Abstract
Eleven reference and 25 clinical isolates of Fusarium were subject to multilocus DNA sequence analysis to determine the species and haplotypes of the fusarial isolates from Beijing and Shandong, China. Seven loci were analyzed: the translation elongation factor 1 alpha gene (EF-1α); the nuclear rRNA internal transcribed spacer (ITS), large subunit (LSU), and intergenic spacer (IGS) regions; the second largest subunit of the RNA polymerase gene (RPB2); the calmodulin gene (CAM); and the mitochondrial small subunit (mtSSU) rRNA gene. We also evaluated an IGS-targeted PCR/reverse line blot (RLB) assay for species/haplotype identification of Fusarium. Twenty Fusarium species and seven species complexes were identified. Of 25 clinical isolates (10 species), the Gibberella (Fusarium) fujikuroi species complex was the commonest (40%) and was followed by the Fusarium solani species complex (FSSC) (36%) and the F. incarnatum-F. equiseti species complex (12%). Six FSSC isolates were identified to the species level as FSSC-3+4, and three as FSSC-5. Twenty-nine IGS, 27 EF-1α, 26 RPB2, 24 CAM, 18 ITS, 19 LSU, and 18 mtSSU haplotypes were identified; 29 were unique, and haplotypes for 24 clinical strains were novel. By parsimony informative character analysis, the IGS locus was the most phylogenetically informative, and the rRNA gene regions were the least. Results by RLB were concordant with multilocus sequence analysis for all isolates. Amphotericin B was the most active drug against all species. Voriconazole MICs were high (>8 μg/ml) for 15 (42%) isolates, including FSSC. Analysis of larger numbers of isolates is required to determine the clinical utility of the seven-locus sequence analysis and RLB assay in species classification of fusaria.
INTRODUCTION
Fusarium species are well-known plant pathogens with significant economic impact (18, 19). In humans, they cause infections ranging from superficial disease (e.g., keratitis and onychomycosis) to life-threatening disseminated infections; the latter occur predominantly in severely immunocompromised patients (7, 18, 19) in whom mortality approaches 100% (10). This high mortality is due in part to frequent delays in diagnosis and to the fact that most Fusarium species, particularly those nested within the Fusarium solani species complex (FSSC), are typically resistant to most antifungal agents (1, 2, 28).
Fusarium is a large, taxonomically complex genus. Molecular phylogenetic studies of medically important fusaria have revealed that the more commonly reported pathogens (i.e., F. solani, F. oxysporum, F. moniliforme, F. incarnatum, F. chlamydosporum, and F. dimerum) actually comprise species complexes that collectively contain at least 70 species (15, 21–26). Since the majority of fusarial isolates cannot be identified to species level by traditional morphological methods only (errors in identification, ≈33 to 50%), which greatly underestimate species diversity (19, 22, 24), DNA sequence-based molecular tools are increasingly used to enable accurate species determination (4, 27).
Molecular identification schemes for Fusarium have focused on the need for the identification of species limits on the basis of multilocus DNA sequence data, or genealogical concordance of phylogenetic species recognition (GCPSR) (35). The concept of GCPSR, already recognized to be necessary for the accurate classification of a number of other fungi, including Aspergillus (4), is particularly relevant for Fusarium since many pathogenic species lack Latin binomials. As such, nomenclature systems based on multilocus sequence typing (MLST) schemes incorporating sequence analysis of 2 to 5 genetic loci have been developed to classify a number of Fusarium species complexes according to genotype or haplotype (7, 9, 12, 15, 23, 24, 27). This approach has further revealed the necessity of developing species and sequence type (ST) nomenclatures to designate cryptic speciation within the FSSC (2, 9, 20, 24, 27, 42), F. oxysporum species complex (FOSC) (22, 26, 27), Gibberella (Fusarium) fujikuroi species complex (GFSC) (21, 27), F. dimerum species complex (FDSC) (23, 27, 33), F. chlamydosporum species complex (FCSC) and F. incarnatum-F. equiseti species complex (FIESC) (25, 27), F. sambucinum species complex (FSAMSC), and F. tricinctum species complex (FTSC) (27). Recently, O'Donnell et al. evaluated a new 3-locus MLST scheme incorporating the partial translation elongation factor 1 alpha gene (EF-1α), the largest subunit of the RNA polymerase gene (RPB1), and the second largest subunit of the RNA polymerase gene (RPB2) loci to identify with good discriminatory capacity 69 Fusarium species within the FCSC, FDSC, FIESC, FOSC, FSAMSC, FSSC, and GFSC (27).
Since species distribution varies with geographic region (21, 40) and different species have different drug susceptibility patterns (19, 24), accurate species assignment is important for epidemiological studies and guiding clinical management. MLST methods, however, are costly and not readily available in all laboratories. The PCR-based reverse line blot (RLB) assay is a simple method to identify microbial pathogens using PCR followed by hybridization with species-specific oligonucleotide probes (39, 41). This has the capacity to analyze simultaneously up to 43 strains against multiple probes (13). Herein, we determined the ability of a 7-locus MLST scheme to differentiate between the major Fusarium species. The EF-1α gene, the internal transcribed spacer (ITS) region, D1 and D2 domains of the large subunit (LSU), and intergenic spacer (IGS) region of the rRNA gene, and the RPB2, calmodulin (CAM), and partial mitochondrial small subunit rRNA (mtSSU) genes were chosen based on results of previous analyses (15, 22–25). We then developed and evaluated an RLB assay employing species-, species complex-, and group-specific probes targeting the Fusarium IGS regions to identify members of this genus. Finally, we determined the susceptibilities of Fusarium species, as identified by multilocus sequence and RLB analyses, to six antifungal agents.
MATERIALS AND METHODS
Fusarium isolates.
Thirty-six Fusarium isolates representing 20 species were studied; 25 clinical strains were obtained from Beijing Tongren Hospital, Beijing, and Shandong Eye Institute, Qingdao, Shandong Province, People's Republic of China (Table 1), and 11 reference strains (10 species) from the China General Microbiological Culture Collection Center (CGMCC), Chinese Academy of Sciences, Beijing, China (Table 1). Isolates were identified to the genus and species complex levels by standard phenotypic methods. All clinical isolates, with the exception of a single strain grown from skin/subcutaneous tissue, were recovered from the eye (Table 1).
Table 1.
Source, species identification, and in vitro susceptibilities of 36 Fusarium isolatesa
| Strain ID no. | Site of isolation | ID by multilocus sequence analysisb | Unique seven-locus haplotypesb,g | ID by IGS-RLB | MIC (μg/ml) |
|
|---|---|---|---|---|---|---|
| AMB | VRC | |||||
| PUF001 | Cornea | FSSC-3+4a(n1)c,e | 1-1-1-1-1-1-1 | FSSC-3+4a(n1/n2) | 1 | 8 |
| PUF002 | Cornea | FSSC-3+4a(n1)c,e | 1-1-1-1-1-1-1 | FSSC-3+4a(n1/n2) | 1 | 8 |
| PUF003 | Cornea | FSSC-3+4a(n2)c,e | 2-1-1-2-2-2-1 | FSSC-3+4a(n1/n2) | 1 | 8 |
| PUF004 | Cornea | FSSC-3+4a(n2)c,e | 2-1-1-2-2-2-1 | FSSC-3+4a(n1/n2) | 1 | 8 |
| PUF005 | Cornea | FSSC-3+4a(n3)c,e | 3-2-2-3-3-3-2 | FSSC-3+4a(n3/n4) | 2 | >16 |
| PUF006 | Cornea | FSSC-3+4a(n4)c,e | 4-2-2-4-3-3-2 | FSSC-3+4a(n3/n4) | 2 | >16 |
| PUF007 | Cornea | FSSC-5a(n1)c,e | 5-3-3-5-4-4-2 | FSSC-5a(n1) | 4 | 16 |
| PUF008 | Eye discharge | FSSC-5a(n1)c,e | 5-3-3-5-4-4-2 | FSSC-5a(n1) | 4 | 16 |
| PUF009 | Cornea | FSSC-5a(n2)c,e | 6-4-4-6-5-5-2 | FSSC-5a(n2) | 4 | 16 |
| PUF010 | Facial lesion | GFSC-F. verticillioides | 7-5-5-7-6-6-3 | F. verticillioides | 2 | 2 |
| PUF011 | Cornea | GFSC-F. verticillioides | 7-5-5-8-6-6-3 | F. verticillioides | 2 | 2 |
| PUF012 | Eye discharge | GFSC-F. verticillioides | 7-5-5-9-6-6-3 | F. verticillioides | 2 | 2 |
| PUF013 | Eye discharge | GFSC-F. verticillioides | 7-5-5-9-6-6-3 | F. verticillioides | 2 | 2 |
| PUF014f | Cornea | GFSC-F. verticillioides | 8-5-5-10-7-6-3 | F. verticillioides | 2 | 2 |
| PUF015 | Unknown, reference strain | GFSC-Fusarium napiforme | 9-5-6-11-8-7-4 | F. napiforme | 2 | 4 |
| PUF016f | Unknown, reference strain | GFSC-Fusarium subglutinans | 10-5-7-12-9-8-5 | F. subglutinans | 4 | 2 |
| PUF017 | Cornea | FOSC-197(n1)d,e | 11-6-8-13-10-9-6 | FOSC-197(n1) | 4 | 8 |
| PUF018f | Unknown, reference strain | GFSC-Fusarium proliferatum | 12-6-9-14-11-10-7 | F. proliferatum | 4 | 8 |
| PUF019f | Cornea | GFSC-F. proliferatum | 12-6-9-14-11-10-7 | F. proliferatum | 4 | 4 |
| PUF020f | Eye discharge | GFSC-F. proliferatum | 13-6-9-15-12-11-7 | F. proliferatum | 2 | 4 |
| PUF021f | Eye discharge | GFSC-F. proliferatum | 13-6-9-15-12-11-7 | F. proliferatum | 4 | 4 |
| PUF022f | Unknown, reference strain | GFSC-Fusarium fujikuroi | 14-6-9-16-13-12-7 | F. fujikuroi | 2 | 2 |
| PUF023 | Cornea | GFSC-F. annulatum | 15-7-9-17-14-13-8 | F. annulatum | 8 | 2 |
| PUF024 | Eye discharge | GFSC-F. thapsinum | 16-8-10-18-15-14-9 | F. thapsinum | 4 | 2 |
| PUF025 | Unknown, reference strain | GFSC-Fusarium nygamai | 17-9-11-19-16-15-10 | F. nygamai | 4 | >16 |
| PUF026 | Plant pathogen; reference strain | Fusarium redolens | 18-10-12-20-17-16-11 | F. redolens | 4 | 4 |
| PUF027f | Cornea | FDSC-F. delphinoides | 19-11-13-21-18-17-12 | F. delphinoides | 0.5 | 8 |
| PUF028f | Cornea | FDSC-F. delphinoides | 19-11-13-21-18-17-12 | F. delphinoides | 0.5 | 8 |
| PUF029 | Cornea | FIESC-6b(n1)c,e | 20-12-14-22-19-18-13 | FIESC-6b(n1) | 8 | 4 |
| PUF030 | Cornea | FIESC-25b(n1)c,e | 21-13-14-23-20-19-14 | FIESC-25b(n1) | 8 | 4 |
| PUF031 | Cornea | FIESC-1cc,e | 22-14-15-24-21-20-15 | FIESC-1c | 8 | 4 |
| PUF032 | Plant pathogen; reference strain | FSAMSC-Fusarium sporotrichioides | 23-15-16-25-22-21-16 | F. sporotrichioides | 4 | >16 |
| PUF033f | Plant pathogen; reference strain | Fusarium graminearum | 24-16-17-26-23-22-17 | F. graminearum | 4 | 4 |
| PUF034f | Unknown, reference strain | FTSC-Fusarium avenaceum | 25-17-18-27-24-23-18 | F. avenaceum | 4 | 4 |
| PUF035f | Plant pathogen; reference strain | FTSC-Fusarium acuminatum | 26-18-19-28-25-24-18 | F. acuminatum | 4 | 4 |
| PUF036f | Plant pathogen; reference strain | FTSC-Fusarium acuminatum | 27-18-19-29-26-24-18 | F. acuminatum | 4 | 4 |
Abbreviations: AMB, amphotericin B; FDSC, F. dimerum species complex; FIESC, F. incarnatum-equiseti species complex; FOSC, F. oxysporum species complex; FSAMSC, F. sambucinum species complex; FSSC, F. solani species complex; FTSC, F. tricinctum species complex; GFSC, Gibberella (Fusarium) fujikuroi species complex; ID, identification; ITS, internal transcribed spacer; IGS, nuclear ribosomal DNA intergenic spacer; RLB, reverse line blot; VRC, voriconazole.
Species complex, species, and unique haplotypes were identified by seven-locus sequence analysis.
Within FSSC and FIESC, Arabic numerals designate species identified and lowercase Roman letters (a, b, c, etc.) represent most identical unique haplotypes in the FUSARIUM-ID database, as previously reported (24, 25).
Within FOSC, Arabic numerals designate most identical multilocus sequence type in the FUSARIUM-ID database as described previously (22).
If strains within FSSC, FIESC, and FOSC in this study were not 100% identical to existing unique haplotypes in the FUSARIUM-ID database, “n1,” “n2,” etc., in parentheses were used to represent new unique haplotypes identified by seven-locus sequences.
Strains whose EF-1α sequences failed to yield an exact match with a known Fusarium species (13 in all). Species assignments for these strains were based on multilocus sequence analysis.
Seven-locus haplotypes in the present study. Haplotypes of each of seven genes for each strain studied are shown, in the order of EF-1α gene, ITS region, LSU rRNA gene, IGS rRNA gene, RPB2 gene, CAM gene, and mtSSU rRNA gene.
DNA extraction.
Organisms were grown on potato dextrose agar (PDA; Sigma, St. Louis, MO) plates for 7 days. Total genomic DNA was extracted according to the protocol previously described, with a few modifications (21). Generally, mycelia (0.20 g) were scraped from a colony growing on a PDA plate, frozen in liquid nitrogen, and ground to a fine powder with a pestle and mortar. DNA was extracted in cetyltrimethylammonium bromide (CTAB) buffer (CTAB, 22 mM; Sarkosyl, 34 mM; sorbitol, 137 mM; EDTA, 22 mM; polyvinylpolypyrolidone [PVPP], 1%; NaCl, 1.2 mM) at 65°C, for 30 min. An equal volume of chloroform/octanol (24:1) was added, mixed, and centrifuged at 14,000 rpm for 10 min. The aqueous phase was removed to a fresh tube, an equal volume of isopropanol was added, and centrifugation as described above followed, to precipitate the DNA. The pellet was then washed in 70% ethanol and dissolved in TE buffer (10 mM Tris-HCl [pH 8], 0.1 mM EDTA). DNA was quantitated and stored at −20°C until required.
Primers for PCR, DNA sequencing, and RLB.
In this study, portions of the seven following fungus-specific genetic loci—EF-1α, ITS, LSU, IGS, RPB2, CAM, and mtSSU—were amplified and subjected to DNA sequencing using published primer pairs (see Table S1 in the supplemental material for primer sequences) (22, 25, 26). For PCR in preparation for the RLB assay, primer CNS1b was 5′-end biotin labeled for amplification of the IGS regions. All primers were synthesized at AuGCT (Beijing AuGCT Biotechnology Co. Ltd., Beijing, People's Republic of China).
PCR and DNA sequencing.
Amplification of the seven genetic loci was performed as previously published (22, 25, 26). Aliquots (8 μl) of each PCR product were visualized by electrophoresis using a 1.5% agarose gel. The PCR products were then sequenced by Tsingke Co. Ltd. (Beijing, People's Republic of China) with the DNA analyzer ABI 3730XL system (Applied Biosystems, Foster City, CA). EF-1α, ITS, LSU, RPB2, CAM, and mtSSU gene fragments were sequenced in both directions using the PCR amplification primer pairs (see Table S1 in the supplemental material). In addition, to obtain whole IGS sequences (amplified by the primer pair CNL12/CNS1, ≈2.1 to 2.7 kbp in length), a “primer walking” strategy was employed as previously described (38).
Species identification and phylogenetic analysis.
All DNA sequences obtained in the study were aligned manually to ensure high-quality sequences. Fusarium isolates were initially identified to the species complex or species level by using the EF-1α gene sequence as a BLAST query against the FUSARIUM-ID database (http://isolate.fusariumdb.org) and the Centraalbureau voor Schimmelcultures (CBS-KNAW) Fungal Biodiversity Center (http://www.cbs.knaw.nl/fusarium) (12, 27). In the absence of an exact match of the EF-1α gene for a known species, the sequences of the remaining six loci (as described above) were examined (12, 27).
For members of the FSSC, FOSC, and FIESC, most of which lack Latin binomials (7, 22, 24, 25), the multilocus haplotype (genotype) nomenclatural system based on the method of O' Donnell et al. was employed for species identification with a few modifications, which follow (22, 24, 25). (i) For strains within the FSSC and FIESC, Arabic numerals (1, 2, 3, and so on) were used to designate isolates to species level and lowercase Roman letters (i.e., a, b, c, and so on) were used to represent a 100% sequence match to the unique haplotype within each species as contained in the FUSARIUM-ID (http://isolate.fusariumdb.org) and CBS-KNAW Fungal Biodiversity Center (http://www.cbs.knaw.nl/fusarium) databases (7, 24, 25). (ii) For the strain belonging to the FOSC, Arabic numerals represented most identical multilocus sequence types in the FUSARIUM-ID and CBS-KNAW Fungal Biodiversity Center databases (22). (iii) For all strains within the FSSC, FIESC, and FOSC, in the absence of 100% sequence identity to a unique haplotype for that species, the letter “n” followed by each successive number within parentheses [i.e., (n1), (n2), and so on] was used to represent novel, unique haplotypes identified by the seven-locus sequence analysis. For example, for strain PUF001 (Table 1), multilocus sequence analysis identified the strain as “FSSC-3+4a(n1)”—i.e., this strain was assigned to the FSSC at the species complex level; “3+4” represents the species represented according to the method of Chang et al. (7), while “a(n1)” represents a new unique haplotype which had 99.5% sequence homology to the species “FSSC-3+4a” (strain NRRL 43441) archived in the FUSARIUM-ID databases but which did not have 100% sequence similarity.
The sequences for each gene were aligned using ClustalW software (36) and then adjusted manually to form the consensus sequences for the 36 isolates. Haplotypes for each of seven genes, as well as unique seven-locus haplotypes were identified using COLLAPSE (version 1.2; http://inbio.byu.edu/Faculty/kac/crandall_lab/Computer.html). Phylogenetic trees were computed with MEGA, version 4 (Molecular Evolutionary Genetic Analysis software, v4.0.2; http://www.megasoftware.net), using maximum-parsimony analysis (34), with all positions containing gaps and missing data eliminated from the data set. On review of published sequences in the GenBank database, no fungal isolate was found that had been characterized by DNA sequences represented by all seven loci examined in this study, except for a single Trichoderma harzianum (Hypocrea lixii) isolate (strain CBS 226.95); the sequences of this isolate were thus used as the outgroups to generate the maximum-parsimony trees (GenBank accession no. AY605833 for EF-1α, AF057606 for ITS, AF399236 for LSU, AY570796 for IGS, AF545549 for RPB2, FJ577684 for CAM, and AF276094 for mtSSU).
Oligonucleotide probe and primer design.
Among those of the seven genes selected for study, IGS sequence polymorphisms were further evaluated as genetic targets for incorporation into the RLB assay. This gene locus was chosen since sequence comparison of known Fusarium IGS sequences identified potentially informative nucleotide polymorphisms for probe design. All probes were designed to achieve maximum specificity according to published protocols (13) after alignment with archived IGS sequences in the GenBank, FUSARIUM-ID, and CBS-KNAW Fungal Biodiversity Center databases.
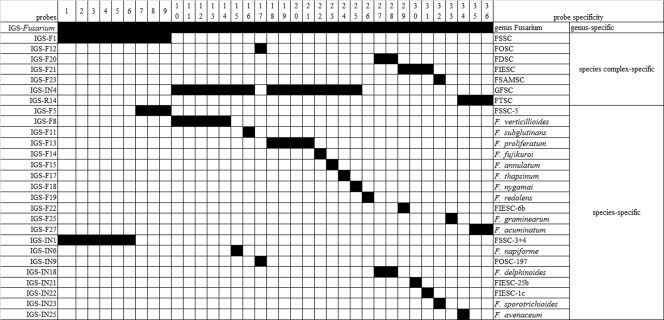
Three heterogeneous regions within the IGS region were used to design IGS-targeted probes, namely, the F region (nucleotide positions 100 to 400), IN region (positions 500 to 800), and R region (positions 2000 and downstream). Although nucleotide positions 800 to 2000 also contained potentially informative polymorphisms, they were not further evaluated as probe targets (see Table S1 in the supplemental material). Eighteen species-, 5 species complex-, and 4 haplotype (or genotype)-specific (targeting a unique haplotype within the FSSC) probes were designed to target the F region, 24 species- and 2 species complex-specific probes the IN region, and 10 species-, 7 group- (6 of which were complex-) specific probes the R region. In addition, one Fusarium genus-specific probe, IGS-Fusarium, was successfully designed to target the F region (see Table S1). All probes were 5′-hexylamine modified and synthesized by Beijing AuGCT Biotechnology Co. Ltd.
RLB hybridization assay.
For PCR in preparation for the RLB assay, primer pair CNL12/CNS1b, with its reverse primer CNS1b, 5′-end biotin labeled (Beijing AuGCT Biotechnology Co. Ltd., Beijing, People's Republic of China), was used to amplify the IGS region (see Table S1 in the supplemental material).
The RLB assay was performed as described previously (13, 41). Briefly, probes (final concentration of 10 μM in 0.5 M NaHCO3, pH 8.4) were covalently linked to a Biodyne C nylon membrane (Pall Life Sciences, Ann Arbor, MI), which was “negatively activated” by incubation in 16% (wt/vol) 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDAC) (Sigma). The membrane was then placed in a MN45 miniblotter (Immunetics, Boston, MA). Amplified PCR product (20 μl) was diluted in 150 μl of 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA; pH 7.7)-0.1% sodium dodecyl sulfate (SDS) denatured at 100°C for 10 min and cooled immediately on ice.
Hybridization was performed by adding 150 μl of diluted denatured PCR product to membrane-bound probes at 60°C for 1 h. The membrane was washed twice (10 min each time) at 60°C with 2× SSPE-0.5% SDS and incubated in peroxidase-labeled streptavidin conjugate (Roche Diagnostics, Mannheim, Germany) at 42°C for 1 h. The membrane was further washed with 2× SSPE buffer-0.5% SDS at 42°C and then at 25°C. Bound PCR products were detected by chemiluminescence using ECL detection liquid (Amersham, GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) and visualized by exposure for 30 min to a hyperfilm ECL X-ray film (Amersham).
Antifungal susceptibility testing.
Antifungal susceptibility testing was performed to determine the in vitro activity of fluconazole (FLC), voriconazole (VRC), itraconazole (ITC), amphotericin B (AMB), caspofungin (CAS), and micafungin (MICA) against all isolates by the use of the Clinical and Laboratory Standards Institute (CLSI) broth microdilution method (M38-A2 protocol) for filamentous fungi (17). Inoculum suspensions were prepared from 7-day-old cultures grown on PDA (Sigma) slants (17). After inoculation, the microtiter plates were incubated at 35°C for 48 h. All isolates produced adequate growth at 48 h. The MIC was defined as the lowest drug concentration at which there was complete inhibition of growth (for AMB) or at which there was prominent reduction of growth (50% growth reduction) compared to that for the drug-free control (for azoles, CAS and MICA) (17, 24).
Nucleotide sequence accession numbers.
The EF-1α, ITS, LSU, IGS, RPB2, CAM, and mtSSU gene sequences of the 36 isolates included in the present study have been deposited in GenBank with accession numbers HQ165832 to HQ165867 (for the EF-1α gene), HQ165904 to HQ165939 (for the ITS region), HQ147569 to HQ147604 (for the LSU rRNA gene), HQ165868 to HQ165903 (for the IGS rRNA gene), HQ423195 to HQ423230 (for RPB2), HQ412311 to HQ412346 (for CAM), and HQ165796 to HQ165831 (for the mtSSU rRNA gene).
RESULTS
Species identification based on seven-locus sequence analysis.
All 36 Fusarium isolates were identified to either the species complex or species level by EF-1α gene sequence analysis by querying the unknown sequence against EF-1α sequences in the FUSARIUM-ID database (http://isolate.fusariumdb.org) (12). For 13 isolates (Table 1) whose EF-1α sequences failed to yield an exact match with a known Fusarium species, species assignment was based on MLST analysis of the remaining six genetic loci; the results are summarized in Table 1. A total of 20 Fusarium species were identified.
Sequence analysis of the seven chosen loci assigned species to all 11 reference strains (10 species) that were concordant with the species identification provided in the CGMCC database (strain information available at the website http://www.cgmcc.net/). Of 25 clinical isolates, the most common species complex was the GFSC (n = 10; 40%), followed by the FSSC (n = 9; 36%) and FIESC (n = 3; 12%). There were two FDSC isolates and a single FOSC strain; 10 species were identified (Table 1).
Fusarium verticillioides was the predominant species (n = 5) within the GFSC and was followed by F. proliferatum (n = 4), with a single isolate for each of the remaining six GFSC species (Table 1). Within the FSSC, six isolates were the species FSSC-3+4 (Latin binomial F. falciforme) (24), and three were assigned to FSSC-5. All three strains within the FIESC were of different species. Three reference strains were assigned within the FTSC, including two F. acuminatum and one F. avenaceum isolate. Both isolates identified within the FDSC (strains PUF027 and PUF028) were the species FDSC-F. delphinoides. There was a single isolate each identified within the FSAMSC and FOSC, which were F. sporotrichioides and FOSC-197, respectively (Table 1).
Phylogenetic diversity and haplotype characteristics of Fusarium isolates.
A phylogenetic analysis of all 36 Fusarium strains was performed by combining the sequence datasets from the sequences of amplified portions of the seven loci: the EF-1α gene (708 bp), the ITS region (493 bp), the LSU rRNA gene (482 bp), the IGS rRNA gene (2,950 bp), the RPB2 gene (1,742 bp), the CAM gene (789 bp), and the mtSSU rRNA gene (720 bp) (Tables 1 and 2).
Table 2.
Tree statistics for the seven individual genetic locia
| Genetic locus | Length (bp) | PIC | PIC/bp | No. of MPTs | MPT length | CI | RI | No. haplotypes |
|---|---|---|---|---|---|---|---|---|
| EF-1α | 708 | 324 | 0.46 | 68 | 518 | 0.65 | 0.88 | 27 |
| ITS | 493 | 128 | 0.26 | 981 | 232 | 0.71 | 0.93 | 18 |
| LSU | 482 | 45 | 0.09 | 2,036 | 67 | 0.76 | 0.95 | 19 |
| IGS | 2,950 | 1,866 | 0.63 | 2 | 2,817 | 0.61 | 0.86 | 29 |
| RPB2 | 1,742 | 577 | 0.33 | 47 | 1,426 | 0.58 | 0.88 | 26 |
| CAM | 789 | 338 | 0.42 | 689 | 630 | 0.66 | 0.90 | 24 |
| mtSSU | 720 | 125 | 0.17 | 1,266 | 69 | 0.83 | 0.97 | 18 |
Abbreviations: CAM, calmodulin; EF-1α, translation elongation factor alpha; IGS, nuclear ribosomal intergenic spacer; ITS, internal transcribed spacer; LSU, the nuclear large subunit; mtSSU, mitochondrial small subunit; CI, consistency index; MPT, maximum parsimony tree; PIC, parsimony informative character; RI, retention index; RPB2, the second largest subunit of RNA polymerase.
A total of 27 EF-1α, 18 ITS, 19 LSU, 29 IGS, 26 RPB2, 24 CAM, and 18 mtSSU haplotypes and 29 unique seven-locus haplotypes were identified (Table 2). Of note, the haplotypes for 24 of 25 clinical strains were novel (the exception was strain PUF031, identified as FIESC-1c; Table 1) after alignment with archived fusarial sequences in the FUSARIUM-ID and CBS-KNAW Fungal Biodiversity Center databases.
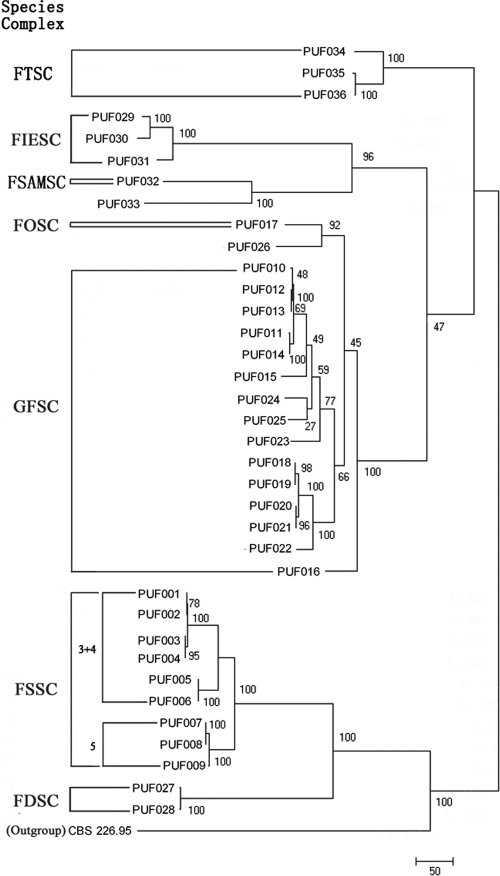
Based on parsimony informative characters (PIC) per bp (Table 2), the IGS locus was the most phylogenetically informative with the greatest nucleotide diversity and was followed by the EF-1α locus. Portions of the amplified rRNA gene regions were the least phylogenetically informative (Table 2). Maximum parsimony trees (MPT) were generated for each locus rooted by the outgroup T. harzianum (H. lixii) strain CBS 226.95. Figure 1 illustrates the MPTs for IGS gene regions. Notably, full-length IGS sequences were able to differentiate all 29 seven-locus haplotypes, including three haplotypes within F. verticillioides that none of the remaining four loci could distinguish between (Tables 1 and 2). Of 17 FOSC/GFSC/F. redolens isolates, two major ITS types were evident (data not shown). However, FOSC-197(n1) and several species of the GFSC (F. verticillioides/F. napiforme/F. subglutinans) in ITS type I could not be further differentiated and neither could the remaining species of the GFSC (F. annulatum/F. fujikuroi/F. proliferatum/F. nygamai/F. thapsinum) and F. redolens in the ITS type II analysis.
Fig. 1.
MPT of nuclear rRNA IGS. IGS sequences of T. harzianum (H. lixii) strain CBS 226.95 were used as outgroup. Five species complexes identified in the present study are marked. Bootstrap values are indicated. The scale bar indicates nucleotide changes (steps).
RLB assay and results.
The primers CNL12/CNS1b amplified the IGS region for all test isolates (see Table S1 in the supplemental material). Table 1 summarizes the species identification of the study isolates obtained by an RLB assay, while Fig. 2 shows the RLB patterns obtained for the study isolates.
Fig. 2.
IGS RLB result of 36 Fusarium isolates studied. The left column shows the probe list (see Table S1 in the supplemental material for details of probes). Lanes 1 to 36: Fusarium strains no. 1 to 36 (see strain information in Table 1).
The Fusarium genus-specific probe, IGS-Fusarium, hybridized with the amplified DNA of the study isolates, with all 36 correctly identified to the species level. Thirteen species complex-specific probes correctly assigned all isolates within the FDSC, FIESC, FSAMSC, FSSC, FTSC, and GFSC to the species complex level using results obtained by multilocus sequence analysis as the “reference” method (Table 1). Fifty-two species-specific probes were able to assign correct species (Fig. 2). One group-specific probe (IGS-R5) (see Table S1 in the supplemental material) hybridized with the DNA of strain PUF017 (FOSC) and closely related F. redolens (strain PUF026). Among members of the FSSC, four haplotype-specific probes correctly identified FSSC-3+4a(n1/n2), FSSC-3+4a(n3/n4), FSSC-5a(n1), and FSSC-5a(n2) (Table 1).
All IGS-targeted probes were 100% specific for their target species/species complex/haplotype/group, with the exception that the F. napiforme-specific probe IGS-F9 showed weak cross-hybridization with F. verticillioides DNA (Fig. 2).
Selection of probes for RLB-based identification of Fusarium species.
On the basis of hybridization patterns obtained with the RLB assay, we propose a set of IGS-targeted probes for species identification of Fusarium. Many of the 71 IGS-targeted probes were functionally redundant (i.e., they hybridized with same set of strains; see Table S1 in the supplemental material). To minimize the number of probes used and for simplicity, probes may be selected for incorporation into the assay according to a clinical requirement. For example, to identify isolates to species complex level, only seven probes are required (see Table S1, in which a set of such probes has been labeled with a superscript “d”). However, to identify all 20 species analyzed in this study, 20 probes are necessary (one appropriate set has been labeled with a superscript “e” in Table S1). Figure 3 shows a set of in silico RLB results using the proposed set of probes labeled with superscripts “d” and “e” in Table S1.
Fig. 3.
The in silico RLB results by proposed set of IGS-targeted genus-, species complex-, and species-specific probes. A set of IGS-targeted probes were proposed for species complex and species identification of Fusarium (labeled with superscripts “d” and “e” in Table S1 in the supplemental material). The black squares represent positive signals on the RLB membrane. Lanes 1 to 36: Fusarium strains no. 1 to 36 (see strain information in Table 1).
Antifungal susceptibility testing.
Based on the molecular phylogenetic results, the susceptibilities of 36 isolates representing 20 species were tested for susceptibility to six antifungal agents in vitro (AMB and VRC MICs are shown in Table 1). AMB was the most active agent, with MICs ranging from 0.5 to 8 μg/ml (MIC90 of 4 μg/ml). FDSC isolates (n = 2) had the lowest AMB MICs (0.5 μg/ml), while three of four FIESC strains had AMB MICs of 8 μg/ml (Table 1). The MICs of AMB among nine members of the FSSC ranged from 1 to 4 μg/ml (geometric mean [GM] MIC of 2.7 μg/ml). CAS and MICA demonstrated little activity against the isolates tested (MICs of >8 μg/ml for all isolates). Among the azoles, FLC MICs were ≥64 μg/ml for 27 of 36 isolates (75%), while the MIC range and MIC90 for ITC were 4 to >16 μg/ml and >16 μg/ml, respectively (data not shown). Corresponding results for VRC were 2 to >16 and 16 μg/ml (Table 1). However, the GM MIC of VRC was lower than that of ITC (5.1 versus 12.9 μg/ml). Isolates belonging to the FSSC, FDSC, and FOSC had high MICs for all three triazoles (FLC, ≥64 μg/ml; VRC, ≥8 μg/ml; ITC, ≥16 μg/ml) (data not shown).
DISCUSSION
The present molecular phylogenetic study using sequence analysis of seven genetic loci is the first of its kind to characterize and improve the understanding of the genetic diversity of circulating clinically relevant Fusarium isolates in China. We also developed and evaluated an IGS-targeted RLB assay as a practical alternative to MLST analysis for the identification of fusarial isolates. Given the inherent resistance of fusaria to antifungal agents, the antifungal susceptibilities of the isolates, based on robust species identification, were determined.
To date, studies focusing on molecular phylogenetic analysis and/or genotyping of Fusarium species have utilized sequence data obtained from the testing of different combinations of genetic loci, known to contain informative species-specific sequence polymorphisms. These have utilized two loci to determine species identification and strain differentiation within the GFSC (EF-1α and RBP2) (15, 21, 23) and for FOSC isolates (EF-1α and IGS) (15, 22). In contrast, four (EF-1α, RPB2, ITS, and LSU) (15, 24) and five (EF-1α, RPB2, CAM, ITS, and LSU) (25) loci have been required to adequately discriminate between cryptic species within the FSSC and within the IESC and FCSC, respectively. Most recently, the largest subunit of the RNA polymerase gene (RPB1) was incorporated as a target in a new three-locus MLST scheme incorporating the EF-1α and RPB2 identification system to identify 69 Fusarium species (27).
The results of the seven-locus DNA typing scheme discussed herein extend and provide additional data on the species/species complex distribution and genetic diversity of major pathogenic Fusarium spp. based on previous sequence analyses that were based on two to five loci (15, 21–25, 27). Of the clinical isolates, members of the GFSC (40%) and FSSC (36%) were the more common pathogens, as has been found elsewhere (7, 15, 23). Conversely, the FOSC was rare (n = 1 isolate). A recent study has reported that among 58 Fusarium isolates from Italy, 20 were of the FOSC; however, these isolates were all recovered from skin/nails from patients with onychomyosis (15). Chang et al., analyzing an outbreak of Fusarium keratitis in the United States, Singapore, and Hong Kong, noted that 77% of 39 isolates were nested within the FSSC and 7 (18%) in the FOSC (7); only one isolate was F. fujikuroi (GFSC). Notably, 10 of 24 isolates recovered from the cornea/eye in our study belonged to the GFSC (Table 1). This suggests that there is regional variation in the species distribution of fusaria and underlines the need to delineate local epidemiological trends and for accurate species determination to guide therapy. All FSSC isolates in the present study were classified within FSSC clade 3, consistent with what has previously been described (7, 20, 24). For three clinical isolates identified as species within the FIESC, two strains (isolates PUF029 and PUF031) were within the Equiseti clade and the other (isolate PUF030) was within the Incarnatum clade, and members of this clade have also been the predominant strains recovered from human Fusarium infections in another study (25). The single FOSC strain PUF017 was classified within FOSC clade 1, in contrast to previous findings that most human FOSC isolates nest within clades 3 and 4 (7, 26).
Of note, there was substantial genetic variation between species and within species (Table 1). Using the combined data set from the seven genetic loci, 29 unique haplotypes were identified among 36 isolates, 28 of which were novel (except strain PUF031, haplotype FIESC-1c). No unique haplotype comprised more than two isolates. Within the FSSC, there were four unique haplotypes, FSSC-3+4a(n1/n2/n3/n4), among six isolates (Table 1); these were genetically most identical to FSSC-3+4a (24). A previous survey of FSSC isolates found that FSSC-1a and FSSC-2d were the most widespread haplotypes (23).
Among the seven loci studied, the ITS region and mtSSU and partial LSU rRNA genes were the least informative (18 or 19 haplotypes) for the differentiation of the 36 Fusarium isolates studied (Table 2). This contrasts with the preferred use of the ITS region as a target for the identification and phylogenetic classification of other fungi (4, 6, 41). Indeed, the ITS locus was the only locus that could not differentiate the GFSC and FOSC from close relative F. redolens. Previous taxonomic research of the FOSC and GFSC has revealed that the close phylogenetic relationship between the ITS regions of these two species complexes may be a result of “switches” between different ITS types during their evolution (24), arguing against the use of single-locus data, especially from nuclear rRNA genes, for inferring species for Fusarium (35). Conversely, the IGS was the most informative locus. All 29 unique seven-locus haplotypes could be correctly identified by the IGS rRNA gene alone. IGS sequence polymorphisms contained much useful data for phylogenetic reconstruction and for inferring species limits with high discriminatory power within Fusarium as also previously noted by O'Donnell et al. (22). The IGS has become one of the more popular loci for investigating genetic diversity or haplotypes nested within the FOSC (5, 11, 14) and, more recently, within the GFSC and other Fusaria spp. (16, 32).
Nonetheless, sequence polymorphisms within other genes assisted in species identification. EF-1α gene sequences were able to assign a species to 23 isolates with 100% sequence similarity to a known Fusarium species in the FUSARIUM-ID database and to assign a species complex level to 12 of the remaining 13 isolates, with the exception of F. graminearum (isolate PUF033). These results support the use of this locus as a useful screen for assigning isolates at least to the species complex level (12, 27). In addition, EF-1α gene data enabled the identification of 27 of 29 haplotypes, with only 3 unique haplotypes (four F. verticillioides strains; isolates PUF010 to PUF013) not identified. However, a single-locus (EF-1α gene) best match at 96 to 98% sequence identity would likely indicate a species that is not represented in the database (27), and sequence data from additional loci are recommended to accurately determine species (27). The CAM and RPB2 genes have also been studied previously as loci for the identification of isolates within the FCSC and FIESC (15, 19, 21, 24, 25, 27). In particular, the RPB2 locus has recently been incorporated along with the EF-1α and RPB1 genes in an internet-accessible platform for the species identification of 69 Fusarium species (27). Both the CAM and RPB2 loci were phylogenetically informative (identified 24 and 26 haplotypes, respectively) (27); in another study of FCSC isolates, sequence polymorphisms within the CAM gene were more informative for species identification than the EF-1α gene (25). We found the RPB2 locus (PIC per bp of 0.33, 26 haplotypes) to be less discriminatory than the IGS and the EF-1α gene loci (PIC per bp of 0.63 and 0.46, 29 and 27 haplotypes, respectively) in the present study.
MLST identification schemes (23–25, 27) undoubtedly provide valuable phylogenetic information. To this end, MLST software incorporating multilocus DNA sequence data of Fusarium is accessible (the FUSARIUM-ID database at http://isolate.fusariumdb.org and CBS-KNAW Fungal Biodiversity Center at http://www.cbs.knaw.nl/fusarium) (12, 27). However, MLST-based approaches are expensive, labor intensive, and not readily adaptable in routine clinical laboratories. The RLB assay described is a useful adjunct to DNA sequencing for species identification of Fusarium, especially if there is a need to simultaneously identify multiple isolates. The IGS-targeted RLB assay correctly identified all isolates studied to species complex or further to species level simply by using the respective species complex- or species-specific probe (Fig. 2). Further, the assay can potentially be used to identify unique haplotypes within species. Although only the IGS region was targeted, and only 20 of >70 known Fusarium species were evaluated in the present study, additional probes may be incorporated into the assay to extend the range of pathogens that can be identified. This flexibility allows customization of the RLB format to meet specific requirements for inclusion of target species according to local epidemiology. A potential limitation of the present assay is time. A working day is required for DNA extraction, PCR (3 h) and product detection and identification using the RLB assay (5 h). However, the RLB membrane can be reused at least 20 times. Running costs associated with the assay are $7 per isolate compared to $14 to $35 for DNA sequencing.
Consistent with the findings of others (1, 2, 24), the six antifungal agents tested in the present study showed poor overall in vitro activities against 20 phylogenetically diverse species. The most significant finding is that AMB was the most active drug against all Fusarium species, including the FSSC, as has been reported by others (2, 24); however, AMB MICs varied with species, and MICs against the FIESC were >8 μg/ml (Table 1). Susceptibility to VRC also varied with species: of note, the agent had poor in vitro activity against the FSSC and FDSC and the single FOSC strain. Both AMB formulations and VRC have been associated with favorable outcomes (response rates 45.5%) in patients with fusariosis (29, 30), and both may be recommended in the treatment of fusariosis, yet the optimal treatment strategy is unclear. (3, 19). As in our study, others have reported the FSSC to be the most resistant members of the genus (37). Finally, studies are needed to evaluate whether and to what extent species- and strain-specific differences in antifungal susceptibility exist within Fusarium. Studies have reported that FOSC isolates are “susceptible” to VRC (8, 28, 31); it is possible that the novel haplotype identified in this study may be relatively more drug resistant.
In conclusion, the present study evaluated a seven-locus sequence analysis scheme for the identification of Fusarium isolates and for their phylogenetic study. Five Fusarium species complexes, 20 species, and 29 unique haplotypes among 36 isolates were identified. AMB was the most active drug in vitro against all the Fusarium species, while the three triazoles FLC, VRC and ITC demonstrated poor activity. Further, we developed a PCR/RLB assay for species identification of Fusarium and its use in a diagnostic laboratory. Evaluation of larger numbers of isolates/species and testing its capability to directly detect Fusarium in clinical specimens are required to position the RLB assay within routine diagnostic algorithms.
Supplementary Material
ACKNOWLEDGMENT
This study work was funded by The Ministry of Health of the People's Republic of China, public service sector fund 200802026.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 9 March 2011.
REFERENCES
- 1. Alastruey-Izquierdo A., Cuenca-Estrella M., Monzon A., Mellado E., Rodriguez-Tudela J. L. 2008. Antifungal susceptibility profile of clinical Fusarium spp. isolates identified by molecular methods. J. Antimicrob. Chemother. 61:805–809 [DOI] [PubMed] [Google Scholar]
- 2. Azor M., Gene J., Cano J., Guarro J. 2007. Universal in vitro antifungal resistance of genetic clades of the Fusarium solani species complex. Antimicrob. Agents Chemother. 51:1500–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Azor M., et al. 2008. In vitro antifungal susceptibility and molecular characterization of clinical isolates of Fusarium verticillioides (F. moniliforme) and Fusarium thapsinum. Antimicrob. Agents Chemother. 52:2228–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balajee S. A., et al. 2009. Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in the clinical mycology laboratory: where are we and where should we go from here? J. Clin. Microbiol. 47:877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balmas V., et al. 2010. Multilocus phylogenetics show high levels of endemic fusaria inhabiting Sardinian soils (Tyrrhenian Islands). Mycologia 102:803–812 [DOI] [PubMed] [Google Scholar]
- 6. Borman A. M., Linton C. J., Miles S. J., Johnson E. M. 2008. Molecular identification of pathogenic fungi. J. Antimicrob. Chemother. 61(Suppl. 1):i7–i12 [DOI] [PubMed] [Google Scholar]
- 7. Chang D. C., et al. 2006. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA 296:953–963 [DOI] [PubMed] [Google Scholar]
- 8. Cuenca-Estrella M., et al. 2006. Head-to-head comparison of the activities of currently available antifungal agents against 3,378 Spanish clinical isolates of yeasts and filamentous fungi. Antimicrob. Agents Chemother. 50:917–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Debourgogne A., et al. 2010. Development of a new MLST scheme for differentiation of Fusarium solani species complex (FSSC) isolates. J. Microbiol. Methods 82:319–323 [DOI] [PubMed] [Google Scholar]
- 10. Dignani M. C., Anaissie E. 2004. Human fusariosis. Clin. Microbiol. Infect. 10(Suppl. 1):67–75 [DOI] [PubMed] [Google Scholar]
- 11. Enya J., et al. 2008. Biological and phylogenetic characterization of Fusarium oxysporum complex, which causes yellows on Brassica spp., and proposal of F. oxysporum f. sp. rapae, a novel forma specialis pathogenic on B. rapa in Japan. Phytopathology 98:475–483 [DOI] [PubMed] [Google Scholar]
- 12. Geiser D. M., et al. 2004. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 110:473–479 [Google Scholar]
- 13. Kong F. R., Gilbert G. L. 2006. Multiplex PCR-based reverse line blot hybridization assay (mPCR/RLB)—a practical epidemiological and diagnostic tool. Nat. Protoc. 1:2668–2680 [DOI] [PubMed] [Google Scholar]
- 14. Mbofung G. Y., Hong S. G., Pryor B. M. 2007. Phylogeny of Fusarium oxysporum f. sp. lactucae inferred from mitochondrial small subunit, elongation factor 1-alpha, and nuclear ribosomal intergenic spacer sequence data. Phytopathology 97:87–98 [DOI] [PubMed] [Google Scholar]
- 15. Migheli Q., et al. 2010. Molecular phylogenetic diversity of dermatologic and other human pathogenic fusarial isolates from hospitals in northern and central Italy. J. Clin. Microbiol. 48:1076–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nalim F. A., Elmer W. H., McGovern R. J., Geiser D. M. 2009. Multilocus phylogenetic diversity of Fusarium avenaceum pathogenic on lisianthus. Phytopathology 99:462–468 [DOI] [PubMed] [Google Scholar]
- 17. NCCLS/CLSI 2009. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi, 2nd ed. Approved standard M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 18. Nucci M., Anaissie E. 2002. Cutaneous infection by Fusarium species in healthy and immunocompromised hosts: implications for diagnosis and management. Clin. Infect. Dis. 35:909–920 [DOI] [PubMed] [Google Scholar]
- 19. Nucci M., Anaissie E. 2007. Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 20:695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O'Donnell K. 2000. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 92:919–938 [Google Scholar]
- 21. O'Donnell K., Cigelnik E., Nirenberg H. I. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465–493 [Google Scholar]
- 22. O'Donnell K., et al. 2009. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet. Biol. 46:936–948 [DOI] [PubMed] [Google Scholar]
- 23. O'Donnell K., et al. 2007. Phylogenetic diversity and microsphere array-based genotyping of human pathogenic fusaria, including isolates from the multistate contact lens-associated U.S. keratitis outbreaks of 2005 and 2006. J. Clin. Microbiol. 45:2235–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Donnell K., et al. 2008. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J. Clin. Microbiol. 46:2477–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Donnell K., et al. 2009. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J. Clin. Microbiol. 47:3851–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Donnell K., et al. 2004. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J. Clin. Microbiol. 42:5109–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O'Donnell K., et al. 2010. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 48:3708–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paphitou N. I., et al. 2002. In vitro activities of investigational triazoles against Fusarium species: effects of inoculum size and incubation time on broth microdilution susceptibility test results. Antimicrob. Agents Chemother. 46:3298–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perfect J. R. 2005. Treatment of non-Aspergillus moulds in immunocompromised patients, with amphotericin B lipid complex. Clin. Infect. Dis. 40:S401–S408 [DOI] [PubMed] [Google Scholar]
- 30. Perfect J. R., et al. 2003. Voriconazole treatment for less-common, emerging, or refractory fungal infections. Clin. Infect. Dis. 36:1122–1131 [DOI] [PubMed] [Google Scholar]
- 31. Pfaller M. A., Diekema D. J. 2004. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J. Clin. Microbiol. 42:4419–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sampietro D. A., et al. 2010. A molecular based strategy for rapid diagnosis of toxigenic Fusarium species associated to cereal grains from Argentina. Fungal Biol. 114:74–81 [DOI] [PubMed] [Google Scholar]
- 33. Schroers H. J., et al. 2009. Taxonomy and phylogeny of the Fusarium dimerum species group. Mycologia 101:44–70 [DOI] [PubMed] [Google Scholar]
- 34. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 35. Taylor J. W., et al. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31:21–32 [DOI] [PubMed] [Google Scholar]
- 36. Thompson J. D., Higgins D. G., Gibson T. J. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tortorano A. M., et al. 2008. Species distribution and in vitro antifungal susceptibility patterns of 75 clinical isolates of Fusarium spp. from northern Italy. Antimicrob. Agents Chemother. 52:2683–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu H., Ma Y., Zhang Y., Zhang H. 2004. Complete sequence of virulence plasmid pEIB1 from the marine fish pathogen Vibrio anguillarum strain MVM425 and location of its replication region. J. Appl. Microbiol. 97:1021–1028 [DOI] [PubMed] [Google Scholar]
- 39. Xiao M., et al. 2010. Identification of pathogenic Nocardia species by reverse line blot hybridization targeting the 16S rRNA and 16S-23S rRNA gene spacer regions. J. Clin. Microbiol. 48:503–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yli-Mattila T., et al. 2009. A novel Asian clade within the Fusarium graminearum species complex includes a newly discovered cereal head blight pathogen from the Russian Far East. Mycologia 101:841–852 [DOI] [PubMed] [Google Scholar]
- 41. Zeng X., et al. 2007. Reverse line blot hybridization assay for identification of medically important fungi from culture and clinical specimens. J. Clin. Microbiol. 45:2872–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang N., et al. 2006. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J. Clin. Microbiol. 44:2186–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.