Abstract
Ciona intestinalis voltage-sensing phosphatase (Ci-VSP) has a transmembrane voltage sensor domain and a cytoplasmic region sharing similarity to the phosphatase and tensin homolog (PTEN). It dephosphorylates phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate upon membrane depolarization. The cytoplasmic region is composed of a phosphatase domain and a putative membrane interaction domain, C2. Here we determined the crystal structures of the Ci-VSP cytoplasmic region in three distinct constructs, wild-type (248–576), wild-type (236–576), and G365A mutant (248–576). The crystal structure of WT-236 and G365A-248 had the disulfide bond between the catalytic residue Cys-363 and the adjacent residue Cys-310. On the other hand, the disulfide bond was not present in the crystal structure of WT-248. These suggest the possibility that Ci-VSP is regulated by reactive oxygen species as found in PTEN. These structures also revealed that the conformation of the TI loop in the active site of the Ci-VSP cytoplasmic region was distinct from the corresponding region of PTEN; Ci-VSP has glutamic acid (Glu-411) in the TI loop, orienting toward the center of active site pocket. Mutation of Glu-411 led to acquirement of increased activity toward phosphatidylinositol 3,5-bisphosphate, suggesting that this site is required for determining substrate specificity. Our results provide the basic information of the enzymatic mechanism of Ci-VSP.
Keywords: Enzyme Mechanisms, Enzyme Structure, Phosphatase, Phosphatidylinositol, Protein Structure, X-ray Crystallography, Voltage Sensor
Introduction
The voltage-sensing phosphatase (VSP)3 was discovered through the survey of the genome of the ascidian, Ciona intestinalis, as a hybrid protein that has a voltage sensor domain (VSD) consisting of four α-helices (S1–S4) for voltage sensing and a cytoplasmic region encoding a phosphatidylinositol phosphatase domain (1). The VSP gene is conserved from sea urchins to humans where it is expressed in the testis (2). In the recent report, C. intestinalis VSP (Ci-VSP) is expressed in the cells of the stomach, intestine, and blood of juveniles detected by whole mount in situ hybridization (3). The VSD of VSP bears homology to the VSD of voltage-gated ion channels and a recently identified voltage-gated proton channel protein that contains only the VSD without pore domain, VSOP or Hv1 (4, 5). The Ci-VSP cytoplasmic region, which consists of a phosphatase domain (PD) and a C2 domain, shares high sequence similarity with the phosphatase and tensin homolog (PTEN) (1, 6). The amino acid sequence of the Ci-VSP cytoplasmic region has similarity to that of PTEN with 36% identity. A unique feature of both invertebrate and vertebrate VSPs is that depolarization induces phosphoinositide phosphatase activity through coupling of VSD to the phosphatase region (7–9). The enzymatic activity of Ci-VSP increases in the range of membrane potentials, from −80 to 100 mV, correlating with the extent of voltage sensor movement (7–8). Ci-VSP most likely operates as a monomer as shown by a study of single molecule imaging in heterologous expression system (10). The linker region between the VSD and the cytoplasmic region has been shown to play a critical role in coupling of membrane potential and phosphatase activity (1). Conservation of the phosphoinositide binding motif (PBM) between Ci-VSP and PTEN motivated recent studies of mutation of this motif to show reduction or elimination of coupling (11, 12). Depletion of phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) diminishes the interaction between the VSD and the phosphatase (12). These findings suggest that coupling between the VSD and phosphatase requires constitutive binding of PtdIns(4,5)P2 to PBM (12). In this scenario, PtdIns(4,5)P2 is not only the substrate but also the regulator for Ci-VSP, and the requirement of PtdIns(4,5)P2 binding to PBM may confer negative feedback mechanism to Ci-VSP activities (12).
The substrate specificity of Ci-VSP is different from that of PTEN; Ci-VSP can dephosphorylate both phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3) and phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) (1, 13), whereas PTEN can dephosphorylate only PtdIns(3,4,5)P3 (14, 15). Ci-VSP is a depolarized-activated PtdIns phosphatase that converts PtdIns(4,5)P2 to PtdIns(4)P and PtdIns(3,4,5)P3 to PtdIns(3,4)P2 as shown by imaging of phosphoinositide-sensitive fluorescent proteins (16). Dephosphorylation of the 5′ site phosphate of PtdIns(4,5)P2 has been proven by in vitro measurements of phosphatase activities with site-specific radiolabeling of phosphate on phosphoinositides (13). This distinct substrate specificity of Ci-VSP from PTEN is partly due to the presence of a Gly-365 in the active site. An alanine residue occupies this position in the PTEN active site. Mutation of Gly-365 to alanine remarkably attenuates phosphatase activity toward PtdIns(4,5)P2 (13). However, detailed molecular mechanisms of how the VSP dephosphorylates both PtdIns(3,4,5)P3 and PtdIns(4,5)P2 are not clear.
In this study we determined the crystal structures of three forms of the cytoplasmic regions of Ci-VSP, residues 248–576 (WT-248), residues 236–576 (WT-236), and residues 248–576 with a G365A mutation (G365A-248). Unlike PTEN, the TI loop, which formed the active pocket with the P loop and WPD loop, was oriented toward the center of active site pocket. This resulted in the side chain of Glu-411 protruding into the active site pocket, reducing the pocket size relative to that of the PTEN pocket. To elucidate the role of Glu-411 in the TI loop, the malachite green phosphatase assay was carried out. The degree of the activity of Glu-411 mutant toward PtdIns(3,5)P2 was higher than that of wild type. The structures of WT-236 and G365A-248 had a disulfide bond between the catalytic residue Cys-363 and the proximate Cys-310. On the other hand, this disulfide bond was not found in the structure of WT-248. The activity of the phosphatase assay toward PtdIns(3,4,5)P3 and PtdIns(4,5)P2 was significantly reduced in the presence of H2O2. These results suggest the possibility of the redox regulation for Ci-VSP. In addition, structural features of the cytoplasmic region of Ci-VSP provide insights into mechanisms of coupling from the VSD to the phosphatase activities.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
For crystallization experiments, recombinant proteins with His-thioredoxin tag were produced in Escherichia coli C43(DE3) or BL21-CodonPlus(DE3)-RIPL (Stratagene) cells from WT-248, WT-236, and G365A-248 genes in pET-32a(+). The cells were grown to optical density of 0.6 at 310 K before inducing protein by the addition of 0.4 mm isopropyl β-d-1-thiogalactopyranoside. Cells were pelleted and resuspended in lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5% (v/v) glycerol, 5 mm β-mercaptoethanol, 40 μg/ml lysozyme) and lysed by sonication, and the lysate was centrifuged at 38,000 × g for 30 min at 277 K. His-thioredoxin-tagged protein was purified on a nickel-nitrilotriacetic acid column (Qiagen). The peak fraction containing the fusion protein was digested with protease overnight by dialysis. The digestion was loaded onto HiTrap SP HP column (GE Healthcare) and Superdex75 column (GE Healthcare) in a buffer containing 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 5 mm DTT.
Crystallization and Data Collection
For the crystallization of WT-248, 1 μl of protein solution (14 mg/ml) was mixed with 0.8 μl of reservoir solution containing 0.1 m Tris-HCl, pH 7.0, 2.1 m ammonium sulfate, and 0.2 μl of 0.5 m NaF solution, and the drop solution was equilibrated against the reservoir solution at 293 K by the hanging drop vapor diffusion method. The proteins WT-236 and G365A-248 were also crystallized under similar conditions. The droplet consisted of 1.0 μl of protein solution, 0.8 μl of reservoir solution (0.1 m Tris-HCl, pH 7.0, 2.1 m ammonium sulfate), 0.2 μl of 0.5 m NaF solution, and 0.2 μl of 1.0 m sodium malonate, pH 7.0, solution. The crystals were cryo-protected with reservoir solution with 25% (v/v) glycerol, and they were flash-cooled in liquid nitrogen. X-ray data were collected at beamline BL44XU of SPring-8 (Hyogo, Japan) equipped with an Image Plate detector DIP6040 (Bruker AXS) at 100 K under a nitrogen gas stream. All of the data were processed using the program HKL2000 (17). All crystals belonged to the P21212 space group, with three molecules in the asymmetric unit. Data collection statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statics
| WT-248 | WT-236 | G365A-248 | |
|---|---|---|---|
| Data collection | |||
| Space group | P21212 | P21212 | P21212 |
| Cell dimensions a, b, c (Å) | 130.9, 176.5, 50.8 | 131.1, 176.5, 51.0 | 131.2, 176.5, 50.9 |
| Resolution (Å) | 50.0-2.80 (2.90-2.80)a | 50.0-2.00 (2.07-2.00) | 50.0-2.40 (2.49-2.40) |
| Rmerge | 0.118 (0.439) | 0.086 (0.494) | 0.113 (0.490) |
| 〈I〉/〈σ(I)〉 | 13.4 (3.8) | 19.6 (2.9) | 18.5 (3.6) |
| Completeness (%) | 98.5 (98.2) | 99.6 (99.6) | 100.0 (99.8) |
| Redundancy | 4.5 (4.5) | 4.8 (4.6) | 6.2 (5.7) |
| Refinement | |||
| Resolution (Å) | 44.13-2.80 | 45.88-2.00 | 48.91-2.40 |
| No. reflections | 30,315 | 81,627 | 47,850 |
| Rwork/Rfree | 0.208/0.253 | 0.179/0.223 | 0.194/0.255 |
| No. atoms | |||
| Protein | 7,397 | 7,490 | 7,407 |
| Ligand/ion | 39/1 | 56/- | 50/- |
| Water | 150 | 535 | 250 |
| B-factors (Å2) | |||
| Protein | 26.75 | 27.44 | 31.70 |
| Ligand/ion | 52.17/11.78 | 48.26/- | 51.83/- |
| Water | 19.1 | 37.08 | 30.83 |
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.019 | 0.025 | 0.022 |
| Bond angles (°) | 1.644 | 1.935 | 1.848 |
| Ramachandran plot | |||
| Favored/allowed/outlier (%) | 94.8/4.9/0.3 | 97.1/2.4/0.5 | 96.6/3.1/0.3 |
a Values in parentheses are for the highest-resolution shell.
Structure Determination of Three Protein Constructs of the Ci-VSP Cytoplasmic Region
The structure of Ci-VSP cytoplasmic region was determined by molecular replacement. The molecular replacement was performed by the program BALBES (18) using the human PTEN (PDB ID 1D5R) as a search model. This search model was selected by the program BALBES (18). The phase was improved by density modification that included non-crystallographic symmetry averaging, as carried out by the program DM (19) from CCP4i in the CCP4 program suite (20). The models with three versions of the Ci-VSP cytoplasmic region in asymmetric unit were manually built by the program Coot (21) and refined using Refmac5 (22) and phenix.refine (23). Stereochemical properties were evaluated by MolProbity (24). Refinement statistics are summarized in Table 1.
In Vitro Phosphatase Assay
For the phosphatase assay, DNA fragments encoding the Ci-VSP cytoplasmic regions (248–576) of E411A, E411Q, and E411T were subcloned from pET-32a(+). His-thioredoxin fusion proteins of each construct were expressed in E. coli C41 (DE3) and purified in the same way as in WT-248. Di-C16 phosphoinositides and 1-palmitoyl-2-oleoyl-phosphatidylserine (10 nmol) were dried together under reduced pressure and resuspended via sonication in 9 μl of assay buffer A (100 mm Tris-HCl, pH 8.0, 10 mm DTT, 0.04% (v/v) Nonidet P-40). In the redox state phosphatase assay, 1 and 10 mm of H2O2 was added into the assay buffer B (100 mm Tris-HCl, pH 8.0, 0.04% (v/v) Nonidet P-40). As a negative control, water without H2O2 was added. Each purified protein (1 μg/tube) was added to the substrate solution and incubated at 23 °C. Reaction was terminated by the addition of N-ethylmaleimide and centrifugation. BIOMOL GREENTM reagent (BIOMOL Research Laboratories, Inc.) was added to the supernatants, and A620 was measured. To determine the kinetics of wild-type and Glu-411 mutants toward PtdIns(3,4,5)P3 and PtdIns(4,5)P2, the cytoplasmic region (248–576) of these proteins was expressed and purified in the same way as for WT-248. Initial rates, V0, of Ci-VSP-catalyzed dephosphorylation of PtdIns(3,4,5)P3 and PtdIns(4,5)P2 were determined from reactions with various concentrations of PtdIns(3,4,5)P3 and PtdIns(4,5)P2. Data were fit by the equation V0 = Vmax[S]/(Km + [S]), where [S] is concentration of substrate, PtdIns(3,4,5)P3, or PtdIns(4,5)P2.
RESULTS
Atomic Structures of the Ci-VSP Cytoplasmic Region
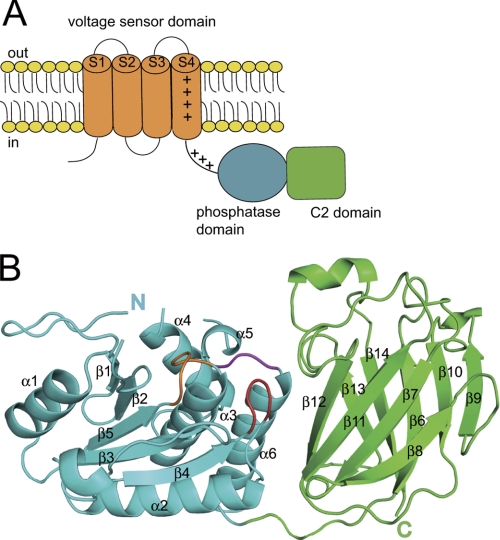
Ci-VSP contains an N-terminal VSD followed by a cytoplasmic region that has a phosphoinositide phosphatase domain preceding a C2 domain (Fig. 1A). We determined the crystal structure of the Ci-VSP cytoplasmic region in three distinct forms; the polypeptide starting from just after the S4 helix region (216–236) of the VSD (WT-236), the polypeptide starting 12 residues downstream of the S4 helix region of the VSD (WT-248), and the polypeptide starting from the 248th residue with mutation of G365A, which is known to eliminate phosphatase activity toward PtdIns(4,5)P2 (G365A-248) (13). All three crystal forms had three molecules in the asymmetric unit (MolA residues 257–279, 287–399, and 404–572; MolB residues 254–279, 287–399, and 405–572; MolC residues 253–279, 287–399, and 405–572 for WT-248; MolA residues 257–279, 287–399, and 404–571; MolB residues 254–279, 286–399, and 405–572; MolC residues 253–280, 287–399, and 404–572 for WT-236; MolA residues 254–279, 287–399, and 405–571; MolB residues 254–279, 287–399, and 406–572; MolC residues 254–279, 287–399, and 404–572 for G365A-248). Each structure in the asymmetric unit was almost the same (r.m.s. deviations of Cα atoms of each model were in the range of 0.094–0.472 Å). The final model contains the N-terminal PD (residues 248–432) and the C-terminal C2 domain (residues 433–576) (Fig. 1B). The crystal structure could not be built for the whole PBM region because this PBM has a high B-factor and is almost disordered (supplemental Fig. S1A).
FIGURE 1.
The structural feature of Ci-VSP. A, a schematic structure of Ci-VSP is shown. Ci-VSP has four transmembrane segments that form the VSD (orange). VSD is connected with the cytoplasmic region by a 16-amino acid linker. The cytoplasmic region is composed of the PD (cyan) and the C2 domain (green). B, the overall structure of Ci-VSP cytoplasmic region is represented in a ribbon diagram. The PD is colored in cyan, and the C2 domain is in green. In the PD, the P loop containing HCKGGK motif is colored in orange, the TI loop in magenta, and the WPD loop in red.
The PD consists of a central four β-strands surrounded by six α-helices as in PTEN (25). The PD contains the protein-tyrosine phosphatases (PTP) signature motif, HCXXGXXR (X is any residue), and this motif makes a loop (P loop, residues 362–369). As in human PTEN (25), the P loop forms an active site pocket with two other elements, WPD loop (residues 330–333) and TI loop (residues 410–412). A previous study showed that the G365A mutant cannot dephosphorylate PtdIns(4,5)P2 both by in vitro phosphatase assay and electrophysiological experiments (13). However, the overall structure of the G365A mutant was similar to that of WT Ci-VSP (r.m.s. deviations of Cα atoms of the each molecules were 0.32–0.33 Å) (supplemental Fig. S2). Any large structural differences among WT-248, WT-236, and G365A-248 were not found.
Structure of C2 Domain
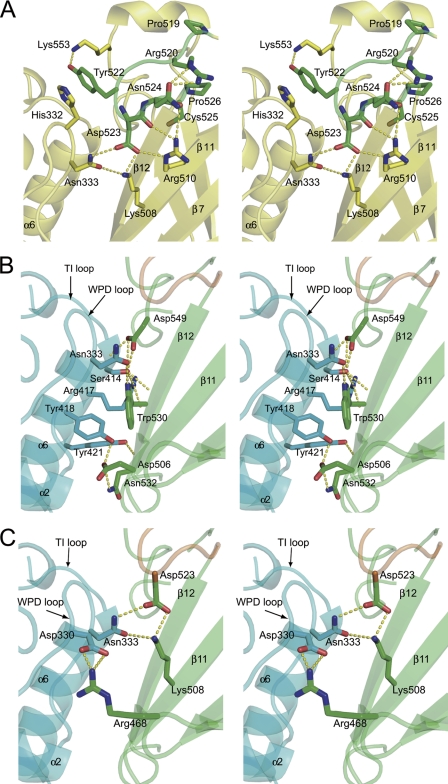
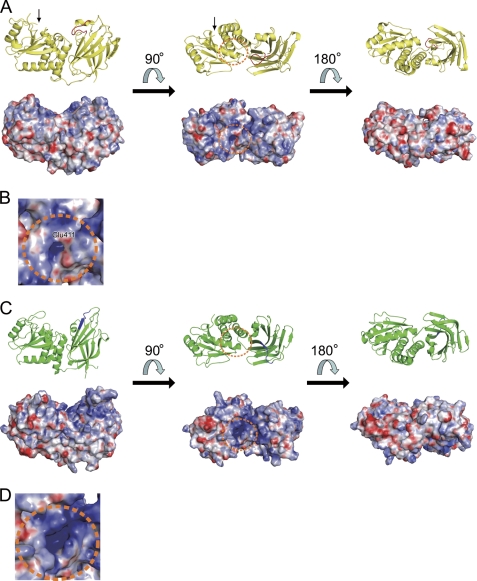
The r.m.s. deviation of Cα atoms of the PD and the C2 domain between Ci-VSP and PTEN were 1.13 and 1.58 Å, respectively. The C2 domain of Ci-VSP consists of nine β-strands and loops, and the overall structure of the C2 domain of Ci-VSP is not remarkably different from that of PTEN. However, VSP showed a different structure at a loop region corresponding to “CBR3 loop” in PTEN that adopts a structure similar to that of phospholipase Cδ subunit and plays a role in association of the C2 domain with membrane (25). The CBR3 loop of PTEN does not bind to calcium, unlike the phospholipase Cδ subunit loop. The CBR3 loop of PTEN is exposed to the solvent region and extends perpendicularly away from the membrane-facing site of the C2 domain (25). The CBR3 loop of PTEN also contains solvent-exposed hydrophobic residues, Met-264 and Leu-265, which are proposed to insert into the lipid bilayer (26, 27). In contrast, the CBR3 loop (residue 519–526) of Ci-VSP does not extend perpendicular to membrane but instead is oriented toward the PD (Fig. 2). It has only one positively charge residue, Arg-520, in contrast to four in PTEN. Therefore, the surface electrostatic potential of the CBR3 loop of Ci-VSP is less positive than that of PTEN (Fig. 3, A–D).
FIGURE 2.
The interaction between the PD and the C2 domain. A, shown is a stereo view of the Ci-VSP CBR3 loop. The yellow dash lines show hydrogen bond contacts in Ci-VSP. The carbon atoms of amino acid residues in the CBR3 loop are represented in green. B, hydrogen bonds that are conserved between Ci-VSP and PTEN are shown. C, hydrogen bonds between PD and the C2 domain that are unique to Ci-VSP, but not conserved in PTEN, are shown. The PD is shown in cyan, the C2 domain is in green, and the CBR3 loop is in orange.
FIGURE 3.
Surface electron potential map of Ci-VSP and PTEN. A, a ribbon diagram (above) and surface electrostatic potential (below) of Ci-VSP are represented in the same orientation. The CBR3 loop of Ci-VSP in ribbon diagram is colored in red. The phosphatase active site is shown in orange dot circle. The black arrow indicates the N terminus of the Ci-VSP cytoplasmic region. B, shown is a close-up view of the active site of the Ci-VSP cytoplasmic region. The color codes are the same as in A. C, a ribbon diagram (above) and surface electrostatic potential (below) of PTEN are represented in the same orientation. The corresponding region of the CBR3 loop of Ci-VSP in ribbon diagram is colored in blue. The phosphatase active site is shown in orange dot circle. D, shown is a close-up view of the active site of PTEN. The color codes are the same as in C. Surface electrostatic potential was calculated by APBS (42).
Moreover, the CBR3 loop of Ci-VSP makes contact with the WPD loop of the PD with residues forming hydrogen bonds. Asp-523 forms hydrogen bond with Asp-333 in the WPD loop and Lys-508 and Arg-510 in the C2 domain (Fig. 2A). Tyr-522 is located near the phosphatase active site and forms a hydrogen bond with Lys-553 and may potentially stack with His-332 from the WPD loop, which is about 3.5 Å away (Fig. 2A). Residues Arg-520, Asn-524, and Pro-526 form a hydrogen bond network within the C2 domain. Asn-333 interacts with Lys-508 and Asp-523 (Fig. 2A), and the Asp-330 interacts with Arg-468 (Fig. 2C). Hydrogen bond networks involving Asp-523 of the CBR3 loop and Arg-468 (Fig. 2C) are not present in PTEN. On the other hand, profiles of hydrogen bonds in the domain interface distant from the CBR3 loop are identical between PTEN and Ci-VSP (Fig. 2B), except that interaction corresponding to the hydrogen bond between Tyr-188 and Asn-276 in PTEN (25), close to the linker between the PD and the C2 domain, is not found in Ci-VSP (not indicated in Fig. 2B). Thus, the C2 domain of Ci-VSP has a more intimate interaction with the WPD loop, mainly through the hydrogen bond network in the CBR3 loop, than in PTEN.
Structure of Phosphatase Domain
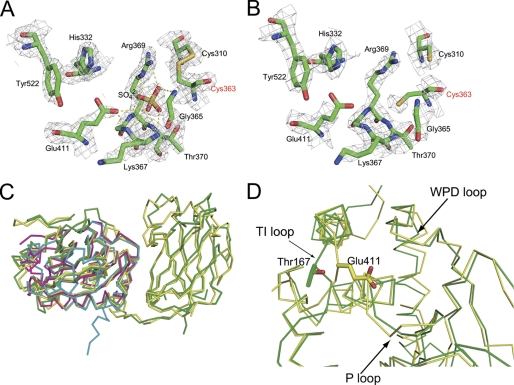
We found that the crystal structures of WT-236 and G365A-248 have a disulfide bond between the catalytic residue Cys-363 and Cys-310 in the phosphatase domain (Fig. 4A). This disulfide bond was not present in the structure of WT-248 in which Cys-363 remained reduced (Fig. 4B). The electron density for the thiol group of the catalytic residue, Cys-363, is well defined in the both oxidized and reduced conditions (Fig. 4, A and B). The distance between Cα of Cys-363 and Cys-310 in the oxidized form is 5.1 Å within the normal range of 4.4–6.8 Å as the Cα-Cα distance of disulfide bonds (28). The residue corresponding to Cys-310 is strictly conserved among VSP orthologs (supplemental Fig. S3). The active site pocket of the oxidized form had a strong tetrahedral-shaped electron density, which was assigned as a sulfate ion in the active site. The sulfate ion is derived from ammonium sulfate that was contained in crystallization mother liquor.
FIGURE 4.
The structural feature of the active site of the PD. A and B, the active site of the PD is shown for the oxidized form and the reduced form. All residues are shown as stick models. In the oxidized form, disulfide bond between Cys-363 and distal Cys-310 is shown. Hydrogen bonds formed by the active site sulfate ion are shown in a yellow dash line. The αA-weighted 2mFo − DFc electron density map of the active site of the PD is shown in gray mesh. The map is contoured at 1σ level. C, shown is superimposition of the cytoplasmic region with known structures of phosphatases, represented in an Cα trace. The structures of the Ci-VSP cytoplasmic region, PTEN (PDB ID 1D5R), VH1-like phosphatase Z (PDB ID 2IMG), and protein-tyrosine phosphatase 1B (PDB ID 2I6I) are shown in yellow, green, pink, and cyan, respectively. D, a close-up view of the active site of the Ci-VSP cytoplasmic region and PTEN represented in Cα trace is shown. The structure of Ci-VSP differs from that of PTEN in TI loop. Glu-411 of Ci-VSP and Thr-167 of human PTEN are shown in a stick model.
The r.m.s. deviation of Cα atoms of the PD of Ci-VSP and PTEN (25), protein-tyrosine phosphatase 1B (29), and VH1-like phosphatase Z (30), dual specific phosphatases (PDB IDs 1D5R, 2I6I, and 2IMG), were 1.13, 1.74, and 1.54 Å, respectively (Fig. 4C). The phosphatase active site pocket of Ci-VSP, which was composed of the side chains and backbone groups from the P loop, the WPD loop, and the TI loop, is similar to that of protein-tyrosine phosphatase 1B but is different from that of PTEN. The TI loop is positioned closer to the WPD loop, and the side chain of the Glu-411 is oriented toward the center of the active site pocket. This results in a smaller active site pocket for Ci-VSP as compared with that of PTEN (Fig. 3D). In addition, the Glu-411 residue of Ci-VSP contributes to making the active site more negative (Figs. 3A and 2D). PTEN has threonine at the site corresponding to Glu-411 of Ci-VSP. Comparison of the surface electrostatic potential between Ci-VSP and PTEN shows that the difference of Glu/Thr in the TI loop makes the active site of PTEN more positively charged than that of Ci-VSP (Figs. 3, C and D).
Phosphatase Assay of the Ci-VSP Cytoplasmic Region in Different Oxidation States
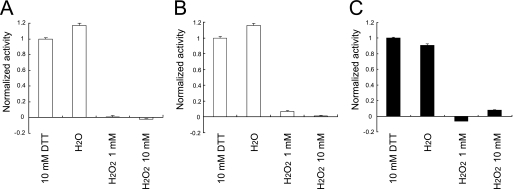
A disulfide bond between the catalytic residue Cys-363 and Cys-310 was present in the structures of WT-236 and G365A-248. This may be caused by the sensitivity of Ci-VSP for the oxidation. To elucidate the sensitivity to oxidation of Ci-VSP cytoplasmic region in vitro, the malachite green phosphatase assay for PtdIns(3,4,5)P3 and PtdIns(4,5)P2 was performed in the different oxidation state. In the malachite green phosphatase assay, WT and G365A mutant proteins had similar enzymatic activity toward PtdIns(3,4,5)P3 and PtdIns(4,5)P2 with 10 mm DTT. In contrast, the presence of 1 or 10 mm H2O2 caused a loss of the activity of WT and G365A mutant for PtdIns(3,4,5)P3 or PtdIns(4,5)P2, respectively (Fig. 5).
FIGURE 5.
Effect of oxidation on Ci-VSP phosphatase activity. A, shown is specific phosphatase activity of Ci-VSP WT toward PtdIns(3,4,5)P3 measured by the malachite green assay in the oxidized and reduced conditions. B, shown are phosphatase activities in various oxidation conditions of G365A toward PtdIns(3,4,5)P3. C, shown are phosphatase activities in various oxidation conditions of WT toward PtdIns(4,5)P2. Values are normalized based on the activity of the condition with 10 mm DTT. Experiments were carried out five times for each condition.
Phosphatase Assay of Wild-type and Glu-411 Mutants
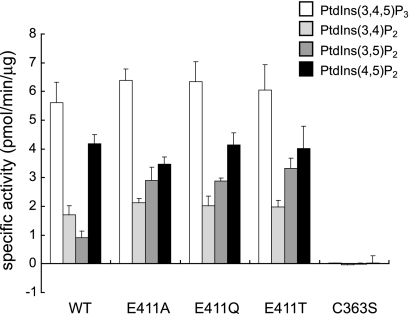
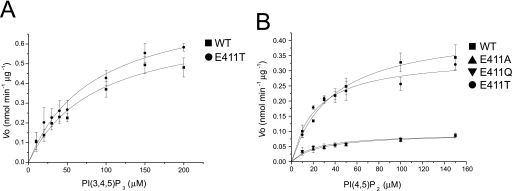
In the active site of the PD, Glu-411 made a smaller pocket size in Ci-VSP. To clarify the role of the residue of Glu-411 for the phosphoinositide activity, we made three mutants, E411T, E411Q, and E411A, and performed the phosphatase assay with PtdIns(3,4,5)P3, PtdIns(3,4)P2, PtdIns(3,5)P2, and PtdIns(4,5)P2 (Fig. 6). For PtdIns(3,4,5)P3 and PtdIns(4,5)P2, the enzymatic activity of WT was similar to that of Glu-411 mutants. The activity toward PtdIns(3,5)P2 was higher in Glu-411 mutants (E411A (2.91 ± 0.46 pmol min−1 μg−1), E411Q (2.87 ± 0.11 pmol min−1 μg−1), and E411T (3.32 ± 0.36 pmol min−1 μg−1)) than WT (0.90 ± 0.25 pmol min−1 μg−1). The degree of enzymatic activity of WT toward PtdIns(3,5)P2 was less than 30% that of Glu-411 mutants. In addition, the enzyme kinetic parameters of the WT and Glu-411 mutants of Ci-VSP cytoplasmic region were examined for PtdIns(3,4,5)P3 and PtdIns(4,5)P2 (Fig. 7, Tables 2 and 3). The Km values of WT Ci-VSP cytoplasmic region for PtdIns(3,4,5)P3 and PtdIns(4,5)P2 were 86.01 and 38.46 μm, respectively. The Vmax values of WT for PtdIns(3,4,5)P3 and PtdIns(4,5)P2 were 0.714 and 0.440 nmol min−1 μg−1, respectively. The Km value of E411T for PtdIns(3,4,5)P3 was almost the same as that of WT. The Vmax of E411T for PtdIns(3,4,5)P3 was slightly larger than that of WT. In the case of PtdIns(4,5)P2, the Km values of three Glu-411 mutant proteins were lower than that of WT. In E411T mutant, the Vmax value was a little lower than WT, whereas Vmax values were significantly lower in E411A and E411Q than that of WT.
FIGURE 6.
The substrate specificities of Ci-VSP WT and Glu-411 mutant proteins. The activity of Ci-VSP dephosphorylation of various phosphoinositides (100 μm) was carried out. After 2 h of incubation, the isolated phosphate from phosphoinositide was detected by a malachite green assay. Experiments of WT, E411A, E411Q, E411T, and C363S were carried out 10, 10, 10, 10, and 5 times each, respectively, for PtdIns(3,4,5)P3. Experiments of WT, E411A, E411Q, E411T, and C363S were carried out 7, 7, 7, 7, and 5 each, respectively, for PtdIns(3,4)P2. Experiments of WT, E411A, E411Q, E411T, and C363S were carried out 7, 7, 7, 7, and 5 each, respectively, for PtdIns(3,5)P2. Experiments of WT, E411A, E411Q, E411T and C363S were carried out 8, 8, 8, 8, and 5 each, respectively, for PtdIns(4,5)P2.
FIGURE 7.
The kinetics of the Ci-VSP cytoplasmic region toward PtdIns(3,4,5)P3 and PtdIns(4,5)P2. The initial rate of WT and Glu-411 mutants of Ci-VSP cytoplasmic region-catalyzed dephosphorylation of PtdIns(3,4,5)P3 (A) and PtdIns(4,5)P2 (B) was determined using various concentrations of PtdIns(3,4,5)P3 and PtdIns(4,5)P2. n = 5.
TABLE 2.
The kinetics toward PtdIns(3,4,5)P3
| Km | Vmax | Kcat | |
|---|---|---|---|
| μm | nmol min−1μg−1 | min−1 | |
| WT | 86.01 ± 17.46 | 0.714 ± 0.067 | 27.1 ± 2.55 |
| E411T | 84.32 ± 15.49 | 0.828 ± 0.070 | 31.5 ± 2.66 |
TABLE 3.
The kinetics toward PtdIns(4,5)P2
| Km | Vmax | Kcat | |
|---|---|---|---|
| μm | nmol min−1μg−1 | min−1 | |
| WT | 38.46 ± 4.40 | 0.440 ± 0.020 | 16.7 ± 0.76 |
| E411A | 27.95 ± 6.90 | 0.094 ± 0.008 | 3.6 ± 0.31 |
| E411Q | 22.79 ± 5.49 | 0.093 ± 0.007 | 3.5 ± 0.27 |
| E411T | 22.79 ± 4.98 | 0.347 ± 0.024 | 13.2 ± 0.92 |
DISCUSSION
We reported the crystal structure of Ci-VSP cytoplasmic region. Although the overall structure of Ci-VSP cytoplasmic region was similar to that of PTEN, our structures showed that the TI loop was different from that of PTEN, and the residue Glu-411 in the TI loop was oriented toward the active center, making the active pocket smaller and more negatively charged than that of PTEN. An in vitro phosphatase assay showed that the mutation of Glu-411 made the substrate specificity broader. These results provide the basic information of the substrate specificity of Ci-VSP cytoplasmic region.
The Substrate Specificity of Ci-VSP Cytoplasmic Region
Ci-VSP can dephosphorylate both PtdIns(3,4,5)P3 and PtdIns(4,5)P2 (1, 13), whereas PTEN does not dephosphorylate PtdIns(4,5)P2 (14, 15). This phosphatase activity of Ci-VSP toward PtdIns(4,5)P2 requires the presence of a Gly-365 instead of an alanine as in PTEN (13). However, comparison of the crystal structure between WT Ci-VSP and the G365A mutant showed small differences, making it difficult for us to discern how this substrate specificity is achieved. It is possible that the glycine residue at 365 allows for more flexibility in the structure of the substrate binding pocket than alanine when PtdIns(4,5)P2 is bound to the active site. The B-factors of atoms in the P loop of Ci-VSP are higher than those of the atoms in surrounding loops (supplemental Fig. S1), suggesting that this local area is flexible. In addition, the residues after Cys-363 and Lys364 are Gly-365 and Gly366, which can adopt a much wider range of conformations than the other residues. Because of the flexibility of the P loop, Ci-VSP can dephosphorylate PtdIns(3,4,5)P3 despite its narrow pocket size. However, we cannot completely exclude that G365A has the same local structure in the PD as in WT, because the enzyme active site was oxidized.
Comparison of the Ci-VSP structure with that of human PTEN shows that the TI loop of Ci-VSP cytoplasmic region is more closely positioned to the active site, thereby creating a narrower active site pocket (Fig. 4D). This depends on the side chain of Glu-411 of the TI loop, located near the catalytic Cys-363 (Fig. 4, A and B). The analysis of enzyme kinetics of Ci-VSP against PtdIns(4,5)P2 showed that the Km and Vmax were 38.46 μm and 0.440 nmol min−1 μg−1. This is the first report of kinetic analysis toward PtdIns(4,5)P2. All Glu-411 mutants exhibited increased affinity for PtdIns(4,5)P2 (Fig. 7 and Table 3), suggesting that the affinity toward PtdIns(4,5)P2 is affected by Glu-411 through its negative charge rather than the size of residue. The lower Vmax of E411A and E411Q than WT and the 80% of WT Vmax for E411T suggest that high turnover rate requires Glu or Thr at the 411th amino acid (Fig. 7 and Table 3). The malachite green phosphatase assay of WT toward various phosphoinositides showed that Ci-VSP can preferably dephosphorylate PtdIns(3,4,5)P3 and PtdIns(4,5)P2 (Fig. 6), consistent with a previous study (13). In the Glu-411 mutants, the degree of phosphatase activity toward PtdIns(3,5)P2 was higher than that of WT (Fig. 6). On the other hand, the activity of Glu-411 mutants toward other phosphoinositides was not much different from that of WT. This depicts that the lower activity of Ci-VSP cytoplasmic region against PtdIns(3,5)P2 is due to the presence of Glu-411. Given the rotational freedom of the inositol ring in phosphoinositide (31) and the near symmetry of the inositol ring along the axis from position 1 (D1) to position 4 (D4), the ring might be able to flip over. In the model of PTEN with Ins(1,3,4,5)P4, the position 5 (D5) phosphate is located near the TI loop to dephosphorylate at position 3 (D3) (25).
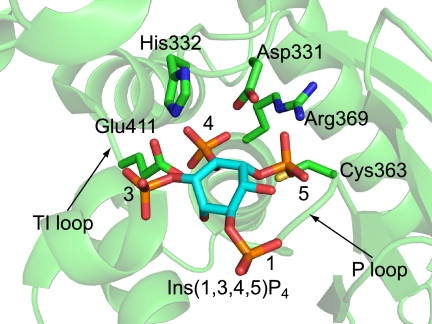
As in a complex model of PTEN and Ins(1,3,4,5)P4 (25), we built a complex model of Ci-VSP and Ins(1,3,4,5)P4 (Fig. 8). When the D5 phosphate group was placed at the active site, which was occupied by sulfate ion in the WT-236 structure, the D3 phosphate positioned near the side chain of the Glu-411 in TI loop. In addition, the D4 phosphate is placed at the active site without any steric hindrance. In this model the side chain of Glu-411 might be placed near the D3 position of Ins(1,3,4,5)P4 (Fig. 8). Glu-411 of Ci-VSP WT may prevent PtdIns(3,5)P2 from binding the active site of Ci-VSP because of charge repulsion between the side chain of Glu-411 and the D3 phosphate. Ci-VSP WT may not prefer PtdIns(3,5)P2 as a substrate. In this model it is unclear why Ci-VSP prefers to dephosphorylate the D5 position phosphate to the D3 position. These suggest that the negative charge of the position 411th amino acid may prevent Ci-VSP cytoplasmic region from dephosphorylating PtdIns(3,5)P2.
FIGURE 8.
Model of Ci-VSP cytoplasmic region and Ins(1,3,4,5)P3 complex. The side chains that may be related to enzymatic activity are shown in a stick model.
On the other hand, the Km value against PtdIns(3,4,5)P3, 86.01 μm, is not consistent with the previously reported value, 38 μm (1). This may be due to the different construction between two studies, the GST tag in the previously report and His-thioredoxin tag in this study. Furthermore, in the case of PtdIns(3,4,5)P3 the Km values of WT and E411T are almost the same. It is unclear at present why activities toward PtdIns(3,4,5)P3 with a more negative charge than PtdIns(4,5)P2 are not affected by the residue Glu-411. Future studies with cocrystals of Ci-VSP and substrate phosphoinositide will be necessary to unravel a more detailed structural basis underlying substrate specificity of VSP.
Regulation of Ci-VSP in Redox State
The P loop containing Cys-363 corresponds to the signature motif, HCXXGXXR, that is the defining feature of the PTP superfamily (32). In the structures of WT-236 and G365A-248, the catalytic residue Cys-363 forms a disulfide bond with the adjacent residue Cys-310 (Fig. 4A). The oxidized form of Ci-VSP in the crystal may be due to a long incubation time in the hanging drop vapor diffusion method we used. The WT and G365A proteins exhibited the substantial reduction of enzyme activity toward PtdIns(3,4,5)P3 and PtdIns(4,5)P2 with H2O2. These results represent that Ci-VSP cytoplasmic region is sensitive to oxidation reagents. In the oxidized form, a sulfate ion was observed in the center of the active site. This may indicate that the phosphate on the inositol ring of phosphoinositides could dock to this site. In the PTPs, P loop conformation generates an unusually low pKa (4.7–5.4) for the active site cysteine that exists as a thiolate anion at neutral pH (32). In PTPs, it is known that this cysteine in the active site is sensitive to oxidation and inactivated by reactive oxygen species (33, 34). In the structural studies of cdc25B, one of the dual specific phosphatases, and lymphoid tyrosine phosphatase that belongs to the proline-, glutamic acid-, serine-, and threonine-rich (PEST) family of PTPs, it was reported that the active site cysteines form disulfide bonds with neighboring cysteines (35, 36). In cdc25B, the disulfide bond state structure is unable to bind the substrate without breaking the disulfide bond (35). In PTEN, Cys-124 reversibly forms a disulfide bond with Cys-71 dependent upon the cellular redox status (37), although the crystal structure of the oxidized form has not been solved. A disulfide bond between Cys-363 and Cys-310 in the two of Ci-VSP structures suggests that the activity of Ci-VSP is potentially regulated by its redox state similar to PTEN (37) and other PTPs (33, 34). We have previously shown that hydrogen peroxide treatment eliminates the voltage-dependent phosphatase activity of zebrafish VSP (7). In sperm, alteration of redox state underlies motility and fertilization events (38). Given that Ci-VSP is expressed in sperm (1), this potential redox-dependent regulation of VSP activities may be involved in sperm functions such as capacitation, motility, and fertilization.
Voltage-dependent Phosphatase Activity of Ci-VSP
Mechanisms of coupling between the VSD and the PD have been studied by biophysical detection of voltage sensor movements and measuring phosphoinositides by several sensor molecules (1, 8, 11, 12). The linker region contains the PBM, and mutation of this region eliminates coupling. However, it remains unknown how the motions of the VSD lead to alteration of enzymatic activity. The surface potential map of Ci-VSP indicates that the side of the active site pocket is positively charged (Fig. 3A, middle). On the other hand, the opposite side of that was highly negatively charged (Fig. 3A, right picture). Thus, the Ci-VSP cytoplasmic region is an electrostatically bipolar protein. This nature seems more remarkable in Ci-VSP than PTEN (Fig. 3C), and positive charges are more broadly distributed on the side of active site of Ci-VSP than in PTEN. In addition, the linker of PBM between the VSD and the cytoplasmic region is on the same side as that of the active site pocket (Fig. 3A). These suggest that the active site pocket faced the membrane. On the activation of the VSD upon depolarization, as a possibility of the mechanism of Ci-VSP voltage-dependent phosphatase activity, the active site pocket of the cytoplasmic region may approach the membrane, or the relative configuration between the PD and the C2 domain may be changed. The electrostatic interaction between the cytoplasmic region and a membrane or the interaction between the PD and the C2 domain might be important for Ci-VSP to change the conformation and to approach the plasma membrane. These processes would be important to bring out the voltage-dependent phosphatase activity.
The C2 domain of PTEN has been reported to be important for high affinity membrane binding (39) and assists the membrane docking of the PD, thereby bringing the PD in close proximity with its substrate, PtdIns(3,4,5)P3 (40). PTEN has several residues that form networks of hydrogen bonds in the interface between the PD and the C2 domain, and some of them are targeted for tumorigenic mutations (25, 41). In a previous report, the bacterial expression level of the PTEN without the C2 domain was extremely low (39). This may suggest that the C2 domain of PTEN plays roles not only in interacting with plasma membrane but also is important for folding.
In Ci-VSP, the C2 domain has more interactions with the PD than in PTEN (Fig. 2). The CBR3 of PTEN was important for interacting with a phospholipid-containing membrane (25). These loops are exposed to the solvent region and have positively charged residues that were involved in interacting with a phospholipid-containing membrane. The CBR3 loop of Ci-VSP is less positively charged than that of PTEN. In contrast with PTEN, the CBR3 loop of Ci-VSP makes contact with the WPD loop of the PD by hydrogen bonds (Fig. 2). In Ci-VSP, the CBR3 loop of C2 domain may contribute to stabilizing the protein through intimate interaction with the PD. When the VSD is in an activated state by depolarization, the cytoplasmic region is recruited to the membrane through binding of the PBM to phosphoinositide. In this process, stability of the cytoplasmic region through interaction between the PD and C2 domain may help conformational change of the substrate binding pocket or orienting the active site of PD toward substrate phosphoinositide.
The crystal structure of the Ci-VSP cytoplasmic region showed two features of the Ci-VSP; that is, the surface electron potential and the CBR3 loop in the C2 domain. Rigorous functional studies in heterologous expressions in the future will address how the voltage-dependent actions of phosphatase are imprinted in these two structural features.
Supplementary Material
Acknowledgments
We thank the laboratory members for helpful discussion. We also thank Dr. Jack E. Dixon and Dr. Carolyn A. Worby for helpful discussion and reading of the manuscript and Dr. Nikola P. Pavletich for providing the structure factor data of human PTEN.
This work has been supported by Targeted Proteins Research Program from the Ministry of Education, Culture, Sports, Science, and Technology (to Y. O. and A. N.), a research grant from the Human Frontier Science Program (to Y. O.), and the Japan Aerospace Exploration Agency-Granada Crystallization Facility High Quality Protein Crystallization Project on the Protein Structure and Function Analysis for Application (to A. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
The atomic coordinates and structure factors (codes 3AWE, 3AWF, and 3AWG) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- VSP
- voltage-sensing phosphatase
- Ci-VSP
- C. intestinalis VSP
- PBM
- phosphoinositide binding motif
- PD
- phosphatase domain
- PTEN
- phosphatase and tensin homolog
- PtdIns(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- Ins(1,3,4,5)P4
- phosphatidylinositol 1,3,4,5-tetrephosphate
- PTP
- protein-tyrosine phosphatase
- r.m.s.
- root mean square
- VSD
- voltage sensor domain.
REFERENCES
- 1. Murata Y., Iwasaki H., Sasaki M., Inaba K., Okamura Y. (2005) Nature 435, 1239–1243 [DOI] [PubMed] [Google Scholar]
- 2. Okamura Y. (2007) Pflugers Arch. 454, 361–371 [DOI] [PubMed] [Google Scholar]
- 3. Ogasawara M., Sasaki M., Nakazawa N., Nishino A., Okamura Y. (2011) Gene Expr. Patterns 11, 233–238 [DOI] [PubMed] [Google Scholar]
- 4. Ramsey I. S., Moran M. M., Chong J. A., Clapham D. E. (2006) Nature 440, 1213–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sasaki M., Takagi M., Okamura Y. (2006) Science 312, 589–592 [DOI] [PubMed] [Google Scholar]
- 6. Okamura Y., Dixon J. E. (2011) Physiology 26, 6–13 [DOI] [PubMed] [Google Scholar]
- 7. Hossain M. I., Iwasaki H., Okochi Y., Chahine M., Higashijima S., Nagayama K., Okamura Y. (2008) J. Biol. Chem. 283, 18248–18259 [DOI] [PubMed] [Google Scholar]
- 8. Murata Y., Okamura Y. (2007) J. Physiol. 583, 875–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Okamura Y., Murata Y., Iwasaki H. (2009) J. Physiol. 587, 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kohout S. C., Ulbrich M. H., Bell S. C., Isacoff E. Y. (2008) Nat. Struct. Mol. Biol. 15, 106–108 [DOI] [PubMed] [Google Scholar]
- 11. Villalba-Galea C. A., Miceli F., Taglialatela M., Bezanilla F. (2009) J. Gen. Physiol. 134, 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kohout S. C., Bell S. C., Liu L., Xu Q., Minor D. L., Jr., Isacoff E. Y. (2010) Nat. Chem. Biol. 6, 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iwasaki H., Murata Y., Kim Y., Hossain M. I., Worby C. A., Dixon J. E., McCormack T., Sasaki T., Okamura Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7970–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maehama T., Dixon J. E. (1998) J. Biol. Chem. 273, 13375–13378 [DOI] [PubMed] [Google Scholar]
- 15. Maehama T., Taylor G. S., Dixon J. E. (2001) Annu. Rev. Biochem. 70, 247–279 [DOI] [PubMed] [Google Scholar]
- 16. Halaszovich C. R., Schreiber D. N., Oliver D. (2009) J. Biol. Chem. 284, 2106–2113 [DOI] [PubMed] [Google Scholar]
- 17. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18. Long F., Vagin A. A., Young P., Murshudov G. N. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cowtan K. D., Zhang K. Y. (1999) Prog. Biophys. Mol. Biol. 72, 245–270 [DOI] [PubMed] [Google Scholar]
- 20. Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 15299374 [Google Scholar]
- 21. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 23. Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. (2005) CCP4 Newsletter 42, contribution 8, Daresbury Laboratory, Daresbury, Warrington, U.K [Google Scholar]
- 24. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee J. O., Yang H., Georgescu M. M., Di Cristofano A., Maehama T., Shi Y., Dixon J. E., Pandolfi P., Pavletich N. P. (1999) Cell 99, 323–334 [DOI] [PubMed] [Google Scholar]
- 26. Bittova L., Sumandea M., Cho W. (1999) J. Biol. Chem. 274, 9665–9672 [DOI] [PubMed] [Google Scholar]
- 27. Chapman E. R., Davis A. F. (1998) J. Biol. Chem. 273, 13995–14001 [DOI] [PubMed] [Google Scholar]
- 28. Cho S. H., Lee C. H., Ahn Y., Kim H., Kim H., Ahn C. Y., Yang K. S., Lee S. R. (2004) FEBS Lett. 560, 7–13 [DOI] [PubMed] [Google Scholar]
- 29. Chu H. M., Wang A. H. (2007) Proteins 66, 996–1003 [DOI] [PubMed] [Google Scholar]
- 30. Agarwal R., Burley S. K., Swaminathan S. (2008) J. Biol. Chem. 283, 8946–8953 [DOI] [PubMed] [Google Scholar]
- 31. Kishore A. I., Prestegard J. H. (2003) Biophys. J. 85, 3848–3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Denu J. M., Dixon J. E. (1998) Curr. Opin. Chem. Biol. 2, 633–641 [DOI] [PubMed] [Google Scholar]
- 33. Salmeen A., Andersen J. N., Myers M. P., Meng T. C., Hinks J. A., Tonks N. K., Barford D. (2003) Nature 423, 769–773 [DOI] [PubMed] [Google Scholar]
- 34. van Montfort R. L., Congreve M., Tisi D., Carr R., Jhoti H. (2003) Nature 423, 773–777 [DOI] [PubMed] [Google Scholar]
- 35. Buhrman G., Parker B., Sohn J., Rudolph J., Mattos C. (2005) Biochemistry 44, 5307–5316 [DOI] [PubMed] [Google Scholar]
- 36. Tsai S. J., Sen U., Zhao L., Greenleaf W. B., Dasgupta J., Fiorillo E., Orrú V., Bottini N., Chen X. S. (2009) Biochemistry 48, 4838–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee S. R., Yang K. S., Kwon J., Lee C., Jeong W., Rhee S. G. (2002) J. Biol. Chem. 277, 20336–20342 [DOI] [PubMed] [Google Scholar]
- 38. Aitken R. J., Paterson M., Fisher H., Buckingham D. W., van Duin M. (1995) J. Cell Sci. 108, 2017–2025 [DOI] [PubMed] [Google Scholar]
- 39. Das S., Dixon J. E., Cho W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7491–7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tonks N. K. (2006) Nat. Rev. Mol. Cell Biol. 7, 833–846 [DOI] [PubMed] [Google Scholar]
- 41. Eng C. (2003) Hum. Mutat. 22, 183–198 [DOI] [PubMed] [Google Scholar]
- 42. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.