Summary
Bacterial stress responses provide them the opportunity to survive hostile environments, proliferate and potentially cause diseases in humans and animals. The way in which pathogenic bacteria interact with host immune cells triggers a complicated series of events that include rapid genetic re‐programming in response to the various host conditions encountered. Viewed in this light, the bacterial host‐cell induced stress response (HCISR) is similar to any other well‐characterized environmental stress to which bacteria must respond by upregulating a group of specific stress‐responsive genes. Post stress, bacteria must resume their pre‐stress genetic program, and, as a consequence, must degrade unnecessary stress responsive transcripts through RNA decay mechanisms. Further, there is a well‐established role for several ribonucleases in the cold shock response whereby they modulate the changing transcript landscape in response to the stress, and during acclimation and subsequent genetic re‐programming post stress. Recently, ribonucleases have been implicated as virulence‐associated factors in several notable Gram‐negative pathogens including, the yersiniae, the salmonellae, Helicobacter pylori, Shigella flexneri and Aeromonas hydrophila. This review will focus on the roles played by ribonucleases in bacterial virulence, other bacterial stress responses, and on their novel therapeutic applications.
Introduction
RNA decay
Initially, mRNA decay was thought to be the result of a random recycling pathway in which salvaged nucleotides could be reused; original thinking also believed mRNA turnover to be a rapid, non‐specific and inevitable end‐point for all transcripts regardless of their length and structure (Deutscher and Li, 2001; Kushner, 2002). Since that time, much progress has been made regarding the understanding of RNA decay, which is now believed to be a series of specific, temporally controlled events in which specific enzymes target the various RNA species including mRNA (Deutscher and Li, 2001; Kushner, 2002). Consequently, our understanding of RNA decay and the enzymes responsible for this metabolic process has broadened greatly. The majority of RNA decay studies have predominantly used E. coli as the model organism. In sharp contrast, RNA decay has not been as well characterized in Gram‐positive bacteria with the exception of preliminary studies characterizing this process in Bacillus subtilis (Even et al., 2005; Mäder et al., 2008), Streptococcus pyogenes (Barnett et al., 2007), S. pneumoniae (Domingues et al., 2009) and Staphylococcus aureus (Huntzinger et al., 2005; Boisset et al., 2007). Common to both Gram‐positive and Gram‐negative organisms, ribonucleases are enzymes that degrade ribonucleotides, and two broad types exist, endo‐ and exo‐ribonucleases. Endoribonucleases cleave RNA molecules endoribonucleolytically, at times in a 5′ end‐dependent manner, while exoribonucleases degrade RNA molecules in a 3′–5′ direction (Table 1).
Table 1.
List of key E. coli ribonucleases, their size and their function.
| Ribonuclease | Size (kDa) | Encoding gene | Function |
|---|---|---|---|
| RNase E | 118 | rnr | Essential endoribonuclease that processes rRNA and degrades mRNA; acts as a scaffolding protein upon which PNPase, enolase and DEAD box RNA helicases associate with to form the multi‐protein complex ‘the degradosome’ which putatively enhances RNA processing and degradation in the cell |
| RNase G | 55 | rng/cafA | Non‐essential endoribonuclease that shares N‐terminal homology with RNase E; has no C‐terminal scaffolding region upon which the degradosome assembles |
| RNase III | 26 | rnc | Endoribonuclease that cleaves double stranded rRNA during rRNA maturation; also involved in degradation of mRNA including the pnp transcript that encodes PNPase |
| RNase II | 72 | rnb | The primary hydrolytic exoribonuclease that degrades mRNA in a 3′–5′ direction but poorly degrades structured mRNA |
| RNase R | 95 | vacB/rnr | The second most abundant hydrolytic exoribonuclease in the cell that easily degrades mRNA with extensive secondary structure, processes rRNA and is cold‐inducible |
| PNPase | 80 | pnp | The primary phosphorolytic exoribonuclease in the cell that is cold inducible and associates with RNase E in the degradosome for cooperative degradation of mRNA. PNPase is required for cold growth (15°C) and, like RNase II, poorly degrades structured RNA |
| RNase PH | 45–50 | rph | The second phosphorolytic exoribonuclease in the cell that shares homology with the catalytic domains of PNPase (which contains two RNase PH catalytic domains); RNase PH has also been shown to physically associate with RNase E and it processes tRNA. |
PNPase, polynucleotide phosphorylase.
Endoribonucleases, RnaseE and the degradosome
So far, at least nine endoribonucleases have been identified in E. coli, including RNase E, G, III, I and P (Kushner, 2002; Anderson and Dunman, 2009). RNase E is the predominant mRNA degrading endoribonuclease in E. coli, and is encoded by the rne gene. Interestingly, Gram‐positive organisms lack RNase E, but instead S. pyogenes possesses endoribonucleases J1 and J2 which, like RNase E in E. coli, were found to be essential for bacterial growth (Bugrysheva and Scott, 2010). A temperature sensitive mutant of RNase E, ams‐1 (altered mRNA stability), displayed a gross mRNA‐decay deficiency (Ono and Kuwano, 1979). RNase E, itself, is autoregulated at the post‐transcriptional level mediated by its own 5′ untranslated region (Mudd and Higgins, 1993; Jain and Belasco, 1995; Sousa et al., 2001). RNase E (Fig. 1) is a 1061‐amino‐acid residue protein (118 kDa) with three distinct domains that form a homo‐tetramer (Callaghan et al., 2005) and has been shown to be involved in rRNA and tRNA maturation as well as mRNA degradation (Kushner, 2002). The first 500 residues at the amino terminus include both the catalytic and the S1 RNA binding domains of RNase E. Residues 597–684 encode an arginine‐rich RNA binding domain, and the carboxy‐terminus ranging from residues 734–1060 serve as a scaffolding region upon which the various components of a multi‐protein complex, the degradosome, bind and include the exo‐ribonuclease polynucleotide phosphorylase (PNPase), a DEAD box (core sequence of eight amino acids including D, E, A and D) helicase RNA helicase B (RhlB), RhlE or CsdA of E. coli (Généreux et al., 2004), and the glycolytic enzyme enolase (Carpousis et al., 1994;Vanzo et al., 1998; Khemici and Carpousis, 2004). There are other minor protein components of the degradosome that have been identified and include DnaK, GroEL and polyphosphate kinase (Miczak et al., 1996; Blum et al., 1997).
Figure 1.
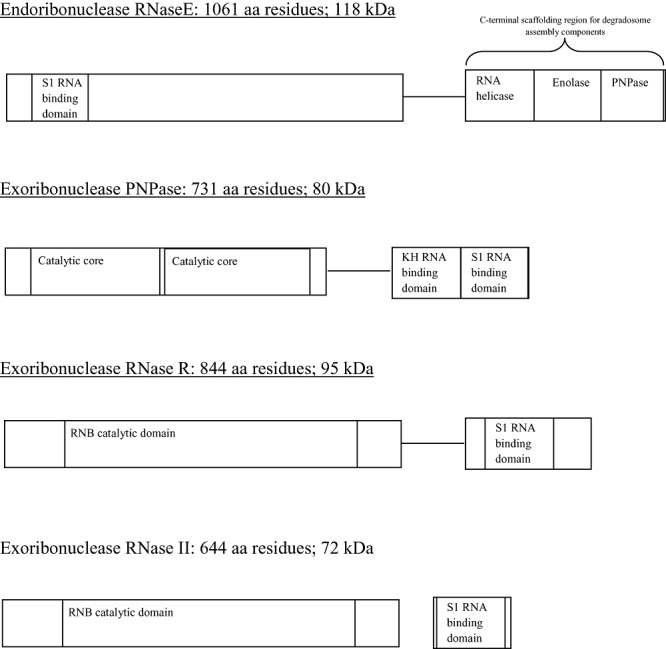
Various E. coli ribonucleases and location of their S1 RNA binding domains. The endoribonuclease RNase E (including its carboxy‐terminus degradosome scaffolding region upon which RhlB helicase, enolase and PNPase associate), the exoribonuclease PNPase, the exoribonuclease RNase R and the exoribonuclease RNase II are depicted. Their S1 RNA binding domains are indicated as well as the KH RNA binding domain that is only present in PNPase.
The degradosome is a large multi‐protein complex thought to patrol the cytoplasm targeting RNA molecules destined for decay and/or processing. Current knowledge describes degradosome‐mediated RNA degradation as a synchronized series of events in which PNPase serves as the RNA trapping molecule by virtue of its S1 RNA binding domain. The PNPase‐bound RNA is then fed to RNase E that attacks the molecule in a 5′ end dependent fashion exposing a free 3′ end. RhlB, known to play a role in rRNA processing, is thought to facilitate the process by unwinding extensively structured RNA which often impedes RNA degradation. The exposed 3′ end or RNA is then fed back into the catalytic (RNase PH) domains of the homo‐trimeric PNPase for final processing/degradation. It is still unclear what role the glycolytic enzyme enolase, DnaK, GroEL, and polyphosphate kinase play in degradosome function (Carpousis, 2002).
In Pseudomonas syringae, a degradosome was discovered containing RNase E, the exoribonuclease RNase R and the DEAD box helicase RhlE (Purusharth et al., 2005). However, the related yeast exosome, which structurally resembles PNPase in its core nine subunits and also contains an RNase II‐family member exo‐ribonuclease in its 10th subunit termed Rrp44 (Mitchell et al., 1997), coordinates endoribonucleolytic and exoribonucleolytic decay (Schaeffer et al., 2008). The presence of PNPase in the E. coli degradosome as well as the presence of PNPase‐like structurally related proteins in the yeast exosome highlights the significance of PNPase in the multi‐protein degradosome and degradosome‐like complexes across taxa.
Despite degradosome assembly within bacteria being demonstrated by various assays including Far Western blot analysis, co‐immunoprecipitation and yeast‐two hybrid experiments, the physiological role played by the degradosome remains currently quite controversial since an E. coli RNase E mutant devoid of its carboxy‐terminus processed and degraded RNA similar to that of its corresponding wild‐type (WT) strain (Kido et al., 1996; Jiang et al., 2000). Additionally, an E. coli homologue of RNase E, RNase G, appears to have some functional overlap with RNase E despite being considerably smaller with only a catalytic domain and no scaffolding region (Umitsuki et al., 2001; Kushner, 2002). Interestingly, PNPase was found to independently associate with enolase in addition to a DEAD box helicase, further complicating the PNPase degradosome model (Portier, 1975; Carpousis et al., 1994; Lin and Lin‐Chao, 2005). Also, RNase PH, a small phosphorolytic ribonuclease resembling one of PNPase's catalytic cores, associated with RNase E in a pull‐down assay, suggests that other degradosomes might exist within E. coli (Durán‐Figueroa et al., 2006). Taken together, the precise role that RNase E plays in RNA decay still remains to be elucidated to determine whether it functions in a degradosome‐dependent or degradosome‐independent fashion in Gram‐negative organisms.
Exoribonucleases, PNPase and their role in RNA metabolism
To date, eight exoribonucleases have been identified in E. coli (Deutscher and Li, 2001; Anderson and Dunman, 2009). RNase II (Fig. 1), the gene product of rnb, is a 72 kDa hydrolytic exoribonuclease that accounts for up to 90% RNA decay and is conserved in eukaryotes (Deutscher and Li, 2001). However, RNase II exhibits impaired catalysis when faced with RNA molecules containing extensive secondary structures (Guarneros and Portier, 1990). RNase R (Fig. 1), the gene product of vacB/rnr, is a 95 kDa hydrolytic exoribonuclease that was identified in an E. coli rnb‐deleted mutant strain and is the second most abundant exoribonuclease. However, unlike RNase II, RNase R is capable of degrading RNA molecules with extensive secondary structures including rRNA (Nikolaev et al., 1976; Kasai et al., 1977). Interestingly, an E. coli RNase R‐RNase II double mutant was viable while either RNase II or RNase R‐PNPase double mutants were non‐viable (Cheng et al., 1998), suggesting some functional overlap among the exoribonucleases, and that PNPase plays some indispensable role when acting alongside either RNase II or R.
PNPase (Fig. 1), the gene product of pnp, is a ubiquitous 80 kDa homo‐trimeric exoribonuclease found in most bacteria with the exception of the Mycoplasma and the archeabacteria. Further, PNPase is also found in yeast, plant chloroplasts and humans (Leszczyniecka et al., 2002). Originally, PNPase was used as a synthetic enzyme (Grunberg‐Manago et al., 1955) to help decipher the genetic code; however, today, its primary physiological role in prokaryotes is believed to be degradative, and like RNase PH, a smaller ribonuclease resembling one of PNPase's two catalytic cores, the mechanism of RNA catalysis is phosphorolysis. Like RNase II, PNPase also poorly degrades structured RNA molecules. Additionally, PNPase coordinately auto‐regulates itself at the post‐transcriptional level together with the endoribonucleases RNase III (Jarrige et al., 2001; 2002). As described earlier, PNPase is a degradosome constituent in E. coli, and exactly what percentage of PNPase's degradative effort is spent while associated with the degradosome has yet to be determined.
Ultimately, many questions in the field of RNA metabolism remain, including why are there so many ribonuclease homologues. Another pressing question is whether the degradosome is physiologically relevant or simply an experimental artifact. Studies to address some of these questions have been conducted in attempts to answer these as well as other RNA metabolism questions. Below, the roles played by each ribonuclease in stress responses important for pathogenesis and bacterial virulence will be discussed and are summarized in Table 2.
Table 2.
List of organisms and ribonucleases that modulate their virulence.
| Organism | Ribonuclease | Role in virulence | Reference |
|---|---|---|---|
| Gram‐negatives | |||
| Shigella flexnari | RNase R | Promotes expression of virulence genes and murine infection | Tobe et al. (1992) |
| Enteroinvasive E. coli | RNase R | Promotes virulence in murine infection | Tobe et al. (1992) |
| Aeromonas hydrophila | RNase R | Promotes cold growth | Erova et al. (2008) |
| Motility and virulence in murine infection | Erova et al. (2008) | ||
| Brucella abortus | RNase R | No role played | Miyoshi et al. (2007) |
| Salmonella enterica | PNPase | Promotes cold growth | Clements et al. (2002) |
| Promotes acute infection and virulence in murine infections | Clements et al. (2002) | ||
| Suppresses Salmonella plasmid virulence genes | Ygberg et al. (2006) | ||
| RNase E | Degrade mgt required from intercellular survival with non‐coding Amg RNA | Lee and Groisman (2010) | |
| Yersinia enterocolitica | PNPase | Promotes cold growth | Goverde et al. (1998) |
| Yersinia psuedotuberculosis | PNPase | Promotes cold growth | Rosenzweig et al. (2005) |
| Promotes virulence in murine and cell culture infections | Rosenzweig et al. (2007) | ||
| Yersinia pestis | PNPase | Promotes virulence in cell culture infection an optimal T3SS function | Rosenzweig et al. (2005) |
| Promotes virulence in murine model of infection | This work (Fig. 2) | ||
| RNase E | Promotes virulence in cell culture infection and optimal T3SS function | Yang et al. (2008) | |
| Dichelobacter nodosus | PNPase | Suppresses virulence and twitching motility | Palanisamy et al. (2010) |
| Campylobacter jejuni | PNPase | Promotes cold growth | Haddad et al. (2009) |
| Legionella pneumophila | RNase R | Promotes cold growth | Söderberg and Cianciotto (2010) |
| Helicobacter pylori | RNase R | Cold inducible; promotes motility and expression of apoptosis genes | Tsao et al. (2009) |
| Gram‐positives | |||
| Staphylococcus aureus | RNase III | Suppress inflammation by degrading spa (encodes Staph protein A) | Huntzinger et al. (2005) |
| Suppresses expression of virulence‐associated genes | Boisset et al. (2007) | ||
| PNPase | Promotes cold growth | Anderson and Dunman (2009) | |
| Streptococcus pyogenes | PNPase | Degrades virulence transcripts during late exponential phase | Barnett et al. (2007) |
| CvfA | Regulate virulence transcripts at stationary phase and promotes virulence | Kang et al. (2010) | |
| Streptococcus mutans | PNPase | Is upregulated during acid‐shock (dental implications) | Len et al. (2004) |
T3SS, type three secretion system; CvfA, a putative ribonuclease.
PNPase as a stress response ribonuclease and virulence‐associated gene product
The role played by PNPase in cold shock/growth and oxidative stress
As alluded to earlier, RNA metabolism can be both ongoing and temporal in nature. In fact, responses to low temperatures are referred to as ‘cold shock responses’, and in E. coli, a small subset of cold inducible genes becomes upregulated (Jones et al., 1987), including a family of six cold shock protein (Csp) encoding genes (Goldstein et al., 1990). In addition to csp transcripts being upregulated in E. coli during a cold shock, other cold inducible genes have been identified including pnp (Jones et al., 1987); the encoded gene product, PNPase, has been shown to play a central role in conferring cold growth capabilities to E. coli, the yersiniae, Salmonella enterica, Campylobacter jejuni, S. aureus and B. subtilis (Jones et al., 1987; Wang and Bechhofer, 1996; Goverde et al., 1998; Clements et al., 2002; Rosenzweig et al., 2005; 2007; Anderson and Dunman, 2009; Haddad et al., 2009). This is of medical significance, since several of the pathogenic yersiniae and C. jejuni are contracted via the oral fecal route, and the fact that these pathogens can persist and grow at refrigerated/cold temperatures poses health threats in the form of yersiniosis and C. jejuni‐induced gastroenteritis.
After cold shock, PNPase is specifically required for the degradation of unnecessary csp transcripts and resuming pre‐stress genetic programming in both E. coli and Yersinia enterocolitica (Goverde et al., 1998; Neuhaus et al., 2000; Yamanaka and Inouye, 2001; Polissi et al., 2003). Further, PNPase serves to degrade csp‐bound (trapped) ribosomes, which would otherwise be prevented from binding non‐cold induced transcripts at the conclusion of a cold shock period. To achieve this mechanistically, PNPase is believed to specifically target and degrade ribosome‐bound csp transcripts, thereby, facilitating ribosome recycling after extricating the captive ribosomes. In the aforementioned model, PNPase is required during the post‐cold shock phase, termed the acclimation phase, in both E. coli and Y. enterocolitica rather than during the cold shock itself (Neuhaus et al., 2000; Polissi et al., 2003). In fact, RNase E, together with PNPase, has also been suggested to play a role in the cold growth adaptation of Y. enterocolitica by virtue of its recognizing and cleaving AGUAA motifs (termed cold‐shock‐cut boxes) in csp transcripts (Neuhaus et al., 2003). In addition, it appeared that the catalytic activity of PNPase and, to a lesser extent, its RNA binding domains were required for conferring cold growth capabilities (Rosenzweig et al., 2005).
Interestingly, PNPase was recently shown to be involved in the oxidative stress response of E. coli. In fact, a PNPase‐deleted mutant exhibited reduced growth in the presence of H2O2 and accumulated oxidatively damaged RNA. These deficiencies were readily complemented by expressing PNPase‐encoding gene in trans. In addition, PNPase appeared to respond to oxidative stress in a degradosome‐independent fashion since an RNase E truncated variant that did not express the degradosome scaffolding region was unable to demonstrate compromised growth when exposed to H2O2 (Wu et al., 2009).
Taken together, PNPase plays a definitive role in several notable Gram‐negative pathogens' ability to grow and persist in the cold. This cold‐growth conferring property provides pathogens with a decisive edge when persisting on various surfaces prior to human infections, thus, forcing us to re‐evaluate low temperature as a growth‐retarding mechanism. Further, the ability of a pathogen to withstand oxidative stress and survive oxidative bursts of neutrophils and/or macrophages is essential for their survival, and PNPase, being required for such an oxidative stress response, could help a pathogen subvert the deleterious effects of host‐cell produced reactive oxygen species.
PNPase and S. enterica virulence
In addition to temperature (cold) and oxidative stress, PNPase has also been shown to be necessary for another stress response of bacterial pathogens, the host cell induced stress response (HCISR). In the aforementioned stress, host immune cells attempting to neutralize and eliminate the pathogen cause the bacteria to respond by expressing numerous anti‐host, virulence genes that enable the pathogen to persist. In S. enterica, PNPase was required for both acute infection as well as lethality in a murine model of infection, as evidenced by a PNPase‐deleted strain's 10‐fold decrease in virulence. However, despite exhibiting a slightly attenuated virulence, the S. enterica PNPase‐deleted strain appeared more effective at actively invading macrophages and generally displaying an increase in Salmonella type three secretion system (T3SS) transcript levels required for active phagocyte invasion (Clements et al., 2002). Further, PNPase was required for repressing Salmonella plasmid virulence gene (spv) transcript levels by acting through the spv activator/regulator spvR; the spv gene products are required for the intracellular survival of this bacterium (Ygberg et al., 2006). Perhaps in S. enterica, PNPase is acting as a molecular switch permitting the pathogen to choose either an acute or persistent infection program. Such decision making would clearly enhance the pathogen's chance for a successful infection, immune evasion, and potential transmission to additional hosts.
PNPase and its role played in yersiniae virulence and cold growth
Like the salmonellae, when the yersiniae are confronted/threatened by innate immune cells, they also experience a HCISR prompting a rapid genetic re‐programming whereby the T3SS needle tip presumably serves as the low intracellular calcium sensor and, together with the 37°C mammalian temperature, signals to de‐repress the T3SS gene arsenal that have anti‐host properties (Cornelis, 2002; Torruellas et al., 2005; Viboud and Bliska, 2005). Following such an encounter (should the bacteria be successful), the yersiniae must resume pre‐stress gene expression profiles, thereby requiring the removal of unwanted and unnecessary T3SS transcripts.
Earlier studies revealed that PNPase was required for cold growth of all three pathogenic yersiniae (Goverde et al., 1998; Neuhaus et al., 2000; Rosenzweig et al., 2005). It appeared that the catalytic activity of PNPase and, to a lesser extent, its RNA binding domain were required for conferring cold growth capabilities (Rosenzweig et al., 2005). Interestingly, PNPase was also found to be needed for optimal activity of the yersiniae T3SS injectisome; however, the requirement of PNPase was independent of its catalytic activity. Instead, only the S1 RNA binding domain of PNPase appeared necessary for optimal T3SS function in cell culture infection assays (Rosenzweig et al., 2005; 2007; Rosenzweig and Schesser, 2007). Ultimately in the yersiniae, PNPase appears to play a role in at least two independent stress responses through distinct mechanisms. Further, the S1 RNA binding domain and the catalytic domain of PNPase mediate unique stress responses in manners independent of one another.
To corroborate the cell culture infections, two independent murine infections were performed in which a Y. psuedotuberculosis pnp‐deleted mutant exhibited a 50‐fold increased LD50 in a BALB/c oragastric model of infection (Rosenzweig et al., 2007). In addition, by using an intraperitoneal (IP) route of infection in Swiss Webster mice, a Y. pestis pnp‐deleted mutant exhibited greatly attenuated virulence as measured by 80% mouse survival when infected with 1 × 107 colony‐forming units (cfu) compared with 100% mortality of mice infected with the corresponding WT strain at the same dose (Fig. 2). Ultimately, PNPase appears to modulate the virulence of S. enterica and the yersiniae in two distinct manners by promoting acute infection in the former while enhancing the T3SS function of the latter.
Figure 2.
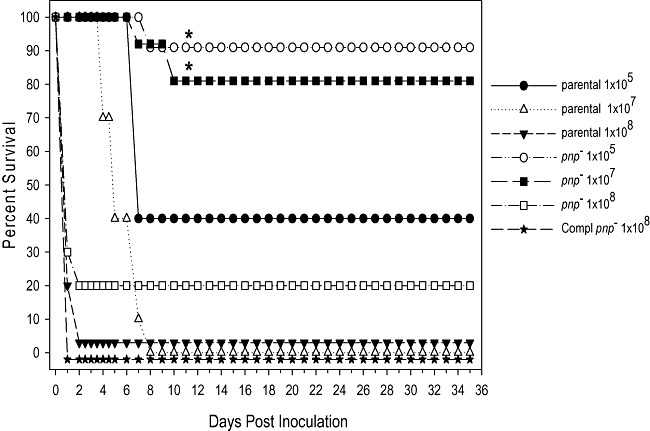
Attenuated virulence of the KIM/D27 Y. pestis pnp deletion mutant in a murine model of infection. Groups of 10 adult, female Swiss Webster mice were infected with a range of infectious doses (105–108) of either Y. pestis KIM/D27 parental (Lindler et al., 1990) or a derived pnp‐deleted mutant (Rosenzweig et al., 2005) strain via the IP route. Additionally, the complemented (compl) pnp mutant strain, in which the pKAK 7‐ plasmid encoded pnp gene was provided in trans, was used at an infectious dose of 1.0 × 108. Per cent survival post infection was monitored and is graphically represented. Yersinia pestis strains were grown overnight in heart infusion broth (Gibco) and the appropriate dilutions were made to achieve the desired infectious dose. Mice were observed up to a period of 35 days post infection. Asterisks represent statistical significance (P < 0.05) as determined by the Kaplan–Meier's survival estimates.
The role played by PNPase in Dichelobacter nodosus virulence and in the pathogenic streptococci
Interestingly, PNPase does not limit itself to involvement with the virulence of human pathogens exclusively. Recently, PNPase was found to repress the virulence of the Gram‐negative, anaerobic, animal pathogen D. nodosus that causes foot rot in sheep. Different isolates of this pathogen result in varying degrees of disease, and PNPase was found to be directly involved in the modulation of virulence. The most virulent isolates exhibited a characteristic phenotype of increased twitching motility. PNPase activity was found to be reduced in the aforementioned isolates. Further, a carboxy‐terminal deleted PNPase variant lacking the S1 RNA binding domain was compromised in its enzymatic activity. More importantly, the S1‐deleted PNPase induced enhanced twitching motility in less virulent/benign isolates (Palanisamy et al., 2010). Previously, it was shown that the PNPase S1 domain was required for optimal virulence of the yersiniae (Rosenzweig et al., 2005; 2007), but in the case of D. nodosus, PNPase acting through its S1 RNA binding domain appears to repress virulence. Thus, PNPase regulates virulence in multiple Gram‐negative pathogens, and that its function varies greatly within the various bacterial species to best accommodate each of their pathogenic strategies.
Interestingly, PNPase potentially mediates the virulence of both S. pyogenes (a group A Streptococci), and S. mutans (the etiological agent of dental caries). In S. pyogenes, PNPase was found to target specific virulence factor encoding transcripts, sagA (encoding a streptolysin precursor) and sda (a DNase that protects S. pyogenes from neutrophilic attack). The two aforementioned transcripts accumulated during late exponential phase but not during stationary phase in the PNPase‐deleted mutant, suggesting a growth phase‐specific role of PNPase in modulating the virulence of a formidable Gram‐positive pathogen (Barnett et al., 2007). Additionally, the proteome of S. mutans was evaluated after an acid shock, and PNPase was found to be upregulated ∼ 4.3‐fold after the pH was shifted from 7.0 to 5.0, suggesting that PNPase might be responsive to other stresses experienced in the human mouth in addition to pH stress (Len et al., 2004).
The diverse roles played by PNPase in modulating/enhancing the virulence of pathogenic bacteria as well as in coping with various environmental stresses (e.g. temperature, oxidative and pH/acid shock) emphasize the importance of this highly conserved exoribonuclease. The multi‐faceted roles played by PNPase make it an attractive protein to study in various model organisms, but the exact mechanism(s) by which PNPase modulates the virulence of the pathogens described above remains largely unknown. Does PNPase bind RNA molecules though its S1 RNA binding domain and then target them for destruction, or through S1 binding do the targeted transcripts protected from degradation? Is PNPase influencing bacterial virulence in a degradosome‐independent or degradosome‐dependent fashion? To date, an exhaustive list of all the target RNA molecules for any one of the above listed pathogens remains undetermined. Perhaps, PNPase targets small regulatory non‐coding RNA molecules that influence the transcript levels of their targeted transcripts. In considering these scenarios, it is clear that beyond describing the phenomenonology of PNPase influencing a pathogen's virulence little is known beyond that.
RNase R as a stress response ribonuclease and virulence‐associated gene product
The role played by RNase R in cold shock/growth
In addition to csp genes and pnp being cold inducible in E. coli, vacB/rnr, which encodes RNase R, was also found to be cold‐inducible and was needed for maturation of SsrA/tmRNA which serves to rescue stalled ribosomes by serving as both a messenger RNA as well as a transfer RNA (Cairrão et al., 2003). However, vacB/rnr is not cold inducible in all organisms. For example, in the psychrotroph P. syringae, vacB/rnr was not cold‐inducible despite RNase R being essential for the organism's cold growth, and the RNase R‐deleted mutant's cold growth deficiency could not be rescued by the overexpression of plasmid‐encoded pnp. Further, in the same P. syringae RNase R‐deleted mutant, impaired processing/maturation of 16 and 5S ribosomal RNA was also observed (Purusharth et al., 2007). These findings suggest that despite having some functional redundancy in gross level RNA decay, each ribonuclease has specific functions and might act in unique stress responses in individual bacterial species.
More recently, RNase R was also found to be essential for 4°C cold‐growth of the human diarrheal pathogen Aeromonas hydrophila; however, growth at the standard 37°C temperature was not impaired (Erova et al., 2008) further demonstrating that PNPase is not the sole ribonuclease required for the cold‐shock response/cold growth of all bacterial pathogens. Additionally, the RNase R encoding gene vacB/rnr in the human pathogen Helicobacter pylori (the causative agent of peptic ulcers) exhibited a 1.7‐fold increase in abundance during a cold shock exposure of 22°C (Tsao et al., 2009). Both RNase R and the cold‐inducible DEAD box helicase CsdA, observed in the E. coli cold shock degradosome (Généreux et al., 2004), were identified as being required for cold growth of the pathogen Legionella pneumophila in a random mutagenesis experiment (Söderberg and Cianciotto, 2010). This is of medical significance since L. pneumophila is an environmental pathogen that survives the cold environment of tainted air conditioners to then cause pneumonia (Legionnaires disease). As was seen with PNPase, RNase R appears to also be cold inducible and essential for growth of several pathogens at cold temperatures. For A. hydrophila and L. pneumophila, RNase R could play an essential role in enabling these pathogens to survive fluctuations in environmental temperatures prior to their introduction into warm hospitable human hosts.
Shigella flexneri, enteroinvasive E. coli and RNase R
In addition to allowing pathogens to withstand colder ambient or refrigerated temperatures prior to infection, RNase R, like PNPase, has also been clearly demonstrated to modulate the virulence of several pathogens. In fact, the first report of a ribonuclease being implicated in bacterial virulence was the gene product encoded by vacB/rnr, later found to be RNase R (Tobe et al., 1992; Cheng et al., 1998). Production of S. flexneri's virulence factors, including IpaB, C and D (invasion associated antigens for the colonic epithelial cells) as well as VirG (a virulence gene product required for intracellular Shigella to recruit host F‐actin), appeared to be dependent upon the presence of the vacB/rnr gene product RNase R (Tobe et al., 1992). The decreased virulence protein levels in the vacB/rnr mutant strain were due to post‐transcriptional modifications since transcript levels of the virulence genes themselves were not significantly different than levels found in the corresponding WT strain. Furthermore, the vacB/rnr mutant strain suffered attenuated virulence as evidenced by decreased invasion of and decreased spreading to neighbouring host cells (Tobe et al., 1992). Interestingly, a vacB/rnr‐deleted enteroinvasive E. coli (EIEC) strain was engineered and similarly suffered attenuated virulence marked by a 90% decrease in host cell invasion (Tobe et al., 1992). To this end, biochemical evaluation of the catalytic activities of RNase R and the other RNase II family member ribonuclease, RNase II, were evaluated in S. Typhimurium and were found to behave in ways similar to their E. coli counterparts. Further, the sole RNase II family member ribonuclease, RNase R in the Gram‐positive pathogen S. pnuemoniae was isolated and, like the E. coli RNase R, was also able to degrade structured RNA molecules (Domingues et al., 2009). Although an important first step in elucidating the mechanism of RNA degradation in these bacterial pathogens, neither RNase II nor RNase R was reported as being necessary for virulence in either of the two aforementioned organisms.
Brucella abortus, Y. pestis, H. pylori and A. hydrophila: the vacB/rnr story thickens
The vacB/rnr‐encoded RNase R has more recently been implicated as playing a role in the pathogenesis of the gastroenteritis causing bacterial pathogen A. hydrophila. This pathogen also leads to septicemic infections in animals and humans. An RNase R‐deficient A. hydrophila mutant was found to be less motile than the corresponding WT strain which could, in part, have contributed to the attenuated virulence observed in a murine infection of Swiss‐Webster mice. Using a lethal dose of 3 × 107 cfu in an IP challenge, 70% of the animals infected with the RNase R‐deficient mutant survived from day 4 until day 16 when the experiment was terminated. This was in sharp contrast to what was observed for both the WT and vacB/rnr complemented strains where by day 3 only 10% and 20% of the animals had survived respectively (Erova et al., 2008). Interestingly, RNase R in H. pylori promoted apoptosis of host cells while enhancing bacterial motility and chemotaxis, all of these features are required for optimal virulence (Tsao et al., 2009).
However, RNase R is not required for virulence in all pathogens. Unlike in A. hydrophila, S. flexneri, H. pylori and EIEC, RNase R was found to be dispensable for B. abortus virulence. Brucella abortus is a zoonotic pathogen found in cattle reservoirs. Expression levels of virulence B operon, virB, genes that encode a type 4 secretion system (essential for intracellular survival) were not significantly different between the RNase R‐deficient mutant and the corresponding WT strain. Further, in a murine model of infection using interferon regulatory factor‐1 knockout BALB/c mice, there was no significant difference in either survival kinetics or number of surviving mice. By day 28, all mice were killed by both strains, while at day 21, all WT‐infected mice had died and two mice infected with the RNase R‐deficient mutant had survived (Miyoshi et al., 2007).
In Y. pestis, an RNase R‐deleted mutant strain was evaluated using various cell culture infection assays. Mirroring what was observed with the B. abortus, the RNase R‐deleted Y. pestis mutant's ability to survive and proliferate in co‐culture with mouse macrophages was indistinguishable from that of the WT strain and showed no signs of attenuation in virulence (Rosenzweig et al., 2005)
Unlike PNPase, which appears to modulate the virulence of all pathogens tested to date in some way, RNase R appears to only play a role in the virulence of several pathogens while not affecting the virulence of either Y. pestis or B. abortus. Considering this RNase R inconsistency among pathogens, perhaps PNPase serves as the global regulator of virulence across prokaryotes despite RNase R being the first identified ribonuclease required for bacterial virulence.
RNase E as a virulence‐associated gene product
The role played by RNase E in yersiniae and salmonellae virulence
Interestingly, in addition to exoribonucleases, several endoribonucleases have also been demonstrated to influence bacterial virulence. For example, RNase E has been shown to play a role in the modulation of yersiniae virulence, suggesting that the yersiniae cope with the aforementioned HCISR in a degradosome‐dependent fashion. RNase E is essential for Gram‐negative bacterial viability. As a result, an RNase E‐deleted mutant strain cannot be engineered. However, using an ectopically expressed, carboxy‐terminal truncated, 465 amino acid residue long RNase E variant (RNase E1‐465– that can no longer form a degradosome), the role of RNase E's C‐terminal can be studied. Production of RNase E1‐465 in Y. pseudotuberculosis resulted in a dominant‐negative effect in which there was a loss of RNase E autoregulation and, more importantly, resulted in attenuated virulence as evidenced by reduced bacterial proliferation during a cell culture infection assay. Further, the resulting attenuation in virulence was shown to be a result of a suboptimally functioning T3SS (Yang et al., 2008). Overall, the general pattern of T3SS dysfunction mirrored the dysfunction observed in the PNPase‐deficient yersiniae (Rosenzweig et al., 2005; 2007). In fact, RNase E was found to co‐purify with PNPase in an immunoprecipitation assay (Yang et al., 2008). This strongly suggests that the yersiniae can potentially respond to the HCISR through the degradosome; however, more experiments need to be carried out confirming the suggestion, including evaluation of the PNPase‐deficient mutant expressing and producing the truncated RNase E1‐465 variant.
Additionally, in S. enterica RNase E was also found to modulate virulence. After the antisense RNA molecule, AmgR, binds to the mgtC portion of the mgtCBR polycistron, RNase E degrades the mgtC transcript (which encodes MgtC, an inner membrane protein important for intracellular survival within macrophages and virulence in a murine model of infection). In an rne temperature‐sensitive mutant (in which the encoded gene product RNase E is greatly reduced at 42°C), levels of mgtC were elevated at various time points tested, and the RNase E‐dependent degradation of the mgtC transcript required the non‐coding regulatory AmgR RNA molecule. This represents one of the first reports elucidating the mechanism by which a ribonuclease, in this case the endoribonuclease RNase E, modulates virulence through the degradation of a particular target transcript and the requirement of a non‐coding antisense regulatory molecule in the process (Lee and Groisman, 2010). In fact, the role played by several of the major RNases (RNase E, G, III and PNPase) in the targeted degradation of four highly conserved small, non‐coding regulatory RNA molecules (CsrB, CsrC, MicA and SraL) in S. Typhimurium has been evaluated. It appeared that the RNase E degradosome and PNPase were required for the decay of all four of the aforementioned small RNA regulatory molecules (Viegas et al., 2007).
RNase III as a virulence‐associated gene product
The role played by RNase III in S. aureus virulence
Importantly, another endoribonuclease has been demonstrated to modulate virulence in a Gram‐positive organism. In S. aureus, RNase III was found to degrade the staphylococcal protein A (SpA) encoding transcript, spa, after the non‐coding regulatory RNAIII RNA molecule binds to the sense spa transcript. As a virulence‐associated protein, SpA typically binds to tumour necrosis factor receptor I and promotes inflammation (Gómez et al., 2004). The initial binding of RNAIII to spa blocks translation initiation; however, after RNase III is recruited, efficient degradation of the transcript ensues eliminating the possibility of spa gene expression (Huntzinger et al., 2005).
Likewise, RNAIII was also found to bind in an antisense manner to a class of virulence factor encoding genes including: a gene encoding a fibrinogen binding protein as well as the staphylocoagulase precursor (both of which are required for an early S. aureus infection) leading to RNase III‐mediated degradation. Additionally, RNAIII was also found to bind to rot in an antisense manner, thereby preventing the expression of Rot, a transcriptional regulatory protein that promotes expression of virulence‐associated genes in S. aureus (Boisset et al., 2007). However, can the S. aureus RNase III function in a manner similar to the better characterized E. coli RNase III? That question was answered when S. aureus RNase III was found to degrade RNA‐mRNA duplexes mirroring the biochemical properties of E. coli RNase III (Chevalier et al., 2008).
The fact that endoribonucleases RNase E and RNase III are emerging as modulators of bacterial virulence suggests that perhaps even other, yet to be characterized ribonucleases, might also play roles in bacterial virulence. Researchers should not restrict their studies to the more ‘usual suspects’ (including RNase R and PNPase) when assessing the roles played by ribonucleases on bacterial virulence; perhaps there remains an as‐of‐yet‐undiscovered world of ribonucleases that influence bacterial pathogenesis.
CvfA as a virulence‐associated gene product
The role played by CvfA in S. pyogenes virulence
As alluded to above, one such novel and putative ribonuclease has been recently implicated as a virulence‐associated factor in S. pyogenes. CvfA, a putative RNase with a KH RNA binding domain that controls various exo‐protein expression in S. pyogenes (including haemolysin), was identified as being required for virulence in a silk worm model of infection (Nagata et al., 2008). Further, growth of a CvfA‐deleted S. pyogenes mutant to stationary phase in carbohydrate poor medium resulted in differentially expressed virulence genes, including streptolysin, streptokinase and SpeB (streptococcal pyrogenic exotoxin); additionally, the CvfA‐deleted S. pyogenes mutant was attenuated in a murine model of infection (Kang et al., 2010).
Taken together, we are learning appreciably more about RNA decay in Gram‐positive organisms, and several studies are addressing the question of whether RNases play a role in these pathogens. However, compared with Gram‐negative organisms, relatively little is known and additional Gram‐positive research on RNA metabolism, RNase activities and RNase influence on virulence needs to be conducted.
RNases and their therapeutic applications
Anti‐bacterial therapeutic application of RNase treatment
Since RNases are being reported as necessary for optimum virulence of multiple pathogens, such bacterial ribonucleases could serve as putative targets for the development of novel antibacterial and anti‐viral therapeutics (Table 3). Additionally, anti‐bacterial human RNases are found on our skin, so further exploitation of these naturally occurring compounds and their inclusion in various hand sanitizers and/or lotions could be a possibility. One such candidate for commercial exploitation is RNase 7. Recently, RNase 7, a member of the RNase A super‐family found in abundance on healthy human skin, was shown to contribute to the innate first line of defence against the Gram‐negative pathogen Enterococcus faecium. Surprisingly, the RNase 7‐specific anti‐E. faecium activity was found to be independent of its catalytic/enzymatic activity. RNase 7 is secreted by keratinocytes at levels and locations on the skin that vary from person to person explaining why the presence of E. faecium is at low levels in the perianal folds (Köten et al., 2009). This finding suggests that RNase 7 could be applied topically as an E. faecium treatment, or that multiple RNases could be applied as a broad spectrum anti‐septic to treat antibiotic resistant cutaneous infections.
Table 3.
List of ribonucleases and their therapeutic applications.
| Ribonuclease | Therapeutic role/potential role | Reference |
|---|---|---|
| hPNPaseold35 | Prevents tumour cell division; drives tumour cells to apoptosis and senescence | Leszczyniecka et al. (2003) |
| Successfully used as a neuroblastoma therapy | Van Maerken et al. (2009) | |
| Onoconase | In clinical trials as an anti‐tumour therapy; promotes apoptosis of tumour cells | Ardelt et al. (2009) |
| RNase 7 | Selectively inhibits Enterococcus faecium on human keratinocytes/skin application | Köten et al. (2009) |
| Dicer like enzymes/RNase III | Antiviral properties in fungal, plant and protozoan models | Sun et al. (2009); Lewsey and Carr (2009); Patrick et al. (2009) |
Dicer/RNaseIII as a eukaryotic viral defence mechanism
In addition to anti‐bacterial roles, two recent reports have indicated a role played by the RNase III eukaryotic homologue Dicer in plant/fungal anti‐viral defence. In the case of chestnut blight fungus Cryphonectria parasitica, dcl2, which encodes Dicer, is regulated by an Argonaute‐like protein Agl2, which allows dcl2 transcript levels to accumulate to high levels. The physiological consequence is antisense silencing of viral transcripts promoting viral integration and fungal host cell survival. Interestingly, viruses have developed a countermeasure in a protein repressor that blocks agl2 promoter activity reducing the agl2 transcript level (Sun et al., 2009). Therefore, it is important to consider viral escape mutants that could acquire resistance to novel RNase‐based therapeutics, and that such cases of resistance could occur more frequently in the more rapidly dividing and error prone viruses than in pathogenic prokaryotes.
Additionally, Arabidopsis thaliana was used as a model organism to determine whether DICER‐like endoribonucleases 2, 3 and 4 were required for the plant's antiviral defence when salicylic acid (which independently induces anti‐viral defences in plants) was used as a pre‐treatment prior to infection with two positive‐sense RNA viruses. Interestingly, in the absence of the DICER proteins, silencing effects and plant antiviral defences were still enabled when salicylic acid was used as a treatment (Lewsey and Carr, 2009). This suggests that perhaps RNase‐based therapies should be coupled with additional compounds possessing known anti‐microbial effects to further combat any cases of resistance to the novel therapies.
Further, DICER‐like enzymes have been shown to patrol the cytoplasm of the eukaryotic human pathogen Trypanasoma brucei (etiological agent of African sleeping sickness) targeting any foreign invading nucleic acids, including viral nucleic acid, thereby, protecting the pathogen (Patrick et al., 2009). Ultimately, anti‐viral/phage roles played by bacterial RNases need to be elucidated in efforts of enhancing anti‐bacterial phage therapies. Through a simultaneous anti‐sense silencing of ribonuclease encoding transcripts identified as being necessary for anti‐phage defences, phage therapy could be greatly augmented providing a therapeutic alternative to the increasingly less effective conventional antibiotics in today's post‐antibiotic era.
Future trend challenges and spin offs
Many bacterial pathogens are exposed to temperature stress and, in order for the pathogen to survive, ribonucleases are required to help modulate the cold‐shock transcriptome during the cold shock, during the acclimation phase, or during the post‐stress reprogramming needed to resume growth under the normal growing temperature. In this vein, both PNPase and RNase R have been shown to either confer cold‐growth or are themselves cold‐inducible in several formidable pathogens. It is important to recall that in order for bacterial virulence to be experienced by a host, the pathogen must survive long enough in the environment to remain viable upon host encounter. For pathogens contracted via the oral–fecal route, survival at refrigerated or near‐refrigerated temperatures in contaminated food products is an essential component of the pathogen's strategy. It appears that ribonucleases empower the pathogen to cope with and adjust to changing climatic conditions at the transcript level. Such growth plasticity is proving to be a serious challenge, and in addition to developing better growth‐control methods for food pathogens, perhaps novel therapeutics that target RNA decay machinery could yield a novel class of anti‐bacterial compounds.
However, ribonucleases appear to respond to a plethora of stresses and are not limited to temperature stress alone. Similar to oxidative, osmolarity, antibiotic and pH‐induced stress, the host cell‐induced stress response can be construed as another type of environmental stress wherein poised phagocytic cells (in the context of the host‐pathogen immune response) trigger bacterial stress signals resulting in the upregulation of virulence‐associated genes. In the case of Gram‐negative enteric pathogens, T3SS genes are rapidly transcribed, and their encoded protein anti‐host factors/proteins are then injected into the targeted host cell through the T3SS injectisome. During this frenzied transcriptional response, PNPase and/or RNase R seemingly play regulatory roles for a subset of transcripts as in the case of the S. flexneri, the yersiniae and S. enterica (Tobe et al., 1992; Clements et al., 2002; Ygberg et al., 2006; Rosenzweig et al., 2007). Alternatively, the aforementioned ribonucleases could directly degrade specific transcripts thereby freeing stalled ribosomes allowing the bacteria to resume ‘normal’ genetic programming post HCISR, as in the case of the cold shock response of E. coli (Jones et al., 1987; Polissi et al., 2003).
Further, endoribonucleases, like RNase E and RNase III, have been shown to modulate the virulence of S. Typhmurium and S. aureus, respectively, in conjunction with the binding of small non‐coding anti‐sense RNA molecules to the mRNA targets (Huntzinger et al., 2005; Boisset et al., 2007; Lee and Groisman, 2010). However, exhaustive studies characterizing potential roles played by the most predominant ribonucleases in the virulence of all notable Gram‐negative and Gram‐positive pathogens have yet to be carried out. For example, to date, there have been no reports of RNase II being a virulence‐associated gene despite being the most abundant hydrolytic exoribonuclease. In addition, new ribonucleases are being discovered and putative ribonucleases, like CvfA in S. pyogenes (Kang et al., 2010), are being implicated in virulence. Current thinking about how ribonucleases respond to stress (including the HCISR) becomes even more ambiguous when considering any potential contributions made by the large multi‐protein complex, the degradosome. More specifically, it remains unclear as to whether the degradosome directly contributes to the HCISR of bacterial pathogens, or whether degradosome constituents function independently in a synchronized degradosome‐ independent fashion.
In as much as we continue to learn about novel small, non‐coding RNAs that serve regulatory functions by working in concert with ribonucleases postrancriptionally, we are reminded of how little we actually know about the complex nature of RNA decay as it relates to changing environmental stresses in Gram‐negative organisms. However, relative to knowledge about Gram‐negative RNA decay and established roles of ribonucleases in virulence, we know even less about Gram‐positive organisms. B. subtilis and S. aureus have been the most characterized with regards to both function of their ribonucleases and their roles in virulence in the case of the latter. Interestingly, RNase II appears to be absent in both of the aforementioned bacteria; however, PNPase and RNAse R homologues are present and fairly well conserved. Further, RNase E, the essential endoribonuclease found in multiple Gram‐negative organisms, is also absent in both B. subtilis and S. aureus (Anderson and Dunman, 2009). However, unique endoribonucleases, RNase J1 and J2 (not found in E. coli), carry out functions similar to RNase E in B. subtilis (Even et al., 2005; Mäder et al., 2008) and are also found in S. aureus and the streptococci.
Ultimately, our understanding of RNA metabolism and how it impacts bacterial virulence is in its infancy with the least known about how RNases impact the virulence of Gram‐positive pathogens. This exciting new dimension deepens our appreciation of the many regulated networks controlling expression of bacterial virulence genes. Perhaps, one of the most exciting possibilities lies in the development of a potential class of novel antimicrobial therapeutics. In our current post‐antibiotic‐era crisis, RNases administered as either stand‐alone treatments or administered in cocktail with other classes of antimicrobial compounds (including anti‐sense RNAs) offers a new hope in combating multiple drug resistant bacterial and viral pathogens as well as treating recalcitrant tumours as anti‐cancer agents.
Acknowledgments
We would like to thank Greg Plano (University of Miami) for his generosity in sharing the Yersinia pestis wild‐type and pnp mutant strains. Also, we would like to thank Ambro van Hoof (University of Texas Health Science Center Houston), Kurt Schesser (Univerity of Miami), Chaitanya Jain (University of Miami) and Murray Deutscher (University of Miami) for stimulating conversation. We would also like to thank Shishir Shishodia, Hector Miranda and Ayodotun Sodipe (Texas Southern University) for their insight. We thank Jian Sha, Elena Kozlova, Sheri Foltz and Michelle Kirtley (University of Texas Medical Branch Galveston) for performing murine infections with Y. pestis and its pnp mutant. Work on this manuscript was supported by the National Aeronautics and Space Administration (NASA) cooperative agreement NNX08B4A47A and NIH/NIAID AI064389 and N01 AI30065 grants awarded to AKC.
References
- Anderson K.L., Dunman P.M. Messenger RNA turnover processes in Escherichia coli, Bacillus subtilis, and emerging studies in Staphylococcus aureus. Int J Microbiol. 2009;2009:525491. doi: 10.1155/2009/525491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardelt W., Ardelt B., Darzynkiewicz Z. Ribonucleases as potential modalities in anticancer therapy. Eur J Pharmacol. 2009;625:181–189. doi: 10.1016/j.ejphar.2009.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett T.C., Bugrysheva J.V., Scott J.R. Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus. J Bacteriol. 2007;189:1866–1873. doi: 10.1128/JB.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum E., Py B., Carpousis A.J., Higgins C.F. Polyphosphate kinase is a component of the Escherichia coli RNA degradosome. Mol Microbiol. 1997;26:387–398. doi: 10.1046/j.1365-2958.1997.5901947.x. [DOI] [PubMed] [Google Scholar]
- Boisset S., Geissmann T., Huntzinger E., Fechter P., Bendridi N., Possedko M. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva J.V., Scott J.R. The ribonucleases J1 and J2 are essential for growth and have independent roles in mRNA decay in Streptococcus pyogenes. Mol Microbiol. 2010;75:731–743. doi: 10.1111/j.1365-2958.2009.07012.x. [DOI] [PubMed] [Google Scholar]
- Cairrão F., Cruz A., Mori H., Arraiano C.M. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol Microbiol. 2003;50:1349–1360. doi: 10.1046/j.1365-2958.2003.03766.x. [DOI] [PubMed] [Google Scholar]
- Callaghan A.J., Marcaida M.J., Stead J.A., McDowall K.J., Scott W.G., Luisi B.F. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- Carpousis A.J. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem Soc Trans. 2002;30:150–155. [PubMed] [Google Scholar]
- Carpousis A.J., Van Houwe G., Ehretsmann C., Krisch H.M. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Cheng Z.F., Zuo Y., Li Z., Rudd K.E., Deutscher M.P. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- Chevalier C., Huntzinger E., Fechter P., Boisset S., Vandenesch F., Romby P., Geissmann T. Staphylococcus aureus endoribonuclease III purification and properties. Methods Enzymol. 2008;447:309–327. doi: 10.1016/S0076-6879(08)02216-7. [DOI] [PubMed] [Google Scholar]
- Clements M.O., Eriksson S., Thompson A., Lucchini S., Hinton J.C., Normark S., Rhen M. Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc Natl Acad Sci USA. 2002;99:8784–8789. doi: 10.1073/pnas.132047099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G.R. Yersinia type III secretion: send in the effectors. J Cell Biol. 2002;158:401–408. doi: 10.1083/jcb.200205077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M.P., Li Z. Exoribonucleases and their multiple roles in RNA metabolism. Prog Nucleic Acid Res Mol Biol. 2001;66:67–105. doi: 10.1016/s0079-6603(00)66027-0. [DOI] [PubMed] [Google Scholar]
- Domingues S., Matos R.G., Reis F.P., Fialho A.M., Barbas A., Arraiano C.M. Biochemical characterization of the RNase II family of exoribonucleases from the human pathogens Salmonella typhimurium and Streptococcus pneumoniae. Biochemistry. 2009;48:11848–11857. doi: 10.1021/bi901105n. [DOI] [PubMed] [Google Scholar]
- Durán‐Figueroa N.V., Piña‐Escobedo A., Schroeder I., Simons R.W., García‐Mena J. Polynucleotide phosphorylase interacts with ribonuclease E through a betabetaalphabetabetaalpha domain. Biochimie. 2006;88:725–735. doi: 10.1016/j.biochi.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Erova T.E., Kosykh V.G., Fadl A.A., Sha J., Horneman A.J., Chopra A.K. Cold shock exoribonuclease R (VacB) is involved in Aeromonas hydrophila pathogenesis. J Bacteriol. 2008;190:3467–3474. doi: 10.1128/JB.00075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even S., Pellegrini O., Zig L., Labas V., Vinh J., Bréchemmier‐Baey D., Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Généreux C., Dehareng D., Devreese B., Van Beeumen J., Frère J.M., Joris B. Mutational analysis of the catalytic centre of the Citrobacter freundii AmpD N‐acetylmuramyl‐L‐alanine amidase. Biochem J. 2004;377:111–120. doi: 10.1042/BJ20030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J., Pollitt N.S., Inouye M. Major cold shock protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez M.I., Lee A., Reddy B., Muir A., Soong G., Pitt A. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat Med. 2004;10:842–848. doi: 10.1038/nm1079. et al. [DOI] [PubMed] [Google Scholar]
- Goverde R.L., Huis in't Veld J.H., Kusters J.G., Mooi F.R. The psychrotrophic bacterium Yersinia enterocolitica requires expression of pnp, the gene for polynucleotide phosphorylase, for growth at low temperature (5 degrees C) Mol Microbiol. 1998;28:555–569. doi: 10.1046/j.1365-2958.1998.00816.x. [DOI] [PubMed] [Google Scholar]
- Grunberg‐Manago M., Ortiz P.J., Ochoa S. Enzymatic synthesis of nucleic acid like polynucleotides. Science. 1955;122:907–910. doi: 10.1126/science.122.3176.907. [DOI] [PubMed] [Google Scholar]
- Guarneros G., Portier C. Different specificities of ribonuclease II and polynucleotide phosphorylase in 3’mRNA decay. Biochimie. 1990;72:771–777. doi: 10.1016/0300-9084(90)90186-k. [DOI] [PubMed] [Google Scholar]
- Haddad N., Burns C.M., Bolla J.M., Prévost H., Fédérighi M., Drider D., Cappelier J.M. Long‐term survival of Campylobacter jejuni at low temperatures is dependent on polynucleotide phosphorylase activity. Appl Environ Microbiol. 2009;75:7310–7318. doi: 10.1128/AEM.01366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E., Boisset S., Saveanu C., Benito Y., Geissmann T., Namane A. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain C., Belasco J.G. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: unusual sensitivity of the rne transcript to RNase E activity. Genes Dev. 1995;9:84–96. doi: 10.1101/gad.9.1.84. [DOI] [PubMed] [Google Scholar]
- Jarrige A., Bréchemier‐Baey D., Mathy N., Duché O., Portier C. Mutational analysis of polynucleotide phosphorylase from Escherichia coli. J Mol Biol. 2002;321:397–409. doi: 10.1016/s0022-2836(02)00645-9. [DOI] [PubMed] [Google Scholar]
- Jarrige A.C., Mathy N., Portier C. PNPase autocontrols its expression by degrading a double‐stranded structure in the pnp mRNA leader. EMBO J. 2001;20:6845–6855. doi: 10.1093/emboj/20.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Diwa A., Belasco J.G. Regions of RNase E important for 5′‐end‐dependent RNA cleavage and autoregulated synthesis. J Bacteriol. 2000;182:2468–2475. doi: 10.1128/jb.182.9.2468-2475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.G., VanBogelen R.A., Neidhardt F.C. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol. 1987;169:2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.O., Caparon M.G., Cho K.H. Virulence gene regulation by CvfA, a putative RNase: the CvfA‐enolase complex in Streptococcus pyogenes links nutritional stress, growth‐phase control, and virulence gene expression. Infect Immun. 2010;78:2754–2767. doi: 10.1128/IAI.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai T., Gupta R.S., Schlessinger D. Exoribonucleases in wild type Escherichia coli and RNase II‐deficient mutants. J Biol Chem. 1977;252:8950–8956. [PubMed] [Google Scholar]
- Khemici V., Carpousis A.J. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP‐stabilizers. Mol Microbiol. 2004;51:777–790. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- Kido M., Yamanaka K., Mitani T., Niki H., Ogura T., Hiraga S. RNase E polypeptides lacking a carboxyl‐terminal half suppress a mukB mutation in Escherichia coli. J Bacteriol. 1996;178:3917–3925. doi: 10.1128/jb.178.13.3917-3925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köten B., Simanski M., Gläser R., Podschun R., Schröder J.M., Harder J. RNase 7 contributes to the cutaneous defense against Enterococcus faecium. PLoS ONE. 2009;4:e6424. doi: 10.1371/journal.pone.0006424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner S.R. mRNA decay in Escherichia coli comes of age. J Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.J., Groisman E.A. An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol Microbiol. 2010;76:1020–1033. doi: 10.1111/j.1365-2958.2010.07161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Len A.C., Harty D.W., Jacques N.A. Stress‐responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology. 2004;150:1339–1351. doi: 10.1099/mic.0.27008-0. [DOI] [PubMed] [Google Scholar]
- Leszczyniecka M., Kang D.C., Sarkar D., Su Z.Z., Holmes M., Valerie K., Fisher P.B. Identification and cloning of human polynucleotide phosphorylase, hPNPase old‐35, in the context of terminal differentiation and cellular senescence. Proc Natl Acad Sci USA. 2002;99:16636–16641. doi: 10.1073/pnas.252643699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka M., Su Z.Z., Kang D.C., Sarkar D., Fisher P.B. Expression regulation and genomic organization of human polynucleotide phosphorylase, hPNPase(old‐35), a Type I interferon inducible early response gene. Gene. 2003;316:143–156. doi: 10.1016/s0378-1119(03)00752-2. [DOI] [PubMed] [Google Scholar]
- Lewsey M.G., Carr J.P. Effects of DICER‐like proteins 2, 3 and 4 on cucumber mosaic virus and tobacco mosaic virus infections in salicylic acid‐treated plants. J Gen Virol. 2009;90:3010–3014. doi: 10.1099/vir.0.014555-0. [DOI] [PubMed] [Google Scholar]
- Lin P.H., Lin‐Chao S. RhlB helicase rather than enolase is the beta‐subunit of the Escherichia coli polynucleotide phosphorylase (PNPase)‐exoribonucleolytic complex. Proc Natl Acad Sci USA. 2005;102:16590–16595. doi: 10.1073/pnas.0500994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindler L.E., Klempner M.S., Straley S.C. Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect Immun. 1990;58:2569–2577. doi: 10.1128/iai.58.8.2569-2577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäder U., Zig L., Kretschmer J., Homuth G., Putzer H. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol Microbiol. 2008;70:183–196. doi: 10.1111/j.1365-2958.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- Miczak A., Kaberdin V.R., Wei C.L., Lin‐Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′‐>5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Miyoshi A., Rosinha G.M., Camargo I.L., Trant C.M., Cardoso F.C., Azevedo V., Oliveira S.C. The role of the vacB gene in the pathogenesis of Brucella abortus. Microbes Infect. 2007;9:375–381. doi: 10.1016/j.micinf.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Mudd E.A., Higgins C.F. Escherichia coli endoribonuclease RNase E: autoregulation of expression and site‐specific cleavage of mRNA. Mol Microbiol. 1993;9:557–568. doi: 10.1111/j.1365-2958.1993.tb01716.x. [DOI] [PubMed] [Google Scholar]
- Nagata M., Kaito C., Sekimizu K. Phosphodiesterase activity of CvfA is required for virulence in Staphylococcus aureus. J Biol Chem. 2008;283:2176–2184. doi: 10.1074/jbc.M705309200. [DOI] [PubMed] [Google Scholar]
- Neuhaus K., Rapposch S., Francis K.P., Scherer S. Restart of exponential growth of cold‐shocked Yersinia enterocolitica occurs after down‐regulation of cspA1/A2 mRNA. J Bacteriol. 2000;182:3285–3288. doi: 10.1128/jb.182.11.3285-3288.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus K., Anastasov N., Kaberdin V., Francis K.P., Miller V.L., Scherer S. The AGUAAA motif in cspA1/A2 mRNA is important for adaptation of Yersinia enterocolitica to grow at low temperature. Mol Microbiol. 2003;50:1629–1645. doi: 10.1046/j.1365-2958.2003.03795.x. [DOI] [PubMed] [Google Scholar]
- Nikolaev N., Folsom V., Schlessinger D. Escherichia coli mutants deficient in exoribonucleases. Biochem Biophys Res Commun. 1976;70:920–924. doi: 10.1016/0006-291x(76)90679-3. [DOI] [PubMed] [Google Scholar]
- Ono M., Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J Mol Biol. 1979;129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- Palanisamy S.K., Fletcher C., Tanjung L., Katz M.E., Cheetham B.F. Deletion of the C‐terminus of polynucleotide phosphorylase increases twitching motility, a virulence characteristic of the anaerobic bacterial pathogen Dichelobacter nodosus. FEMS Microbiol Lett. 2010;302:39–45. doi: 10.1111/j.1574-6968.2009.01831.x. [DOI] [PubMed] [Google Scholar]
- Patrick K.L., Shi H., Kolev N.G., Ersfeld K., Tschudi C., Ullu E. Distinct and overlapping roles for two Dicer‐like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc Natl Acad Sci USA. 2009;106:17933–17938. doi: 10.1073/pnas.0907766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polissi A., De Laurentis W., Zangrossi S., Briani F., Longhi V., Pesole G., Dehò G. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res Microbiol. 2003;154:573–580. doi: 10.1016/S0923-2508(03)00167-0. [DOI] [PubMed] [Google Scholar]
- Portier C. Quaternary structure of polynucleotide phosphorylase from Escherichia coli: evidence of a complex between two types of polypeptide chains. Eur J Biochem. 1975;55:573–582. doi: 10.1111/j.1432-1033.1975.tb02194.x. [DOI] [PubMed] [Google Scholar]
- Purusharth R.I., Klein F., Sulthana S., Jäger S., Jagannadham M.V., Evguenieva‐Hackenberg E. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J Biol Chem. 2005;280:14572–14578. doi: 10.1074/jbc.M413507200. et al. [DOI] [PubMed] [Google Scholar]
- Purusharth R.I., Madhuri B., Ray M.K. Exoribonuclease R in Pseudomonas syringae is essential for growth at low temperature and plays a novel role in the 3′ end processing of 16 and 5 S ribosomal RNA. J Biol Chem. 2007;282:16267–16277. doi: 10.1074/jbc.M605588200. [DOI] [PubMed] [Google Scholar]
- Rosenzweig J.A., Schesser K. Polynucleotide phosphorylase and the T3SS. Adv Exp Med Biol. 2007;603:217–224. doi: 10.1007/978-0-387-72124-8_19. [DOI] [PubMed] [Google Scholar]
- Rosenzweig J.A., Weltman G., Plano G.V., Schesser K. Modulation of yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J Biol Chem. 2005;280:156–163. doi: 10.1074/jbc.M405662200. [DOI] [PubMed] [Google Scholar]
- Rosenzweig J.A., Chromy B., Echeverry A., Yang J., Adkins B., Plano G.V. Polynucleotide phosphorylase independently controls virulence factor expression levels and export in Yersinia spp. FEMS Microbiol Lett. 2007;270:255–264. doi: 10.1111/j.1574-6968.2007.00689.x. et al. [DOI] [PubMed] [Google Scholar]
- Schaeffer D., Tsanova B., Barbas A., Reis F.P., Dastidar E.G., Sanchez‐Rotunno M. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2008;16:56–62. doi: 10.1038/nsmb.1528. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg M.A., Cianciotto N.P. Mediators of lipid A modification, RNA degradation, and central intermediary metabolism facilitate the growth of Legionella pneumophila at low temperatures. Curr Microbiol. 2010;60:59–65. doi: 10.1007/s00284-009-9502-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa S., Marchand I., Dreyfus M. Autoregulation allows Escherichia coli RNase E to adjust continuously its synthesis to that of its substrates. Mol Microbiol. 2001;42:867–878. doi: 10.1046/j.1365-2958.2001.02687.x. [DOI] [PubMed] [Google Scholar]
- Sun Q., Choi G.H., Nuss D.L. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc Natl Acad Sci USA. 2009;106:17927–17932. doi: 10.1073/pnas.0907552106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe T., Sasakawa C., Okada N., Honma Y., Yoshikawa M. vacB, a novel chromosomal gene required for expression of virulence genes on the large plasmid of Shigella flexneri. J Bacteriol. 1992;174:6359–6367. doi: 10.1128/jb.174.20.6359-6367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torruellas J., Jackson M.W., Pennock J.W., Plano G.V. The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol Microbiol. 2005;57:1719–1733. doi: 10.1111/j.1365-2958.2005.04790.x. [DOI] [PubMed] [Google Scholar]
- Tsao M.Y., Lin T.L., Hsieh P.F., Wang J.T. The 3′‐to‐5′ exoribonuclease (encoded by HP1248) of Helicobacter pylori regulates motility and apoptosis‐inducing genes. J Bacteriol. 2009;191:2691–2702. doi: 10.1128/JB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umitsuki G., Wachi M., Takada A., Hikichi T., Nagai K. Involvement of RNase G in in vivo mRNA metabolism in Escherichia coli. Genes Cells. 2001;6:403–410. doi: 10.1046/j.1365-2443.2001.00430.x. [DOI] [PubMed] [Google Scholar]
- Van Maerken T., Sarkar D., Speleman F., Dent P., Weiss W.A., Fisher P.B. Adenovirus‐mediated hPNPase(old‐35) gene transfer as a therapeutic strategy for neuroblastoma. J Cell Physiol. 2009;219:707–715. doi: 10.1002/jcp.21719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo N.F., Li Y.S., Py B., Blum E., Higgins C.F., Raynal L.C. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud G.I., Bliska J.B. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu Rev Microbiol. 2005;59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- Viegas S.C., Pfeiffer V., Sittka A., Silva I.J., Vogel J., Arraiano C.M. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–7664. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Bechhofer D.H. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J Bacteriol. 1996;178:2375–2382. doi: 10.1128/jb.178.8.2375-2382.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Jiang Z., Liu M., Gong X., Wu S., Burns C.M., Li Z. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry. 2009;48:2012–2020. doi: 10.1021/bi801752p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Inouye M. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J Bacteriol. 2001;183:2808–2816. doi: 10.1128/JB.183.9.2808-2816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Jain C., Schesser K. RNase E regulates the Yersinia type 3 secretion system. J Bacteriol. 2008;190:3774–3778. doi: 10.1128/JB.00147-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ygberg S.E., Clements M.O., Rytkönen A., Thompson A., Holden D.W., Hinton J.C., Rhen M. Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect Immun. 2006;74:1243–1254. doi: 10.1128/IAI.74.2.1243-1254.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


