Abstract
Internalization of Shigella into host epithelial cells, where the bacteria replicates and spreads to neighboring cells, requires a type 3 secretion system (T3SS) effector coined IpaA. IpaA binds directly to and activates the cytoskeletal protein vinculin after injection in the host cell cytosol, and this was previously thought to be directed by two amphipathic α-helical vinculin-binding sites (VBS) found in the C-terminal tail domain of IpaA. Here, we report a third VBS, IpaA-VBS3, that is located N-terminal to the other two VBSs of IpaA and show that one IpaA molecule can bind up to three vinculin molecules. Biochemical in vitro Shigella invasion assays and the 1.6 Å crystal structure of the vinculin·IpaA-VBS3 complex showed that IpaA-VBS3 is functionally redundant with the other two IpaA-VBSs in cell invasion and in activating the latent F-actin binding functions of vinculin. Multiple VBSs in IpaA are reminiscent of talin, which harbors 11 VBSs. However, most of the talin VBSs have low affinity and are buried in helix bundles, whereas all three of the VBSs of IpaA are high affinity, readily available, and in close proximity to each other in the IpaA structure. Although deletion of IpaA-VBS3 has no detectable effects on Shigella invasion of epithelial cells, deletion of all three VBSs impaired bacterial invasion to levels found in an ipaA null mutant strain. Thus, IpaA-directed mimicry of talin in activating vinculin occurs through three high affinity VBSs that are essential for Shigella pathogenesis.
Keywords: Bacterial Toxins, Cell Adhesion, Crystal Structure, Cytoskeleton, Protein-Protein Interactions, Signal Transduction
Introduction
Vinculin is a cytoskeletal helix bundle protein that strengthens and stabilizes focal adhesion complexes (1). This occurs through its ability to bind to actin and to force-activate talin that is tethered to the cytoplasmic tail of integrin receptors (2, 3). The N-terminal seven-helix bundle vinculin head domain, Vh1, contains binding sites for talin and α-actinin (4–6), whereas the five-helix bundle tail domain, Vt, contains binding sites for F-actin (7) and raver1 (8–10). The activity of vinculin is inhibited by intramolecular Vh1-Vt interactions that are severed by the binding of vinculin binding sites (VBSs) to the Vh1 domain (11), which triggers remarkable changes in the conformation of Vh1 and displaces Vt (6, 12, 13). VBSs are amphipathic α-helices of about 19 residues found in vinculin activators such as talin and α-actinin (8, 9). VBSs are also found in the IpaA invasin, a T3SS2 effector of Shigella (14, 15). Crystal structures of Vh1 with various VBS complexes have shown that these VBSs usually insert into and contort the N-terminal four-helix bundle subdomain of Vh1 (6, 12, 15, 16). However, binding to the C-terminal four-helix bundle subdomain of Vh1 has also been observed with some VBSs, and this occurs via a helix addition mechanism where the VBS simply binds to the four-helix bundle without altering its structure (15).
Shigella spp. are Gram-negative, facultative anaerobic, intracellular pathogens that are the principal cause of bacterial dysentery in humans (17). Indeed, dysentery provoked by Shigella causes 1.1 million deaths a year, mostly in children in underdeveloped countries (18). To enter into intestinal epithelial cells of the host, Shigella requires extensive remodeling of the actin cytoskeleton that is directed by virulence determinants that are transferred through the T3SS (19). Four invasins, IpaA, IpaC, IpgB1, and IpgB2, function in a coordinated manner to direct host cell entry (20–23).
IpaA is a 633-residue protein that harbors an N-terminal chaperone binding domain, a disordered central region, and an all-helical C-terminal tail domain. The last two C-terminal helices are tandem VBSs through which IpaA binds the Vh1 domain of vinculin (14). We have shown that these VBSs are required for maximal entry of Shigella into host cells and that they activate vinculin by acting as very efficient mimics of talin (14). Furthermore, the vinculin·IpaA complex also regulates actin polymerization by modulating barbed end capping activity (24). Although talin contains 11 VBSs, all of these appear to be buried within helix bundles in the talin rod, and to activate vinculin they must be released from these bundles by mechanical force, for example following integrin binding to matrix components (25, 26). By contrast, IpaA does not require pre-activation to bind to vinculin (20), where its high affinity, tandem VBSs are available to bind to and activate vinculin (14).
The two C-terminal VBSs of IpaA do not, however, fully account for the activity of full-length IpaA, as Shigella expressing an IpaA mutant lacking these VBSs can still recruit vinculin to bacterial entry sites and is only partially compromised in bacterial invasion (14). Thus, we posited that yet another VBS harbored by IpaA provides this function. Here, we report a third IpaA VBS (residues 488–512), which also binds to and activates vinculin. The crystal structure of this IpaA-VBS3 in complex with vinculin as well as biochemical and biological studies support a model where the three juxtaposed high affinity VBSs of IpaA endow Shigella with the ability to rapidly and efficiently activate vinculin at sites of pathogen entry.
EXPERIMENTAL PROCEDURES
Protein Purification and Crystallization
For biochemical studies, the newly identified VBS of IpaA (residues 488–512; IpaA-VBS3) was amplified by PCR from a virulence plasmid derived from Shigella flexneri ΔIpaBC strain (27). The PCR product was cloned into NdeI/EcoRI sites of the pET28a vector (Novagen) and NheI/EcoRI sites of pMAL-c2e (New England Biolabs) to generate pET28a-IpaA-VBS3 and pMAL-IpaA-VBS3, producing IpaA-VBS3 with an N-terminal hexahistidine tag and a maltose-binding protein, respectively. Furthermore, pMAL-IpaA-VBS3 was mutated at Val-499 and Leu-503 using PCR mutagenesis technique (28) to produce pMAL-IpaA-VBS3 V499E/L503Q.
For crystallographic studies, the human vinculin head domain (residues 1–258; Vh1) followed by an internal ribosome-binding site and a start codon was PCR-amplified and inserted into the EcoRI site of pET28a-IpaA-VBS3 to generate the bicistronic expression vector pET28a-IpaA-VBS3-Vh1. Plasmids were transformed into the Escherichia coli BL21(DE3) strain (Novagen). The binary complex of Vh1·IpaA-VBS3 was expressed in E. coli cells grown overnight in auto-induction media (29) containing kanamycin at 30 °C.
The Vh1·IpaA-VBS3 complex was purified by passage over an affinity nickel column and size exclusion chromatography column on an Äkta FPLC system (GE Healthcare). In brief, cells were harvested by centrifugation and resuspended in sonication buffer (10 mm Tris-HCl (pH 8), 300 mm NaCl, and 10 mm imidazole), and the supernatant was loaded onto a HisTrap HP affinity chromatography column (GE Healthcare). The column was then washed with a linear gradient from 10 to 250 mm imidazole to elute adsorbed proteins. The elutions were pooled and concentrated with a 30-kDa cutoff ultrafiltration unit (Amicon), and the buffer was exchanged to phosphate-buffered saline (PBS) for thrombin digestion. A ratio of 0.1 unit of thrombin (Sigma-Aldrich) to 1 mg of Vh1·IpaA-VBS3 was used, and samples were incubated at room temperature for 4 h. Digestion by thrombin was confirmed by SDS-PAGE. Vh1 without the affinity tag was then loaded onto a Superdex 200 26/60 sizing chromatography column that was equilibrated with 10 mm Tris-HCl (pH 7.5), 20 mm NaCl, 1 mm EDTA, and 1 mm DTT. The fractions containing Vh1·IpaA-VBS3 were pooled and concentrated to 30 mg/ml.
Crystal screening on 96-well plates using commercial screening solutions and the TTP LabTech Mosquito crystallization robot identified several crystallization hits. Best diffracting crystals were obtained from a reservoir containing 0.1 m sodium citrate, 20% PEG-3350, and 0.1 m cacodylate (pH 7.2).
For biochemical studies, the human Vh1 domain, full-length human vinculin, and the human Vt domain were purified as described previously (14). Wild type IpaA-VBS3 and mutant IpaA-VBS3 V499E/L503Q, having N-terminal maltose-binding protein tags, were purified by a single step amylose affinity chromatography column (New England Biolabs) following the manufacturer's instructions.
Data Collection and Structure Determination
Crystals of the Vh1·IpaA-VBS3 complex were briefly soaked in a step gradient of cryoprotectant solutions (the reservoir solution supplemented with 5, 10, and 20% of ethylene glycol) and flash-frozen in liquid nitrogen. A complete x-ray diffraction data set to 1.61-Å Bragg spacings was collected at 100 K using a wavelength of 0.9795 Å on the SER-CAT beamline 22ID at the Advanced Photon Source, Argonne National Laboratory. The data were processed with AutoPROC (30), which utilizes the XDS program (31) as the data-processing engine. The structure of the Vh1·IpaA-VBS3 complex was determined by molecular replacement using BALBES (32), a component of the CCP4 programs suite version 6.1.3 (33), using the Vh1 from the Vh1·IpaA-VBS1 structure (14) as a search model.
Crystallographic refinement of the Vh1·IpaA-VBS3 structure was performed using the program BUSTER (34) applying both translation, libration, and screw-motion and the local structure similarity restraint approach to enforcing noncrystallographic symmetry (NCS) (35), interspersed by cycles of manual rebuilding with Coot (36). The model was analyzed with the CCP4 (33) program WATNCS to identify NCS-related water molecules, showing that 600 of the 804 water molecules obeyed the NCS. Chain identity and the residue number of these water molecules were reassigned to the related protein chains so that the NCS-related water molecules have the same residue number. These were then subsequently included in the NCS definitions for the final rounds of refinement. One ordered cacodylate per heterodimer was identified, modeled, and refined under NCS restraints; each led to the ordering of the Lys-199 side chain through water-mediated electrostatic interactions. Structure validation was carried out with MolProbity (37). Detailed crystal parameters, data reduction statistics, and refinement parameters are provided in Tables 1 and 2, respectively.
TABLE 1.
Data reduction statistics for the Vh1·IpaA-VBS3 complex
| Space group | P212121 |
| Unit cell dimensions | |
| a | 53.58 ± 0.17 Å |
| b | 85.32 ± 0.17 Å |
| c | 137.90 ± 0.22 Å |
| α = β = γ | 90° |
| Resolution | 137 to 1.61 Å |
| Last shell | 1.7 to 1.61 Å |
| Total measurements | 558,840 |
| No. of unique reflections | 82,474 |
| Last shell | 11,885 |
| Wavelength | 0.9795 Å |
| R-mergea | 0.037 |
| Last shell | 0.5 |
| I/σ(I) | 25.2 |
| Last shell | 3.7 |
| Completeness | 0.999 |
| Last shell | 1 |
| Redundancy | 6.8 |
| Last shell | 6.7 |
a R-merge = ΣhklΣi|Ii(hkl) − Ī (h̄k̄l̄)|/ΣhklΣiIi(hkl).
TABLE 2.
Crystallographic refinement statistics for the Vh1·IpaA-VBS3 complex
| Resolution | 53 to 1.61 Å |
| Last shell | 1.65 to 1.61 Å |
| No. of reflections (working set) | 78,259 |
| No. of reflections (test set) | 4,129 |
| R-factora | 0.1849 |
| Last shell | 0.1956 |
| R-freeb | 0.1954 |
| Last shell | 0.2496 |
| No. of protein atoms | 8,871 |
| No. of water molecules | 804 |
| No. of cacodylic anions | 2 |
| Average B-factor | |
| Vh1 | 31.65 Å2 |
| IpaA-VBS3 | 29.61 Å2 |
| Solvent | 32.92 Å2 |
| r.m.s.d. from ideal geometry | |
| Bond lengths | 0.01 Å |
| Bond angles | 1.02° |
a R-factor = Σhkl‖Fobs(hkl)|−|Fcalc(hkl)|, where 〈|Fcalc|〉 denotes the expectation of |Fcalc(hkl)| used in defining the likelihood refinement target.
b The free R-factor is a cross-validation residual calculated by using about 5% reflections, which were randomly chosen and excluded from the refinement.
Native PAGE Assays
For binding studies, native PAGE was performed with a PhastGel system (GE Healthcare) according to the manufacturer's instructions. 10 μm of Vh1 was incubated with IpaA-VBS3 in 5:1, 5:3, and 1:1 molar ratios at room temperature for 10 min before analysis on an 8–25% gradient native gel. Vh1·Vt displacement assays with IpaA-VBS3 were performed as described previously (14).
Surface Plasmon Resonance
Binding studies of wild type IpaA-VBS3 and mutant IpaA-VBS3 A495K to Vh1 were performed at 25 °C using a Biacore 2000 (Biacore AB, Uppsala, Sweden) equipped with a carboxymethyl dextran-coated gold surface (CM5 sensor chip; Biacore AB, Uppsala, Sweden). The carboxymethyl groups on the chip were activated with 1-ethyl-3-[(3-(dimethylamino)propyl]carbodiimide and N-hydroxysuccinimide to form the N-hydroxysuccinimide ester of carboxymethyl dextran. Vh1 (0.75 μm to about 1.5 μm) in 10 mm sodium acetate (pH 4.5) was immobilized at densities of about 300–2700 resonance units on the surface of several CM5 sensor chips through amine coupling. The remaining reactive sites on the surface were blocked by reaction with ethanolamine. A reference surface was prepared as for Vh1 without adding Vh1. Association rate constants of the binding was measured by flowing wild type or mutant IpaA-VBS3 in 10 mm HEPES buffer (pH 7.4), 150 mm NaCl, 0.5 mm DTT, 0.01% Tween 20, and 3% DMSO at a flow rate of 20 μl/min over reference or sample surfaces. After each injection, the dissociation rate constant of the IpaA-VBSs was measured by flowing buffer only over the surfaces. The Vh1 surfaces were regenerated with 20 mm phosphoric acid after each binding cycle. Data fitting was performed using BIAEvaluation 4.1 (Biacore AB, Uppsala, Sweden). The binding dissociation constants were calculated from the observed on- and off-rates for the interactions.
Plasmid Constructs
The deletion of IpaA-VBS3 was generated using a PCR-based deletion strategy (38) on plasmid pCR2.1:ipaA (10), using the primers 5′-CGCGCAGACTAATGCGGCCATGGTCAGAAGTGTTGAGGCAC-3′ and 5′-GTGCCTCAACACTTCTGACCATGGCCGCATTAGTCTGCGCG-3′, which generate a deletion of residues 489–511 (AΔVBS3). The AΔCterm construct deleted for residues 550–633, including IpaA-VBS1 and IpaA-VBS2, was described previously (10). To generate the AΔVBS3ΔCterm construct, pCR2.1:ipaAΔVBS3 was digested with SnaBI and ApaI, subjected to end-filling using T4 DNA polymerase, and ligated to generate deletions of residues 550–633. All constructs were verified by DNA sequencing.
Secretion Assay
Shigella strains were grown to mid-exponential phase, and Congo Red was added at 100 μg/ml to induce type III secretion as described previously (39). Secreted proteins from the bacterial supernatants were precipitated and analyzed by Western blotting with antibody specific for IpaA. As a control, immunoprecipitation-Western blotting was also performed with antibody specific for SepA (40), a Shigella protein that is secreted independently of the T3SS.
Gentamicin Protection Assay
Bacterial internalization was performed using the gentamicin protection assay as described previously (39). Semi-confluent HeLa cells were challenged with bacteria grown to mid-log phase at a multiplicity of infection of 200 bacteria per cell for 30 min at 37 °C in RPMI 1640 medium containing 50 mm HEPES buffer (pH 7.3). Samples were washed twice with PBS buffer and incubated for another 30 min at 37 °C in RPMI 1640 medium containing 10% fetal calf serum and 50 mg/ml gentamicin. Samples were washed three times with PBS buffer containing 1 mm MgCl2 and 0.5 mm CaCl2, lysed in 0.5% sodium deoxycholate, and plated onto trypticase soy plate for colony-forming unit determination.
Actin Co-sedimentation Assay
Full-length human vinculin protein was filtered using a 0.02-μm syringe filter and diluted with PBS to a final concentration of 15 μm. Each IpaA-VBS dissolved in PBS was then added to full-length vinculin at a 1:2 (vinculin:IpaA) molar ratio and incubated for 20 min at room temperature. G-actin was purchased from Cytoskeleton Inc. and polymerized following the manufacturer's instructions. The final concentration of F-actin was adjusted to 60 μm in PBS. 7 μl of F-actin and 33 μl of each vinculin·IpaA complex were mixed and incubated for 1 h at room temperature and then centrifuged at 100,000 × g for 25 min. Supernatants and pellets were resolved on SDS-PAGE using a PhastGel system (GE Healthcare).
RESULTS
Identification of Novel VBS in IpaA
We identified residues 488–512 as a predicted amphipathic α-helix using the Phyre server (41) that has 47% and 42% sequence identity with IpaA-VBS1 and IpaA-VBS2, respectively. Helical wheel comparison and sequence alignment suggested that this predicted α-helix might function as a third VBS of IpaA (IpaA-VBS3; Fig. 1). We therefore performed several binding and crystallographic studies to test if our newly identified IpaA-VBS3 is a bona fide third VBS for IpaA.
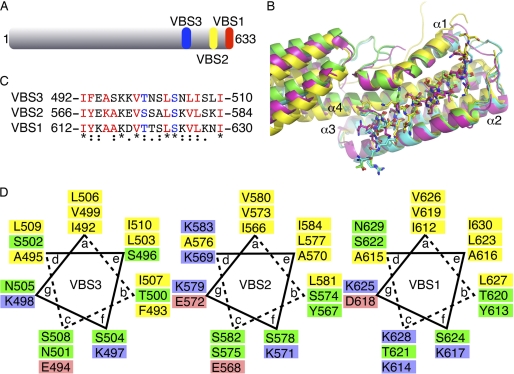
FIGURE 1.
Vh1·I paA-VBS3 crystal structure. A, schematic of IpaA. Full-length IpaA is shown in gray, IpaA-VBS1 in red, IpaA-VBS2 in yellow, and the newly identified IpaA-VBS3 in blue. B, superposition of the three crystal structures of IpaA-VBSs in their Vh1-bound states (Vh1·IpaA-VBS1, PDB entry 2gww, magenta; Vh1·IpaA-VBS2, PDB entry 2hsq, yellow; Vh1·IpaA-VBS3, cyan and green for the two heterodimers in the asymmetric unit, respectively). Vh1 is shown as a cartoon, and the IpaA-VBSs is shown in a ball-and-stick representation. α-Helices α1 through α4 of Vh1 are labeled. The 20 central residues of the IpaA-VBSs (IpaA-VBS1, residues 611–630; IpaA-VBS2, residues 565–584) were aligned with IpaA-VBS3 (residues 491–510) with r.m.s.d. of 0.37 Å and 0.38 Å, respectively, resulting in almost identical Vh1-bound subdomain structures except for the C-terminal region of α-helix α1 that unfurls upon VBS binding. The C-terminal four-helix Vh1 subdomain, which is only partially shown, adopts several relative orientations with respect to the N-terminal four-helix bundle subdomain (shown) due to the flexibility between the two Vh1 subdomains. This panel was prepared with PyMol. C, structure-based sequence alignment of the three IpaA-VBSs. The asterisk, colon, and period symbols below the sequence each represent identity, and high and low conservation, respectively. The most C-terminal VBS is IpaA-VBS1, and the most N-terminal VBS is IpaA-VBS3, a nomenclature that reflects the timing of their discoveries rather than their position in the polypeptide chain. IpaA-VBS residues in red are those that are involved in major hydrophobic interactions with Vh1, and those in blue have hydrogen bonding interactions with Vh1. D, helical wheel analysis of IpaA-VBS3 (left), IpaA-VBS2 (middle), and IpaA-VBS1 (right) indicates that all three VBSs are indeed amphipathic α-helices. The hydrophobic residues, charged residues, and polar residues are colored yellow, red and blue, and green, respectively.
Crystal Structure of the Vh1·IpaA-VBS3 Complex
Vinculin binds to IpaA with its N-terminal seven-helix bundle domain coined Vh1. We therefore crystallized Vh1 in complex with IpaA-VBS3. The crystal structure of Vh1·IpaA-VBS3 complex was determined to 1.61 Å resolution (Tables 1 and 2). This resolution represents by far the highest resolution Vh1·VBS structure to date, where eight Vh1·talin-VBS structures were determined at 3.3 Å to 1.8 Å resolution (6, 12, 42, 43), respectively, and Vh1·IpaA-VBS1 and Vh1·IpaA-VBS2 structures were solved to 2.7 Å (14) and 3.2 Å (15) resolution, respectively. The final model consists of two Vh1·IpaA-VBS3 heterodimers, two cacodylate ions, and 804 water molecules in the asymmetric unit. The model has no protein geometry violations according to MolProbity results (37). The Ramachandran plot analysis shows that all residues fall in allowed regions. MolProbity (37) and clash scores are 0.79 (99th percentile) and 0.75 (100th percentile), respectively, when compared with 1.6 Å resolution structures or worse that are available in the PDB. Electron density is missing for the last IpaA-VBS3 residue in one heterodimer in the asymmetric unit and for the last two IpaA-VBS3 residues in the other heterodimer in the asymmetric unit, with temperature factors increased (from under 20 Å2 to over 50 Å2) for the last two modeled residues in either IpaA-VBS3 (supplemental Fig. S1).
The structure of the Vh1·IpaA-VBS3 complex closely resembles the crystal structures of Vh1 in complex with other IpaA-VBSs (Fig. 1B). Indeed, the Vh1·IpaA-VBS3 complex structure superimposes well with the other Vh1·IpaA-VBS structures (PDB entries 2gww, Vh1·IpaA-VBS1; and 2hsq, Vh1·IpaA-VBS2) with r.m.s.d. of 1.9 Å and 2.46 Å, respectively, for 241 Cα positions. Furthermore, the Vh1 N-terminal four-helix bundle subdomain (residues 8–126) that binds to the IpaA-VBSs superimposed with r.m.s.d. of 1.37 Å and 1.46 Å, respectively. The superposition of one subdomain alone is better due to the relative movement of the two four-helix bundle subdomains relative to each other (Fig. 1B).
Vh1-IpaA-VBS3 Interactions
The 1.6 Å resolution Vh1·IpaA-VBS3 crystal structure allows a clear definition of the vinculin-IpaA interaction. As first seen in the Vh1·talin-VBS3 structure (12), the majority of the Vh1-IpaA-VBS3 interactions are hydrophobic and contributed by eight residues: IpaA residues Ile-492 and Phe-493 interact with Vh1 residues Thr-8, Ile-12, Val-57, Gly-58, Leu-123, and Phe-126; IpaA residue Ala-495 with Vh1 Leu-54 and Val-57; IpaA residue Val-499 with Vh1 Ala-50, Val-51, Leu-54, and Ile-115; IpaA residue Leu-503 with Vh1 Val-16, Ile-20, Ser-112, and Ile-115; IpaA residues Leu-506 and Ile-507 with Vh1 Val-16, Ile-20, Leu-23, Leu-40, Pro-43, Val-44, and Leu-88; and IpaA residue Ile-510 with Vh1 Leu-108. These interactions contribute up to 72% (835 Å2) of the total interface (1,152 Å2) between IpaA-VBS3 and Vh1.
Three interactions between IpaA-VBS3 and Vh1 are mediated by ordered water molecules, and all of these involve Vh1 residues residing on α-helices α1 and α2 as follows: the main chain carbonyl of Vh1 Ile-12 and side chain of IpaA Asn-501; the main-chain carbonyls of Vh1 Pro-43 and IpaA Ser-502; and main chain carbonyls of Vh1 Ala-50 and IpaA Ala-495. Additionally, a few notable hydrogen bonding interactions are formed by IpaA residues Arg-489, Glu-490, Lys-497, Thr-500, Ser-504, and Ser-508, as described below.
Comparison of Vh1 Binding to IpaA-VBS3 Versus Other VBSs
Comparison of the IpaA-VBSs bound to Vh1 shows that a highly conserved 19-residue core α-helix directs interactions of IpaA with vinculin (Fig. 1B and supplemental Figs. S2 and S3). Previous mutagenesis studies (14) have shown that equivalent Tyr residues of IpaA-VBS1 (Tyr-613) and IpaA-VBS2 (Tyr-567) are important for high affinity binding to vinculin compared with the talin VBSs (14). IpaA-VBS3 has a Phe at this position that interacts with Vh1 similar to that seen for the other two IpaA-VBSs.
There are two conserved hydrogen bonds between the carbonyl group of Vh1 residue Ile-12 in all three IpaA-VBS complex crystal structures, with the IpaA-VBS side chain (Thr-500 in IpaA-VBS3, Ser-574 in IpaA-VBS2, and Thr-620 in IpaA-VBS1) and the side chain of Vh1 residue Gln-19 with the hydroxyl of the IpaA-VBS serine (504, 578, and 624, respectively). In contrast, equivalent residues are not strictly conserved in the VBSs of talin and are often substituted by Ala and polar amino acids, respectively. Additionally, Lys-497 of IpaA-VBS3 engages in an additional hydrogen bonding interaction with Ser-11 and Glu-14 of Vh1, and the equivalent residues have high temperature factors in the IpaA-VBS1 (∼120 Å2 for Lys-617, PDB entry 2gww) and IpaA-VBS2 (over 150 Å2 for Lys-571, PDB entry 2hsq) structures. Moreover, the aliphatic side chain of IpaA Lys-497 also engages in hydrophobic interactions with vinculin Ile-12. Importantly, IpaA-VBS3 residue Lys-497 is conserved among the three IpaA-VBSs (Lys-571 and Lys-617 in VBS2 and VBS1, respectively). The interaction between Ser-496 of IpaA-VBS3 and Vh1 is less favorable compared with the equivalent interaction in the other two VBSs of IpaA (Ala-570 and Ala-616) due to burial of the polar side chain. However, two electrostatic interactions are formed by residues outside of the 19-residue VBS core that further stabilize the binding of Vh1 with IpaA-VBS3 as follows: Arg-489 and Glu-490 interact with Glu-60 and Arg-56 of Vh1, respectively (supplemental Figs. S2 and S3).
IpaA Harbors Three VBSs
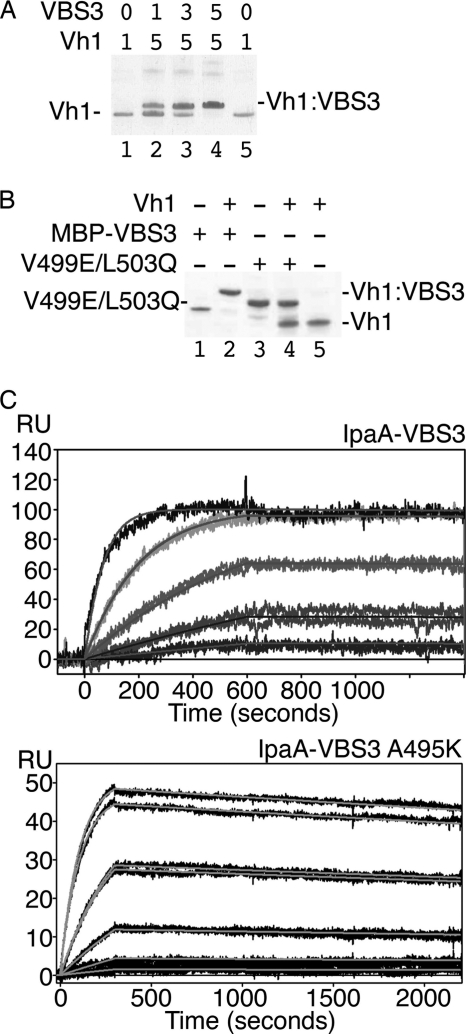
We used native gel shift mobility assays to test whether IpaA-VBS3 binds to the vinculin Vh1 domain in solution (Fig. 2A). Indeed, the Vh1·IpaA-VBS3 complex formed under physiological conditions and binding was saturated at equimolar ratios. Mutation of conserved small hydrophobic residues of VBSs to polar or charged residues disrupts Vh1 binding (25). Thus, we tested the binding of the IpaA-VBS3 V499E/L503Q mutant and found this double mutant did not bind to Vh1 (Fig. 2B).
FIGURE 2.
IpaA-VBS3 binds to vinculin in solution. A, binding of IpaA-VBS3 to vinculin. Native gel shift mobility assays were run on an 8–25% gradient gel. Vh1 (lanes 1 and 5) migrates faster than the Vh1·IpaA-VBS3 complex (lanes 2–4). Equimolar concentrations (indicated at the top of each lane) of IpaA-VBS3 and Vh1 (lane 4) result in saturation of Vh1 with IpaA-VBS3. VBS3, IpaA-VBS3. B, IpaA-VBS3 V499E/L503Q mutant fails to bind to vinculin. Native gel shift mobility assays were run on an 8–25% gradient gel. Migration of MBP-IpaA-VBS3 (lane 1) and Vh1 (lane 5) is retarded upon Vh1·MBP-IpaA-VBS3 complex formation (lane 2). However, the V499E/L503Q MBP-IpaA-VBS3 mutant fails to bind to Vh1 (lane 4; lane 3, MBP-IpaA-VBS3 V499E/L503Q mutant alone). MBP-VBS3, MBP-IpaA-VBS3; V499E/L503Q, MBP-IpaA-VBS3 V499E/L503Q mutant. C, binding affinity of IpaA-VBS3 for Vh1. Biacore surface plasmon resonance was used to measure binding affinity by amine coupling Vh1 on a carboxymethyl dextran-coated gold surface (Biacore CM-5 chip). Wild type IpaA-VBS3 at 100 nm, 33.3 nm, 11.1 nm, 3.7 nm, and 1.23 nm for the traces from top to bottom, respectively (top panel), and mutant IpaA-VBS3 A495K at 100 nm, 100 nm, 33.3 nm, 33.3 nm, 11.1 nm, 3.7 nm, and 1.23 nm for the traces from top to bottom, respectively (bottom panel), were injected in 3-fold dilution series over a reference or Vh1-immobilized surface, and the binding rate constants of IpaA were measured. The results obtained from duplicate injections of each sample over three Vh1 surfaces were normalized by subtracting the reference value. Representative sensograms are shown. The calculated Kd was 0.054 nm (wild type, top) and 1 nm (A495K mutant, bottom), respectively. RU, resonance units.
To determine the binding affinity of IpaA-VBS3 for Vh1, we used surface plasmon resonance (Fig. 2C). In agreement with the native gel shift mobility assays (Fig. 2A), the stoichiometry was 1:1. The binding affinity Kd of IpaA-VBS3 for Vh1 was 0.054 nm (Fig. 2C), which is significantly higher than that of IpaA-VBS2 (6.61 nm) (14) but similar to that of IpaA-VBS1 (0.11 nm) (14). The lower binding affinity of IpaA-VBS2 is accounted for Lys-569, as mutating this residue to an alanine (as seen in IpaA-VBS1; Ala-615) increased its affinity about 12-fold to 0.53 nm (14). We therefore tested if the equivalent alanine residue in IpaA-VBS3 (Ala-495) was also responsible for its tighter binding. Indeed, mutant IpaA-VBS3 (A495K) lowered the affinity about 20-fold to 1 nm. Thus, the higher affinity of IpaA-VBS3 is in part due to Ala-495 (Fig. 2C).
Finally, to determine the total number of VBSs present in IpaA, we determined the stoichiometry of the Vh1·IpaA complex (supplemental Figs. S4 and S5). Indeed, three molecules of Vh1 bound one molecule of IpaA. Thus, IpaA harbors three VBSs, and a single IpaA molecule can potentially bind to and activate three molecules of vinculin.
Deletion of All Three IpaA VBSs Impairs IpaA Function during Bacterial Invasion of Epithelial Cells
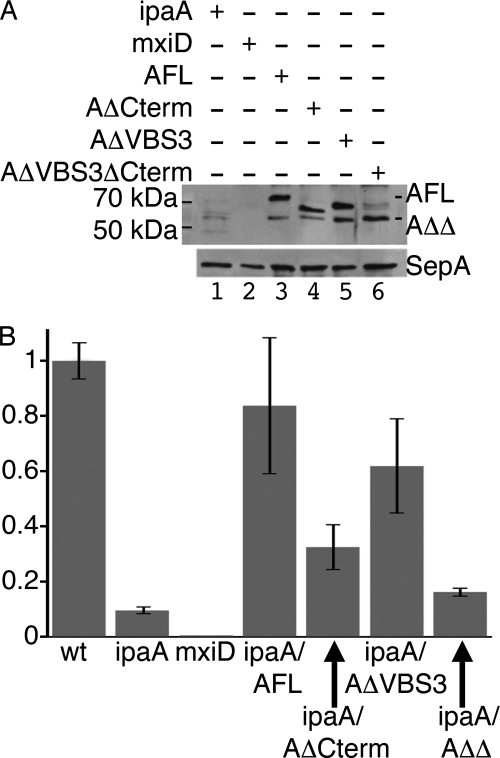
To test the role of the IpaA VBSs in the invasion of Shigella into epithelial cells, we generated the IpaA strains lacking residues 488–512 (AΔVBS3) or residues 488–512 and 550–633 corresponding to the deletion of all three VBSs (AΔVBS3ΔCterm). These mutants were subcloned into a bacterial expression vector and introduced into an ipaA Shigella mutant strain. As shown in Fig. 3A, type III secretion of AΔVBS3 and AΔVBS3ΔCterm was readily detected, at levels that were at least equivalent to that of the AΔCterm mutant lacking both IpaA-VBS1 and IpaA-VBS2 (residues 550–633). Gentamicin protection assays (Fig. 3B) indicated that expression of the AΔVBS3 mutant restored invasion functions to levels similar to those of full-length IpaA. As described previously (10), complementation with the AΔCterm containing a deletion of the previously identified IpaA-VBS1 and IpaA-VBS2 sites resulted in partial complement bacterial invasion (14). The AΔVBS3ΔCterm mutant, however, failed to complement the ipaA mutant Shigella (Fig. 3B). Thus, deletion of all three IpaA-VBSs severely impairs IpaA invasion functions.
FIGURE 3.
Deletion of all three of the VBSs of IpaA impairs IpaA function in bacterial invasion. A, secretion of the IpaA derivatives in Shigella. Shigella strains were grown to mid-exponential phase, and Congo Red was added at 100 μg/ml to induce type III secretion. Secreted proteins from the bacterial supernatants were precipitated and analyzed by anti-IpaA Western blotting (top panel) or with an antibody that recognizes SepA (40), a Shigella protein whose secretion does not depend on the T3SS (lower panel). ipaA, Shigella ipaA deletion mutant (lane 1); mxiD, isogenic Shigella strain deficient for type III secretion (lane 2); AFL, ipaA deletion mutant strain complemented with full-length IpaA (lane 3) or IpaA derivatives containing deletions of residues 550–633 (AΔCterm, lane 4), 488–512 (AΔVBS3, lane 5), or 488–512 and 550–633 (AΔVBS3ΔCterm, AΔΔ, lane 6). AFL and AΔΔ bands are indicated, with AΔCterm bands below AFL and AΔVBS3 above AΔΔ (not indicated for clarity). B, invasion efficiencies of Shigella ipaA deletion mutant complemented with the indicated IpaA derivatives. HeLa cells were challenged with bacterial strains, and bacterial invasion was determined using the gentamicin protection assay. The results are expressed as the average of the percentage of invasion relative to wild type Shigella of three independent experiments performed in triplicate. ipaA, Shigella ipaA deletion mutant; mxiD, isogenic Shigella strain deficient for type III secretion; ipaA/AFL, ipaA mutant strain complemented with full-length IpaA or IpaA derivatives containing deletions of residues 550–633 (ipaA/AΔCterm), 488–512 (ipaA/AΔVBS3), or 488–512 and 550–633 (ipaA/AΔΔ where AΔΔ is AΔVBS3ΔCterm abbreviated).
Activation of Vinculin by IpaA-VBS3
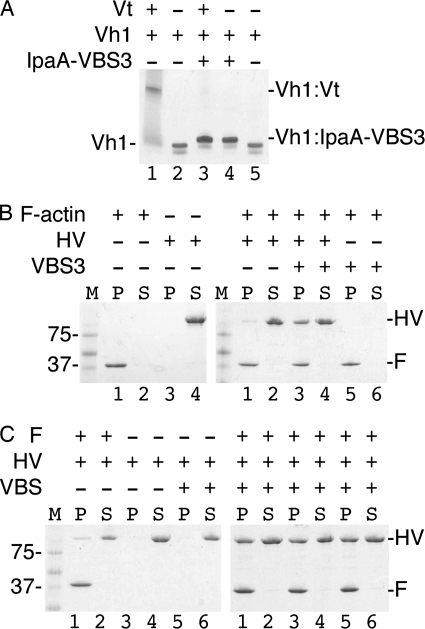
The similar binding mode of IpaA-VBS3 with the other IpaA-VBSs suggested that IpaA-VBS3 can activate vinculin, which requires severing of the intramolecular head-tail (Vh1-Vt) interaction that clamps vinculin in its inactive conformation (7). To test this, we incubated IpaA-VBS3 with the pre-formed Vh1·Vt complexes. The Vt domain was efficiently displaced by IpaA-VBS3 (Fig. 4A). Thus, IpaA-VBS3, like IpaA-VBS1 and IpaA-VBS2, is sufficient to trigger vinculin activation.
FIGURE 4.
IpaA-VBS3 is sufficient to trigger activation of vinculin. A, IpaA-VBS3 is sufficient to sever the vinculin head·tail complex that clamps vinculin in its inactive conformation. Native gel mobility shift assays are shown. IpaA-VBS3 displaces the Vt domain from pre-formed Vh1·Vt complexes (lane 1) to form Vh1·IpaA-VBS3 complexes (lanes 3 and 4), which migrate slower compared with unbound Vh1 (lanes 2 and 5). Free Vt does not migrate into the gel due to its basic pI. B, actin co-sedimentation experiments. M, molecular weight markers; P, pellet; S, supernatant; HV, human vinculin; F, F-actin. Left gel, F-actin pellets (lanes 1 and 2) and full-length vinculin remain in the supernatant in the absence of IpaA and F-actin (lanes 3 and 4). Right gel, native unbound full-length vinculin remains mainly in the supernatant in the presence of F-actin (i.e. in the absence of IpaA; lanes 1 and 2), but vinculin co-sediments with F-actin in the presence of IpaA-VBS3 (lanes 3 and 4). The behavior of F-actin is not affected by IpaA-VBS3 alone (lanes 5 and 6). Thus, IpaA-VBS3 is sufficient to activate the latent F-actin binding functions of vinculin. C, activation efficiency of vinculin by the VBSs of IpaA were compared using F-actin-vinculin co-sedimentation assays. M, molecular weight marker; P, pellet; S, supernatant; HV, human vinculin; F, F-actin. Left gel, vinculin remains in the supernatant in the absence of IpaA-VBS and F-actin (lanes 1–4) or in the presence of a mixture of IpaA-VBSs (1:2 molar ratio of each vinculin:IpaA-VBS) without F-actin (lanes 5 and 6). Right gel, vinculin co-sediments with F-actin if it is pre-bound by IpaA-VBS3 (lanes 5 and 6), and this is comparable with vinculin activated by IpaA-VBS1 (lanes 1 and 2) or IpaA-VBS2 (lanes 3 and 4).
A hallmark of activated vinculin is its ability to bind to F-actin, which is mediated by the Vt domain (7). Thus, we tested whether IpaA-VBS3 was sufficient to activate the latent F-actin binding functions of full-length human vinculin by performing F-actin pulldown assays. Notably, vinculin that was pre-activated by incubating with IpaA-VBS3 readily bound to F-actin (Fig. 4B). In contrast, only minute amounts of native vinculin bound to F-actin. Finally, the extent of vinculin activation by IpaA-VBS3 as measured by F-actin binding was similar to that triggered by the binding of IpaA-VBS1 or IpaA-VBS2 (Fig. 4C). Thus, IpaA-VBS3, like IpaA-VBS1 and IpaA-VBS2, can trigger the latent F-actin binding functions of vinculin.
DISCUSSION
Our studies revealed that the IpaA invasin of Shigella harbors three tandem high affinity binding sites that bind to and activate vinculin. Specifically, our biochemical and structural studies demonstrated that a third VBS of this invasin is located N-terminal to the two C-terminal VBSs, and they demonstrated that all three VBSs of IpaA can function in a redundant manner to sever the head-tail interaction that normally locks vinculin in its inactive conformation and that all three IpaA-VBSs are capable of activating the latent F-actin binding functions of full-length vinculin, a hallmark of activated vinculin at focal adhesions (7). Finally, our 1.6 Å crystal structure of IpaA-VBS3 in complex with vinculin, by far the highest resolution structure of any vinculin·VBS complex to date, with the resolution for previously determined Vh1·IpaA complexes ranging from 2.7 Å to 4 Å, revealed that this VBS also functions in an analogous manner to the two other VBSs of IpaA, by acting as a talin-VBS mimic that avidly inserts into the N-terminal four-helix bundle subdomain of Vh1 to generate an entirely new helix bundle structure.
Vinculin is required for the efficient entry of Shigella into the host cell (39). The fact that IpaA harbors three likely functionally redundant VBSs explains the paradox that an IpaA C-terminal deletion mutant that lacks IpaA-VBS1 and IpaA-VBS2 is competent to rescue an ipaA deletion mutant for Shigella invasion (14). Here, we posit that IpaA-VBS3 provides these functions, as it can bind to and activate vinculin at sites of pathogen entry. Accordingly, deletion of all three VBSs in IpaA compromised bacterial invasion, similar to those observed in an ipaA-deficient Shigella strain.
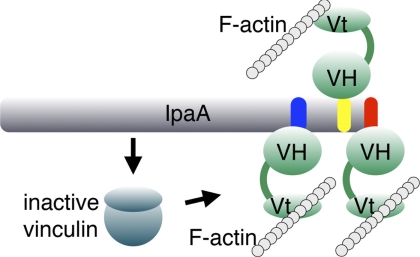
Finally, it is quite striking that the three high affinity VBSs of IpaA appear in tandem in the C-terminal tail of IpaA. Tandem repeats of the IpaA VBSs are somewhat comparable with those seen in talin, where its 11 VBSs bind vinculin upon force-induced activation of the talin rod domain (26). However, unlike talin-VBSs, the IpaA-VBSs are readily available for binding without activation (supplemental Fig. S4). Indeed, IpaA-VBS1 (residues 611–633), IpaA-VBS2 (residues 565–587), and IpaA-VBS3 (residues 488–512) are separated by linkers of only 24 or 53 residues, respectively (Fig. 5). The high local concentration of these VBSs would also ensure tight binding to vinculin. Indeed, our binding studies suggest that IpaA can simultaneously bind up to three Vh1 molecules, a scenario that would stabilize the focal adhesion-like complexes seen at pathogen entry sites (Fig. 5) (44).
FIGURE 5.
Vinculin activation by IpaA. IpaA has three VBSs that can activate vinculin by the binding of any one of its three VBSs (IpaA-VBS1, red; IpaA-VBS2, yellow; and IpaA-VBS3, blue) to the vinculin head domain (VH) by helix bundle conversion, which severs vinculin head-tail interactions (indicated by the change in color). Activated vinculin can then bind to F-actin through Vt-mediated interactions. The three VBSs of IpaA serve to amplify the response and/or promote simultaneous binding of IpaA to several vinculin molecules, which would promote actin cross-linking at sites of Shigella entry.
Supplementary Material
Acknowledgments
We are indebted to John Cleveland (Scripps, Florida) for discussions and critical review of the manuscript and Gerard Bricogne (Global Phasing Ltd.) for significant help with crystallography. We thank Philippe Bois (Scripps, Florida) for helpful discussions, Rebecca Rich and David Myszka (University of Utah) for surface plasmon resonance, and Zhen Wu and Philippe Bois (Scripps Florida) for sequencing. We are grateful to the staff at the Advanced Photon Source, SER-CAT, for synchrotron support.
This work was supported, in whole or in part, by National Institutes of Health grants (to T. I.). This work was also supported by start-up funds provided to Scripps Florida from the State of Florida (to T. I.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
The atomic coordinates and structure factors (code 3rf3) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- T3SS
- type 3 secretion system
- NCS
- noncrystallographic symmetry
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean squares deviation
- VBS
- vinculin binding site
- Vh1
- N-terminal seven-helix bundle head domain of vinculin
- Vt
- C-terminal five-helix bundle tail domain of vinculin.
REFERENCES
- 1. Ziegler W. H., Liddington R. C., Critchley D. R. (2006) Trends Cell Biol. 16, 453–460 [DOI] [PubMed] [Google Scholar]
- 2. Zamir E., Geiger B. (2001) J. Cell Sci. 114, 3583–3590 [DOI] [PubMed] [Google Scholar]
- 3. Critchley D. R. (2004) Biochem. Soc. Trans. 32, 831–836 [DOI] [PubMed] [Google Scholar]
- 4. Borgon R. A., Vonrhein C., Bricogne G., Bois P. R., Izard T. (2004) Structure 12, 1189–1197 [DOI] [PubMed] [Google Scholar]
- 5. Rangarajan E. S., Lee J. H., Yogesha S. D., Izard T. (2010) PLoS One 5, e10679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Izard T., Vonrhein C. (2004) J. Biol. Chem. 279, 27667–27678 [DOI] [PubMed] [Google Scholar]
- 7. Johnson R. P., Craig S. W. (1995) Nature 373, 261–264 [DOI] [PubMed] [Google Scholar]
- 8. Lee S. W., Wulfkuhle J. D., Otto J. J. (1992) J. Biol. Chem. 267, 16355–16358 [PubMed] [Google Scholar]
- 9. Kroemker M., Rüdiger A. H., Jockusch B. M., Rüdiger M. (1994) FEBS Lett. 355, 259–262 [DOI] [PubMed] [Google Scholar]
- 10. Hüttelmaier S., Illenberger S., Grosheva I., Rüdiger M., Singer R. H., Jockusch B. M. (2001) J. Cell Biol. 155, 775–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bois P. R., O'Hara B. P., Nietlispach D., Kirkpatrick J., Izard T. (2006) J. Biol. Chem. 281, 7228–7236 [DOI] [PubMed] [Google Scholar]
- 12. Izard T., Evans G., Borgon R. A., Rush C. L., Bricogne G., Bois P. R. (2004) Nature 427, 171–175 [DOI] [PubMed] [Google Scholar]
- 13. Bois P. R., Borgon R. A., Vonrhein C., Izard T. (2005) Mol. Cell. Biol. 25, 6112–6122 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 14. Izard T., Tran Van Nhieu G., Bois P. R. (2006) J. Cell Biol. 175, 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nhieu G. T., Izard T. (2007) EMBO J. 26, 4588–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Papagrigoriou E., Gingras A. R., Barsukov I. L., Bate N., Fillingham I. J., Patel B., Frank R., Ziegler W. H., Roberts G. C., Critchley D. R., Emsley J. (2004) EMBO J. 23, 2942–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Niyogi S. K. (2005) J. Microbiol. 43, 133–143 [PubMed] [Google Scholar]
- 18. Kotloff K. L., Winickoff J. P., Ivanoff B., Clemens J. D., Swerdlow D. L., Sansonetti P. J., Adak G. K., Levine M. M. (1999) Bull. World Health Organ. 77, 651–666 [PMC free article] [PubMed] [Google Scholar]
- 19. Sansonetti P. J., Egile C. (1998) Antonie Van Leeuwenhoek 74, 191–197 [DOI] [PubMed] [Google Scholar]
- 20. Bourdet-Sicard R., Rüdiger M., Jockusch B. M., Gounon P., Sansonetti P. J., Nhieu G. T. (1999) EMBO J. 18, 5853–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Niebuhr K., Jouihri N., Allaoui A., Gounon P., Sansonetti P. J., Parsot C. (2000) Mol. Microbiol. 38, 8–19 [DOI] [PubMed] [Google Scholar]
- 22. Alto N. M., Shao F., Lazar C. S., Brost R. L., Chua G., Mattoo S., McMahon S. A., Ghosh P., Hughes T. R., Boone C., Dixon J. E. (2006) Cell 124, 133–145 [DOI] [PubMed] [Google Scholar]
- 23. Mounier J., Popoff M. R., Enninga J., Frame M. C., Sansonetti P. J., Van Nhieu G. T. (2009) PLoS Pathog. 5, e1000271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramarao N., Le Clainche C., Izard T., Bourdet-Sicard R., Ageron E., Sansonetti P. J., Carlier M. F., Tran Van Nhieu G. (2007) FEBS Lett. 581, 853–857 [DOI] [PubMed] [Google Scholar]
- 25. Gingras A. R., Ziegler W. H., Frank R., Barsukov I. L., Roberts G. C., Critchley D. R., Emsley J. (2005) J. Biol. Chem. 280, 37217–37224 [DOI] [PubMed] [Google Scholar]
- 26. del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P. (2009) Science 323, 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ménard R., Sansonetti P. J., Parsot C. (1993) J. Bacteriol. 175, 5899–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Picard V., Ersdal-Badju E., Lu A., Bock S. C. (1994) Nucleic Acids Res. 22, 2587–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 30. Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., Bricogne G. (2011) Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 32. Long F., Vagin A. A., Young P., Murshudov G. N. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Collaborative Computational Project No 4. (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 15299374 [Google Scholar]
- 34. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek P., Roversi P., Sharff A., Smart O., Vonrhein C., Womack T. (2010) BUSTER, version 2.9, Global Phasing Ltd., Cambridge, United Kingdom [Google Scholar]
- 35. Smart O. S., Brandl C., Flensburg P., Keller W., Paciorek C., Vonrhein C., Womack T., Bricogne G. (2008) in Abstracts of Annual Meeting American Crystallographic Association, Abstract no. TP139, American Crystallographic Association, Buffalo, NY [Google Scholar]
- 36. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang W., Malcolm B. A. (1999) BioTechniques 26, 680–682 [DOI] [PubMed] [Google Scholar]
- 39. Tran Van Nhieu G., Ben-Ze'ev A., Sansonetti P. J. (1997) EMBO J. 16, 2717–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tran Van Nhieu G., Caron E., Hall A., Sansonetti P. J. (1999) EMBO J. 18, 3249–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 42. Fillingham I., Gingras A. R., Papagrigoriou E., Patel B., Emsley J., Critchley D. R., Roberts G. C., Barsukov I. L. (2005) Structure 13, 65–74 [DOI] [PubMed] [Google Scholar]
- 43. Gingras A. R., Vogel K. P., Steinhoff H. J., Ziegler W. H., Patel B., Emsley J., Critchley D. R., Roberts G. C., Barsukov I. L. (2006) Biochemistry 45, 1805–1817 [DOI] [PubMed] [Google Scholar]
- 44. Nhieu G. T., Sansonetti P. J. (1999) Curr. Opin. Microbiol. 2, 51–55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.