Abstract
The increased worldwide spread of carbapenem-resistant Enterobacteriaceae (CRE) emphasizes the need for a sensitive screening procedure to identify these microorganisms. Gastrointestinal carriers may serve as the reservoir for cross-transmission in the health care setting, and thus active surveillance is a key part in preventing the spread of such strains. Three agar-based methods for direct CRE detection from rectal swabs were compared: CHROMagar-KPC (Chrom); MacConkey agar with imipenem at 1 μg/ml (MacI); and MacConkey plates with imipenem, meropenem, and ertapenem disks (MacD). First, we compared the levels of detection (LODs) of 10 molecularly characterized carbapenemase-producing Enterobacteriaceae strains by the three methods. Second, we compared their performance in a surveillance study using rectal swabs (n = 139). The LODs of carbapenemase-producing Enterobacteriaceae strains were influenced by their MICs to carbapenems and were best for MacI, followed by Chrom. The MacD method was able to detect only the strains exhibiting MICs of ≥32 μg/ml to at least ertapenem. In the surveillance study, both Chrom and MacI had greater sensitivity (85%) than MacD (76%). However, MacI was the most specific method. In conclusion, MacI appears to be most appropriate medium for the detection of CRE in settings in which multiclonal CRE strains with various MICs to carbapenems are circulating.
INTRODUCTION
Carbapenem-resistant Enterobacteriaceae (CRE) have emerged globally and have become a major threat to public health (1, 17). Carbapenem resistance may be caused by a variety of mechanisms and has been identified in a variety of Enterobacteriaceae species (1, 20). In 2006, an epidemic strain of KPC-3-producing Klebsiella pneumoniae, exhibiting resistance to nearly all antimicrobial agents, spread in all major Israeli hospitals (11, 16). This strain, identified as sequence type (ST) 258, is identical to the epidemic strain that had spread across the United States (9, 12) and is characterized by high MIC values of carbapenem antibiotics (16).
As gastrointestinal carriage may serve as a reservoir for CRE cross-transmission in health care settings, active surveillance among high-risk patients has been deemed important for controlling this epidemic in acute-care facilities (2, 23). The implementation of a reliable and sensitive method for detection of this strain as well as other CRE is therefore critical to the success of infection control measures. Although PCR-based methods have been proven to be highly sensitive and reliable for rapid diagnosis (8, 22), these methods require expertise that is not readily available in many centers. Moreover, as the emergence and spread of other types of CRE are increasingly reported (7, 19), culture-based methods are still essential for the initial detection of these strains.
In our center, we have been using MacConkey agar supplemented with imipenem at 1 μg/ml (HyLabs, Rehovot, Israel) as the main screening agar plate for the detection of CRE from rectal swabs. In the present study, we compared this method to two other culture-based methods, namely, CHROMagar-KPC (HyLabs, Rehovot, Israel) and MacConkey plates with imipenem, meropenem, and ertapenem disks. This paper reports the laboratory and clinical evaluation of these screening media.
MATERIALS AND METHODS
Setting, patient selection, and collection of surveillance specimens.
The study was conducted as part of an ongoing surveillance program that had been implemented at the Tel Aviv Sourasky Medical Center, a 1,200-bed tertiary care hospital in Tel Aviv, Israel. From August 2008 through April 2009, rectal specimens were collected from known CRE carriers and from contacts of patients newly discovered to be harboring CRE, as previously described (22). A nylon flocked swab system with liquid Amies medium was used according to the manufacturer's instructions (Eswab, Copan, Brescia, Italy), immediately transferred to the laboratory following sampling, and processed.
Analysis of LODs of the CRE screening plates.
To analyze the limit of detection (LOD) of CRE, we compared the three type of screening plates used in our study for their ability to detect 10 distinct, well-characterized strains (Table 1). Strains were stored in LB broth with 25% glycerol at −80°C, thawed, and subcultured onto MacConkey agar plates before use. All strains were isolated in Israel, except for strains 2565 and 2577, which were isolated in Europe (provided by Marek Gniadkowski, Department of Molecular Microbiology, National Medicines Institute, Warsaw, Poland). Isolates were suspended in saline to the density of a 0.5 McFarland standard, followed by serial 10-fold dilutions. An aliquot of 100 μl from each dilution (0.5 McFarland standard,10 and lower) of each study strain was plated on each of the three screening plates evaluated in this study, as well as on Muller-Hinton agar for determination of viable colony counts. The following selective agar plates were used: (i) CHROMagar-KPC (Chrom); (ii) MacConkey agar with imipenem at 1 μg/ml (MacI); and (iii) MacConkey agar plates with standard imipenem, meropenem, and ertapenem 10-μg paper disks (Oxoid) sterilely applied at the 4-, 8-, and 12-o'clock positions (MacD). Plates were incubated overnight at 35°C in ambient air and then read. The LOD was determined based on the minimal colony count allowing detection on the respective screening plate. The cost of each method per one sample was as follows: Chrom, 7.27 Israeli new shekels (NIS 7.27); MacI, NIS 2.1; and MacD, NIS 1.95 ($1 = NIS 3.6).
Table 1.
Levels of detection for the three CRE screening plates for detection of carbapenem-resistant Enterobacteriaceae
| Bacterial strain | Species | bla type | MIC (μg/ml) |
Level of detection (CFU/ml) on the following screening platea: |
Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| Imipenem | Meropenem | Ertapenem | MacI | MacD | Chrom | ||||
| 490 (ST 258) | K. pneumoniae | KPC-3 | 32 | 32 | 64 | 1.1 × 102 | 1.1 × 103 | 1.1 × 102 | 11 |
| 9 | K. pneumoniae | KPC-3 | 0.5 | 1 | 1.5 | 4.1 × 102 | ND | 4.1 × 103 | 16 |
| 14 | K. pneumoniae | KPC-3 | >32 | >32 | >32 | 1 × 102 | 1 × 102 | 1 × 102 | 16 |
| 533 | Enterobacter cloacae | KPC-2 | 4 | 2 | 12 | 1.1 × 102 | ND | 1.1 × 102 | 3 |
| 360 | Escherichia coli | KPC-2 | 1 | 1.5 | 2 | 9.7 × 101 | ND | 9.7 × 104 | 6 |
| 2438 | E. coli | KPC-2 | 12 | 12 | >32 | 6.5 × 101 | 2 × 106 | 6.5 × 101 | 6 |
| 1679 | E. coli | KPC-2 | 4 | 2 | 12 | 8.9 × 101 | ND | 8.9 × 101 | 6 |
| 2112 | E. coli | KPC-3 | 1 | 1.5 | 0.75 | 8.3 × 106 | ND | ND | 5 |
| 2565 | K. pneumoniae | IMP-1 | 2 | 32 | 4 | 1.4 × 104 | ND | 1.4 × 106 | MOSAR1144 |
| 2577 | K. pneumoniae | VIM-1 | 32 | 32 | 32 | 1.1 × 102 | 1.1 × 105 | 1.1 × 102 | MOSAR1156 |
Chrom, CHROMagar-KPC plates; MacI, MacConkey agar with imipenem at 1 μg/ml; MacD, MacConkey agar plates with standard imipenem, meropenem, and ertapenem 10-μg paper disks; ND, no detection.
Detection and identification of CRE from rectal swabs.
We compared the three agar-based methods for the detection of CRE directly from rectal swabs. Swabs were vortexed for 10 s, and 100-μl aliquots were plated onto the three different selective agar plates in parallel. Plates were then processed as described above. Following incubation, plates were visualized for suspected CRE growth by two different observers.
CRE colonies on Chrom were identified according to the manufacturer's instructions (Klebsiella and Enterobacter species, medium-size dark metallic blue colonies; E. coli, medium to large pink/dark rose colonies). CRE colonies on MacI and MacD were identified as any typical growth of lactose-fermenting pink colonies on the plate or within a 21-mm diameter of at least one of the carbapenem disks, respectively. Suspected CRE colonies were subcultured from the respective screening plate onto standard MacConkey plates. Identification and antimicrobial susceptibility testing (AST) of bacterial strains were then performed with the Vitek-2 system using GN-ID and GN09 cards (bioMérieux, Marcy l'Etoile, France). Imipenem and meropenem MICs were verified with the Etest (AB Biodisk, Solna, Sweden). Susceptibility was determined using the 2010 MIC breakpoint criteria of the Clinical and Laboratory Standards Institute (CLSI) (4).
Isolates nonsusceptible to either imipenem or meropenem were defined as CRE positive and were subjected to PCR for the blaKPC gene according to a previously described protocol (22). PCR-negative isolates were further tested by the modified Hodge test (MHT) according to CLSI recommendations (4).
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and overall accuracy were calculated for each of the methods. A true positive was defined as growth with phenotypic features compatible with CRE diagnosed as CRE by confirmatory testing. A false positive was defined as growth with phenotypic features compatible with CRE diagnosed as non-CRE by confirmatory testing. We also calculated the turnaround time (TAT) as the time elapsed from sample receipt in the laboratory to communication of the final result to the clinician.
RESULTS
Analytical sensitivities of the three screening plates for identification of different CRE strains.
The reference strains used, MICs of different carbapenems, and growth performance with the studied screening methods are summarized in Table 1. Strains exhibited various MICs to different carbapenems. Any growth was detected for 10/10 strains on MacI, 9/10 on Chrom, and 4/10 on MacD. With MacI, the LOD for detected strains was <104 CFU/ml in 8/10. The rates were 2/4 for MacD and 7/9 for Chrom. All three CRE screening plates successfully detected all the strains with MICs of 32 μg/ml or higher for all carbapenems. These included the epidemic K. pneumoniae ST 258 (strain 490) and strains 14 and 2577 (Table 1). These strains were detected at a lower inoculum by Chrom and MacI but only at a higher inoculum (10-fold) by MacD. For the 9 strains that grew on both MacI and Chrom, the LOD was lower with MacI for 3 and similar with both media for 6. The colony morphologies and colors of the various K. pneumoniae strains were indistinguishable.
Performance of screening agar plates in recovery of CRE from rectal swabs.
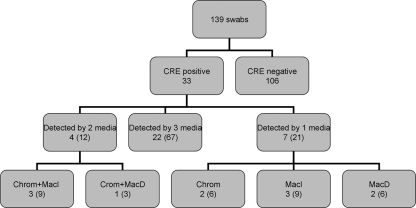
A total of 139 rectal swabs were collected; CRE were identified in 33 (24%) of the samples (31 patients) on at least one of the screening plates (Fig. 1). All isolates were K. pneumoniae except for 2 (Klebsiella oxytoca and Enterobacter aerogenes). The MIC10, MIC50, and MIC90 of the isolates, respectively, as determined by Etest, were as follows: imipenem, 6, >32, and > 32 μg/ml; meropenem, 8, >32, and >32 μg/ml. All isolates but one tested positive by blaKPC PCR. The PCR-negative isolate was K. pneumoniae; this isolate tested negative by the MHT and had the lowest MIC values (1 and 6 μg/ml to imipenem and meropenem, respectively).
Fig. 1.
Recovery of carbapenem-resistant Enterobacteriaceae (CRE) from rectal swabs and performance of screening agar plates. Numbers in parentheses show the percentage of the total number of positive samples (n = 33). MacI, MacConkey agar with imipenem (1-μg/ml plates); Chrom, CHROMagar-KPC plates; MacD, MacConkey agar plates with ertapenem, imipenem, and meropenem disks; ND, not detected.
The performances of different screening media are summarized in Table 2 and Fig. 1. Chrom and MacI detected 28 of 33 CRE strains, while MacD detected only 25. Of the CRE isolates, 67% were detected by all media, 12% by two media, and 21% by only one type of medium. There were 12, 6, and 11 samples in which growth on Chrom, MacI, and MacD, respectively, was initially mistaken for CRE, resulting in unnecessary laboratory work-up. The implicated false-positive isolates were mainly carbapenem-susceptible Enterobacteriaceae and also Acinetobacter baumannii. Chrom and MacI showed similar sensitivities and negative predictive values, but MacI had superior specificity and positive predictive value and thus greater overall accuracy. The turnaround times were comparable. MacD had clearly inferior sensitivity, and its specificity was similar to that of Chrom.
Table 2.
Summary of CRE screening plate performances
| Methoda | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | Mean turnaround time (range), days |
|---|---|---|---|---|---|---|
| MacI | 84.9 | 94.3 | 82.3 | 95.2 | 92.1 | 2.8 (2–4) |
| MacD | 75.8 | 89.6 | 69.5 | 92.2 | 86.3 | 2.8 (2–4) |
| Chrom | 84.9 | 88.7 | 70 | 95 | 87.8 | 3.0 (2–4) |
Chrom, CHROMagar-KPC plates; MacI, MacConkey agar with imipenem (1 μg/ml); MacD, MacConkey agar plates with standard imipenem, meropenem, and ertapenem 10-μg paper disks.
DISCUSSION
In this study, we compared the performances of three culture-based screening methods for the detection of CRE from surveillance rectal swabs. In the clinical evaluation study, we found that Chrom and MacI screening plates had comparable sensitivities and negative predictive values in detection of the highly resistant blaKPC-producing CRE strain that had spread in our hospital and exhibits high-level carbapenem resistance (11). MacI had higher specificity and positive predictive value than Chrom. Both screening plates were superior to MacD, in line with previous reports (21). Notably, MacI performed slightly better than Chrom during the laboratory evaluation, and both were superior to MacD (Table 1). Only strains that were highly resistant to at least ertapenem (Etest MIC value, ≥32 μg/ml) were detected by all three methods. Two strains with an Etest MIC of 12 μg/ml (strains 1679 and 533) were not detected by the MacD method. This might be explained by the fact that we used inocula that were at least 10-fold lower than the 0.5 McFarland standard, which better resemble physiologic conditions. The difference in sensitivity between MacI and Chrom was not apparent during the surveillance study, as the dominant CRE strain at the time of study in our hospital was the blaKPC-producing K. pneumoniae strain that is characterized by a high level of resistance to carbapenems (11). However, with the increase in the diversity of CRE strains from different genera with variable MIC values (5–7, 14, 15), the higher sensitivity of the MacI plates is likely to become important for adequate detection of CRE carriage.
The MacI medium had higher specificity; i.e., it was the least likely to require additional work-up following growth of non-CRE strains (either carbapenem-susceptible Enterobacteriaceae or A. baumannii). This medium is considerably less expensive than Chrom, which had similar performance during clinical evaluation. Combining these factors, in our hands, MacI appeared to be superior to the other two methods as the primary method for CRE surveillance.
Several studies have compared the performances of different culture-based methods, either to direct detection by PCR (8, 21, 22) or to other culture-based methods (10, 13). A summary of these studies is presented in Table 3. The differences in studied populations, laboratory methods, and study design make a head-to-head comparison difficult, but several conclusions can be made: (i) among agar-based methods, the use of carbapenem disks on MacConkey plates appears to be the least sensitive, and (ii) unlike screening for vancomycin-resistant enterococci (18), enrichment in broth was not superior to direct plating (13). Our study adds to the current literature the direct comparison of MacI with two previously described agar-based methods, along with an examination of the analytic sensitivity of these methods in detecting various types of CRE strains. Although direct detection by PCR has the advantage of rapid identification of CRE carriers, it is limited to the detection of CRE that harbor the target β-lactamase gene, e.g., blaKPC, and will inevitably miss non-carbapenemase-producing CRE or CRE strains that carry other genes, such as blaVIM-1 or blaNDM-1 (New Delhi metallo-β-lactamase 1). Moreover, the recovery of CRE strains is essential in order to perform molecular epidemiology studies (e.g., pulsed-field gel electrophoresis), especially in an outbreak situation, in order to better direct infection control measures.
Table 3.
Summary of published studies on detection of CRE from rectal swabs
| Design (n) | Method evaluateda | Results (%)b | Reference |
|---|---|---|---|
| Surveillance rectal swabs (187) | MAC plate + ERT, IMI, and MER disks; cutoff not stated | SN = 87, SP = 100 | 8 |
| blaKPC qPCR from swabs following extraction method A | SN = 100, SP = 95 | ||
| blaKPC qPCR from swabs following extraction method B | SN = 97.9, SP = 96.4 | ||
| LOD analysis of CRE strains; surveillance rectal swabs (51) | TSB + IMI disk, subcultured to MAC plates | Comparable analytical LOD, SN = 100 | 10 |
| TSB, subcultured to MAC plates + IMI disk; cutoff, <16 mm | Comparable analytical LOD, SN = 50 | ||
| Surveillance rectal swabs (149) | TSB + IMI disk, subcultured to MAC plates | SN = 65.6, SP = 49.6 | 13 |
| MAC plate + ERT disks; cutoff, <27 mm | SN = 97, SP = 90.5 | ||
| Surveillance rectal swabs (phenotypic methods compared to blaKPC PCR) (122) | MAC plate + ERT, IMI, and MER disks; cutoff not stated | SN = 92.7, SP = 95.9 | 21 |
| CHROMagar KPC | SN = 100, SP = 98.4 | ||
| Surveillance rectal swabs (755) | Inoculated BHI broth subjected to blaKPC PCR | SN = 92.2, SP = 99.4 | 22 |
| MAC + IMI (1 μg/ml) | SN = 87.5, SP = 99.6 | ||
| Surveillance rectal swabs (139) | MAC plate + ERT, IMI, and MER disks; cutoff, <22 mm | SN = 75.8, SP = 89.6 | Current study |
| MAC + IMI (1 μg/ml) | SN = 84.9, SP = 94.3 | ||
| CHROMagar KPC | SN = 84.9, SP = 88.7 |
TSB, tryptic soy broth; IMI, imipenem; ERT, ertapenem; MER, meropenem; MAC, MacConkey; qPCR, quantitative PCR; BHI, brain heart infusion.
SN, sensitivity; SP, specificity.
In conclusion, in geographic regions such as Israel, where CRE of various genera and with a wide range of MICs to carbapenems are being discovered (5–7, 14, 15), the use of MacConkey agar supplemented with imipenem at 1 μg/ml is the most appropriate for detection of CRE carriage. This screening plate offers a sensitive, convenient, and relatively low-cost method for identifying CRE species, and it is able to detect even CRE species with relatively low carbapenem MICs.
ACKNOWLEDGMENTS
We acknowledge Marek Gniadkowski, Department of Molecular Microbiology, National Medicines Institute, Warsaw, Poland, for providing us with the metallo-β-lactamase control strains characterized during the MOSAR project.
This work was supported in part by European Commission Research grant FP7: SATURN—Impact of Specific Antibiotic Therapies on the Prevalence of Human Host Resistant Bacteria grant no. 241796.
Footnotes
Published ahead of print on 6 April 2011.
REFERENCES
- 1. Bilavsky E., Schwaber M. J., Carmeli Y. 2010. How to stem the tide of carbapenemase-producing enterobacteriaceae?: proactive versus reactive strategies. Curr. Opin. Infect. Dis. 23:327–331 [DOI] [PubMed] [Google Scholar]
- 2. Calfee D., Jenkins S. G. 2008. Use of active surveillance cultures to detect asymptomatic colonization with carbapenem-resistant Klebsiella pneumoniae in intensive care unit patients. Infect. Control Hosp. Epidemiol. 29:966–968 [DOI] [PubMed] [Google Scholar]
- 3. Chmelnitsky I., Navon-Venezia S., Strahilevitz J., Carmeli Y. 2008. Plasmid-mediated qnrB2 and carbapenemase gene bla(KPC-2) carried on the same plasmid in carbapenem-resistant ciprofloxacin-susceptible Enterobacter cloacae isolates. Antimicrob. Agents Chemother. 52:2962–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. Approved standard MS100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Goren M. G., et al. 2010. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patients. Emerg. Infect. Dis. 16:1014–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goren M. G., Navon-Venezia S., Chmelnitsky I., Carmeli Y. 2010. Carbapenem-resistant KPC-2-producing Escherichia coli in a Tel Aviv medical center, 2005 to 2008. Antimicrob. Agents Chemother. 54:2687–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goren M. G., Chmelnitsky I., Carmeli Y., Navon-Venezia S. 2011. Plasmid-encoded OXA-48 carbapenemase in Escherichia coli from Israel. J. Antimicrob. Chemother. 66:672–673 [DOI] [PubMed] [Google Scholar]
- 8. Hindiyeh M., et al. 2008. Rapid detection of blaKPC carbapenemase genes by real-time PCR. J. Clin. Microbiol. 46:2869–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kitchel B., et al. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53:3365–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Landman D., Salvani J. K., Bratu S., Quale J. 2005. Evaluation of techniques for detection of carbapenem-resistant Klebsiella pneumoniae in stool surveillance cultures. J. Clin. Microbiol. 43:5639–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leavitt A., Navon-Venezia S., Chmelnitsky I., Schwaber M. J., Carmeli Y. 2007. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob. Agents Chemother. 51:3026–3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leavitt A., et al. 2010. Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel. Antimicrob. Agents Chemother. 54:3002–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lolans K., Calvert K., Won S., Clark J., Hayden M. K. 2010. Direct ertapenem disk screening method for identification of KPC-producing Klebsiella pneumoniae and Escherichia coli in surveillance swab specimens. J. Clin. Microbiol. 48:836–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marchaim D., Navon-Venezia S., Schwaber M. J., Carmeli Y. 2008. Isolation of imipenem-resistant Enterobacter species: emergence of KPC-2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob. Agents Chemother. 52:1413–1418 inelevel0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Navon-Venezia S., et al. Plasmid-mediated imipenem-hydrolyzing enzyme KPC-2 among multiple carbapenem-resistant Escherichia coli clones in Israel. Antimicrob. Agents Chemother. 050:3098–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Navon-Venezia S., et al. 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob. Agents Chemother. 53:818–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nordmann P., Cuzon G., Naas T. 2009. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9:228–236 [DOI] [PubMed] [Google Scholar]
- 18. Novicki T. J., et al. 2004. Convenient selective differential broth for isolation of vancomycin-resistant enterococcus from fecal material. J. Clin. Microbiol. 42:1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poirel L., Lagrutta E., Taylor P., Pham J., Nordmann P. 2010. Emergence of metallo-β-lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob. Agents Chemother. 54:4914–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Queenan A. M., Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Samra Z., et al. 2008. Evaluation of CHROMagar KPC for rapid detection of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 46:3110–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schechner V., et al. 2009. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J. Clin. Microbiol. 47:3261–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schwaber M., and TASMC CRE Working Group Abstr. 19th Annu. Sci. Meet. Soc. Healthcare Epidemiol. Am., abstr. 484 [Google Scholar]