Abstract
Tumor necrosis factor receptor 2 (TNFR2) activates transcription factor κB (NF-κB) and c-Jun N-terminal kinase (JNK). The mechanisms mediating these activations are dependent on the recruitment of TNF receptor-associated factor 2 (TRAF2) to the intracellular region of the receptor. TNFR2 also induces TRAF2 degradation. We show that in addition to the well characterized TRAF2 binding motif 402-SKEE-405, the human receptor contains another sequence located at the C-terminal end (amino acids 425–439), which also recruits TRAF2 and activates NF-κB. In addition to that, human TNFR2 contains a conserved region (amino acids 338–379) which is responsible for TRAF2 degradation and therefore of terminating NF-κB signaling. TRAF2 degradation and the lack of NF-κB activation when both TNFR1 and TNFR2 are co-expressed results in an enhanced ability of TNFR1 to induce cell death, showing that the cross-talk between both receptors is of a great biological relevance. Induction of TRAF2 degradation appears to be independent of TRAF2 binding to the receptor. Amino acids 343-TGSSDSS-349 are essential for inducing TRAF2 degradation because deletion mutants of this region or point mutations at serine residues 345 and 346 or 348 and 349 obliterate the ability of TNFR2 to induce TRAF2 degradation.
Keywords: Proteasome, Protein Degradation, Signal Transduction, TRAF, Tumor Necrosis Factor (TNF)
Introduction
Tumor necrosis factor receptor 2 (TNFR2)5 is one of the two receptors known to bind TNF, this cytokine can be found as a transmembrane (mTNF) or a soluble (sTNF) form. Whereas both mTNF and sTNF activate TNFR1, TNFR2 is mainly activated by mTNF (1, 2), which upon binding to the receptor, causes its trimerization and activation. The fact that mTNF is the optimal activator of TNFR2 has implied serious limitations in the study of this receptor. Because the activation of TNFR2 depends on its aggregation, receptor activation can be forced by its overexpression or by the use of specific antibodies against the receptor (3, 4).
TNFR2 lacks any intrinsic catalytic activity within its cytoplasmic tail, thus any signal emerging from the receptor depends on the recruitment of adaptor proteins. TNFR2 can bind directly TRAF2 and through this interaction signals for NF-κB and JNK activation, as the expression of a dominant negative form of the adaptor protein (TRAF2dn) can suppress both the activation of NF-κB and JNK (5).
Seven different TRAF proteins have been identified so far (6). All of them share a highly conserved TRAF domain at the protein C terminus and, with the exception of TRAF1, a N-terminal-RING finger domain followed by five to seven zinc-finger motifs (7, 8). TRAF proteins were initially considered as adaptor proteins between TNFRs and the kinases implicated in the activation of JNK or IκB kinase IKK (9). It was then described that TRAF proteins, because of their RING finger domain, might act as E3 ubiquitin ligases, which catalyze K63-linked ubiquitination (10). In the case of TRAF2 this requires the interaction with cellular inhibitor of apoptosis 1 (cIAP1) and 2 (cIAP2) (11). More recently, it has been suggested that TRAF2 is by itself unable to act as a E3 ligase (12) and that sphingosine 1-phosphate might act as a cofactor required for the E3 ligase activity associated to TRAF2 (13) to catalyze K63-linked ubiquitination.
As for the interaction of TRAF2 and TNFR2, by using sequential truncations of the receptor cytoplasmic tail (amino acids 266–439), a sequence comprising amino acids 384 to 423 was proposed to be responsible of TRAF2 recruitment (14). Further analysis of this sequence limited the TRAF2 binding motif to amino acids 402-SKEE-405 (15, 16). More recently, another TRAF2 binding site with a suggested negative regulatory role has been identified on the murine TNFR2 (17). This site comprises amino acids 433–437, and its sequence does not fit with any of the consensus motifs known for TRAF2 interaction with TNFRs.
Recently it has been described that TNFR2 activation induces changes in TRAF2 subcellular localization (18, 19). Upon TNFR2 stimulation, TRAF2 translocates to a Triton X-100 insoluble compartment (20, 21) where the adaptor protein is K48-linked ubiquitinated and finally degraded by the proteasome (19, 22). In this process, cIAP1 interacts with the E2 protein Ubc6 thus leading to ubiquitination and proteasomal degradation of TRAF2 (11, 19, 22). As it has been previously mentioned, TNFR2 has no enzymatic activity within its cytoplasmic tail. Nevertheless it has been reported the activation of serine-threonine kinases and tyrosine kinases by TNFR2. Thus, it has been established the association between p80TRAK (p80 TNF receptor-associated kinase) and a region of TNFR2 comprising amino acids 354–397 (23). The role of this kinase in TNFR2 signaling remains to be investigated. The role of tyrosine kinases in TNFR2 signaling has not been clarified either. It has been described that TNFR2 can recruit the tyrosine kinase ETK, which interacts with the 15 C-terminal amino acids of TNFR2 (amino acids 425–439). Its activation seems to be independent of TRAF2, because overexpression of the mutant TRAF2dn does not affect ETK activation by the receptor (24).
Our work has focused on the study of NF-κB signaling pathway considering both its triggering and its termination. We have found two regions within the receptor involved in NF-κB activation. Both of them recruit TRAF2 upon receptor activation. One of these regions contains the well characterized TRAF2 binding motif 402-SKEE-405. The second region identified is located on the C terminus, comprises amino acids 425–439 of the receptor and its sequence does not fit with known TRAF2 binding motifs. This region overlaps with the receptor's ETK binding site, pointing to a possible connection between tyrosine kinases and the new NF-κB activation site. We have also found that human TNFR2 contains a conserved region localized between amino acids 338 and 379 of the receptor in which amino acids 343–349 are essential for inducing TRAF2 degradation and therefore for terminating NF-κB signaling by TNFR2, also having important consequences in the signaling by TNFR1. This region contains several serine residues which are potential phosphorylation sites and partially overlaps with the p80TRAK binding region (amino acids 354–397), suggesting a possible connection between this kinase activity and the functions here assigned to the 338–379 region of TNFR2.
EXPERIMENTAL PROCEDURES
Plasmids
Expression plasmids encoding human Myc-tagged TRAF2 and its mutant version Myc-TRAF2dn (lacking amino acids 1–87, corresponding to the Ring Finger) have been described elsewhere (25). The NF-κB reporter construct pNF-κB-SEAP encodes a thermoresistant secretable alkaline phosphatase under the control of a promoter where its minimal promoter region was complemented with four tandem copies of the NF-κB binding site (GGGGACTTTCCC) (26). Plasmid encoding NIK-mut, in which lysines 429 and 430 are both mutated to alanines, and plasmid encoding IκBαdn, lacking amino acids 1 to 36, were a gift from David Wallace (The Weizmann Institute of Science, Rehovot, Israel). Plasmid encoding TNFR1 was a gift from Bharat B. Aggarwal (MD Anderson, Houston, Texas). Expression plasmids encoding human FLAG-tagged TNFR2 (pCMV1-FLAG-TNFR2) and its deletion R2-Δ379 were a gift from B.B. Aggarwal (MD Anderson Cancer Center, Houston, Texas). Unless otherwise indicated, TNFR2 constructs used in this work were generated by PCR using standard methods and the primers indicated in supplemental Table S1. To generate point mutations by PCR mutagenesis, the Quick Change Site-directed Mutagenesis kit of Stratagene was used together with the primers indicated in supplemental Table S1. The sequences of all plasmids generated in this work were verified by automated DNA sequencing.
Cell Lines and Transient Transfection
Human embryonic kidney 293 (HEK 293) cell line was obtained from the American Type Culture Collection and cultured in minimal essential medium supplemented with 10% fetal bovine serum, 1% glutamine, 0.13% bicarbonate, and antibiotics (100 units/ml penicillin, 50 mg/ml streptomycin). For transient transfections, HEK 293 cells (2 × 105 cells/well on 6-well plates) were seeded and transfected with Lipofectamine Reagent Plus (Gibco BRL) following manufacturer's instructions. Cells and the conditioned supernatants were harvested 24–36 h after transfection.
Western Blotting and Antibodies
Proteins were separated by SDS-PAGE, electroblotted onto PVDF membranes (Bio-Rad), blocked for 1 h in 5% nonfat milk in TBS-0.1% Tween and incubated with the indicated primary antibodies (at 1:1.000 dilution in TBS-0.1% Tween) and the appropriate secondary antibody (at 1:10.000 dilution in 5% nonfat milk in TBS-0.1% Tween).
Primary antibody to TRAF2 (c-20) was obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and anti-FLAG (F3165) and anti-β-actin (A5441) from Sigma. Secondary antibodies anti-rabbit IgG and anti-mouse IgG conjugated horseradish peroxidase were purchased from Sigma. Blots were developed with an enhanced chemiluminescence detection system (GE Healthcare).
TNFR2 and TRAF2 Co-immunoprecipitation Assay
HEK 293 cells were cotransfected with 1 μg of pCMV1-FLAG-TNFR2 or its deletions and/or mutants together with 0.3 μg of pRK-Myc-TRAF2. 24 h after transfection, cells were harvested and lysed in 200 μl of Lysis buffer (20 mm Tris pH 8.0, 150 mm NaCl, 1 mm DTT, 2 mm EDTA, 1% Triton X-100, 1 μg/μl leupeptine, 1 μg/μl aprotinine, 2 mm sodium orthovanadate). Two 20-μl fractions were collected as inputs for TRAF2 and TNFR2 or its variants as expression controls. The remaining 160 μl fraction was incubated with 1 μg of anti-FLAG antibody at 4 °C in rotation for 1 h and then with 20 μl of protein G-Sepharose beads in rotation at 4 °C overnight. After three washes with lysis buffer and an additional one with low salt buffer (20 mm Tris pH 8.0, 25 mm NaCl, 1 mm DTT, 1 μg/μl leupeptin, 1 μg/μl aprotinin, 2 mm sodium orthovanadate), the immunoprecipitate was resuspended in SDS-sample buffer and subjected, together with inputs, to SDS-PAGE and Western blot with anti-TRAF2 (TRAF2) and anti-FLAG (TNFR2 and its variants).
NF-κB-SEAP Reporter Assay
HEK 293 cells were transiently cotransfected with pNF-κB-SEAP (0.5 μg) and the expression plasmids specified in each case. 36 h after transfection, the conditioned medium was removed and assayed for SEAP activity as previously described (26). The activity of SEAP was assayed on a 96-well fluorescent plate reader (Cytofluor TM 2350, Millipore). The average (± S.D.) number of relative fluorescent light units for each transfection was then determined and reported as fold activation with respect to control vector-transfected cells.
Sequence Analysis
The multiple sequence alignment of the intracellular region of TNFR2 from different species was generated using ClustalW (27). The alignment was edited using GENEDOC. Intracellular TNFR2 protein sequences were obtained from NCBI protein data base and corresponds to the following access numbers : gi 339758 gb AAA36755.1 (Homo sapiens); gi 55591327 ref XP_525188.1 (Pan troglodytes); gi 108997205 ref XP_001105753.1 (Macaca mulata); gi 199828 gb AAA39752.1 (Mus musculus); gi 27662550 ref XP_233644.1 (Rattus norvegicus); gi 281349276 gb EFB24860.1 (Ailuropoda melanoleuca); gi 76674315 ref XP_582626.2 (Bos taurus); gi 157427685 ref NP_001090910.2 (Sus scrofa); gi 194208037 ref XP_001489894.2 (Equus caballus); gi 73950773 ref XP_544562.2 (Canis familiaris). TNFR2 protein sequence was analyzed with NetPhos (28) that produces neural network predictions for serine, threonine, and tyrosine phosphorylation sites in eukaryotic proteins.
Quantification of the Hypodiploid Population
HEK293 cells were transfected with the plasmids of interest together with 0.2 μg of pEGFP-F, a plasmid expressing a farnesylated version of GFP used as a reporter of transfection. After 36 h, the cells were collected together with any floating cells in the culture media, washed with PBS first, and then with 500 μl of buffer H (20 mm Hepes pH 7.2, 160 mm NaCl and 1 mm EGTA). After these washes the cells were incubated for 5 min at room temperature in 500 μl of 0.04% digitonin in buffer H, pelleted and resuspended in 1 ml of buffer H with 200 μg/ml of RNase A and 10 μg/ml of propidium iodide for a final incubation of 30 min in the dark. The cells were analyzed in a Cytomics FC500 (Beckman Coulter) cytometer to quantify variations in the sub G0/G1 population of the transfected cells.
RESULTS
TRAF2 Depletion Induced by TNFR2 Correlates with a Decrease in TNFR2-mediated NF-κB Activation
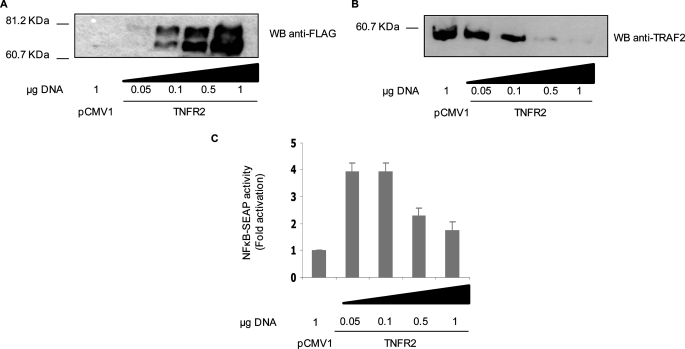
Signaling via TNFR2 causes TRAF2 degradation (22). To investigate the relationship between this phenomenon and the activation of NF-κB, we analyzed the activation of this transcription factor in several conditions in which the extent of TNFR2-induced TRAF2 degradation was controlled. Thus, increasing amounts of a mammalian expression vector encoding FLAG-tagged human TNFR2 was transfected into HEK 293 cells together with either a TRAF2 encoding plasmid or a NF-κB-dependent reporter construct (pNF-κB-SEAP). As shown in Fig. 1, A and B, as the expression of the receptor increased there was a depletion of the adaptor protein so that at the highest dose assayed TRAF2 was undetectable. On the other hand, TRAF2 was easily detected in the absence of TNFR2. As for NF-κB, the highest activation was observed at low doses of the receptor, which correlates with elevated levels of TRAF2; then the extent of NF-κB activation decreased as the amount of expressed receptor increased (Fig. 1C) and again, this correlates with the lowering in the levels of the adaptor protein. With respect to receptor overexpression, two different molecular weight bands corresponding to TNFR2 were detected by Western blot. The different electrophoretic mobilities may be the consequence of post-translational modifications (N-glycosylation and O-glycosylation) experimented by the receptor (29).
FIGURE 1.
TRAF2 depletion induced by TNFR2 correlates with the inhibition of TNFR2-mediated NF-κB activation. HEK 293 cells were transiently transfected with the indicated amounts of pFLAG-TNFR2 either with 0.3 μg of pRK-Myc-TRAF2 or with 0.5 μg of pNF-κB-SEAP reporter vector. To analyze TRAF2 protein levels, cells were harvested 36 h after transfection, and cell extracts subjected to SDS-PAGE and Western blot analysis with anti-FLAG antibody to detect receptor expression (A) or with anti-TRAF2 antibody (B). For the analysis of NF-κB activation (C), conditioned medium was removed 36 h after transfection and assayed for SEAP activity as described under “Experimental Procedures.” The results in C represent the mean ± S.D. of three independent transfection experiments.
TNFR2 Activates NF-κB in the Absence of the Canonical 402-SKEE-405 TRAF2 Binding Site
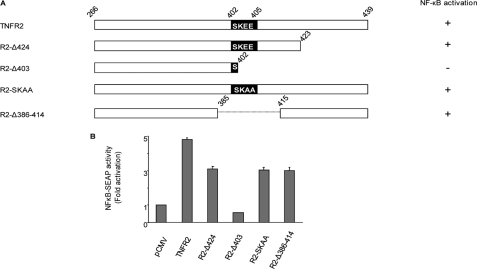
We wanted to test whether the abrogation of TRAF2 recruitment by the receptor had the same consequences in signaling than depletion of TRAF2. With this purpose we tested TNFR2 capability to activate NF-κB when the canonical 402-SKEE-405 TRAF2 binding site was abolished either by mutation (R2-SKAA) or by internal deletion (R2-Δ386–414). As shown in Fig. 2B, both the double point mutation and the deletion mutant were still able to mediate NF-κB activation although to a lower extent than the wild-type receptor. On the other hand the mutant receptor R2-Δ424 (lacking the 15 C-terminal amino acids but still carrying the intact SKEE TRAF2 binding motif) showed a reduced ability to activate NF-κB. Moreover, the mutant receptor R2-Δ403 (in which amino acids 403–439 were deleted) was the only one unable to activate NF-κB, indicating that the complete ability to activate NF-κB lies in the C-terminal half of the receptor (Fig. 2, A and B). All this led us to consider the possible existence of a novel region of the receptor capable of binding TRAF2.
FIGURE 2.
TNFR2 activates NF-κB in the abscence of the canonical 402-SKEE-405 TRAF2 binding site. A, schematic diagram of TNFR2 and its mutants or deletions. Diagrammatic representation of TNFR2 intracellular region (amino acids 266–439) and its mutants or deletions. Numbers indicate the amino acid position in the full-length human receptor sequence. In deletions, the last residue retained is numbered. Point mutations are indicated in black boxes. The ability to activate NF- κB is also indicated on the right. B, NF-κB activation induced by TNFR2 and its mutants or deletions. HEK 293 cells were transiently transfected with 0.1 μg of expression vectors encoding TNFR2, R2-Δ424, R2-Δ403, R2-SKAA, or R2-Δ386–414 together with 0.5 μg of pNF-κB-SEAP. Conditioned medium was removed 36 h after transfection and assayed for SEAP activity. The results represent the mean ± S.D. of three independent transfection experiments.
TNFR2 Binds TRAF2 through a Region Comprising the 15 C-terminal Amino Acids of the Receptor
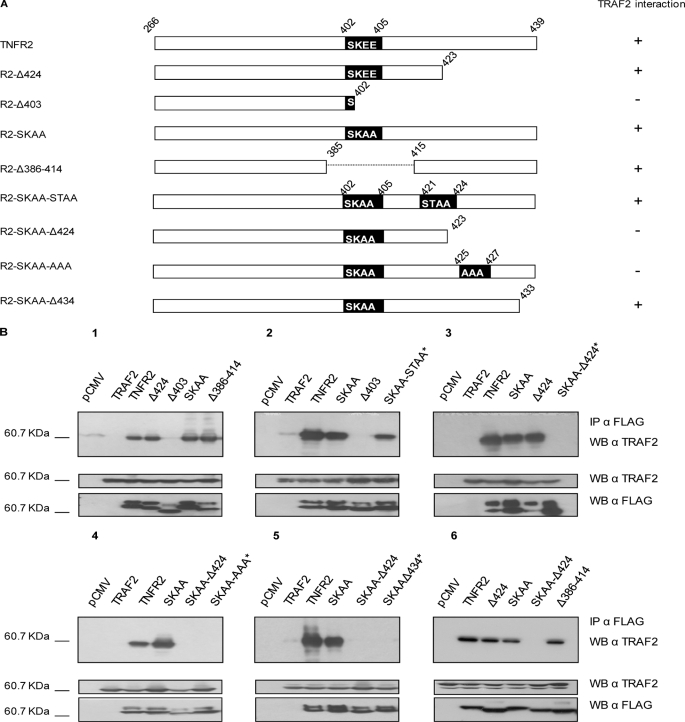
The results reported above raised the question of whether the R2-SKAA mutant or the R2-Δ386–414 deletion mutant were able of recruiting TRAF2 and whether these alterations were not sufficient to obliterate the binding of TRAF2 to the receptor, thus providing an explanation to the activation of NF-κB induced by the receptors carrying the double mutation or the deletion. For this purpose, we carried out co-immunoprecipitation assays of TRAF2 with TNFR2 and some receptor variants affecting the 402-SKEE-405 binding site (represented in Fig. 3A). As shown in Fig. 3B, panel 1, TNFR2 and R2-Δ424 (both carrying an intact canonical 402-SKEE-405 TRAF2 binding motif) bound TRAF2. Mutant receptors R2-Δ386–414 and R2-SKAA, both with alterations in the canonical TRAF2 binding site, also bound TRAF2, indicating the existence of a novel TRAF2 binding site in the receptor. From the binding of TRAF2 to the mutant receptor R2-Δ386–414 and the lack of binding to the deletion mutant R2-Δ403 it can be inferred that an alternative TRAF2 binding module should be located in the region comprising the C-terminal amino acids 415–439. In this region, which sequence is 415-PETLLGSTEEKPLPLGVPDAGMKPS-439, it is included the sequence 421-STEE-424 that fits to the major TRAF2 binding consensus motif (P/S/T/A)X(Q/E)E (30). To analyze the implication of this sequence in the novel TNFR2-TRAF2 interaction detected, we constructed and assayed the mutant receptor R2-SKAA-STAA (containing the sequences 402-SKAA-405 and 421-STAA-424). This receptor still bound TRAF2, (Fig. 3B, panel 2), indicating that the sequence 421-STEE-424 is not responsible for the novel interaction detected. Therefore, we ruled out that the sequence 421-STEE-424 within the C-terminal end of the receptor was responsible for binding TRAF2. We therefore focused our studies on the sequences 415-PETLLG-420 and 425-KPLPLGVPDAGMKPS-439. To test whether 415-PETLLG-420 was able to interact with TRAF2, we assayed R2-SKAA-Δ424, a receptor, which lacks amino acids 424–439 and carries the mutation 402-SKAA-405. This receptor did not bind TRAF2, (Fig. 3B, panel 3) indicating that the second TRAF2 binding site should be located within amino acids 425-KPLPLGVPDAGMKPS-439. To test the function of the most N-terminal amino acids of this sequence, we constructed the mutant R2-SKAA-AAA (which contains the mutations 402-SKAA-405 and 425-AAA-427) and to test the most C-terminal amino acids of the sequence we generated a mutant receptor containing the double mutation 402-SKAA-405 and deleted from amino acid A434 to the C-terminal residue (R2-SKAA-Δ434). As shown in Fig. 3B, panels 4 and 5, neither of these two mutant receptors bound TRAF2, indicating the implication of amino acids at both ends of this C-terminal sequence in the binding of the adaptor protein. We also carried out experiments studying the interaction between wild-type TNFR2 or key mutant receptors described above with endogenous TRAF2, rather than with the ectopically expressed adaptor protein. The results confirmed all the observations made with the ectopically expressed TRAF2 protein (Fig. 3B, panel 6). From our study it can be concluded the existence of a novel TRAF2 binding region in TNFR2, located at the 15 C-terminal amino acids of the receptor which fits neither the major (P/S/T/A)X(Q/E)E nor the minor PXQXT TRAF2 binding consensus motifs (30).
FIGURE 3.
TNFR2 binds TRAF2 through a region comprising the 15 C-terminal amino acids of the receptor. A, schematic diagram of TNFR2 and its mutants or deletions. Diagrammatic representation is as in Fig. 2. The ability to bind TRAF2 is also indicated on the right. B, TNFR2-TRAF2 co-immunoprecipitation. HEK 293 cells were cotransfected with 1 μg of expression plasmid encoding FLAG-tagged TNFR2 or its variants as indicated and 0.3 μg of pRK-Myc-TRAF2 (1 to 5) or without pRK-Myc-TRAF2 (6) to test the interaction with endogenous TRAF2. 24 h after transfection, cells were harvested and lysed. Inputs were analyzed to determine receptor and TRAF2 expression levels. Immunoprecipitation was carried out with anti-FLAG antibody. Immunoprecipitates were resolved by SDS-PAGE and Western blotted with anti-TRAF2 antibody as indicated. Novel constructs tested in each panel are marked with an asterisk.
The Novel TRAF2 Binding Region Signals for NF-κB Activation in a TRAF2-, NIK-, and IκBα-dependent Fashion
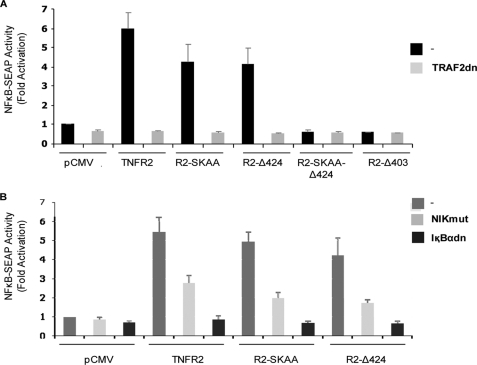
We next addressed the question of whether the new TRAF2 binding site was responsible for the activation of NF-κB detected in the absence of 402-SKEE-405 TRAF2 binding motif (see Fig. 2B). With this goal we transiently cotransfected TNFR2 (both TRAF2 binding sites intact), R2-SKAA (without a functional canonical TRAF2 binding site), R2-Δ424 (lacking the C-terminal TRAF2 binding site), R2-SKAA-Δ424 (with the canonical TRAF2 binding motif mutated and the C-terminal one deleted) and R2-Δ403 (with both TRAF2 binding sites deleted) either alone or together with a expression plasmid encoding mutant inactive versions of TRAF2 (TRAF2dn) and assayed them for the activation of NF-κB using the NF-κB dependent reporter plasmid pNF-κB-SEAP. As expected from our previous results (see Fig. 3), TNFR2, R2-SKAA and R2-Δ424, each one carrying at least one functional TRAF2 binding site, activated NF-κB whereas R2-SKAA-Δ424 and R2-Δ403, with no functional TRAF2 binding sites, were unable to activate the transcription factor (Fig. 4A), indicating again that TRAF2 recruitment by TNFR2 is a pre-requisite for the activation of NF-κB. This idea is further stressed because TRAF2dn abolished signaling from either TNFR2 region (Fig. 4A). It is remarkable that each region implicated in NF-κB activation signals with similar strength. The wild-type TNFR2, however, displayed an activation power that was not additive of those two components, indicating that both sites were not completely independent. We also wanted to test the functional implication of NIK and IκBα, two signal transducers classically related to NF-κB signaling pathway, in the activation of NF-κB by the C-terminal region of the receptor. For this purpose, we transiently cotransfected the expression plasmids encoding TNFR2, R2-SKAA, or R2-Δ424 with either a plasmid encoding a catalytically inactive version of NIK (NIK-mut) or a plasmid encoding a non degradable version of IκBα (IκBαdn) and assayed them for activation of NF-κB as described above. As shown in Fig, 4B, both NIK-mut and IκBαdn obliterated NF-κB activation induced by both TRAF2 binding regions in TNFR2. We therefore concluded that there are no differences between the two TRAF2 binding sites as for the pathways they use to activate NF-κB. It can also be concluded that TNFR2 activates NF-κB by both the classical and the alternative pathways.
FIGURE 4.
The novel TRAF2 binding site activates NF-κB in a TRAF2, NIK, and IκBα-dependent fashion. A, analysis of TRAF2 implication in NF-κB activation mediated by the C-terminal TRAF2 binding site. HEK 293 cells were transiently transfected with 0.1 μg of expression vectors encoding TNFR2, R2-SKAA, R2-Δ424, R2-SKAA-Δ424, or R2-Δ403 as indicated, together with 0.5 μg of pNF-κB-SEAP and, when indicated, with 0.2 μg of a plasmid encoding TRAF2dn. Conditioned medium was removed 36 h after transfection and assayed for SEAP activity. B, analysis of the functional relevance of NIK and IκBα in TNFR2-dependent NF-κB activation. HEK 293 cells were transiently transfected with 0.1 μg of expression vectors encoding TNFR2, R2-SKAA, R2-Δ424, or R2-SKAA-Δ424 as indicated, together with 0.5 μg of pNF-κB-SEAP and, when indicated, with 0.2 μg of an expression vector encoding NIK-mut or IκBαdn. Conditioned medium was removed 36 h after transfection and assayed for SEAP activity.
The Novel NF-κB Activation/TRAF2 Binding Site Is Highly Conserved
To explore the conservation status of the novel NF-κB activation/TRAF2 binding site, we made a multiple amino acid sequence alignment of TNFR2 intracellular regions from different species. The result indicates that the novel NF-κB activation/TRAF2 binding site, comprising amino acids 425 to 439 of the human receptor, is highly conserved among the species analyzed (supplemental Fig. S1). This comparison revealed another three modules which are highly conserved in addition to the NF-κB activation/TRAF2 binding sites. These three conserved modules comprise amino acids 266–285 (module I), amino acids 291–321 (module II) and amino acids 338–379 (module III). We therefore aimed at studying the possible implication of these modules in the degradation of TRAF2 triggered by TNFR2.
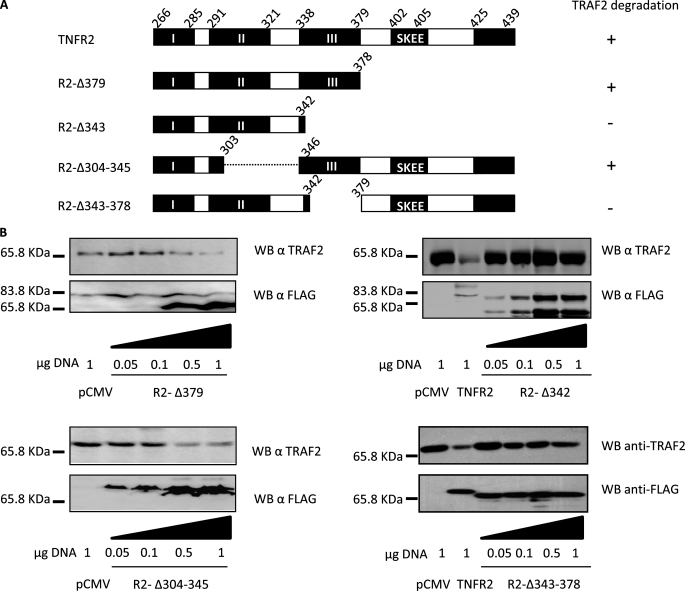
The Region Comprising Amino Acids 338–379 of TNFR2 Is Involved in TRAF2 Degradation
The observations reported above led us to generate some new variants of TNFR2 with some internal deletions lacking amino acids 304–345 (R2-Δ304–345) in which module II has been partially eliminated or lacking amino acids 343–378 (R2-Δ343–378) in which module III has been eliminated, as well as others containing partial deletions from the C terminus (R2-Δ379 and R2-Δ343) as shown in Fig. 5A. We cotransfected the plasmids encoding such mutant receptors together with TRAF2 expression vector and examined their ability to trigger TRAF2 degradation. As shown in Fig. 5B, R2-Δ379 and R2-Δ304–345 were able to induce TRAF2 degradation. By contrast, R2-Δ343 and R2-Δ343–379 failed to induce TRAF2 degradation. These results indicate that the region comprised between amino acids 343 and 378 is involved in the induction of TRAF2 degradation. These amino acids are comprised within the conserved module III of the receptor mentioned above, which may therefore have an important role in TNFR2 signal transduction.
FIGURE 5.
The region comprising amino acids 343–378 of TNFR2 is involved in TRAF2 degradation. A, schematic diagram of TNFR2 and its deletions. Diagrammatic representation is as in Fig. 2. Black boxes indicate the conserved modules and, when required, the amino acid sequence is indicated. The ability to induce degradation of TRAF2 is also indicated on the right. B, analysis of TRAF2 degradation induced by different TNFR2 deletion mutants. HEK 293 cells were transiently transfected with 0.3 μg of pRK-Myc-TRAF2 and the indicated amounts of pTNFR2, pR2-Δ379, pR2-Δ343, pR2-Δ304–345, or pR2-Δ344–379, all FLAG tagged. 36 h after transfection, cells were harvested and lysed. Equal aliquots were resolved by SDS-PAGE and Western blotted with anti-TRAF2 or anti-FLAG (receptor) antibodies.
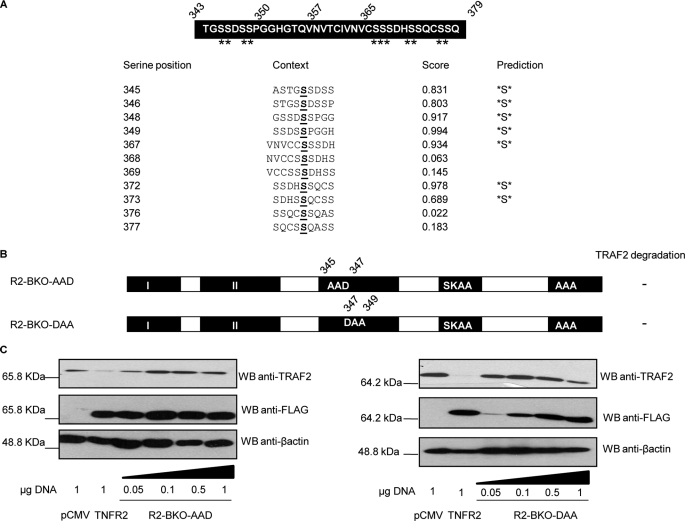
Amino Acids 345-SS-346 and 348-SS-349 of TNFR2 Are Responsible for Receptor-induced TRAF2 Depletion
As mentioned above, amino acids 343–378 of TNFR2 appear to be responsible for the degradation of TRAF2 induced by the receptor. We therefore focused on the search of the specific residues responsible of this activity. The region 343–378 of TNFR2 contains 11 serine residues (Fig. 6A) which were analyzed with the NetPhos software to predict possible phosphorylation sites. As shown in Fig. 6A, 7 of the 11 serine residues were predicted as phosphorylation sites with a score higher than 0.8. We therefore generated new mutant receptors (represented in supplemental Fig. S2A) with internal deletions within this region and also with the TRAF2 binding sites (402-SKEE-405 and 425-KPL-427) mutated (402-SKAA-405 and 425-AAA-427) so that they were unable to interact with the adaptor protein (and therefore named R2 BKO, from TRAF2 binding knock out). The analysis of their ability to induce TRAF2 degradation showed that the region comprising amino acids 343–349 is responsible of this activity (supplemental Fig. S2B). In this sequence of 6 amino acids, 4 of them are serines which are potential phosphorylation sites. We therefore generated two new mutant receptors in which serines 345 and 346 or serines 348 and 349 were mutated. In these receptors the binding of TRAF2 was also knocked out and were therefore named R2-BKO-AAD and R2-BKO-DAA, respectively (see Fig. 6B). The Western blot analysis of cell extracts in which increasing amounts of the expression plasmids coding for these receptors were cotransfected with the expression plasmid coding for TRAF2 showed that neither receptor was able to induce TRAF2 degradation (Fig. 6C). We can therefore conclude that any of these 4 serine residues appear to be essential for inducing TRAF2 degradation, thus providing the receptor site in which NF-κB signal termination is initiated. It is important to stress that all the receptors used in this study were unable to bind TRAF2, thus showing that the induction of TRAF2 degradation by TNFR2 appears to be independent of TRAF2 binding.
FIGURE 6.
Amino acids 345-SS-346 and 348-SS-349 of TNFR2 are responsible for receptor-induced TRAF2 degradation. A, sequence analysis of region 343–379 to detect potential phosphorylation sites. The amino acid sequence between residues 343 and 379 is indicated. The 11 serine residues within this sequence are marked with asterisks. The sequence was analyzed with the NetPhos software to identify serine residues that could be phosphorylated. 7 of the serine residues show a score higher than 0.8. B, schematic representation of the mutations of region 343–349 of TNFR2. Diagrammatic representation of TNFR2 mutants is as in Fig. 5. The ability to induce TRAF2 degradation is also indicated on the right. C, mutation of serines 345- 346 or 348–349 abrogates TRAF2 degradation. HEK 293 cells were transiently transfected with 0.3 μg of pRK-Myc-TRAF2 and increasing amounts of pR2-BKO-AAD or pR2-BKO-DAA (all FLAG tagged) as indicated. 36 h after transfection cells were harvested and lysed. Equal aliquots of the lysates were resolved by SDS-PAGE and Western blotted with anti-TRAF2, anti-FLAG (receptors) or anti-β-actin antibodies.
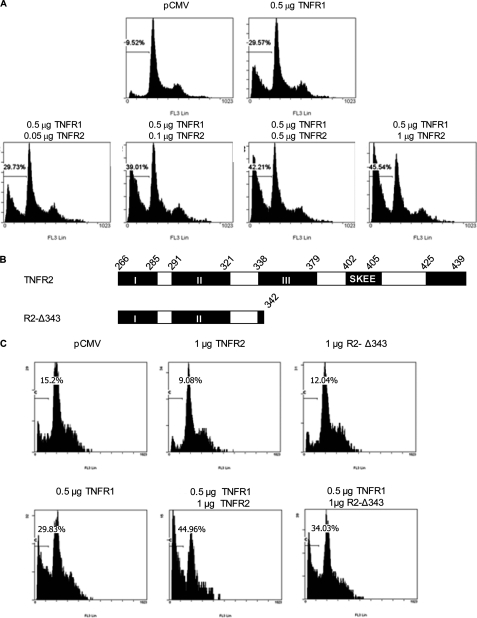
TNFR2 Enhances the Ability of TNFR1 to Induce Cell Death
Both TNF receptors can trigger NF-κB activation (5, 31), and it is known that TNFR2 can potentiate the cytotoxicity of TNFR1 (32, 33). To test if the TRAF2 degradation module identified in the cytoplasmic region of TNFR2 is necessary for the cooperation between the two TNF receptors to induce cell death, HEK293 cells were transfected with a fixed amount of TNFR1 and increasing amounts of wild-type TNFR2 or with R2- Δ 343, a TNFR2 deletion mutant lacking the two TRAF2 binding sites and the module necessary to induce the degradation of TRAF2 (see Fig. 7B). As shown in Fig. 7A the percentage of hypodiploid cells induced by TNFR1 increased in a dose-dependent manner with respect to the amount of TNFR2 that was cotransfected with it. On the other hand, when the mutant receptor R2- Δ343 was cotransfected with TNFR1, no enhancement of TNFR1 cytotoxicity was observed.
FIGURE 7.
TNFR2 enhances the ability of TNFR1 to induce cell death. A, quantification of the sub G0/G1 population induced by coexpression of TNFR1 and TNFR2.HEK 293 cells were transfected with 0.5 μg of pTNFR1 alone or together with increasing amounts of pTNFR2 as indicated, as well as with 0.2 μg of pGEGF-F. Thirty-six hours later, cells were harvested, stained with propidium iodide, and analyzed by flow cytometry to quantify the sub Go/G1 population in the transfected cells. B, schematic diagram of TNFR2 and its deletion R2-Δ343. The receptors are represented as in Fig. 5. C, quantification of the sub-G0/G1 population induced by coexpression of TNFR1 and R2-Δ343. HEK 293 cells were transfected as in A and the percentage of hypodiploid cells in the transfected cells was quantified by flow cytometry.
DISCUSSION
It was initially described that TNFR2 initiates its signal transduction through TRAF2 recruitment to the receptor cytoplasmic tail, leading to NF-κB (5, 14) and JNK activation (4). To identify the binding site for TRAF2, sequential deletions of the intracellular tail of TNFR2 lacking the 16 (TNFR2–16), 37 (TNFR2–37), and 59 (TNFR2–59) C-terminal amino acids were used (14). It was shown that both full-length TNFR2 and TNFR2–16-bound TRAF2, while TNFR2–37 and further deletions did not associate with the adaptor protein. From these experiments it was concluded that the 16 C-terminal amino acids of TNFR2 were not relevant for TRAF2 binding, and consequently, for NF-κB activation. This first approach has been generally accepted, and no interest has been focused in this region as a potential NF-κB activator module. In our study we show that in addition to the well characterized TRAF2 binding motif 402-SKEE-405, TNFR2 contains a second region implicated in the activation of NF-κB. Our results show that this novel TRAF2 binding site regulates NF-κB activation positively, as its deletion decreases the ability of the receptor to activate NF-κB. This novel binding site, comprising amino acids 425–439 partially overlaps with the one described by Grech et al. (17) for the murine receptor for which a negative regulatory role in NF-κB signaling was suggested. In this regard, it should be mentioned that the murine receptor has 13 extra amino acids in its C-terminal region which could account for this functional difference. Moreover, we show that NF-κB activation by both TRAF2 binding sites occurs via the classical and the alternative pathways. From our experiments it is not possible to discern if TRAF2 association to this new binding site is direct or indirect. The sequence of this region fits neither into the major TRAF2 binding motif nor into the minor one and, on the other hand, this region was shown to directly associate with other proteins such as the tyrosine kinase ETK (24). It is therefore likely an indirect association between this new binding site and TRAF2. This novel NF-κB activator/TRAF2 association site is highly conserved among different species. When the phenomenon of TRAF2 degradation was described (11, 19, 22, 34) TNFR2 signal transduction became more complex. The present study brings TRAF2 degradation into consideration and opens a new aspect of TNFR2 signal transduction.
In our study NF-κB is activated with different efficiency as increasing amounts of TNFR2 are expressed. This correlates with changes in TRAF2 protein level and agrees with the well known implication of TRAF2 in NF-κB activation (5). We show that a novel region of the receptor comprising amino acids 343–349 is responsible for the induction of TRAF2 degradation, a process that negatively regulates TNFR2-induced NF-κB activation. TRAF2 is the target of TNF-induced ubiquitination, a modification that has been linked to a proteasome dependent degradation (11, 19) through K48-linked polyubiquitin chains. In addition, it has also been reported that TNF, through TNFR1, can induce another TRAF2 ubiquitination pattern, implicating K63-linked polyubiquitin chains, a modification not related to degradation but to JNK activation (35). In this regard it is interesting the identification of TRAF2 interaction with ubiquitin proteases such as USP31, which shows specificity for K63-linked polyubiquitin chains (36) or CYLD (37). It is therefore conceivable that TRAF2 can be initially modified through K63-linked polyubiquitin chains, converted into an active signal transducer and then, through K48-linked polyubiquitin chains, tagged for the proteasomal machinery, bringing to an end TRAF2-dependent signaling process. It is well known that TRAF2 interacts with cIAP1 and cIAP2 and that these interactions cause TRAF2 degradation by the proteasome (19). In this context, several authors have identified different intermediates that might be implicated in the interaction TRAF2-cIAPs, such as AIMP2 (38) or sphingosine 1-phosphate (13), which has been described as a cofactor for E3 ubiquitin ligase activity of TRAF2 (39). On the other hand, it has been recently described the role of A20 on TRAF2 lysosomal degradation in conditions in which TNF triggers cell death. So, in these conditions TRAF2 should be degraded both by the proteasome and the lysosome (40).
The connection between region 343–349 and TRAF2 ubiquitination is probably mediated through a phosphorylation event. Such post-translational modification has been classically related as the initiator step in the ubiquitination process. In this regard it is interesting that the conserved module III, in which amino acids 343–349 are included, partially overlaps with the TNFR2 p80TRAK (p80 TNF Receptor Associated Kinase) binding motif, which is located between amino acids 338 and 379 (23). Thus, it is possible that region 343–378, through its association with p80TRAK or another kinase as yet unidentified, mediates TRAF2 phosphorylation and subsequent ubiquitination, a process necessary both for NF-κB activation and TRAF2 degradation. In summary, region 338–379 could act as an “on-off” switch in TNFR2-induced NF-κB activation. In this regard, it is particularly interesting that while the “on” signal requires TRAF2 binding to the receptor, the “off” signal appears to be independent of TRAF2 recruitment since R2-Δ379, a receptor lacking both TRAF2 binding sites, is able to induce TRAF2 degradation. TNFR2 does not have a death domain in its intracellular region, so it is unable to induce cell death by itself. However, we found that it is able to increase the cytotoxic effect mediated by TNFR1. This crosstalk between the two receptors seems to relay on the ability of TNFR2 to induce TRAF2 degradation. In this context, TRAF2 could be considered as a central player in the cytotoxic effect mediated by TNFR1, since TRAF2 interaction with TNFR1 and the anti-apoptotic proteins cIAP1 and cIAP2 reduced the ability of this receptor to induce cell death (33). On the other hand, when TNFR2 and TNFR1 are coactivated the degradation of TRAF2 induced by TNFR2 abrogates the interaction between TNFR1 and the anti-apoptotic proteins, thus enhancing the cell death induced by TNFR1. This observation could be envisaged as a particular situation of the classical phenomenom in which the balance between the apoptotic and antiapoptotic signals triggered by TNF determines whether the final output will be either cell death or cell survival.
Supplementary Material
This work was supported by Grant SAF2009-07227 from the Spanish Ministry of Science and Technology (MICINN) and by Grant IB09-066 from the Plan Regional de Ciencia y Tecnología del Principado de Asturias (PCTI). The IUOPA is supported by Obra Social Cajastur.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- TNFR2
- tumor necrosis factor receptor 2
- TRAF2
- TNF receptor-associated factor 2
- cIAP
- cellular inhibitor of apoptosis
- TRAK
- TNF receptor-associated kinase.
REFERENCES
- 1. Grell M., Douni E., Wajant H., Löhden M., Clauss M., Maxeiner B., Georgopoulos S., Lesslauer W., Kollias G., Pfizenmaier K., Scheurich P. (1995) Cell 83, 793–802 [DOI] [PubMed] [Google Scholar]
- 2. Wajant H., Pfizenmaier K., Scheurich P. (2003) Cell Death Differ. 10, 45–65 [DOI] [PubMed] [Google Scholar]
- 3. Espevik T., Shalaby R., Liabakk N. B., Waage A., Sundan A. (1990) Scand. J. Clin. Lab. Invest. Suppl. 202, 160 [PubMed] [Google Scholar]
- 4. Reinhard C., Shamoon B., Shyamala V., Williams L. T. (1997) EMBO. J. 16, 1080–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rothe M., Sarma V., Dixit V. M., Goeddel D. V. (1995) Science 269, 1424–1427 [DOI] [PubMed] [Google Scholar]
- 6. Bouwmeester T., Bauch A., Ruffner H., Angrand P. O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., Hopf C., Huhse B., Mangano R., Michon A. M., Schirle M., Schlegl J., Schwab M., Stein M. A., Bauer A., Casari G., Drewes G., Gavin A. C., Jackson D. B., Joberty G., Neubauer G., Rick J., Kuster B., Superti-Furga G. (2004) Nat. Cell Biol. 6, 97–105 [DOI] [PubMed] [Google Scholar]
- 7. Bradley J. R., Pober J. S. (2001) Oncogene 20, 6482–6491 [DOI] [PubMed] [Google Scholar]
- 8. Wajant H., Henkler F., Scheurich P. (2001) Cell Signal. 13, 389–400 [DOI] [PubMed] [Google Scholar]
- 9. Häcker H., Karin M. (2006) Sci. STKE. 2006, re13. [DOI] [PubMed] [Google Scholar]
- 10. Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. (2000) Cell 103, 351–361 [DOI] [PubMed] [Google Scholar]
- 11. Li X., Yang Y., Ashwell J. D. (2002) Nature 416, 345–347 [DOI] [PubMed] [Google Scholar]
- 12. Yin Q., Lamothe B., Darnay B. G., Wu H. (2009) Biochemistry 48, 10558–10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alvarez S., Harikumar K., Hait N., Allegood J., Strub G., Kim E., Maceyka M., Jiang H., Luo C., Kordula T., Milstein S., Spiegel S. (2010) Nature 465, 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rothe M., Wong S. C., Henzel W. J., Goeddel D. V. (1994) Cell 78, 681–692 [DOI] [PubMed] [Google Scholar]
- 15. Boucher L. M., Marengère L. E., Lu Y., Thukral S., Mak T. W. (1997) Biochem. Biophys. Res. Commun. 233, 592–600 [DOI] [PubMed] [Google Scholar]
- 16. Park Y. C., Burkitt V., Villa A. R., Tong L., Wu H. (1999) Nature 398, 533–538 [DOI] [PubMed] [Google Scholar]
- 17. Grech A. P., Gardam S., Chan T., Quinn R., Gonzales R., Basten A., Brink R. (2005) J. Biol. Chem. 280, 31572–31581 [DOI] [PubMed] [Google Scholar]
- 18. Arch R. H., Gedrich R. W., Thompson C. B. (2000) Biochem. Biophys. Res. Commun. 272, 936–945 [DOI] [PubMed] [Google Scholar]
- 19. Wu C. J., Conze D. B., Li X., Ying S. X., Hanover J. A., Ashwell J. D. (2005) EMBO. J. 24, 1886–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feng X., Gaeta M. L., Madge L. A., Yang J. H., Bradley J. R., Pober J. S. (2001) J. Biol. Chem. 276, 8341–8349 [DOI] [PubMed] [Google Scholar]
- 21. Wicovsky A., Henkler F., Salzmann S., Scheurich P., Kneitz C., Wajant H. (2009) Oncogene 28, 1769–1781 [DOI] [PubMed] [Google Scholar]
- 22. Chan F. K., Lenardo M. J. (2000) Eur. J. Immunol. 30, 652–660 [DOI] [PubMed] [Google Scholar]
- 23. Darnay B. G., Singh S., Aggarwal B. B. (1997) FEBS. Lett. 406, 101–105 [DOI] [PubMed] [Google Scholar]
- 24. Pan S., An P., Zhang R., He X., Yin G., Min W. (2002) Mol. Cell. Biol. 22, 7512–7523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Darnay B. G., Haridas V., Ni J., Moore P. A., Aggarwal B. B. (1998) J. Biol. Chem. 273, 20551–20555 [DOI] [PubMed] [Google Scholar]
- 26. Darnay B. G., Ni J., Moore P. A., Aggarwal B. B. (1999) J. Biol. Chem. 274, 7724–7731 [DOI] [PubMed] [Google Scholar]
- 27. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blom N., Gammeltoft S., Brunak S. (1999) J. Mol. Biol. 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 29. Pennica D., Lam V. T., Weber R. F., Kohr W. J., Basa L. J., Spellman M. W., Ashkenazi A., Shire S. J., Goeddel D. V. (1993) Biochemistry 32, 3131–3138 [DOI] [PubMed] [Google Scholar]
- 30. Ye H., Park Y. C., Kreishman M., Kieff E., Wu H. (1999) Mol. Cell. 4, 321–330 [DOI] [PubMed] [Google Scholar]
- 31. Tartaglia L. A., Goeddel D. V. (1992) Immunol Today 13, 151–153 [DOI] [PubMed] [Google Scholar]
- 32. Fotin-Mleczek M., Henkler F., Samel D., Reichwein M., Hausser A., Parmryd I., Scheurich P., Schmid J. A., Wajant H. (2002) J. Cell Sci. 115, 2757–2770 [DOI] [PubMed] [Google Scholar]
- 33. Weiss T., Grell M., Hessabi B., Bourteele S., Müller G., Scheurich P., Wajant H. (1997) J. Immunol. 158, 2398–2404 [PubMed] [Google Scholar]
- 34. Duckett C. S., Thompson C. B. (1997) Genes Dev. 11, 2810–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shi C. S., Kehrl J. H. (2003) J. Biol. Chem. 278, 15429–15434 [DOI] [PubMed] [Google Scholar]
- 36. Tzimas C., Michailidou G., Arsenakis M., Kieff E., Mosialos G., Hatzivassiliou E. G. (2006) Cell Signal. 18, 83–92 [DOI] [PubMed] [Google Scholar]
- 37. Kovalenko A., Chable-Bessia C., Cantarella G., Israël A., Wallach D., Courtois G. (2003) Nature 424, 801–805 [DOI] [PubMed] [Google Scholar]
- 38. Choi J. W., Kim D. G., Park M. C., Um J. Y., Han J. M., Park S. G., Choi E. C., Kim S. (2009) J. Cell Sci. 122, 2710–2715 [DOI] [PubMed] [Google Scholar]
- 39. Alvarez S. E., Harikumar K. B., Hait N. C., Allegood J., Strub G. M., Kim E. Y., Maceyka M., Jiang H., Luo C., Kordula T., Milstien S., Spiegel S. (2010) Nature 465, 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li L., Soetandyo N., Wang Q., Ye Y. (2009) Biochim. Biophys. Acta 1793, 346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.