Abstract
Lipomannan (LM) and lipoarabinomannan (LAM) are key Corynebacterineae glycoconjugates that are integral components of the mycobacterial cell wall, and are potent immunomodulators during infection. LAM is a complex heteropolysaccharide synthesized by an array of essential glycosyltransferase family C (GT-C) members, which represent potential drug targets. Herein, we have identified and characterized two open reading frames from Corynebacterium glutamicum that encode for putative GT-Cs. Deletion of NCgl2100 and NCgl2097 in C. glutamicum demonstrated their role in the biosynthesis of the branching α(1→2)-Manp residues found in LM and LAM. In addition, utilizing a chemically defined nonasaccharide acceptor, azidoethyl 6-O-benzyl-α-D-mannopyranosyl-(1→6)-[α-D-mannopyranosyl-(1→6)]7-D-mannopyranoside, and the glycosyl donor C50-polyprenol-phosphate-[14C]-mannose with membranes prepared from different C. glutamicum mutant strains, we have shown that both NCgl2100 and NCgl2097 encode for novel α(1→2)-mannopyranosyltransferases, which we have termed MptC and MptD respectively. Complementation studies and in vitro assays also identified Rv2181 as a homologue of Cg-MptC in Mycobacterium tuberculosis. Finally, we investigated the ability of LM and LAM from C. glutamicum, and C. glutamicumΔmptC and C. glutamicumΔmptD mutants, to activate Toll-like receptor 2. Overall, our study enhances our understanding of complex lipoglycan biosynthesis in Corynebacterineae and sheds further light on the structural and functional relationship of these classes of polysaccharides.
Introduction
The Corynebacterineae, a sub-order of the Actinobacteria, represent an unusual group within Gram-positive bacteria, with a distinctive cell wall architecture. Prominent members are the human pathogens, Corynebacterium diphtheriae, Mycobacterium tuberculosis and Mycobacterium leprae (Bloom and Murray, 1992). Typically, the cell walls of Corynebacterineae contain mycolic acids (m), arabinogalactan (AG) and peptidoglycan (P), which are covalently linked to each other to form the mycolyl-arabinogalactan-peptidoglycan (mAGP) complex (Daffe et al., 1990; McNeil et al., 1990; 1991; Besra et al., 1995; 1997; Brennan, 2003; Dover et al., 2004). In addition, specialized glycophospholipids, phosphatidyl-myo-inositol mannosides (PIMs) and lipoglycans, lipomannan (LM) and lipoarabinomannan (LAM), are found in the outer leaflet of mAGP (Hill and Ballou, 1966; Brennan and Ballou, 1967; 1968; Brennan and Nikaido, 1995; Besra et al., 1997; Morita et al., 2004). The non-pathogenic bacillus, Corynebacterium glutamicum, also belongs to this class of bacteria, and is widely used in the industrial production of amino acids (Eggeling and Bott, 2005). Use of this easily cultivatable bacterium, together with the fact that the cell walls of Corynebacterineae share identical basic structures and building blocks, have contributed significantly to decipher the biosynthesis of their complex cell walls. For instance, in both C. glutamicum and M. tuberculosis the orthologous acyl-CoA carboxylase genes (Gande et al., 2007) together with Pks (Gande et al., 2004), are key to mycolic acid biosynthesis, and the glycosyltransferases AftA (Alderwick et al., 2006a), and AftB (Seidel et al., 2007a) introduce specific arabinofuranose (Araf) residues into AG, and AftC in both AG and LAM of Mycobacterium smegmatis (Birch et al., 2008; 2010;). A speciation-specific difference is the presence of the glycosyltransferase RptA in C. glutamicum that introduces decorating rhamnose residues into AG (Birch et al., 2009), whereas a structurally very similar protein introduces galactosamine residues into AG of M. tuberculosis (Skovierova et al., 2010).
A particular interesting group of cell wall components are the lipoglycans LM and LAM that are based on PIMs. They play poorly defined roles in Corynebacterineae but mycobacterial LAM has been implicated in many of the key aspects of the pathogenesis of tuberculosis and leprosy, such as induction of phagocytosis, phagosomal alteration and acquired T cell-mediated immunity (Briken et al., 2004). Mycobacterial LAM has attached to the PIM base, an elongated α(1→6) linear, α(1→2) branched mannan ‘core’, of approximately 30 mannopyranose (Manp) residues, and linked to its terminus a branched d-arabinan domain. In mycobacteria, this large arabinan domain consists of approximately 70 arabinofuranose (Araf) residues and is assembled through several distinct motifs (Chatterjee et al., 1991; Alderwick et al., 2007; Birch et al., 2010), formed via specific α(1→5), α(1→3) and β(1→2) arabinofuranosyltransferases (Besra et al., 1997; Zhang et al., 2003; Alderwick et al., 2005; Birch et al., 2010), and can be capped to various degrees with either short α(1→2) mannopyranosyl chains, as is the case in M. tuberculosis (Chatterjee et al., 1992), or in M. smegmatis by inositol phosphate (Khoo et al., 1995), or completely devoid of caps as found in Mycobacterium chelonae (Guerardel et al., 2002). There is very limited structural information on LAM in other Corynebacterineae, except for C. glutamicum (Tatituri et al., 2007a,b;). In this organism the core α(1→6) linear α(1→2) branched LM (see Fig. 9), which is structurally akin to mycobacterial LM, is substituted by t-Araf residues to form a modified LAM-like molecule (Tatituri et al., 2007a,b;).
Fig. 9.
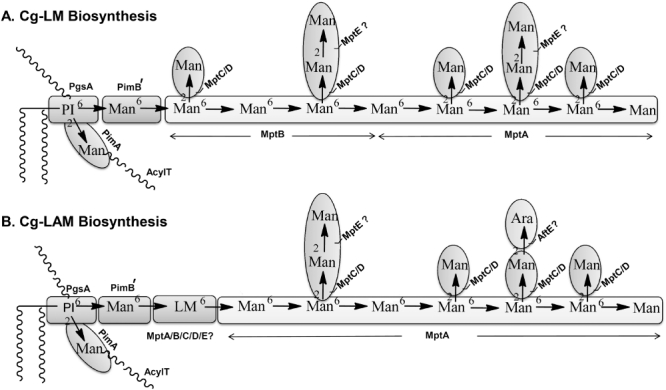
Biosynthetic pathway of LM (A) and LAM (B) biogenesis in Corynebacterineae. Cg-LM biosynthesis initially involves the formation of the α(1→6) mannan backbone by MptB, followed by α(1→2)-Manp branching by MptC (or MptD). This biosynthetic precursor would then be the substrate for further α(1→6)-Manp elongation by MptA, followed by α(1→2)-Manp branching by MptD (or MptC) to afford initially Cg-LM (A). Further extension by MptA along with additional GT-C glycosyltransferases further modify the α(1→6)-mannan backbone. These GT-Cs include MptC/D and MptE (formation of the dimannoside side-chain), and AftE/MptC/D and formation of Araf-Manp side-chains (B).
The current model of lipoglycan biosynthesis supported by biochemical and genetic studies, follows a linear pathway from PI→PIM2→LM→LAM (Besra and Brennan, 1997; Besra et al., 1997). As with AG synthesis selected enzymatic and structural features are in part shared within the Corynebacterineae. Glycosylation of phosphatidyl-myo-inositol (PI) by different α-mannopyranosyltransferases and acylation by acyltransferase(s) results in the synthesis of mono- and di-acylated PIMs (Kordulakova et al., 2002; 2003; Kremer et al., 2002; Morita et al., 2006; Lea-Smith et al., 2008; Mishra et al., 2009). Previous studies have identified the enzymes involved in the early and late stages of LM and LAM biosynthesis in Corynebacterineae. For instance, NCgl1505/Rv1459c (MptB) and NCgl2093/Rv2174 (MptA), synthesize the proximal and distal α(1→6) mannan backbone in LM and LAM (Kaur et al., 2007; Mishra et al., 2007; 2008a;). However, the enzymatic steps involved in the biosynthesis of mannan branching of LM and LAM, still remain to be studied in detail in Corynebacterineae (Kaur et al., 2006; 2008; Sena et al., 2010).
In the current study we have examined two open reading frames from C. glutamicum, which encode for putative glycosyltransferase family C (GT-C) members (Liu and Mushegian, 2003). On the basis of mutant studies, chemical and enzymatic data we report that NCgl2100 (Cg-MptC) and its M. tuberculosis homologue Rv2181 function as α(1→2)-mannopyranosyltransferases, and NCgl2097 functions as a second α(1→2)-mannopyranosyltransferase (Cg-MptD), both of which are required for complete mannose-branching found in LM and LAM in Corynebacterineae.
Results
Genome locus and structural features of MptC and MptD
We previously identified the α(1→6)-mannopyranosyltransferases, MptA and MptB, which catalyse the transfer of Manp residues from the glycosyl donor C50-polyprenol-phosphate mannose, to the distal and proximal ends of the mannan backbone of LM and LAM respectively (Mishra et al., 2007; 2008a;). In search of further GT-Cs, which synthesize the elaborated structures of LM and LAM, we interrogated the genome of C. glutamicum for additional GT-C family members (Liu and Mushegian, 2003). Located within a 16 kb genomic region containing mptA, which has been demonstrated to encode an α(1→2)-mannopyranosyltransferase in M. tuberculosis (Kaur et al., 2008), as does its orthologue MSMEG_4247 in M. smegmatis (Kaur et al., 2006; Sena et al., 2010), are two additional GT-Cs (NCgl2100 [Cg-mptC], and NCgl2097 [Cg-mptD]) (Fig. 1A). In both Corynebacterium and Mycobacterium additional genes are retained at this locus: a serine/threonine protein kinase (pknL), hypothetical proteins (NCgl2099, Rv2179c; NCgl2094, Rv2175c) and a 3-deoxy-7-phosphoheptulonate synthase (aroG) (Fig. 1A).
Fig. 1.
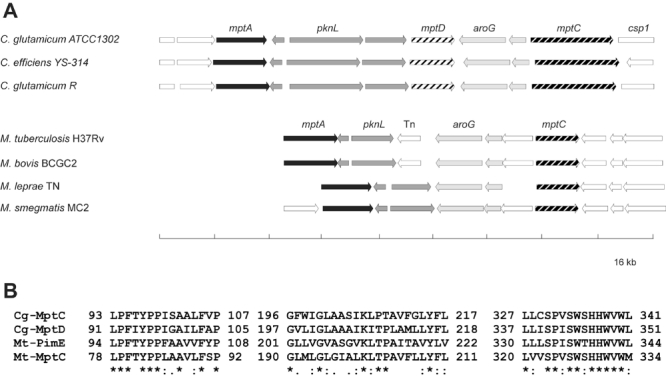
Comparative gene relatedness of α(1→2) and α(1→6) mannopyranosyltransferases.
A. The genomic region in selected Corynebacterianeae containing the GT-C mannosyltransferases MptA, MptC and MptD.
B. Partial sequence alignment of the strongly related α(1→2) mannosyltransferases Cg-MptC, Cg-MptD, Mt-PimE and Mt-MptC illustrating conserved residues.
The Cg-mptC encoded protein is a long hydrophobic polytopic membrane protein of 812 amino acid (aa) residues. The first half of the protein (1–417 aa), shares 37% identity (55% similarity) with Mt-MptC and 38% identity (57% similarity) to Cg-MptD, whereas the second half (417–812 aa), has no counterpart in various Mycobacterium species. The protein appears to be a fusion of two membrane proteins, functional as an α(1→2)-mannopyranosyltransferase as described below, and termed in this study Cg-MptC. Cg-MptD is a hydrophobic polytopic membrane protein of 436 amino acid residues with 11 transmembrane helixes (TMH). After TMH-1 and TMH-7, larger loop regions are present probably localized in the periplasm. Cg-MptC, Cg-MptD and Mt-MptC, together with Mt-PimE, an α(1→2)-mannopyranosyltransferase demonstrated to be involved in PIM6 synthesis (Morita et al., 2006), share a similar size, degree of hydrophobicity and regions of high identity (Fig. 1B), suggesting closely related functions.
Construction of deletion mutants and growth phenotype
The construct pK19mobsacBΔCg-mptC was made containing 12 nucleotides (nt) of the 3′ end of NCgl2100 together with the genomic upstream sequences, and 36 nt of the 5′ end together with genomic downstream sequences (Table S1). This non-replicative vector was used to transform C. glutamicum to kanamycin-resistance (Kanr) indicating chromosomal integration. Sucrose-resistant (Sucr) clones were selected in a second round of positive selection, indicating loss of the vector-encoded sacB function (Schäfer et al., 1994). From 12 Kans and Sucr clones analysed via PCR, seven had lost NCgl2100, whereas in the remaining five the genomic wild type situation was restored. One clone with deleted NCgl2100 was selected and will be referred to as C. glutamicumΔmptC (Fig. S1) In an analogous manner, pK19mobsacBΔCg-mptD was constructed and used to select from the wild type of C. glutamicum by double cross-over events as described above for deletion of Cg-mptD, yielding C. glutamicumΔmptD (Fig. S1). Starting from C. glutamicumΔmptC and using pK19mobsacBΔCg-mptD the double deletion mutant C. glutamicumΔmptCΔmptD was also constructed (Fig. S1).
The mutants were inoculated into liquid BHI and CGXII medium and their growth patterns studied. There was no statistical difference observed in three independent cultivations for each of the strains. The growth rates in CGXII were 0.39 ± 0.02 h−1 for wild-type C. glutamicum, C. glutamicumΔmptC and C. glutamicumΔmptD, and 0.37 ± 0.02 h−1 for C. glutamicumΔmptCΔmptD (data not shown). The growth rates on BHI medium were 0.66 ± 0.07 h−1 for all three strains and illustrate that the genes are not essential for growth as observed for other GT-Cs involved in mAGP synthesis like, Cg-emb, Cg-aftA and Cg-aftB (Alderwick et al., 2005; 2006a; Seidel et al., 2007a).
Purification and general characteristic features of lipoglycans from C. glutamicumΔmptC and C. glutamicumΔmptD
Lipoglycans from wild-type C. glutamicum, C. glutamicumΔmptC and C. glutamicumΔmptD with complemented strains C. glutamicumΔmptC pEKEx2-Cg-mptC, C. glutamicumΔmptC pEKEx2-Rv2181, C. glutamicumΔmptD pEKEx2-Cg-mptD and C. glutamicumΔmptD pEKEx2-Rv2181, were extracted from delipidated cells by ethanol/water extraction followed by hot-phenol water treatment and enzymatic degradation. The crude lipoglycan extract was subjected to hydrophobic interaction and gel permeation chromatography allowing the separation of Cg-LM (Cg-LM-A and Cg-LM-B) and Cg-LAM (Tatituri et al., 2007b; Lea-Smith et al., 2008; Mishra et al., 2008b). Because the mannan backbone structures of Cg-LM-A and Cg-LM-B are similar as revealed through previous matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS), and gas chromatography-mass spectrometry (GC-MS) analyses of partially methylated alditol acetates (Mishra et al., 2008b), subsequent analyses focused on the Cg-LM as a mixture. The extracted lipoglycans were examined for their size and mobility on 15% SDS-PAGE (Fig. 2A). The crude extracts from wild-type C. glutamicum showed the presence of Cg-LM and Cg-LAM (Tatituri et al., 2007a; Lea-Smith et al., 2008; Mishra et al., 2008b) (Fig. 2A), while the crude lipoglycans from mutant strains, C. glutamicumΔmptC and C. glutamicumΔmptD, showed subtle differences in their migration on SDS-PAGE (Fig. 2A). Purified Cg-LAM from C. glutamicumΔmptC and C. glutamicumΔmptD (Fig. 2A) migrated faster on SDS-PAGE in comparison with those from wild-type C. glutamicum, indicating that these lipoglycans were smaller in size with low molecular weights. In addition, fractionation using gel permeation chromatography also revealed a degree of heterogeneity in Cg-LM and Cg-LAM, as evident when fractions were pooled. Complementation of C. glutamicumΔmptC and C. glutamicumΔmptD by pEKEx2-Cg-mptC and pEKEx2-Cg-mptD, respectively, restored the wild-type phenotype, as illustrated by the slower and co-migration on SDS-PAGE in comparison with wild-type C. glutamicum (Fig. 2A). In contrast, cross-complementation experiments, C. glutamicumΔmptC by pEKEx2-Cg-mptD, and C. glutamicumΔmptD by pEKEx2-Cg-mptC, failed to restore a wild-type phenotype and reversion to a slower migration of products on SDS-PAGE (Fig. S2), as observed in our previous experiments described above (Fig. 2A).
Fig. 2.
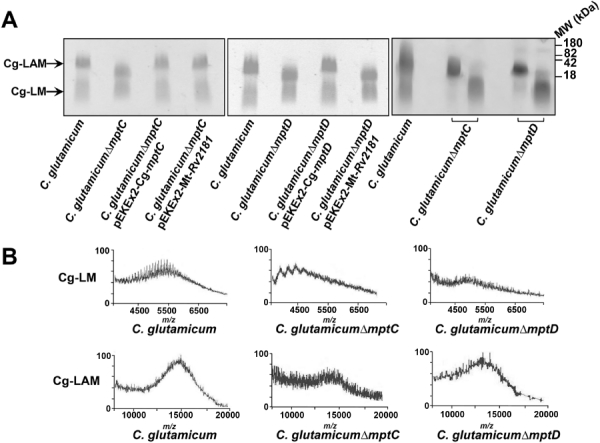
Lipoglycan profiles of wild-type and mutant strains of C. glutamicum.
A. The left-hand and middle panels illustrate lipoglycans extracted from wild-type, mutant and complemented strains analysed using SDS-PAGE and visualized using a Pro-Q emerald glycoprotein stain (Invitrogen). The right-hand panel illustrates purified Cg-LAM and Cg-LM from C. glutamicumΔmptC and C. glutamicumΔmptD, respectively, in comparison with wild-type C. glutamicum lipoglycans. Lipoglycan profiles are represented with standard molecular weight markers of glycoproteins of 180, 82, 42 and 18 kDa.
B. MALDI-TOF-MS spectra of Cg-LM and Cg-LAM from different strains of C. glutamicum. MALDI-TOF-MS spectra were acquired in the linear negative mode with delayed extraction using 2,5-dihydrobenzoic acid as a matrix.
The molecular weights of the lipoglycans were investigated by MALDI-TOF-MS. The Cg-LAM from wild-type C. glutamicum exhibited a broad unresolved peak centred at m/z 15 000 (approximately 85 Manp/Araf glycosyl residues), and Cg-LM at m/z 5500 (approximately 25 Manp residues), indicating a molecular weight of approximately 15 and 5.5 kDa for the major molecular species of these lipoglycans (Fig. 2B). Interestingly, Cg-LAM from C. glutamicumΔmptC (Fig. 2A) peaked at m/z 13 800 (approximately 78 Manp/Araf glycosyl residues), indicating a decrease of around 1.2 kDa for the Cg-LAM isolated from C. glutamicumΔmptC as compared with wild-type Cg-LAM (Fig. 2B). In addition, Cg-LM from C. glutamicumΔmptC (Fig. 2A) exhibited a broad unresolved peak centred at m/z 4400 (approximately 20 Manp glycosyl residues), indicating a molecular weight of approximately 4.4 kDa, a decrease of 1.1 kDa in comparison with wild-type C. glutamicum (Fig. 2B). Similarly, the lipoglycans, Cg-LM and Cg-LAM from C. glutamicumΔmptD (Fig. 2A) are smaller and centred around m/z 13 000 (approximately 73 Manp/Araf glycosyl residues), and 4700 (approximately 20 Manp glycosyl residues) respectively (Fig. 2B). The differences in size of Cg-LAM and Cg-LM from the mutant strains, suggest that there is a difference in a common component of both mutant strains, which also reflects the earlier observation on SDS-PAGE (Fig. 2A).
Glycosyl composition of purified lipoglycans from C. glutamicumΔmptC and C. glutamicumΔmptD
Purified lipoglycans from wild-type C. glutamicum, C. glutamicumΔmptC and C. glutamicumΔmptD were analysed as alditol acetates and their glycosyl composition was determined by gas chromatography (GC) (Fig. S3). GC analysis of alditol acetates prepared from wild-type Cg-LAM revealed a molar ratio of Araf : Manp of 0.47:1.0. The mutant Cg-LAM from C. glutamicumΔmptC and C. glutamicumΔmptD yielded a significant reduction in Manp content concomitant with a relative increase in the amount of Araf. The C. glutamicumΔmptC yielded a mutant Cg-LAM with an Araf : Manp ratio of 0.59:1.0, while Cg-LAM from C. glutamicumΔmptD, yielded an Araf : Manp ratio of 0.7:1.0. The data suggest that both Cg-MptC and Cg-MptD are involved in the synthesis of the mannan portion of Cg-LAM together with Cg-MptA and Cg-MptB (Mishra et al., 2007; 2008a;).
Glycosyl linkage of purified lipoglycans from C. glutamicumΔmptC and C. glutamicumΔmptD
The glycosyl linkages present in the lipoglycans from wild-type and mutant strains of C. glutamicum were analysed by GC-MS of per-O-methylated alditol acetate derivatives prepared from purified lipoglycans. Cg-LM from C. glutamicum indicated a normal profile of glycosidic linkages corresponding to t-Manp (49%), 6-Manp (15%), 2-Manp (6%) and 2,6-Manp (30%) (Fig. 3A). However, the relative ratios of different linkages in Cg-LM from C. glutamicumΔmptC[t-Manp (23%), 6-Manp (58%), 2-Manp (6%) and 2,6-Manp (13%)] and C. glutamicumΔmptD[(t-Manp (25%), 6-Manp (45%), 2-Manp (8%) and 2,6-Manp (22%)] indicated subtle changes in the mannan domain in Cg-LM from these strains consistent with the earlier MALDI-TOF-MS data indicating a reduction in size (Fig. 2B). The relative abundance of t-Manp and 2,6-Manp were reduced with a concomitant increase in the abundance of 6-Manp in Cg-LM from these strains (Fig. 3A). Similarly, per-O-methylated alditol acetate derivatives of Cg-LAM from these strains showed a net reduction in relative abundance of t-Manp and 2,6-Manp and increase in 6-Manp (Fig. 3B). For instance, Cg-LAM from C. glutamicum indicated a normal profile of glycosidic linkages corresponding to t-Araf (40%), t-Manp (14%), 6-Manp (6%), 2-Manp (11%) and 2,6-Manp (29%) (Fig. 3A). Whereas the relative ratios of different linkages in Cg-LAM from C. glutamicumΔmptC were t-Araf (40%), t-Manp (6%), 6-Manp (30%), 2-Manp (11%) and 2,6-Manp (13%) and C. glutamicumΔmptD were t-Araf (40%), t-Manp (8%), 6-Manp (26%), 2-Manp (12%) and 2,6-Manp (14%). The complementation of C. glutamicumΔmptC and C. glutamicumΔmptD with pEKEx2-Cg-mptC and pEKEx2-Cg-mptD, respectively, restored the wild-type phenotype in these strains (data not shown). Altogether, the data together suggest that Cg-LM and Cg-LAM from C. glutamicumΔmptC and C. glutamicumΔmptD have reduced 2,6-Manp branching residues indicating that MptC and MptD are involved in the synthesis of α(1→2)-Manp residues in Cg-LM and Cg-LAM in C. glutamicum.
Fig. 3.
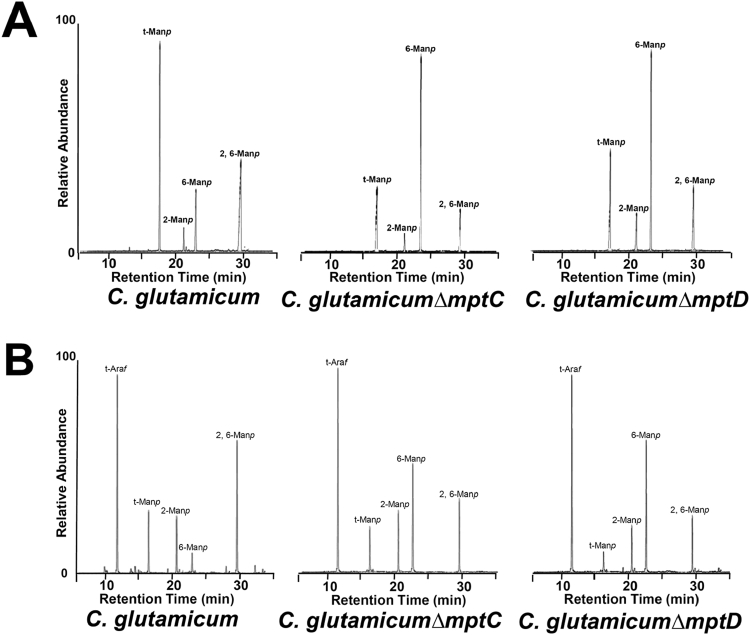
Glycosyl linkage analysis of Cg-LM and Cg-LAM from different strains of C. glutamicum.
A. Cg-LM per-O-methylated samples were hydrolysed using 2 M TFA, reduced and per-O-acetylated. The resulting partially per-O-methylated, per-O-acetylated alditol acetates were analysed using GC-MS.
B. Cg-LAM per-O-methylated samples were hydrolysed using 2 M TFA, reduced and per-O-acetylated. The resulting partially per-O-methylated, per-O-acetylated alditol acetates were analysed using GC-MS. The partially per-O-methylated, per-O-acetylated alditol acetates were identified based on their characteristic alditol cleavage acetal profiles: t-Araf (m/z 118, 161), t-Manp (m/z 118, 102, 145, 161, 162, 205), 2-Manp (m/z 129, 130, 161, 190), 6-Manp (m/z 118, 129, 162, 189, 233) and 2,6-Manp (m/z 129, 130, 189, 190).
Characterization of Cg-LAM from C. glutamicumΔmptCΔmptD
Because the data in Fig. 3A,B suggest that deletion of mptC or mptD results in only partial removal of 2,6-Manp branching residues, we constructed a C. glutamicumΔmptCΔmptD double mutant strain (as described earlier). The extracted lipoglycans were examined for their size and mobility on 15% SDS-PAGE (Fig. 4A). The crude lipoglycan extracts of C. glutamicumΔmptCΔmptD in comparison with wild-type C. glutamicum, C. glutamicumΔmptC and C. glutamicumΔmptD, migrated faster on SDS-PAGE, indicating that these lipoglycans were smaller in size (Fig. 4A). The purified Cg-LAM from C. glutamicumΔmptCΔmptD was analysed by 1H-13C-NMR HMQC (Fig. 4C) and compared with our previously reported 1H-13C-NMR HMQC (Fig. 4B) assignment of anomeric resonances of Cg-LAM from C. glutamicum (Tatituri et al., 2007b). Briefly, correlations at δH1C1 5.20/112.2 and 5.13/112.0 were attributed to t-α-Araf units; 5.06/105.2 to t-α-Manp units; 5.12/101.4, 5.07/10.1.7, and 5.04/101.9 to 2,6-α-Manp units; 4.93/102.6 to 6-α-Manp units; and 5.00/104.9 to 2-α-Manp units, which were consistent with our above glycosyl linkage data for Cg-LAM from C. glutamicum (Fig. 2B). In contrast, the 1H-13C-NMR HMQC (Fig. 4C) of Cg-LAM from C. glutamicumΔmptCΔmptD was dominated by a single anomeric resonance at δH1C1 4.92/102.5 and assigned to 6-α-Manp units, consistent with a complete removal of 2,6-α-Manp branching residues. GC-MS of per-O-methylated alditol acetate derivatives from Cg-LAM purified from C. glutamicumΔmptCΔmptD confirmed these results and the identification of 2,3,4-tri-O-CH3-1,5,6-tri-O-COCH3-mannitol with the characteristic fragment alditol cleavage ions m/z 102, 118, 129, 162, 189 and 233 (Fig. S4).
Fig. 4.

Lipoglycan analysis of C. glutamicumΔmptCΔmptD.
A. Lipoglycans were extracted from wild-type and mutant strains and were analysed using SDS-PAGE and visualized using a Pro-Q emerald glycoprotein stain (Invitrogen). Lipoglycan profiles are represented with standard molecular weight markers of glycoproteins of 180, 82, 42 and 18 kDa.
B. 2D 1H-13C-NMR HMQC characterization of Cg-LAM from C. glutamicum as described in Tatituri et al. (2007b).
C. 2D 1H-13C-NMR HMQC characterization of Cg-LAM from C. glutamicumΔmptCΔmptD. NMR spectra of Cg-LAMs were recorded in D2O at 313 K.
In vitroα(1→2)-mannopyranosyltransferase assay utilizing a novel blocked nonasaccharide acceptor
For characterization of α(1→2)-mannopyranosyltransferase activities of Cg-MptC and Cg-MptD a nonasaccharide acceptor, azidoethyl 6-O-benzyl-α-D-mannopyranosyl-(1→6)-[α-D-mannopyranosyl-(1→6)]7-D-mannopyranoside, Acc-Man9 (Fig. 5A, compound 1) was chemically synthesized (Fig. 5A,B). The synthesis of the target nonamannopyranoside 1 was planned starting from the previously synthesized 2 (Aqueel et al., 2008) via synthesis of oligosaccharide 3 (Fig. 5A). For the synthesis of 3, donor thioglycoside 4 was synthesized starting from the reported 5,6-O-benzylidene protected thioglycoside 5 (Cumpstey et al., 2004) (Fig. 5B). In a straight forward reaction sequence, thioglycoside 5 was treated with benzoyl chloride in pyridine to access the thioglycoside 6, which on regioselective ring opening with Et3SiH gave 6-OBn thioglycoside 7. The 1H NMR and 13C NMR spectral studies confirmed the structure of 7. An acetate protecting group was installed at the 4-OH position in thioglycoside 7 by reacting with acetic anhydride in pyridine resulting in the thioglycoside donor 4. An acetate group was specifically chosen to easily characterize the reaction product during coupling of 4 and 2 by NMR. The donor thioglycoside 4 and 2 were coupled in the presence of NIS and triflic acid at −20°C, resulting in nonasaccharide 3 after chromatographic purification. The observed singlet signal in the 1H NMR spectrum of 3 at δ 1.88 ppm was assigned to the three protons of the acetyl protecting group, and the resonances at δ 4.48 and 4.28 ppm (doublets, J = 11.9 Hz), for one proton each to the benzyl protecting group, supporting the structure of 3. The 13C NMR spectrum further confirmed the structure of 3, with the presence of signals at 73.45 and 20.81 ppm, which were assigned to the CH2 of the benzyl and CH3 of the acetyl carbons respectively. The ester protecting groups in oligosaccharide 3 were removed in an overnight reaction using sodium methoxide in CH3OH. The reaction was neutralized using H+ Amberlite resin and the reaction product purified using Bio-Beads SM-4 using a CH3OH-H2O mobile phase and 0–60% gradient resulting in the desired nonomannoside 1. The 1H NMR spectrum of 1 showed the presence of benzyl signals at δ 4.64 and 4.61 ppm (doublet, J = 11.7 Hz). In the 13C NMR spectrum, the benzyl carbon was observed at δ 73.32 ppm and CH2N3 at δ 51.79 ppm. Finally, the structure of 1 was further supported by HR-MS, which showed a [M + Na]+ peak at 1685.55.
Fig. 5.
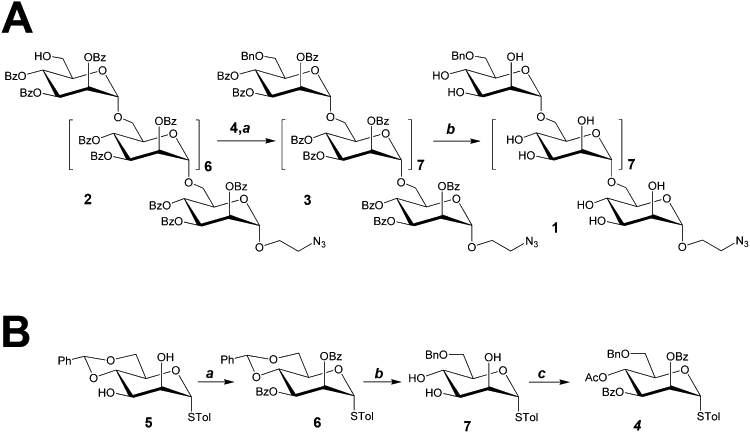
Chemical synthesis of the nonasaccharide acceptor azidoethyl
6-O-benzyl-α-D-mannopyranosyl-(1→6)-[α-D-mannopyranosyl-(1→6)]7-D-mannopyranoside (1) and key intermediate 4. Reagents and conditions.
A. (a) NIS, triflic acid, CH2Cl2, −20°C–0°C, 1 h, 82%; (b) Sodium methoxide, CHCl3-CH3OH (1:2), overnight, 35%;
B. (a) BzCl, pyridine, room temperature, 4 h, 94%; (b) Et3SiH, triflic acid, CH2Cl2, −78°C, 1 h, 80%; (c) acetic anydride, pyridine, room temperature, 5 h, 94%.
The structural features of the acceptor allowed its specific isolation and purification of in vitro reaction products synthesized by distinct α(1→2)-mannopyranosyltransferases using a strong anion exchange cartridge, thus removing from the in vitro assay unused C50-polyprenol-phospho-[14C]-mannose (PP-[14C]-M) and other in vitro synthesized products, such as higher [14C]-PIMs, [14C]-LM and [14C]-LAM. In addition, the non-reducing sugar of 1 (which we have abbreviated as Acc-Man9) is blocked at the O-6 position by a benzyl ether, thus preventing further elongation by endogenous activities through MptA and MptB via α(1→6)-mannopyranosyltransferases. Overall, the utilization of Acc-Man9 and the in vitro assay provides a novel route to evaluate distinct α(1→2)-mannopyranosyltransferases in Corynebacterineae (Fig. 6A). The in vitro transfer of [14C]-Manp from PP-[14C]-M onto the Acc-Man9 acceptor was examined using membrane preparations from wild-type and mutant strains of C. glutamicum (Fig. 6B). In this improved assay, membrane preparations from wild-type C. glutamicum transferred 3072 ± 132 cpm of [14C]-Manp from PP-[14C]M onto the synthetic Acc-Man9 via an α(1→2)-mannopyranosyltransferase (Fig. 6B and inset). Assays repeated in the absence of Acc-Man9 afforded background activity 102 ± 12 cpm, demonstrating the efficiency of the assay (Fig. 6B). Membrane preparations from both C. glutamicumΔmptC and C. glutamicumΔmptD used in above assays afforded α(1→2)-mannopyranosyltransferase activities, but in each case the relative level of activity in comparison with wild-type C. glutamicum membranes was reduced (Fig. 6B).
Fig. 6.
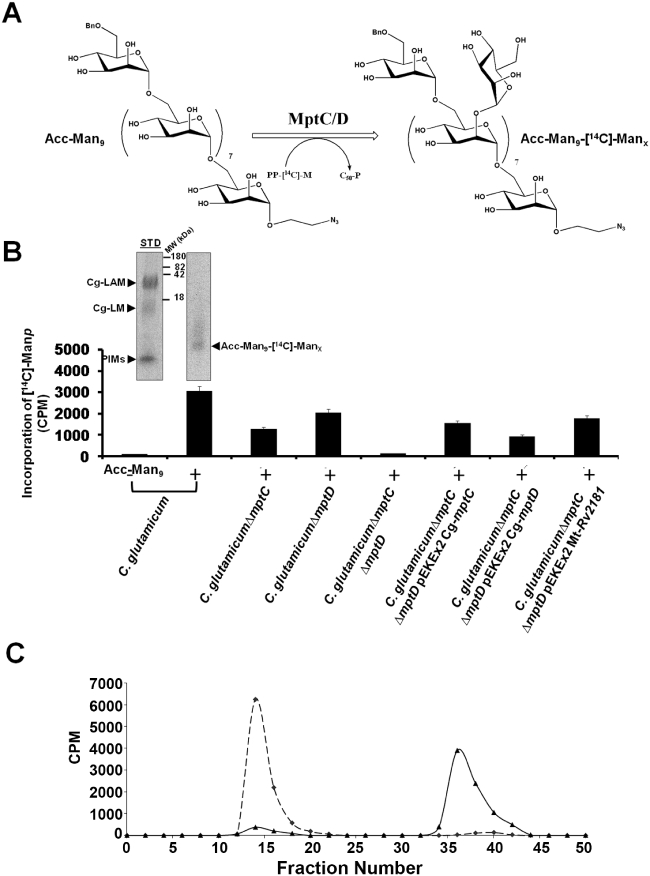
In vitroα(1→2)-mannopyranosyltransferase assay utilizing a nonasaccharide acceptor.
A. Reaction scheme illustrating in vitro assays performed with the chemically synthesized nonasaccharide acceptor, azidoethyl 6-O-benzyl-α-D-mannopyranosyl-(1→6)-[α-D-mannopyranosyl-(1→6)]7-D-mannopyranoside, Acc-Man9, and either MptC or MptD, resulting in the synthesis of Acc-Man9-[14C]-Manx.
B. The reactions were set up with and without Acc-Man9 (+ or −), PP-[14C]-M, and membrane preparations from different strains of C. glutamicum as described in the Experimental procedures.
C. The Acc-Man9-[14C]-Manx product from the use of C. glutamicum membranes was dried and subsequently incubated with an α(1→2)-mannosidase. The reaction mixture was then fractionated on a Bio-Gel P-2 gel filtration column. The control Acc-Man9-[14C]-Manx (dotted line) product before α(1→2)-D-mannosidase treatment eluted from the Bio-Gel P-2 column at fractions 11–13, and the digested product, released [14C]-Man (solid line), was retained and eluted in later fractions 33–39, co-eluting with a [14C]-mannose standard (Cooper et al., 2002).
The analysis of [14C] reaction products was simplified further by utilizing membrane preparations from the branching deficient double mutant, C. glutamicumΔmptCΔmptD, lacking Cg-MptC and Cg-MptD. The C. glutamicumΔmptCΔmptD mutant possessed background (149 ± 15 cpm) incorporation of [14C]-Manp (Fig. 6B), demonstrating the complete absence of α(1→2)-mannopyranosyltransferase activity in this double mutant. Therefore, the double mutant, C. glutamicumΔmptCΔmptD was used to analyse the enzymatic activities of Cg-MptC and Cg-MptD. Cg-mptC and Cg-mptD were cloned into pEKEx2 and introduced into C. glutamicumΔmptCΔmptD resulting in C. glutamicumΔmptCΔmptD pEKEx2-Cg-mptC and C. glutamicumΔmptCΔmptD pEKEx2-Cg-mptD. C. glutamicumΔmptCΔmptD pEKEx2-Cg-mptC resulted in 1570 ± 83 cpm of Acc-Man9-[14C]-Manx, while C. glutamicumΔmptCΔmptD pEKEx2-Cg-mptD resulted in 953 ± 24 cpm of Acc-Man9-[14C]-Manx.
Acc-Man9-[14C]-Manx selective cleavage by an α(1→2)-D-mannosidase
The extracted Acc-Man9-[14C]-Manx reaction product from the above assays performed with C. glutamicum membranes (Fig. 6B) were dried and subsequently treated with a previously characterized α(1→2)-D-mannosidase from Trichoderma reesei (Maras et al., 2000) as described (Ichishima et al., 1981; Yokoyama and Ballou, 1989) to determine the α(1→2) addition of Manp. The digested mixtures were fractionated on a gel filtration column. The control Acc-Man9-[14C]-Manx reaction product, before α(1→2)-D-mannosidase treatment eluted from the Bio-Gel P-2 column at fractions 13–16, and this completely enzymatically digested product (measured through the release of [14C]-Manp), was retained and eluted in later fractions 35–42 with a [14C]-mannose standard (Fig. 6C; Cooper et al., 2002). The results demonstrate that the addition of [14C]-Manp to Acc-Man9 is via an α(1→2)-mannopyranosyltransferase, and combined with the data from Fig. 6B and the use of the C. glutamicumΔmptCΔmptD mutant strain, has shown that MptC and MptD are solely responsible for this α(1→2)-mannopyranosyltransferase activity.
Functional identification of the mycobacterial homologue of Cg-MptC
As described above (Fig. 1), Cg-MptC showed highest similarity of 55% to Rv2181 of M. tuberculosis, while Cg-MptD showed weaker, but significant similarity of 47.9% to Rv2181. To assay for a complementary function of Rv2181 in C. glutamicum, plasmid pEKEx2-Mt-Rv2181 was constructed. Upon introduction into C. glutamicumΔmptC, the wild-type phenotype of C. glutamicum was restored (Fig. 2A) illustrating that Mt-MptC is functional and utilizes the corynebacterial substrates. In addition, Rv2181 was very specific for C. glutamicumΔmptC as complementation of the mutant phenotype in C. glutamicumΔmptD was not obtained (Fig. 2A). The complementation of C. glutamicumΔmptC with pEKEx2-Mt-Rv2181 also restored the wild-type glycosyl linkage phenotype in this strain (Fig. 7, and see Fig. 3A and B). Furthermore, use of membrane preparations from C. glutamicumΔmptCΔmptD pEKEx2-Mt-Rv2181 and the synthetic Acc-Man9 (Fig. 6A) resulted in the incorporation of 1811 ± 87 cpm of [14C]-Manp into the [14C]-mannan product (Fig. 6B). Altogether, these data strongly suggest that Rv2181 is a homologue of Cg-mptC and both encode for α(1→2)-mannopyranosyltransferase activities, involved in the synthesis of the α(1→2)-Manp branches of LM and LAM in Corynebacterineae.
Fig. 7.
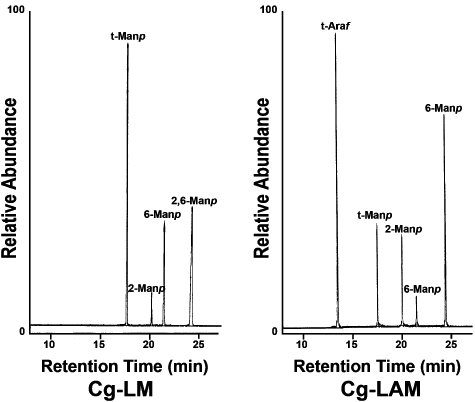
Characterization of the mycobacterial homologue Rv2181 of Cg-MptC. Lipoglycans were extracted from C. glutamicumΔmptC pEKEx2-Mt-Rv2181, and the resulting Cg-LM and Cg-LAM products purified, per-O-methylated, and hydrolysed using 2 M TFA, reduced and per-O-acetylated. The resulting partially per-O-methylated, per-O-acetylated alditol acetates were analysed by GC-MS and products identified based on their characteristic alditol cleavage acetal profiles: t-Araf (m/z 118, 161), t-Manp (m/z 118, 102, 145, 161, 162, 205), 2-Manp (m/z 129, 130, 161, 190), 6-Manp (m/z 118, 129, 162, 189, 233) and 2,6-Manp (m/z 129, 130, 189, 190).
Activation of Toll-like receptor 2 (TLR2) by wild-type and mutant Cg-LM and LAM
Both LM and LAM are bioactive molecules thought to be involved in modulating the host immune response (Briken et al., 2004). Bacterial lipoglycans are recognized by a variety of host immune receptors including TLR2. In fact, mycobacterial LMs are well known to be potent activators of this latter receptor hence providing them strong pro-inflammatory activity (Vignal et al., 2003; Quesniaux et al., 2004; Gilleron et al., 2006). On the other hand, LAMs are only poorly active; a feature attributed to the presence of the arabinan domain that masks the ‘bioactive’ mannan core (Vignal et al., 2003; Nigou et al., 2008; Birch et al., 2010). To investigate the ability of Cg-LM and Cg-LAM to activate TLR2 to assess possible consequences of Cg-mptC and Cg-mptD inactivation in this process, purified LM and LAM from C. glutamicumΔmptC and C. glutamicumΔmptD, were used to stimulate HEK293 cells expressing TLR2. To prevent interference of co-purified lipopeptides, the glycolipids were first pretreated with hydrogen peroxide (Morr et al., 2002; Geurtsen et al., 2009; Birch et al., 2010). As shown in Fig. 8, wild-type Cg-LAM was unable to activate TLR2. This result is consistent with an earlier study in which it was shown that LAMs substituted with single arabinosyl residues were unable to induce the production of tumour necrosis factor-α in human THP-1 monocytes (Nigou et al., 2008). Interestingly, LAMs isolated from the C. glutamicumΔmptC and C. glutamicumΔmptD mutant strains demonstrated an increased activity as evidenced by the enhanced TLR2-dependent IL-8 production by HEK293 cells (Fig. 8). This suggests that partial removal of α(1→2)-Manp units, increases the ability of Cg-LAM to activate TLR2. In contrast to mycobacterial LMs, wild-type Cg-LM did not show any activity towards TLR2. Furthermore, mutation of Cg-mptC and Cg-mptD did not alter this behaviour and the mutant LMs remained inactive (Fig. 8). These results demonstrate that wild-type Cg-LAM and Cg-LM, are both TLR2 inactive. Furthermore, partial removal of α(1→2)-Manp branching may promote TLR2 activation, but this is only the case for Cg-LAM.
Fig. 8.
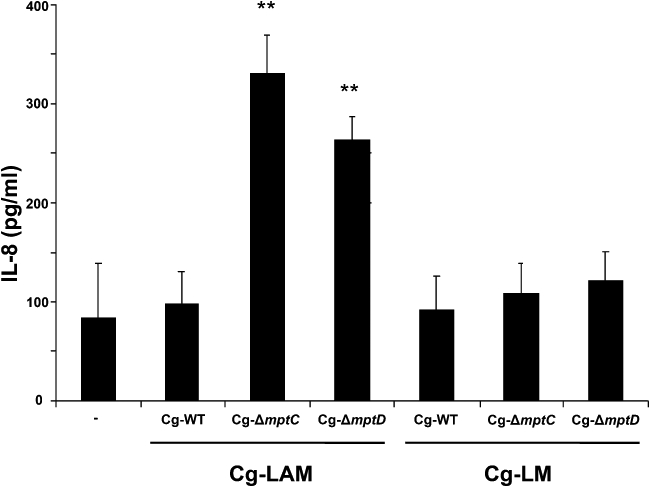
TLR-2 stimulation by wild-type and mutant Cg-LM and Cg-LAM. HEK293 cells transfected with TLR2 were stimulated with 50 µg ml−1 of wild-type (WT) or mutant (ΔmptC/ΔmptD) Cg-LAM or Cg-LM. Unstimulated cells (−) served as a control. After 24 h of stimulation (37°C), supernatants were harvested and analysed for IL-8 content using ELISA. The results represent the mean IL-8 production ± SD from three independent experiments (each experiment was performed in triplicate). Double asterisks indicate highly significant (P < 0.001) differences as compared to the unstimulated control cells.
Discussion
Apart from sharing a similar cell wall architecture, C. glutamicum possesses similar genetic loci responsible for cell wall biosynthesis like M. tuberculosis. As a model, it has proven extremely useful in the study of essential genetic elements of M. tuberculosis (Alderwick et al., 2006a,b;). In particular, it has enabled us to use it as a suitable model for the identification and functional study of mycobacterial genes involved in mycolic acid (Gande et al., 2004; 2007;), arabinogalactan (Alderwick et al., 2005; 2006b; 2007; Seidel et al., 2007a,b; Birch et al., 2008; 2009;) and LAM biosynthesis (Gibson et al., 2003; Mishra et al., 2007; 2008a,b; 2009; Tatituri et al., 2007a,b;). Previously, we reported the identification of enzymes involved in early and late stages of LM and LAM biosynthesis in Corynebacterineae (Mishra et al., 2007; 2008a,b; 2009;).
SDS-PAGE and MALDI-TOF-MS analysis of different lipoglycan species, including Cg-LM and Cg-LAM from C. glutamicumΔmptC and C. glutamicumΔmptD, suggested that there is a difference in a similar component of all these species. Two-dimensional thin-layer chromatography (2D-TLC) analysis supplemented with negative MALDI-TOF-MS of extracted glycolipids from mutants failed to show any differences in the PIM precursors (data not shown), and the presence of MptA and MptB in these mutants, negates the possibility of a change in the α(1→6) mannan backbone of these lipoglycans, leaving the only possibility of a change in α(1→2)-Manp branching, suggesting that MptC and MptD are involved in synthesis of these α(1→2)-Manp units in these lipoglycans. In addition, glycosyl linkage analysis revealed a reduction in the relative abundance of t-Manp and 2,6-Manp with a concomitant increase in the abundance of 6-Manp in Cg-LM and Cg-LAM from these strains. The cumulative chemical analysis of C. glutamicumΔmptC and C. glutamicumΔmptD agrees with the view that Cg-MptC and Cg-MptD are involved in synthesis of α(1→2)-Manp units in Cg-LM and Cg-LAM in C. glutamicum. This view was strengthened by the analysis of a C. glutamicumΔmptCΔmptD double mutant and the specificity of MptC and MptD in cross-complementation studies (Fig. S2). In summary, the data clarified the occurrence of residual t-α-Manp, α(1→2)-Manp and t-α-Araf units in the Cg-MptC and Cg-MptD mutants, as the singular mutants only partially removed 2,6-Manp branching. The absence of these units in the C. glutamicumΔmptCΔmptD mutant provides evidence that these units are present as t-α-Manp-(α1→2)-Manp and t-α-Araf-(α1→2)-Manp motifs (Fig. 9). Furthermore, specific in vitro assays utilizing a chemically synthesized nonasaccharide acceptor demonstrate that both Cg-mptC and Cg-mptD encode for α(1→2)-mannopyranosyltransferases, responsible for the synthesis of the α(1→2)-Manp units on the mannan backbone of LM and LAM. From the earlier studies with Rv2181 (Kaur et al., 2008), and the fact that Cg-MptC and Rv2181 are homologues, it is tempting to speculate that Cg-MptC adds α(1→2)-Manp residues to the proximal backbone of Cg-LM, or one could imply that Rv2181 in vitro has MptC function. A plasmid encoding Rv2181 was used to transform the C. glutamicum mutants, and Rv2181 was able to restore the wild-type phenotype in only the C. glutamicumΔmptC mutant, and possessed α(1→2)-mannopyranosyltransferase activity in the defined in vitro acceptor assay reported in this study.
Whereas the LM/LAM backbone is potentially rather similar within the Corynebacterineae, as indicated also from the relatedness of the α(1→6)-mannopyranosyltransferases (Fig. 1), it is clear from structural, immunological and genetic evidence, that the more elaborated structures and decorations of these glycolipids are variable. Known structural variations include large arabinan domains in M. tuberculosis (Chatterjee et al., 1991; McNeil et al., 1994), Man-capping of arabinan domains in M. tuberculosis (Chatterjee et al., 1992) and their PI capping in M. smegmatis (Khoo et al., 1995). Accordingly, also immunological properties of LM/LAM are different (Briken et al., 2004). This variability is naturally reflected at the genetic level. An example is the capping GT-C transferase of M. tuberculosis encoded by Rv1635c (Appelmelk et al., 2008), which is not present in C. glutamicum. The fact that MptC in C. glutamicum is a fusion protein carrying a second domain with unknown function, also illustrates that mature LM and LAM glycolipids are species-specific and accordingly the corresponding biosynthesis genes more prone to rapid evolution. Duplication or fusion of genes may indicate that the types of enzymes involved in the later steps of LM and LAM synthesis are highly dynamic and of evolutionary change within the Corynebacterineae (Xie et al., 2003).
A key question in lipoglycan biosynthesis is the division of LM and LAM abundance in the cell wall. The separation of LM and LAM in C. glutamicum, biosynthetically probably occurs after the initial formation of the α(1→6) mannan backbone by MptB, followed by α(1→2)-Manp branching by MptC (or MptD). The product of these two enzymes would then be the preferred substrate for further α(1→6)-Manp elongation by MptA, followed by α(1→2)-Manp branching by MptD (or MptC) (Fig. 9). Additional GT-C glycosyltransferases remain to be characterized involved in further modification of the α(1→6)-mannan backbone. These include how the dimanoside (via MptE?) and Araf-Manp side-chains are synthesized (AftE?) and are summarized in Fig. 9.
Both LAM and LM are immunomodulatory molecules (Briken et al., 2004). Whereas LAM has an important role during intracellular survival, for instance by preventing phagosome-lysosome fusion (Fratti et al., 2003), LM has strong pro-inflammatory activity through its ability to activate TLR2 (Vignal et al., 2003; Quesniaux et al., 2004; Gilleron et al., 2006). Previous research has demonstrated that the pro-inflammatory activity of LM is at least partly based upon its acylation status (Gilleron et al., 2006; Doz et al., 2007). PI-based LAM and LM species can differ in their degree of acylation, each carrying between one and four acyl chains. Investigation of the immune activating properties of differentially acylated LM species demonstrated that LM should at least be tri-acylated to be pro-inflammatory, with species carrying less acyl chains exhibiting immunosuppressive activity in a mechanism independent of TLR2 (Doz et al., 2007).
Here we investigated different Cg-LAM and LM species for their ability to activate TLR2. Similar to mycobacterial LAMs, wild-type Cg-LAM was found to be inactive (Fig. 8). This finding is consistent with previous observations that LAM species carrying only t-Araf substitutions, such as LAMs isolated from Rhodococcus ruber and Turicella otitidis, are largely inactive (Nigou et al., 2008). Comparison of wild-type Cg-LAM to the LAMs isolated from the C. glutamicumΔmptC and C. glutamicumΔmptD mutant strains demonstrated that the mutant LAMs had gained the ability to activate TLR2 (Fig. 8). The explanation for this increased activity is currently not known. Interestingly, wild-type Cg-LM, like wild-type Cg-LAM, but unlike mycobacterial LM was found to be completely TLR2-inactive (Fig. 8). The reason for the discrepancy is unclear, but may be related to structural differences in their membrane anchors. In mycobacteria, LAMs and LMs share a common PI anchor, on which further assembly occurs to give mature LM and LAM. However, in C. glutamicum, although Cg-LAM contains a normal PI anchor, two different pools of Cg-LM (Cg-LM-A and Cg-LM-B) dominate the majority of lipoglycan composition of the corynebacterial cell wall (Tatituri et al., 2007a; Lea-Smith et al., 2008; Mishra et al., 2008b). The large majority of the Cg-LM species (∼95%), Cg-LM-B, contains an anchor based upon a di-acylated α-D-glucopyranosyluronic acid-(1→3)-glycerol moiety, and only a minor fraction contains the more classical anchor based on PI, Cg-LM-A (Mishra et al., 2008b). Despite the different lipid anchors, the mannan backbone structures of Cg-LM-A and Cg-LM-B are similar as revealed through GC-MS analysis of partially methylated alditol acetates (Mishra et al., 2008b). This means that the majority of Cg-LM species will contain a di-acylated GlcAGroAc2 anchor, which is a feature that has been demonstrated to be incompatible with activation of TLR2 (Doz et al., 2007).
Furthermore, a complete investigation of the structural and functional relationship of these lipoglycans during host–pathogen interaction requires the availability of mutants defective in their respective biosynthetic pathways. The availability of complete genome sequences of several mycobacteria and related actinomycetes, like C. glutamicum, and the development of novel tools for genetic manipulation has opened up these possibilities (Cole et al., 1998).
Experimental procedures
Bacterial strains, cell culture and growth conditions
Corynebacterium glutamicum and E. coli DH5αmcr were grown in LB broth at 30°C and 37°C respectively. For C. glutamicum complex medium BHI and minimal medium CGXII was used (Eggeling and Bott, 2005). Kanamycin and ampicillin were used at a concentration of 50 µg ml−1 for selection of recombinants. Samples for lipid analysis were prepared by harvesting cells at an OD of 10–15, followed by a saline wash and freeze drying. HEK293 cells transfected with TLR2 (Flo et al., 2002) were kept in DMEM (Invitrogen) containing 10% FCS, 100 U ml−1 penicillin, 100 µg ml−1 streptomycin, 0.5 mg ml−1 G418, 2 mM L-glutamine and 110 mg l−1 pyruvate.
Construction of plasmids and strains
To delete Cg-mptC and Cg-mptD the deletion vectors pK19mobsacBΔCg-mptC, and pK19mobsacBΔCg-mptD were constructed. In each case cross-over PCR was used with genomic DNA as template and two different PCR's with primer pairs AB and CD (Table S1). The resulting PCR products served as template for primer pairs AD. The PCR product contained 12 nucleotides (nt) of the 3′ end of the respective gene together with genomic upstream sequences, and 36 nt of the 5′ end together with genomic downstream sequences. The inserts of all plasmids used in this work were confirmed by sequencing. Genes were deleted in by first introducing plasmids prepared from E. coli via electroporation into C. glutamicum and then selection for sucrose resistance in a procedure as described (Schäfer et al., 1994). Starting from C. glutamicumΔmptC and using pK19mobsacBΔCg-mptD the double deletion mutant C. glutamicumΔmptCΔmptD was made. The starting strain used was the type strain C. glutamicum ATCC13032. Chromosomal deletions were confirmed using primer pairs AD, as well as the additional new primer pairs EF, hybridizing outside of the regions used for plasmid constructions.
To assay for complementation of chromosomal deletions four expression plasmids were used. To construct pEKEx2-Cg-mptC primer pairs Ex-2100-RBS-for and Ex-2100-rev were used to amplify the 5′ half of NCgl2100, the PCR product treated with SalI/EcoRI and ligated with similar treated pEKEx2. To construct pEKEx2-Cg-mptD primer pairs Ex-2097-RBS-for and Ex-2097-rev were used to amplify NCgl2097, the PCR product treated with SalI/EcoRI and ligated with similar treated pEKEx2. To construct pEKEx2-Mt-Rv2181, chromosomal DNA of M. tuberculosis served as template, and Rv2181 was amplified using primer pairs Ex-Rv2181-for and Ex-Rv2181-rev, the former providing the sequence CTGCAG as a ribosome binding site. The amplified product was treated with PstI and EcoRI and ligated with pEKEx2. Plasmids were used to transform the respective mutants via electroporation to kanamycin resistance, and recombinants confirmed by plasmid preparation.
Lipid extraction and analysis
Polar and apolar lipids were extracted as described by Dobson et al. (1985). Briefly, 6 g of dry C. glutamicum cells was treated in 220 ml of methanolic saline (20 ml 0.3% NaCl and 200 ml CH3OH) and 220 ml of petroleum ether for 2 h (Dobson et al., 1985). The suspension was centrifuged and the upper layer containing apolar lipids was separated. An additional 220 ml of petroleum ether was added, mixed and centrifuged as described above. The two upper petroleum ether fractions were combined and dried. For polar lipids, 260 ml CHCl3/CH3OH/0.3% NaCl (9:10:3, v/v/v) was added to the lower aqueous phase and stirred for 4 h. The mixture was filtered and the filter cake re-extracted twice with 85 ml of CHCl3/CH3OH/0.3% NaCl (5:10:4, v/v/v). Equal amounts of CHCl3 and 0.3% NaCl (145 ml each) were added to the combined filtrates and stirred for 1 h. The mixture was allowed to settle, and the lower layer containing the polar lipids recovered and dried. The polar lipid extract was examined by 2D-TLC on aluminium backed plates of silica gel 60 F254 (Merck 5554), using CHCl3/CH3OH/H2O (65:25:4, v/v/v) in the first direction and CHCl3/CH3COOH/CH3OH/H2O (40:25:3:6, v/v/v/v) in the second direction. The thin-layer chromatographic plates sprayed with the appropriate staining solution to detect the presence of lipids, glycolipids or phospholipids as described (Dobson et al., 1985).
Extraction and purification of lipoglycans
Lipoglycans were extracted from delipidated cells as previously described (Ludwiczak et al., 2001). Briefly, cells were broken by sonication (MSE Soniprep 150, 12 micron amplitude, 60 s ON, 90 s OFF for 10 cycles, on ice) and the cell debris refluxed five times with 50% C2H5OH at 68°C, for 12 h intervals. The cell debris was removed by centrifugation and the supernatant containing lipoglycans, neutral glycans and proteins dried. This dried extract was then treated with hot phenol–H2O. The aqueous phase was dialysed and dried, followed by extensive treatments with α-amylase, DNase, RNase chymotrypsin and trypsin. This fraction was dialysed to remove the low MW break-down products formed after the enzyme treatment, thus yielding the crude lipoglycan fraction.
The crude lipoglycan extract was dried and resuspended in buffer A (50 mM ammonium acetate and 15% propan-1-ol) and subjected to Octyl Sepharose CL-4B chromatography (2.5 × 50 cm). The column (2.5 × 50 cm) was washed initially with four column volumes of buffer A to ensure removal of neutral glycans followed by buffer B (50 mM ammonium acetate and 50% propan-1-ol). The eluent was collected and concentrated to approximately 1 ml and precipitated using 5 ml of C2H5OH. The sample was dried using a Savant Speedvac and then resuspended in buffer C [0.2 M NaCl, 0.25% sodium deoxycholate (w/v), 1 mM EDTA and 10 mM Tris-HCl, pH 8] to a final concentration of 20 mg ml−1. The sample was gently mixed and left to incubate for 48 h at room temperature. The sample was then loaded onto a Sephacryl S-200 (2.5 cm × 50 cm) column previously equilibrated with buffer C. The sample was eluted with 400 ml of buffer C at a flow rate of 3 ml h−1, collecting 1.5 ml fractions. The fractions were monitored by SDS-PAGE using either a silver stain or a Pro-Q emerald glycoprotein stain and individual fractions pooled and dialysed extensively against buffer D (10 mM Tris-HCl, pH 8, 0.2 M NaCl, 1 mM EDTA) for 72 h with frequent changes of buffer. The samples were further dialysed against deionized water for 48 h with frequent changes of water, lyophilized and stored at −20°C.
MALDI-TOF-MS and NMR analysis
The matrix used was 2,5-dihydroxybenzoic acid at a concentration of 10 µg µl−1, in a mixture of H2O/C2H5OH (1:1, v/v), 0.1% trifluoroacetic acid (TFA). Samples (0.5 µl) at a concentration of 10 µg µl−1 were mixed with 0.5 µl of the matrix solution. Analyses were performed on a Voyager DE-STR MALDI-TOF instrument (PerSeptive Biosystems, Framingham, MA, USA) using linear mode detection. Mass spectra were recorded in the negative mode using a 300 ns time delay with a grid voltage of 80% of full accelerating voltage (25 kV) and a guide wire voltage of 0.15%. The mass spectra were mass assigned using external calibration. NMR spectra of LAM samples were recorded on a Bruker DMX-500 equipped with a double-resonance (1H/X)-BBi z-gradient probe head. All samples were exchanged in D2O, with intermediate lyophilization, and then dissolved in 0.7 ml D2O and analysed at 313 K. The 1H and 13C NMR chemical shifts were referenced relative to internal acetone at 2.225 and 34.00 ppm respectively. All details concerning NMR sequences and the experimental procedures were described previously (Tatituri et al., 2007b).
Glycosyl compositional and linkage analysis
Lipoglycans were hydrolysed using 250 µl of 2 M TFA at 120°C for 2 h, reduced with 100 µl of NaBD4 (10 mg ml−1 in C2H5OH:1 M NH4OH), and the resultant alditols per-O-acetylated using acetic anhydride (100 ml) at 120°C for 1 h, before examination by GC (Tatituri et al., 2007b). GC was performed using a Thermoquest Trace GC 2000 and a DB225 column (Supelco) with samples injected in the split-less mode. The oven was programmed to hold at an isothermal temperature of 275°C for a run time of 15 min. Glycosyl linkage analyses were performed as described previously (Tatituri et al., 2007b). Briefly, lipoglycan samples were per-O-methylated using dimethyl sulfinyl carbanion, hydrolysed using 2 M TFA, reduced using NaB2H4, and per-O-acetylated before analysis by GC-MS (Tatituri et al., 2007b). GC-MS was carried out on a Finnigan Polaris GCQ PlusTM using a BPX5 (Supleco) column. The injector temperature was set at 50°C, held for 1 min and then increased to 110°C at 20°C min−1. The oven was held at 110°C then ramped to 290°C at 8°C min−1 and held for 5 min to ensure all the products had eluted from the column. All the data were collected and analysed using Xcaliber (v.1.2) software.
General methods for chemical synthesis of Acc-Man9 (compound 1)
Reactions were performed under a dry argon atmosphere. Whenever necessary, compounds and starting materials were dried by azeotropic removal of water with toluene under reduced pressure. The reactions were monitored by TLC on pre-coated silica gel (60F254) plates (0.25 mm) from E. Merck and visualized using UV light (254 nm) and/or heating after spraying with (NH4)2SO4 solution (150 g ammonium sulphate, 30 ml H2SO4, 750 ml H2O). Flash column chromatography was carried out on Isco Teledyne CombiFlash Rf200 chromatographic system using pre-packed columns. 1H and 13C NMR spectra were recorded at 400 MHz and 100 MHz, respectively, on a Varian 400-MR spectrometer. Coupling constants (J) are reported in Hz and chemical shifts (δ) in ppm relative to a residual solvent peak or an internal standard (TMS). The HR-MS spectra were recorded on a Agilent 6140 liquid chromatography mass spectrometry instrument.
p-Tolyl 2,3-Di-O-benzoyl-4,5-O-benzylidene-1-thio-α-D-mannopyranoside (6)
Compound 5 (230 mg, 0.61 mmol) was dissolved in 5 ml of anhydrous pyridine, and benzoyl chloride (179 µl, 1.54 mmol) was added dropwise. The reaction mixture was stirred for 4 h at room temperature, poured into ice-water (15 ml) and extracted with CHCl3 (2 × 10 ml). The combined CHCl3 layers were washed with brine, dried over Na2SO4 and concentrated. Flash chromatographic purification of the crude product using cyclohexane/ethyl acetate as eluent gave pure 6 (335 mg, 94%) as a colourless solid (mp 66°C). Rf = 0.44 (cyclohexane/ ethyl acetate, 2:1). 1H NMR (CDCl3, 400 MHz): δ 8.06 (2H, m, Ar), 7.92 (2H, m, Ar), 7.61 (1H, m, Ar), 7.46 (8H, m, Ar), 7.33 (4H, m, Ar), 7.15 (2H, d, J = 6.3 Hz, Ar), 5.95 (1H, dd, J = 0.9, 2.7 Hz, H-2), 5.82 (1H, dd, J = 2.7, 7.7 Hz, H-3), 5.68 (1H, s, CHPh), 5.59 (1H, d, J = 0.9 Hz, H-1), 4.64 (1H, ddd, J = 3.9, 6.9, 7.5, Hz, H-5), 4.39 (1H, dd, J = 7.2, 7.5 Hz, H-4), 4.33 (1H, dd, J = 3.6, 7.8 Hz, H-6a), 3.96 (1H, dd, J = 7.5, 7.8 Hz, H-6b), 2.34 (3H, s, CH3). 13C NMR (100 MHz, CDCl3): δ 165.40, 165.29 (2 × C = O), 138.52, 138.03, 133.54, 133.11, 133.00, 130.07, 129.98, 129.84, 129.79, 129.57, 129.48, 129.10, 128.59, 128.27, 128.25, 126.19 (Ar), 101.96 (CHPh), 87.31 (C-1), 76.95 (C-4), 72.44 (C-2), 69.22 (C-3), 68.61 (C-6), 65.21 (C-5), 21.17 (CH3). HR-MS: m/z 605.1604 [M + Na]+ calculated for C34H30O7SNa, found 605.1611.
p-Tolyl 2,3-Di-O-benzoyl-6-O-benzyl-1-thio-α-D-mannopyranoside (7)
To an anhydrous CH2Cl2 (10 ml) solution of compound 6 (300 mg, 0.52 mmol) was added 200 mg of 4 Å molecular sieves under an atmosphere of argon and cooled to −78°C. Et3SiH (266 µl, 1.66 mmol) was added dropwise to the cooled reaction mixture followed by addition of triflic acid (115 µl, 1.30 mmol). The reaction mixture was then stirred at −78°C for 1 h. The reaction was quenched by the addition of triethylamine (0.25 ml) and CH3OH (0.5 ml), diluted with CH2Cl2 (20 ml), and washed with saturated aqueous NaHCO3 solution (10 ml), H2O (10 ml) and brine (10 ml). The organic layer was dried on Na2SO4, filtered and the filtrate concentrated to a syrup. Flash chromatographic purification of the crude product using cyclohexane/ethyl acetate as eluent gave pure 7 (241 mg, 80% yield) as colourless foam. Rf = 0.38 (cyclohexane/ethyl acetate, 2:1). 1H NMR (CDCl3, 400 MHz): δ 8.03 (2H, m, Ar), 7.94 (2H, m, Ar), 7.58 (1H, m, Ar), 7.52 (1H, m, Ar), 7.43 (3H, m, Ar), 7.34 (8H, m, Ar), 7.08 (2H, d, J = 6.0 Hz, Ar), 5.85 (1H, dd, J = 0.9, 2.2 Hz, H-2), 5.61 (1H, d, J = 0.9 Hz, H-1), 5.57 (1H, dd, J = 2.2, 7.2 Hz, H-3), 4.69 (1H, d, J = 9.0 Hz, CH2Pha), 4.59 (1H, d, J = 9.0 Hz, CH2Phb), 4.53 (1H, m, H-5), 4.40 (1H, ddd, J = 2.7, 7.2, 7.5 Hz, H-4), 3.98 (1H, dd, J = 3.5, 7.8 Hz, H-6a), 3.89 (1H, dd, J = 2.7, 7.8 Hz, H-6b), 2.81 (1H, d, J = 3.0 Hz, 4-OH), 2.31 (3H, s, CH3). 13C NMR (100 MHz, CDCl3): δ 166.51, 165.37 (2 × C = O), 138.18, 138.04, 133.43, 133.38, 132.68, 129.93, 129.88, 129.85, 129.53, 129.41, 129.32, 128.53, 128.41, 128.39, 127.68, 127.60 (Ar), 86.42 (C-1), 73.76 (CH2Ph), 73.18 (C-3), 72.44 (C-5), 72.01 (C-2), 68.99 (C-6), 67.80 (C-4), 21.14 (CH3). HR-MS: m/z 607.1761 [M + Na]+ calculated for C34H32O7SNa, found 607.1757.
p-Tolyl 4-O-Acetyl-2,3-di-O-benzoyl-6-O-benzyl-1-thio-α-D-mannopyranoside (4)
Compound 7 (200 mg, 0.34 mmol) was dissolved in 3 ml of dry pyridine, and acetic anhydride (49 µl, 0.51 mmol) was added under an argon atmosphere. The reaction mixture was stirred for 5 h at room temperature, poured into an ice-water mixture (10 ml) and extracted with CHCl3 (2 × 15 ml). The combined CHCl3 layers were washed with brine, dried over Na2SO4 and concentrated. Flash chromatographic purification of the crude product using cyclohexane/ethyl acetate as eluent gave pure 8 (202 mg, 94% yield) as colourless solid (m.p. 93.7°C). Rf = 0.45 (cyclohexane/ethyl acetate, 2:1). 1H NMR (CDCl3, 400 MHz): δ 8.02 (2H, m, Ar), 7.91 (2H, m, Ar), 7.53 (2H, m, Ar), 7.45 (2H, m, Ar), 7.38 (2H, m, Ar), 7.32 (7H, m, Ar), 7.07 (2H, d, J = 6.0 Hz, Ar), 5.87 (1H, dd, J = 1.5, 2.2 Hz, H-2), 5.81 (1H, t, J = 7.5 Hz, H-4), 5.65 (1H, dd, J = 2.2, 7.3 Hz, H-3), 5.64 (1H, d, J = 1.5 Hz, H-1), 4.69 (1H, m, H-5), 4.63 (1H, d, J = 8.7 Hz, CH2Pha), 4.51 (1H, d, J = 8.7 Hz, CH2Phb), 3.75 (1H, dd, J = 3.4, 8.2 Hz, H-6a), 3.70 (1H, dd, J = 2.2, 8.2 Hz, H-6b), 2.30 (3H, s, PhCH3), 1.93 (3H, s, COCH3). 13C NMR (100 MHz, CDCl3): δ 169.76 (COCH3), 165.47, 165.40 (2 × COPh), 138.28, 137.99, 133.42, 133.34, 132.74, 129.94, 129.91, 129.81, 129.31, 129.20, 129.12, 128.53, 128.46, 128.33, 127.80, 127.60 (Ar), 86.24 (C-1), 73.62 (CH2Ph), 71.88 (C-2), 70.95 (C-5), 70.68 (C-3), 68.93 (C-6), 66.85 (C-4), 21.14 (PhCH3), 20.76 (COCH3). HR-MS: m/z 649.1867 [M + Na]+ calculated for C36H34O8SNa, found 649.1852.
1-Azidoethyl 4-O-Acetyl-2,3-di-O-benzoyl-6-O-benzyl-α-D-mannopyranosyl-(1→6)-α-D-mannopyranosyl-(1→6)-α-D-mannopyranosyl-(1→6)-2,3,4-tri-O-benzoyl-α-D-mannopyranosyl-2,3,4-tri-O-benzoyl-α-D-mannopyranosyl-(1→6)-2,3,4-tri-O-benzoyl-α-D-mannopyranosyl-(1→6)-2,3,4-tri-O-benzoyl-α-D-mannopyranosyl-(1→6)-2,3,4-tri-O-benzoyl-α-D-mannopyranosyl-(1→6)-2,3,4-tri-O-benzoyl-α-D-mannopyranoside (3)
To a solution of thiooctasaccharide 2 (150 mg, 0.04 mmol) in dry CH2Cl2 (5 ml) were added the donor thioglycoside 4 (47 mg, 0.08 mmol) and 100 mg of 4 Å molecular sieves. The mixture was cooled to −20°C and NIS (11.0 mg, 0.048 mmol) and triflic acid (2 µl, 0.02 mmol) added and the reaction was stirred for 15 min at −20°C. The temperature of the reaction mixture was raised to room temperature and after 1 h, 5 ml of saturated aqueous NaHCO3 was added. The reaction mixtures were extracted with 2 × 10 ml of CHCl3 and the organic layer was washed with saturated aqueous sodium thiosulfate solution (5 ml). The organic layer was dried over Na2SO4 and concentrated to a syrup. Flash chromatographic purification of the crude product was performed using cyclohexane/ethyl acetate as eluent and gave pure nonasaccharide 3 (169 mg, 82% yield) as colourless solid (m.p. 202°C). Rf = 0.43 (cyclohexane/ethyl acetate, 1:1). 1H NMR (CDCl3, 400 MHz): δ 8.16 (15H, m, Ar), 8.05 (18H, m, Ar), 7.87 (17H, m, Ar), 7.47 (50H, m, Ar), 7.26 (35H, m, Ar), 6.18 (8H, m), 6.02 (2H, m), 5.98 (7H, m), 5.85 (3H, m), 5.26 (1H, s), 5.22 (1H, d, J = 1.2 Hz), 4.96 (6H, m), 4.48 (1H, d, J = 11.9 Hz, CH2Pha), 4.57 (1H, dd, J = 1.8, 7.5 Hz), 4.38 (1H, dd, J = 2.4, 7.2 Hz), 4.28 (1H, d, J = 11.9 Hz, CH2Phb), 4.13 (8H, m), 3.86 (9H, m), 3.66 (1H, dd, J = 2.1, 8.4 Hz), 3.57 (4H, m), 3.37 (7H, m), 1.88 (3H, s, CH3). 13C NMR (100 MHz, CDCl3): δ 166.79 (COCH3), 165.72, 165.56, 165.54, 165.52, 165.45, 165.37, 165.35, 165.33, 165.25, 165.23, 165.11 (COPh), 133.72, 133.51, 133.44, 133.29, 133.05, 132.96, 130.06, 130.01, 129.86, 129.67, 129.48, 129.42, 129.40, 129.37, 129.28, 129.21, 129.18, 129.15, 129.05, 128.82, 128.78, 128.70, 128.60, 128.51, 128.38, 128.27 (Ar), 98.28, 98.09, 97.95, 97.87 (C-1's), 73.45 (OCH2Ph), 70.88, 70.61, 70.53, 70.39, 70.29, 70.25, 70.17, 69.90, 69.71, 69.63, 69.54, 69.47, 69.40, 69.30, 67.19, 67.12, 66.74, 66.41, 66.32, 66.24, 66.03, 65.96, 65.75, 50.48 (CH2N3), 20.81 (COCH3).
1-Azidoethyl 6-O-Benzyl-α-D-mannopyranosyl-(1→6)-2,3,4-tri-O-benzoyl-α-D-mannopyranosyl-(1→6)-α-D-mannopyranosyl-(1→6)-α-D-mannopyranosyl-α-D-mannopyranosyl-(1→6)-α-D-mannopyranosyl-(1→6)-α-D-mannopyranosyl-(1→6)-α-D-mannopyranosyl-(1→6)-α-D-mannopyranoside (1)
To an ice-cooled solution of compound 3 (150 mg, 0.03 mmol) in dry CH2Cl2–CH3OH (1:2, 10 ml) was added a sodium methoxide solution in CH3OH (25% w/v, 2.0 ml). The reaction mixture was allowed to stir overnight at room temperature and H+ Amberlite resin (200 mg) was added. It was filtered, solvent was evaporated and the residue was washed with CHCl3 (3 × 10 ml). An aqueous solution (2 ml) crude nonasaccharide was passed through a small column of Bio-Beads SM-4 (15 g, 20–50 mesh) and eluted with 0–60% CH3OH–H2O. The aqueous solution that remained after brief evaporation under vacuum was frozen and lyophilized to a colourless foam (19 mg, 35%). 1H NMR (CDCl3, 400 MHz): δ 7.68 (1H, m), Ar), 7.38 (4H, m, Ar), 4.89 (1H, s), 4.64 (1H, d, J = 11.6 Hz, OCH2Ph), 4.61 (1H, d, J = 11.7 Hz, OCH2Ph), 3.99–3.60 (63H, m), 2.68 (2H, m, CH2N3). 13C NMR (100 MHz, CDCl3): δ 138.04, 128.39, 128.37, 127.01 (Ar), 101.77, 101.04, 100.96, 100.72, 100.55 (C-1's), 74.48, 74.37, 73.32 (OCH2Ph), 73.18, 72.86, 72.77, 72.62, 72.56, 72.42, 72.04, 70.97, 68.84, 68.71, 68.56, 67.86, 67.45, 67.31, 67.00, 51.79 (CH2N3). HR-MS: m/z 1658.5548 [M + Na]+ calculated for C63H101N3O46Na, found 1658.5503.
In vitroα(1→2)-mannopyranosyltransferase assay utilizing a novel nonasaccharide acceptor, Acc-Man9
Corynebacterium glutamicum, C. glutamicumΔmptC, C. glutamicumΔmptD, complemented strains, and a double mutant devoid of Manp branching activities (MptC and MptD) were used to characterize the enzymatic activity of Cg-MptC and Cg-MptD, and Mt-Rv2181. Membranes from all strains of C. glutamicum were prepared as described previously (Mishra et al., 2008a) and resuspended in 100 mM sodium acetate (pH 6.0), containing 5 mM β-mercaptoethanol and 7 mM MgCl2 (Buffer E) to a final concentration of 10–15 mg ml−1. The chemically synthesized nonasaccharide acceptor, azidoethyl 6-O-benzyl-α-D-mannopyranosyl-(1→6)-[α-D-mannopyranosyl-(1→6)]7-D-mannopyranoside (Acc-Man9), and PP-[14C]-M [Gurcha et al., 2002; stored in CHCl3/CH3OH, 2:1, v/v], were aliquoted into 1.5 ml Eppendorf tubes to a final concentration of 2 mM and 250 000 cpm (0.305 Ci mmol−1), respectively, and dried under nitrogen. IgePal CA-630 (8 µl, Sigma Aldrich) was added and the tubes sonicated to resuspend the lipid linked components, and 1 mM ATP, 1 mM NADP, 5 mM MnCl2, and membrane protein (1 mg) added to a final volume of 100 µl. The reaction mixture incubated at 37°C for 2 h as described (Yokoyama and Ballou, 1989) and then terminated by the addition of 5 ml of 50% C2H5OH. The supernatant was recovered, dried and resuspended in 700 µl of water and loaded onto a 1 ml strong anion exchange SepPak cartridge (Supelco). The cartridge washed with H2O (5 ml), which was collected, dried and resuspended in 200 µl of H2O and 10% of the reaction product was quantified by scintillation counting using 5 ml of Ecolume (ICN Biomedicals, Costa Mesa, CA, USA) and the remaining material analysed by 15% SDS-PAGE-/autoradiography as described previously (Mishra et al., 2008a).
Acc-Man9-[14C]-Manx and selective cleavage by an α(1→2)-D-mannosidase
The extracted Acc-Man9-[14C]-Manx products from the assays performed with C. glutamicum membranes were dried and subsequently incubated at 30°C for 24 h in 0.1 M acetate buffer, pH 5.0, with 5 mU of α(1→2)-D-mannosidase (Maras et al., 2000) as described previously (Ichishima et al., 1981; Yokoyama and Ballou, 1989). The reaction mixture was fractionated on a 50 ml Bio-Gel P-2 gel filtration column (30 cm × 1.5 cm; Bio-Rad) in H2O. The control Acc-Man9-[14C]-Manx before α(1→2)-D-mannosidase treatment eluted form the Bio-Gel P-2 column at fractions 11–13, and digested products (release of [14C]Manp) was retained and eluted in later fractions 33–39 based on a [14C]-mannose standard (Cooper et al., 2002).
Cell stimulation assays
Trypsine-released HEK293 cells were washed with and resuspended in culture medium at 1.11 × 106 cells ml−1. Cells (1 × 105, 90 µl) were transferred to a 96-well U-bottom plate (Greiner) and left for 2 h to let the cells re-adhere. After this, cells were stimulated (in triplicate) with 50 µg ml−1 wild-type or mutant Cg-LM and Cg-LAM (pretreated with 1% hydrogen peroxide for 96 h as described (Birch et al., 2010) in a final stimulation volume of 100 µl. Unstimulated cells served as a control. Culture supernatants were harvested after 24 h by centrifugation and stored at −80°C for cytokine measurements using enzyme-linked immunosorbent assay (ELISA) against human IL-8 (Invitrogen).
Acknowledgments
AKM is a Darwin Trust sponsored student. JG is financially supported by the Netherlands Organization for Scientific Research (NWO) through a VENI research Grant (016.101.001). GSB acknowledges support in the form of a Personal Research Chair from Mr James Bardrick, Royal Society Wolfson Research Merit Award, as a former Lister Institute-Jenner Research Fellow, the Medical Research Council and The Wellcome Trust (081569/Z/06/Z). We are thankful to Dr Nico Callewaert (University of Ghent, Belgium) for his generous gift of T. reeseiα(1→2)-D-mannosidase. Furthermore, we want to thank Dr D. Golenbock (University of Massachusetts Medical School, Worcester, MA, USA) for kindly providing the HEK293 TLR2 cell line. Figure 4B were originally published in Journal of Biological Chemistry. Tatituri et al. (2007b); © the American Society for Biochemistry and Molecular Biology.
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alderwick LJ, Radmacher E, Seidel M, Gande R, Hitchen PG, Morris HR, et al. Deletion of Cg-emb in corynebacterianeae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an arabinan-deficient mutant with a cell wall galactan core. J Biol Chem. 2005;280:32362–32371. doi: 10.1074/jbc.M506339200. [DOI] [PubMed] [Google Scholar]
- Alderwick LJ, Seidel M, Sahm H, Besra GS, Eggeling L. Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J Biol Chem. 2006a;281:15653–15661. doi: 10.1074/jbc.M600045200. [DOI] [PubMed] [Google Scholar]
- Alderwick LJ, Dover LG, Seidel M, Gande R, Sahm H, Eggeling L, et al. Arabinan-deficient mutants of Corynebacterium glutamicum and the consequent flux in decaprenylmonophosphoryl-D-arabinose metabolism. Glycobiology. 2006b;16:1073–1081. doi: 10.1093/glycob/cwl030. [DOI] [PubMed] [Google Scholar]
- Alderwick LJ, Birch HL, Mishra AK, Eggeling L, Besra GS. Structure, function and biosynthesis of the Mycobacterium tuberculosis cell wall: arabinogalactan and lipoarabinomannan assembly with a view to discovering new drug targets. Biochem Soc Trans. 2007;35:1325–1328. doi: 10.1042/BST0351325. [DOI] [PubMed] [Google Scholar]
- Appelmelk BJ, den Dunnen J, Driessen NN, Ummels R, Pak M, Nigou J, et al. The mannose cap of mycobacterial lipoarabinomannan does not dominate the Mycobacterium-host interaction. Cell Microbiol. 2008;10:930–944. doi: 10.1111/j.1462-5822.2007.01097.x. [DOI] [PubMed] [Google Scholar]
- Aqueel MS, Pathak V, Pathak AK. Concise assembly of linear α(1→6)-linked octamannan fluorescent probe. Tetrahedron Lett. 2008;49:7157–7160. doi: 10.1016/j.tetlet.2008.09.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besra GS, Brennan PJ. The mycobacterial cell wall: biosynthesis of arabinogalactan and lipoarabinomannan. Biochem Soc Trans. 1997;25:845–850. doi: 10.1042/bst0250845. [DOI] [PubMed] [Google Scholar]
- Besra GS, Khoo KH, McNeil MR, Dell A, Morris HR, Brennan PJ. A new interpretation of the structure of the mycolyl-arabinogalactan complex of Mycobacterium tuberculosis as revealed through characterization of oligoglycosylalditol fragments by fast-atom bombardment mass spectrometry and 1H nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34:4257–4266. doi: 10.1021/bi00013a015. [DOI] [PubMed] [Google Scholar]
- Besra GS, Morehouse CB, Rittner CM, Waechter CJ, Brennan PJ. Biosynthesis of mycobacterial lipoarabinomannan. J Biol Chem. 1997;272:18460–18466. doi: 10.1074/jbc.272.29.18460. [DOI] [PubMed] [Google Scholar]
- Birch HL, Alderwick LJ, Bhatt A, Rittmann D, Krumbach K, Singh A, et al. Biosynthesis of mycobacterial arabinogalactan: identification of a novel α(1→3) arabinofuranosyltransferase. Mol Microbiol. 2008;69:1191–1206. doi: 10.1111/j.1365-2958.2008.06354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch HL, Alderwick LJ, Rittmann D, Krumbach K, Etterich H, Grzegorzewicz A, et al. Identification of a terminal rhamnopyranosyltransferase (RptA) involved in Corynebacterium glutamicum cell wall biosynthesis. J Bacteriol. 2009;191:4879–4887. doi: 10.1128/JB.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch H, Alderwick LJ, Appelmelk BJ, Maaskant J, Bhatt A, Singh A, et al. A truncated lipoglycan from mycobacteria with altered immunological properties. Proc Natl Acad Sci USA. 2010;107:2634–2639. doi: 10.1073/pnas.0915082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom BR, Murray CJ. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- Brennan PJ. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2003;83:91–97. doi: 10.1016/s1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- Brennan PJ, Ballou CE. Biosynthesis of mannophosphoinositides by Mycobacterium phlei. The family of dimannophosphoinositides. J Biol Chem. 1967;242:3046–3056. [PubMed] [Google Scholar]
- Brennan PJ, Ballou CE. Biosynthesis of mannophosphoinositides by Mycobacterium phlei. Enzymatic acylation of the dimannophosphoinositides. J Biol Chem. 1968;243:2975–2984. [PubMed] [Google Scholar]
- Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- Briken V, Porcelli SA, Besra GS, Kremer L. Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol Microbiol. 2004;53:391–403. doi: 10.1111/j.1365-2958.2004.04183.x. [DOI] [PubMed] [Google Scholar]
- Chatterjee D, Bozic CM, McNeil M, Brennan PJ. Structural features of the arabinan component of the lipoarabinomannan of Mycobacterium tuberculosis. J Biol Chem. 1991;266:9652–9660. [PubMed] [Google Scholar]
- Chatterjee D, Lowell K, Rivoire B, McNeil MR, Brennan PJ. Lipoarabinomannan of Mycobacterium tuberculosis. Capping with mannosyl residues in some strains. J Biol Chem. 1992;267:6234–6239. [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Cooper HN, Gurcha SS, Nigou J, Brennan PJ, Belisle JT, Besra GS, et al. Characterization of mycobacterial protein glycosyltransferase activity using synthetic peptide acceptors in a cell-free assay. Glycobiology. 2002;12:427–434. doi: 10.1093/glycob/cwf051. [DOI] [PubMed] [Google Scholar]
- Cumpstey I, Chayajarus K, Fairbanks AJ, Redgrave AJ, Seward CMP. Allyl protecting group mediated intramolecular aglycon delivery: optimisation of mixed acetal formation and mechanistic investigation. Tetrahedron Asymmetry. 2004;15:3207–3221. [Google Scholar]
- Daffe M, Brennan PJ, McNeil M. Predominant structural features of the cell wall arabinogalactan of Mycobacterium tuberculosis as revealed through characterization of oligoglycosyl alditol fragments by gas chromatography/mass spectrometry and by 1H and 13C NMR analyses. J Biol Chem. 1990;265:6734–6743. [PubMed] [Google Scholar]
- Dobson G, Minnikin DE, Minnikin SM, Parlett JH, Goodfellow M, Ridell M, et al. Systematic analysis of complex mycobacterial lipids. In: Goodfellow M, Minnikin DE, editors. Chemical Methods in Bacterial Systematics. London: Academic Press; 1985. pp. 237–265. [Google Scholar]
- Dover LG, Cerdeno-Tarraga AM, Pallen MJ, Parkhill J, Besra GS. Comparative cell wall core biosynthesis in the mycolated pathogens, Mycobacterium tuberculosis and Corynebacterium diphtheriae. FEMS Microbiol Rev. 2004;28:225–250. doi: 10.1016/j.femsre.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Doz E, Rose S, Nigou J, Gilleron M, Puzo G, Erard F, et al. Acylation determines the toll-like receptor (TLR)-dependent positive versus TLR2-, mannose receptor- and SIGNR1-independent negative regulation of pro-inflammatory cytokines by mycobacterial lipomannan. J Biol Chem. 2007;282:26014–26025. doi: 10.1074/jbc.M702690200. [DOI] [PubMed] [Google Scholar]
- Eggeling L, Bott M. Handbook of Corynebacterium Glutamicum. Boca Raton, FL: CRC Press, Taylor Francis Group; 2005. [Google Scholar]
- Flo TH, Ryan L, Latz E, Takeuchi O, Monks BG, Lien E, et al. Involvement of toll-like receptor (TLR) 2 and TLR4 in cell activation by mannuronic acid polymers. J Biol Chem. 2002;277:35489–35495. doi: 10.1074/jbc.M201366200. [DOI] [PubMed] [Google Scholar]
- Fratti RA, Chua J, Vergne I, Deretic V. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc Natl Acad Sci USA. 2003;100:5437–5442. doi: 10.1073/pnas.0737613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gande R, Gibson KJ, Brown AK, Krumbach K, Dover LG, Sahm H, et al. Acyl-CoA carboxylases (accD2 and accD3), together with a unique polyketide synthase (Cg-pks), are key to mycolic acid biosynthesis in Corynebacterianeae such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J Biol Chem. 2004;279:44847–44857. doi: 10.1074/jbc.M408648200. [DOI] [PubMed] [Google Scholar]
- Gande R, Dover LG, Krumbach K, Besra GS, Sahm H, Oikawa T, et al. The two carboxylases of Corynebacterium glutamicum essential for fatty acid and mycolic acid synthesis. J Bacteriol. 2007;189:5257–5264. doi: 10.1128/JB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurtsen J, Chedammi S, Mesters J, Cot M, Driessen NN, Sambou T, et al. Identification of mycobacterial alpha-glucan as a novel ligand for DC-SIGN: involvement of mycobacterial capsular polysaccharides in host immune modulation. J Immunol. 2009;183:5221–5231. doi: 10.4049/jimmunol.0900768. [DOI] [PubMed] [Google Scholar]
- Gibson KJ, Eggeling L, Maughan WN, Krumbach K, Gurcha SS, Nigou J, et al. Disruption of Cg-Ppm1, a polyprenyl monophosphomannose synthase, and the generation of lipoglycan-less mutants in Corynebacterium glutamicum. J Biol Chem. 2003;278:40842–40850. doi: 10.1074/jbc.M307988200. [DOI] [PubMed] [Google Scholar]
- Gilleron M, Nigou J, Nicolle D, Quesniaux V, Puzo G. The acylation state of mycobacterial lipomannans modulates innate immunity response through toll-like receptor 2. Chem Biol. 2006;13:39–47. doi: 10.1016/j.chembiol.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Guerardel Y, Maes E, Elass E, Leroy Y, Timmerman P, Besra GS, et al. Structural study of lipomannan and lipoarabinomannan from Mycobacterium chelonae. Presence of unusual components with α1,3-mannopyranose side chains. J Biol Chem. 2002;277:30635–30648. doi: 10.1074/jbc.M204398200. [DOI] [PubMed] [Google Scholar]
- Gurcha SS, Baulard AR, Kremer L, Locht C, Moody DB, Muhlecker W, et al. Ppm1, a novel polyprenol monophosphomannose synthase from Mycobacterium tuberculosis. Biochem J. 2002;365:441–450. doi: 10.1042/BJ20020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DL, Ballou CE. Biosynthesis of mannophospholipids by Mycobacterium phlei. J Biol Chem. 1966;241:895–902. [PubMed] [Google Scholar]
- Ichishima E, Arai M, Shigematsu Y, Kumagai H, Sumida-Tanaka R. Purification of an acidic alpha-D-mannosidase from Aspergillus saitoi and specific cleavage of 1,2-α-D-mannosidic linkage in yeast mannan. Biochim Biophys Acta. 1981;658:45–53. doi: 10.1016/0005-2744(81)90248-5. [DOI] [PubMed] [Google Scholar]
- Kaur D, Berg S, Dinadayala P, Gicquel B, Chatterjee D, McNeil MR, et al. Biosynthesis of mycobacterial lipoarabinomannan: role of a branching mannosyltransferase. Proc Natl Acad Sci USA. 2006;103:13664–13669. doi: 10.1073/pnas.0603049103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur D, McNeil MR, Khoo KH, Chatterjee D, Crick DC, Jackson M, et al. New insights into the biosynthesis of mycobacterial lipomannan arising from deletion of a conserved gene. J Biol Chem. 2007;282:27133–27140. doi: 10.1074/jbc.M703389200. [DOI] [PubMed] [Google Scholar]
- Kaur D, Obregon-Henao A, Pham H, Chatterjee D, Brennan PJ, Jackson M. Lipoarabinomannan of Mycobacterium: mannose capping by a multifunctional terminal mannosyltransferase. Proc Natl Acad Sci USA. 2008;105:17973–17977. doi: 10.1073/pnas.0807761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo KH, Dell A, Morris HR, Brennan PJ, Chatterjee D. Inositol phosphate capping of the nonreducing termini of lipoarabinomannan from rapidly growing strains of Mycobacterium. J Biol Chem. 1995;270:12380–12389. doi: 10.1074/jbc.270.21.12380. [DOI] [PubMed] [Google Scholar]
- Kordulakova J, Gilleron M, Mikusova K, Puzo G, Brennan PJ, Gicquel B, et al. Definition of the first mannosylation step in phosphatidylinositol mannoside synthesis. PimA is essential for growth of mycobacteria. J Biol Chem. 2002;277:31335–31344. doi: 10.1074/jbc.M204060200. [DOI] [PubMed] [Google Scholar]
- Kordulakova J, Gilleron M, Puzo G, Brennan PJ, Gicquel B, Mikusova K, et al. Identification of the required acyltransferase step in the biosynthesis of the phosphatidylinositol mannosides of mycobacterium species. J Biol Chem. 2003;278:36285–36295. doi: 10.1074/jbc.M303639200. [DOI] [PubMed] [Google Scholar]
- Kremer L, Gurcha SS, Bifani P, Hitchen PG, Baulard A, Morris HR, et al. Characterization of a putative α-mannosyltransferase involved in phosphatidylinositol trimannoside biosynthesis in Mycobacterium tuberculosis. Biochem J. 2002;363:437–447. doi: 10.1042/0264-6021:3630437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea-Smith DJ, Martin KL, Pyke JS, Tull D, McConville MJ, Coppel RL, et al. Analysis of a new mannosyltransferase required for the synthesis of phosphatidylinositol mannosides and lipoarbinomannan reveals two lipomannan pools in Corynebacterineae. J Biol Chem. 2008;283:6773–6782. doi: 10.1074/jbc.M707139200. [DOI] [PubMed] [Google Scholar]
- Liu J, Mushegian A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003;12:1418–1431. doi: 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwiczak P, Brando T, Monsarrat B, Puzo G. Structural characterization of Mycobacterium tuberculosis lipoarabinomannans by the combination of capillary electrophoresis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 2001;73:2323–2330. doi: 10.1021/ac001368h. [DOI] [PubMed] [Google Scholar]
- McNeil M, Daffe M, Brennan PJ. Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J Biol Chem. 1990;265:18200–18206. [PubMed] [Google Scholar]
- McNeil M, Daffe M, Brennan PJ. Location of the mycolyl ester substituents in the cell walls of mycobacteria. J Biol Chem. 1991;266:13217–13223. [PubMed] [Google Scholar]
- McNeil MR, Robuck KG, Harter M, Brennan PJ. Enzymatic evidence for the presence of a critical terminal hexa-arabinoside in the cell walls of Mycobacterium tuberculosis. Glycobiology. 1994;4:165–173. doi: 10.1093/glycob/4.2.165. [DOI] [PubMed] [Google Scholar]
- Maras M, Callewaert N, Piens K, Claeyssens M, Martinet W, Dewaele S, et al. Molecular cloning and enzymatic characterization of a Trichoderma reesei 1,2-α-D mannosidase. J Biotechnol. 2000;77:255–263. doi: 10.1016/s0168-1656(99)00222-9. [DOI] [PubMed] [Google Scholar]
- Mishra AK, Alderwick LJ, Rittmann D, Tatituri RV, Nigou J, Gilleron M, et al. Identification of an α(1→6) mannopyranosyltransferase (MptA), involved in Corynebacterium glutamicum lipomanann biosynthesis, and identification of its orthologue in Mycobacterium tuberculosis. Mol Microbiol. 2007;65:1503–1517. doi: 10.1111/j.1365-2958.2007.05884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Alderwick LJ, Rittmann D, Wang C, Bhatt A, Jacobs WR, Jr, et al. Identification of a novel α(1→6) mannopyranosyltransferase MptB from Corynebacterium glutamicum by deletion of a conserved gene, NCgl1505, affords a lipomannan- and lipoarabinomannan-deficient mutant. Mol Microbiol. 2008a;68:1595–1613. doi: 10.1111/j.1365-2958.2008.06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Klein C, Gurcha SS, Alderwick LJ, Babu P, Hitchen PG, et al. Structural characterization and functional properties of a novel lipomannan variant isolated from a Corynebacterium glutamicum pimB' mutant. Antonie Van Leeuwenhoek. 2008b;94:277–287. doi: 10.1007/s10482-008-9243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Batt S, Krumbach K, Eggeling L, Besra GS. Characterization of the Corynebacterium glutamicum deltapimB' deltamgtA double deletion mutant and the role of Mycobacterium tuberculosis orthologues Rv2188c and Rv0557 in glycolipid biosynthesis. J Bacteriol. 2009;191:4465–4472. doi: 10.1128/JB.01729-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita YS, Patterson JH, Billman-Jacobe H, McConville MJ. Biosynthesis of mycobacterial phosphatidylinositol mannosides. Biochem J. 2004;378:589–597. doi: 10.1042/BJ20031372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita YS, Sena CB, Waller RF, Kurokawa K, Sernee MF, Nakatani F, et al. PimE Is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of phosphatidylinositol mannoside in Mycobacteria. J Biol Chem. 2006;281:25143–25155. doi: 10.1074/jbc.M604214200. [DOI] [PubMed] [Google Scholar]
- Morr M, Takeuchi O, Akira S, Simon MM, Mühlradt PF. Differential recognition of structural details of bacterial lipopeptides by toll-like receptors. Eur J Immunol. 2002;32:3337–3347. doi: 10.1002/1521-4141(200212)32:12<3337::AID-IMMU3337>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Nigou J, Vasselon T, Ray A, Constant P, Gilleron M, Besra GS, et al. Mannan chain length controls lipoglycans signalling via and binding to TLR2. J Immunol. 2008;180:6696–6702. doi: 10.4049/jimmunol.180.10.6696. [DOI] [PubMed] [Google Scholar]
- Quesniaux VJ, Nicolle DM, Torres D, Kremer L, Guérardel Y, Nigou J, et al. Toll-like receptor 2 (TLR2)-dependent-positive and TLR2-independent-negative regulation of proinflammatory cytokines by mycobacterial lipomannans. J Immunol. 2004;172:4425–4434. doi: 10.4049/jimmunol.172.7.4425. [DOI] [PubMed] [Google Scholar]
- Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Seidel M, Alderwick LJ, Birch HL, Sahm H, Eggeling L, Besra GS. Identification of a novel arabinofuranosyltransferase AftB involved in a terminal step of cell wall arabinan biosynthesis in Corynebacterianeae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J Biol Chem. 2007a;282:14729–14740. doi: 10.1074/jbc.M700271200. [DOI] [PubMed] [Google Scholar]
- Seidel M, Alderwick LJ, Sahm H, Besra GS, Eggeling L. Topology and mutational analysis of the single Emb arabinofuranosyltransferase of Corynebacterium glutamicum as a model of Emb proteins of Mycobacterium tuberculosis. Glycobiology. 2007b;17:210–219. doi: 10.1093/glycob/cwl066. [DOI] [PubMed] [Google Scholar]
- Sena CB, Fukuda T, Miyanagi K, Matsumoto S, Kobayashi K, Murakami Y, et al. Controlled expression of branch-forming mannosyltransferase is critical for mycobacterial lipoarabinomannan biosynthesis. J Biol Chem. 2010;285:13326–13336. doi: 10.1074/jbc.M109.077297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovierova H, Larrouy-Maumus G, Pham H, Belanova M, Barilone N, Dasgupta A, et al. Biosynthetic origin of the galactosamine substituent of arabinbogalactan in Mycobacterium tuberculosis. J Biol Chem. 2010;285:41348–41355. doi: 10.1074/jbc.M110.188110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatituri RV, Alderwick LJ, Mishra AK, Nigou J, Gilleron M, Krumbach K, et al. Structural characterization of a partially arabinosylated lipoarabinomannan variant isolated from a Corynebacterium glutamicum ubiA mutant. Microbiology. 2007a;153:2621–2629. doi: 10.1099/mic.0.2007/008078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatituri RV, Illarionov PA, Dover LG, Nigou J, Gilleron M, Hitchen PG, et al. Inactivation of Corynebacterium glutamicum NCgl0452 and the role of MgtA in the biosynthesis of a novel mannosylated glycolipid involved in lipomannan biosynthesis. J Biol Chem. 2007b;282:4561–4572. doi: 10.1074/jbc.M608695200. [DOI] [PubMed] [Google Scholar]
- Vignal C, Guérardel Y, Kremer L, Masson M, Legrand D, Mazurier J, et al. Lipomannans, but not lipoarabinomannans, purified from Mycobacterium chelonae and Mycobacterium kansasii induce TNF-alpha and IL-8 secretion by a CD14-toll-like receptor 2-dependent mechanism. J Immunol. 2003;171:2014–2023. doi: 10.4049/jimmunol.171.4.2014. [DOI] [PubMed] [Google Scholar]
- Xie G, Keyhani NO, Bonner CA, Jensen RA. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol Mol Biol Rev. 2003;67:303–342. doi: 10.1128/MMBR.67.3.303-342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, Ballou CE. Synthesis of α(1→6)-mannooligosaccharides in Mycobacterium smegmatis. Function of beta-mannosylphosphoryldecaprenol as the mannosyl donor. J Biol Chem. 1989;264:21621–21628. [PubMed] [Google Scholar]
- Zhang N, Torrelles JB, McNeil MR, Escuyer VE, Khoo KH, Brennan PJ, Chatterjee D. The Emb proteins of mycobacteria direct arabinosylation of lipoarabinomannan and arabinogalactan via an N-terminal recognition region and a C-terminal synthetic region. Mol Microbiol. 2003;50:69–76. doi: 10.1046/j.1365-2958.2003.03681.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


