Abstract
The RAD3 gene of Saccharomyces cerevisiae is required for excision repair and is essential for cell viability. RAD3 encoded protein possesses a single stranded DNA-dependent ATPase and DNA and DNA.RNA helicase activities. Mutational studies have indicated a requirement for the RAD3 helicase activities in excision repair. To examine the extent of conservation of structure and function of RAD3 during eukaryotic evolution, we have cloned the RAD3 homolog, rhp3+, from the distantly related yeast Schizosaccharomyces pombe. RAD3 and rhp3+ encoded proteins are highly similar, sharing 67% identical amino acids. We show that like RAD3, rhp3+ is indispensable for excision repair and cell viability, and our studies indicate a requirement of the putative rhp3+ DNA helicase activity in DNA repair. We find that the RAD3 and rhp3+ genes can functionally substitute for one another. The level of complementation provided by the rhp3+ gene in S.cerevisiae rad3 mutants or by the RAD3 gene in S.pombe rhp3 mutants is remarkable in that both the excision repair and viability defects in both yeasts are restored to wild type levels. These observations suggest a parallel evolutionary conservation of other protein components with which RAD3 interacts in mediating its DNA repair and viability functions.
Full text
PDF

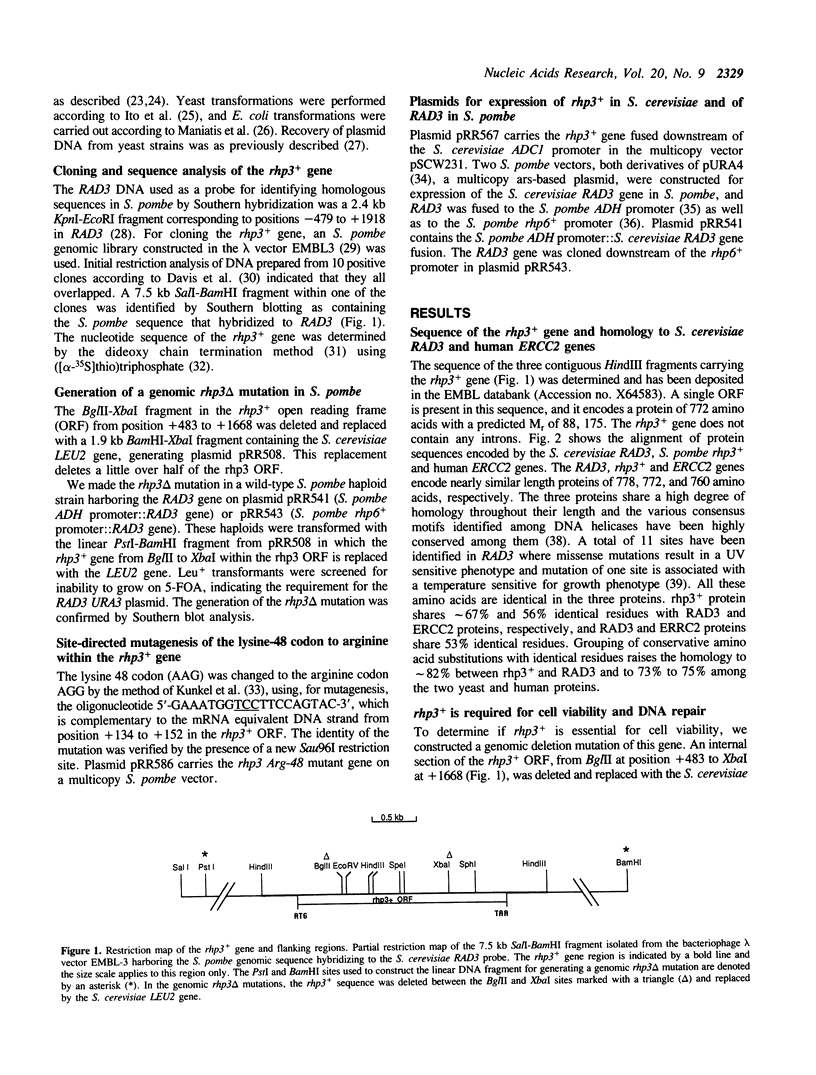

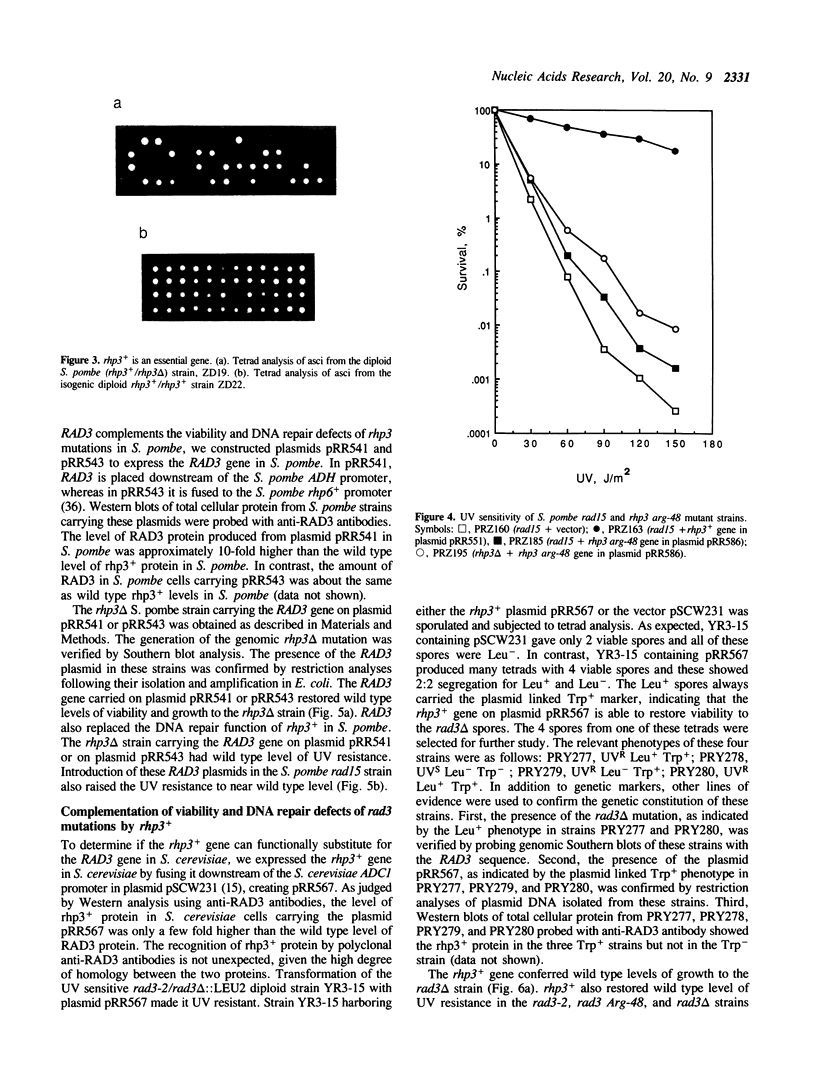


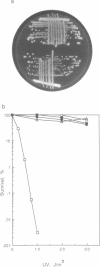
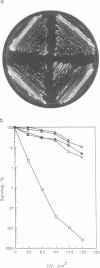
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailly V., Sung P., Prakash L., Prakash S. DNA.RNA helicase activity of RAD3 protein of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9712–9716. doi: 10.1073/pnas.88.21.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankmann M., Prakash L., Prakash S. Yeast RAD14 and human xeroderma pigmentosum group A DNA-repair genes encode homologous proteins. Nature. 1992 Feb 6;355(6360):555–558. doi: 10.1038/355555a0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootsma D., Keijzer W., Jung E. G., Bohnert E. Xeroderma pigmentosum complementation group XP-I withdrawn. Mutat Res. 1989 Sep;218(2):149–151. doi: 10.1016/0921-8777(89)90021-9. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Kohli J., Murray J., Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet. 1988 Dec;215(1):81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- Higgins D. R., Prakash S., Reynolds P., Polakowska R., Weber S., Prakash L. Isolation and characterization of the RAD3 gene of Saccharomyces cerevisiae and inviability of rad3 deletion mutants. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5680–5684. doi: 10.1073/pnas.80.18.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Huysmans E., Dams E., Vandenberghe A., De Wachter R. The nucleotide sequences of the 5S rRNAs of four mushrooms and their use in studying the phylogenetic position of basidiomycetes among the eukaryotes. Nucleic Acids Res. 1983 May 11;11(9):2871–2880. doi: 10.1093/nar/11.9.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. T., Elliott G. C., Squires S., Joysey V. C. Lack of complementation between xeroderma pigmentosum complementation groups D and H. Hum Genet. 1989 Feb;81(3):203–210. doi: 10.1007/BF00278989. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Miller R. D., Prakash L., Prakash S. Defective excision of pyrimidine dimers and interstrand DNA crosslinks in rad7 and rad23 mutants of Saccharomyces cerevisiae. Mol Gen Genet. 1982;188(2):235–239. doi: 10.1007/BF00332681. [DOI] [PubMed] [Google Scholar]
- Miller R. D., Prakash L., Prakash S. Genetic control of excision of Saccharomyces cerevisiae interstrand DNA cross-links induced by psoralen plus near-UV light. Mol Cell Biol. 1982 Aug;2(8):939–948. doi: 10.1128/mcb.2.8.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim A., Smith B. P. Genetic control of radiation sensitivity in Schizosaccharomyces pombe. Genetics. 1975 Apr;79(4):573–582. doi: 10.1093/genetics/79.4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Friedberg E. C. A DNA repair gene required for the incision of damaged DNA is essential for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4818–4821. doi: 10.1073/pnas.80.15.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975 Aug 14;256(5518):547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- Reynolds P., Higgins D. R., Prakash L., Prakash S. The nucleotide sequence of the RAD3 gene of Saccharomyces cerevisiae: a potential adenine nucleotide binding amino acid sequence and a nonessential acidic carboxyl terminal region. Nucleic Acids Res. 1985 Apr 11;13(7):2357–2372. doi: 10.1093/nar/13.7.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Koken M. H., Hoeijmakers J. H., Prakash S., Prakash L. The rhp6+ gene of Schizosaccharomyces pombe: a structural and functional homolog of the RAD6 gene from the distantly related yeast Saccharomyces cerevisiae. EMBO J. 1990 May;9(5):1423–1430. doi: 10.1002/j.1460-2075.1990.tb08258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R. J., Friedberg E. C. Molecular mechanisms of pyrimidine dimer excision in Saccharomyces cerevisiae: incision of ultraviolet-irradiated deoxyribonucleic acid in vivo. J Bacteriol. 1981 May;146(2):692–704. doi: 10.1128/jb.146.2.692-704.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P., Moreno S., Reed S. I. Conservation of mitotic controls in fission and budding yeasts. Cell. 1989 Apr 21;57(2):295–303. doi: 10.1016/0092-8674(89)90967-7. [DOI] [PubMed] [Google Scholar]
- Russell P., Nurse P. Schizosaccharomyces pombe and Saccharomyces cerevisiae: a look at yeasts divided. Cell. 1986 Jun 20;45(6):781–782. doi: 10.1016/0092-8674(86)90550-7. [DOI] [PubMed] [Google Scholar]
- Russell P., Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986 Apr 11;45(1):145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. M., Montelone B. A., Siede W., Friedberg E. C. Effects of multiple yeast rad3 mutant alleles on UV sensitivity, mutability, and mitotic recombination. J Bacteriol. 1990 Dec;172(12):6620–6630. doi: 10.1128/jb.172.12.6620-6630.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Higgins D., Prakash L., Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988 Oct;7(10):3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Prakash L., Matson S. W., Prakash S. RAD3 protein of Saccharomyces cerevisiae is a DNA helicase. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8951–8955. doi: 10.1073/pnas.84.24.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Prakash L., Weber S., Prakash S. The RAD3 gene of Saccharomyces cerevisiae encodes a DNA-dependent ATPase. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6045–6049. doi: 10.1073/pnas.84.17.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L. H., Brookman K. W., Dillehay L. E., Mooney C. L., Carrano A. V. Hypersensitivity to mutation and sister-chromatid-exchange induction in CHO cell mutants defective in incising DNA containing UV lesions. Somatic Cell Genet. 1982 Nov;8(6):759–773. doi: 10.1007/BF01543017. [DOI] [PubMed] [Google Scholar]
- Thompson L. H., Shiomi T., Salazar E. P., Stewart S. A. An eighth complementation group of rodent cells hypersensitive to ultraviolet radiation. Somat Cell Mol Genet. 1988 Nov;14(6):605–612. doi: 10.1007/BF01535314. [DOI] [PubMed] [Google Scholar]
- Weber C. A., Salazar E. P., Stewart S. A., Thompson L. H. ERCC2: cDNA cloning and molecular characterization of a human nucleotide excision repair gene with high homology to yeast RAD3. EMBO J. 1990 May;9(5):1437–1447. doi: 10.1002/j.1460-2075.1990.tb08260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda G., van Ham R. C., Vermeulen W., Bootsma D., van der Eb A. J., Hoeijmakers J. H. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne's syndrome. Cell. 1990 Aug 24;62(4):777–791. doi: 10.1016/0092-8674(90)90122-u. [DOI] [PubMed] [Google Scholar]
- Wilcox D. R., Prakash L. Incision and postincision steps of pyrimidine dimer removal in excision-defective mutants of Saccharomyces cerevisiae. J Bacteriol. 1981 Nov;148(2):618–623. doi: 10.1128/jb.148.2.618-623.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin M., de Wit J., Odijk H., Westerveld A., Yasui A., Koken M. H., Hoeijmakers J. H., Bootsma D. Molecular characterization of the human excision repair gene ERCC-1: cDNA cloning and amino acid homology with the yeast DNA repair gene RAD10. Cell. 1986 Mar 28;44(6):913–923. doi: 10.1016/0092-8674(86)90014-0. [DOI] [PubMed] [Google Scholar]