Abstract
As human pituitary tumor transforming gene (hPTTG1) is upregulated in endocrine tumors, we studied regulatory mechanisms for hPTTG1 expression. We identified Oct-1-binding motifs in the hPTTG1 promoter region and show Oct-1-specific binding to the hPTTG1 promoter using chromatin immunoprecipitation. We overexpressed Oct-1 and observed ~2.5-fold activation of hPTTG1 promoter luciferase constructs (−2642/−1 and −1717/−1). Transcriptional activation was abrogated by co-transfection of an inactive Oct-1 form lacking the POU domain or by utilizing mutated hPTTG1 promoters or mutants devoid of two Oct-1-binding motifs (−1717/−1mut, −637/−1 or −433/−1). Using biotin–streptavidin pull-down assays, we confirmed Oct-1 binding to the two octamer motifs in the hPTTG1 promoter (−1669/−1631 and −1401/−1361). Endogenous hPTTG1 mRNA and protein increased up to approximately fourfold in Oct-1 transfectants, as measured by real-time PCR and western blot. In contrast, siRNA-mediated suppression of endogenous Oct-1 attenuated both the hPTTG1 mRNA and protein levels. Using confocal immunofluorescence imaging, Oct-1 and hPTTG1 were concordantly upregulated in pituitary (57 and 62%, n=79, P<0.01) and breast tumor specimens (57 and 42%, n=77, P<0.05) respectively. The results show that Oct-1 transactivates hPTTG1, and both proteins are concordantly overexpressed in endocrine tumors, thus offering a mechanism for endocrine tumor hPTTG1 abundance.
Introduction
Pituitary tumor transforming gene (PTTG1), the index mammalian securin, was isolated from rat pituitary tumor cells (Pei & Melmed 1997), and the human homolog (hPTTG1) was identified as a proto-oncogene (Zhang et al. 1999b). hPTTG1 facilitates cell cycle progression, maintains chromosomal stability, and mediates in vitro transformation and in vivo tumorigenesis (Zou et al. 1999, Jallepalli et al. 2001, Yu et al. 2003). hPTTG1 abundance (Yu et al. 2003) or loss of function (Jallepalli et al. 2001, Wang et al. 2001, 2003) both result in abnormal mitosis and chromosomal instability. In contrast to restricted normal tissue expression, hPTTG1 is upregulated in pituitary (Zhang et al. 1999a, Filippella et al. 2006), thyroid (Kim et al. 2006), breast (Solbach et al. 2004), esophageal (Shibata et al. 2002), and colorectal tumors (Heaney et al. 2000). The hPTTG1 overexpression correlates with tumor invasiveness, differentiation, tumor recurrence, and prognosis. Notably, hPTTG1 has been identified as a key signature gene associated with tumor metastases (Ramaswamy et al. 2003). hPTTG1 activates c-myc (Pei 2001) and CCND3 (Tong et al. 2007) as a co-transcription factor enhancing cell proliferation. hPTTG1 interacts with Ku and represses its activity (Romero et al. 2001, Kim et al. 2007b) and modulates p53 (Bernal et al. 2002) participating in DNA damage/repair and apoptosis (Yu et al. 2000, Zhou et al. 2003, Kim et al. 2007b). Pttg deletion results in pituitary gland senescence and is associated with DNA damage pathway activation (Chesnokova et al. 2007). hPTTG1 increases b-fibroblast growth factor (b-FGF) and vascular endothelial growth factor (VEGF) expression and induces tumor angiogenesis (Kim et al. 2007a). Several factors induce hPTTG1, including estrogen, b-FGF, epidermal growth factor (EGF), and insulin (Vlotides et al. 2007). In pituitary folliculostellate TtT/GF cells, EGF-induced PTTG1 is mediated by phosphoinositide-3-kinase (PI3K), protein kinase C (PKC), and mitogen activated protein kinase (MAPK) pathways (Vlotides et al. 2006). hPTTG1 is also regulated as a target gene of the β-catenin/transcription factor (TCF) pathway in esophageal and colon cancers (Zhou et al. 2005, Hlubek et al. 2006). Nevertheless, proximal regulatory mechanisms for hPTTG1 expression are still unclear. Understanding mechanisms regulating hPTTG1 expression could elucidate the process of tumor progression and help identify subcellular antitumor targets.
The transcription factor Oct-1, a member of the POU homeodomain family, is ubiquitously expressed in adult tissues and, unlike other POU family members, is not expressed in a specific temporal or spatial pattern. The Oct-1 protein mid-region POU domain is a conserved bipartite DNA-binding domain consisting of two subdomains, a POU-specific domain and a homeodomain, separated by a flexible linker. Oct-1 binds to a specific promoter octamer sequence (5′-ATGCAAAT-3′) and activates transcription of several genes including those for histone H2B (Fletcher et al. 1987), small nuclear RNA (Murphy et al. 1989), and gonadotrophin-releasing hormone (Eraly et al. 1998). Oct-1 also represses transcription including that of von Willebrand factor (Schwachtgen et al. 1998) and vascular cell adhesion molecule (Iademarco et al. 1992). In addition to these actions, Oct-1 binds cofactors that interact with the DNA-binding (POU) domain to regulate target genes. Oct-1 protein is induced in a p53-independent manner after treatment with DNA-damaging and therapeutic agents (Zhao et al. 2000). Accordingly, Oct-1-deficient mouse fibroblasts exhibit altered expression of cellular stress-induced genes (Tantin et al. 2005). Oct-1 might also play a role in tumorigenesis, as Oct-1 expression is enhanced in intestinal metaplasia and gastric carcinomas (Almeida et al. 2005).
In this study, we detected two putative Oct-1-binding motifs in the hPTTG1 promoter region and show that Oct-1 specifically binds and transactivates the hPTTG1 promoter. Moreover, endogenous hPTTG1 mRNA and protein levels were elevated in HEK293 cells overexpressing Oct-1. In contrast, siRNA-mediated Oct-1 suppression attenuated both hPTTG1 mRNA and protein expression. Concordant overexpression of both immunoreactive Oct-1 and hPTTG1 was observed in human pituitary (P<0.01) and breast tumors (P<0.05), and also in colon cancers (P<0.05). The results indicate a mechanism underlying tumor hPTTG1 abundance.
Materials and methods
Cell culture and transfections
HEK293 cells were obtained from the American tissue culture collection (ATCC) and cultured according to conditions of the ATCC manual. Transfections were performed in 70–80% confluent cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
Chromatin immunoprecipitation (ChIP)
Using a ChIP kit (Active Motif, Carlsbad, CA, USA), about 107 cells were cross-linked and lysed. Chromatin was sonicated to ~500–700 bp length fragments with four rounds of 10 s pulses using 25% power. Sheared chromatin DNA mixture (normalized inputs) was incubated with 3 μg Oct-1 antibody (Oct-1E-8 sc-28334X, Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4 °C. Negative control IgG and positive control transcription factor IIB (TFIIB) antibody were added at 10 μl (4 μg) per ChIP reaction. Real-time PCR were amplified using precipitated immunocomplexes as template and the following hPTTG1 promoter primers. Primer 1, forward: 5′-TTGGGCCGCGAGTTGT-3′, reverse: 5′-CACTCACGCAGGTCTTAACAGC-3′; Primer 2, forward: 5′-GAGAATGACTCAAACGCTGCTG-3′, reverse: 5′-TTGGGTCTAAAGAATACTAGCAGAGAAG-3′; Primer 3, forward: 5′-CAATATTGGTCCTGAAATGCCA-3′, reverse: 5′-AAAGGCTTGACACTATACCTGACATAGA-3′; Primer 4, forward: 5′-GGAATCAGAAAGCGCAGGAC-3′, reverse: 5′-CCCGGCTTTCCTAGACCTTC-3′; Primer 5, forward: 5′-GGTGCTGTGAGAACATAAGGCAT-3′, reverse: 5′-AAGGGATGGGATAGCATAGATACTGT-3′. These five primer pairs were designed in different promoter regions, with primer 1 the closest to ATG and primer 5 the furthest. The TFIIB-positive control was amplified with glyceraldehyde-3-phosphatase dehydrogenase (GAPDH) primers. Negative primers, not expected to be enriched, were used as negative controls.
Plasmids and siRNA
Four different hPTTG1 promoter fragments were amplified from human genomic DNA (Roche Company) using TaKaRa LA Taq, and inserted into the pGL3-Basic luciferase reporter vector (Promega). The following primers were used for PCR of hPTTG1 promoters −2642/−1, −1717/−1, −637/−1, and −433/−1: forward 1 (from −2642): 5′-GGGGTACCATCTATGCTATCCCATCCCT-3′; forward 2 (from −1717): 5′-GGGGTACCGATACAGGAGGAAATAAAGGATGGGGATA-3′; forward 3 (from −637): 5′-GGGGTACCTCTTCCCAGAAAACGTGCCACAAAG-3′; forward 4 (from −433): 5′-GGGGTACCGTTAAGACCTGCGTGAGTGAATGGG-3′; reverse (to −1): 5′-GAAGATCTTCTGGATTATTCTAAGAATG-3′. Promoter −1717/−1mut was mutated by using Quik-Change multi site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) with primers 5′-GTGAAGGGACAATAAAGGTATATTTTTCTGTCTTTCCTC-3′ and 5′-GCTGATCTGCAGCTTAAAATATATGGTGAACTGTGGG-3′. Artificial wild-type and mutant promoters containing six repeat wild-type or mutant Oct-1-binding motifs (designated WT-Oct or mut-Oct) were ligated with annealed complementary oligonucleotides and linear pGL3-Basic vector. Synthesized oligonucleotide sequences were: WT-Oct, sense: 5′-CGTATGCATATGTATGCATATGTATGCATATGTATGCATATGTATGCATATGTATGCATATA-3′, antisense: 5′-GATCTATATGCATACATATGCATACATATGCATACATATGCATACATATGCATACATATGCATACGGTAC-3′; mut-Oct, sense: 5′-CGTATGCAGGCGTATGCAGGCGTATGCAGGCGTATGCAGGCGTATGCAGGCGTATGCAGGCA-3′, anti-sense: 5′-GATCTGCCTGCATACGCCTGCATACGCCTGCATACGCCTGCATACGCCTGCATACGCCTGCATACGGTAC-3′. Complementary oligos were annealed in annealing buffer (10 mM Tris–HCl (pH 7.5), 50 mM NaCl, and 1 mM EDTA).
Three Oct-1 expression plasmids were amplified from a human pre-made pituitary cDNA library (Invitrogen) using TaKaRa LA Taq and cloned into the pcDNA3.1/myc-his vector (Invitrogen). The following primers were used for PCR of Oct-1 (full length), Oct-1 POU (shorter form with the POU domain), and Oct-1delPOU (shortest form lacking the POU domain). Forward: 5′-CGGGATCCACCATGAACAATCCGTCAGAAACC-3′; reverse Oct-1: 5′-CCGCTCGAGCTGTGCCTTGGAGGCGGTGG-TG-3′; reverse Oct-1 POU: 5′-CCGCTCGAGACTGCTTGGTGGGTTGATTCTTTT-3′; reverse Oct-1delPOU: 5′-CCGCTCGAGTGGTGTTGACTGGCTCTGTGG AAG-3′. Plasmids were sequenced by Sequetech. Pre-designed Oct-1 (ID#114253) and negative control siRNAs (Cat#4611) were obtained from Ambion (Austin, TX, USA).
Reporter assay
Cells were split into 24-well plates and incubated at 37 °C overnight. For Oct-1-induced hPTTG1 promoter reporter assays, each well was co-transfected with 1) 200 ng luciferase promoter, pGL3-Basic as control, hPTTG1 promoter −2642/−1, −1717/−1, −1717/−1mut, −637/−1, or −433/−1; and 2) 800 ng expression plasmid, empty pcDNA3.1 vector as control, full length Oct-1, Oct-1 POU, or Oct-1delPOU. For artificial Oct-1 luciferase reporter assays, each well was co-transfected with 1) 200 ng luciferase promoter, pGL3-Basic as control, WT-Oct or mut-Oct; and 2) 800 ng expression plasmid, empty pcDNA3.1 vector as control, full length Oct-1 or Oct-1delPOU. pRL-TK (Promega) encoding Renilla luciferase was used as an internal control (5 ng/well) to assess transfection efficiency. After 24 h, whole-cell lysates were collected for reporter detection by the Dual Luciferase Reporter System (Promega) according to the manufacturer’s protocol. Reactions were measured using an Orion Microplate Luminometer (Berthold Detection System, Pforzheim, Germany). Transfections were performed in triplicate and repeated four times to assure reproducibility.
Biotin–streptavidin pull-down assay
Four oligonucleotides-labeled biotin on the 5′-nucleotide of the sense strand were used in the pull-down assay. These oligonucleotides are synthesized as follows: 1) WT Oct-1 −1655 sense: 5′-GGGACAATAAAGGTATGCATATTTTTCTGTCTTTCCTCTT-3′, which corresponds to positions −1669 to −1631 of the human PTTG promoter; 2) Mut Oct-1 −1655 sense: 5′-GGGACAATAAAGGTATGCAGGCTTTTCTGTCTTTCCTCTT-3′, in which GGC nucleotides replaced TAT; 3) WT Oct-1 −1384 sense: 5′-CTGATCTGCAGCTTAAAATGCATATATGGTGAACTGTGGG-3′, which corresponds to positions −1401 to −1361 of the human PTTG promoter; 4) Mut Oct-1 −1384 sense: 5′-CTGATCTGCA GCTTAAAATGCAGGCATGGTGAACTGTGGG-3′. Oligonucleotides were annealed to their respective complementary oligonucleotides as follows: 1) WT Oct-1 −1655 anti-sense: 5′-AAGAGGAAAGACAGAAAAATATGCATACCTTTATTGTCCC-3′; 2) Mut Oct-1−1655 anti-sense: 5′-AAGAGGAAAGACAGAAAAGCCTGCATACCTTTATTGTCCC-3′; 3) WT Oct-1 −1384 anti-sense: 5′-CCCACAGTTCACCATATATGCATTTTAAGCTGCAGATCAG-3′; 4) Mut Oct-1 −1384 anti-sense: 5′-CCCACAGTTCACCATGCCTGCATTTTAAGCTGCAGATCAG-3′. HEK293 nuclear protein was extracted using a nuclear extract kit (Active motif). Protein concentration was measured by Coomassie Plus Assay Kit (Pierce, Rockford, IL, USA). One microgram of each oligonucleotide was incubated with 1 mg nuclear protein for 20 min at room temperature in binding buffer containing 12% glycerol, 12 mM HEPES (pH 7.9), 4 mM Tris (pH 7.9), 150 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, and 10 μg poly(dI-dC) competitor. Following this reaction, 30 μl streptavidin–agarose (Sigma) was added and incubated at 4 °C for 4 h. Prior to this step, 300 μl original streptavidin–agarose bead preparation was preabsorbed with 500 μg BSA, 50 μg poly(dI-dC) (Sigma), and 50 μg sheared salmon sperm DNA (Sigma) for 20 min at 25 °C; beads were washed three times and resuspended in 300 μl binding buffer. The protein–DNA–streptavidin–agarose complex was washed three times with binding buffer and loaded onto NuPAGE Novex Bis-Tris Gel (Invitrogen). Oct-1 protein was detected by western blot as described below.
RNA extraction and real-time PCR
Total RNA was isolated using TRIZOL Reagent (Invitrogen). Three micrograms of total RNA were used to synthesize cDNA with SuperScript II Reverse Transcriptase (Invitrogen). Real-time PCR was amplified in 20 μl reaction mixtures (100 ng template, 0.5 μM of each primer, 10 μl 2× SYBR GREEN Master Mix (Applied Biosystems, Foster City, CA, USA)) using the following parameters: 95 °C for 1 min, followed by 40 cycles of 95 °C for 20 s, 60 °C for 40 s. β-actin was used as internal control. Real-time PCR primers were designed as follows: hPTTG1 forward: 5′-TGATCCTTGACGAGGAGAGAG-3′, hPTTG1 reverse: 5′-GGTGGCAATTCAACATCCAGG-3′; β-actin forward: 5′-CATGTACGTTGCTATCCAGGC-3′, β-actin reverse: 5′-CTCCTTAATGTCACGCACGAT-3′.
Cell lysis and Western blot analysis
Total cell lysate was prepared in RIPA buffer (Sigma) containing Protease Inhibitor Cocktail (Sigma). Protein concentrations were measured by Coomassie Plus Assay Kit (Pierce) using BSA as standard. Equal amounts (100 μg) of proteins were separated by NuPAGE Novex Bis-Tris Gels (Invitrogen) and transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membrane was incubated in blocking solution (TBS buffer containing 5% nonfat dry milk (Bio-Rad)) for 1 h at room temperature, followed by incubation with primary antibody (Oct-1 ab15112, 1:200, Abcam (Cambridge, MA, USA); Securin DCS-280, 1:200, Abcam; β-actin, 1:5000, Sigma; Anti-myc tag clone 9E10,1:1000, Upstate (Lake Placid, NY, USA) at 4 °C overnight. After washes with TTBS (0.5% Tween-20 in TBS), membranes were subsequently incubated with horseradish peroxidase-linked secondary antibody (GE Healthcare, Piscataway, NJ, USA) for 1 h at room temperature and developed using ECL Western Blotting Detection Reagents (GE Healthcare).
Immunofluorescence
Tissue-array slides of human breast and colorectal tumor were obtained from US Biomax (BR801, BR802, BR804, CO801, and CO803). Each tumor specimen with matched normal adjacent tissue was pathologically confirmed (http://www.biomax.us/tissue-arrays). Seventy nine human pituitary tumor samples and three normal tissues were obtained from the Pathology department of Cedars-Sinai Medical Center. The protocol was approved by the Institutional Review Board. Slides were deparaffinized in xylene, hydrated in graded ethanol, and heated in Target Retrieval Solution (DakoCytomation, Carpinteria, CA, USA) to retrieve antigen at 95 °C for 40 min. Slides were permeabilized with 1% triton-X-100 in PBS for 30 min, and incubated in blocking buffer (10% goat serum, 1% BSA, and 0.1% triton-X-100 in PBS) for 1 h. After washing with PBS, slides were hybridized with antibody against Oct-1 (E-8 sc-28334X, 1:500, Santa Cruz) and hPTTG1 (Securin DCS-280, 1:50, Abcam) at 4 °C overnight. Mouse IgG was used as negative controls. Alexa Fluor anti-mouse antibody with green fluorescence (Molecular Probes, Carlsbad, CA, USA) was used as secondary antibody and incubated at room temperature avoiding light for 1 h. Slides were mounted with ProLong Gold Antifade Reagent with DAPI (Invitrogen), and nuclei dyed by DAPI with blue fluorescence.
Confocal microscopy
Samples were imaged with a Leica TCS/SP spectral confocal scanner (Leica Microsystems, Mannheim, Germany) in dual emission mode to separate autofluorescence from specific staining. A spectral window from 500 to 550 nm wavelength detected emission of Alexa 488. A second window from 560 to 620 nm detected the autofluorescence contribution to the signal. In the final images, Alexa 488 appears green and autofluorescence appears red. The two images were merged, so that all autofluorescence appears yellow, and any true signal appears green.
Evaluation and statistical analysis
Immunofluorescence slides were independently examined by two observers who were blinded to the pathological results and to each other’s records. Human pituitary tumor specimens staining hPTTG1 were calculated as a ratio of immunoreactive cell numbers per field (200×magnification). Three to thirty different fields were counted on each slide according to the tissue size. The average score of all three normal tissues (scaled as 0.02) was used as basal control and compared with all tumors. Tumor hPTTG1 expression was evaluated as follows. 1) No change, if score is ≤0.2; 2) Over-expression, if score is >0.2. Pituitary, breast, and colorectal tumors samples stained with Oct-1, and breast and colorectal tumors samples stained with hPTTG1 were scaled as a percentage of positively stained cells. Tumor specimens were compared with normal tissues and were scored as follows: 1) No change, if positive tumor epithelial cell immunofluorescence was increased <15% than in matched normal tissue; 2) Overexpression, if positive tumor epithelial cell immunofluorescence was increased ≥15% than in matched normal tissue. Oct-1 and hPTTG1 expression were correlated using Pearson’s χ2-test with statistical software SPSS10.0. P<0.05 was considered significant.
Results
Oct-1 protein binds the hPTTG1 promoter
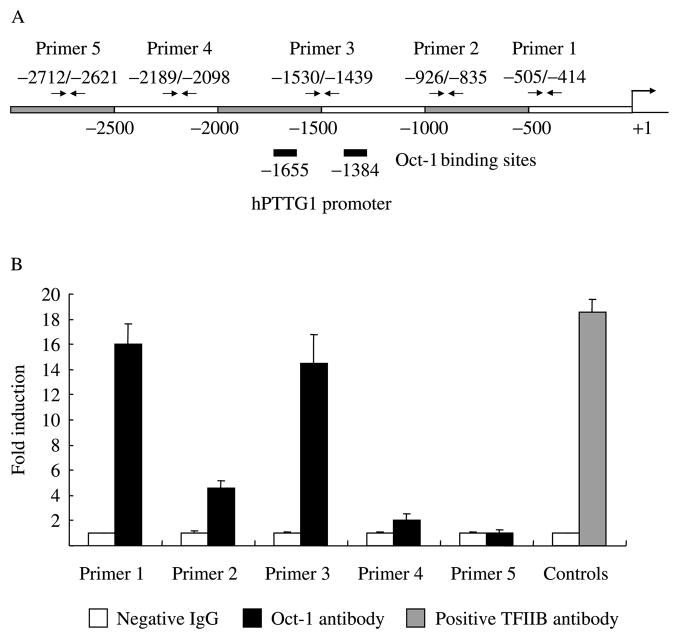
We screened the human PTTG1 promoter with Genomatix MatInspector (http://www.genomatix.de/) and detected Oct-1-binding motifs. We therefore performed ChIP to identify Oct-1 binding to the hPTTG1 promoter. About 107 HEK293 cells were fixed and sonicated into ~600 bp length chromatin DNA. Equal amounts of chromatin DNA (normalized inputs) were incubated separately with negative IgG control, Oct-1 antibody or positive control TFIIB antibody. The antibody captured specific chromatin DNA fragments, and chromatin DNA pulled down by protein G beads detected as template using real-time PCR. Five hPTTG1 promoter primer pairs were designed for real-time PCR (Fig. 1A). Primer 1 is the closest to the ATG translation initiation site and primer 5 the furthest. As shown in Fig. 1B, anti-Oct-1-immunoprecipitated DNA with the enriched Oct-1 locus was amplified by primers 1–4, indicating specific Oct-1 binding to the hPTTG1 promoter. Amplifications obtained from primers 1 and 3 were higher than others, implying stronger Oct-1 protein-binding affinity to hPTTG1 promoter regions around these primers.
Figure 1.
Oct-1 binds the hPTTG1 promoter. (A). Schematic representation of five chromatin immunoprecipitation primers designed for hPTTG1 promoter. (B). Real-time PCR result of chromatin immunoprecipitation. Immunocomplexed DNA pulled down by Oct-1 antibody and negative IgG were amplified as template using real-time PCR with specific hPTTG1 promoter primers 1–5. TFIIB antibody was used as positive control for the ChIP. White Bar: incubated with negative control IgG; Black Bar: incubated with Oct-1 antibody (primers 1–5); Grey Bar: incubated with positive control TFIIB antibody (controls). Each negative IgG control was normalized to Unit ‘1’. Each Real-time PCR was performed in triplicate and ChIP experiments were repeated twice.
Oct-1 activates hPTTG1 transcription
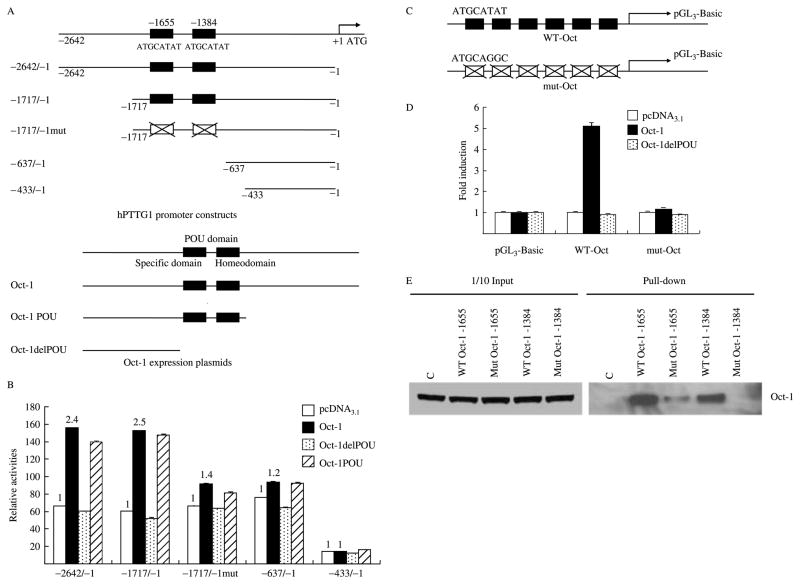
Genomatix MatInspector identified two high scoring Oct-1-binding motifs 5′-ATGCATAT-3′ located at −1655 and −1384 bp upstream of the translation initiation site (ATG) and which were similar to the consensus octamer-binding motifs 5′-ATGCAAAT-3′ (Verrijzer et al. 1992). To ascertain whether Oct-1 regulates hPTTG1 transcription, we cloned and mutated hPTTG1 promoter constructs −2642/−1, −1717/−1, −1717/−1mut, −637/−1, and −433/−1. The −2642/−1 and −1717/−1 fragments contain two Oct-1-binding sites, while the −1717/−1mut contains two mutant sites and both the −637/−1 and −433/−1 fragments are devoid of the motifs (Fig. 2A). We measured hPTTG1 promoter activity in response to Oct-1 co-transfection with a luciferase reporter assay in HEK293 cells exhibiting high transfection efficiency. The DNA-binding POU domain is located in the mid-region of the Oct-1 protein. We cloned three Oct-1 expression plasmids: Oct-1 full length (Oct-1), a shorter form of Oct-1 with the POU domain (Oct-1 POU), and the shortest form lacking the POU domain (Oct-1delPOU) that lacks DNA-binding ability (Fig. 2A). Figure 2B shows that Oct-1 induced hPTTG1 promoter transcriptional activity. Co-transfection of the hPTTG1 promoter (−2642/−1 or −1717/−1) with Oct-1 or Oct-1 POU resulted in ~2.5-fold induction of promoter activity compared with empty vector pcDNA3.1. Results of experimental inductions were normalized to pGL3-Basic as background control. Co-transfected Oct-1delPOU abrogated hPTTG1 promoter activation but did not alter basal transcription levels. However, the transcriptional response to Oct-1 was mostly attenuated (~80% induction) with the mutant −1717/−1mut promoter or truncated −637/−1 promoter, and totally abolished by the shortest −433/−1 promoter. The results indicate that the two Oct-1-binding motifs located at −1655 and −1384 bp are mainly required for Oct-1-induced hPTTG1 promoter activity.
Figure 2.
Oct-1 activates hPTTG1 promoter transcriptional activity. (A) Schematic representation of hPTTG1 promoter constructs and Oct-1 expression plasmids. Upper figure: −2642/−1: hPTTG1 promoter construct from −2642 to −1 which contains two Oct-1-binding motifs located at −1655 and −1384 bp; −1717/−1: from −1717 to −1 containing two WT Oct-1-binding motifs; −1717/−1: from −1717 to −1 containing two mutant Oct-1-binding motifs; −637/−1: truncated promoter from −637 to −1; −433/−1: truncated promoter from −433 to −1. Black boxes represent the Oct-1-binding motifs and white boxes represent the mutant motifs. Lower figure: Oct-1: full length Oct-1 with POU domain; Oct-1 POU: shorter form of Oct-1 with POU domain; Oct-1delPOU: shortest form of Oct-1 without POU domain. Black boxes depict the POU domain. (B) Oct-1 activates hPTTG1 transcriptional activity. HEK293 cells (each well of 24-well plate) were co-transfected 200 ng pGL3-Basic, hPTTG1 promoter −2642/−1, −1717/−1, −1717/−1mut, −637/−1, or −433/−1, together with 800 ng empty vector pcDNA3.1 (white bar), Oct-1 (black bar), Oct-1delPOU (dotted bar), or Oct-1 POU (hatched bar). Relative activities were normalized to empty vector pGL3-Basic as background control. Compared with induction of empty pGL3-Basic vector, pcDNA3.1, Oct-1, Oct-1delPOU, and Oct-1 POU induced hPTTG1 promoter −2642/−1 activity 66.3±0.02-, 155.9±0.09-, 60.2±0.13-, and 140±0.14-fold respectively; promoter −1717/−1 activity was induced 60.2±0.39-, 152.8±0.11-, 52.1±0.69-, and 147.4±0.82-fold respectively; promoter −1717/− 1mut activity was induced 66.4±0.59-, 91.4±0.53+-, 63.4±0.25-, and 81.4±0.85-fold respectively; promoter −637/−1 activity was induced 75.9±0.06-, 93.8±0.56-, 64.6±0.10-, and 92.6±0.12-fold respectively; promoter −433/−1 activity was induced 14.2±0.02-, 14.5±0.06-, 12.1±0.009-, and 16.2±0.006-fold respectively. Compared with pcDNA3.1-induced transcriptional response (as unit ‘1’, white bar), Oct-1-activated hPTTG1 promoters −2642/−1, −1717/−1, −1717/−1mut, −637/−1, or −433/−1 transcription 2.4-fold (P<0.01), 2.5-fold (P<0.01), 1.4-fold (P<0.01), 1.2-fold (P<0.05), and 1-fold respectively (t-test). (C). Schematic representation of artificial Oct-1 luciferase reporter constructs. WT-Oct: construct including six repeat wild-type Oct-1-binding motif (ATGCATAT) in pGL3-Basic vector; mut-Oct: construct including six repeated mutant Oct-1-binding motif (ATGCAGGC) in pGL3-Basic vector. (D) Two Oct-1-binding sites located at −1655 and −1384 bp were involved in hPTTG1 promoter transactivation response to Oct-1. HEK293 cells were co-transfected with 200 ng pGL3-Basic, artificial WT-Oct, or mut-Oct, together with 800 ng empty vector pcDNA3.1 (white bar), Oct-1 (black bar), or Oct-1delPOU (dotted bar). Relative activities were normalized to empty vector pGL3-Basic as background control. Oct-1- and Oct-1delPOU-induced transactivition were normalized to pcDNA3.1. Compared with the pcDNA3.1 response (control as unit ‘1’, white bar), Oct-1 or Oct-1delPOU induced WT-Oct promoter transcription 5.1±0.04- (P<0.01) and 0.9±0.001-fold respectively and induced mut-Oct promoter transcription 1.2±0.009- and 0.9±0.004-fold respectively (t-test). Five ng pRL-TK plasmid was co-transfacted to normalize transfection efficiency. Assays were performed 24 h after transfection. Transfections were performed in triplicate and repeated four times. (E). Western blot of biotin–streptavidin pull-down assay. Equal amounts of HEK293 nuclear extract were incubated with one of the following: no oligonucleotide; WT Oct-1 −1655 or −1384, intact sequences containing wild-type Oct-1-binding motif; Mut Oct-1 −1655 or −1384, in which GGC nucleotides replaced TAT. Binding proteins were precipitated by streptavidin–agarose and detected by Oct-1 antibody. The left 1/10 input lanes confirmed equal extract aliquots added to each reaction. C: no oligonucleotide control.
Next, to confirm roles of the two octamer-binding motifs (−1655 and −1384 bp) involved in Oct-1 activated hPTTG1 transcription, we constructed two artificial luciferase reporter plasmids, WT-Oct and mut-Oct, composed of six repeat wild-type Oct-1-binding motifs (ATGCATAT) or mutant sequences (ATG-CAGGC) (Fig. 2C). As shown in Fig. 2D, co-transfected Oct-1 increased activation (~5.1-fold) of WT-Oct luciferase reporters but not that of mut-Oct luciferase reporters in HEK293 cells. The WT-Oct luciferase reporter did not respond to Oct-1delPOU. Furthermore, we carried out a biotin–streptavidin pull-down assay to verify the interaction between Oct-1 protein and octamer motifs at −1655 and −1384 bp of the hPTTG1 promoter. Equal amounts of Oct-1 transfected HEK293 nuclear extract (normalized inputs) were incubated with one of the following: no oligo, WT Oct-1 −1655, Mut Oct-1 −1655, WT Oct-1 −1384, or Mut Oct-1 −1384. Oligonucleotides captured specific proteins and were then precipitated with steptavidin beads by biotin-labeled 5′-ends. The pulled down DNA–protein complex was detected with antibody directed against Oct-1 by western blot. As shown in Fig. 2E, wild-type Oct-1 −1655 and −1384 both specifically bound Oct-1 protein. Oct-1 −1655 binding was abrogated ~95% and −1384 binding completely abrogated by mutant Oct-1 oligonucleotides. Inputs confirmed equal amounts of nuclear proteins in each reaction (Fig. 2E). These results indicated that the two Oct-1-binding motifs in the PTTG promoter (−1655 and −1384) participate in hPTTG1 transcriptional regulation by Oct-1, and directly bind the Oct-1 protein.
Oct-1 induces hPTTG1 mRNA and protein expression
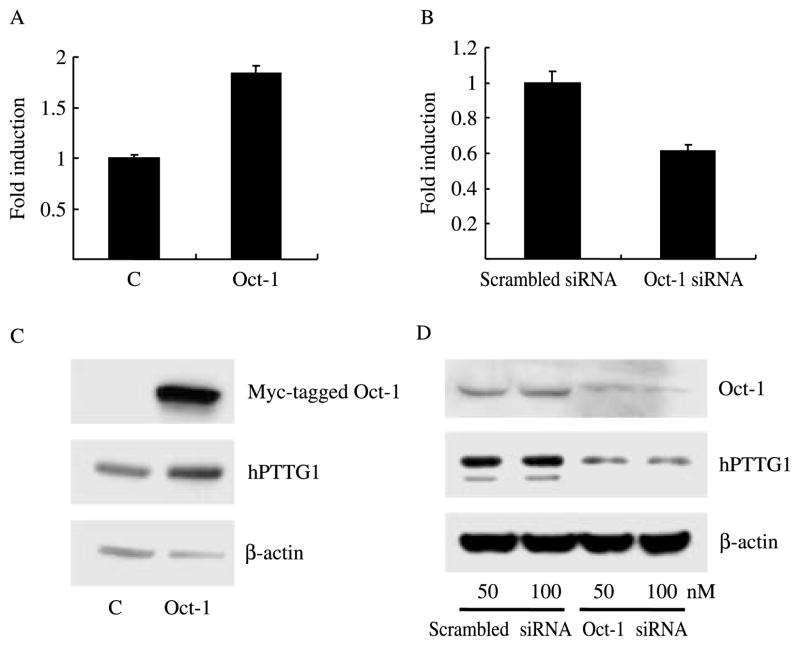
As Oct-1 binds the hPTTG1 promoter and activates transcription, we tested Oct-1 regulation of endogenous hPTTG1 expression. We transiently transfected empty vector pcDNA3.1 or Oct-1 expression plasmids into HEK293 cells. mRNA expression was measured by real-time PCR using β-actin as an internal control. As shown in Fig. 3A, overexpressed Oct-1 increased hPTTG1 mRNA levels approximately twofold. Transfection of Oct-1 siRNA (100 nM) suppressed endogenous Oct-1 mRNA and also inhibited hPTTG1 mRNA expression by ~40% (Fig. 3B). Cell lysates analyzed by western blot verified that hPTTG1 protein expression followed the same pattern as mRNA levels, i.e. overexpressed Oct-1 enhanced hPTTG1 protein expression about fourfold (measured by BandScan), and transfected Oct-1 siRNA (both 50 nM and 100 nM) attenuated hPTTG1 protein levels by ~70% (measured by BandScan) in HEK293 cells (Fig. 3C and D).
Figure 3.
Oct-1 induces hPTTG1 mRNA and protein expression. HEK 293 cells were transfected with empty vector pcDNA3.1 or Oct-1 (A and C), or transfected with negative control siRNA or Oct-1 siRNA (B and D). (A) Compared with pcDNA3.1 control, Oct-1 induced hPTTG1 mRNA expression 1.8±0.08-folds (P<0.01). (B) Compared with negative control siRNA, Oct-1 siRNA attenuated hPTTG1 mRNA expression by 39±4% (P<0.01). C: empty vector pcDNA3.1; Scrambled siRNA: negative control siRNA. hPTTG1 mRNA levels were detected by real-time PCR (A and B); Oct-1 and hPTTG1 protein levels were measured by western blot (C and D).
Oct-1 and hPTTG1 are concordantly expressed in endocrine tumors and colon cancers
hPTTG1 is overexpressed in multiple human tumors including pituitary, breast, and colorectal tumors (Zhang et al. 1999a, Heaney et al. 2000, Solbach et al. 2004, Filippella et al. 2006). High Oct-1 protein expression was detected in gastric carcinomas (Almeida et al. 2005). Since our results show that overexpressed Oct-1 induces hPTTG1 expression in vitro, we assessed Oct-1 and hPTTG1 protein levels in human pituitary, breast, and colorectal tumor tissues by confocal immunofluorescent imaging.
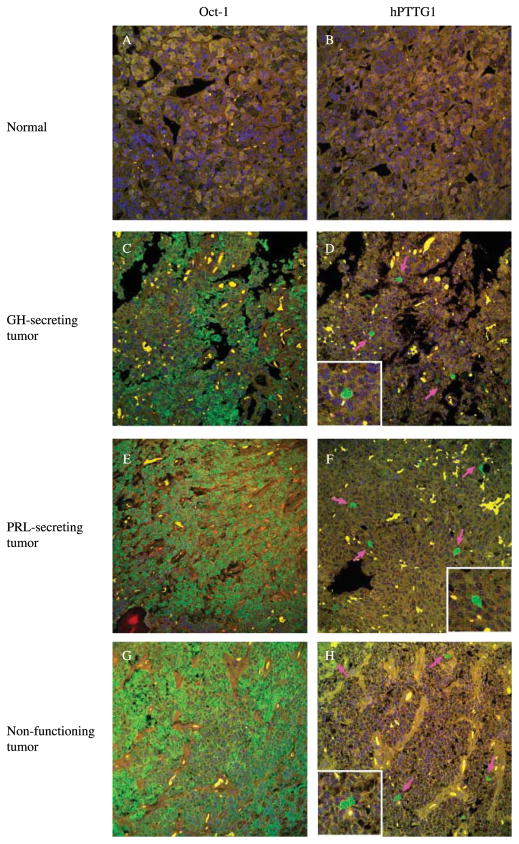
We studied 3 normal pituitary tissue specimens and 79 pituitary tumor specimens including 1 adrenocorticotrophin-secreting, 16 growth hormone-secreting, 13 prolactin-secreting, and 49 non-functioning tumors. Normal non-tumorous pituitary tissue was not immunoreactive for hPTTG1 (Fig. 4B). Weak Oct-1 staining was detected in up to 10% of normal pituitary cells (Fig. 4A), whereas abundant Oct-1 staining was observed in up to 85% of pituitary tumor cells (Fig. 4C, E and G). Upregulated immunoreactive Oct-1 and hPTTG1 were detected in 57% (45 of the 79) and 62% (49 of the 79) of pituitary tumors respectively (Fig. 4, Table 1). Thirty seven tumors with high Oct-1 expression were also immunoreactive for hPTTG1 (Fig. 4C–H), and twenty-two cases with unchanged Oct-1 levels did not exhibit enhanced hPTTG1 signals. Taken together, 75% of tumor samples exhibited a concordant pattern of Oct-1 and hPTTG1 expression. Oct-1 and hPTTG1 expression correlated positively (P<0.01, Pearson’s χ2-test) (Table 1). hPTTG1 and concordant Oct-1/hPTTG1 overexpression correlate with proliferation. The frequency of hPTTG1 and concordant Oct-1/hPTTG1 overexpression increased with elevated Ki-67 staining in pituitary tumors. Either hPTTG1 or both Oct-1/hPTTG1 overexpression correlate with Ki-67 expression (P<0.01 and P<0.05 respectively, Pearson’s χ2-test).
Figure 4.
Oct-1 and hPTTG1 immunoreactivity in pituitary tumor specimens. Oct-1 and hPTTG1 protein immunoreactivity were detected using fluorescence immunohistochemistry. Panel A and B show negative staining of both Oct-1 and hPTTG1 in normal tissues; compared with normal tissue, panels C–H show high Oct-1 and hPTTG1 expression in GH-secreting, PRL-secreting, and non-functioning pituitary tumors respectively. Pink arrows indicate positive hPTTG1 staining cells. Size: 375×375 μM. Green signal: staining of Oct-1 or hPTTG1; blue signal: DAPI nuclear staining; yellow signal: autofluorescence background.
Table 1.
Oct-1 and pituitary tumor transforming gene (hPTTG1) expression in 79 human pituitary tumors
| hPTTG1
|
Total | ||
|---|---|---|---|
| Overexpression | No change | ||
| Oct-1 | |||
| Overexpression | 37 | 8 | 45 |
| No change | 12 | 22 | 34 |
| Total | 49 | 30 | 79 |
Oct-1 and hPTTG1 immunoreactivity were assessed by confocal microscopy in 79 human pituitary tumors (Fig. 4). Correlation was analyzed using Pearson’s χ2-test, P<0.01.
In 77 human breast tumors and matched normal breast tissues, undetectable or low Oct-1 and hPTTG1 immunoreactivity were observed in normal breast epithelium (Fig. 5C, D, G and H), whereas high Oct-1 and hPTTG1 expression was observed in tumors (Fig. 5A and B). Compared with normal tissue, 57% (44 of the 77) and 42% (32 of the 77) of breast tumors exhibited upregulated Oct-1 and hPTTG1 (Table 2). Sixty one percent of cases (47 of 77), including 23 cases showing both overexpressed (Fig. 5A–D) and 24 cases showing both unchanged (Fig. 5E–H), exhibited concordant expression of the two immunoreactive proteins (P<0.05, Pearson’s χ2-test) (Table 2).
Figure 5.
Oct-1 and hPTTG1 immunoreactivity in breast tumor specimens. Case 1 (A–D) shows concordantly abundant Oct-1 (A) and hPTTG1 (B) expression in breast tumor (A and B) compared with undetectable expression in matched normal tissue (C and D). Case 2 (E–H) shows concordantly unchanged Oct-1 and hPTTG1 expression in tumor (E and F) compared with normal tissue (G and H). Picture size: 750×750 μM; corner inserts size: 110×110 μM. Green signal: staining of Oct-1 or hPTTG1; blue signal: DAPI nuclear staining; yellow signal: autofluorescence background.
Table 2.
Oct-1 and pituitary tumor transforming gene (hPTTG1) expression in 77 human breast tumors
| hPTTG1
|
Total | ||
|---|---|---|---|
| Overexpression | No change | ||
| Oct-1 | |||
| Overexpression | 23 | 21 | 44 |
| No change | 9 | 24 | 33 |
| Total | 32 | 45 | 77 |
Oct-1 and hPTTG1 immunoreactivity were assessed by confocal microscopy in 77 paried human breast tumor specimens (Fig. 5). Correlation was analyzed using Pearson’s χ2-test, P<0.05.
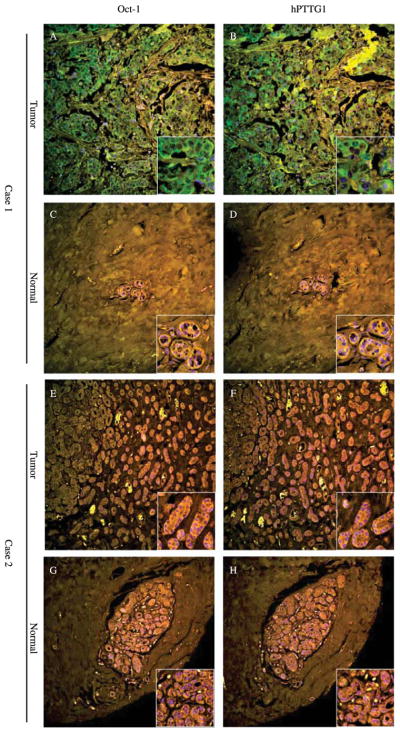
Since Oct-1 protein is ubiquitously expressed in adult tissues, in addition to endocrine tumors, we also analyzed 71 colorectal tumors with paired normal tissues. Discrete Oct-1 and hPTTG1 staining were detected in a small percentage of normal tissues (Fig. 6C, D, G and H); in contrast, most tumor tissues stained strongly with a high percentage of positive cells (Fig. 6A and B). Interestingly, in normal colorectal mucosal samples, hPTTG1 was expressed predominantly at the bottom of the crypts, where vigorous cell proliferation occurs (Fig. 6D). Upregulated Oct-1 and hPTTG1 expression were observed in 44% and 42% of tumor tissues (Table 3). Remarkably, about 65% of tumor samples (46 of the 71) exhibited a concordant pattern of Oct-1 and hPTTG1 expression (P<0.05, Pearson’s χ2-test) (Table 3), i.e. 18 of the 46 cases show both proteins overexpressed (Fig. 6A–D) and 28 of the 46 cases show both unchanged (Fig. 6E–H). Oct-1, hPTTG1, and concordant Oct-1/hPTTG1 overexpression all correlated with gender (P<0.05, Pearson’s χ2-test). Oct-1, hPTTG1, and concordant Oct-1/hPTTG1 are over-expressed more often in males.
Figure 6.
Oct-1 and hPTTG1 immunoreactivity in colon cancer specimens. Case 1 (A–D) shows concordantly upregulated Oct-1 (A) and hPTTG1 (B) expression in colon cancer (A and B) compared with low expression in matched normal tissue (C and D). hPTTG1 is expressed predominantly at the bottom of crypts in normal tissue (D). Case 2 (E–H) shows concordantly unchanged Oct-1 and hPTTG1 expressions in tumor (E and F) compared with normal tissue (G and H). Size: 750×750 μM; corner inserts size: 110× 110 μM. Green signal: staining of Oct-1 or hPTTG1; blue signal: DAPI nuclear staining; yellow signal: autofluorescence background.
Table 3.
Oct-1 and pituitary tumor transforming gene (hPTTG1) expression in 71 human colon cancers
| hPTTG1
|
Total | ||
|---|---|---|---|
| Overexpression | No change | ||
| Oct-1 | |||
| Overexpression | 18 | 13 | 31 |
| No change | 12 | 28 | 40 |
| Total | 30 | 41 | 71 |
Oct-1 and hPTTG1 immunoreactivity were assessed by confocal microscopy in 71 paried human colon cancer specimens (Fig. 6). Correlation was analyzed using Pearson’s χ2-test, P<0.05.
Thus, Oct-1 and hPTTG1 are concordantly over-expressed in ex vivo tumor specimens, suggesting that increased Oct-1 expression participates in abundant tumor hPTTG1 expression.
Discussion
hPTTG1 is upregulated in human tumors and is involved in multiple steps of tumor progression including tumorigenesis, invasiveness, metastasis, and angiogenesis (Vlotides et al. 2007). Therefore, understanding regulatory mechanisms of hPTTG1 expression is important to develop novel approaches for endocrine tumor therapy. We showed that EGF-induced PTTG1 is mediated by PI3K, PKC, and MAPK pathways (Vlotides et al. 2006). hPTTG1 is also a target gene of the β-catenin/TCF pathway (Zhou et al. 2005). hPTTG1 expression is increased by 17-β-estradiol in rat pituitary tumors (Heaney et al. 1999) and in MCF7 breast cancer cells (Yen-hao Chen unpublished). In this study, we present evidence that the transcription factor Oct-1 transactivates hPTTG1 and induces hPTTG expression both in vitro and in ex vivo tumor tissues. Two octamer-binding motifs on the hPTTG1 promoter respectively located at −1655 and −1384 bp upstream of ATG directly bind Oct-1 and are required for Oct-1-induced activation (Fig. 2). Mutation or deletion of the two Oct-1-binding sites abrogated binding and the transactivation response. In addition to binding octamer motifs, Oct-1 also promotes target gene transactivation by association with co-activators (Gstaiger et al. 1995, Strubin et al. 1995). Oct-1 protein forms complexes with nuclear factor Y, signal transducer and activator of transcription 5 (STAT5), or nuclear factor κB (NF-κB), and drives target gene promoters via other elements such as CAAT box, STAT5 motif, CREB, or NF-κB-binding sites (Boulon et al. 2002, Fan et al. 2002, Magne et al. 2003, Kam et al. 2005, dela Paz et al. 2007). By scanning the hPTTG1 promoter region, several CAAT boxes and STAT5 motifs are located at −640 to −490 bp upstream of ATG, implying that other motifs may act as complementary signaling motifs. Real-time PCR results of the ChIP assay showed higher amplification of primers 1 and 3, indicating higher binding affinity of Oct-1 protein around these areas. The area of primer 3 approximates the two Oct-1-binding motifs (−1655 and −1384 bp), and the area of primer 1 overlaps the CAAT- and STAT5-binding sites located at −640 to −490 bp. Deletion of both Oct-1-binding sites (−1655 and −1384 bp) together with CAAT and STAT5 motifs abrogated Oct-1 stimulation and basal transcriptional activity (Fig. 2B, −433/−1).
Oct-1 is a transcriptional activator involved in H2B and snRNA gene regulation (Fletcher et al. 1987, Murphy et al. 1989), both important for rapid cell division; furthermore, Oct-1 promotes the oncogene cyclin D1 transcription (Boulon et al. 2002, Magne et al. 2003) and represses tumor suppressor gene p15INK4b transcription (Hitomi et al. 2007). Oct-1 antisense transformants reduced cell growth rates (Palubin et al. 2006). We show here that Oct-1 overexpression transactivated hPTTG1 and induced its expression. In contrast, Oct-1 knockdown by siRNA suppressed hPTTG1 mRNA and protein levels. hPTTG1, as a human securin, is a key component of the spindle checkpoint pathway that facilitates cell proliferation and tumor progression (Zhang et al. 1999b, Zou et al. 1999). Identification of hPTTG1 as an Oct-1 target gene provides evidence linking Oct-1 function to cell proliferation and growth control.
Oct-1 is ubiquitously expressed in adult tissues. Most studies of Oct-1 have focused on transcriptional regulatory functions, and the Oct-1 expression status in human tumors has been described in a report of enhanced Oct-1 expression in gastric carcinomas (Almeida et al. 2005). This universal transcriptional factor may therefore be abnormally elevated in human tumors. We now identify overexpressed Oct-1 in endocrine-related pituitary and breast tumors, and also in colon cancers (Fig. 4–6) by confocal immunofluorescent imaging. Notably, a concordant Oct-1 and hPTTG1 expression pattern was detected in these tumors. The results strongly support our in vitro results that hPTTG1 acts as an Oct-1 target and may explain at least in part, abundant tumor hPTTG expression.
Cellular stress responses include cell cycle growth arrest, apoptosis, and DNA repair, and may ultimately result in genomic instability and malignant cell transformation. Oct-1 functions as a stress sensor (Tantin et al. 2005). Oct-1 is induced in response to cellular stress, and DNA-binding affinity is enhanced to regulate downstream genes transcription via the octamer-binding sequence (Zhao et al. 2000, Jin et al. 2001, Schild-Poulter et al. 2003, Tantin et al. 2005, Duggan et al. 2006). hPTTG1, as a downstream gene of Oct-1, participates in events critical to the cellular stress response, including cell cycle control, DNA repair (Kim et al. 2007b), and apoptosis (Yu et al. 2000). After genotoxic stress, hPTTG1 deficiency is associated with compromised cell survival and proliferation (Bernal et al. 2008). Since both proteins are concordantly overexpressed in endocrine tumors, cellular stress may result in Oct-1-induced tumor hPTTG1 upregulation, thus enabling genomic instability and tumorigenesis.
Acknowledgments
We thank Shiri Gutman for assistance with immunofluorescent staining.
Funding
NIH Grant CA75979 (S Melmed) and the Doris Factor Molecular Endocrinology Laboratory.
Footnotes
Declaration of interest
There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Almeida R, Almeida J, Shoshkes M, Mendes N, Mesquita P, Silva E, Van Seuningen I, Reis CA, Santos-Silva F, David L. OCT-1 is over-expressed in intestinal metaplasia and intestinal gastric carcinomas and binds to, but does not transactivate, CDX2 in gastric cells. Journal of Pathology. 2005;207:396–401. doi: 10.1002/path.1861. [DOI] [PubMed] [Google Scholar]
- Bernal JA, Luna R, Espina A, Lazaro I, Ramos-Morales F, Romero F, Arias C, Silva A, Tortolero M, Pintor-Toro JA. Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nature Genetics. 2002;32:306–311. doi: 10.1038/ng997. [DOI] [PubMed] [Google Scholar]
- Bernal JA, Roche M, Mendez-Vidal C, Espina A, Tortolero M, Pintor-Toro JA. Proliferative potential after DNA damage and non-homologous end joining are affected by loss of securin. Cell Death and Differentiation. 2008;15:202–212. doi: 10.1038/sj.cdd.4402254. [DOI] [PubMed] [Google Scholar]
- Boulon S, Dantonel JC, Binet V, Vie A, Blanchard JM, Hipskind RA, Philips A. Oct-1 potentiates CREB-driven cyclin D1 promoter activation via a phospho-CREB- and CREB binding protein-independent mechanism. Molecular and Cellular Biology. 2002;22:7769–7779. doi: 10.1128/MCB.22.22.7769-7779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokova V, Zonis S, Rubinek T, Yu R, Ben-Shlomo A, Kovacs K, Wawrowsky K, Melmed S. Senescence mediates pituitary hypoplasia and restrains pituitary tumor growth. Cancer Research. 2007;67:10564–10572. doi: 10.1158/0008-5472.CAN-07-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan SP, Gallagher WM, Fox EJ, Abdel-Latif MM, Reynolds JV, Kelleher D. Low pH induces co-ordinate regulation of gene expression in oesophageal cells. Carcinogenesis. 2006;27:319–327. doi: 10.1093/carcin/bgi211. [DOI] [PubMed] [Google Scholar]
- Eraly SA, Nelson SB, Huang KM, Mellon PL. Oct-1 binds promoter elements required for transcription of the GnRH gene. Molecular Endocrinology. 1998;12:469–481. doi: 10.1210/mend.12.4.0092. [DOI] [PubMed] [Google Scholar]
- Fan W, Jin S, Tong T, Zhao H, Fan F, Antinore MJ, Rajasekaran B, Wu M, Zhan Q. BRCA1 regulates GADD45 through its interactions with the OCT-1 and CAAT motifs. Journal of Biological Chemistry. 2002;277:8061–8067. doi: 10.1074/jbc.M110225200. [DOI] [PubMed] [Google Scholar]
- Filippella M, Galland F, Kujas M, Young J, Faggiano A, Lombardi G, Colao A, Meduri G, Chanson P. Pituitary tumour transforming gene (PTTG) expression correlates with the proliferative activity and recurrence status of pituitary adenomas: a clinical and immunohistochemical study. Clinical Endocrinology. 2006;65:536–543. doi: 10.1111/j.1365-2265.2006.02630.x. [DOI] [PubMed] [Google Scholar]
- Fletcher C, Heintz N, Roeder RG. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 1987;51:773–781. doi: 10.1016/0092-8674(87)90100-0. [DOI] [PubMed] [Google Scholar]
- Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens CM. AB-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- Heaney AP, Horwitz GA, Wang Z, Singson R, Melmed S. Early involvement of estrogen-induced pituitary tumor transforming gene and fibroblast growth factor expression in prolactinoma pathogenesis. Nature Medicine. 1999;5:1317–1321. doi: 10.1038/15275. [DOI] [PubMed] [Google Scholar]
- Heaney AP, Singson R, McCabe CJ, Nelson V, Nakashima M, Melmed S. Expression of pituitary-tumour transforming gene in colorectal tumours. Lancet. 2000;355:716–719. doi: 10.1016/S0140-6736(99)10238-1. [DOI] [PubMed] [Google Scholar]
- Hitomi T, Matsuzaki Y, Yasuda S, Kawanaka M, Yogosawa S, Koyama M, Tantin D, Sakai T. Oct-1 is involved in the transcriptional repression of the p15(INK4b) gene. FEBS Letters. 2007;581:1087–1092. doi: 10.1016/j.febslet.2007.01.092. [DOI] [PubMed] [Google Scholar]
- Hlubek F, Pfeiffer S, Budczies J, Spaderna S, Jung A, Kirchner T, Brabletz T. Securin (hPTTG1) expression is regulated by beta-catenin/TCF in human colorectal carcinoma. British Journal of Cancer. 2006;94:1672–1677. doi: 10.1038/sj.bjc.6603155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iademarco MF, McQuillan JJ, Rosen GD, Dean DC. Characterization of the promoter for vascular cell adhesion molecule-1 (VCAM-1) Journal of Biological Chemistry. 1992;267:16323–16329. [PubMed] [Google Scholar]
- Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, Kinzler KW, Vogelstein B, Lengauer C. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- Jin S, Fan F, Fan W, Zhao H, Tong T, Blanck P, Alomo I, Rajasekaran B, Zhan Q. Transcription factors Oct-1 and NF-YA regulate the p53-independent induction of the GADD45 following DNA damage. Oncogene. 2001;20:2683–2690. doi: 10.1038/sj.onc.1204390. [DOI] [PubMed] [Google Scholar]
- Kam KY, Jeong KH, Norwitz ER, Jorgensen EM, Kaiser UB. Oct-1 and nuclear factor Y bind to the SURG-1 element to direct basal and gonadotropin-releasing hormone (GnRH)-stimulated mouse GnRH receptor gene transcription. Molecular Endocrinology. 2005;19:148–162. doi: 10.1210/me.2004-0025. [DOI] [PubMed] [Google Scholar]
- Kim DS, Franklyn JA, Boelaert K, Eggo MC, Watkinson JC, McCabe CJ. Pituitary tumor transforming gene (PTTG) stimulates thyroid cell proliferation via a vascular endothelial growth factor/kinase insert domain receptor/inhibitor of DNA binding-3 autocrine pathway. Journal of Clinical Endocrinology and Metabolism. 2006;91:4603–4611. doi: 10.1210/jc.2006-1291. [DOI] [PubMed] [Google Scholar]
- Kim CS, Ying H, Willingham MC, Cheng SY. The pituitary tumor-transforming gene promotes angiogenesis in a mouse model of follicular thyroid cancer. Carcinogenesis. 2007a;28:932–939. doi: 10.1093/carcin/bgl231. [DOI] [PubMed] [Google Scholar]
- Kim DS, Franklyn JA, Smith VE, Stratford AL, Pemberton HN, Warfield A, Watkinson JC, Ishmail T, Wakelam MJ, McCabe CJ. Securin induces genetic instability in colorectal cancer by inhibiting double-stranded DNA repair activity. Carcinogenesis. 2007b;28:749–759. doi: 10.1093/carcin/bgl202. [DOI] [PubMed] [Google Scholar]
- Magne S, Caron S, Charon M, Rouyez MC, Dusanter-Fourt I. STAT5 and Oct-1 form a stable complex that modulates cyclin D1 expression. Molecular and Cellular Biology. 2003;23:8934–8945. doi: 10.1128/MCB.23.24.8934-8945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S, Pierani A, Scheidereit C, Melli M, Roeder RG. Purified octamer binding transcription factors stimulate RNA polymerase III – mediated transcription of the 7SK RNA gene. Cell. 1989;59:1071–1080. doi: 10.1016/0092-8674(89)90763-0. [DOI] [PubMed] [Google Scholar]
- Palubin KM, Goodwin BL, Niesen MI, Le EA, Osborne AR, Blanck G. A direct mechanistic link between growth control and a tumor cell immune function: increased interleukin-8 secretion accounts for elimination of Oct-1 antisense transformants from scid mice. Anti-cancer Research. 2006;26:1733–1738. [PubMed] [Google Scholar]
- dela Paz NG, Simeonidis S, Leo C, Rose DW, Collins T. Regulation of NF-κB-dependent gene expression by the POU domain transcription factor Oct-1. Journal of Biological Chemistry. 2007;282:8424–8434. doi: 10.1074/jbc.M606923200. [DOI] [PubMed] [Google Scholar]
- Pei L. Identification of c-myc as a down-stream target for pituitary tumor-transforming gene. Journal of Biological Chemistry. 2001;276:8484–8491. doi: 10.1074/jbc.M009654200. [DOI] [PubMed] [Google Scholar]
- Pei L, Melmed S. Isolation and characterization of a pituitary tumor-transforming gene (PTTG) Molecular Endocrinology. 1997;11:433–441. doi: 10.1210/mend.11.4.9911. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nature Genetics. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Romero F, Multon MC, Ramos-Morales F, Dominguez A, Bernal JA, Pintor-Toro JA, Tortolero M. Human securin, hPTTG, is associated with Ku heterodimer, the regulatory subunit of the DNA-dependent protein kinase. Nucleic Acids Research. 2001;29:1300–1307. doi: 10.1093/nar/29.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild-Poulter C, Shih A, Yarymowich NC, Hache RJ. Down-regulation of histone H2B by DNA-dependent protein kinase in response to DNA damage through modulation of octamer transcription factor 1. Cancer Research. 2003;63:7197–7205. [PubMed] [Google Scholar]
- Schwachtgen JL, Remacle JE, Janel N, Brys R, Huylebroeck D, Meyer D, Kerbiriou-Nabias D. Oct-1 is involved in the transcriptional repression of the von willebrand factor gene promoter. Blood. 1998;92:1247–1258. [PubMed] [Google Scholar]
- Shibata Y, Haruki N, Kuwabara Y, Nishiwaki T, Kato J, Shinoda N, Sato A, Kimura M, Koyama H, Toyama T, et al. Expression of PTTG (pituitary tumor transforming gene) in esophageal cancer. Japanese Journal of Clinical Oncology. 2002;32:233–237. doi: 10.1093/jjco/hyf058. [DOI] [PubMed] [Google Scholar]
- Solbach C, Roller M, Fellbaum C, Nicoletti M, Kaufmann M. PTTG mRNA expression in primary breast cancer: a prognostic marker for lymph node invasion and tumor recurrence. Breast. 2004;13:80–81. doi: 10.1016/j.breast.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Strubin M, Newell JW, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- Tantin D, Schild-Poulter C, Wang V, Hache RJ, Sharp PA. The octamer binding transcription factor Oct-1 is a stress sensor. Cancer Research. 2005;65:10750–10758. doi: 10.1158/0008-5472.CAN-05-2399. [DOI] [PubMed] [Google Scholar]
- Tong Y, Tan Y, Zhou C, Melmed S. Pituitary tumor transforming gene interacts with Sp1 to modulate G1/S cell phase transition. Oncogene. 2007;26:5596–5605. doi: 10.1038/sj.onc.1210339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer CP, Alkema MJ, van Weperen WW, Van Leeuwen HC, Strating MJ, van der Vliet PC. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO Journal. 1992;11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlotides G, Cruz-Soto M, Rubinek T, Eigler T, Auernhammer CJ, Melmed S. Mechanisms for growth factor-induced pituitary tumor transforming gene-1 expression in pituitary folliculostellate TtT/GF cells. Molecular Endocrinology. 2006;20:3321–3335. doi: 10.1210/me.2006-0280. [DOI] [PubMed] [Google Scholar]
- Vlotides G, Eigler T, Melmed S. Pituitary tumor-transforming gene: physiology and implications for tumorigenesis. Endocrine Reviews. 2007;28:165–186. doi: 10.1210/er.2006-0042. [DOI] [PubMed] [Google Scholar]
- Wang Z, Yu R, Melmed S. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Molecular Endocrinology. 2001;15:1870–1879. doi: 10.1210/mend.15.11.0729. [DOI] [PubMed] [Google Scholar]
- Wang Z, Moro E, Kovacs K, Yu R, Melmed S. Pituitary tumor transforming gene-null male mice exhibit impaired pancreatic beta cell proliferation and diabetes. PNAS. 2003;100:3428–3432. doi: 10.1073/pnas.0638052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Heaney AP, Lu W, Chen J, Melmed S. Pituitary tumor transforming gene causes aneuploidy and p53-dependent and p53-independent apoptosis. Journal of Biological Chemistry. 2000;275:36502–36505. doi: 10.1074/jbc.C000546200. [DOI] [PubMed] [Google Scholar]
- Yu R, Lu W, Chen J, McCabe CJ, Melmed S. Overexpressed pituitary tumor-transforming gene causes aneuploidy in live human cells. Endocrinology. 2003;144:4991–4998. doi: 10.1210/en.2003-0305. [DOI] [PubMed] [Google Scholar]
- Zhang X, Horwitz GA, Heaney AP, Nakashima M, Prezant TR, Bronstein MD, Melmed S. Pituitary tumor transforming gene (PTTG) expression in pituitary adenomas. Journal of Clinical Endocrinology and Metabolism. 1999a;84:761–767. doi: 10.1210/jcem.84.2.5432. [DOI] [PubMed] [Google Scholar]
- Zhang X, Horwitz GA, Prezant TR, Valentini A, Nakashima M, Bronstein MD, Melmed S. Structure, expression, and function of human pituitary tumor-transforming gene (PTTG) Molecular Endocrinology. 1999b;13:156–166. doi: 10.1210/mend.13.1.0225. [DOI] [PubMed] [Google Scholar]
- Zhao H, Jin S, Fan F, Fan W, Tong T, Zhan Q. Activation of the transcription factor Oct-1 in response to DNA damage. Cancer Research. 2000;60:6276–6280. [PubMed] [Google Scholar]
- Zhou Y, Mehta KR, Choi AP, Scolavino S, Zhang X. DNA damage-induced inhibition of securin expression is mediated by p53. Journal of Biological Chemistry. 2003;278:462–470. doi: 10.1074/jbc.M203793200. [DOI] [PubMed] [Google Scholar]
- Zhou C, Liu S, Zhou X, Xue L, Quan L, Lu N, Zhang G, Bai J, Wang Y, Liu Z, et al. Overexpression of human pituitary tumor transforming gene (hPTTG), is regulated by β-catenin/TCF pathway in human esophageal squamous cell carcinoma. International Journal of Cancer. 2005;113:891–898. doi: 10.1002/ijc.20642. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]