7b VCH, Weinheim, Germany [Google Scholar]
- 35. Blau N., Niederwieser A. 1985. GTP-cyclohydrolases: a review. J. Clin. Chem. Clin. Biochem. 23:169–176 [PubMed] [Google Scholar]
- 36. Blount K. F., Breaker R. R. 2006. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 24:1558–1564 [DOI] [PubMed] [Google Scholar]
- 37. Bonomi H. R., et al. 2010. An atypical riboflavin pathway is essential for Brucella abortus virulence. PLoS One 5:e9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boretsky Y. R., et al. 2005. Positive selection of mutants defective in transcriptional repression of riboflavin synthesis by iron in the flavinogenic yeast Pichia guilliermondii. FEMS Yeast Res. 5:829–837 [DOI] [PubMed] [Google Scholar]
- 39. Boretsky Y. R., et al. 2007. Mutations and environmental factors affecting regulation of riboflavin synthesis and iron assimilation also cause oxidative stress in the yeast Pichia guilliermondii. J. Basic Microbiol. 47:371–377 [DOI] [PubMed] [Google Scholar]
- 40. Boretsky Y. R., et al. 2007. Development of a transformation system for gene knock-out in the flavinogenic yeast Pichia guilliermondii. J. Microbiol. Methods 70:13–19 [DOI] [PubMed] [Google Scholar]
- 41. Boretsky Y., et al. 1999. Identification of an ARS element and development of a high efficiency transformation system for Pichia guilliermondii. Curr. Genet. 36:215–221 [DOI] [PubMed] [Google Scholar]
- 41a. Boretsky Y. R., et al. 2011. Identification of the genes affecting regulation of riboflavin synthesis in the flavinogenic yeast Pichia guilliermondii using insertion mutagenesis. FEMS Yeast Res. 11:307–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bornemann S. 2002. Flavoenzymes that catalyse reactions with no net redox change. Nat. Prod. Rep. 19:761–772 [DOI] [PubMed] [Google Scholar]
- 43. Boshoff H. I., Barry C. E., III 2006. Is the mycobacterial cell wall a hopeless drug target for latent tuberculosis? Drug Disc. Today Dis. Mech. 3:237–245 [Google Scholar]
- 44. Bowling K. M., et al. 2008. Direct binding of GTP cyclohydrolase and tyrosine hydroxylase: regulatory interactions between key enzymes in dopamine biosynthesis. J. Biol. Chem. 283:31449–31459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brachat S., et al. 2003. Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol. 4:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bracher A., et al. 1998. Biosynthesis of pteridines. NMR studies on the reaction mechanisms of GTP cyclohydrolase I, pyruvoyltetrahydropterin synthase, and sepiapterin reductase. J. Biol. Chem. 273:28132–28141 [DOI] [PubMed] [Google Scholar]
- 47. Braden B. C., Velikovsky C. A., Cauerhff A. A., Polikarpov I., Goldbaum F. A. 2000. Divergence in macromolecular assembly: X-ray crystallographic structure analysis of lumazine synthase from Brucella abortus. J. Mol. Biol. 297:1031–1036 [DOI] [PubMed] [Google Scholar]
- 48. Bresler S. E., Glazunov E. A., Chernik T. P., Perumov D. A. 1973. Study of the operon of riboflavin biosynthesis in Bacillus subtilis. V. Flavin mononucleotide and flavin adenine dinucleotide as effectors in the operon of riboflavin biosynthesis. Genetika 9:84–91 (In Russian.)4619996 [Google Scholar]
- 49. Bresler S. E., Glazunov E. A., Gorinchuk G. F., Chernik T. P., Perumov D. A. 1978. Riboflavin biosynthesis operon of Bacillus subtilis. XIV. Operator-constitutive mutants. Genetika 14:1530–1538 [PubMed] [Google Scholar]
- 50. Bresler S. E., Glazunov E. A., Perumov D. A. 1972. Study of the operon of riboflavin biosynthesis in Bacillus subtilis. IV. Regulation of the synthesis of riboflavin synthetase. Study of riboflavin transport through cell envelope. Genetika 8:109–117 (In Russian.) [PubMed] [Google Scholar]
- 51. Bresler S. E., Glazunov E. A., Perumov D. A., Chernik T. P. 1977. Riboflavin biosynthesis operon of Bacillus subtilis. XIII. Genetic and biochemical study of mutants with regard to intermediate stages of biosynthesis. Genetika 13:2006–2016 (In Russian.) [PubMed] [Google Scholar]
- 52. Bresler S. E., Kalinin V. L., Kriviskiy A. S., Perumov D. A. 1969. The mutant of Bacillus subtilis producing large amounts of riboflavin. Genetika 5:133–138 (In Russian.) [Google Scholar]
- 53. Briggs W. R., Christie J. M. 2002. Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 7:204–210 [DOI] [PubMed] [Google Scholar]
- 54. Brizio C., et al. 2006. Over-expression in Escherichia coli and characterization of two recombinant isoforms of human FAD synthetase. Biochem. Biophys. Res. Commun. 344:1008–1016 [DOI] [PubMed] [Google Scholar]
- 55. Brooke A. G., Dijkhuizen L., Harder W. 1986. Regulation of flavin biosynthesis in the methylotrophie yeast Hansenula polymorpha. Arch. Microbiol. 145:62–70 [Google Scholar]
- 56. Brown J. A., Marsh M. M. 1952. Paper chromatographic separation and determination of some water-soluble vitamins. Anal. Chem. 24:1952–1956 [Google Scholar]
- 57. Burgess C. M., et al. 2006. The riboflavin transporter RibU in Lactococcus lactis: molecular characterization of gene expression and the transport mechanism. J. Bacteriol. 188:2752–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Burgess C. M., Smid E. J., Rutten G., van Sinderen D. 2006. A general method for selection of riboflavin-overproducing food grade micro-organisms. Microb. Cell Fact. 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Burkholder P. R. 1943. Synthesis of riboflavin by yeast. Proc. Natl. Acad. Sci. U. S. A. 29:166–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Burrows R. B., Brown G. M. 1978. Presence of Escherichia coli of a deaminase and a reductase involved in biosynthesis of riboflavin. J. Bacteriol. 136:657–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Callahan S. M., Dunlap P. V. 2000. LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J. Bacteriol. 182:2811–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Callahan S. M., et al. 2008. Controlled inactivation of recombinant viruses with vitamin B2. J. Virol. Methods 148:132–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Campbell G. R. O., et al. 2006. Sinorhizobium meliloti bluB is necessary for production of 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc. Natl. Acad. Sci. U. S. A. 103:4634–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Campuzano V., et al. 1996. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271:1423–1427 [DOI] [PubMed] [Google Scholar]
- 65. Cecchini G., Perl M., Lipsick J., Singer T. P., Kearney E. B. 1979. Transport and binding of riboflavin by Bacillus subtilis. J. Biol. Chem. 254:7295–7301 [PubMed] [Google Scholar]
- 66. Cerletti P., Strom R., Giordano M. G., Barra D., Giovenco S. 1965. Flavin coenzymes, flavinogenesis and reproduction in Ashbya gossypii. J. Biochem. 57:773–786 [DOI] [PubMed] [Google Scholar]
- 67. Chatwell L., et al. 2006. Biosynthesis of riboflavin: structure and properties of 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5′-phosphate reductase of Methanocaldococcus jannaschii. J. Mol. Biol. 359:1334–1351 [DOI] [PubMed] [Google Scholar]
- 68. Chen S. C., Chang Y. C., Lin C. H., Liaw S. H. 2006. Crystal structure of a bifunctional deaminase and reductase from Bacillus subtilis involved in riboflavin biosynthesis. J. Biol. Chem. 281:7605–7613 [DOI] [PubMed] [Google Scholar]
- 69. Cheng V. W., Ma E., Zhao Z., Rothery R. A., Weiner J. H. 2006. The iron-sulfur clusters in Escherichia coli succinate dehydrogenase direct electron flow. J. Biol. Chem. 281:27662–27668 [DOI] [PubMed] [Google Scholar]
- 70. Christie J. M., Briggs W. R. 2001. Blue light sensing in higher plants. J. Biol. Chem. 276:11457–11460 [DOI] [PubMed] [Google Scholar]
- 71. Chu S. Y., Henderson J. F. 1972. Inhibition of the phosphoribosyl-formylglycineamidine synthetase of Ehrlich ascites tumor cells by glutamine analogues. Biochem. Pharmacol. 21:401–406 [DOI] [PubMed] [Google Scholar]
- 72. Ciptadjaya C. G., Guo W., Angeli J. M., Obare S. O. 2009. Controlling the reactivity of chlorinated ethylenes with flavin mononucleotide hydroquinone. Environ. Sci. Technol. 43:1591–1597 [DOI] [PubMed] [Google Scholar]
- 73. Clarebout G., Villers C., Leclercq R. 2001. Macrolide resistance gene mreA of Streptococcus agalactiae encodes a flavokinase. Antimicrob. Agents Chemother. 45:2280–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Coats J. H., Li G. P., Kuo M. S., Yurek D. A. 1989. Discovery, production, and biological assay of an unusual flavenoid cofactor involved in lincomycin biosynthesis. J. Antibiot. (Tokyo). 42:472–474 [DOI] [PubMed] [Google Scholar]
- 75. Cochrane J. C., Strobel S. A. 2008. Riboswitch effectors as protein enzyme cofactors. RNA 14:993–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cooperman J. M., Lopez R. 1991. Riboflavin, p. 283–310 In Macklin J. (ed.), Handbook of vitamins. Marcel Dekker, New York, NY [Google Scholar]
- 77. Coursolle, Baron D. D. B., Bond D. R., Gralnick J. A. 2010. The Mtr respiratory pathway is essential for reducing flavins and electrodes in Shewanella oneidensis. J. Bacteriol. 192:467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Crossley R. A., et al. 2007. Riboflavin biosynthesis is associated with assimilatory ferric reduction and iron acquisition by Campylobacter jejuni. Appl. Environ. Microbiol. 73:7819–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Davidson A. L., Dassa E., Orelle C., Chen J. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72:317–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Decker K., Brandsch R. 1997. Determining covalent flavinylation. Methods Enzymol. 280:413–423 [DOI] [PubMed] [Google Scholar]
- 81. De Colibus L., Mattevi A. 2006. New frontiers in structural flavoenzymology. Curr. Opin. Struct. Biol. 16:722–728 [DOI] [PubMed] [Google Scholar]
- 82. Delbrück M., Ootaki T. 1979. An unstable nuclear gene in Phycomyces. Genetics 92:27–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Demain A. L. 1972. Riboflavin oversynthesis. Annu. Rev. Microbiol. 26:369–388 [DOI] [PubMed] [Google Scholar]
- 84. Demain A. L. 2007. The business of biotechnology. Ind. Biotechnol. 3:269–283 [Google Scholar]
- 85. Diamond L. S., Cunnick C. C. 2007. A serum-free, partly defined medium, PDM-805, for axenic cultivation of Entamoeba histolytica Schaudinn, 1903 and other Entamoeba. J. Eukaryot. Microbiol. 38:211–216 [DOI] [PubMed] [Google Scholar]
- 86. Dietrich F. S., et al. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304–307 [DOI] [PubMed] [Google Scholar]
- 87. Dikanskaia E. M. 1971. Yeast variants producing riboflavin and resistant to cobalt. Mikrobiologiia 40:1077–1083 (In Russian.) [PubMed] [Google Scholar]
- 88. Dikanskaia E. M., Gorobtsova T. A. 1975. Increased flavin synthesis in yeasts utilizing hydrocarbons. Mikrobiologiia 44:784–790 (In Russian.) [PubMed] [Google Scholar]
- 89. Dikanskaia E. M., Gorobtsova T. A., Kulikova V. P. 1976. Flavinogenesis in methylotrophic yeasts. Mikrobiologiia 45:955–959 (In Russian.) [PubMed] [Google Scholar]
- 90. Dmytruk K. V., et al. 2004. Cloning of structural genes involved in riboflavin synthesis of the yeast Candida famata. Ukr. Biokhim. Zhurn. 76:78–87 [PubMed] [Google Scholar]
- 91. Dmytruk K. V., Voronovsky A. Y., Sibirny A. A. 2006. Insertion mutagenesis of the yeast Candida famata (Debaryomyces hansenii) by random integration of linear DNA fragments. Curr. Genet. 50:183–191 [DOI] [PubMed] [Google Scholar]
- 92. Dmytruk K. V., Yatsyshyn V. Y., Fedorovych D. V., Sibirny A. A. 2011. Metabolic engineering and classic selection of the yeast Candida famata (Candida flareri) for construction of the strains with enhanced riboflavin production. Metab. Eng. 13:82–88 [DOI] [PubMed] [Google Scholar]
- 93. Dobler W., Eggersdorfer M., Paust J. January 1991. Purification of salts of riboflavin 5′-phosphate, in particular of monosodium riboflavin 5′-phosphate. U.S. patent 4,987,229
- 94. Dong H., Beer S. V. 2000. Riboflavin induces disease resistance in plants by activating a novel signal transduction pathway. Phytopathology 90:801–811 [DOI] [PubMed] [Google Scholar]
- 95. Drepper T., et al. 2007. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25:443–445 [DOI] [PubMed] [Google Scholar]
- 96. Duan Y. X., Chen T., Chen X., Zhao X. M. 2010. Overexpression of glucose-6-phosphate dehydrogenase enhances riboflavin production in Bacillus subtilis. Appl. Microbiol. Biotechnol. 85:1907–1914 [DOI] [PubMed] [Google Scholar]
- 97. Duurkens R. H., Tol M. B., Geertsma E. R., Permentier H. P., Slotboom D. J. 2007. Flavin binding to the high affinity riboflavin transporter RibU. J. Biol. Chem. 282:10380–10386 [DOI] [PubMed] [Google Scholar]
- 98. Eberhardt S., Korn S., Lottspeich F., Bacher A. 1997. Biosynthesis of riboflavin: an unusual riboflavin synthase of Methanobacterium thermoautotrophicum. J. Bacteriol. 179:2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Echt S., et al. 2004. Potential anti-infective targets in pathogenic yeasts: structure and properties of 3,4-dihydroxy-2-butanone 4-phosphate synthase of Candida albicans. J. Mol. Biol. 341:1085–1096 [DOI] [PubMed] [Google Scholar]
- 100. Edwards A. M. 2006. General properties of flavins, p. 1–11 In Silva E., Edwards A. M. (ed.), Comprehensive series in photochemical and photobiological sciences, vol. 6 Flavins: photochemistry and photobiology RSC, Cambridge, United Kingdom [Google Scholar]
- 101. Efimov I., Kuusk V., Zhang X., McIntire W. S. 1998. Proposed steady-state kinetic mechanism for Corynebacterium ammoniagenes FAD synthetase produced by Escherichia coli. Biochemistry 37:9716–9723 [DOI] [PubMed] [Google Scholar]
- 102. Eggeling L., Sahm H., Wagner F. 1977. Induction of FMN adenylyltransferase in the methanol utilizing yeast Candida boidinii. FEMS Microbiol. Lett. 1:205–211 [Google Scholar]
- 103. Eker A. P., Hessels J. K., Dekker R. H. 1986. Photoreactivating enzyme from Streptomyces griseus. VI. Action spectrum and kinetics of photoreactivation. Photochem. Photobiol. 44:197–205 [DOI] [PubMed] [Google Scholar]
- 104. Eker A. P. M., Kooiman P., Hessels J. K., Yasui A. 1990. DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. J. Biol. Chem. 265:8009–8015 [PubMed] [Google Scholar]
- 105. Eker A. P., Quayle C., Chaves I., van der Horst G. T. 2009. DNA repair in mammalian cells: direct DNA damage reversal: elegant solutions for nasty problems. Cell Mol. Life Sci. 66:968–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Enary T. M. 1955. Effect of cobalt and iron on riboflavin by Candida guilliermondii. Acta Chem. Scand. 9:1726–1729 [Google Scholar]
- 107. Exinger F., Lacroute F. 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 22:9–11 [DOI] [PubMed] [Google Scholar]
- 108. Fassbinder F., Kist M., Bereswill S. 2000. Structural and functional analysis of the riboflavin synthesis genes encoding GTP cyclohydrolase II (ribA), DHBP synthase (ribBA), riboflavin synthase (ribC), and riboflavin deaminase/reductase (ribD) from Helicobacter pylori strain P1. FEMS Microbiol. Lett. 191:191–197 [DOI] [PubMed] [Google Scholar]
- 109. Fedorovich D. V., Kityk I. V., Dzhala V. I., Protchenko O. V., Shavlovskii G. M. 1997. Accumulation and redox transformations of iron on the yeast Pichia guilliermondii and its flavinogenic mutants. Mikrobiologiia 66:60–64 (In Rusian.) [Google Scholar]
- 110. Fedorovich D., Protchenko O., Lesuisse E. 1999. Iron uptake by the yeast Pichia guilliermondii. Flavinogenesis and reductive iron assimilation are co-regulated processes. Biometals 12:295–300 [DOI] [PubMed] [Google Scholar]
- 111. Fedorovych D., Kszeminska H., Babjak L., Kaszycki P., Koloczek H. 2001. Hexavalent chromium stimulation of riboflavin synthesis in flavinogenic yeast. Biometals 14:23–31 [DOI] [PubMed] [Google Scholar]
- 112. Fischer M., Bacher A. 2005. Biosynthesis of flavocoenzymes. Nat. Prod. Rep. 22:324–350 [DOI] [PubMed] [Google Scholar]
- 113. Fischer M., Bacher A. 2006. Biosynthesis of vitamin B2 in plants. Physiol. Plant 126:304–318 [Google Scholar]
- 114. Fischer M., Bacher A. 2008. Biosynthesis of vitamin B2: structure and mechanism of riboflavin synthase. Arch. Biochem. Biophys. 474:252–265 [DOI] [PubMed] [Google Scholar]
- 115. Fischer M., Bacher A. 2010. Riboflavin biosynthesis, p. 3–36 In Mander L., Liu H. W. (ed.), Comprehensive natural products. II. Chemistry and biology, vol. 7 Cofactors Elsevier, Philadelphia, PA [Google Scholar]
- 116. Fischer M., et al. 2002. Biosynthesis of riboflavin: 6,7-dimethyl-8-ribityllumazine synthase of Schizosaccharomyces pombe. Eur. J. Biochem. 269:519–526 [DOI] [PubMed] [Google Scholar]
- 117. Fischer M., et al. 2005. Evolution of vitamin B2 biosynthesis: riboflavin synthase of Arabidopsis thaliana and its inhibition by riboflavin. Biol. Chem. 386:417–428 [DOI] [PubMed] [Google Scholar]
- 118. Fischer M., et al. 2003. Enzyme catalysis via control of activation entropy: site-directed mutagenesis of 6,7-dimethyl-8-ribityllumazine synthase. J. Mol. Biol. 326:783–793 [DOI] [PubMed] [Google Scholar]
- 119. Fischer M., et al. 2004. Evolution of vitamin B2 biosynthesis: structural and functional similarity between pyrimidine deaminases of eubacterial and plant origin. J. Biol. Chem. 279:36299–36308 [DOI] [PubMed] [Google Scholar]
- 120. Fischer M., et al. 2002. Biosynthesis of riboflavin in archaea: studies on the mechanism of 3,4-dihydroxy-2-butanone-4-phosphate synthase of Methanococcus jannaschii. J. Biol. Chem. 277:41410–41416 [DOI] [PubMed] [Google Scholar]
- 121. Fischer M., et al. 2004. Evolution of vitamin B2 biosynthesis. A novel class of riboflavin synthase in Archaea. J. Mol. Biol. 343:267–278 [DOI] [PubMed] [Google Scholar]
- 122. Food Nutrition Board, Institute of Medicine, National Academy of Sciences, 1998. Riboflavi, n, p. 87–122 In Dietary reference intakes: thiamin, riboflavin, niacin, vitamin B6, vitamin B12, pantothenic acid, biotin, folate and choline. The National Academies Press, Washington, DC: [PubMed] [Google Scholar]
- 123. Foor F., Brown G. M. 1975. Purification and properties of guanosine triphosphate cyclohydrolase II from Escherichia coli. J. Biol. Chem. 250:3545–3551 [PubMed] [Google Scholar]
- 124. Foor F., Brown G. M. 1980. GTP cyclohydrolase II from Escherichia coli. Methods Enzymol. 66:303–307 [DOI] [PubMed] [Google Scholar]
- 125. Foraker A. B., Khantwal C. M., Swaan P. W. 2003. Current perspectives in cellular uptake and trafficking of riboflavin. Adv. Drug Deliv. Rev. 55:1467–1483 [DOI] [PubMed] [Google Scholar]
- 126. Forouhar F., et al. 2008. Molecular insights into the biosynthesis of the F420 coenzyme. J. Biol. Chem. 283:11832–11840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Förster C., Revuelta J. L., Krämer R. 2001. Carrier-mediated transport of riboflavin in Ashbya gossypii. Appl. Microbiol. Biotechnol. 55:85–89 [DOI] [PubMed] [Google Scholar]
- 128. Förster C., Santos M. A., Ruffert S., Krämer R., Revuelta J. L. 1999. Physiological consequence of disruption of the VMA1 gene in the riboflavin overproducer Ashbya gossypii. J. Biol. Chem. 274:9442–9448 [DOI] [PubMed] [Google Scholar]
- 129. Foy H., Mbaya V. 1977. Riboflavin, Prog. Food Nutr. Sci. 2:357–394 [PubMed] [Google Scholar]
- 130. Fraaije M. V., Mattevi A. 2000. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci. 25:126–132 [DOI] [PubMed] [Google Scholar]
- 131. Fraaije M. W., van den Heuvel R. H., van Berkel W. J., Mattevi A. 1999. Covalent flavinylation is essential for efficient redox catalysis in vanillyl-alcohol oxidase. J. Biol. Chem. 274:35514–35520 [DOI] [PubMed] [Google Scholar]
- 132. Fraga A. A., Reddy C. A. 1982. Nutritional requirements of Corynebacterium pyogenes. J. Clin. Microbiol. 16:334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Frago S., Martínez-Júlvez M., Serrano A., Medina M. 2008. Structural analysis of FAD synthetase from Corynebacterium ammoniagenes. BMC Microbiol. 8:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Frago S., Velázquez-Campoy A., Medina M. 2009. The puzzle of ligand binding to Corynebacterium ammoniagenes FAD synthetase. J. Biol. Chem. 284:6610–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Froehlich A. C., Liu Y., Loros J. J., Dunlap J. C. 2002. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297:815–819 [DOI] [PubMed] [Google Scholar]
- 136. Fujio T., Maruyama A. 1997. Enzymatic production of pyrimidine nucleotides using Corynebacterium ammoniagenes cells and recombinant Escherichia coli cells: enzymatic production of CDP-choline from orotic acid and choline chloride (part I). Biosci. Biotechnol. Biochem. 61:956–959 [DOI] [PubMed] [Google Scholar]
- 137. García-Ramírez J. J., Santos M. A., Revuelta J. L. 1995. The Saccharomyces cerevisiae RIB4 gene codes for 6,7-dimethyl-8-ribityllumazine synthase involved in riboflavin biosynthesis. Molecular characterization of the gene and purification of the encoded protein. J. Biol. Chem. 270:23801–23807 [DOI] [PubMed] [Google Scholar]
- 138. Gelfand M. S., Mironov A. A., Jomantas J., Kozlov Y. I., Perumov D. A. 1999. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 15:439–442 [DOI] [PubMed] [Google Scholar]
- 139. Gerhardt S., et al. 2002. The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase. J. Mol. Biol. 318:1317–1329 [DOI] [PubMed] [Google Scholar]
- 140. Gerhardt S., et al. 2002. Studies on the reaction mechanism of riboflavin synthase: X-ray crystal structure of a complex with 6-carboxyethyl-7-oxo-8-ribityllumazine. Structure 10:1371–1381 [DOI] [PubMed] [Google Scholar]
- 141. Gershanovich V. N., Kukanova A. I., Galushkina Z. M., Stepanov A. I. 2000. Transketolase mutation in riboflavin-synthesizing strains of Bacillus subtilis. Mol. Gen. Mikrobiol. Virusol. 3:3–7 (In Russian.) [PubMed] [Google Scholar]
- 142. Ghisla S., Mayhew S. G. 1976. Identification and properties of 8-hydroxyflavin-adenine dinucleotide in electron-transferring flavoprotein from Peptostreptococcus elsdenii. Eur. J. Biochem. 63:373–390 [DOI] [PubMed] [Google Scholar]
- 143. Giancaspero T. A., Locato V., de Pinto M. C., De Gara L., Barile M. 2009. The occurrence of riboflavin kinase and FAD synthetase ensures FAD synthesis in tobacco mitochondria and maintenance of cellular redox status. FEBS J. 276:219–231 [DOI] [PubMed] [Google Scholar]
- 144. Giancaspero T. A., Wait R., Boles E., Barile M. 2008. Succinate dehydrogenase flavoprotein subunit expression in Saccharomyces cerevisiae—involvement of the mitochondrial FAD transporter, Flx1p. FEBS J. 275:1103–1117 [DOI] [PubMed] [Google Scholar]
- 145. Giri K. V., Krishnaswamy P. W., Rao N. A. 1957. Occurrence of flavokinase activity in plants. Nature 179:1134–1135 [DOI] [PubMed] [Google Scholar]
- 146. Glas A. F., et al. 2009. The archaeal cofactor F0 is a light-harvesting antenna chromophore in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 106:11540–11545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Gliszczynska A., Koziolowa A. 1998. Chromatographic determination of flavin derivatives in baker's yeast. J. Chromatogr. A 822:59–66 [DOI] [PubMed] [Google Scholar]
- 148. Gomelsky M., Klug G. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27:497–500 [DOI] [PubMed] [Google Scholar]
- 149. Gonzalez-Cabo P., Ros S., Palau F. 2010. Flavin adenine dinucleotide rescues the phenotype of frataxin deficiency. PLoS One 5:e8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Goodwin T. W., McEvoy D. 1959. Studies on the biosynthesis of riboflavin. 5. General factors controlling flavinogenesis in the yeast Candida flareri. Biochem. J. 71:742–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Graham D. E., White R. H. 2002. Elucidation of methanogenic coenzyme biosyntheses: from spectroscopy to genomics. Nat. Prod. Rep. 19:133–147 [DOI] [PubMed] [Google Scholar]
- 152. Graham D. W., Brown J. E., Ashton W. T., Brown R. D., Rogers E. F. 1977. Anticoccidial riboflavine antagonists. Experientia 33:1274–1276 [DOI] [PubMed] [Google Scholar]
- 153. Graupner M., Xu H., White R. H. 2002. The pyrimidine nucleotide reductase step in riboflavin and F420 biosynthesis in Archaea proceeds by the eukaryotic route to riboflavin. J. Bacteriol. 184:1952–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Gray M. J., Escalante-Semerena J. C. 2007. Single-enzyme conversion of FMNH2 to 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc. Natl. Acad. Sci. U. S. A. 104:2921–2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Grill S., Busenbender S., Pfeiffer M., Köhler U., Mack M. 2008. The bifunctional flavokinase/flavin adenine dinucleotide synthetase from Streptomyces davawensis produces inactive flavin cofactors and is not involved in resistance to the antibiotic roseoflavin. J. Bacteriol. 190:1546–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Grill S., et al. 2007. Identification and characterization of two Streptomyces davawensis riboflavin biosynthesis gene clusters. Arch. Microbiol. 188:377–387 [DOI] [PubMed] [Google Scholar]
- 157. Grininger M., Staudt H., Johansson P., Wachtveitl J., Oesterhelt D. 2009. Dodecin is the key player in flavin homeostasis of archaea. J. Biol. Chem. 284:13068–13076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Grininger M., Zeith K., Oesterhelt D. 2006. Dodecins: a family of lumichrome binding proteins. J. Mol. Biol. 357:842–857 [DOI] [PubMed] [Google Scholar]
- 159. Grochowski L. L., Xu H., White R. H. 2008. Identification and characterization of the 2-phospho-l-lactate guanylyltransferase involved in coenzyme F420 biosynthesis. Biochemistry 47:3033–3037 [DOI] [PubMed] [Google Scholar]
- 160. Grochowski L. L., Xu H., White R. H. 2009. An iron(II) dependent formamide hydrolase catalyzes the second step in the archaeal biosynthetic pathway to riboflavin and 7,8-didemethyl-8-hydroxy-5-deazariboflavin. Biochemistry 48:4181–4188 [DOI] [PubMed] [Google Scholar]
- 161. Groom K. R., Heyman H. C., Steffen M. C., Hawkins L., Martin N. C. 1998. Kluyveromyces lactis SEF1 and its Saccharomyces cerevisiae homologue bypass the unknown essential function, but not the mitochondrial RNase P function, of the S. cerevisiae RPM2 gene. Yeast 14:77–87 [DOI] [PubMed] [Google Scholar]
- 162. Guilliermond A., Fontaine M., Raffy A. 1935. Sur l'existence dans l'Eremothecium ashbyii d'un pigment jaune se rapportant au groupe des flavines. C. R. Hebd. Seances Acad. Sci. 201:1077–1080 [Google Scholar]
- 163. Gusarov I. I., et al. 1997. Primary structure and functional activity of the Bacillus subtilis ribC gene. Mol. Biol. (Moscow) 31:820–825 [PubMed] [Google Scholar]
- 164. Hagihara T., Fujio T., Aisaka K. 1995. Cloning of FAD synthetase gene from Corynebacterium ammoniagenes and its application to FAD and FMN production. Appl. Microbiol. Biotechnol. 42:724–729 [DOI] [PubMed] [Google Scholar]
- 165. Harvey R. A., Plaut G. W. 1966. Riboflavin synthetase from yeast. Properties of complexes of the enzyme with lumazine derivatives and riboflavin. J. Biol. Chem. 241:2120–2136 [PubMed] [Google Scholar]
- 166. Haupt W. 1999. Chloroplast movement: from phenomenology to molecular biology. Prog. Bot. 60:3–35 [Google Scholar]
- 167. Hebbeln P., Rodionov D. A., Alfandega A., Eitinger T. 2007. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc. Natl. Acad. Sci. U. S. A. 104:2909–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Heefner, Boyts D. L. A., Burdzinski L., Yarus M. July 1993. Efficient riboflavin production with yeast. U.S. patent 5,231,007
- 169. Heefner D., et al. December 1988. Riboflavin producing strains of microorganisms, method for selecting, and method for fermentation. Patent WO 88/09822
- 170. Heefner D. L., Weaver C. A., Yarus M. J., Burdzinski L. A. November 1992. Method for producing riboflavin with Candida famata. U.S. patent 5,164,303
- 171. Hemmerich P., Massey V., Fenner H. 1977. Flavin and 5-deazaflavin: a chemical evaluation of ‘modified’ flavoproteins with respect to the mechanisms of redox biocatalysis. FEBS Lett. 84:5–21 [DOI] [PubMed] [Google Scholar]
- 172. Henderson G. B., Zevely E. M., Huennekens G. M. 1979. Coupling of energy to folate transport in Lactobacillus casei. J. Bacteriol. 139:552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Herguedas B., Martínez-Júlvez M., Frago S., Medina M., Hermoso J. A. 2010. Oligomeric state in the crystal structure of modular FAD synthetase provides insights into its sequential catalysis in prokaryotes. J. Mol. Biol. 400:218–230 [DOI] [PubMed] [Google Scholar]
- 174. Herz S., Eberhardt S., Bacher A. 2000. Biosynthesis of riboflavin in plants. The ribA gene of Arabidopsis thaliana specifies a bifunctional GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase. Phytochemistry 53:723–731 [DOI] [PubMed] [Google Scholar]
- 175. Hickey R. J. 1945. The inactivation of iron by 2,2′-bipyrimidine and its effect on riboflavin synthesis by Clostridium acetobutylicum. Arch. Biochem. 8:439–447 [PubMed] [Google Scholar]
- 176. Higa A., Miyamoto E., ur Rahman L., Kitamura Y. 2008. Root tip-dependent, active riboflavin secretion by Hyoscyamus albus hairy roots under iron deficiency. Plant Physiol. Biochem. 46:452–460 [DOI] [PubMed] [Google Scholar]
- 177. Higashitsuji Y., Angerer A., Berghaus S., Hobl B., Mack M. 2007. RibR, a possible regulator of the Bacillus subtilis riboflavin biosynthetic operon, in vivo interacts with the 5′-untranslated leader of rib mRNA. FEMS Microbiol. Lett. 274:48–54 [DOI] [PubMed] [Google Scholar]
- 178. Hohmann H. P., Stahmann K. P. 2010. Biotechnology of riboflavin production, p. 115–139 In Mander L., Liu H. W. (ed.), Comprehensive natural products. II. Chemistry and biology, vol. 7 Cofactors Elsevier, Philadelphia, PA [Google Scholar]
- 179. Hollander I., Brown G. M. 1979. Biosynthesis of riboflavin: reductase and deaminase of Ashbya gossypii. Biochem. Biophys. Res. Commun. 89:759–763 [DOI] [PubMed] [Google Scholar]
- 180. Horwitt M. K. 1972. Riboflavine. XII. Requirements and factors influencing them, p. 73–88 In Sebrell W. H., Harris R. S. (ed.), The vitamins, vol. 5 Academic Press, New York, NY [Google Scholar]
- 181. Howell D. M., White R. H. 1997. d-Erythro-neopterin biosynthesis in the methanogenic archaea Methanococcus thermophila and Methanobacterium thermoautotrophicum deltaH. J. Bacteriol. 179:5165–5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Huang S. N., Swaan P. W. 2001. Riboflavin uptake in human trophoblast-derived BeWo cell monolayers: cellular translocation and regulatory mechanisms. J. Pharmacol. Exp. Ther. 298:264–271 [PubMed] [Google Scholar]
- 183. Huang Y. F., Liu S. Y., Yen C. L., Yang P. W., Shieh C. C. 2009. Thapsigargin and flavin adenine dinucleotide ex vivo treatment rescues trafficking-defective gp91phox in chronic granulomatous disease leukocytes. Free Radic. Biol. Med. 47:932–940 [DOI] [PubMed] [Google Scholar]
- 184. Huerta C., Borek D., Machius M., Grishin N. V., Zhang H. 2009. Structure and mechanism of a eukaryotic FMN adenylyltransferase. J. Mol. Biol. 389:388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Hümbelin M., et al. 1999. GTP cyclohydrolase II and 3,4-dihydroxy-2-butanone 4-phosphate synthase are rate-limiting enzymes in riboflavin synthesis of an industrial Bacillus subtilis strain used for riboflavin production. J. Ind. Microbiol. Biotechnol. 22:1–7 [Google Scholar]
- 186. Hustad S., Ueland P. M., Schneede J. 1999. Quantification of riboflavin, flavin mononucleotide, and flavin adenine dinucleotide in human plasma by capillary electrophoresis and laser-induced fluorescence detection. Clin. Chem. 45:862–868 [PubMed] [Google Scholar]
- 187. Ifergan I., Goler-Baron V., Assaraf Y. G. 2009. Riboflavin concentration within ABCG2-rich extracellular vesicles is a novel marker for multidrug resistance in malignant cells. Biochem. Biophys. Res. Commun. 380:5–10 [DOI] [PubMed] [Google Scholar]
- 188. Iino M. 2001. Phototropism in higher plants, p. 659–811 In Hader D. P., Lebert M. (ed.), Photomovement. Elsevier, Amsterdam, Netherlands [Google Scholar]
- 189. Ikeno S., Aoki D., Hamada M., Hori M., Tsuchiya K. S. 2006. DNA sequencing and transcriptional analysis of the kasugamycin biosynthetic gene cluster from Streptomyces kasugaensis M338-M1. J. Antibiot. (Tokyo) 59:18–28 [DOI] [PubMed] [Google Scholar]
- 190. Illarionov B., Eisenreich W., Schramek N., Bacher A., Fischer M. 2005. Biosynthesis of vitamin B2: diastereomeric reaction intermediates of archaeal and non-archaeal riboflavin synthases. J. Biol. Chem. 280:28541–28546 [DOI] [PubMed] [Google Scholar]
- 191. Imaizumi T., Tran H. G., Swartz T. E., Briggs W. R., Kay S. A. 2003. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426:302–306 [DOI] [PubMed] [Google Scholar]
- 192. International Union of Biochemistry, 1984. Nomenclature of electron-transfer proteins. In Enzyme nomenclature, recommendations. Academic Press, Orlando, FL [Google Scholar]
- 193. Ishchuk O. P., et al. 2006. Construction of the flavinogenic yeast Candida famata strains with high riboflavin kinase activity using gene engineering. Ukr. Biokhim. Zh. 78:63–69 (In Ukrainian.) [PubMed] [Google Scholar]
- 194. Ishchuk O. P., et al. 2008. Development of a promoter assay system for the flavinogenic yeast Candida famata based on the Kluyveromyces lactis β-galactosidase LAC4 reporter gene. Enzyme Microb. Technol. 42:208–215 [Google Scholar]
- 195. Jiménez A., Santos M. A., Pompejus M., Revuelta J. L. 2005. Metabolic engineering of the purine pathway for riboflavin production in Ashbya gossypii. Appl. Environ. Microbiol. 71:5743–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Jiménez A., Santos M. A., Revuelta J. L. 2008. Phosphoribosyl pyrophosphate synthetase activity affects growth and riboflavin production in Ashbya gossypii. BMC Biotechnol. 8:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Jin C., Barrientos A., Tsagoloff A. 2003. Yeast dihydroxybutanone phosphate synthase, an enzyme of the riboflavin biosynthetic pathway, has a second unrelated function in expression of mitochondrial respiration. J. Biol. Chem. 278:14698–14703 [DOI] [PubMed] [Google Scholar]
- 198. Johnson J. L., Bastian N. R., Rajagopalan K. V. 1990. Molybdopterin guanine dinucleotide: a modified form of molybdopterin identified in the molybdenum cofactor of dimethyl sulfoxide reductase from Rhodobacter sphaeroides forma specialis denitrificans. Proc. Natl. Acad. Sci. U. S. A. 87:3190–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Jordan D. B., Bacot K. O., Carlson T. J., Kessel M., Viitanen P. V. 1999. Plant riboflavin biosynthesis. Cloning, chloroplast localization, expression, purification, and partial characterization of spinach lumazine synthase. J. Biol. Chem. 274:22114–22121 [DOI] [PubMed] [Google Scholar]
- 200. Juárez O., Nilges M. J., Gillespie P., Cotton J., Barquera B. 2008. Riboflavin is an active redox cofactor in the Na+-pumping NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. J. Biol. Chem. 283:33162–33167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Juri N., et al. 1987. Formation of roseoflavin from 8-amino- and 8-methylamino-8-demethyl-d-riboflavin. J. Biochem. 101:705–711 [DOI] [PubMed] [Google Scholar]
- 202. Jusko W. J., Levy G. 1970. Pharmacokinetic evidence for saturable renal tubular reabsorption of riboflavin. J. Pharm. Sci. 59:765–772 [DOI] [PubMed] [Google Scholar]
- 203. Kaesler B., et al. January 1997. Riboflavin production process by means of microorganisms with modified isocitrate lyase activity. Patent WO 9703208-A
- 204. Kainosho M., Kyogoku Y. 1972. High-resolution proton and phosphorus nuclear magnetic resonance spectra of flavin-adenine dinucleotide and its conformation in aqueous solution. Biochemistry 11:741–752 [DOI] [PubMed] [Google Scholar]
- 205. Kaiser J., et al. 2007. A high-throughput screening platform for inhibitors of the riboflavin biosynthesis pathway. Anal. Biochem. 365:52–61 [DOI] [PubMed] [Google Scholar]
- 206. Kaiser J., et al. 2002. Biosynthesis of vitamin B2. Eur. J. Biochem. 269:5264–5270 [DOI] [PubMed] [Google Scholar]
- 207. Kalingan A. E., Krishnan M. R. V. 1997. Application of agro-industrial by-products for riboflavin production by Eremothecium ashbyii NRRL 1363. Appl. Microbiol. Biotechnol. 47:226–230 [Google Scholar]
- 208. Kanehisa M., Goto S., Kawashima S., Nakaya A. 2002. The KEGG databases at GenomeNet. Nucleic Acids Res. 30:42–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Karos M., Vilariño C., Bollschweiler C., Revuelta J. L. 2004. A genome-wide transcription analysis of a fungal riboflavin overproducer. J. Biotechnol. 113:69–76 [DOI] [PubMed] [Google Scholar]
- 210. Karrer P., Schopp K., Benz F. 1935. Synthesen von Flavinen. Helv. Chim. Acta 18:426–429 [Google Scholar]
- 211. Karthikeyan S., Zhou Q., Osterman A. L., Zhang H. 2003. Ligand binding-induced conformational changes in riboflavin kinase: structural basis for the ordered mechanism. Biochemistry 42:12532–12538 [DOI] [PubMed] [Google Scholar]
- 212. Kasai S., et al. 1978. Anti-riboflavin activity of 8N-alkyl analogues of roseoflavin in some Gram-positive bacteria. J. Nutr. Sci. Vitaminol. (Tokyo) 24:339–350 [DOI] [PubMed] [Google Scholar]
- 213. Kasai S., Yamanaka S., Wang S. C., Matsui K. 1979. Anti-riboflavin activity of 8-O-alkyl derivatives of riboflavin in some Gram-positive bacteria. J. Nutr. Sci. Vitaminol. (Tokyo) 25:289–298 [DOI] [PubMed] [Google Scholar]
- 214. Kashchenko V. E., Shavlovskii G. M. 1976. Purification and properties of the riboflavin kinase of the yeast Pichia guilliermondii. Biokhimiia 41:376–383 (In Russian.) [PubMed] [Google Scholar]
- 215. Kashchenko V. E., Preobrazhenskaya E. N., Sibirny A. A. November 1991. The method for determination of riboflavin kinase activity. USSR author's certificate no. 1631089 [Google Scholar]
- 216. Katagiri H., Yamada H., Imai K. 1959. Biosynthesis of flavin coenzymes by microorganisms. II. Enzymatic synthesis of flavin adenine dinucleotide in Escherichia coli. J. Vitaminol. (Kyoto) 5:307–311 [DOI] [PubMed] [Google Scholar]
- 217. Kato T., Park E. Y. 2006. Expression of alanine:glyoxylate aminotransferase gene from Saccharomyces cerevisiae in Ashbya gossypii. Appl. Microbiol. Biotechnol. 71:46–52 [DOI] [PubMed] [Google Scholar]
- 218. Kearney E. B., Englard S. 1951. The enzymatic phosphorylation of riboflavin. J. Biol. Chem. 193:821–834 [PubMed] [Google Scholar]
- 219. Kearney E. B., Goldenberg J., Lipsick J., Perl M. 1979. Flavokinase and FAD synthetase from Bacillus subtilis specific for reduced flavins. J. Biol. Chem. 254:9551–9557 [PubMed] [Google Scholar]
- 220. Kelly M. J., et al. 2001. The NMR structure of the 47-kDa dimeric enzyme 3,4-dihydroxy-2-butanone-4-phosphate synthase and ligand binding studies reveal the location of the active site. Proc. Natl. Acad. Sci. U. S. A. 98:13025–13030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Kil Y. V., Mironov V. N., Gorishin I. Y., Kreneva R. A., Perumov D. A. 1992. Riboflavin operon of Bacillus subtilis: unusual symmetric arrangement of the regulatory region. Mol. Gen. Genet. 233:483–486 [DOI] [PubMed] [Google Scholar]
- 222. Kim J. K., Ezaki J., Himeno M., Kato K., Kim S. 1993. Purification and characterization of flavine-adenine dinucleotide phosphohydrolase from rat liver lysosomal membranes. J. Biochem. 114:126–131 [DOI] [PubMed] [Google Scholar]
- 223.Reference deleted.
- 224. Kis K., Bacher A. 1995. Substrate channeling in the lumazine synthase/riboflavin synthase complex of Bacillus subtilis. J. Biol. Chem. 270:16788–16795 [DOI] [PubMed] [Google Scholar]
- 225. Kis K., Kugelbrey K., Bacher A. 2001. Biosynthesis of riboflavin. The reaction catalyzed by 6,7-dimethyl-8-ribityllumazine synthase can proceed without enzymatic catalysis under physiological conditions. J. Org. Chem. 66:2555–2559 [DOI] [PubMed] [Google Scholar]
- 226. Klinke S., et al. 2007. Structural and kinetic properties of lumazine synthase isoenzymes in the order Rhizobiales. J. Mol. Biol. 373:664–680 [DOI] [PubMed] [Google Scholar]
- 227. Knight S. A., Lesuisse E., Stearman R., Klausner R. D., Dancis A. 2002. Reductive iron uptake by Candida albicans: role of copper, iron and the TUP1 regulator. Microbiology 148:29–40 [DOI] [PubMed] [Google Scholar]
- 228. Knusel F. 1957. Biosynthesis of riboflavin in Candida guilliermondii (A. Cast) Langeron and Guerra and some related species; influence of trace elements, especially iron and zinc. Arch. Mikrobiol. 27:219–259 (In German.) [PubMed] [Google Scholar]
- 229. Kobayashi M., et al. 1998. Potential of Escherichia coli GTP cyclohydrolase II for hydrolyzing 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 273:26394–26399 [DOI] [PubMed] [Google Scholar]
- 230. Kobayashi T., Suzue T. 1961. Flavin adenine dinucleotide-synthesizing enzyme in Eremothecium ashbyii. J. Vitaminol. (Kyoto) 7:42–47 [DOI] [PubMed] [Google Scholar]
- 231. Koch M., et al. 2004. Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase. Eur. J. Biochem. 271:3208–3214 [DOI] [PubMed] [Google Scholar]
- 232. Koh Y. S., Choih J., Lee J. H., Roe J. H. 1996. Regulation of the ribA gene encoding GTP cyclohydrolase II by the soxRS locus in Escherichia coli. Mol. Gen. Genet. 251:591–598 [DOI] [PubMed] [Google Scholar]
- 233. Koh Y. S., Chung W. H., Lee J. H., Roe J. H. 1999. The reversed SoxS-binding site upstream of the ribA promoter in Escherichia coli. Mol. Gen. Genet. 261:374–380 [DOI] [PubMed] [Google Scholar]
- 234. Koizumi S., Teshiba S. 1998. Riboflavin biosynthetic genes of Corynebacterim ammoniagenes. J. Ferment. Bioeng. 86:130–133 [Google Scholar]
- 235. Koizumi S., Yonetani Y., Maruyama A., Teshiba S. 2000. Production of riboflavin by metabolically engineered Corynebacterium ammoniagenes. Appl. Microbiol. Biotechnol. 53:674–679 [DOI] [PubMed] [Google Scholar]
- 236. Koka P., Lee J. 1979. Separation and structure of the prosthetic group of the blue fluorescence protein from the bioluminescent bacterium Photobacterium phosphoreum. Proc. Natl. Acad. Sci. U. S. A. 76:3068–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Kolinsky M. A., Gross S. S. 2004. The mechanism of potent GTP cyclohydrolase I inhibition by 2,4-diamino-6-hydroxypyrimidine: requirement of the GTP cyclohydrolase I feedback regulatory protein. J. Biol. Chem. 279:40677–40682 [DOI] [PubMed] [Google Scholar]
- 238. Kolonne S., Seviour R. J., McDougall B. M. 1994. Effect of pH on exocellular riboflavin production by Eremothecium ashbyii. Biotechnol. Lett. 16:79–84 [Google Scholar]
- 239. Koltun L. V., Shavlovskii G. M., Kashchenko V. E., Trach V. M. 1984. Changes in the enzyme activity of flavinogenesis in the process of culturing the fungus Eremothecium ashbyii. Mikrobiologiia 53:43–47 (In Russian.) [PubMed] [Google Scholar]
- 240. Konings E. J. M. 2006. Water-soluble vitamins. J. AOAC Intern. 89:285–288 [PubMed] [Google Scholar]
- 241. Koser S. A. 1968. Vitamin requirements of bacteria and yeasts. Charles C. Thomas, Springfield, IL [Google Scholar]
- 242. Kovárová-Kovar K., et al. 2000. Application of model-predictive control based on artificial neural networks to optimize the fed-batch process for riboflavin production. J. Biotechnol. 79:39–52 [DOI] [PubMed] [Google Scholar]
- 243. Kreneva R. A., et al. 2000. Study of the phenotypic occurrence of ypaA gene inactivation in Bacillus subtilis. Genetika 36:1166–1168 (In Russian.) [PubMed] [Google Scholar]
- 244. Kuhn R., Weygand F. 1934. Synthetic vitamin B2. Berichte 67B:2084–2085 [Google Scholar]
- 245. Kutsiaba V. I., Stenchuk N. N., Fedorovich D. V. 2002. Riboflavin overproduction in 4-aminopyrazole[3,4-d]pyrimidine-treated yeast Pichia guilliermondii. Prikl. Biokhim. Mikrobiol. 38:268–272 (In Russian.) [PubMed] [Google Scholar]
- 246. Kuwada S., Masuda T., Kishi T., Asai M. 1958. On the biosynthesis of riboflavin in the culture of Eremothecium ashbyii. J. Vitaminol. (Tokyo) 4:217–225 [DOI] [PubMed] [Google Scholar]
- 247. Ladenstein R., Ritsert K., Huber R., Richter G., Bacher A. 1994. The lumazine synthase/riboflavin synthase complex of Bacillus subtilis. X-ray structure analysis of hollow reconstituted beta-subunit capsids. Eur. J. Biochem. 223:1007–1017 [DOI] [PubMed] [Google Scholar]
- 248. Ladenstein R., et al. 1988. Heavy riboflavin synthase from Bacillus subtilis. Crystal structure analysis of the icosahedral beta 60 capsid at 3.3 A resolution. J. Mol. Biol. 203:1045–1070 [DOI] [PubMed] [Google Scholar]
- 249. Lambooy J. P. 1975. Biological activities of analogs of riboflavin. Plenum, New York, NY [Google Scholar]
- 250. Lambooy J. P., Shaffner C. S. 1977. Utilization of analogues of riboflavin by the riboflavin-deficient chick embryo. J. Nutr. 107:245–250 [DOI] [PubMed] [Google Scholar]
- 251. Lee C. Y., Meighen E. A. 1992. The lux genes in Photobacterium leiognathi are closely linked with genes corresponding in sequence to riboflavin synthesis genes. Biochem. Biophys. Res. Commun. 186:690–697 [DOI] [PubMed] [Google Scholar]
- 252. Lee C. Y., O'Kane D. J., Meighen E. A. 1994. Riboflavin synthesis genes are linked with the lux operon of Photobacterium phosphoreum. J. Bacteriol. 176:2100–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Lee E. R., Blount K. F., Breaker R. R. 2009. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 6:187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Lee J. 1993. Lumazine protein and the excitation mechanism in bacterial bioluminescence. Biophys. Chem. 48:149–158 Rev. [DOI] [PubMed] [Google Scholar]
- 255. Lee J., O'Kane D. J., Visser A. J. 1985. Spectral properties and function of two lumazine proteins from Photobacterium. Biochemistry 24:1476–1483 [DOI] [PubMed] [Google Scholar]
- 256. Lee R. S.-F., Ford H. C. 1997. Purification and characterization of 5′-nucleotidase/FAD pyrophosphatase from human placenta. Methods Enzymol. 280:424–436 [DOI] [PubMed] [Google Scholar]
- 257. Lehmann M., et al. 2009. Biosynthesis of riboflavin. Screening for an improved GTP cyclohydrolase II mutant. FEBS J. 276:4119–4129 [DOI] [PubMed] [Google Scholar]
- 258. Lessner D. J., et al. 2006. An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. U. S. A. 103:17921–17926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Leulliot N., et al. 2010. Crystal structure of yeast FAD synthetase (Fad1) in complex with FAD. J. Mol. Biol. 398:641–646 [DOI] [PubMed] [Google Scholar]
- 260. Levine H., Oyaas J. E., Wasserman L., Hoogerheide J. C., Stern R. M. 1949. Riboflavin production by Candida yeasts. Ind. Eng. Chem. 41:1665–1668 [Google Scholar]
- 261. Li H., Graupner M., Xu H., White R. H. 2003. CofE catalyzes the addition of two glutamates to F420-0 in F420 coenzyme biosynthesis in Methanococcus jannaschii. Biochemistry 42:9771–9778 [DOI] [PubMed] [Google Scholar]
- 262. Li Z., Wakao S., Fischer B. B., Niyogi K. K. 2009. Sensing and responding to excess light. Annu. Rev. Plant Biol. 60:239–260 [DOI] [PubMed] [Google Scholar]
- 263. Liao D. I., Calabrese J. C., Wawrzak Z., Viitanen P. V., Jordan D. B. 2001. Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase of riboflavin biosynthesis. Structure 9:11–18 [DOI] [PubMed] [Google Scholar]
- 264. Liao D. I., Viitanen P. V., Jordan D. B. 2000. Cloning, expression, purification and crystallization of dihydroxybutanone phosphate synthase from Magnaporthe grisea. Acta Crystallogr. D Biol. Crystallogr. 56:1495–1497 [DOI] [PubMed] [Google Scholar]
- 265. Liao D. I., Wawrzak Z., Calabrese J. C., Viitanen P. V., Jordan D. B. 2001. Crystal structure of riboflavin synthase. Structure 9:399–408 [DOI] [PubMed] [Google Scholar]
- 266. Liauta-Teglivets O., Hasslacher M., Boretskii I. R., Kohlwein S. D., Shavlovskii G. M. 1995. Molecular cloning of the GTP-cyclohydrolase structural gene RIB1 of Pichia guilliermondii involved in riboflavin biosynthesis. Yeast 11:945–952 [DOI] [PubMed] [Google Scholar]
- 267. Liesegang A., Straube G., Fritsche W., Reinbothe H. 1974. Correlation between growth and uptake of excreted riboflavin in Candida guilliermondii. Z. Allg. Mikrobiol. 14:691–699 (In German.) [DOI] [PubMed] [Google Scholar]
- 268. Lim S. H., Choi J. S., Park E. Y. 2001. Microbial production of riboflavin uasing riboflavin overproducers, Ashbya gossypii, Bacillus subtilis and Candida famata: an overview. Biotechnol. Bioprocess Eng. 6:75–88 [Google Scholar]
- 269. Lin C., et al. 1995. Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269:968–970 [DOI] [PubMed] [Google Scholar]
- 270. Lin J. W., Chao Y. F., Weng S. F. 2001. Riboflavin synthesis genes ribE, ribB, ribH, ribA reside in the lux operon of Photobacterium leiognathi. Biochem. Biophys. Res. Commun. 284:587–595 [DOI] [PubMed] [Google Scholar]
- 271. Liu X. D., Mazumdar T., Xu Y., Getzoff E. D., Eissa N. T. 2009. Identification of a flavin mononucleotide module residue critical for activity of inducible nitrite oxide synthase. J. Immunol. 183:5977–5982 [DOI] [PubMed] [Google Scholar]
- 272. Logvinenko E. M., Shavlovskiĭ G. M., Koltun L. V., Ksheminskaia G. P. 1975. Nature of riboflavin precursors in Pichia guilliermondii yeast. Mikrobiologiia 44:48–54 (In Russian.) [PubMed] [Google Scholar]
- 273. Logvinenko E. M., Shavlovskii G. M., Kontorovskaya N. Y. 1987. On biochemical functions of the products of genes RIB5 and RIB6 involved in riboflavin biosynthesis in the yeast Pichia guilliermondii. Genetika 23:1699–1701 (In Russian.) [PubMed] [Google Scholar]
- 274. Logvinenko E. M., Shavlovskii G. M., Trach V. M., Sibirnyi V. A. 1973. Role of flavins in regulating riboflavin synthetase synthesis in Pichia guilliermondii and Candida utilis. Mikrobiologiia 42:1008–1014 (In Russian.) [PubMed] [Google Scholar]
- 275. Logvinenko E. M., Shavlovskii G. M., Zakalskii A. E., Samarskii V. A. 1989. Regulation of the activity and synthesis of enzymes participating in the formation of 6,7-dimethyl-8-ribityllumazine, a riboflavin precursor in yeast. Ukr. Biokhim. Zhurn. 61:28–32 (In Russian.) [PubMed] [Google Scholar]
- 275a. Logvinenko E. M., Shavlovskii G. M., Zakalskii A. E., Seniuta E. Z. 1980. Detection of phosphorylated pyrimidine precursors of riboflavin in yeasts. Biokhimiia 45:1284–1292 (In Russian.) [PubMed] [Google Scholar]
- 276. Logvinenko E. M., Shavlovskii G. M., Zakalskii A. E., Zakhodylo I. V. 1982. Biosynthesis of 6,7-dimethyl-8-ribityllumazine in the extracts of the yeast Pichia guilliermondii. Biokhimiia 47:931–936 (In Russian.) [PubMed] [Google Scholar]
- 277. Logvinenko E. M., et al. 1993. Cloning of the RIB7 gene encoding the riboflavin synthase of the yeast Pichia guilliermondii. Genetika 29:922–927 (In Russian.) [PubMed] [Google Scholar]
- 278. Long Q., Ji L., Wang H., Xie J. 2010. Riboflavin biosynthetic and regulatory factors as potential novel anti-infective drug targets. Chem. Biol. Drug Des. 75:339–347 [DOI] [PubMed] [Google Scholar]
- 279. Mack M., Grill S. 2006. Riboflavin analogs and inhibitors of riboflavin biosynthesis. Appl. Microbiol. Biotechnol. 71:265–275 [DOI] [PubMed] [Google Scholar]
- 280. Mack M., van Loon A. P., Hohmann H. P. 1998. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J. Bacteriol. 180:950–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Magalhães M. L., Argyrou A., Cahill S. M., Blanchard J. S. 2008. Kinetic and mechanistic analysis of the Escherichia coli ribD-encoded bifunctional deaminase-reductase involved in riboflavin biosynthesis. Biochemistry 47:6499–6507 [DOI] [PubMed] [Google Scholar]
- 282. Mailänder B., Bacher A. 1976. Biosynthesis of riboflavin. Structure of the purine precursor and origin of the ribityl side chain. J. Biol. Chem. 251:3623–3628 [PubMed] [Google Scholar]
- 283. Maita N., Hatakeyama K., Okada K., Hakoshima T. 2004. Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein. J. Biol. Chem. 279:51534–51540 [DOI] [PubMed] [Google Scholar]
- 284. Maley G. F., Plaut G. W. 1959. The isolation, synthesis, and metabolic properties of 6,7-dimethyl-8-ribityllumazine. J. Biol. Chem. 234:641–647 [PubMed] [Google Scholar]
- 285. Mansour N. M., Sawhney M., Tamang D. G., Vogl C., Saier M. H., Jr 2007. The bile/arsenite/riboflavin transporter (BART) superfamily. FEBS J. 274:612–629 [DOI] [PubMed] [Google Scholar]
- 286. Manstein D. J., Pai E. F. 1986. Purification and characterization of FAD synthetase from Brevibacterium ammoniagenes. J. Biol. Chem. 261:16169–16173 [PubMed] [Google Scholar]
- 287. Marsili E., et al. 2008. Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U. S. A. 105:3968–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Marx H., Mattanovich D., Sauer M. 2008. Overexpression of the riboflavin biosynthetic pathway in Pichia pastoris. Microb. Cell Fact. 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Mashhadi Z., Zhang H., Xu H., White R. H. 2008. Identification and characterization of an archaeon-specific riboflavin kinase. J. Bacteriol. 190:2615–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Massey V. 2000. The chemical and biological versality of riboflavin. Biochem. Soc. Trans. 28:283–296 [PubMed] [Google Scholar]
- 291. Masuda T. 1955. Application of chromatography. XXVIII. On the formation of FAD in the culture of Eremothecium ashbyii. Pharm. Bull. 3:434–440 [DOI] [PubMed] [Google Scholar]
- 292. Mateos L., Jiménez A., Revuelta J. L., Santos M. A. 2006. Purine biosynthesis, riboflavin production, and trophic-phase span are controlled by a Myb-related transcription factor in the fungus Ashbya gossypii. Appl. Environ. Microbiol. 72:5052–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Mathes T., Vogl C., Stolz J., Hegemann P. 2009. In vivo generation of flavoproteins with modified cofactors. J. Mol. Biol. 385:1511–1518 [DOI] [PubMed] [Google Scholar]
- 294. Matsui H., Sato K., Enei H., Hirose Y. 1977. Mutation of an inosine-producing strain of Bacillus subtilis to dl-methionine sulfoxide resistance for guanosine production. Appl. Environ. Microbiol. 34:337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Matsui H., Sato K., Enei H., Hirose Y. 1979. Production of guanosine by psicofuranine and decoyinine resistant mutants of Bacillus subtilis. Agric. Biol. Chem. 43:1739–1744 [Google Scholar]
- 296. Matsui K., Juri N., Kubo Y., Kasai S. 1979. Formation of roseoflavin from guanine through riboflavin. J. Biochem. 86:167–175 [PubMed] [Google Scholar]
- 297. Matsui K., Kasai S. 1996. Identification of nektoflavin as 7-alpha-hydroxyriboflavin. J. Biochem. 119:441–447 [DOI] [PubMed] [Google Scholar]
- 298. Mattevi A. 2006. To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes. Trends Biochem. Sci. 31:276–283 [DOI] [PubMed] [Google Scholar]
- 299. McCormick J. R. D., Morton G. O. 1982. Identity of cosynthetic factor 1 of Streptomyces aureofaciens and fragment F0 from coenzyme F420 of Methanobacterium sp. J. Am. Chem. Soc. 104:4014–4015 [Google Scholar]
- 300. McCormick D. B., Oka M., Bowers-Komro D. M., Yamada Y., Hartman H. A. 1997. Purification and properties of FAD synthetase from liver. Methods Enzymol. 280:407–413 [DOI] [PubMed] [Google Scholar]
- 301. McCullough J. L., Maren T. H. 1973. Inhibition of dihydropteroate synthetase from Escherichia coli by sulfones and sulfonamides. Antimicrob. Agents Chemother. 3:665–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Meighen E. A. 1993. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 7:1016–1022 [DOI] [PubMed] [Google Scholar]
- 303. Meining W., et al. 2000. The atomic structure of pentameric lumazine synthase from Saccharomyces cerevisiae at 1.85 A resolution reveals the binding mode of a phosphonate intermediate analogue. J. Mol. Biol. 299:181–197 [DOI] [PubMed] [Google Scholar]
- 304. Messner R., Prillinger H. J., Ibl M., Himmler G. 1995. Sequences of ribosomal genes and internal transcribed spacers move three plant parasitic fungi, Eremothecium ashbyi, Ashbya gossypii, and Nematospora coryli, towards Saccharomyces cerevisiae. J. Gen. Appl. Microbiol. 41:31–42 [Google Scholar]
- 305. Miersch J., Logvinenko E. M., Zakalsky A. E., Shavlovsky G. M., Reinbothe H. 1978. Origin of the ribityl side-chain of riboflavin from the ribose moiety of guanosine triphosphate in Pichia guilliermondii yeast. Biochim. Biophys. Acta 543:305–312 [DOI] [PubMed] [Google Scholar]
- 306. Miramar M. D., et al. 2001. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J. Biol. Chem. 276:16391–16398 [DOI] [PubMed] [Google Scholar]
- 307. Mironov A. S., et al. 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111:747–756 [DOI] [PubMed] [Google Scholar]
- 308. Mironov V. N., et al. 1994. Functional organization of the riboflavin biosynthesis operon from Bacillus subtilis SHgw. Mol. Gen. Genet. 242:201–208 [DOI] [PubMed] [Google Scholar]
- 309. Mironov V. N., Perumov D. A., Kraev A. S., Stepanov A. I., Skriabin K. G. 1990. Unusual structure of the regulatory region of the riboflavin biosynthesis operon in Bacillus subtilis. Mol. Biol. (Moscow) 24:256–261 [PubMed] [Google Scholar]
- 310. Mitsuda H., Tomozawa Y., Kawai F. 1963. Studies on plant flavokinase. II. The purification and some properties of bean flavokinase. J. Vitaminol. (Tokyo) 66:142–148 [PubMed] [Google Scholar]
- 311. Miura R. 2001. Versatility and specificity in flavoenzymes: control mechanisms of flavin reactivity. Chem. Rec. 1:183–194 [DOI] [PubMed] [Google Scholar]
- 312. Monschau N., Sahm H., Stahmann K. P. 1998. Threonine aldolase overexpression plus threonine supplementation enhanced riboflavin production in Ashbya gossypii. Appl. Environ. Microbiol. 64:4283–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Moore H. N., de Becze G., Schraffenberger E. 1947. Studies on Eremothecium ashbyii. J. Bacteriol. 53:502. [PubMed] [Google Scholar]
- 314. Morgunova E., et al. 2006. Structural and thermodynamic insights into the binding mode of five novel inhibitors of lumazine synthase from Mycobacterium tuberculosis. FEBS J. 273:4790–4804 [DOI] [PubMed] [Google Scholar]
- 315. Morgunova E., et al. 2005. Crystal structure of lumazine synthase from Mycobacterium tuberculosis as a target for rational drug design: binding mode of a new class of purinetrione inhibitors. Biochemistry 44:2746–2758 [DOI] [PubMed] [Google Scholar]
- 316. Morgunova E., et al. 2007. Lumazine synthase from Candida albicans as an anti-fungal target enzyme: structural and biochemical basis for drug design. J. Biol. Chem. 282:17231–17241 [DOI] [PubMed] [Google Scholar]
- 317. Mori H., Iida A., Fujio T., Teshiba S. 1997. A novel process of inosine 5′-monophosphate production using overexpressed guanosine/inosine kinase. Appl. Microbiol. Biotechnol. 48:693–698 [DOI] [PubMed] [Google Scholar]
- 318. Mörtl S., et al. 1996. Biosynthesis of riboflavin. Lumazine synthase of Escherichia coli. J. Biol. Chem. 271:33201–33327 [DOI] [PubMed] [Google Scholar]
- 319. Naik M. S., Das N. B. 1964. Effect of copper and zinc deficiency on the synthesis of protein and riboflavin by Aspergillus niger. Indian J. Exp. Biol. 2:59–64 [Google Scholar]
- 320. Nakagawa S., et al. 1995. Nucleotide sequence of the FAD synthetase gene from Corynebacterium ammoniagenes and its expression in Escherichia coli. Biosci. Biotechnol. Biochem. 59:694–702 [DOI] [PubMed] [Google Scholar]
- 321. Natraj U., Kumar R. A., Kadam P. 1987. Termination of pregnancy in mice with antiserum to chicken riboflavin-carrier protein. Biol. Reprod. 36:677–685 [DOI] [PubMed] [Google Scholar]
- 322. Nguyen H. V., Gaillardin C., Neuvéglise C. 2009. Differentiation of Debaryomyces hansenii and Candida famata by rRNA gene intergenic spacer fingerprinting and reassessment of phylogenetic relationships among D. hansenii, C. famata, D. fabryi, C. flareri (=D. subglobosus) and D. prosopidis: description of D. vietnamensis sp. nov. closely related to D. nepalensis. FEMS Yeast Res. 9:641–662 [DOI] [PubMed] [Google Scholar]
- 323. Nielsen P., Bacher A. 1981. Biosynthesis of riboflavin. Characterization of the product of the deaminase. Biochim. Biophys. Acta 662:312–317 [DOI] [PubMed] [Google Scholar]
- 324. Nielsen P., Rauschenbach P., Bacher A. 1983. Phosphates of riboflavin and riboflavin analogs: a reinvestigation by high-performance liquid chromatography. Anal. Biochem. 130:359–368 [DOI] [PubMed] [Google Scholar]
- 325. Nudler E., Mironov A. S. 2004. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 29:11–17 [DOI] [PubMed] [Google Scholar]
- 326. Ohkawa H., Ohnishi N., Yagi K. 1983. New metabolites of riboflavin appear in human purine. J. Biol. Chem. 258:5623–5628 [PubMed] [Google Scholar]
- 327. Oka M., McCormick D. B. 1987. Complete purification and general characterization of FAD synthetase from rat liver. J. Biol. Chem. 262:7418–7422 [PubMed] [Google Scholar]
- 328. O'Kane D. J., Woodward B., Lee J., Prasher D. C. 1991. Borrowed proteins in bacterial bioluminescence. Proc. Natl. Acad. Sci. U. S. A. 88:1100–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Olczyk C. 1978. n-Alkanes as a substratum for riboflavin production. I. Investigations of the dynamics of the flavinogenesis in chosen yeasts of the genus Candida. Pol. J. Pharmacol. Pharm. 30:83–88 [PubMed] [Google Scholar]
- 330. Oltmanns O., Bacher A. 1972. Biosynthesis of riboflavine in Saccharomyces cerevisiae: the role of genes rib1 and rib7. J. Bacteriol. 110:818–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Oltmanns O., Bacher A., Lingens F., Zimmermann F. K. 1969. Biochemical and genetic classification of riboflavine deficient mutants of Saccharomyces cerevisiae. Mol. Gen. Genet. 105:306–313 [DOI] [PubMed] [Google Scholar]
- 332. O'Neill H., Mayhew S. G., Butler G. 1998. Cloning and analysis of the genes for a novel electron-transferring flavoprotein from Megasphaera elsdenii. Expression and characterization of the recombinant protein. J. Biol. Chem. 273:21015–21024 [DOI] [PubMed] [Google Scholar]
- 333. Otani S. 1976. Studies on roseoflavin: isolation, physical, chemical and biological properties, p. 323–327 In Singer T. P. (ed.), Flavins and flavoproteins. Elsevier Scientific Publishing Co., Amsterdam, Netherlands [Google Scholar]
- 334. Otani S., Takatsu M., Nakano M., Kasa S., Miura R. 1974. Roseoflavin, a new antimicrobial pigment from Streptomyces. J. Antibiot. (Tokyo) 27:86–87 [PubMed] [Google Scholar]
- 335. Ott, Stolz E. J., Lehmann M., Mack M. 2009. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol. 6:276–280 [DOI] [PubMed] [Google Scholar]
- 336. Özbas T., Kutsal T. 1986. Riboflavin production by Eremothecium ashbyii in a batch stirred tank fermentor. Biotechnol. Lett. 8:441–444 [Google Scholar]
- 337. Özbas T., Kutsal T. 1986. Comparative study of riboflavin production by two organisms Eremothecium ashbyii and Ashbya gossypii. Enzyme Microb. Technol. 3:593–596 [Google Scholar]
- 338. Ozimek P., Veenhuis M., van der Klei I. J. 2005. Alcohol oxidase: a complex peroxisomal, oligomeric flavoprotein. FEMS Yeast Res. 5:975–983 [DOI] [PubMed] [Google Scholar]
- 339. Park E. Y., Zhang J. H., Tajima S., Dwiarti L. 2007. Isolation of Ashbya gossypii mutant for an improved riboflavin production targeting for biorefinery technology. J. Appl. Microbiol. 103:468–476 [DOI] [PubMed] [Google Scholar]
- 340. Paterson T., Wood H. C. 1972. The biosynthesis of pteridines. VI. Studies of the mechanism of riboflavin biosynthesis. J. Chem. Soc. Perkin 1 8:1051–1056 [DOI] [PubMed] [Google Scholar]
- 341. Perkins J. B., Pero J. 2002. Vitamin biosynthesis, p. 271–286 In Sonenshein A., Hoch J., Losick R. (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington DC [Google Scholar]
- 342. Perkins J. B., Pero J. G., Sloma A. January 1991. Riboflavin overproducing strains of bacteria. European patent EP 0 405 370 B2
- 343. Perkins J. B., et al. 1999. Genetic engineering of Bacillus subtilis for the commercial production of riboflavin. J. Ind. Microbiol. Biotechnol. 22:8–18 [Google Scholar]
- 344. Perkins J. B., et al. July 1999. Bacterial strains which overproduce riboflavin. U.S. patent 5,925,538
- 345. Perl M., Kearney E. B., Singer T. P. 1976. Transport of riboflavin into yeast cells. J. Biol. Chem. 251:3221–3228 [PubMed] [Google Scholar]
- 346. Persson K., Schneider G., Jordan D. B., Viitanen P. V., Sandalova T. 1999. Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly. Protein Sci. 8:2355–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Peschke U., Schmidt H., Zhang H. Z., Piepersberg W. 1995. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol. Microbiol. 16:1137–1156 [DOI] [PubMed] [Google Scholar]
- 348. Pett T. 1936. Lactoflavin in microorganisms. Biochem. J. 30:143816746176 [Google Scholar]
- 349. Petushkov V. N., Lee J. 1997. Purification and characterization of flavoproteins and cytochromes from the yellow bioluminescence marine bacterium Vibrio fischeri strain Y1. Eur. J. Biochem. 245:790–796 [DOI] [PubMed] [Google Scholar]
- 350. Philippsen P., Kaufmann A., Schmitz H. P. 2005. Homologues of yeast polarity genes control the development of multinucleated hyphae in Ashbya gossypii. Curr. Opin. Microbiol. 8:370–377 [DOI] [PubMed] [Google Scholar]
- 351. Pickering W. R., Woods R. A. 1973. Genetics of resistance to 4-aminopyrazolo-(3,4-d)-pyrimidine in Saccharomyces cerevisiae. Mol. Gen. Genet. 122:231–242 [DOI] [PubMed] [Google Scholar]
- 352. Plaut G. W. 1960. Studies on the stoichiometry of the enzymic conversion of 6,7-dimethyl-8-ribityllumazine to riboflavin. J. Biol. Chem. 235:PC41–PC42 [PubMed] [Google Scholar]
- 353. Plaut G. W. 1963. Studies on the nature of the enzymic conversion of 6,7-dimethyl-8-ribityllumazine to riboflavin. J. Biol. Chem. 238:2225–2243 [PubMed] [Google Scholar]
- 354. Plaut G. W. E. 1971. The biosynthesis of riboflavin, p. 11–45 In Florkin M., Stolz E. H. (ed.), Elsevier, Amsterdam, Netherlands [Google Scholar]
- 355. Plaut G. W., Beach R. L., Aogaichi T. 1970. Studies on the mechanism of elimination of protons from the methyl groups of 6,7-dimethyl-8-ribityllumazine by riboflavin synthetase. Biochemistry 9:771–785 [DOI] [PubMed] [Google Scholar]
- 356. Powers H. J. 2003. Riboflavin (vitamin B-2) and health. Am. J. Clin. Nutr. 77:1352–1360 [DOI] [PubMed] [Google Scholar]
- 357. Protchenko O. V., Boretsky Y. R., Romanyuk T. M., Fedorovych D. V. 2000. Oversynthesis of riboflavin by yeast Pichia guilliermondii in response to oxidative stress. Ukr. Biokhim. Zh. 72:19–23 [PubMed] [Google Scholar]
- 358. Protchenko O., Rodriguez-Suarez R., Androphy R., Bussey H., Philpott C. C. 2006. A screen for genes of heme uptake identifies the FLC family required for import of FAD into the endoplasmic reticulum. J. Biol. Chem. 281:21445–21457 [DOI] [PubMed] [Google Scholar]
- 359. Purwantini E., Daniels L. 1996. Purification of a novel coenzyme F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J. Bacteriol. 178:2861–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Purwantini E., Daniels L. 1998. Molecular analysis of the gene encoding F420-dependent glucose-6-phosphate dehydrogenase from Mycobacterium smegmatis. J. Bacteriol. 180:2212–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. Pynyaha Y. V., et al. 2009. Deficiency in frataxin homologue YFH1 in the yeast Pichia guilliermondii leads to missregulation of iron acquisition and riboflavin biosynthesis and affects sulfate assimilation. Biometals 22:1051–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Ramsperger A., et al. 2006. Crystal structure of an archaeal pentameric riboflavin synthase in complex with a substrate analog inhibitor: stereochemical implications. J. Biol. Chem. 281:1224–1232 [DOI] [PubMed] [Google Scholar]
- 363. Ravindranath S. D., Rao N. A. 1971. Nucleotidases in plants. V. Purification & properties of an enzyme hydrolysing flavin adenine dinucleotide at acid pH value from Phaseolus radiatus. Indian J. Biochem. 8:219–226 [PubMed] [Google Scholar]
- 364. Reihl P., Stolz J. 2005. The monocarboxylate transporter homolog Mch5p catalyzes riboflavin (vitamin B2) uptake in Saccharomyces cerevisiae. J. Biol. Chem. 280:39809–39817 [DOI] [PubMed] [Google Scholar]
- 365. Ren J., et al. 2005. GTP cyclohydrolase II structure and mechanism. J. Biol. Chem. 280:36912–36919 [DOI] [PubMed] [Google Scholar]
- 366. Renz P. 1999. Biosynthesis of the 5,6-dimethylbenzimidazole moiety of cobalamin and of the other bases found in natural corrinoids, p. 557–575 In Banerjee R. (ed.), Chemistry and Biochemistry of B12. Wiley, New York, NY [Google Scholar]
- 367. Renz P. 2007. Investigations on the biosynthesis of the 5,6-dimethylbenzimidazole moiety of vitamin B12, p. 119–130 In Kräutler B., Arigoni D., Golding B. T. (ed.), Vitamin B12 and B12 proteins. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 368. Reuke B., Korn S., Eisenreich W., Bacher A. 1992. Biosynthetic precursors of deazaflavins. J. Bacteriol. 174:4042–4049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Rhodes P. M., Winskill N., Friend E. J., Warren M. 1981. Biochemical and genetic characterisation of Streptomyces rimosus mutants impaired in oxytetracycline biosynthesis. J. Gen. Microbiol. 124:329–338 [Google Scholar]
- 370. Richter G., et al. 1992. Biosynthesis of riboflavin: cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase of Escherichia coli. J. Bacteriol. 174:4050–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371. Richter G., et al. 1993. Biosynthesis of riboflavin: cloning, sequencing, mapping, and expression of the gene coding for GTP cyclohydrolase II in Escherichia coli. J. Bacteriol. 175:4045–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Richter G., et al. 1997. Biosynthesis of riboflavin: characterization of the bifunctional deaminase-reductase of Escherichia coli and Bacillus subtilis. J. Bacteriol. 179:2022–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Ritz H., et al. 2001. Biosynthesis of riboflavin: studies on the mechanism of GTP cyclohydrolase II. J. Biol. Chem. 276:22273–22277 [DOI] [PubMed] [Google Scholar]
- 374. Rivlin R. S. (ed.) 1975. Riboflavin. Plenum Press, New York, NY [Google Scholar]
- 375. Roberts G. A., et al. 2003. A self-sufficient cytochrome P450 with a primary structural organization that includes a flavin domain and a [2Fe-2S] redox center. J. Biol. Chem. 278:48914–48920 [DOI] [PubMed] [Google Scholar]
- 376. Rodionov D. A., et al. 2009. A novel class of modular transporters for vitamins in prokaryotes. J. Bacteriol. 191:42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377. Römisch-Margl W., Eisenreich W., Haase I., Bacher A., Fischer M. 2008. 2,5-Diamino-6-ribitylamino-4(3H)-pyrimidinone 5′-phosphate synthases of fungi and archaea. FEBS J. 275:4403–4414 [DOI] [PubMed] [Google Scholar]
- 378. Roth A., Breaker R. R. 2009. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 78:305–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379. Roughead Z. K., McCormick D. B. 1990. Qualitative and quantitative assessment of flavins in cow's milk. J. Nutr. 120:382–388 [DOI] [PubMed] [Google Scholar]
- 380. Rowan T., Wood H. C. 1968. The biosynthesis of pteridines. V. The synthesis of riboflavin from pteridine precursors. J. Chem. Soc. Perkin 1 4:452–458 [DOI] [PubMed] [Google Scholar]
- 381. Rühl M., Zamboni N., Sauer U. 2010. Dynamic flux responses in riboflavin overproducing Bacillus subtilis to increasing glucose limitation in fed-batch culture. Biotechnol. Bioeng. 105:795–804 [DOI] [PubMed] [Google Scholar]
- 382. Said H. M., Arianas P. 1991. Transport of riboflavin in human intestinal brush border membrane vesicles. Gastroenterology 100:82–88 [DOI] [PubMed] [Google Scholar]
- 383. Sakai T., Watanabe T., Chibata I. 1973. Selection of microorganism producing flavin-adenine dinucleotide from FMN and adenine (AMP) and production of flavin-adenine dinucleotide by Sarcina lutea. Agric. Biol. Chem. 37:849–856 [Google Scholar]
- 384. Sancar A. 2000. Cryptochrome: the second photoactive pigment in the eye and its role in circadian photoreception. Annu. Rev. Biochem. 69:31–67 [DOI] [PubMed] [Google Scholar]
- 385. Sancar A. 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103:2203–2237 [DOI] [PubMed] [Google Scholar]
- 386. Sancar A. 2004. Photolyase and cryptochrome blue-light photoreceptors. Adv. Protein Chem. 69:73–100 [DOI] [PubMed] [Google Scholar]
- 387. Sandoval F. J., Roje S. 2005. An FMN hydrolase is fused to a riboflavin kinase homolog in plants. J. Biol. Chem. 280:38337–38345 [DOI] [PubMed] [Google Scholar]
- 388. Sandoval F. Y., Zhang Y., Roje S. 2008. Flavin nucleotide metabolism in plants: monofunctional enzymes synthesize FAD in plastids. J. Biol. Chem. 283:30890–30900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Santos M. A., García-Ramírez J. J., Revuelta J. L. 1995. Riboflavin biosynthesis in Saccharomyces cerevisiae. Cloning, characterization, and expression of the RIB5 gene encoding riboflavin synthase. J. Biol. Chem. 270:437–444 [DOI] [PubMed] [Google Scholar]
- 390. Santos M. A., Jimenez A., Revuelta J. L. 2000. Molecular characterization of FMN1, the structural gene for the monofunctional flavokinase of Saccharomyces cerevisiae. J. Biol. Chem. 275:28618–28624 [DOI] [PubMed] [Google Scholar]
- 391. Santos M. A., Mateos L., Stahmann K. P., Revuelta J. L. 2006. Insertional mutagenesis in the vitamin B2 producer fungus Ashbya gossypii, p. 283–300 In Barredo J. L. (ed.), Methods in biotechnology, vol. 18 Microbial processes and products Humana Press Inc., Totowa, NJ [Google Scholar]
- 392. Santos R., et al. 2004. Candida albicans lacking the frataxin homologue: a relevant yeast model for studying the role of frataxin. Mol. Microbiol. 54:507–519 [DOI] [PubMed] [Google Scholar]
- 393. Sato Y., et al. 2010. Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1′-d-ribityl) lumazine, and its analogues, riboflavin and flavin mononucleotide, at high resolution. J. Bacteriol. 192:127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394. Sauer U., Cameron D. C., Bailey J. E. 1998. Metabolic capacity of Bacillus subtilis for the production of purine nucleosides, riboflavin, and folic acid. Biotechnol. Bioeng. 59:227–238 [PubMed] [Google Scholar]
- 395. Saxild H. H., Nygaard P. 1987. Genetic and physiological characterization of Bacillus subtilis mutants resistant to purine analogs. J. Bacteriol. 169:2977±2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396. Schlösser T., Schmidt G., Stahmann K. P. 2001. Transcriptional regulation of 3,4-dihydroxy-2-butanone 4-phosphate synthase. Microbiology 147:3377–3386 [DOI] [PubMed] [Google Scholar]
- 397. Schlösser T., et al. 2007. Growth stress triggers riboflavin overproduction in Ashbya gossypii. Appl. Microbiol. Biotechnol. 76:569–578 [DOI] [PubMed] [Google Scholar]
- 398. Schlüpen C., et al. 2003. Disruption of the SHM2 gene, encoding one of two serine hydroxymethyltransferase isoenzymes, reduces the flux from glycine to serine in Ashbya gossypii. Biochem. J. 369:263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399. Schmidt G., Stahmann K. P., Kaesler B., Sahm H. 1996. Correlation of isocitrate lyase activity and riboflavin formation in the riboflavin overproducer Ashbya gossypii. Microbiology 142:419–426 [DOI] [PubMed] [Google Scholar]
- 400. Schmidt G., Stahmann K. P., Sahm H. 1996. Inhibition of purified isocitrate lyase identified itaconate and oxalate as potential antimetabolites for the riboflavin overproducer Ashbya gossypii. Microbiology 142:411–417 [DOI] [PubMed] [Google Scholar]
- 401. Schott K., Kellermann J., Lottspeich F., Bacher A. 1990. Riboflavin synthases of Bacillus subtilis. Purification and amino acid sequence of the alpha subunit. J. Biol. Chem. 265:4204–4209 [PubMed] [Google Scholar]
- 402. Schramek N., Bracher A., Bacher A. 2001. Biosynthesis of riboflavin. Single turnover kinetic analysis of GTP cyclohydrolase II. J. Biol. Chem. 276:44157–44162 [DOI] [PubMed] [Google Scholar]
- 403. Schrecker A. W., Kornberg A. 1950. Reversible enzymatic synthesis of flavin-adenine dinucleotide. J. Biol. Chem. 182:795–803 [PubMed] [Google Scholar]
- 404. Serganov A., Huang L., Patel D. J. 2009. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 458:233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405. Serganov A., Patel D. J. 2007. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 8:776–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406. Serganov A., Patel D. J. 2008. Towards deciphering the principles underlying an mRNA recognition code. Curr. Opin. Struct. Biol. 18:120–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407. Shavlovskii G. M., Babiak L. I., Sibirnyi A. A., Logvinenko E. M. 1985. Genetic control of riboflavin biosynthesis in Pichia guilliermondii yeasts. The detection of a new regulator gene RIB81. Genetika 21:368–374 [PubMed] [Google Scholar]
- 408. Shavlovskii G. M., Fedorovich D. V. 1977. The activity of enzymes involved in synthesis and hydrolysis of flavin adenine dinucleotide in Pichia guilliermondii: studies at different levels of flavinogenesis. Mikrobiologiia 46:904–911 (In Russian.) [PubMed] [Google Scholar]
- 409. Shavlovskii G. M., Fedorovich, Babiak L. Y. 1993. Effects of rib81 mutation on riboflavin biosynthesis and iron transport in the yeast Pichia guilliermondii. Mikrobiologiia 62:897–904 (In Russian.) [Google Scholar]
- 410. Shavlovskii G. M., Fedorovich D. V., Kutsiaba V. I., Babiak L. Y., Stenchuk N. N. 1992. Involvement of gene RIB80 in regulation of riboflavin biosynthesis and iron transport in Pichia guilliermondii. Genetika 28:25–32 (In Russian.) [Google Scholar]
- 411. Shavlovskii G. M., Fedorovich D. V., Logvinenko E. M., Koltun L. V. 1985. Isolation and characterization of the flavinogenic strains of Pichia guilliermondii bearing regulatory mutation rib80 (ribR). Mikrobiologiia 54:919–925 (In Russian.) [Google Scholar]
- 412. Shavlovskii G. M., Koltun L. V., Kashchenko V. E. 1978. Regulation of the activity of GTP-cyclohydrolase, the enzyme of the first step of flavinogenesis in yeasts. Biokhimiia 43:2074–2081 (In Russian.) [PubMed] [Google Scholar]
- 413. Shavlovskii G. M., Koltun L. V., Kshanovskaya B. V., Logvinenko E. M., Stenchuk N. N. 1989. Regulation of biosynthesis of riboflavin by elements of positive control in Pichia guilliermondii yeast. Genetika 25:250–258 (In Russian.) [Google Scholar]
- 414. Shavlovskii G. M., Logvinenko E. M. 1985. Flavin biogenesis in yeasts. Ukr. Biokhim. Zh. 57:98–112 (In Russian.) [PubMed] [Google Scholar]
- 415. Shavlovskii G. M., Logvinenko E. M. 1988. Supersynthesis of flavins in microorganisms and its molecular mechanism (review of the literature). Prikl. Biokhim Mikrobiol. 24:435–447 (In Russian.) [PubMed] [Google Scholar]
- 416. Shavlovskii G. M., Logvinenko E. M., Kashchenko V. E., Koltun L. V., Zakalskii A. E. 1976. Detection in Pichia guilliermondii of GTP-cyclohydrolase, an enzyme involved in the 1st stage of flavinogenesis. Dokl. Akad. Nauk SSSR 230:1485–1487 (In Russian.) [PubMed] [Google Scholar]
- 417. Shavlovskii G. M., Logvinenko E. M., Sibirnyi A. A., Fedorovich D. V., Zakalskii A. E. 1981. Activity of the enzyme of the 2d step of flavinogenesis, 2,5-diamino-6-hydroxy-4-ribosylaminopyrimidine-5′-phosphate reductase, in Pichia guilliermondii yeasts. Mikrobiologiia 50:1008–1011 (In Russian.) [PubMed] [Google Scholar]
- 418. Shavlovskii G. M., Logvinenko E. M., Zakalskii A. E. 1983. Purification and some properties of GTP cyclohydrolase of the yeast Pichia guilliermondii. Biokhimia 48:837–843 (In Russian.) [PubMed] [Google Scholar]
- 419. Shavlovskii G. M., Sibirnyi A. A., Fedorovich D. V., Seniuta E. Z. 1982. Selection and mutant properties of Pichia guilliermondii yeasts with derepressed GTP cyclohydrolase, the enzyme of the 1st step in flavinogenesis. Mikrobiologiia 51:96–101 (In Russian.) [PubMed] [Google Scholar]
- 420. Shavlovskii G. M., Sibirnyi A. A., Kshanovskaia B. V., Koltun L. V., Logvinenko E. M. 1979. Genetic classification of riboflavin-dependent mutants of Pichia guilliermondii yeasts. Genetika 15:1561–1568 (In Russian.) [PubMed] [Google Scholar]
- 421. Shavlovskii G. M., Sibirnyi A. A., Ksheminskaya G. P., Pinchuk G. E. 1980. Overproduction of riboflavin in mutants of Pichia guilliermondii yeasts resistant to 7-methyl-8-trifluoromethyl-10-(1′-d-ribityl)isoalloxazine. Mikrobiologiia 49:702–707 (In Russian.) [PubMed] [Google Scholar]
- 422. Shavlovskii G. M., Tesliar G. E., Strugovshchikova L. P. 1982. Flavinogenesis regulation in riboflavin-dependent Escherichia coli mutants. Mikrobiologiia 51:986–992 (In Russian.) [PubMed] [Google Scholar]
- 423. Shavlovskii G. M., et al. 1978. Flavinogenic activity of natural strains of the yeast Pichia guilliermondii. Prikl. Biokhim. Mikrobiol. 14:184–189 (In Russian.) [Google Scholar]
- 424. Shavlovsky G. M., Kashchenko V. E. 1975. Determination of riboflavin kinase activity in yeast. Ukr. Biokhim. Zh. 47:536–541 (In Ukrainian.) [PubMed] [Google Scholar]
- 425. Schavlovsky G. M., et al. 1980. First reaction of riboflavin biosynthesis: catalysis by a guanosine triphosphate cyclohydrolase from yeast. Arch. Microbiol. 124:255–259 [Google Scholar]
- 426. Shavlovsky G. M., Sibirny A. A. 1985. Riboflavin transport in yeasts and its regulation, p. 385–392 In Kulaev I. S., Tempest D. W., Dawes E. A. (ed.), Environmental regulation of microbial metabolism. Academic Press, London, United Kingdom [Google Scholar]
- 427. Shavlovsky G. M., Sibirny A. A., Ksheminskaya G. P. 1977. Permease and “excretase” for riboflavin in the mutants of Pichia guilliermondii yeast. Biochem. Physiol. Pflanzen. 171:139–145 [Google Scholar]
- 428. Shenton W., Mann S., Cölfen H., Bacher A., Fischer M. 2001. Synthesis of nanophase irone oxide in lumazine synthase capsids. Angew Chem. Intern. Edit. Engl. 40:442–445 [DOI] [PubMed] [Google Scholar]
- 429. Shi S., Chen T., Zhang Z., Chen X., Zhao X. 2009. Transcriptome analysis guided metabolic engineering of Bacillus subtilis for riboflavin production. Metab. Eng. 11:243–252 [DOI] [PubMed] [Google Scholar]
- 430. Shimizu S. 2001. Vitamins and related compounds: microbial production, p. 322–326 In Rehm H. (ed)., Biotechnology, vol. 10 Wiley-VCH, Weinheim, Germany [Google Scholar]
- 431. Shimizu S., Ishida M., Kato N., Tani Y., Ogata K. 1977. Derepression of FAD pyrophosphorylase and flavin changes during growth of Kloeckera sp. no. 2201 on methanol. Agric. Biol. Chem. 41:2215–2220 [Google Scholar]
- 432. Shimizu S., Ishida M., Tani Y., Ogata K. 1977. Flavin changes of Kloeckera sp. no. 2201 during adaptation to methanol. Agric. Biol. Chem. 41:423–424 [Google Scholar]
- 433. Shimizu S., Yamane K., Tani Y., Yamada H. 1983. Enzymatic synthesis of flavin adenine dinucleotide. Appl. Biochem. Biotechnol. 8:237–247 [DOI] [PubMed] [Google Scholar]
- 434. Shinkai S., et al. 1986. Coenzyme models. 40. Spectral and reactivity studies of roseoflavin analogs: correlation between reactivity and spectral parameters. Bioorg. Chem. 14:119–133 [Google Scholar]
- 435. Shukla J. P., Prabku K. A. 1961. Studies on the production of riboflavin by Aspergillus niger. J. Sci. Ind. Res. 40:20–24 [Google Scholar]
- 436. Sibirny A. A. 1996. Pichia guilliermondii, p. 255–272 In Wolf K. (ed.), Nonconventional yeasts in biotechnology. Springer Verlag, Berlin, Germany [Google Scholar]
- 437. Sibirny A. A., Boretsky Y. R. 2009. Pichia guilliermondii, p. 113–134 In Satyanarayana T., Kunze G. (ed.), Yeast biotechnology: diversity and applications. Springer Science, New York, NY [Google Scholar]
- 438. Sibirny A. A., Fedorovych D. V., Boretsky Y. R., Voronovsky A. Y. 2006. Microbial synthesis of flavins. Naukova Dumka, Kiev, Ukraine: (In Ukrainian.) [Google Scholar]
- 439. Sibirny A. A., Dmytruk K. V., Fedorovych D. V. May 2010. The yeast strain Candida famata IMB Y-5034, the riboflavin producer. UA patent no. 90754
- 440. Sibirny A. A., Shavlovsky G. M. 1984. Identification of regulatory genes of riboflavin permease and α-glucosidase in the yeast Pichia guilliermondii. Curr. Genet. 8:107–114 [DOI] [PubMed] [Google Scholar]
- 441. Sibirny A. A., Voronovsky A. Y. 2009. Candida famata (Debaryomyces hansenii), p. 85–111 In Satyanarayana T., Kunze G. (ed.), Yeast biotechnology: diversity and applications. Springer Science, New York, NY [Google Scholar]
- 442. Sibirnyi A. A., Ksheminskaya G. P., Orlovskaya A. G., Shavlovskii G. M. 1981. Riboflavin transport in the mutant of Pichia guilliermondii yeast defective in glucose uptake. Biokhimiia 46:1761–1763 (In Russian.) [PubMed] [Google Scholar]
- 443. Sibirnyi A. A., Shavlovskii G. M. 1978. On the inhibition of alkaline phosphatase I of the yeast Pichia guilliermondii in vitro and in vivo. Ukr. Biokhim. Zhurn. 50:226–231 (In Russian.) [PubMed] [Google Scholar]
- 444. Sibirnyi A. A., Shavlovskii G. M. 1981. Increase in yeast and bacterial sensitivity to inhibitors and riboflavin as affected by high sulfate and phosphate concentrations. Mikrobiologiia 50:242–248 (In Russian.) [PubMed] [Google Scholar]
- 445. Sibirnyi A. A., Shavlovskii G. M. August 1984. Mutants of the yeast Pichia guilliermondii accumulating high amounts of riboflavin in the cells. USSR author's certificate no. 207914 [Google Scholar]
- 446. Sibirnyi A. A., Shavlovskii G. M. January 1984. The method of the production of 6,7-dimethyl-8-ribityllumazine. USSR author's certificate no. 1092952 [Google Scholar]
- 447. Sibirnyi A. A., Shavlovskii G. M., Goloshchapova G. V. 1977. The mutants of the yeast Pichia guilliermondii with multiple sensitivity to antibiotics and antimetabolites. Selection and some properties of the mutants. Genetika 13:872–879 (In Russian.) [PubMed] [Google Scholar]
- 448. Sibirnyi A. A., Shavlovskii G. M., Ksheminskaya G. P., Naumov G. I. 1977. Hybridization and meiotic segregation in the paraffin-utilizing yeast Pichia guilliermondii Wickerham. Genetika 13:314–321 (In Russian.) [Google Scholar]
- 449. Sibirnyi A. A., Shavlovskii G. M., Ksheminskaya G. P., Orlovskaya A. G. 1977. Active riboflavin transport in the yeast Pichia guilliermondii. Finding and properties of the cryptic riboflavin permease. Biokhimiia 42:1841–1851 (In Russian.) [PubMed] [Google Scholar]
- 450. Sibirnyi A. A., Shavlovskii G. M., Ksheminskaya G. P., Orlovskaya A. G. 1977. On riboflavin transport in the cells of riboflavinless yeast mutants. Mikrobiologiia 46:376–378 (In Russian.) [PubMed] [Google Scholar]
- 451. Sibirnyi A. A., Shavlovskii G. M., Ksheminskaya G. P., Orlovskaya A. G. 1978. The influence of glucose and some its derivates on the systems for uptake and excretion of riboflavin in yeast Pichia guilliermondii. Biokhimiia 43:1414–1422 (In Russian.) [PubMed] [Google Scholar]
- 452. Sibirnyi A. A., Shavlovskii G. M., Ksheminskaya G. P., Orlovskaya A. G. 1979. Coordinate regulation of riboflavin permease and α-glucosidase synthesis in the yeast Pichia guilliermondii. Biokhimiia 44:1558–1568 (In Russian.) [PubMed] [Google Scholar]
- 453. Sibirnyi A. A., Trach V. M., Shavlovskii G. M. September 1984. The method for riboflavin sorption from solutions. USSR author's certificate no. 209240 [Google Scholar]
- 454. Sibirnyi A. A., Zharova V. P., Kshanovskaia B. V., Shavlovskii G. M. 1977. Selection of a genetic strain of Pichia guilliermondii yeasts capable of forming a significant quantity of spores. Tsitol. Genet. 11:330–333 (In Russian.) [PubMed] [Google Scholar]
- 455. Siddiqi R., Khan M. A. 1982. Vitamin and nitrogen base requirements for Listeria monocytogenes and haemolysin production. Zentralbl. Bakteriol. Mikrobiol. Hyg. A. 253:225–235 [PubMed] [Google Scholar]
- 456. Snell E. E., Strong F. M. 1939. A microbiological assay for riboflavin. Ind. Eng. Chem. Anal. ed. 11:346–350 [Google Scholar]
- 457. Solovieva I. M., Kreneva R. A., Errais Lopez L., Perumov D. A. 2005. The riboflavin kinase encoding gene ribR of Bacillus subtilis is a part of a 10 kb operon, which is negatively regulated by the yrzC gene product. FEMS Microbiol. Lett. 243:51–58 [DOI] [PubMed] [Google Scholar]
- 458. Solovieva I. M., Kreneva R. A., Leak D. J., Perumov D. A. 1999. The ribR gene encodes a monofunctional riboflavin kinase which is involved in regulation of Bacillus subtilis riboflavin operon. Mikrobiologiia 145:67–73 [DOI] [PubMed] [Google Scholar]
- 459. Spector R. 1982. Riboflavin transport by rabbit kidney slices: characterization and relation to cyclic organic acid transport. J. Pharmacol. Exp. Ther. 221:394–398 [PubMed] [Google Scholar]
- 460. Spitzner A., Perzlmaier A. F., Geillinger K. E., Reihl P., Stolz J. 2008. The proline-dependent transcription factor Put3 regulates the expression of the riboflavin transporter MCH5 in Saccharomyces cerevisiae. Genetics 180:2007–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461. Spoonamore J. E., Dahlgran A. L., Jacobsen N. E., Bandarian V. 2006. Evolution of new function in the GTP cyclohydrolase II proteins of Streptomyces coelicolor. Biochemistry 45:12144–12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462. Stahmann K. P., Revuelta J. L., Seulberger H. 2000. Three biotechnical processes using Ashbya gossypii, Candida famata, or Bacillus subtilis compete with chemical riboflavin production. Appl. Microbiol. Biotechnol. 53:509–516 [DOI] [PubMed] [Google Scholar]
- 463. Stahmann K. P., et al. 2001. Riboflavin, overproduced during sporulation of Ashbya gossypii, protects its hyaline spores against ultraviolet light. Environ. Microbiol. 3:545–550 [DOI] [PubMed] [Google Scholar]
- 464. Stasyk O. V., et al. 2004. A hexose transporter homologue controls glucose repression in the methylotrophic yeast Hansenula polymorpha. J. Biol. Chem. 279:8116–8125 [DOI] [PubMed] [Google Scholar]
- 465. Steinbacher S., Schiffmann S., Bacher A., Fischer M. 2004. Metal sites in 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii in complex with the substrate ribulose 5-phosphate. Acta Crystallogr. D Biol. Crystallogr. 60:1338–1340 [DOI] [PubMed] [Google Scholar]
- 466. Steinbacher S., et al. 2003. Structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii in complex with divalent metal ions and the substrate ribulose 5-phosphate: implications for the catalytic mechanism. J. Biol. Chem. 278:42256–42265 [DOI] [PubMed] [Google Scholar]
- 467. Steiner S., Philippsen P. 1994. Sequence and promoter analysis of the highly expressed TEF gene of the filamentous fungus Ashbya gossypii. Mol. Gen. Genet. 242:263–271 [DOI] [PubMed] [Google Scholar]
- 468. Steiner S., Wendland J., Wright M. C., Philippsen P. 1995. Homologous recombination as the main mechanism for DNA integration and cause of rearrangements in the filamentous ascomycete Ashbya gossypii. Genetics 140:973–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469. Stenchuk M. M., Kapustyak K. E. 1999. The new gene RED1 controlling biosynthesis of riboflavin and ferrireductase activity in the yeast Pichia guilliermondii. Biopolymers Cell 15:522–528 (In Ukrainian.) [Google Scholar]
- 470. Stenchuk N. N., Kapustyak K. E. 2003. The red mutations impair regulation of flavinogenesis and metal homeostasis in the yeast Pichia guilliermondii. Genetika 39:1026–1032 (In Russian.) [PubMed] [Google Scholar]
- 471. Stenchuk N. N., Kutsiaba V. I., Kshanovskaia B. V., Fedorovich D. V. 2001. Effect of rib83 mutation on riboflavin biosynthesis and iron assimilation in Pichia guilliermondii. Mikrobiologiia 70:753–758 (In Russian.) [PubMed] [Google Scholar]
- 472. Stenchuk N. N., Protchenko O. V., Fedorovich D. V., Shavlovskii G. M. 1991. The mutants of Pichia guilliermondii with enhanced ability to reduce iron ions and riboflavin. Genetika 27:561–564 (In Russian.) [Google Scholar]
- 473. Stenmark P., Moche M., Gurmu D., Nordlund P. 2007. The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism. J. Mol. Biol. 373:48–64 [DOI] [PubMed] [Google Scholar]
- 474. Stepanov A. I., Beburov M. Y., Zhdanov V. G. 1974. Mutants of Eremothecium ashbyii resistant to 8-azaguanine. Communication I. Isolation of mutants and study of the level of riboflavin biosynthesis. Sov. Genet. 8:729–733 [PubMed] [Google Scholar]
- 475. Stepanov A. I., Zhdanov V. G. 1974. The use of mutagenic factors in the selection of the riboflavin producer Eremothecium ashbyii. Sov. Genet. 8:745–749 [PubMed] [Google Scholar]
- 476. Stover C. K., et al. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405:962–966 [DOI] [PubMed] [Google Scholar]
- 477. Straube G. 1980. Riboflavin accumulation by cells of the yeast Pichia (Candida) guilliermondii. Z. Allg. Mikrobiol. 20:215–218 [PubMed] [Google Scholar]
- 478. Stripp B. 1975. Intestinal absorption of riboflavin in man. Acta Pharmacol. Toxicol. 22:353–362 [DOI] [PubMed] [Google Scholar]
- 479. Strugovshchikova L. P., Shavlovskiĭ G. M., Fedorovich I. P., Kucheras R. V. 1973. Purification and some properties of the alkaline phosphatase I of Pichia guilliermondii yeasts. Ukr. Biokhim. Zhurn. 45:312–317 (In Ukrainian.) [PubMed] [Google Scholar]
- 480. Sugimoto T., Kanamasa S., Kato T., Park E. Y. 2009. Importance of malate synthase in the glyoxylate cycle of Ashbya gossypii for the efficient production of riboflavin. Appl. Microbiol. Biotechnol. 83:529–539 [DOI] [PubMed] [Google Scholar]
- 481. Sugimoto T., Morimoto A., Nariyama M., Kato T., Park E. Y. 2010. Isolation of an oxalate-resistant Ashbya gossypii strain and its improved riboflavin production. J. Ind. Microbiol. Biotechnol. 37:57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482. Surribas A., Resina D., Ferrer P., Valero F. 2007. Riboflavin may interfere with on-line monitoring of secreted green fluorescence protein fusion proteins in Pichia pastoris. Microb. Cell Fact. 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483. Survase S. A., Bajaj I. B., Singhal R. S. 2006. Biotechnological production of vitamins. Food Technol. Biotechnol. 44:381–396 [Google Scholar]
- 484. Susin S., et al. 1993. Riboflavin 3′-sulfate and 5′-sulfate, two novel flavins accumulating in the roots of iron-deficient sugar beet (Beta vulgaris). J. Biol. Chem. 268:20958–20965 [PubMed] [Google Scholar]
- 485. Suzuki Y., Katagiri H. 1963. Studies on dextransucrase. II. Factors affecting the formation of riboflavinylglucoside in growing cultures of Leuconostoc mesenteroides. J. Vitaminol. (Kyoto) 10:293–298 [PubMed] [Google Scholar]
- 486. Sybesma W., Burgess C., Starrenburg M., Sinderen D. V., Hugenholtz J. 2004. Multivitamin production in Lactococcus lactis using metabolic engineering. Metab. Eng. 6:109–115 [DOI] [PubMed] [Google Scholar]
- 487. Sydorovych I. B., Fedorovych D. V. 2002. Influence of manganese on iron accumulation and flavinogenesis in yeast Debaryomyces hansenii. Mikrobiol. Z. 64:47–52 (In Ukrainian.) [PubMed] [Google Scholar]
- 488. Tachibana S., Murakami T., Ninomiya T. 1975. Identification of the chemical structures of schizoflavins as 7,8-dimethyl-10-(2,3,4-trihydroxy-4-formylbutyl)isoalloxazine and 7,8-dimethyl-10-(2,3,4-trihydroxy-4-carboxybutyl)isoalloxazine. J. Nutr. Sci. Vitaminol. 21:347–353 [DOI] [PubMed] [Google Scholar]
- 488a. Taga M. E., Larsen N. A., Howard-Jones A. R., Walsh C. T., Walker G. C. 2007. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature 446:449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489. Taheri P., Tarighi S. 2010. Riboflavin induces resistance in rice against Rhizoctonia solani via jasmonate-mediated priming of phenylpropanoid pathway. J. Plant Physiol. 167:201–208 [DOI] [PubMed] [Google Scholar]
- 490. Tajima S., Itoh Y., Sugimoto T., Kato T., Park E. Y. 2009. Increased riboflavin production from activated bleaching earth by a mutant strain of Ashbya gossypii. J. Biosci. Biotechnol. 108:325–329 [DOI] [PubMed] [Google Scholar]
- 491. Talukdar A., et al. 2009. Discovery and development of a small molecule library with lumazine synthase inhibitory activity. J. Org. Chem. 74:5123–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492. Tanner F. W., Jr., Voinovich C., Van Lanen J. M. 1945. Riboflavin production by Candida species. Science 101:180–181 [DOI] [PubMed] [Google Scholar]
- 493. Tännler S., Zamboni N., Kiraly C., Aymerich S., Sauer U. 2008. Screening of Bacillus subtilis transposon mutants with altered riboflavin production. Metab. Eng. 10:216–226 [DOI] [PubMed] [Google Scholar]
- 494. Terrade N., Mira de Orduña R. 2009. Determination of the essential nutrient requirements of wine-related bacteria from the genera Oenococcus and Lactobacillus. Int. J. Food Microbiol. 133:8–13 [DOI] [PubMed] [Google Scholar]
- 495. Tesliar G. E., Shavlovskii G. M. 1983. Localization of the genes coding for GTP cyclohydrolase II and riboflavin synthase on the chromosome of Escherichia coli K-12. Tsitol. Genet. 17:54–56 (In Russian.) [PubMed] [Google Scholar]
- 496. Tielker D., Eichhof I., Jaeger K. E., Ernst J. F. 2009. Flavin mononucleotide-based fluorescent protein as an oxygen-independent reporter in Candida albicans and Saccharomyces cerevisiae. Eukaryot. Cell 8:913–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497. Torchetti E. M., Brizio C., Coletta M., Galuccio M., Giancaspero T. A. 2010. Mitochondrial localization of human FAD synthetase isoform 1. Mitochondrion 10:263–273 [DOI] [PubMed] [Google Scholar]
- 498. Ubiivovk V. M., Sibirnyi A. A. 1991. Biochemical investigation of the properties of a mutant of the methylotrophic yeast Hansenula polymorpha with reduced FAD and alcohol oxidase content. Biochemistry (Moscow) 56:1570–1576 [Google Scholar]
- 499. Uyakul D., Isobe M., Goto T. 1990. Lampteroflavin, the first riboflavinyl alpha ribofuranoside as light emitter in the luminous mushroom Lampteromyces japonicus. Tetrahedron 46:1367–1378 [Google Scholar]
- 500. van der Klei I. J., Harder W., Veenhuis M. 1991. Biosynthesis and assembly of alcohol oxidase, a peroxisomal matrix protein in methylotrophic yeasts: a review. Yeast 7:195–209 [DOI] [PubMed] [Google Scholar]
- 501. van Herwaarden A. E., et al. 2007. Multidrug transporter ABCG2/breast cancer resistance protein secretes riboflavin (vitamin B2) into milk. Mol. Cell. Biol. 27:1247–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502. Vardja T., Vardja R., Pudersell K., Tohver A. 2004. Riboflavin excretion from the excised roots of Hyoscyamus niger. Pharm. Biol. 42:353–359 [Google Scholar]
- 503. Vinas P., Balsalobre N., Lopez-Erroz C., Hernandez-Cordoba M. 2004. Liquid chromatographic analysis of riboflavin vitamers in foods using fluorescence detection. J. Agric. Food Chem. 52:1789–1794 [DOI] [PubMed] [Google Scholar]
- 504. Vitreschak A. G., Rodionov D. A., Mironov A. A., Gelfand M. S. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30:3141–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505. Vogl C., et al. 2007. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J. Bacteriol. 189:7367–7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506. Volk R., Bacher A. 1988. Biosynthesis of riboflavin. The structure of the four-carbon precursor. J. Am. Chem. Soc. 110:3651–3653 [Google Scholar]
- 507. Volk R., Bacher A. 1990. Studies on the 4-carbon precursor in the biosynthesis of riboflavin. Purification and properties of L-3,4-dihydroxy-2-butanone-4-phosphate synthase. J. Biol. Chem. 265:19479–19485 [PubMed] [Google Scholar]
- 508. Volk R., Bacher A. 1991. Biosynthesis of riboflavin. Studies on the mechanism of l-3,4-dihydroxy-2-butanone 4-phosphate synthase. J. Biol. Chem. 266:20610–20618 [PubMed] [Google Scholar]
- 509. von Canstein H., Ogawa J., Shimizu S., Lloyd J. R. 2008. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 74:615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510. Voronovsky A. Y., et al. 2004. Candida famata (Debaryomyces hansenii) DNA sequences containing genes involved in riboflavin synthesis. Yeast 21:1307–1316 [DOI] [PubMed] [Google Scholar]
- 511. Voronovsky A. Y., et al. 2002. Development of a transformation system for the flavinogenic yeast Candida famata. FEMS Yeast Res. 2:381–388 [DOI] [PubMed] [Google Scholar]
- 512. Vorwieger A., et al. 2007. Iron assimilation and transcription factor controlled synthesis of riboflavin in plants. Planta 226:147–158 [DOI] [PubMed] [Google Scholar]
- 513. Wacker H., Harvey R. A., Winestock C. H., Plaut G. W. 1964. 4-(1′-d-Ribitylamino)-5-amino-2,6-dihydroxypyrimidine, the second product of the riboflavin synthetase reaction. J. Biol. Chem. 239:3493–3497 [PubMed] [Google Scholar]
- 514. Walsh C. 1986. Naturally occurring 5-deazaflavin coenzymes: biological redox roles. Acc. Chem. Res. 19:216–221 [Google Scholar]
- 515. Walsh C., et al. 1978. Chemical and enzymatic properties of riboflavin analogues. Biochemistry 17:1942–1951 [DOI] [PubMed] [Google Scholar]
- 516. Wang L., et al. 2008. Isolation and characterization of Candida membranifaciens subsp. flavinogenie W14-3, a novel riboflavin-producing marine yeast. Microbiol. Res. 163:255–266 [DOI] [PubMed] [Google Scholar]
- 517. Wang W., Kim R., Jancarik J., Yokota H., Kim S. H. 2003. Crystal structure of a flavin-binding protein from Thermotoga maritima. Proteins 52:633–635 [DOI] [PubMed] [Google Scholar]
- 518. Wang W., Kim R., Yokota H., Kim S. H. 2005. Crystal structure of flavin binding to FAD synthetase of Thermotoga maritima. Proteins 58:246–248 [DOI] [PubMed] [Google Scholar]
- 519. Warren M. J. 2006. Finding the final pieces of the vitamin B12 biosynthetic jigsaw. Proc. Natl. Acad. Sci. U. S. A. 103:4799–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520. Watanabe T., Uchida T., Kato J., Chibata I. 1974. Production of flavine-adenine dinucleotide from riboflavine by a mutant of Sarcina lutea. Arch. Microbiol. 27:531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521. Weinstein L. H., Purvis E. R., Meiss A. N., Uhler R. L. 1954. Chelates, absorption and translocation of ethylenediaminetetraacetic acid by sunflower plants. J. Agric. Food Chem. 2:421–424 [Google Scholar]
- 522. Wendland J., Walther A. 2005. Ashbya gossypii: a model for fungal developmental biology. Nat. Rev. Microbiol. 3:421–429 [DOI] [PubMed] [Google Scholar]
- 523. White H. B., III, Merrill A. H., Jr 1988. Riboflavin-binding proteins. Annu. Rev. Nutr. 8:279–299 [DOI] [PubMed] [Google Scholar]
- 524. White R. H. 2001. Biosynthesis of methanogenic cofactors. Vitamins Hormones 61:299–339 [DOI] [PubMed] [Google Scholar]
- 525. Wickerham L. J., Flickinger M. H., Johnston R. M. 1946. The production of riboflavin by Ashbya gossypii. Arch. Biochem. 9:95–98 [PubMed] [Google Scholar]
- 525a. Wiley, 1984. Kirk-Othmer encyclopedia of chemical technology, p. 108–124 Wiley, New York, NY [Google Scholar]
- 526. Wilson A. C., Pardee A. B. 1962. Regulation of flavin synthesis by Escherichia coli. J. Gen. Microbiol. 28:283–303 [DOI] [PubMed] [Google Scholar]
- 527. Winkler A., Hartner F., Kutchan T. M., Glieder A., Macheroux P. 2006. Biochemical evidence that berberine bridge enzyme belongs to a novel family of flavoproteins containing a bi-covalently attached FAD cofactor. J. Biol. Chem. 281:21276–21285 [DOI] [PubMed] [Google Scholar]
- 528. Winkler W. C., Cohen-Chalamish S., Breaker R. R. 2002. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. U. S. A. 99:15908–15913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529. Worst D. J., Gerrits M. M., Vandenbrouke-Grauls C. M., Kusters J. G. 1998. Helicobacter pylori ribBA-mediated riboflavin production is involved in iron acquisition. J. Bacteriol. 180:1473–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530. Wright M. C., Philippsen P. 1991. Replicative transformation of the filamentous fungus Ashbya gossypii with plasmids containing Saccharomyces cerevisiae ARS elements. Gene 109:99–105 [DOI] [PubMed] [Google Scholar]
- 531. Wu M., Repetto B., Glerum D. M., Tzagoloff A. 1995. Cloning and characterization of FAD1, the structural gene for flavin adenine dinucleotide synthetase of Saccharomyces cerevisiae. Mol. Cell. Biol. 15:264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532. Wu Q. L., Chen T., Gan Y., Chen X., Zhao X. M. 2007. Optimization of riboflavin production by recombinant Bacillus subtilis RH44 using statistical designs. Appl. Microbiol. Biotechnol. 76:783–794 [DOI] [PubMed] [Google Scholar]
- 533. Yagi K., Matsuoka Y. 1955. Separating determination of riboflavin nucleotides by paper electrophoresis. J. Biochem. 42:757–762 [Google Scholar]
- 534. Yagi K., Matsuoka Y., Kuyama S., Tada M. 1956. Preparation of flavin adenine dinucleotide from Eremothecium ashbyii. J. Biochem. 43:93–100 [Google Scholar]
- 535. Yagi K., Nagase F. 1975. Flavins in chick embryo. J. Nutr. Sci. Vitaminol. 21:27–30 [DOI] [PubMed] [Google Scholar]
- 536. Yagi K., Okuda J., Matsuoka Y. 1955. Separation of flavins by ion-exchange resins. Nature 175:555–556 [DOI] [PubMed] [Google Scholar]
- 537. Yamada Y., Merrill A. H., Jr., McCormick D. B. 1990. Probable reaction mechanisms of flavokinase and FAD synthetase from rat liver. Arch. Biochem. Biophys. 278:125–130 [DOI] [PubMed] [Google Scholar]
- 538. Yamamoto S., et al. 2009. Identification and functional characterization of rat riboflavin transporter 2. J. Biochem. 145:437–443 [DOI] [PubMed] [Google Scholar]
- 539. Yamasaki I., Yositome W. 1938. Über die Flavingarung der Aceton-Butylalkoholbakterien. Biochem. Z. 297:398 [Google Scholar]
- 540. Yatsyshyn V. Y., Fedorovych D. V., Sibirny A. A. 2009. The microbial synthesis of flavin nucleotides: a review. Appl. Biochem. Microbiol. 45:115–124 [PubMed] [Google Scholar]
- 541. Yatsyshyn V. Y., Fedorovych D. V., Sibirny A. A. 2010. Medium optimization for production of flavin mononucleotide by the recombinant strain of the yeast Candida famata using statistical designs. Biochem. Eng. J. 49:52–60 [Google Scholar]
- 542. Yatsyshyn V. Y., Ishchuk O. P., Voronovsky A. Y., Fedorovych D. V., Sibirny A. A. 2009. Production of flavin mononucleotide by metabolically engineered yeast Candida famata. Metab. Eng. 11:163–167 [DOI] [PubMed] [Google Scholar]
- 543. Yazdanpanah B., et al. 2009. Riboflavin kinase couples TNF receptor 1 to NADPH oxidase. Nature 460:1159–1163 [DOI] [PubMed] [Google Scholar]
- 544. Yonezawa A., Masuda S., Katsura T., Inui K. 2008. Identification and functional characterization of a novel human and rat riboflavin transporter, RFT1. Am. J. Physiol. Cell Physiol. 295:C632–C641 [DOI] [PubMed] [Google Scholar]
- 545. Yuasa, Hirobe H. M., Tomei S., Watanabe J. 2000. Carrier-mediated transport of riboflavin in the rat colon. Biopharm. Drug Dispos. 21:77–82 [DOI] [PubMed] [Google Scholar]
- 546. Zakalskii A. E., et al. 1990. Cloning of the RIB1 gene coding for the enzyme of the first stage of flavinogenesis in the yeast Pichia guilliermondi, GTP cyclohydrolase, in Escherichia coli cells. Genetika 26:614–620 (In Russian.) [PubMed] [Google Scholar]
- 547. Zamboni N., Maaheimo H., Szyperski T., Hohmann H. P., Sauer U. 2004. The phosphoenolpyruvate carboxykinase also catalyzes C3 carboxylation at the interface of glycolysis and the TCA cycle of Bacillus subtilis. Metab. Eng. 6:277–284 [DOI] [PubMed] [Google Scholar]
- 548. Zamboni N., Mouncey N., Hohmann H. P., Sauer U. 2003. Reducing maintenance metabolism by metabolic engineering of respiration improves riboflavin production by Bacillus subtilis. Metab. Eng. 5:49–55 [DOI] [PubMed] [Google Scholar]
- 549. Zandomeneghi M., Carbonaro L., Zandomeneghi G. 2007. Biochemical fluorometric method for the determination of riboflavin in milk. J. Agric. Food Chem. 55:5990–5994 [DOI] [PubMed] [Google Scholar]
- 550. Zhang S., et al. 2009. Riboflavin-induced priming for pathogen defense in Arabidopsis thaliana. J. Integr. Plant Biol. 51:167–174 [DOI] [PubMed] [Google Scholar]
- 551. Zhang X., et al. 2003. A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus. J. Mol. Biol. 328:167–182 [DOI] [PubMed] [Google Scholar]
- 552. Zhang X., Meining W., Fischer M., Bacher A., Ladenstein R. 2001. X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons. J. Mol. Biol. 306:1099–1114 [DOI] [PubMed] [Google Scholar]
- 553. Zhao Y., et al. 2009. Discovery and development of the covalent hydrates of trifluoromethylated pyrazoles as riboflavin synthase inhibitors with antibiotic activity against Mycobacterium tuberculosis. J. Org. Chem. 74:5297–5303 [DOI] [PubMed] [Google Scholar]
- 554. Zhu Y., Chen X., Chen T., Zhao X. 2007. Enhancement of riboflavin production by overexpression of acetolactate synthase in a pta mutant of Bacillus subtilis. FEMS Microbiol. Lett. 266:224–230 [DOI] [PubMed] [Google Scholar]
- 555. Zielińska-Dawidziak M., Grajek K., Olejnik A., Czaczyk K., Grajek W. 2008. Transport of high concentration of thiamin, riboflavin and pyridoxine across intestinal epithelial cells Caco-2. J. Nutr. Sci. Vitaminol. (Tokyo) 54:423–429 [DOI] [PubMed] [Google Scholar]
- 556. Zylberman V., et al. 2006. Evolution of vitamin B2 biosynthesis: 6,7-dimethyl-8-ribityllumazine synthases of Brucella. J. Bacteriol. 188:6135–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]