Abstract
Deficiency in repair of nuclear and mitochondrial DNA damage has been linked to several neurodegenerative disorders. Many recent experimental results indicate that the post-mitotic neurons are particularly prone to accumulation of unrepaired DNA lesions potentially leading to progressive neurodegeneration. Nucleotide excision repair is the cellular pathway responsible for removing helix-distorting DNA damage and deficiency in such repair is found in a number of diseases with neurodegenerative phenotypes, including Xeroderma Pigmentosum and Cockayne syndrome. The main pathway for repairing oxidative base lesions is base excision repair, and such repair is crucial for neurons given their high rates of oxygen metabolism. Mismatch repair corrects base mispairs generated during replication and evidence indicates that oxidative DNA damage can cause this pathway to expand trinucleotide repeats, thereby causing Huntington’s disease. Single-strand breaks are common DNA lesions and are associated with the neurodegenerative diseases, ataxia-oculomotor apraxia-1 and spinocerebellar ataxia with axonal neuropathy-1. DNA double-strand breaks are toxic lesions and two main pathways exist for their repair: homologous recombination and non-homologous end-joining. Ataxia telangiectasia and related disorders with defects in these pathways illustrate that such defects can lead to early childhood neurodegeneration. Aging is a risk factor for neurodegeneration and accumulation of oxidative mitochondrial DNA damage may be linked with the age-associated neurodegenerative disorders Alzheimer’s disease, Parkinson’s disease and amyotrophic lateral sclerosis. Mutation in the WRN protein leads to the premature aging disease Werner syndrome, a disorder that features neurodegeneration. In this article we review the evidence linking deficiencies in the DNA repair pathways with neurodegeneration.
Keywords: DNA repair, Genomic instability, Reactive oxidative species, Neurodegeneration, Aging, Mitochondria, Cockayne syndrome, Alzheimer’s disease, Parkinson’s disease, Werner syndrome
1. Introduction
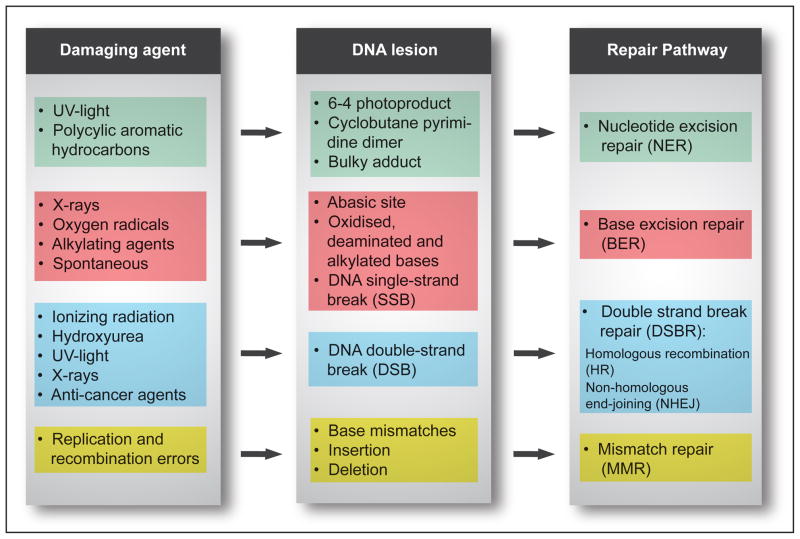
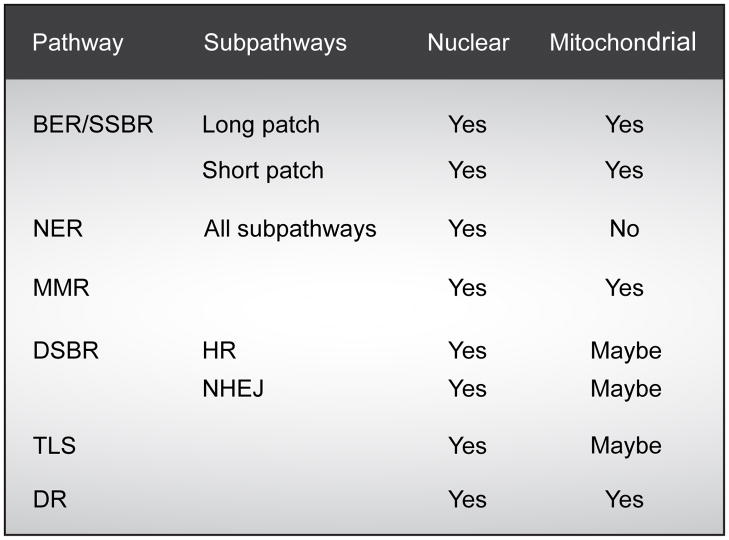
Amongst the fundamental processes, crucial for viability of organisms, including humans, are appropriate cellular signaling responses to DNA damage and the ability to repair such damage. Our cells are constantly exposed to DNA damage caused by endogenous sources such as reactive oxygen species and exogenous sources such as mutagens and radiation. To protect against this damage all cells have various DNA repair pathways. The four major pathways for repairing damage to bases are nucleotide excision repair (NER), base excision repair (BER), mismatch repair (MMR) and double-strand break repair (DSBR) (Fig. 1). NER excises bulky helix-distorting DNA lesions and BER repairs damage to a single nucleotide base, whereas MMR corrects mismatches of the normalbases; such as failure to maintain normal Watson-Crick base pairing. Breakage of the DNA backbone also occurs, either in the form of a single-strand break (SSB) or a double-strand break (DSB). SSBs are handled by the BER pathway. The repair of DNA DSBs involves one of two mechanisms: non-homologous end-joining (NHEJ) or homologous recombination (HR). NHEJ directly joins the broken ends, whereas HR uses the intact sister chromatid as a template for repair. In addition, a type of repair termed direct reversal (DR) can reverse some forms of base damage without removing the base. Translesion DNA synthesis (TLS) uses specialized DNA polymerases to replicate past lesions in the DNA, which although more error-prone than BER, NER and MMR, may reduce the immediate danger of DSBs (Prakash and Prakash, 2002).
Fig. 1. DNA lesions and their repair by the four major DNA repair pathways in higher eukaryotes.
Cells have multiple DNA repair pathways that provide the capacity to repair many different types of DNA lesions. The figure provides an overview of DNA damaging agents, the lesions they cause and the four main pathways responsible for removing and repairing the DNA lesions.
Deficiencies in DNA repair pathways can result in reduced stability of the cellular chromosomes which in turn can lead to mutagenesis, cellular dysfunction and aberrant phenotypes. Such genomic instability would be expected to potentially increase the risk of cancer, and indeed several hereditary DNA repair deficiency diseases (e.g. Xeroderma Pigmentosumare associated with increased cancer risk. Another major clinical feature of such deficiencies is neurological disease, and accordingly, DNA repair deficiencies are implicated in various diseases that feature progressive neurodegeneration. In the central nervous system (CNS), higher levels of DNA damage either due to increased exposure to damaging agentsand/or defective repair of DNA, can lead to pronounced neuropathology. The brain consists largely of non-proliferative neuronal cells and is therefore particularly vulnerable to defective DNA repair that would lead to “accumulation” (more accurately, a greater steady-state level) of unrepaired DNA lesions. These DNA lesions have been proposed to be the cause of the neuropathology observed in several neurodegenerative disorders.
Progressive neurodegeneration occurs when the loss of neuronal structure or function leads to a decline in the number of neurons due to apoptotic cell death. The most consistent risk factor for developing a progressive neurodegenerative disease is aging. With age often comes a decline in brain volume and function, which similarly to neurodegenerative disease can be attributable to the permanent loss of neurons (Brazel and Rao, 2004). The ”free radical theory of aging” hypothesizes that accumulation of unrepaired oxidative damage leads to the cellular decline and associated age-related deterioration (Harman, 1981). Considerable circumstantial evidence supports the role of oxidative damage in the aging process (Balaban et al., 2005; Bokov et al., 2004; Golden et al., 2002; Sinclair, 2005), and neurons have very high rates of oxygen metabolism. In view of this it has been suggested that deficiencies in the repair of oxidative DNA damage with aging, correlates with the cognitive decline and neurodegenerative diseases that are more prominent in the aged population(Weissman et al., 2007a). Mitochondria, the main cellular energy generators, are vital for proper neuronal function and survival, and their dysfunction have been linked to neurodegeneration. In addition, it has been suggested by the “mitochondrial theory of aging” that accumulation of mitochondrial damage is the cause of the normal aging process (Harman, 1972).
In this review, we present an overview of the current understanding of the molecular basis for neuronal DNA repair deficiencies associated with neurodegeneration. This will be done by exploring the evidence gained from the study of both inherited and age-associated neurodegenerative diseases. Included are brief descriptions with illustrations of various pathways of DNA repair that we hope will be helpful to readers not already intimately familiar with these important cellular pathways.
2. Nucleotide excision repair deficiency
2.1. Nucleotide excision repair (NER)
Damage from ultraviolet (UV) radiation and reactive oxygen species can generate helix-distorting DNA lesions. The DNA repair process responsible for removing such lesions is the nucleotide excision repair (NER) pathway. NER is a highly conserved and versatile multistep pathway capable of repairing lesions such as UV-induced cyclobutane pyrimidine dimers and 6-4 photoproducts, intra-strand crosslinks (Niedernhofer et al., 2004), DNA-protein crosslinks (Nouspikel, 2008) and some DNA adducts caused by oxidative damage (D’Errico et al., 2006; Satoh et al., 1993). In human cells, recognition of these helix distortions leads to the removal of a short single-stranded DNA segment which holds the lesion (de Boer and Hoeijmakers, 2000). This creates a single-strand gap in the DNA which subsequently is filled during repair synthesis by a DNA polymerase using the undamaged strand as a template. NER can be divided into two subpathways, global genome NER (GG-NER) and transcription-coupled NER (TC-NER) that differ in the recognition of the DNA lesion, but subsequently uses the same excision mechanism (Fig. 2). GG-NER recognizes and repairs DNA lesions anywhere in the genome whereas TC-NER only resolves lesions in the actively transcribed strand of genes. Recent reviews describe GG-NER (Shuck et al., 2008) and TC-NER in detail (Fousteri and Mullenders, 2008). The NER pathway is active in the post-mitotic neurons though the activity is lower than in fibroblasts (Yamamoto et al., 2007). Two NER-associated disorders, Xeroderma Pigmentosum and Cockayne syndrome, both feature progressive neurodegeneration and will be discussed below.
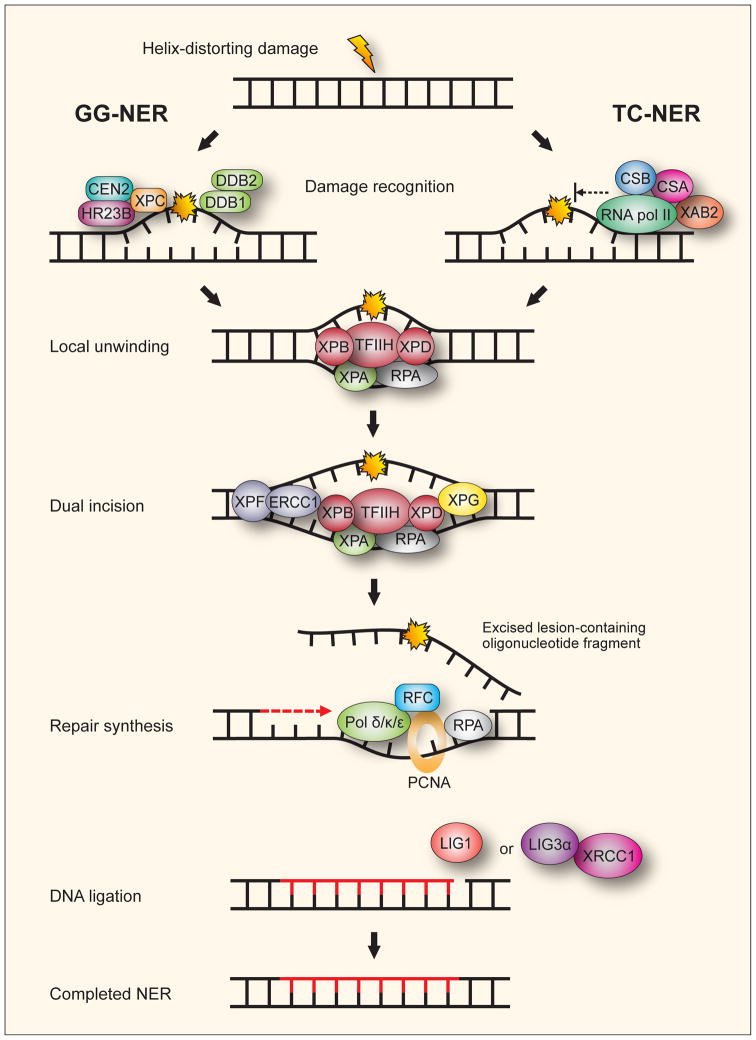
Fig. 2. The two subpathways of mammalian nucleotide excision repair.
In global genome nucleotide excision repair (GG-NER) helix distorting DNA damage anywhere in the genome is recognized by the XPC-HR23B-CEN2 complex. The DDB complex consisting of the two subunits DDB1 and DDB2 (XPE) can facilitate recognition of lesions that by themselves cause little distortion of the helix. In transcription-coupled nucleotide excision repair (TC-NER) recognition is by the stalling of RNA pol II at DNA lesions on the transcribed strand of active genes facilitated by CSB, CSA and XAB2. Either XPC in GG-NER or CSB and CSA in TC-NER recruit TFIIH to the repair site followed by converging of the subpathways. The XPB and XBD subunits of TFIIH are DNA helicases that unwind the DNA in the immediate vicinity of the lesion. RPA and XPA bind to keep the DNA strands apart. For the dual incision, XPA recruits the XPF-ERCC1 endonuclease to incise the damaged DNA strand 5′ to the lesion while XPG incises 3′ to it. The lesion is thus excised in an oligonucleotide fragment leaving behind a single-strand gap. Repair synthesis is performed by DNA polymerase δ and κ, or ε (Pol δ/κ/ε) with the help of the accessory proteins RFC, PCNA and RPA. The remaining nick in the DNA backbone is sealed with ligation by either LIG1 or LIG3α-XRCC1.
2.2. Molecular mechanism of NER
For NER to be initiated the two prerequisites are: the presence of a lesion in the DNA, and a resulting helix-distorting disruption of the duplex DNA structure. For convenience, the NER pathway can be described as a process of five sequential steps:
damagerecognition of the base lesion,
local unwinding of the DNA in the vicinity of the lesion,
dualincision of the DNA strand on the 3′ and 5′ side of the base damage site leading to the excision of a single-stranded lesion-containing oligonucleotide fragment,
repairsynthesis of DNA to fill the nucleotide gap,
DNAligation to seal the nick, restoring covalent integrity.
The first step, recognition, differs between GG-NER and TC-NER.
NER does not recognize the DNA lesion or its nature as such, but rather recognizes the distortion in the structure of the DNA double helix, caused by the lesion. In GG-NER, recognition of helix-distortion is facilitated by XPC, suggested by many studies to be the first protein factor to arrive at the lesion. XPC is complexed with HR23B (most often) or HR23A, two orthologs of the yeast protein Rad23, and also CEN2 (Sugasawa et al., 1997; Sugasawa, 2006; Wood, 1999). XPC is a DNA binding protein that preferentially binds to damaged DNA with distorting structures that are substrates for NER (Wood, 1999). Poly-ubiquitination of XPC occurs upon DNA damage and this post-translational modification increases its affinity for DNA. The function of HR23B is not known but as it is an ortholog of Rad23 it is most likely involved in the ubiquitination of XPC. While not absolutely required, CEN2 is usually present and serves to stabilize the protein complex (Araki et al., 2001). The UV-induced cyclobutane pyrimidine dimersand 6-4 photoproductslesions cause little distortion of the helix by themselves. The DDB complex, consisting of the two subunits DDB1 and DDB2 (XPE), can facilitate recognition of such photo lesions by binding to the lesion and inducing a stronger distortion, thereby enhancing recognition by the XPC-HR23B-CEN2 complex (Sugasawa, 2006). The DDB complex is also part of the E3 ubiquitin ligase responsible for attaching ubiquitin monomers to XPC. In TC-NER, recognition is facilitated by CSB, CSA and XAB2. These are recruited to RNA polymerase II (RNA pol II) to stabilize it when the polymerase is stalled at a DNA lesion in the transcribed strand of a gene during active transcription (Laine and Egly, 2006; Tsutakawa and Cooper, 2000) and to recruit other NER proteins.
For the local unwinding of the DNA duplex, the multi-subunit transcription factor TFIIH is recruited to the site of damage by either XPC (in GG-NER) or CSB and CSA (in TC-NER). XPG binds to TFIIH and stabilizes the complex (Ito et al., 2007). The XPB and XBD subunits of TFIIH are 3′-5′ and 5′-3′ DNA helicases, respectively, unwinding the DNA duplex in the immediate vicinity of the lesion (Winkler et al., 2000). The short stretches of single-stranded DNA (ssDNA) created by the unwinding facilitates binding of the XPA complex consisting of XPA and the ssDNA binding protein RPA. RPA and XPA stabilize the open structure (Missura et al., 2001; Patrick and Turchi, 2002).
For the dual incisions, the heterodimeric XPF-ERCC1 endonuclease protein is recruited by XPA to incise the damaged strand 5′ to the lesion and then the endonuclease activity of XPG incises the damaged strand 3′ to the lesion (Staresincic et al., 2009). The incisions flanking the damaged site generate a single-stranded oligonucleotide fragment 27–30 nucleotides in length which includes the damaged base. The fragment is thus excised from the genome leaving behind a single-stranded gap. No additional specific factors appear to be needed for the excision in mammalian NER, at least invitro(Riedl et al., 2003).
Repair synthesis to fill the gap is performed by DNA polymerase holoenzyme complexes, consisting of DNA polymerase δ and κ, or ε (Pol δ, κ, ε) and accessory proteins, using the undamaged strand as template. RPA appears to recruit the clamp loader RFC and the PCNA clamp to the repair site. Pol δ is then recruited by unmodified PCNA, the classical RFC1-RFC replication factor complex and p66. Pol ε recruitment is dependent on the CTF18-RFC clamp loader. Pol κ appears to be recruited by ubiquitinated PCNA and XRCC1. The clamp loader proteins, RFC1-RFC and CTF18-RFC, catalyze the loading of PCNA on to DNA so it can serve as a processivity factor in DNA synthesis by Pol δ, ε and κ (Ogi et al., 2010; Ogi and Lehmann, 2006). (v): To restore the integrity of the DNA backbone, the remaining nick in the DNA backbone is sealed by DNA ligase I (LIG1) during the S phase of the cell cycle, or by the DNA ligase IIIα (LIG3α)-XRCC1 complex throughout the cell cycle (Moser et al., 2007).
2.3. Xeroderma pigmentosum
Xeroderma Pigmentosum (XP) is an autosomal recessive hereditary disease characterized by marked photosensitivity, hyperpigmentation and a >1000-fold increased risk of skin cancer, mainly basal and squamous cell carcinomas (Kraemer et al., 1987). An additional feature of XP in about 30% of patients (Kraemer et al., 2007) is neurological abnormalities, referred to as XP neurological disease (Rapin et al., 2000; Robbins et al., 1991). Typical neurological symptoms include abnormal motor control, ataxia (uncoordinated movements), peripheral neuropathy, dementia, brain and spinal cord atrophy, microcephaly and sensorineural deafness (Kraemer et al., 1987; Robbins et al., 1991). The severity of the symptoms varies as does the age of diagnosis, and the disease is progressive with increasing severity of symptoms over time, including cognitive decline and dementia. Most of the symptoms in XP neurological disease have been attributed to progressive neurodegeneration by apoptotic neuronal cell death. This is supported at the histological level by the observation that loss of neurons occur in several different regions of the brain (Rapin et al., 2000).
In the late 1960s it was shown that cells from XP patients are defective in NER (Cleaver, 1968; Setlow et al., 1969). XP patients fall into seven complementation groups, XP-A to XP-G, corresponding to mutations in the NER genes XPA-G (Kraemer et al., 2007). An eighth group, XP-V, of patients does not have defective NER, but rather have mutated polymerase η (Masutani et al., 1999). The severity and nature of the symptoms is determined by the specific mutation and reflects the role of the protein in the NER process. XP complementation group A patients most frequently feature neurodegeneration (Hentati et al., 1992). Neurodegeneration may also occur in XP-B and XP-D patients (Hentati et al., 1992) and milder, typically adult onset, neurodegeneration can occur in XP-C and XP-F (Robbins et al., 1991; Sijbers et al., 1998). Neurodegeneration has not been reported so far in patients with the least severe form of the disorder, those from group XP-E (Rapic-Otrin et al., 2003). XPA, XPB, XPD and XPF are required for TC-NER while XPC and XPE (DDB2) are dispensable for this subpathway (Fig. 2). Thus it would seem that TC-NER is the most important pathway for protecting neurons while GG-NER deficiency (XP-C and XP-E patients) leads at most to mild neurodegeneration. This view is supported by the fact that GG-NER activity is attenuated in the post-mitotic neurons relative to mitotic cells while the TC-NER activity is the same (Nouspikel and Hanawalt, 2000). In the case of XP-F there is in vitro evidence that mutations in the XPF part of the XPF-ERRC1 endonuclease are responsible for at least part of the DNA repair defect and symptoms by causing cytoplasmic rather than nuclear cellular localization of XPF-ERCC1 (Ahmad et al., 2010). It was not possible, however, to predict the disease severity of XP-F patients by the level of cytoplasmic XPF-ERCC1.
As UV radiation does not reach the human brain and most chemical adducts cannot cross the blood-brain barrier, it has been hypothesized that the neurodegeneration in XP patients is caused by accumulation of endogenous DNA lesions that would normally be repaired by NER (Andrews et al., 1978). Oxidative damage is considered the main form of endogenous DNA lesion in the brain (Weissman et al., 2007a) and such lesions are mostly repaired by the BER pathway as discussed in Section 3. However, it has been shown that oxygen radicals can generate DNA lesions that are repaired by NER, in vitro(Yamamoto et al., 2007). One such class of oxidative lesions, 8,5′-cyclopurine-2′-deoxynucleosides (cyclopurines), have an extra, second bond between the base and the DNA backbone, and are generated specifically by hydroxyl radicals (•OH) (Brooks, 2007). Cyclopurines fulfill several criteria that can reasonably be applied to DNA lesions responsible for causing the progressive neurodegeneration in XP: they are substrates for NER, but not other DNA repair pathways (Brooks et al., 2000; Kuraoka et al., 2000), they are chemically stable (Brooks, 2007), they are endogenous lesions (Dizdaroglu et al., 2001; Randerath et al., 2001) and they may block transcription by RNA pol II (Brooks et al., 2000). It has not, however, been determined whether cyclopurines accumulate in cells from XP patients (Brooks, 2007). Other potential neurodegenerative DNA lesions are the bulky propane-deoxyguanosine (PdG) adducts that are generated when lipid peroxidation products such as malodialdehyde react with DNA (Burcham, 1998). PdG lesions are endogenous (Marnett, 1999), repaired by NER (Johnson et al., 1997) and can block RNA pol II (Cline et al., 2004). However, PdG adducts are not as likely as cyclopurines to be stable over the period of time when neurodegeneration progresses in XP patients (Brooks, 2007).
Whatever the responsible DNA lesion, cyclopurine, PdG adduct or some as yet uncharacterized lesion, neurodegeneration is thought to partially result from the blocked transcription that occurs as lesions stall RNA pol II. This inactivation of genes could decrease the level of essential protein components of the neurons to the point that cell death becomes inevitable (Andrews et al., 1978) or the blockage could be a more direct trigger for apoptosis pathways due to the presence of stalled RNA pol II (Kohji et al., 1998; Ljungman and Lane, 2004). Also, there are multiple signaling pathways that result from the presence of lesions or stalled polymerase or repair complexes. It should be noted that many of the neurological symptoms in XP also feature in normal aging including peripheral neuropathy, dementia and hearing loss. This deterioration of neurological function has, in both aging and XP, been linked to a permanent loss of neurons that leaves the glial cell population largely unchanged. As such, XP neurological disease can be seen to resemble a form of accelerated aging of the nervous system.
2.4. Cockayne syndrome
Cockayne syndrome (CS) is a rare autosomal recessive disease featuring progressive childhood neurological impairment. The neurological symptoms include demyelination in the cerebral and cerebellar cortex, calcification in basal ganglia and cerebral cortex, neuronal loss, sensorineural hearing loss and decreased nerve conduction. Neurodegeneration is most pronounced in the cerebellum with severe loss of Purkinje and granule neurons and the second most affected areas of pathology is the basal ganglia nuclei and thalamus (Weidenheim et al., 2009). Other symptoms include cachectic dwarfism, retardation of growth and development after birth, and often photosensitivity and cataracts (Licht et al., 2003). The disease does not seem to confer an increased risk of cancer. The life expectancy of CS patients is 12.5 years, and as many of the disease features resemble normal aging, it has been classified as a premature aging syndrome. The cause of CS is in 62% of cases mutations in the CSB gene (CS-B patients) (Laugel et al., 2010), while most of the rest result from mutations in CSA (CS-A patients), though some XP-B, XP-D and XP-G patients can have a combined XP/CS phenotype (de Boer and Hoeijmakers, 2000; Rapin et al., 2000). The symptoms of CS-B patients cannot be distinguished from those of CS-A. The CSB and CSA proteins are active in TC-NER and in CS patients this pathway is defective for the repair of UV-induced cyclobutane pyrimidine dimers and 6-4 photoproducts lesions while the GG-NER pathway functions normally (Licht et al., 2003; Venema et al., 1990). Consequently, one of the hallmarks of CS-B cells is hypersensitivity to UV radiation-induced DNA damage in active genes. The 168 kDa CSB protein belongs to the SWI/SNF2 family of chromatin remodelers. The protein has an SNF2-like ATPase domain consisting of seven conserved motifs (I, Ia and II–VI) and the DNA-dependent ATPase activity is crucial for recovery from cellular UV-sensitivity, RNA synthesis restart after oxidative damage and chromatin remodeling (Stevnsner et al., 2008). CSB plays a key role in the initiation of TC-NER by recruiting the histone acetyltransferase p300 to the damaged site for chromatin remodeling and recruiting other NER proteins to the stalled RNA pol II (Fousteri et al., 2006). CSB association with RNA pol II and TFIIH forms the basis of several different models that have been suggested to explain the neurological symptoms in CS:
The “transcription defect” model proposes that the symptoms are caused by a subtle defect in transcription, and that the reduction in the process stems from mutations in the proteins involved. In support of this model are the findings that CSB associates with RNA pol II in vivo(van Gool et al., 1997) and stimulates elongation in vitro(Selby and Sancar, 1997), and the observation of reduction of transcription in CS-B cells in vivo(Balajee et al., 1997) and in CS-B and XP-B extracts in vitro(Dianov et al., 1997). CS symptoms are also often seen in XP-B and XP-D, and given that XPB and XPD are subunits of TFIIH, it is conceivable that mutations in these proteins could reduce transcription. The neurological symptoms are then in this model explained by high neuronal sensitivity to transcription defects, given that a high proportion of the neuronal (but not glial) genome is transcribed (Lein et al., 2007). This model, however, still leaves the severe white matter degeneracy in CS patients, unexplained.
The “recycling” model proposes that TFIIH shifts between two different conformations, transcription and repair, and that CSB is required to switch TFIIH back to transcription. CS symptoms would then occur when TFHII is stuck in repair conformation because of the mutated CSB, reducing transcription. CSA is part of an E3 ubiquitin ligase complex (Groisman et al., 2003). As CSB is ubiquitinated by the E3 complex and rapidly degraded by the proteasome (Groisman et al., 2006), CSA could be required for removal of CSB allowing TFHII to switch back to transcription mode. A variant of this model postulates that a stalled RNA pol II blocks transcription and impairs TC-NER by preventing the repair enzymes from gaining access to the DNA lesion. TC-NER cannot occur before RNA pol II has been removed (Svejstrup, 2002). Supporting this view is the fact that yeast deficient in RAD26 (CSB homolog) displays slower NER of the transcribed strand than the non-transcribed strand (Tijsterman and Brouwer, 1999), and finding a “footprint” of a stalled RNA pol II (Tornaletti et al., 1999). One way to resolve this blockage would be for RNA pol II to change conformation to reveal the DNA lesion, and there is in vitro evidence that CSB and TFIIH are vital for such remodeling activity (Sarker et al., 2005). A mutated CSB would then presumably impair this RNA pol II remodeling thereby contributing to CS pathology. Another way for the blockage to be resolved would be for CSB to remove stalled RNA pol II from the DNA altogether (Svejstrup, 2002), and mutated CSB would fail to do this. Yet another possibility is for RNA pol II to be ubiquitin-tagged for degradation, controlled by CSA and CSB (Bregman et al., 1996).
It is perhaps not likely that the pleiotropic nature of CS can be explained by just one of the above models, and several different mechanisms may give rise to the CS pathology. Also, like for XP neurological disease, symptom-causing DNA damage may potentially be oxidatively induced lesions. The brains of knockout CSB−/− mice are devoid of CSBand have been observed to contain higher endogenous levels of the cyclopurine (5′S)-8,5′-cyclo-2′-deoxyadenosine, which is a helix-distorting form of DNA lesion (Kirkali et al., 2009). Like the other cyclopurines, this moiety is a form of oxidative DNA damage that due to the presence of a C8-C′5 covalent bond requires resolution by the NER pathway. If such lesions are indeed accumulating in the brains of CS-B patients it implies a role for CSB in their repair in vivo either linked to transcription in TC-NER, globally in the genome by GG-NER, or both.
Lipid peroxidation has been implicated in numerous human diseases particularly many of which have disease progression associated with aging including cancer and atherosclerosis, and lipid peroxidation levels are increased in at least some cell types of aged humans and rats. Peroxidation of omega-3 and omega-6 polyunsaturated fatty acids can give rise to the major product trans-4-hydroxy-2-nonenal (HNE). While the primary targets of HNE in the cell are proteins and thiols, it can also, following oxidation, form an epoxide that readily reacts with DNA bases to form HNE-DNA adducts recognized by mammalian NER proteins (Chung et al., 2003). While toxic to both human wild-type and CSB-deficient cells, the sensitivity to even low physiological levels of HNE was observed to be significantly greater for the CSB-deficient cells (Maddukuri et al., 2009). Induction of HNE-DNA adducts was found to inhibit transcription by T7 RNA polymerase as well as transcription by HeLa cell-free extracts. Interestingly, cells with a CSB ATPase motif II-mutant with no ATPase activity and presumably defective in TC-NER, were just as sensitive to HNE treatment as CSB-null cells. These data present the case for bulky HNE-DNA lesions as transcription inhibitors that block the progress of RNA pol II necessitating active CSB protein for their resolution by TC-NER.
As CSB interacts with proteins of the BER pathway, deficiency in this DNA repair pathway may also be involved in the CS phenotype, a possibility reviewed in Section 3.3. Such an involvement of the CSB protein may extend to the mitochondria as mutated CSB cells also appear to be deficient in mitochondrial BER and display dysfunctional mitochondria, as discussed in Section 7.3. In both the CS and XP diseases neurological tissue have pronounced susceptibility to the effects of NER deficiency, and there may be several reasons for this: neurons may suffer more DNA damage, neurons may also be more sensitive to changes in transcription, and both neurons and glial cells have lower NER activity than other cell types (Yamamoto et al., 2007). The answer to how a defective NER pathway might affect the neurons of the brain shielded as they are from the damaging effect of UV radiation may lie in the endogenously generated cyclopurines. These oxidative lesions are potent blocks for transcription as well as the progress of DNA polymerase δ and the translesion synthesis DNA polymerase η. Brain tissue is very rich in lipids with a particularly high content of the easily oxidized omega-3 and omega-6 polyunsaturated fatty acids while simultaneously having very high rates of oxygen metabolism and a low content of antioxidant enzymes (Barzilai, 2007). Thus compared to other cell types neurons may have to contend with a generation of lipid peroxidation-DNA products, such as PdG and HNE-DNA adducts, that is significantly greater that most tissues, and accordingly, may be highly vulnerable to a cellular NER deficiency. Relevant to this is the observed accumulation of HNE in the cerebrum of Alzheimer’s, Parkinson’s, Huntington’s diseases and amyotrophic lateral sclerosis patients (Zarkovic, 2003). While these diseases are not associated with known defects in NER, the accumulation of HNE is considered a biomarker for oxidative stress. This highlights the potential great impact of oxidative damage in the pathology of many neurodegenerative disorders whether they are linked to a deficiency in NER or other pathways of DNA repair. It is worth keeping in mind, however, that it has so far not been possible to determine whether the neurodegeneration and apparent premature aging in CS are in fact primarily due to a NER deficiency, a transcription deficiency, or a result of a more complex array of defects in the neuronal cells.
3. Base excision repair deficiency
3.1. Base excision repair (BER)
DNA is inherently unstable due to spontaneous hydrolytic decay and due to modification by both endogenous and exogenous alkylating agents (Lindahl, 1993). In addition reactive oxygen species (ROS), such as the highly reactive hydroxyl radical (•OH), superoxide anion (O2•−) and hydrogen peroxide (H2O2) are generated as a result of normal cellular metabolism. ROS is genotoxic and capable of damaging DNA by generating various oxidative DNA lesions with base or sugar damage (Evans et al., 2004; Lindahl, 1993). One such lesion, 7,8-dihydro-8-oxoguanine (8-oxo-dG) is a commonly used cellular biomarker to indicate the extent of oxidative stress (Klaunig and Kamendulis, 2004). A comprehensive description of the various types of oxidative DNA damage can be found in a recent review (Evans et al., 2004). Such DNA lesions can present mutagenic and/or cytotoxic challenges to the cell by blocking replication and transcription. The major pathway responsible for eliminating spontaneous hydrolytic, alkylation and oxidative DNA damage, and thereby restoring genomic integrity, is base excision repair (BER). BER is an evolutionarily conserved DNA repair process responsible for correcting most common forms of DNA damage by recognizing, excising and replacing a broad spectrum of specific forms of DNA modifications (Hoeijmakers, 2001; Krokan et al., 2000). The BER pathway proteins are also involved in repair of DNA single-strand breaks as reviewed in Section 5. The following will describe the process of nuclear BER while mitochondrial BER (mtBER) will be discussed in Section 7.2.
3.2. Molecular mechanism of BER
BER is initiated by a distinct lesion-specific mono- or bifunctional DNA glycosylase and completed by either of two subpathways: short-patch BER (SP-BER) that replaces one nucleotide or long-patch BER (LP-BER) that replaces 2–13 nucleotides (Fig. 3) (Fan and Wilson, 2005; Fortini et al., 2003; Hegde et al., 2008; Sweasy et al., 2006; Wilson and Bohr, 2007). For convenience, the BER pathway can be described as a process of five sequential steps:
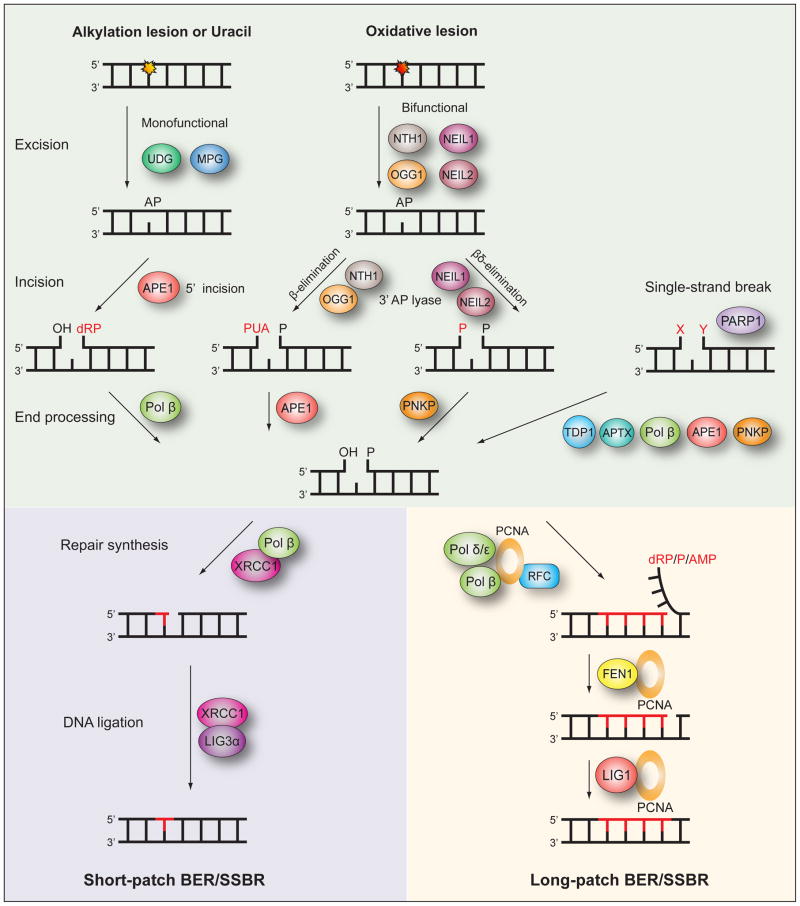
Fig. 3. The mammalian base excision repair and single-strand break repair pathways.
Base excision repair (BER) is initiated by removal of the modified base by either a monofunctional or bifunctional DNA glycosylase to leave an abasic site (AP). If excision is by either one of the monofunctional DNA glycosylases UDG or MPG, the following incision of the DNA backbone 5′ to the AP site is by APE1. Excision by one of the bifunctional DNA glycosylases NTH1, OGG1, NEIL1 or NEIL2 is followed by incision 3′ to the AP site via β- or βδ-elimination facilitated by the intrinsic 3′ AP lyase activity of these enzymes. The resulting single-strand break will contain either a 3′ or 5′ obstructive termini. End processing is then performed by Pol β, APE1 or PNKP depending on the specific nature of the terminus. Single-strand breaks do not only occur as intermediates of BER but also by other means and can contain simultaneous 3′ and 5′ obstructive termini. PARP1 recognizes these breaks and the end processing may utilize the additional factors TDP1 and APTX. When end processing has produced the necessary 3′-OH and 5′-P termini the following BER and single-strand break repair (SSBR) steps diverge into two subpathways, short-patch and long-patch. In short-patch BER/SSBR repair synthesis of the single nucleotide gap is by Pol β aided by the XRCC1 scaffold, and subsequent ligation by LIG3α finishes the repair. In long-patch BER/SSBR repair synthesis of the 2–13 nucleotide gap is by Pol β, and/or Pol δ/ε aided by PCNA and RFC. A resulting 5′ flap is removed by FEN1 and the the final ligation step is by LIG1.
recognition and excision of the inappropriate base moiety (e.g. 8-oxo-dG),
incision of the DNA backbone adjacent to the resulting abasic site,
end processing of the DNA termini to generate a 3′-hydroxyl group (3′-OH) and a 5′-phosphate moiety (5′-P),
repair synthesis to replace the missing nucleotide(s), and
DNAligation to seal the remaining nick.
The initiating step in BER is performed by a distinct DNA glycosylase which recognizes and excises a specific base substrate by catalyzing hydrolysis of the N-glycosylic bond (Dizdaroglu, 2005; Huffman et al., 2005; Stivers and Jiang, 2003). The result of this is an abasic (AP) site with an intact DNA phosphodiester backbone. DNA glycosylases can be either monofunctional or bifunctional. Monofunctional DNA glycosylases, such asUDG andMPG, have only the glycosylase activity. In contrast, bifunctional DNA glycosylases, such as 8-oxoguanine DNA glycosylase (OGG1), NTH1 and NEIL1, have an intrinsic 3′ AP lyase activity in addition to the glycosylase activity.
After excision of the substrate base, the next step is to incise the DNA backbone adjacent to the AP site. The major protein responsible for incision in mammalian BER is APE1 which incises the DNA backbone immediately 5′ to the AP site, leaving a 5′-deoxyribose-5-phosphate (5′-dRP) product (Demple and Sung, 2005; Wilson and Barsky, 2001). The bifunctional DNA glycosylases incise the DNA backbone immediately 3′ to the AP site via β- or βδ-elimination, leaving a DNA SSB with a 3′-phospho-α,β-unsaturated aldehyde (3′-PUA) or a 3′-phosphate (3′-P), respectively.
The third step in BER is end processing of obstructive 3′- and 5′-termini to generate the 3′-OH and 5′-P termini in the gap at the strand break, which is the normal substrate for a DNA polymerase. DNA polymerase β (Pol β) is responsible for removing the 5′-dRP moiety via its 5′-dRP lyase activity (Bennett et al., 1997a; Mol et al., 2000; Wilson, 1998), while APE1 removes the 3-PUA residue generated by β-elimination via its 3′-phosphodiesterase activity. The 3′-P moiety generated by βδ-elimination is a poor substrate for APE1 (Wilson, 2003), so in vivo, such blocking groups are excised primarily by the phosphatase activity of PNKP (Rasouli-Nia et al., 2004; Wiederhold et al., 2004). PNKP is also associated with the resolution of 3′-TOP1-SSB obstructive termini in single-strand break repair (SSBR) (see Section 5.3).
The next step, repair synthesis to replace nucleotide(s), can proceed by one of two subpathways, short-patch (SP) or long-patch (LP). The choice of pathway may depend on several factors (Horton et al., 2000). When the 5′-dRP intermediate can be efficiently removed by Pol β in the previous step (iii), SP-BER is usually favored (Sobol et al., 1996). LP-BER is utilized in cases where the 5′-moiety is refractory to the Pol β AP lyase activity (Gary et al., 1999), for example during repair of a reduced AP site. The majority of BER events is currently thought to proceed via the short-patch pathway (Almeida and Sobol, 2007). In SP-BER, Pol β performs repair synthesis to fill the single nucleotide gap. Pol β interacts with XRCC1, a scaffold protein involved in promoting SP-BER by recruiting other proteins (Gryk et al., 2002; Kubota et al., 1996). In LP-BER, the repair synthesis of 2–13 nucleotides is performed by Pol β, and/or Pol δ/ε coupled with the PCNA clamp in cooperation with the loading factor RFC (Fan and Wilson, 2005). The resulting 5′-flap structure formed during the repair synthesis is removed by the flap endonuclease, FEN1 (Fan and Wilson, 2005; Levin et al., 2004), the activity of which is stimulated by PCNA (Gary et al., 1997; Wu et al., 1996). Poly(ADP-ribose) polymerase-1 (PARP1) may help to facilitate LP-BER (Frouin et al., 2003; Prasad et al., 2001).
The final step in BER is ligation to seal the nick containing a 3′-OH terminus and a 5′-P terminus. In SP-BER the ligation is believed to be performed by the ligase activity of LIG3α present in a LIG3α-XRCC1 complex, where XRCC1 is believed to serve as a scaffold and to stabilize LIG3αin vivo(Caldecott et al., 1994; Caldecott et al., 1995; Cappelli et al., 1997). PARP1 is apredominantly nuclear enzyme that responds to oxidative DNA damage and facilitates DNA repair by BER. It associates with LIG3α and slightly enhances the ligase activity (Caldecott et al., 1996; Leppard et al., 2003; Schreiber et al., 2002). It is thought that PARP1 senses the DNA SSB created at BER intermediates, helping XRCC1 to recruit the end-processing enzymes used in step (iii) (Ziegler and Oei, 2001). Binding of the PARP1 to DNA breaks activates the protein to transiently modify itself and target proteins with branched chains of ADP-ribose units. PARP2 also associates with the LIG3α-XRCC1 complex and seems to be required for efficient BER (Schreiber et al., 2002). In LP-BER, ligation is performed by LIG1 (Levin et al., 2000). LIG1 is physically associated with PCNA (Levin et al., 2000; Montecucco et al., 1998) and this association may be critical for effective ligation (Levin et al., 2000). APE1 can associate with LIG1 and stimulate its activity when the DNA nick has a 5′-P terminus (Ranalli et al., 2002).
3.3. Association of BER deficiency with neurodegeneration and aging
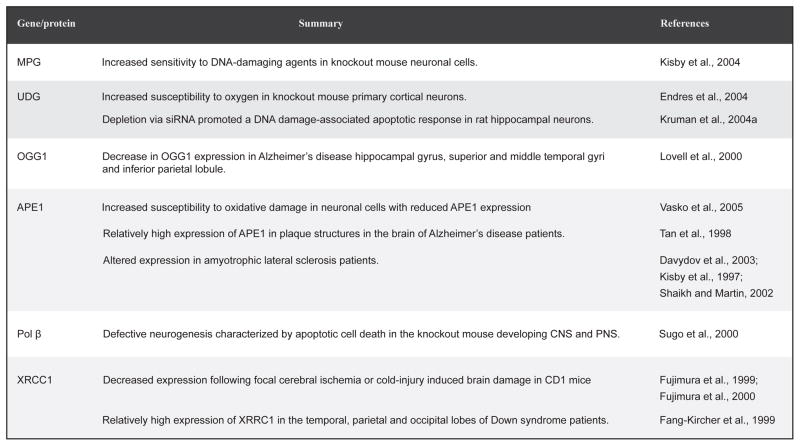
The human brain represents only 2% of the body weight but it extracts aproximatey 50 % of the oxygen and 10% of the glucose from the arterial blood with glucose representingthe obligatory energy substrate utilized by the brain (Magistretti and Pellerin, 1996). Neurons, and particularly their mitochondria, have very high rates of oxygen metabolism due tothe high glucose requirement of neurons and the dependence on aerobic oxidation of glucose as their source of energy (Bell et al., 1993; Bruckner et al., 1999; Ly and Verstreken, 2006). Neurons are not only highly energetic but also long-lived. Combined with the low level of antioxidant enzymes in the brain, the type of DNA damage most likely to occur in the neuronal cells is ROS-induced oxidative DNA lesions(Nouspikel and Hanawalt, 2000; Viswanathan et al., 1999). In fact, the rate of ROS production in the CNS has been proposed to be inversely proportional to life-span in vertebrates(Barja, 2004a; Hinerfeld et al., 2004). As it is believed that accumulation of oxidative DNA damage may lead to neurodegeneration (Barja, 2004b), and since BER is the major pathway for correcting this damage, it has been hypothesized that BER deficiencies may be a cause for neurodegenerative disorders(Yang et al., 2008). The BER pathway is known to be active in the neurons of the CNS. Induction of the BER proteins XRCC1, LIG3α and Pol β is seen in ischemically preconditioned rat brains, and the total BER capacity of nuclear extracts prepared from such brains is increased, most likely due to increased gene expression of these proteins (Li et al., 2007b). The increased ability to repair oxidative DNA base damage probably accounts for the attenuation of neuronal cell death observed in the ischemically preconditioned rat brains after reperfusion and consequent oxidative stress. In aged rats the activity of APE, the major enzyme responsible for incision of the DNA backbone in BER, is reduced in the frontal/parietal cortex, cerebellum, brainstem, midbrain and hypothalamus compared to young rats (Kisby et al., 2010). While APE activity declined with age, there was no change in the protein levels of APE, Pol β and LIG3 in the rat frontal/parietal cortex perhaps suggesting that the reduced APE activity could be due to altered post-translational modification. Interestingly, the decline in APE activity was significantly smaller in rats subjected to caloric restriction at all ages and brain regions. An overview of BER gene expression in the CNS can be found in various reviews (Weissman et al., 2007a; Wilson and Bohr, 2007). Fig. 4 summaries some of the studies (using mostly knockout animals and siRNA techniques) that have been performed to examine the link between BER and neurodegeneration. These studies provide evidence implicating BER in maintaining the genomic stability of neurons.
Fig. 4.
Overview of studies linking BER and neurodegeneration.
CS patients, as described in Section 2.4., are characterized by progressive neurological impairment and features resembling accelerated aging. CSB, the protein mutated in most patients with CS, may also be involved in BER. CSB-deficient cells seem to be deficient in BER of some oxidative lesions (Tuo et al., 2003), which could be due to either reduced transcription of the BER genes or a direct interaction of CSB with BER proteins. CSB mutant cells are defective in both the repair of 8-oxo-dG (Dianov et al., 1999) and 8-oxoA (Tuo et al., 2002) DNA base damage, indicating that CSB may be important for repair of these abundant lesions. The brains of CSB−/− mice accumulate endogenous oxidative formamidopyrimidine DNA lesions (FapyG and FapyA) (Muftuoglu et al., 2009). These lesions are the product of the imidazole ring-opened purines guanine and adenine, respectively, and NEIL1 is the DNA glycosylase responsible for their resolution by BER in both mice and humans. CSB was found to stimulate both the incision and AP lyase activity of NEIL1 on both FapyG and FapyA lesions in vitro, apparently not requiring the ATPase function of CSB. NEIL1 and CSB were found to co-localize in vivo, and additionally, endogenous NEIL1 and CSB could be co-immunoprecipitated(Muftuoglu et al., 2009). These results suggest that CSB can function in a complex with NEIL1 to resolve oxidative FapyG and FapyA lesions. The CSB protein has been shown to physically interact with the BER proteins PARP1 (Thorslund et al., 2005) and APE1 (Wong et al., 2007). PARP1 binds DNA SSBs, such as those generated during BER, and thereby becomes activated (see Section 5.1.). CSB is a substrate for PARP1 by interaction with the N-terminal part of CSB that lacks the ATPase domain, and PARP1-CSB complexes have been shown to relocate to sites of DNA damage in vivo after oxidative stress(Thorslund et al., 2005). The poly(ADP-ribosyl)ation of CSB by PARP1 (Thorslund et al., 2005) accelerates DNA repair (Flohr et al., 2003) suggesting that CSB functions in the PARP1 stimulation of BER. The interaction of CSB with APE1 stimulates incision of the DNA backbone by APE1 in a manner dependent on ATP (Wong et al., 2007). When functional CSB is absent the processing of 8-oxo-dG base modifications by the DNA glycosylase OGG1 appears to cause a strong transcriptional inactivation of the damaged gene (Khobta et al., 2009). Additional OGG1 deficiency can attenuate this effect making it likely that the SSB and AP site repair intermediates generated by OGG1 are the reason for gene inactivation. CSB is then responsible for preventing such intermediates from causing transcriptional inactivation possibly by facilitating and enhancing transcriptional functions relating to the DNA repair process. This would be consistent with an association of CSB with RNA pol II, as described in Section 2.4.
The interactions of CSB with components of the BER pathway described here imply that impaired BER may contribute to the CS phenotype and that CSB may play a general role in processing of BER substrates. Further evidence linking the BER pathway to repair of neuronal DNA is found in studies looking at BER enzymes in aging neurons. These studies have generally found decreased BER activity and BER protein abundance with age (Xu et al., 2008). One study, using an in vitro UDG-initiated BER assay, found a large decline in nuclear BER activity in brain extracts from old mice compared with 6-day-old mice (Intano et al., 2003). Another study found a reduction in Pol β activity and protein abundance in brain nuclear extracts from old mice compared to young mice (Cabelof et al., 2002). It has been known for some time that oxidative DNA damage accumulates in the mammalian brain with aging (Hamilton et al., 2001; Hirano et al., 1996; Kaneko et al., 1996; Nakae et al., 2000; Sai et al., 1992; Shen et al., 2001). This accumulation is considered a possible cause for the progressive loss of neurons associated with aging (Barja, 2004b), and therefore potentially associates BER-deficiency with age-related neurodegeneration.
The BER proteins are also responsible for repairing DNA SSBs. Such breaks accumulate with age, lead to neuronal cell death, and deficient SSB repair can cause neurodegenerative diseases (see Section 5.), again linking BER deficiency with neurodegeneration. It is also possible that mtBER is more important for protection of neurons, as mitochondria are the primary source of endogenous ROS and mtBER deficiency has been implicated in various neurodegenerative disorders such as Alzheimer’s disease.
4. Mismatch repair defects
4.1. Mismatch repair (MMR)
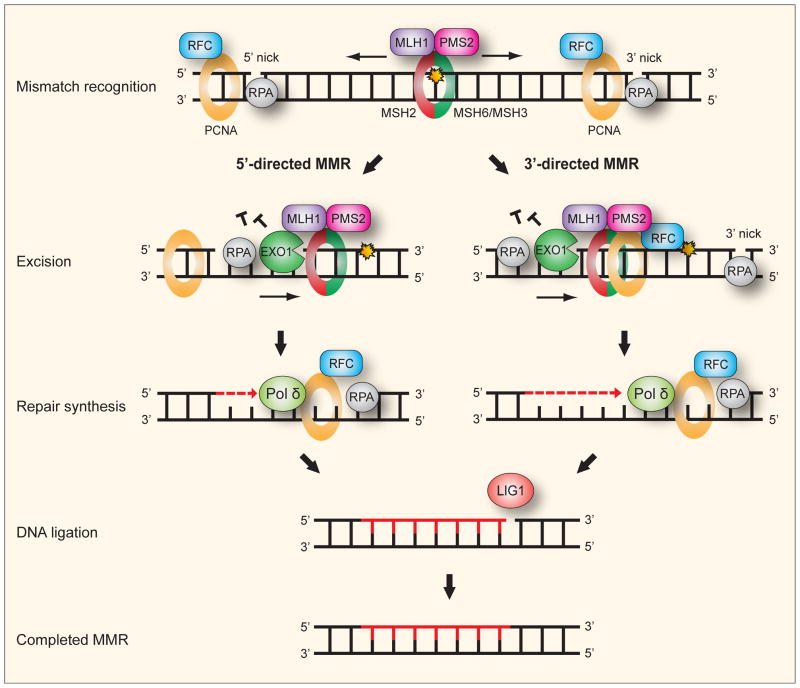
DNA mismatch repair (MMR) is a highly conserved pathway that removes base-base mismatches and insertion-deletion loops that arise during DNA replication and recombination, thereby improving the fidelity of replication 50–1000 fold (Hsieh and Yamane, 2008; Jiricny, 2006). Base-base mismatches are created when errors escape from the proofreading function of DNA polymerases. Insertion-deletion loops arise when primer and template strand in a microsatellite dissociate and re-anneal incorrectly, causing the number of microsatellite-repeat units in the template and in the newly synthesized strand to differ (Kunkel, 1993). Failure of MMR greatly elevates the rate of mutagenesis, increases genomic instability, and is associated with human cancers (Li, 2008). The human MMR system (Fig. 5) has recently been reconstituted with purified proteins, though many aspects remain unclear, and there are several different working models for mammalian MMR. Reviews are available for in depth discussion of MMR (Jiricny, 2006; Li, 2008).
Fig. 5. Human mismatch repair.
For convenience, the mechanism of human mismatch repair (MMR) can be seen as consisting of five consecutive steps: (i) Recognition and binding of a mismatch is by either a MSH2-MSH6 or MSH2-MSH3 heterodimeric ATPase complex. MSH2-MSH6 preferentially recognizes base-base mismatches and insertion deletion loops of 1–2 nucleotides while MSH2-MSH3 has preference for larger insertion-deletion loops. The mismatch-bound MSH2-MSH6 (or MSH2-MSH3) recruits the MLH1-PMS2 complex, a molecular matchmaker with weak ATPase activity, to form a ternary complex. The PCNA clamp recruits MMR proteins to the replication fork while the clamp loader RFC loads PCNA. A strand-specific nick or gap, which may reside either 5′ or 3′ to the mismatch, is sufficient to direct repair in 5′- and 3′-directed MMR, respectively. PCNA appears essential for 3′-directed but not 5′-directed MMR. (ii) Excision is apparently by the 5′ to 3′ exonuclease EXO1 in both 3′- and 5′-directed MMR. For 5′-directed MMR the excision is straightforward by the 5′ to 3′ exonuclease activity of EXO1. For 3′-directed MMR, the endonuclease function of PMS2 is activated by presence of the 3′ nick, and stimulated by RFC, PCNA and ATP, to introduce a necessary second nick 5′ to the mismatch. Excision can then follow by EXO1. RPA binds to protect the single-stranded DNA during the excision and to facilitate the following DNA repair synthesis. (iv) Repair synthesis is accurately performed by Pol δ. (v) Ligation of the remaining nicks after synthesis is by LIG1.
4.2. Huntington’s disease
Huntington’s disease (HD) is an autosomal dominant progressive neurodegenerative disorder. The HD phenotype is characterized by loss of medium spiny neurons, cognitive deterioration and motor dysfunction, with onset of the disease usually in the fourth or fifth decade of life (van Dellen et al., 2005). The mutation in HD is a progressively expanding CAG repeat in the N-terminal coding region of the HD gene. CAG encodes glutamine in the normal gene product named huntingtin, whereas the mutant huntingtin has a growing polyglutamine tract that alters its interaction with huntingtin-binding proteins (Harjes and Wanker, 2003; Li and Li, 2004; Mirkin, 2007). The onset and severity of HD is determined by the length of the repeat. There is evidence to suggest that mutant huntingtin causes mitochondrial dysfunction (Beal, 2005) and elevated mitochondrial DNA damage levels have been found in HD patients (Polidori et al., 1999). Surprisingly, it has been found that the MSH2-MSH3 complex that functions in MMR recognition causes the CAG expansion in HD (Kovtun and McMurray, 2001; Manley et al., 1999; Owen et al., 2005). hHD/MSH2−/− mice displayed abolition of CAG repeat expansion (Kovtun and McMurray, 2001; Manley et al., 1999) and loss of MSH3 abrogated the expansion in transgenic mice harboring the CAG tract in the hHD transgene (Owen et al., 2005). Loss of MSH6 did not prevent expansion (Owen et al., 2005). MSH2-MSH3 has strong repair specificity for small heteroduplex loops, but can also repair single base mismatches (Palombo et al., 1996). CAG can form a stable stem loop structure with a repeat unit of two GC pairs and a mismatched pair (Gacy et al., 1995), and such a heteroduplex loop might conceivably be recognized by MSH2-MSH3, but then fail to be excised in the MMR process (Moore et al., 1999). Since the CAG expansion in HD occurs in post-mitotic neurons (Kovtun et al., 2007), another mechanism is likely to be responsible. One model proposes that expansion occurs as a result of BER of oxidative damage: OGG1 excises an oxidized guanine (8-oxo-dG) to create a single strand nick followed by gap-filling synthesis by a polymerase. CAG repeat hairpins then form by strand displacement together with slippage during synthesis. Removal of the flap by FEN1 is inhibited as the 5′-end is inaccessible, while MSH2-MSH3 stabilizes its hairpin substrate, allowing the slipped-stranded intermediate to be converted to an expansion. This model is supported by several lines of evidence: 8-oxo-dG accumulation in the brain of HD mice correlates with degree of CAG expansion, and that expansion is suppressed by loss of OGG1 (Kovtun et al., 2007). Expansion is suppressed in MSH2−/− mice in the presence of OGG1 (Kovtun and McMurray, 2001) and in OGG1−/− mice in the presence of MSH2 (Kovtun et al., 2007), thus indicating that OGG1 and MSH2 physically or functionally interact together to cause expansion. A role for MMR in also repairing oxidative DNA damage is supported by the literature (Pitsikas et al., 2007). FEN1 have been shown to be inhibited by secondary structures at trinucleotide repeats (Spiro et al., 1999). A model that ties oxidation to expanding CAG repeats is consistent with the progressive nature of HD, and supported by evidence that age-dependent expansion occurs together with oxidative DNA damage accumulation in HD mice (Kovtun et al., 2007).
5. Single-strand break repair deficiency
5.1. Single-strand break repair (SSBR)
SSBs are some of the most common lesions found in chromosomal DNA and they can arise in two different ways: (i) indirectly, via enzymatic cleavage of the phosphodiester backbone. Cleavage occurs during BER of oxidative base damage generated by the attack of ROS (Connelly and Leach, 2004), and also during DNA topoisomerase I (TOP1) activity (Pommier et al., 2003). (ii) Directly, induced by the oxidative damage generated by the attack of ROS such as •OH, O2•− and H2O2, or by ionizing radiation. The repair of direct and indirect SSBs has been termed SSBR. In SSBR, PARP1 is believed to function as a SSB sensor that binds to the break and attracts the LIG3α-XRCC1 complex (Chalmers, 2004). After this, SSBR utilizes many of the same proteins, and follows essentially the same procedure, as BER (Fig. 3). SSBR has two subpathways, short-patch (SP) and long-patch (LP) similar to BER (Caldecott, 2003). Any SSB event leaving an intact 5′-P and 3′-OH group should be easily ligatable, but the DNA termini at SSBs frequently have altered 3′- and 5′ termini that will be obstructions to normal polymerization by Pol β and ligation by LIG3α. Examples of 3′-obstructive termini are 3′-topoisomerase I (3′-TOP1), 3′-phosphate (3′-P), 3′-phosphoglycolate (3′-PG) and 3′-unsaturated aldehyde (3′-PUA). Examples of 5′-obstructive termini are 5′-adenosine monophosphate (5′-AMP), 5′-hydroxyl (5′-OH), 5′-aldehyde (5′-Ade) and 5′-deoxyribose-5-phosphate (5′-dRP) (Wilson and Mattson, 2007).
Two neurodegenerative diseases characterized by cerebellar ataxia, ataxia-oculomotor apraxia-1 (AOA1) and spinocerebellar ataxia with axonal neuropathy-1 (SCAN1), have been found to be associated with defects in the repair of SSBs, specifically defects in the end processing of obstructive termini (Ahel et al., 2006; El-Khamisy et al., 2005). AOA1 and SCAN1 are both hereditary autosomal recessive diseases where the patients lack the non-neurological phenotypes of hypersensitivity to ionizing radiation, increased genetic instability and cancer incidence seen in a number of DNA repair disorders. This may imply that the nervous system is particularly sensitive to defects in the repair of DNA SSBs. Elucidating the molecular basis of these diseases offers insight into the mechanisms by which defective SSBR can cause neurodegeneration, and how the normal SSBR process helps to maintain the genetic integrity of post-mitotic neurons.
5.2. Ataxia-oculomotor apraxia-1
Ataxia-oculomotor apraxia-1 (AOA1) is an autosomal recessive neurodegenerative syndrome associated with progressive cerebellar atrophy, late axonal peripheral motor neuropathy, ataxia, oculomotor apraxia (limited eye movement control), and variable age of onset (1–16 years, mean age is 5) (Aicardi et al., 1988; Le Ber et al., 2003). AOA1 cells are sensitive to the SSB-inducing agents H2O2 and methyl methanesulfonate(Clements et al., 2004; Gueven et al., 2004). Accumulation of SSBs under conditions of oxidative stress has been reported and post-mortem brain sections from AOA1 patients display elevated levels of oxidative DNA damage(Hirano et al., 2007a)as did fibroblasts from a AOA1 patient (Harris et al., 2009) but evidence is conflicting. Recently it was reported that neither lymphoblastoid nor primary fibroblasts from AOA1 patients have reduced rates of SSBR (Reynolds et al., 2009) nor do AOA1-modelAPTX−/− mouse astrocytes (El-Khamisy et al., 2009).
AOA1 results from mutations in the gene (APTX) which encodes the protein aprataxin (Date et al., 2001; Moreira et al., 2001). The gene is a member of the histidine triad superfamily of nucleotide hydrolases and transferases (Brenner, 2002). The aprataxin protein (APTX) can remove 5′-AMP obstructive termini from DNA strand breaks in cell free assays (Ahel et al., 2006). APTX contains three conserved domains: an N-terminal forkhead associated domain that allows interaction with the XRCC1 scaffold protein part of the LIG3α-XRCC1 complex(Clements et al., 2004) that plays a crucial role in short-patch SSBR and BER of chromosomal DNA (Thompson and West, 2000; Whitehouse et al., 2001), a histidine triaddomain that possesses AMP-lysine hydrolysis activity (Ahel et al., 2006; Seidle et al., 2005) and a DNA-binding C2H2 zinc-finger motif (Date et al., 2001). APTX is recruited to DNA SSBs by XRCC1 (Hirano et al., 2007a). There it is believed to function as a nick sensor to scan the SSBs for the 5′-AMP obstructive termini that are intermediates in failed DNA ligase reactions when adenylation occurs prematurely, before a 3′-OH terminus is present (Ahel et al., 2006; Rass et al., 2008). When encountered such obstructions are resolved by excision of the AMP residues in a two-step catalytic reaction (Rass et al., 2008). This deadenylation restores the DNA 5′-P terminus that is necessary for the subsequent repair steps which are believed to mainly be by the short-patch pathway. This is consistent with the function of the three conserved domains and implicates APTX as having a direct role in DNA repair. Most of the disease-causing mutations that so far have been observed in AOA1 patients have been in the catalytic histidine triad domain. The zinc finger domain have been shown to provide stabilizing contacts that lock the enzyme onto its high affinity AMP-DNA target site, something that is necessary for efficient hydrolysis of the DNA adenylate (Rass et al., 2007a). Furthermore, it was shown that APTX can deadenylate DSBR intermediates in addition to BER/SSBR intermediates. APTX also interacts with PCNA, a LP-SSBR scaffold protein (Hirano et al., 2007a). But it has been an open question whether SSBR by the long-patch pathway plays a significant role in the post-mitotic neuronal cells. The main observations that suggest it does not are: long-patch repair seems to functions mainly during the S-phase (Caldecott, 2001) and the LP-SSBR components (PCNA, Pol δ/ε, LIG1 and FEN1) all function during DNA replication. However, a recent study has further examined the involvement of SSBR in AOA1 cells (Reynolds et al., 2009). The presence of 5′-AMP termini caused a failure of short-patch SSBR at the final step of DNA ligation and accumulation of adenylated DNA SSBs in AOA1 lymphoblastoid extracts but not wild-type cells. The end-processing by PNKP and DNA repair synthesis by Pol β in the preceding repair steps did not appear to be affected by the presence of 5′-AMP and the absence of APTX in the AOA1 cells. Despite observing a failure of SP-SSBR, this study, however, did not observe reduced rates of chromosomal SSBR in AOA1 cells nor in quiescent primary APTX−/− mouse neural astrocytes. One explanation that may reconcile these findings is a channeling of adenylated DNA nicks into the long-patch SSBR pathway where the 5′-AMP termini in the absence of APTX presumably would be removed by the combination of Pol δ/ε repair synthesis and FEN1 endonuclease activities (Fig. 3). To examine this possibility, mouse quiescent APTX−/− astrocytes and wild-type astrocytes were treated with H2O2 or methyl methanesulfonate in the presence and absence of the Pol δ/ε inhibitor aphidicolin, and the level of SSBs assessed. This chemical inhibition of LP-SSBR reduced the rates of SSBR in the APTX−/− astrocytes but not the wild-type astrocytes suggesting that compensatory long-patch repair may explain the normal rates of chromosomal SSBR observed in AOA1 cells.
APTX may also process specific 3′-obstructive termini of SSBs in order restore them to the 3′-OH termini that are required for subsequent repair. This has been demonstrated by one study using in vitro assays to show 3′-phosphatase and 3′-PG hydrolase activities for APTX to resolve the 3′-P and 3′-PG obstructive termini, respectively (Takahashi et al., 2007). This study also confirmed that the C-terminal region (histidine triad and zinc finger domains) is responsible for the removal activity while the N-terminal region containing the forkhead associated domain allows interaction with XRCC1, which may enhance the catalytic activities. If AOA1 cells are indeed defective in SP-SBBR and APTX responsible for, or contribute to, processing of some 3′-obstructive SSB termini, this would represent a potential problem for efficient repair. LP-SSBR would be unable to compensate for the deficient short-patch repair as the 3′-OH required for DNA repair synthesis is unavailable. The findings presented above and the lack of a non-neurological phenotype in AOA1, suggests that APTX functions primarily in the repair of SSBs. However, it has been speculated that APTX may have a general proofreading function during ligation in DNA repair, given its ability to deadenylate both BER/SSBR and DSBR intermediates. This view is supported by its ability to associate with both the BER/SSBR scaffold XRCC1, and XRCC4, an indispensable protein in the NHEJ pathway of DSBR (Clements et al., 2004). Also indicative of a potential role for APTX in DSBR is the finding that MDC1, a protein that amplifies ATM-dependent DNA damage signaling in the DSB response, co-localizes and interacts with APTX after induction of DNA breaks (Becherel et al., 2010). MDC1 was found not to be required for SSBR suggesting that its interaction with APTX is in response to DSBs. However, in APTX-deficient cells the repair of ionizing radiation-induced DSBs was found to be normal but as suggested above this may indicate APTX involvement specifically in proofreading DNA ligation and resolution of abortive ligation events at DSBs and not a general role for APTX in DSBR. APTX is considered to be a nuclear protein, present in both the nucleoplasm and the nucleolus, and interestingly, APTX is associated with the nucleolar proteins nucleolin, nucleophosmin and UPF1, and also partially co-localizes with nucleolin (Becherel et al., 2006; Gueven et al., 2004). It is possible that this association serves to ensure effective and efficient processing of obstructive 3′- and 5′ DNA termini to prevent SSBs from reducing gene expression by stalling RNA polymerases at the sites of elevated transcription.
The accumulated data is thus suggestive of a potentially multifaceted cellular role for APTX. It is not yet clear which specific SSBs with obstructive termini, if any, are the neurodegenerative agents in AOA1 patients though 5′-AMP SSBs appear to be the most likely candidate. And while it appears that LP-SSBR can compensate for deficient SP-SSBR in APTX−/− mouse astrocytes it is by no means certain that this is the case for AOA1 neurons given that long-patch repair appears to be deficient in some types of terminally differentiated cells (Narciso et al., 2007).
5.3. Spinocerebellar ataxia with axonal neuropathy-1
Spinocerebellar ataxia with axonal neuropathy-1 (SCAN1) is a rare autosomal recessive neurodegenerative disease associated with progressive cerebellar atrophy and peripheral neuropathy, and with 15 years being the mean age of onset (Takashima et al., 2002). Similar to AOA1, the syndrome is not associated with genomic instability nor is there an increased predisposition to cancer.
SCAN1 is caused by a homozygous mutation in the TDP1 gene resulting in the substitution of histidine 493 for an arginine residue (H493R) (Takashima et al., 2002). TDP1 encodes tyrosyl-DNA phosphodiesterase 1 (TDP1), a member of the phospholipase D superfamily, and as such it contains two HKD motifs which interacts to form a single symmetrical active site. The symmetry of this active site is broken in the H493R SCAN1 mutation, resulting in a decrease of enzyme activity by about 25-fold (Interthal et al., 2005). TDP1 is involved in the repair of DNA strand breaks associated with various obstructive DNA termini.
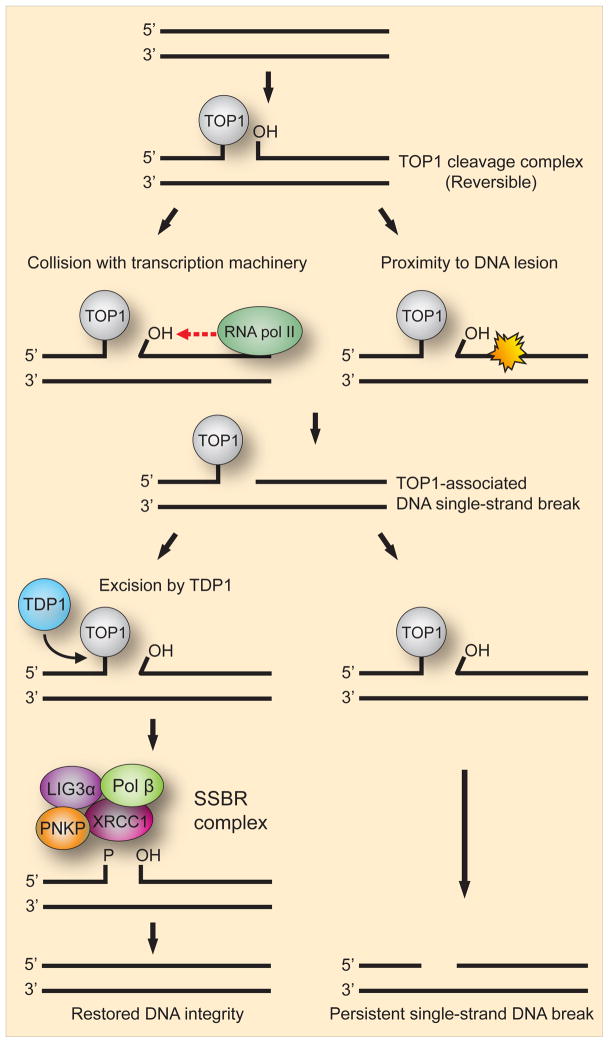
During repair, replication, transcription, recombination and chromatin condensation, DNA topoisomerase I (TOP1) relaxes superhelical tension by nicking DNA followed by controlled rotation of the broken DNA strand around the intact strand and resealing of the nick (Champoux, 2001; Wang, 2002). During this process, reversible and transient 3′-TOP1-DNA intermediates known as TOP1 cleavage complexes (TOP1cc) are formed in which TOP1 is linked, by its active site tyrosine, to the 3′-teminus of the single-stranded nick. However the TOP1cc can become irreversibly “trapped” by endogenous DNA lesions such as base mismatches, DNA breaks or abasic sites that displace the 5′-OH terminus. The TOP1cc can also become unduly long lived due to enhanced binding of TOP1 to oxidative base lesions such as 8-oxo-dG(Lesher et al., 2002) and consequently SCAN1 cells are hypersensitive to the TOP1-inhibitor camptothecin (CPT) that prolong the half-life of TOP1cc (Interthal et al., 2005). The TOP1cc can then be converted into abortive 3′-TOP1-associated DNA SSBs (3′-TOP1-SSBs) by collision with the transcription machinery or proximity to endogenous or exogenous DNA lesions (Pommier et al., 2003) (Fig. 6). TDP1 is well documented to be the primary end-processing enzyme responsible for excision of the covalently linked 3′-TOP1-SSBs (Katyal et al., 2007; Plo et al., 2003; Pouliot et al., 1999; Yang et al., 1996) (Fig. 6). SCAN1 cells are defective for the repair of these SSBs and they have been found to accumulate in TDP1−/− mice(Katyal et al., 2007; Miao et al., 2006). The resolution activity of TDP1 leaves a 3′-P terminus that is converted to 3′-OH by the phosphatase action of PNKP. The kinase activity of PNKP phosphorylates the 5′-OH terminus, allowing gap filling by Pol β and finally the DNA nick is sealed by LIG3α aided by the XRCC1 scaffold.
Fig. 6. Model for the generation of single-strand breaks from TOP1 cleavage complexes.
During various processes of DNA metabolism the enzymatic activity of DNA topoisomerase I (TOP1) generates reversible 3′-TOP1-DNA intermediates known as TOP1 cleavage complexes. Such complexes can, however, become unduly long lived and collision with RNA pol II or the proximity of a DNA lesion creates a TOP1-associated DNA single-strand break. The enzyme responsible for cleaving the link between TOP1 and the 3′-teminus of the single-stranded break is TDP1. If excision is successful the remaining strand break can then be repaired by a SSBR complex consisting of PNKP, XRCC1, Pol β and LIG3α thereby restoring DNA integrity. If excision by TDP1 fails, a persistent DNA single-strand break will be generated.
SCAN1 cells have a reduced ability to repair H2O2-induced oxidative SSBs (Takashima et al., 2002). Human TDP1 has been shown to be able to repair oxidation-induced 3′-PG obstructive termini (El-Khamisy et al., 2005; Zhou et al., 2005). However, another study, using in vitro assays mimicking oxidative SSBs, suggests that APE1 could possibly be responsible for most of the 3′-PG-resolving activity in human cells (Parsons et al., 2004). It is also possible that APTX could be the major contributor to this activity, as it was reported to have a Kcat of 3′-PG hydrolase activity about 20-fold higher than APE1 (Takahashi et al., 2007). APE1 is associated with removal of 3′-phospho-α,β-unsaturated aldehyde (3′-PUA) termini and endonucleolytic cleavage of AP sites. This would suggest that that TDP1 does not make a major contribution to resolving 3′-PG in SSBs. TDP1 does however appear to be required for end processing of PG-terminated 3′ overhangs on DNA DSBs as indicated by the complete inability of SCAN1 cell extract to resolve these substrates (Zhou et al., 2005). A similar result was found in a more recent study using cell extract from TDP1−/− mouse embryonic fibroblasts on protruding 3′-PG DSB substrates (Hawkins et al., 2009). In addition, these investigators found that the processing of 3′-PG on blunt-end DSB substrates was partially deficient in the absence of TDP1 indicating that other enzymes can resolve such blunt-ended substrates although less efficiency than TDP1. This potential role in DSBR is examined in more mechanistic detail in Section 6.5.
TDP1 also facilitates the repair of SSBs induced by ionizing radiation although the nature of these SSBs is unclear. They could be direct breaks, producing obstructive termini such as 3′-PG that TDP1 can resolve, or they could be TOP1-associated SSBs (Parsons et al., 2004). There is evidence linking TDP1 directly to components of the SSB multi-protein repair complex. Thus, yeast two-hybrid and co-immunoprecipitation experiments were used to show a direct interaction between TDP1 and LIG3α mediated by the N-terminal domain of TDP1, and the formation of a complex with LIG3α, XRCC1 and PNKP (El-Khamisy et al., 2005). This complex was found to repair model SSB substrates with 3′-tyrosyl termini. Further strengthening the link, another study showed co-immunoprecipitation of TDP1 with XRCC1 from a rodent cell extract (Plo et al., 2003). These findings are reconciled with the observation that LIG3α-XRCC1 stimulated TDP1 activity on TOP1-associated SSBs in vitro (El-Khamisy et al., 2007).
The N-terminal regions of the TDP1 orthologs from different species of lower eukaryotes are poorly conserved and vary substantially in length (Interthal et al., 2001). Budding yeast species lack the XRCC1 and LIG3α components of the human SSBR complex, possibly because only the much larger mammalian genome requires these factors to enhance the rate of SSBR to avoid accumulation of SSBs. The mammalian nervous system appears to be particularly vulnerable to the effect of a high steady-state level of SSBs. Given the above, it can be speculated that the large and crucial mammalian CNS requires TDP1 to be linked to the SSBR complex, thereby enhancing the ability to rapidly repair SSBs, particularly those associated with TOP1 activity. However, as TDP1 may also process obstructive DSB termini, it appears likely that the physiological role of TDP1, like APTX, is multifaceted.
5.4. Single-strand breaks, neurodegeneration and aging
AOA1 and SCAN1 are ataxias that lack the genomic instability and increased cancer incidence phenotypes seen with many DNA repair deficiency syndromes. Both are however, associated with particular defects in DNA SSBR, indicating the critical role such repair plays in protecting the nervous system against SSBs with 3′ and 5′-obstructive termini. The notion that both APTX and TDP1 probably are required for efficient in vivo SSBR in neural cells was underscored by the latest experiments on a double knockout TDP1−/−/APTX−/− mouse model (El-Khamisy et al., 2009). The level of H2O2-induced SSBs were similar for single knockout TDP1−/− and APTX−/−, and double knockout TDP1−/−/APTX−/− quiescent primary astrocytes, but the global rate of chromosomal repair of these oxidatively induced SSBs was synergistically decreased in the double knockout compared to the single knockouts. A similar result was reported for cerebellar granule neurons. The rate of SSBR was likewise reduced synergistically in double knockouts after treatment with the alkylating agent methyl methanesulfonate. The additional deletion of APTX−/− did not affect the SSBR rate ofTDP1−/− astrocytes treated with CPT probably because these SSBs are associated with 3′-TOP1 but not 5′-AMP obstructive terminus intermediates.
Neuronal cells are terminally differentiated post-mitotic cells that also have unusually high rates of oxygen metabolism compared to other non-dividing cells while also possessing a low level of antioxidant enzymes. This would be expected to lead to an increased accumulation of SSBs with obstructive termini, necessitating effective repair. The dependency of the neurons on SSBR is also increased by the high transcriptional demand of these cells (Flangas and Bowman, 1970; Morris and Geller, 1996; Sarkander and Uthoff, 1976; Sarkander and Dulce, 1978), and their limited cellular regenerative capacity compared to other terminally differentiated cells (Nouspikel and Hanawalt, 2002; Vierck et al., 2000; Yan, 2000). The failure to prevent the accumulation of SSBs in SSBR-deficient neurons can lead to stalled transcription as the break represents an impediment to RNA polymerase progression, particularly when an obstructive terminus is present (Zhou and Doetsch, 1994). Blocked transcription will eventually cause neuronal cell death by depriving the cell of vital transcripts, and perhaps also by more direct induction of apoptosis (Ljungman and Lane, 2004). It is also possible that merely a reduction in the level of transcription resulting from defective SSBR can contribute to neurodegeneration, as suggested by the progressive neurodegeneration in Cockayne syndrome that is associated with impaired transcription-coupled NER (see Section 2.4.). Another possibility is that a high steady-state level of unrepaired SSBs encourage depletion of NAD+ and ATP in cells due to the continuously excessive activation of the SSB sensor PARP1 (Heeres and Hergenrother, 2007), thereby promoting the death of the highly ATP-dependent neuronal cells.
AOA1 features the loss of cerebellar Purkinje neurons (Sugawara et al., 2008) but no post-mortem studies on SCAN1 patients have been published so far. However, several observations make it appear likely that the SCAN1 disease also features loss of these neurons: TOP1 is particularly highly expressed in Purkinje neurons of adolescence human brain (Gorodetsky et al., 2007; Holden et al., 1997), the toxicity of the yeast SCAN1 TDP1 mutation is dependent on high levels of TOP1 (He et al., 2007) and several other ataxia syndromes (e.g. ataxia telangiectasia) feature neurodegeneration of cerebellar Purkinje cells.
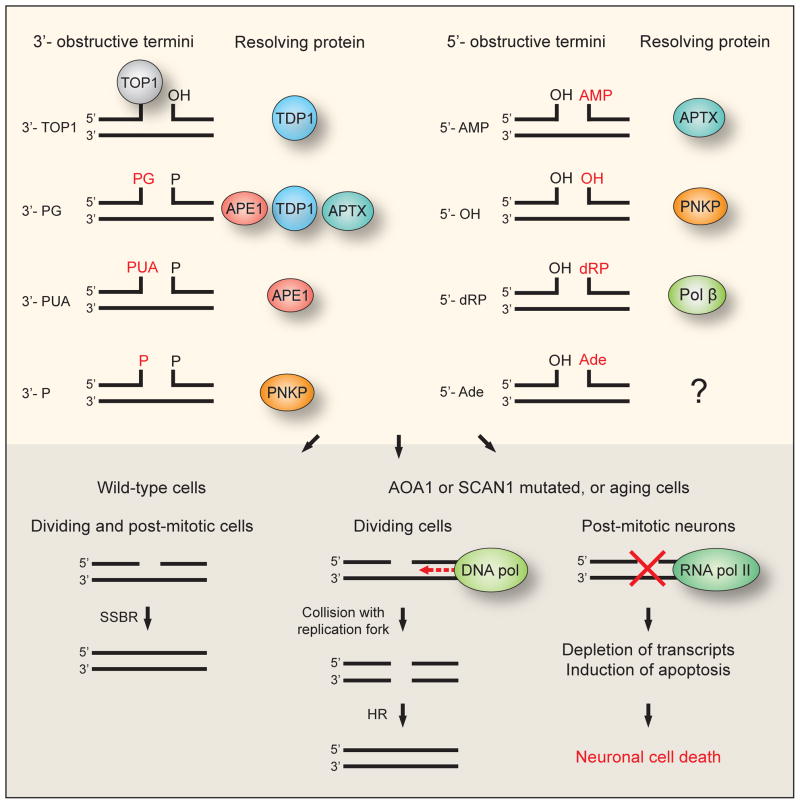
The susceptibility of neurons to unrepaired SSBs compared to dividing cells, and the absence of increased genetic instability and cancer in AOA1 and SCAN1 patients are probably due to alternative processing in dividing cells (Fig. 7). In dividing cells that lack SSBR, the unrepaired SSBs are converted to DSBs when they collide with DNA replication forks during the S phase. Accurate and efficient DSBR is then performed by nuclease-dependent HR during DNA replication. For the 3′-obstructions, the XPF-ERCC1 complex is a possible nuclease and for the 5′-obstuctions FEN1 or EXO1 are candidates. The lack of HR in the post-mitotic neurons will lead to slow accumulation of unresolved 3′ and 5′-obstructive termini over the years, which would be expected to lead to increased levels of SSBs with age. Indeed, an age-related accumulation of DNA SSBs has been demonstrated in certain types of neurons that are not reduced in number during aging in the mouse brain (Rutten et al., 2007). These include cerebellar granule neurons, which were shown to be dependent on TDP1, as such cells derived from TDP1−/− mice displayed SCAN1-like DNA repair deficiencies (Katyal et al., 2007). They furthermore showed that loss of TDP1 resulted in gradual age-related cerebellar atrophy in mice. Deficient SSBR of oxidative damage has been linked to another ataxia disorder, ataxia with oculomotor apraxia 2 (AOA2), a disease caused by mutation in the helicase SETX but the role of this helicase is presently unclear(Rass et al., 2007b). These findings further strengthen the association between DNA damage, DNA repair defects, SSBs, neurodegeneration and aging.
Fig. 7. Model for the differential impact of single-strand breaks on dividing cells and post-mitotic neurons caused by AOA1 or SCAN1 mutations, or aging.
The upper part of the figure shows the various 3′- and 5′-obstructive termini and the proteins responsible for resolving them. The lower part of the figure shows the impact of single-strand breaks on wild-type, AOA1 and SCAN1 cells. In both dividing and post-mitotic wild-type cells efficient single-strand break repair (SSBR) will ensure resolution of the break. In dividing AOA1, SCAN1 or aging cells SSBR is deficient, but the single-strand breaks may be converted to double-strand breaks and subsequently repaired by homologous recombination (HR). In the post-mitotic neurons of AOA1 or SCAN1 patients, or aging individuals HR is not available and persistent unrepaired single-strand breaks in these cells lead to neuronal cell death.
6. Double-strand break repair deficiency
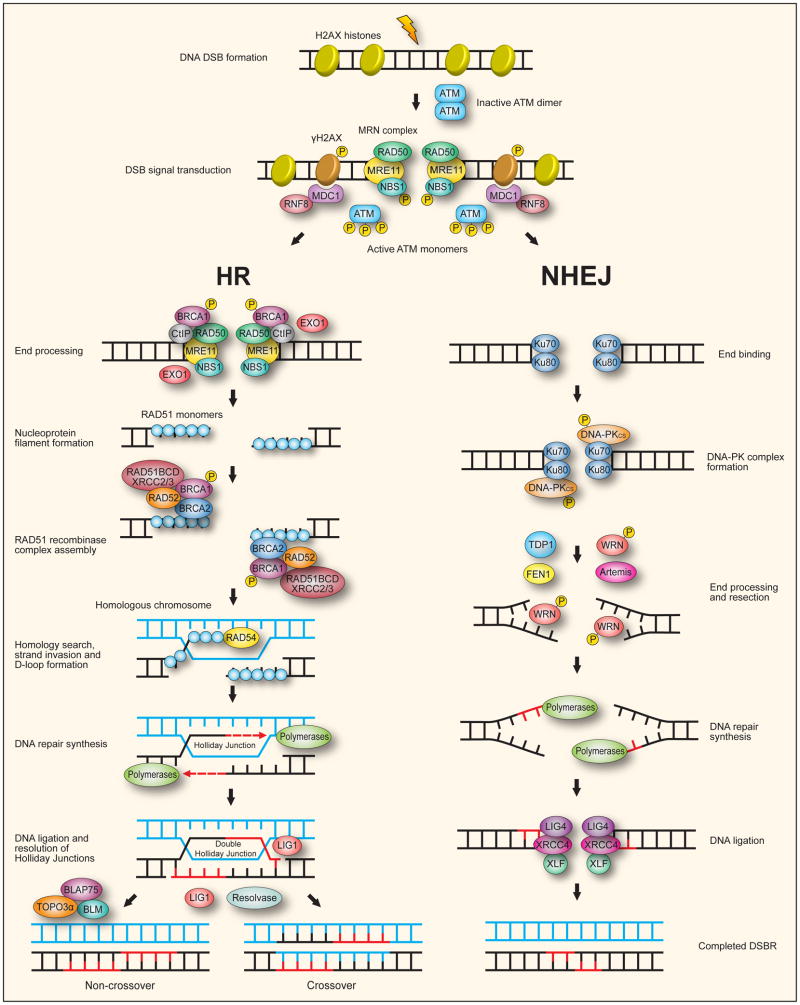
6.1. Double-strand break repair (DSBR)
One of the most toxic and mutagenic lesions is the DNA double-strand break (DSB), as chromosomal breakage may result in an extreme loss of genetic integrity. DSBs can be induced by exogenous sources, such as ionizing radiation and exposure to genotoxic compounds that directly or indirectly damage DNA. DSBs can also be induced by endogenous sources, such as the ROS generated by cellular metabolism, replication fork collapse during DNA replication and repair events, and during meiotic recombination. The result of DSBs in the nervous system is initiation of a defined signaling process leading to cell cycle arrest which allows repair or elimination of the damaged cell by apoptosis.
Two major mechanistically distinct pathways exist for DSB repair (DSBR) in mammalian cells: homologous recombination (HR) and non-homologous end-joining (NHEJ) (Fig. 8). HR allows the error-free repair of DSBs associated with DNA replication by using the other intact sister chromatid, which is in close proximity, as a template. This restricts HR to the late S to G2/M phase of the cell cycle when a sister chromatid is available in proliferating cells. In addition, HR is essential for the preservation of replication forks, telomere maintenance, and chromosome segregation in meiosis I. HR is known to be the predominant DSB repair pathway during the early proliferative stage of embryonic nervous system development (Orii et al., 2006). Recent detailed reviews describe error-free DNA DSBR via HR (Helleday et al., 2007; Lee and McKinnon, 2007; West, 2003; Wyman et al., 2004). NHEJ modifies and ligates the two DNA termini of a DSB, allowing for the error-prone repair of DSBs without the need for an undamaged template. NHEJ operates throughout the cell cycle and can repair differentiated cells. The mature nervous system consists largely of post-mitotic cells therefore NHEJ is the main pathway for repair of DNA DSBs in the post-natal brain. A detailed description of error-prone DNA DSBR via NHEJ can be found in recent reviews (Helleday et al., 2007; Lee and McKinnon, 2007; Lees-Miller and Meek, 2003; Lieber et al., 2003; O’Driscoll and Jeggo, 2006). The choice of which of the two pathways that becomes activated may be linked to the cell cycle or be regulated by the levels of available specific components of the two pathways (Rothkamm et al., 2003). The two pathways can cooperate in the repair of DSBs by being active simultaneously (Richardson and Jasin, 2000).
Fig. 8. The two major mammalian pathways for double-strand break repair.
Repair of a DNA double-strand break (DSB) is usually accomplished by either homologous recombination (HR) when a homologous chromosome is available in the form of a sister chromatid, or by non-homologous end-joining (NHEJ) throughout the cell cycle. Following formation of a DSB, the MRE11-RAD50-NBS1 (MRN) complex is recruited to the break where it binds to the DNA ends through MRE11. A coiled-coil region of RAD50 reaches across the break and through coordination of a central Zn2+ ion, tethers the two broken DNA ends together. The serine/threonine protein kinase ATM is usually present as an inactive dimer but is recruited to the break site by NBS1. This causes ATM to autophosphorylate (P) and monomerize, activating the kinase. The activated ATM phosphorylates NBS1 and a multitude of other proteins that participates in the DNA DSB response, including BRCA1 and phosphorylation of nearby histone H2AX to generate γH2AX. The γH2AX at repair foci acts as a signal to recruit other repair proteins in order to assemble DSBR complexes. One such protein, MDC1, binds to γH2AX which recruits RNF8to the break site where it initiates an ubiquitylation cascade of histones H2A and H2AX causing chromatin restructuring and generation of binding sites for further protein factors. The HR pathway proceeds via the end processing of damaged DNA termini with initial nucleolytic 5′ resection performed by CtIP, a function dependent on CtIP recruitment of BRCA1. Further resection by EXO1 generates single DNA strands with 3′ overhangs upon which RAD51 monomers attach to form nucleoprotein filaments. A RAD51 recombinase complex is then assembled containing the accessory proteins BRCA1, BRCA2, RAD52 and the RAD51 paralogs: RAD51B/C/D and XRCC2/3. This recombinase complex, with the aid of an additional factor, RAD54, facilitates homology search and strand invasion of the homologous chromosome to form displacement loops (D-loops). DNA repair synthesis by DNA polymerase using the homologous strand as template extends the 3′ invading strand allowing branch migration of the Holliday junction, a cruciform intermediate. The repair synthesis allows capture of the other DNA end to create a double Holliday junction intermediate. DNA ligation by LIG1 seals the remaining nicks and resolution of the double Holliday junction by structure-specific endonucleases generate either non-crossover or crossover products depending on where the junctions are cut by the resolvase. The BLM RecQ helicase can cooperate with TOPO3α and BLAP75 to resolve DHJs with generation of exclusively non-crossover products.
The NHEJ pathway first involves binding of Ku70-80 heterodimer to the two DNA termini. This then recruits DNA-PKCS to form the DNA-PK complex bringing the two DNA termini close together. Because of associated lesions not all DNA termini are readily ligatable and must be processed to generate the 5′-P and 3′-OH termini that are necessary for ligation. Autophosphorylation of DNA-PKCS can make the obstructive termini accessible to end processing enzymes such as TDP1. DNA-PKCS also mediates a regulatory phosphorylation of the WRN RecQ helicase. End processing and resection, while not fully understood, may involve the exonuclease activity of FEN1, WRN and Artemis. DNA polymerases then perform any necessary DNA repair synthesis. Finally, the DNA-PK complex, in a Ku70-80 mediated fashion, recruits the LIG4-XRCC4-XLF complex to perform the ligation of the DNA termini via the ligase activity of LIG4.
A variety of human syndromes resulting from defects in the molecular machinery that responds to DNA DSBs are characterized by neuropathology, some of them including progressive neurodegeneration. The following will focus on two neurodegenerative diseases, ataxia telangiectasia and ataxia telangiectasia-like disorder, resulting from a defective response to DNA DSBs. Furthermore, the DSB repair deficiency-associated disorder Nijmegen breakage syndrome, characterized by microcephaly, will be touched upon as it features a defect in the same molecular machinery.
6.2. Ataxia telangiectasia
Ataxia telangiectasia (AT) is a rare autosomal recessive neurodegenerative disease characterized by ataxic movements and telangiectasia (dilated blood vessels), resulting from mutations in the ataxia telangiectasia mutated (ATM) gene (Chun and Gatti, 2004; Gatti et al., 2001; McKinnon, 2004). The onset of AT is in early childhood and by age 2-3 years ataxia becomes visible, followed by progressive neurodegeneration such that by age 10 the patients are confined to a wheelchair (Chun and Gatti, 2004). Neurodegeneration of granule and Purkinje cells in the cerebellum is the hallmark of AT but other features include increased predisposition to lymphoid and breast cancer (Angele et al., 2003; Gumy-Pause et al., 2004; Thorstenson et al., 2003), immunodeficiency (Nowak-Wegrzyn et al., 2004), sterility and extreme cellular and chromosomal sensitivity to ionizing radiation(McKinnon, 2004).
ATM is a member of the PIKK family of serine/threonine protein kinases (Shiloh, 2003). It autophosphorylates after DNA damage and plays a crucial role in the DNA DSB signaling cascade by phosphorylating a multitude of different proteins that all participate in the DSB response including DSBR proteins (Fig. 8). DNA DSBs in the nervous system initiate rapid signaling transduction processes which have been characterized, both from human neurological diseases and in vivo mouse models (Lee and McKinnon, 2007), and is similar to the process for cultured cells, described by numerous in vitro studies(Bakkenist and Kastan, 2003; Bakkenist and Kastan, 2004; Kastan and Bartek, 2004; Kobayashi et al., 2002). The DSB signal response involves a series of post-translational modifications that facilitate signal transduction via the protein-protein interactions of the MRN-ATM signaling pathway, resulting in cell cycle arrest to allow time for repair or apoptosis. Failure to activate ATM in response to DSBs is believed to attenuate repair and prevent apoptosis. This would then cause the damaged cells to escape apoptosis and remain in the maturing brain leading to neurodegeneration in AT as the cells later succumb to the damage and die (Lee et al., 2001).
The MRN complex is a DNA DSB sensor known to be required in both NHEJ and HR (Chen et al., 2001; Tauchi et al., 2002), whereas the requirement for ATM is less clear. ATM together with the MRN complex and Artemis was shown to participate in the repair of a subset of DSBs via the NHEJ pathway (Riballo et al., 2004). ATM together with the MRN complex may also function in DSBR via HR (Kobayashi, 2004; Morrison et al., 2000). However, one study found no observable effect of ATM deficiency in mouse embryos with inactivated HR (XRCC2−/− mice) (Orii et al., 2006). This would suggest that HR only requires ATM in some tissues or later in development. Alternatively, another ATM-related protein, ATR, could be functioning in place of ATM in the DNA DSB signaling during HR (Brown and Baltimore, 2003; Wang et al., 2004). Whreas it was generally thought that ATM was involved in DSBR and ATR in SSBR, it is now evident that functions of these two proteins overlap. ATM signaling in the nervous system seems to function predominantly in recently post-mitotic neural cells rather than proliferative precursor cells where the ATR or DNA-PK kinases may predominate (Lee et al., 2001; Orii et al., 2006). This would also indicate that ATM is involved in DNA DSB repair utilizing NHEJ, as HR is restricted to proliferative cells. ATM is required for DNA damage-induced apoptosis in the nervous system arising from LIG4 deficiency (Lee et al., 2000; Sekiguchi et al., 2001), indicating that ATM directly responds to endogenous DNA damage in the nervous system. When ATM is absent the level of ROS is increased and at least in vitro, treatment with antioxidants can increase the survival of Purkinje cells that does not express ATM(Biton et al., 2008). Reducing the ROS level by increasing the antioxidative capacity can reduce cerebellar atrophy in AT patients (Russo et al., 2009). In fact, very recent data shows that conditions of oxidative stress directly oxidize and activate ATM by the formation of disulfide-crosslinked ATM dimers in the absence of DSBs and the MRN complex (Guo et al., 2010). There is thus strong evidence that ATM is an important direct sensor of oxidative stress.
Recently, post-mortem examination of the brain tissue of AT patients revealed a dramatic age-dependent increase in cerebellar DSBs (Iourov et al., 2009). These breaks were not randomly distributed but instead accumulated at specific chromosomal loci, primarily chromosomal region 14q12. Interestingly, this locus contains two genes, FOXG1B and NOVA1, which are highly expressed in the cerebellum. Thus, the cerebellar-specific nature of the neurodegeneration in AT may be due to improper gene regulation in the presence of DSBs at defined chromosomal loci.
6.3. Ataxia telangiectasia-like disorder
Ataxia telangiectasia-like disorder (ATLD) (Stewart et al., 1999; Taylor et al., 2004) is a very rare disease in which the patient phenotype is similar to the AT phenotype in showing a progressive cerebellar ataxia, genomic instability and hypersensitivity to ionizing radiation. However, ATLD can be distinguished from AT by the absence of telangiectasia, normal immunoglobulin levels, and a later onset and slower progression of the disease. In addition, no increased predisposition to cancer has been observed so far in ATLD patients in contrast to AT patients. The disease is caused by mutations of MRE11, one of the proteins in the MRN complex (Fig. 8). MRE11 interacts with both of the other MRN components NBS1 and RAD50. Additionally, binding of the MRN complex to DNA is through the two DNA-binding motifs of MRE11. Disease-causing mutations generate a hypomorphic allele resulting in either a truncated protein or a full-length mutant. The result of the truncated protein is reduced levels of all three components of the MRN complex. The specific mutation present in cells from ATLD patients with full-length mutant protein determines the strength of interaction with NBS1 and RAD50; both weaker and stronger interactions have been observed (Taylor et al., 2004). Complete loss of function is embryonic lethal (Friedberg and Meira, 2006; Xiao and Weaver, 1997). A neuropathological study of a pair of male siblings with the same heterozygous mutations of the MRE11 gene found both cerebellar atrophy and the absence of MRE11 in the neurons of the cerebellar cortex, cerebral cortex, basal ganglia and midbrain (Oba et al., 2010). Cerebellar atrophy in the two siblings was already present at infancy as indicated by magnetic resonance imaging studies. The similarity of symptoms in AT and ATLD may suggest interrelationship between ATM and the MRN complex, and a connection between DNA DSBs and neurodegeneration.
6.4. Nijmegen breakage syndrome
Nijmegen breakage syndrome (NBS) (Carney et al., 1998; Digweed and Sperling, 2004) is a rare autosomal recessive disorder caused by hypomorphic mutations of NBS1, a component of the MRN complex (Fig. 8). In contrast to AT and ATLD, NBS is characterized by microcephaly. Otherwise, the extra-neurological symptoms are quite similar to AT, including the predisposition to cancer (Valerie and Povirk, 2003). The most common (>90%) mutation of NBS1, 657Δ5, leads to cells with a 26 kDa N-terminal protein fragment, which is not physically associated with the MRN complex as it lacks the C-terminal MRE11-binding domain (Maser et al., 2001; Varon et al., 1998). In the same cells a 70 kDa C-terminal fragment is produced by internal translation initiation within the NBS1 mRNA using an open reading frame generated by the 657Δ5 frameshift. This 70 kDa fragment contains the MRE11 binding domain and can interact with the other two components of the MRN complex, MRE11 and RAD50, with sufficient function to allow embryonic development (Maser et al., 2001). The C-terminal fragment also contains a conserved domain responsible for recruitment of ATM. Complete loss of function of NBS1 is embryonic lethal, as is complete loss of function of MRE11 or RAD50 (Friedberg and Meira, 2006; Xiao and Weaver, 1997). The requirement of human NBS1 for efficient HR and NHEJ was demonstrated in a study in which introduction of full length NBS1 cDNA could restore repair proficiency in NBS1-deficient cells (Howlett et al., 2006).
6.5. Double-strand breaks, neurodegeneration and aging
The three syndromes, AT, ATLD and NBS all result from defective DSBR and share many characteristics, though they also differ in some aspects. The cause of the different clinical phenotypes between the syndromes is unclear. However, the hypomorphic nature of the MRE11 and NBS mutations could be responsible for some of the differences as the proteins are partly functional. It is intriguing that ATLD and NBS cause two different kinds of neuropathology, neurodegeneration and microcephaly, respectively, given that they both stem from mutations in components of the MRN complex. This suggests that despite their presence in the same complex, MRE11 and NBS1 have distinct functions. One possible explanation would be for the mutations to differentially affect the DNA repair function and apoptotic function of the MRN complex. The MRE11 mutations would then interfere with the apoptotic function, leading to the subsequent neurodegeneration in ATLD patients, as cells containing DSBs erroneously are utilized in the development of the nervous system. Conversely, the NBS1 mutations probably interferes with the DNA repair function of the MRN complex causing hypersensitivity to DSBs but without affecting apoptosis, possibly resulting in the microcephaly of NBS patients due to the increased neuronal cell loss during development. This would be consistent with the interaction of NBS1 with γH2AX at sites of DSBs and the requirement of NBS1 for localization of activated ATM at sites of DSBs (Kitagawa et al., 2004; Lukas et al., 2003). Results from a recent study (Shull et al., 2009) on mice lend support to such an interpretation. The nervous system of MRE11ATLD1/ATLD1 mice with truncated MRE11 protein displayed pronounced attenuation of ATM activation and subsequent resistance to apoptosis after damage induced by ionizing radiation. Mutant NBS1ΔB/ΔB mice with truncated NBS1 on the other hand, displayed a normal apoptotic response and a higher degree of ATM activation compared to the MRE11ATLD1/ATLD1 mice after ionizing radiation treatment. Another protein linking potential deficiency in DSBR to neurodegeneration is the end processing enzyme TDP1 that can resolve protruding 3′-PG, and with less efficiency, blunt end 3′-PG DSB termini. This protein is known to be mutated in the ataxia disease SCAN1 and the processing of protruding 3′-PG is completely deficient in SCAN1 cells (see Section 5.3.). Even though TDP1 is an established protein in the repair of SSBs it has been observed to be more active at DSB termini (Raymond et al., 2005). The data on the reported sensitivity of TDP1-deficient cells to DNA damaging agents expected to produce large amounts of 3′-PG-DSB termini, is conflicting. Human SCAN1 cells do not seem to be particularly sensitive to ionizing radiation and a deficiency in the repair of DSBs in these cells were not found after such treatment (Katyal et al., 2007). Also, no defect in DSBR after IR was found in TDP1−/− and TDP1−/−/APTX−/− mouse astrocytes as measured by the disappearance of DSB marker γH2AX foci (El-Khamisy et al., 2009). However, TDP1−/− mice are hypersensitive to the blunt end 3′-PG DSB inducing agent bleomycin (Hirano et al., 2007b). New data has been published on the interplay of TDP1 with the abundant Ku70/80 heterodimer and DNA-PKCS components of the NHEJ pathway (Fig. 8) (Zhou et al., 2009). Ku70/80 and DNA-PKCS are rapidly recruited to DNA ends and in the absence of ATP this was found to render both protruding and blunt 3′-PG termini highly refractory to TDP1. This inhibition was abrogated by addition of ATP to strongly restore TDP1 end processing of 3′-PG. This effect of ATP is most likely due to increased phosphorylation of DNA-PKCS as previous work suggests that the autophosphorylation of DNA-PKCS modulate the accessibility of DNA ends (Reddy et al., 2004). This rearrangement of DNA ends appears to be vital for allowing TDP1 and other factors to accomplish the processing necessary for eventual end joining. SCAN1 lymphoblastoid cells displayed chromosomal hypersensitivity to calicheamicin, which induces DSBs with obstructive 3′-termini on both strands, one protruding 3′-PG and one protruding 3′-P. End processing and resection in NHEJ may involve the exonuclease activities of WRN, FEN1 and Artemis, but such activity could be hindered by the presence of obstructive 3′-termini. The WRN exonuclease activity does not excise 3′-PG and 3′-P (Harrigan et al., 2007) and although the Artemis nuclease together with DNA-PK can process protruding 3′-PG, it does so only slowly and very inefficiently (Povirk et al., 2007). If a DSB is blocked by a 3′-PG in one end and a 3′-P in the other end, such a double block may then present a significant challenge. Thus, TDP1 may serve to process DSBs with 3′-obstructive termini on both strands that are blocks for the end processing exonuclease activities in NHEJ, and consequently the TDP1-deficient SCAN1 cells would lack this function.
DSBR is essential for neural homeostasis as defective DSBR can lead to severe neurodegeneration, particularly of granule and Purkinje cells in the cerebellum. Purkinje cells are some of the largest neurons in the human brain, featuring an elaborate dendritic arbor and a large number of dendritic spines. Granule cells are tiny neurons, with cerebellar granule cells accounting for nearly half of the neurons in the CNS. Cerebellar granule cells send fibers through Purkinje cell dendritic arbors where they synapse onto the dendrites. Several features of Purkinje cells may explain why loss of these neurons is particularly prevalent in ataxia syndromes: high metabolic activity and levels of transcription, the dendritic tree complexity and the storage of relatively large amounts of Ca2+. It can be speculated that DNA repair deficiency may impact Purkinje cells more because of Ca2+ involvement in signaling processes or greater susceptibility to disrupted Ca2+ homeostasis present in these cells.
It is clear that DSBR capability is critical for normal neurogenesis during development. This is illustrated by AT and ATLD where low or no ATM activation is believed to allow damaged cells to escape apoptosis and to incorporate into the developing brain, leading to neurodegeneration in early life(Lee et al., 2001; O’Driscoll and Jeggo, 2008). It is less clear, however, what role the DSBs arising spontaneously in the mature brain due to ROS generated by metabolic activity, might play in the neurodegeneration associated with aging. There is evidence to support ROS as the cause of unrepaired DNA DSBs in NHEJ-deficient neurons (Karanjawala et al., 2002). That unrepaired DSBs could contribute to aging is supported by studies showing accelerated aging in mice defective in NHEJ (Li et al., 2007a; Vogel et al., 1999). One study that analyzed DSBR in aging have found that cells from old mice contain more DSBs than cells from young mice (Singh et al., 2001). A role for DSBs in aging is also supported by studies of the premature aging syndrome, Werner syndrome (WS). WS is caused by a mutation inthe WRN gene that encodes a helicase believed to be involved in DSBR by both the HR and NHEJ pathways, as discussed in Section 8.2.2. There is also evidence that the efficiency and fidelity of NHEJ declines significantly during cellular senescence. A more than four-fold reduction in efficiency of end joining have been found in senescent cells and this process was furthermore associated with extended deletions (Seluanov et al., 2004). This decline and increasingly aberrant function may be hypothesized to contribute to age-related genomic instability and aging, as senescent cells are known to accumulate in older organisms (Choi et al., 2000; Ding et al., 2001). It is therefore plausible that unrepaired DNA DSBs contribute to the neurodegeneration associated with aging. However, as DNA SSBs are far more frequent than DSBs, although less toxic and mutagenic, it is also possible that the mature nervous system relies mainly on SSBR for neuronal homeostasis.
7. Mitochondrial dysfunction and DNA repair
7.1. Mitochondria and neurons
Mitochondria are membrane-enclosed organelles that generate most of the ATP supply in eukaryotic cells, including neurons. To generate ATP high-energy electrons must be transported through the electron transport chain at the inner mitochondrial membrane. This process leads to the generation of ROS when high energy electrons react with O2 to form O2•, which in turn leads to generation of H2O2 and •OH. While the mitochondria have various antioxidant enzymes to deactivate these highly reactive molecules they do not constitute a perfect defense. Therefore damage to DNA will inevitably occur and require repair. The mammalian brain consumes large amounts of oxygen due to its reliance on glucose for production of ATP and this energy production is largely dependent on the neuronal mitochondria. The density of mitochondria is particularly high at synapses to power the energy-intensive synaptic activities (Ly and Verstreken, 2006). The mitochondrial theory of aging presupposes that ROS generated during the lifespan damages DNA, proteins and lipids, and ultimately leads to the aging of the organism. The brain may be highly vulnerable to mitochondrial dysfunction with age due to its high energy demand with concurrent production of ROS, the age-dependent decline of antioxidant defensive mechanisms, and the inability to generally replace lost post-mitotic neurons.
The generation of large amounts of ROS inevitably exposes the mitochondrial DNA (mtDNA) to high levels of oxidative stress. Mitochondria have their own circular genome with each mitochondrion containing approximately 4–10 DNA molecules, and this mtDNA is located in close proximity to the inner mitochondrial membrane. The location of the mtDNA and the lack of protective histones is believed to make this DNA particular vulnerable to oxidative damage (Ames et al., 1993). Consistent with this, levels of oxidative base damage in mtDNA are 2–3 fold higher compared to nuclear DNA (nDNA) (Hudson et al., 1998; Richter et al., 1988), and mtDNA damage is more extensive and persists longer than nDNA damage in human cells (Yakes and Van Houten, 1997). Mitochondrial DNA damage can lead to mitochondrial dysfunction and apoptosis (Kruman et al., 2004b; Linnane et al., 1989). The mitochondrial dysfunction will promote the production of ROS which in turn increase the level of mtDNA damage and conceivably creating a self-amplifying cycle of dysfunction until neuronal cell death occurs by apoptosis. The existence of such a “mitochondrial viscious cycle” is still being debated (Sanz et al., 2006). Accumulation of mtDNA damage in the brain is suggested to be inversely related to maximum mammalian life span (Barja and Herrero, 2000). The level of oxidative DNA lesions is increased in the aging human brain. Starting by middle age accumulation of DNA damage has been found in promoters of age-downregulated genes involved in mitochondrial and synaptic function leading to transcriptional repression (Lu et al., 2004). Such repression/silencing of genes could allow a cell to stay alive with persistent DNA lesions, although with reduced function. This may represent a worthwhile alternative to complete removal of an irreplaceable post-mitotic neuron by apoptosis. Whether damage to promoter regions leading to transcriptional repression also occurs in mtDNA as well as to nDNA involved in mitochodrial function is currently unknown and thus speculative. The transcriptional repression of genes within damaged regions of DNA is a potential contributor to the gradual cognitive decline seen with age. The importance of mitochondria suggest that mitochondrial dysfunction due to unrepaired neuronal mtDNA damage could be involved in the pathogenesis of age-related neurological diseases and there is indeed evidence for this in major neurodegenerative disorders (Lin and Beal, 2006), as discussed below.
7.2. Mitochondrial DNA repair pathways
A number of DNA repair activities and pathways that function in the nucleus have also been identified and characterized in mammalian mitochondria, though the proteins involved and details of the pathway can differ (Fig. 9). The absence of mtDNA NER in mammalian cell was suggested in 1974 (Clayton et al., 1974) and later confirmed by subsequent studies (LeDoux et al., 1992; Pascucci et al., 1997). Mammalian MMR activity for mtDNA has been reported though it may be different from the corresponding activity in the nucleus (Mason et al., 2003). Indeed, very recently, robust MMR activity was found in human mitochondria though key nuclear MMR factors were not detected (de Souza-Pinto et al., 2009). Specifically, evidence was found to suggest that the mitochondrial MMR pathway does not utilize MSH2 as the key mismatch-binding protein but rather the multifunctional YB-1 protein, thus making the nuclear and mitochondrial pathways distinct. There is some evidence for HR in mammalian mitochondria (Kajander et al., 2001; Thyagarajan et al., 1996), but this does not necessarily mean that HR repairs DSBs and there is conflicting reports on repair activity (Cullinane and Bohr, 1998; Cullinane et al., 2000; LeDoux et al., 1992). Some findings indicate that NHEJ may function in mammalian mitochondrial DSBR. Mitochondria seems capable of rejoining both cohesive and blunt-ended linearized plasmid DNA (Lakshmipathy and Campbell, 1999), and proteins that specifically bind to double-stranded DNA ends as well as substrates with 3′ or 5′ overhangs were identified in mitochondrial extracts (Coffey et al., 1999). While some of the DSBR functions have been detected in mitochondria, the proteins, as discussed in the previous section, have not been detected in mitochondria.
Fig. 9.
Overview of DNA repair pathways active in the nucleus and mitochondria of mammalian cells.
No in vivo TLS activity in mitochondria have been shown so far, though human mitochondrial DNA polymerase γ can perform synthesis past benzo[a]pyrene adducts and insert a random nucleotide, in vitro(Graziewicz et al., 2004). DR activity has been shown to remove O6-methyl-2′-deoxyguanosine (Myers et al., 1988) and O6-ethyl-2′-deoxyguanosine (Satoh et al., 1988) from mammalian mtDNA, and consistent with this a study showed repair of methylation but not complex alkylation damage (LeDoux et al., 1992).
By far the most studied and well documented mammalian DNA repair pathway present in mitochondria is BER (mtBER). The mtBER proteins are not encoded by the mitochondrial genome; rather they are mitochondrial versions of nuclear-encoded proteins. Many DNA glycosylases have been found to have mitochondrial forms. OGG1β is a splice variant that localizes to the mitochondria, but has no glycosylase activity, while the OGG1α isomform possesses a strong nuclear localization signal but may in fact be responsible for excision activity at 8-oxo-dG in the mitochondria as well (Hashiguchi et al., 2004). MYH can excise adenine opposite 8-oxo-dG and it is currently unclear, which of the many human splice variants is the mitochondrial MYH (Ohtsubo et al., 2000). NTH1 can excise pyrimidine lesions, though the human NTH1 mainly localizes to the nucleus possibly due to a weak mitochondrial targeting sequence (Ikeda et al., 2002). UNG1, an isoform of UDG, can excise uracil and oxidized cytosines from DNA (Dizdaroglu et al., 1996; Nilsen et al., 1997). NEIL1 excises oxidatively modified pyrimidine bases (Jaruga et al., 2004; Vartanian et al., 2006). In addition, it is possible that MPG can repair alkylation base damage (LeDoux et al., 1993; Myers et al., 1988; Pettepher et al., 1991; Satoh et al., 1988). After excision of the base, the AP endonuclease APE1 performs incision of the abasic site (Tomkinson et al., 1988). The human DNA polymerase γ in mitochondria then performs gap-filling and processing of the termini (Kaguni, 2004). Finally, nick ligation is performed by the mitochondrial variant of DNA ligase III (LIG3β) encoded by the human LIG3 gene (Lakshmipathy and Campbell, 1999), which unlike the nuclear variant does not seem to require XRCC1 (Lakshmipathy and Campbell, 2000). It now appears that the damage-sensing protein PARP1 is also present in the mitochondria and can be found in a complex with LIG3β and mtDNA (Rossi et al., 2009). It seems that many of the mtBER proteins are localized to the inner mitochondrial membrane and are therefore not freely soluble (Stuart et al., 2005). There have been several recent findings which indicate that mtBER also can occur by long patch repair DNA synthesis. Uracil and AP sites was found to be repaired via LP-BER by mitochondrial extracts free of nuclear BER proteins from human cells (Akbari et al., 2008). Two-deoxyribonolactone, a type of oxidized AP site, is known to be almost entirely repaired by LP-BER and dependent on the flap endonuclease activity of FEN1. Such repair activity has now been found in human mitochondrial extract along with FEN1 (Liu et al., 2008). TDP1, the enzyme responsible for resolution of TOP1cc and other 3′ obstructive termini (see Section 5.3.) is encoded by the nuclear TDP1 gene, a fraction of which localizes to the mitochondria (Das et al., 2010). TDP1 appears to be required for efficient repair of oxidative mtDNA damage and mtBER/SSBR was found to be dependent on both TDP1 and LIG3β for resolution of 3′-TOP1 and 3′-PG obstructive termini. Bohr and coworkers have recently discovered that another end processing protein, APTX (see section 5.2.) is present in mitochondria (in press). One of the essential component of mitochondrial nucleoids, mitochondrial transcription factor A (TFAM) is important for transcription and replication in these organelles. The role of TFAM in mtBER has recently been examined and the results indicate that TFAM can modulate mtBER by binding DNA and by protein interactions. TFAM preferentially bound to DNA containing 8-oxo-dG in vitro, and lysates from TFAM knockdown cells had higher 8-oxo-dG incision activity but increased mtDNA damage accumulation compared to the control cells (Canugovi et al., 2010).
7.3. Deficient mitochondrial BER in Cockayne syndrome
While the significance of mtDNA MMR, DSBR and DR in neurodegenerative processes is still unclear, there is substantial evidence, as described below, supporting a link to deficient mtDNA BER. Thus, the pathology of CS may in part be attributable to the role of CSB in the mtBER of oxidative base damage. Levels of the mitochondrial isoform of OGG1 in CSB-deficient cells are low as is the 8-oxo-dG incision activity (Stevnsner et al., 2002). Mutant CSBm/m mouse embryo fibroblasts exhibit hypersensitivity to the mitochondrial oxidant paraquat (de Waard et al., 2003). Mutant CSBm/m mice additionally contain increased levels of 8-oxo-dG in their mtDNA and the cellular hypersensitivity extends to another mitochondrial oxidant, menadione (Osenbroch et al., 2009). CSBm/m cells also display altered organization of respiratory complexes in the inner mitochondrial membrane and consistent with this are slow to recover from ATP depletion induced by menadione. Very recently two different groups have reported mitochondrial localization of CSB and further evidence of CSB involvement in mtBER. CSB was found in mitochondria complexed with mtDNA and mitochondrial OGG1upon oxidative stress (Kamenisch et al., 2010). In addition to increased CSB distribution after oxidative stress, reduced 8-oxo-dG, uracil and 5-hydroxy-uracil incision activities, linked to the mitochondrial inner membrane in CSB-deficient cells, was reported (Aamann et al., 2010). Altogether these findings strongly imply that CSB mutated cells have deficient mtBER as well as dysfunctional mitochondria. Deficient mtBER has also been linked to several age-associated neurodegenerative diseases as described below.
7.4. Alzheimer’s disease
Alzheimer’s disease (AD), a progressive neurodegenerative disease, is the most common age-associated severe dementia. The clinical symptoms of AD are progressive memory impairment, cognitive decline, and behavioral changes. At later stages of the disease, mobility and coordination ability is impaired with slowness, rigidity, tremors, and gait problems. Neuropathologically, AD is characterized by deposition of amyloid β-peptide (Aβ) plaques, intraneuronal neurofibrillary tangles, synaptic degeneration and neuronal death (Mattson, 2004). Aβ accumulates in mitochondria and can impair the function of electron transport enzymes (Caspersen et al., 2005), cause cellular calcium overload (Canevari et al., 2004) and may increase the level of H2O2 in the brain (Manczak et al., 2006), which is consistent with the finding that Aβ can cause oxidative stress such as lipid peroxidation (Bruce-Keller et al., 1998; Butterfield et al., 1994). The neurons lost in AD are the poorly myelinated projection neurons of the cerebral cortex, a population of cells that may be highly vulnerable to mitochondrial dysfunction due to their dependence on a high level of ATP production to enable energy-costly ion fluxes and axonal transport. The lack of myelin is associated with high metabolic demand, high oxygen consumption, and high baseline oxidative stress.
Brains from AD patients have elevated levels of oxidative base damage in both nDNA and mtDNA and interestingly, the mtDNA have approximately 10-fold higher levels than nDNA (Wang et al., 2005). Different brain regions show different levels of DNA damage and this pattern correlates with the pathology of AD brains, with the most affected regions having the highest level of DNA damage (Wang et al., 2005). AD brain cells have increased fragmentation of mtDNA, reduced mtDNA content and display evidence of apoptosis, indicating that mtDNA damage gives rise to the neuronal cell loss in AD (de la Monte et al., 2000). BER deficiency has been found in brain tissue from post mortem sporadic AD patients, with both an affected brain region (inferior parietal lobule) and the least affected region (cerebellum) showing similar alteration in BER function (Weissman et al., 2007b). Specifically, in the inferior parietal lobule, reduced levels of the UDG and DNA polymerase β proteins were observed as well as reduced uracil incision, 8-oxodG incision and DNA repair synthesis. In the cerebellum, both UDG protein levels and uracil incision was found to be reduced as was 8-oxodG incision activity. DNA repair synthesis and Pol β protein levels were likewise significantly reduced although less so than for the inferior parietal lobule. Interestingly, this study also examined patients with the amnestic subtype of mild cognitive impairment, the earliest clinically detectable phase of AD. Samples from these subjects displayed a significant linear decline in uracil incision activity and DNA repair synthesis that was correlated with the severity of clinical symptoms. The finding that BER is impaired is not limited to neuropathological brain regions but extends to the cerebellum, a region with no neuronal cell death in AD, suggests that BER deficiency may be a general characteristic of human AD brains. Furthermore, this defect may be present at the earliest stage of AD as it was also found in amnestic mild cognitive impairment patients. It is likely that the BER deficiency in AD brains described above is even more pronounced when specifically measured in mitochondria, but it is important to note that this remains to be carefully investigated. One study have found decreased neuronal cytoplasmic levels of OGG1, the DNA glycosylase responsible for incision of 8-oxodG, in some AD brain regions (Iida et al., 2002). A more recent investigation adds further evidence for deficient BER in early AD, with reported significant decreases in nuclear and mitochondrial OGG1 activities in the neocortex of mild cognitive impairment patients similar to late-stage AD (Shao et al., 2008). The mtDNA damage in AD patients increases the production of ROS (Swerdlow et al., 1997) resulting in production of more mtDNA damage, and this combined with deficient mtBER may create an increasing cycle of destruction culminating in neuronal death. Polymorphisms in TFAM, a modulator of mtBER (see above), can influence the risk of sporadic late-onset AD (Zhang et al., 2011).
Mouse models of AD allow examination of the earlier stages of AD and its progression while the study of human AD is generally limited by the availability of post-mortem tissue samples. AD mice, unlike human AD patients, do not display neurodegeneration even though both Aβ plaques and neurofibrillary tangles accumulate. A possible explanation is suggested by recent findings. Transgenic mice with AD-like symptoms did not display a generally significant decrease in 8-oxodG, and uracil incision activity, and DNA repair synthesis by Pol β appeared normal in the symptomatic mice (Weissman et al., 2009). An investigation into in mtBER of the synaptic fraction from whole mouse brains during normal aging and in a model of AD found that significant age-related decline of BER capacity occur specifically at the synapses, due to a reduction in the level of BER proteins and reduced mitochondrial axonal transport(Gredilla et al., 2010). However, there was no difference in repair capacity between wild-type and AD-model mice in any age group. A difference in the mechanisms involved in progressions of disease pathology between species may therefore complicate the use of mouse models for research into human AD.
The presence of oxidative damage in the brains from AD patients with Aβ plaques and neurofibrillary tangles led to the hypothesis that such structures represented the main source of oxidative stress in AD brains. However, recent findings indicate that the role of Aβ peptides may be complex. Oxidative damage has been found in brains of individuals with mild cognitive impairment, a state preceding AD (Keller et al., 2005), and oxidative stress precedes Aβ plaque formation in the pathogenesis of AD. A newer, alternative hypothesis therefore proposes that while the Aβ peptides are initially produced as an antioxidant defense mechanism, the later deposition of aggregated Aβ plaques represent pro-oxidant structures. The presence of pro-oxidant Aβ plaques in specific areas of the brain combined with the general deficiency in repair of the oxidative base lesions in AD described above, potentially explains why cell death is limited to particular neuronal cell populations in the AD brain. Also, the apparent absence of a BER deficiency in mouse AD brains may explain why neuronal cell death is absent even though Aβ plaques are present.
7.5. Parkinson’s disease
Parkinson’s disease (PD) is the second most prevalent neurodegenerative disease, affecting approximately 2% of the population over the age of 65 (de Rijk et al., 1997). PD is clinically characterized by tremor, muscular rigidity, bradykinesia, and pathologically by massive loss of dopaminergic neurons in the substantia nigra (SN). The loss of neurons is associated with Lewy bodies, inclusion bodies that contain aggregated α-synuclein protein in large amounts (Mouradian, 2002).
Several studies implicate DNA damage in PD neurodegeneration: oxidatively damagednDNA and mtDNA were found in SN neurons (Alam et al., 1997; Zhang et al., 1999), increased 8-oxo-dG was found in SN mitochondria of PD patients (Shimura-Miura et al., 1999), and the PD mitochondria produce more ROS and have more mtDNA damage than normal mitochondria (Swerdlow et al., 1996). DNA deletions can be caused by oxidative DNA damage (Dumont et al., 2000; Hayakawa et al., 1992) and several brain regions of PD patients display large numbers of mtDNA deletions (Gu et al., 2002), including the substantia nigra (Bender et al., 2006; Kraytsberg et al., 2006). mtDNA point mutations accumulate in SN neurons of PD brains (Cantuti-Castelvetri et al., 2005). The SN of PD patients also displays elevated expression of OGG1β (Fukae et al., 2005). These studies have led to the hypothesis that mtDNA lesions trigger the apoptosis of SN dopaminergic neurons in PD. Exposing neuroblastoma cells to a mitochondrial complex I inhibitor led to increased 8-oxo-dG levels in cells, many of which also displayed morphology characteristics of apoptosis and increased α-synuclein levels (Sherer et al., 2002). Transgenic mice with mutated α-synuclein displayed accumulation of oxidative mtDNA damage in brain cells that also featured increased levels of apoptotic markers (Tanaka et al., 2001). Mutation in α-synuclein have been associated with familial PD (Thomas and Beal, 2007) and altogether these results suggest that neuronal apoptosis induced by α-synuclein mutations is linked to mtDNA lesions.
The exact cause of the selective loss of dopaminergic neurons in PD remains unclear. But it is worth considering whether there are characteristics of dopaminergic neurons that may make them more vulnerable to defects in DNA repair. It has been put forth that these neuronal cells could be under oxidative stress because dopamine at physiological pH is easily oxidized to produce ROS(Izumi et al., 2005). Against this, it is argued that dopamine is normally stored in synaptic vesicles where the acidic conditions prevent auto-oxidation but it is also noted that under conditions of stress, such as exposure to glutamate, extravesicular dopamine will be present and therefore subject to oxidation(Izumi et al., 2009). The excitatory neurotransmitter glutamate has previously been implicated in PD (Choi, 1988)and it has been suggested that excessive stimulation of glutamate receptors on dopaminergic neurons may be involved in the progression of PD(Blandini et al., 1996; Rodriguez et al., 1998). There is recent evidence that dopaminergic neurons are more vulnerable to continuous exposure to low concentrations of glutamate than non-dopaminergic neurons (Izumi et al., 2009). This is intriguing in the context of DNA repair because low, transient, physiological levels of glutamate causes an influx of calcium in the neuron, which in turn induces damage to mitochondrial ROS production resulting in oxidative DNA damage (Yang et al., 2010). Such exposure to low and transient doses of glutamate was found to be followed by increased DNA repair activity and increased mRNA and protein levels of the essential BER protein APE1 resulting in rapid and efficient repair of neuronal DNA. In light of these findings, it seems possible that a prolonged or excessive stimulation of glutamate receptors in PD could cause a mitochondrial ROS production that exceeds the DNA repair capacity of the more vulnerable dopaminergc neurons. Or that any potential deficiency in the repair of oxidative DNA damage in PD patients may adversely affect the capacity of the dopaminergic neurons to deal with the normal mitochondrial ROS production from glutamate receptor stimulation.
7.6. Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS) is the most common adult-onset motor neuron disease, and is characterized by progressive degeneration and death of neurons in the spinal cord, brain stem and cerebral cortex (Boillee et al., 2006). The disorder is clinically characterized by muscle weakness, fasciculations and atrophy that results from the denervation associated with the death of motor neurons. Approximately 90% of cases are sporadic (SALS) and the rest are familial (FALS). The protein ALS2 may protect neurons against oxidative stress, and a rare mutation in the ALS2 gene can cause an inherited infantile/juvenile onset FALS (Hadano et al., 2001; Hadano et al., 2007). SALS patients exhibit mitochondrial dysfunction and higher levels of mtDNA point mutations in spinal cord cells (Wiedemann et al., 2002). Mutations in the Cu/Zn-superoxide dismutase 1 (SOD1) antioxidant enzyme that converts O2•− to H2O2 may be the cause of about 20% of FALS cases (Bruijn et al., 2004) and the SOD1 version from a mouse model of human FALS increases the vulnerability of mitochondria (Jaiswal and Keller, 2009). ALS patients have increased levels of 8-oxo-dG in the spinal cord (Kikuchi et al., 2002), as do transgenic SOD1 mice spinal cord motor neuron mtDNA (Warita et al., 2001). Both SSBs and DSBs of mtDNA were found in the motor neurons of SOD1 mutant mice, and the DSBs increased with age (Martin et al., 2007). The mtDNA repair enzyme DNA polymerase γ is decreased in SOD1 mutant mice spinal cord neurons (Murakami et al., 2007). Taken together, these results suggest that the motor neurons in ALS may have deficient mtDNA repair of oxidative lesions, and suffer from both mtDNA damage and mitochondrial dysfunction, potentially contributing to neurodegeneration.
8. RecQ helicase disorders
8.1. Human RecQ helicases
Helicases are ATP-hydrolysis powered motor proteins that separate the two complementary strands of nucleic acid duplexes. Humans posses five distinct DNA helicases of the RecQ family: WRN, BLM, RECQ1, RECQ4 and RECQ5 (van Brabant et al., 2000). Human RecQ helicases are active in replication, recombination, DNA repair and possibly transcription, chromatin structure regulation and telomere maintenance (Bohr, 2008). These functions provide the RecQ helicases with a key role in maintaining genomic integrity by protecting against deleterious changes to the DNA, and perhaps accordingly, mutations in these enzymes can cause rare diseases characterized by genomic instability (van Brabant et al., 2000). The RecQ helicases interact with various proteins of the DNA repair pathways as discussed in recent reviews (Brosh and Bohr, 2007; Cobb and Bjergbaek, 2006; Hanada and Hickson, 2007; Kusumoto et al., 2007; Ouyang et al., 2008; Sharma et al., 2006). Important cellular functions of human RecQ helicases are: unwinding of DNA duplexes by the 3′ to 5′ helicase activity (Sharma et al., 2006), strand annealing of complementary ssDNA molecules (Sharma et al., 2006), and in the case of WRN, 3′ to 5′ exonuclease activity (Huang et al., 1998; Kamath-Loeb et al., 1998; Shen et al., 1998). Mutations in three of the helicases, BLM, WRN and RECQ4, result in the disorders Bloom syndrome, Werner syndrome and Rothmund-Thomson syndrome, respectively. One of these diseases, Werner syndrome, features both progressive neurodegeneration and premature aging as discussed below.
8.2. Werner syndrome
8.2.1. Characteristics of Werner syndrome and the WRN protein
Werner syndrome (WS) is a rare autosomal recessive disease characterized by genetic instability, premature aging and elevated cancer risk (Epstein et al., 1966; Goto, 1997; Schellenberg, 2001). WS mimics many changes seen with normal aging such as wrinkling of the skin, graying of the hair, cataracts, diabetes, osteoporosis, and progressive neurodegeneration (Huang et al., 1998). From microarray studies it has been observed that transcriptional changes seen in WS closely resemble those seen in normal aging (Kyng et al., 2005). WS cells are characterized by high levels of chromosomal aberrations and hypersensitivity to some DNA damaging agents (Brosh and Bohr, 2007). The WRN gene encodes a 1432 amino acid protein with a 3′ to 5′ DNA helicase domain and a 3′ to 5′ exonuclease domain (Bohr et al., 2000). WS is caused by loss-of-function mutations in the WRN gene and both the helicase and exonuclease activities must be lost for the disease to develop (Huang et al., 2006). WRN has been shown to physically interact with proteins in several different DNA repair pathways including DSBR, BER/SSBR, MMR and DNA interstrand cross-link (ICL) repair, in addition to proteins involved in telomere maintenance (Brosh and Bohr, 2007; Saydam et al., 2007).
8.2.2. WRN in DSBR
WRN interacts with components of both the HR and NHEJ pathways of DSBR. For HR, WRN interaction with RAD52 either stimulates or inhibits the helicase activity depending on DNA structure, and increases the efficiency of strand annealing mediated by RAD52 (Baynton et al., 2003). This implicates WRN in the rescue of replication forks after DNA damage. WRN also interacts with BRCA1 (Cheng et al., 2006), and an interaction with the MRN complex through NSB1 stimulates the helicase activity (Cheng et al., 2004). RAD51 associates with WRN and RAD54 co-localizes strongly with WRN after replication arrest (Otterlei et al., 2006). In the NHEJ pathway, Ku70-80 heterodimer interaction with WRN strongly stimulates the 3′ to 5′ exonuclease activity (Cooper et al., 2000; Li and Comai, 2000) and DNA-PKCS dependent phosphorylation of WRN regulates the helicase and exonuclease activities (Karmakar et al., 2002). Certain oxidative DNA lesions with OH groups at the C8 position in the major groove of DNA can block the exonuclease activity (Machwe et al., 2000). However, the presence of Ku70-80 can stimulate the WRN exonuclease to bypass these lesions (Bukowy et al., 2008). The interaction between Ku70-80 may therefore be important in the context of DSBs induced by both ionizing radiation and radiomimetic drugs, which are known to generate oxidatively induced base lesions in addition to DNA strand breaks. WRN thus appears to be an important accessory protein for increasing the fidelity of NHEJ. It is, however, not necessarily an essential component, as deficiency in core components of NHEJ causes hypersensitivity to ionizing radiation while the cells of WS patients appear to have only mildly increased sensitivity. It has been reported that the exonuclease activity of WRN is stimulated by the physical interaction with the LIG4-XRCC4-XLF complex, involved in NHEJ, and that this complex can ligate a substrate processed by the WRN exonuclease (Kusumoto et al., 2008). In light of the evidence for WRN activity in the DSBR pathways, the chromosomal aberrations seen in WS cells may partly result from unrepaired DSBs.
8.2.3. WRN in BER
WS cells are sensitive to DNA damaging agents repaired by the BER pathway (Blank et al., 2004; Harrigan et al., 2006) and accumulate 8-oxo-dG lesions (Von Kobbe et al., 2004b), indicating a role for WRN in BER. Furthermore, WRN is required for PARP1 ribosylation (von Kobbe et al., 2003), and physical interaction between WRN and PARP1 regulates the helicase and exonuclease activity of WRN (von Kobbe et al., 2004a), which implicates WRN in SSBR given that PARP1 is a sensor of SSBs. WRN may in fact be important in BER/SSBR as it interacts functionally with many of the proteins in these pathways. APE1 inhibits WRN helicase activity possibly to prevent undesired unwinding of the BER intermediates before repair processing by downstream enzymes in the BER pathway (Ahn et al., 2004). WRN stimulates Pol β strand displacement synthesis (Harrigan et al., 2003). The FEN1 cleavage of the 5′ flap created by strand displacement synthesis is greatly stimulated (Brosh et al., 2001) and the NEIL1 DNA glycosylase is also stimulated by WRN (Das et al., 2007). WRN can protect non-dividing cells from oxidative DNA damage and depletion of the protein produce accelerated cellular senescence in normal fibroblasts (Szekely et al., 2005). Taken together these results indicate that the neurodegenerative and premature aging phenotype of WS patients may relate to oxidative damage and SSB accumulation.
8.2.4. WRN in MMR
It has been suggested by an earlier study showing deficient base-base mismatch and insertion-deletion loop repair in WS fibroblast cells, that WRN might have a role in MMR (Bennett et al., 1997b). WRN can physically interact with the MSH2-MSH6, MSH2-MSH3 and MLH1-PMS2 complexes involved in initiation of MMR (Saydam et al., 2007). The helicase activity of WRN was also found to be strongly stimulated by MSH2-MSH6 and MSH2-MSH3.
8.2.5. WRN in ICL repair
DNA ICLs are rare events formed by covalent binding of the complementary strands of the double helix that can prevent DNA strand separation and hence represent formidable blocks to replication and transcription. While they occur relatively infrequently, they are highly cyto- and genotoxic with an estimated 40 unrepaired ICLs being enough to induce cell death (Akkari et al., 2000). Accumulation of ICLs in the genome over time has been proposed to contribute to genomic instability and the aging process (Mitchell et al., 2003).
ICL repair in mammals is not yet well understood but may involve proteins from HR and the NER pathways as well as error-prone TLS polymerases (Hinz, 2010). When considering models for a mammalian ICL repair pathway, the current data argues for choice of subpathway and repair proteins that depend on cell cycle status (McHugh and Sarkar, 2006). In the DNA of non-replicating cells or outside of the S-phase, ICL repair is thought to rely primarily on NER proteins as well as TLS polymerase activity (Fig. 10A). In the context of actively replicating cells ICL repair appears significantly more complex involving coordination of the S-phase checkpoint, the Fanconi anemia (FA) pathway, TLS, NER and HR (Fig. 10B). Excellent reviews on ICL repair and the FA pathway have been published recently and are recommended for interested readers (Andreassen and Ren, 2009; de Winter and Joenje, 2009).
Fig. 10. Model for mammalian inter-strand crosslink repair.
The ICL repair process is influenced by cell-cycle status. (A) In ICL repair in non-replicating cells the crosslink is recognized by the XPC-HR23B-CEN2 complex or RNA pol II. The XPF-ERCC1 complex makes incisions in the DNA on either side of the ICL leaving a gap in the opposing strand and the incised oligonucleotide still attached to the intact strand. TLS polymerases fill the gap while the flipped out crosslinked base is recognized by the DBB1-DDB2 complex, triggering the completion of repair by NER. (B) In actively replicating cells ICL repair at stalled replication forks is initiated when the arrested fork activates ATR and its downstream kinase CHK1. Facilitated by RPA and the MRN complex, ATR and CHK1 phosphorylates many proteins of the FA-BRCA network. The FANCM-FAAP24 complex enables access of other repair proteins, and subsequently becomes part of the FA core complex. The FA core complex monoubiquitylates the FANCD2-FANCI complex, which is then retained at the damaged region by BRCA1. The ICL is unhooked by XPF-ERCC1 and MUS81-EME1, leaving a DSB. The monoubiquitinated FANCD2-FANCI complex releases the replicative polymerase Pol δ and loads an error-prone TLS polymerase to perform bypass synthesis to repair the gap, possibly assisted by FANCJ. The crosslinked base is removed by NER. After 5′ end resection, possibly by the MRN complex, the BRCA2-FANCN-RAD51 complex initiates the reconstruction of the replication fork by HR.
There is considerable evidence linking WRN to ICL repair. WS cells are more sensitive to ICL-inducing compounds than any other genotoxic compounds (Liu et al., 2009; Poot et al., 2001; Poot et al., 2002). Triggering of the intra-S checkpoint after exposure to DNA damaging agents is necessary to inhibit DNA synthesis and allow for repair. It has been reported that WRN is required for this S phase checkpoint as well as the damage signaling in response to ICL-induced DNA lesions (Cheng et al., 2008). The helicase but not exonuclease activity of WRN is required for the repair of at least some types of DNA ICLs in vivo (Cheng et al., 2006). This study also found evidence to suggest WRN cooperates with BRCA1 in the HR steps of ICL repair. The interaction of WRN with BRCA1 was found to increase in cells treated with ICL-inducing agents. WRN also participates in a complex with the ATR and RAD51 in response to ICL-induced replication arrest and interacts directly with RAD51 (Otterlei et al., 2006).
8.2.6. WRN in TLS
For TLS to bypass DNA lesions, it is necessary for the PCNA clamp to be activated by mono-ubiquitination by Rad18 (Lehmann, 2006). Very recently it has been suggested that WRN can participate in TLS through interaction with NBS1 of the MRN complex (Kobayashi et al., 2010). DNA damage induced the dissociation of PCNA from WRN in an NBS1-dependent manner leading to the ubiquitination of PCNA. Furthermore, WS cells exhibited increase in spontaneous formation of Rad18 foci, an increase in the interaction between PCNA and Rad18 and spontaneous mono-ubiquitination of PCNA.
8.2.7. WRN in maintenance of telomeres
The ends of linear eukaryotic chromosomes have specialized nucleoprotein complexes called telomeres that function to protect against events that may lead to genome instability. During the normal aging process a progressive shortening of the telomeres is observed. Non-functional short telomeres are believed to contribute to the aging process. Fibroblast cells from WS patients have higher rates of telomere shortening than those of normal fibroblasts (Baird et al., 2004) and display symptoms of telomere dysfunction including premature senescence (Davis et al., 2003; Schulz et al., 1996). WRN interacts with telomeric DNA structures and telosome proteins, including TRF1 and TRF2, regulators of telomere length (Brosh and Bohr, 2007; Opresko et al., 2002). TRF2 stimulates the helicase activity of WRN (Machwe et al., 2004; Opresko et al., 2002) and it has been proposed that WRN plays a role in capping of the telomeres (Multani and Chang, 2007). TRF2 also binds to BER proteins, and both TRF1 and TRF2 interact with proteins that function in DSBR (Brosh and Bohr, 2007), possibly to recruit BER and DSBR to maintain the telomeres. It is worth noting that because of the high guanine content in telomeric DNA, the level of 8-oxo-dG base lesions is higher than in non-telomeric DNA, and the BER activity on this lesion is less effective in some telomere structures(Rhee et al., 2011). Very recently, an interesting study was published that links location (genomic vs. telomeric DNA) and chromosomal aberration frequencies with premature cellular senescence (Hagelstrom et al., 2010). In telomerase-negative WRN-deficient cells, the frequencies of telomeric but not genomic chromosomal abnormalities are significantly increased compared to control cells. It was furthermore predicted that in cells with increased telomeric chromosomal aberrations, the onset of replicative senescence will be dramatically accelerated, a factor that may be contributing to the accelerated aging observed in WS. These findings suggest that telomere dysfunction that activates apoptotic pathways may be involved in the neurodegenerative and premature aging phenotype of WS patients. Telomere dysfunction adversely affects many tissues including more quiescent systems and intriguingly, a very recent investigation has linked dysfunctional telomeres with impaired mitochondrial biogenesis and function as well as increased production of ROS (Sahin et al., 2011). Conceivably, this could have repercussions also for neuronal tissue.
8.2.8. Neurodegeneration in Werner syndrome
WS is often referred to as a segmental progeroid syndrome due to the phenotypic similarity to some symptoms of normal aging including progressive neurodegeneration. This similarity has prompted much research into the functions of the WRN gene as it may hold the promise of illuminating some of the cellular functions that can become deficient during the normal aging process. As described above, WRN is involved in many DNA metabolic pathways notably those involved in upholding the integrity of the genome. Failure to properly repair DNA is therefore a likely reason for the neurodegenerative symptoms seen in WS patients but what defective pathway(s) are responsible may be difficult to determine. Defective HR during neurogenesis may lead to damaged cells being incorporated into the brain, or alternatively, a defective NHEJ pathway results in DSBs in the mature non-dividing neurons. If BER/SSBR is compromised by a mutated WRN protein oxidative lesions will remain unrepaired with a rapidly rising steady-state level that resembles an accelerated form of the observed accumulation in normal aging brains. Finally, it is possible that the change in transcriptional pattern seen with this disease is simply not compatible with the high transcriptional demand of neurons. The study of the WS disorder has linked deficient DNA repair to the aging phenotype but the question still remains whether such deficiency is a cause for aging per se or rather a reflection of increasing cellular dysfunction with age.
9. Conclusion
The neurodegenerative diseases examined in this review highlight the importance of DNA repair in maintaining genomic integrity in the CNS, and implicates DNA damage and DNA repair deficiency in the aging of the brain. Much work still needs to be done to better understand the role of DNA repair enzymes and pathways in neurons. In particular, it would be of great interest to determine and clarify if certain neuronal subpopulations are more vulnerable to the effects of DNA repair deficiencies given that some neurodegenerative disorders involve specific types of neurons. Neuronal DNA repair is one of the most exciting areas for future DNA repair investigations as understanding neurodegenerative processes will be of increasing concern in aging populations with the preservation of cognitive function as a key component of healthy aging. The study of the SSBR and DSBR syndromes so far may suggest that SSBs have the greater impact on the aging brain, but the extent to which the aging brain depends on the repair of the two types of strand breaks is not yet clear. Given the evidence that mitochondrial dysfunction is implicated in several common neurodegenerative diseases associated with aging, it will be of great interest to further investigate the nature and extent of neuronal mtDNA repair. More insight is also needed for potential future therapeutic strategies. While this review has presented evidence linking DNA repair defects and aging with neurodegeneration, we would like to end on a positive note by highlighting the fact that neurodegenerative disease is not an inevitable consequence of long life, not even in 115-year olds (den Dunnen et al., 2008).
Article Highlights.
Neurological disease is a major symptom of genomic instability disorders
Deficient DNA repair is linked to progressive neurodegeneration
Several hereditary ataxia disorders are characterized by defective DNA repair
Mitochondrial DNA damage is implicated in the age-associated Alzheimer’s disease
Neurodegeneration is a feature of the premature aging disorder Werner syndrome
Acknowledgments
We thank members of the Laboratory for DNA repair and Aging, Dept Molecular Biology, Aarhus University, Denmark and members of the Laboratory of Molecular Gerontology, NIA-NIH (Baltimore, MD) - especially Deborah Croteau for critically reading the manuscript.
D.K. Jeppesen was supported by the Danish Cancer Society and part of this work was supported by a grant from The Velux Foundation to the Danish Aging Research Center.
Abbreviations
- 3′-OH
3′-hydroxyl
- 3′-P
3′-phosphate
- 3′-PG
3′-phosphoglycolate
- 3′-PUA
3′-phospho-α,β-unsaturated aldehyde
- 3′-TOP1
3′-topoisomerase I
- 5′-Ade
5′-aldehyde
- 5′-AMP
5′-adenosine monophosphate
- 5′-dRP
5′-deoxyribose-5-phosphate
- 5′-OH
5′-hydroxyl
- 5′-P
5′-phosphate
- 8-oxo-dG
7,8-dihydro-8-oxoguanine
- Aβ
amyloid β-peptide
- AD
Alzheimer’s disease
- ALS
amyotrophic lateral sclerosis
- AOA1
ataxia-oculomotor apraxia-1
- AP
abasic
- APTX
aprataxin
- AT
ataxia telangiectasia
- ATLD
ataxia telangiectasia-like disorder
- ATM
ataxia telangiectasia mutated
- BER
base excision repair
- CNS
central nervous system
- CS
Cockayne syndrome
- DR
direct reversal
- DSB
double-strand break
- DSBR
double-strand break repair
- GG-NER
global genome nucleotide excision repair
- HD
Huntington’s disease
- HNE
trans-4-hydroxy-2-nonenal
- H2O2
hydrogen peroxide
- HR
homologous recombination
- ICL
interstrand cross-link
- LIG1
DNA ligase I
- LIG3α
DNA ligase IIIα
- LIG4
DNA Ligase IV
- LP-BER
long-patch base excision repair
- MMR
mismatch repair
- mtBER
mitochondrial BER
- mtDNA
mitochondrial DNA
- NBS
Nijmegen breakage syndrome
- nDNA
nuclear DNA
- NER
nucleotide excision repair
- NHEJ
non-homologous end-joining
- OGG1
8-oxoguanine DNA glycosylase
- PARP1
poly(ADP-ribose) polymerase-1
- PD
Parkinson’s disease
- PdG
propane-deoxyguanosine
- Pol β
DNA polymerase β
- Pol δ/ε
DNA polymerase δ or ε
- RNA pol II
RNA polymerase II
- ROS
reactive oxygen species
- SCAN1
spinocerebellar ataxia with axonal neuropathy-1
- SN
substantia nigra
- SOD1
superoxide dismutase 1
- SP-BER
short-patch base excision repair
- SSB
single-strand break
- SSBR
single-strand break repair
- ssDNA
single-stranded DNA
- TC-NER
transcription-coupled nucleotide excision repair
- TDP1
tyrosyl-DNA phosphodiesterase 1
- TLS
Translesion DNA synthesis
- TOP1
DNA topoisomerase I
- TOP1cc
TOP1 cleavage complexes
- UV
ultraviolet
- WS
Werner syndrome
- XP
Xeroderma Pigmentosum
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–6. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- Ahmad A, Enzlin JH, Bhagwat NR, Wijgers N, Raams A, Appledoorn E, Theil AF, JHJH, Vermeulen W, NGJJ, Scharer OD, Niedernhofer LJ. Mislocalization of XPF-ERCC1 nuclease contributes to reduced DNA repair in XP-F patients. PLoS Genet. 2010;6:e1000871. doi: 10.1371/journal.pgen.1000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn B, Harrigan JA, Indig FE, Wilson DM, 3rd, Bohr VA. Regulation of WRN helicase activity in human base excision repair. J Biol Chem. 2004;279:53465–74. doi: 10.1074/jbc.M409624200. [DOI] [PubMed] [Google Scholar]
- Aicardi J, Barbosa C, Andermann E, Andermann F, Morcos R, Ghanem Q, Fukuyama Y, Awaya Y, Moe P. Ataxia-ocular motor apraxia: a syndrome mimicking ataxia-telangiectasia. Ann Neurol. 1988;24:497–502. doi: 10.1002/ana.410240404. [DOI] [PubMed] [Google Scholar]
- Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7:605–16. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol. 2000;20:8283–9. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam ZI, Jenner A, Daniel SE, Lees AJ, Cairns N, Marsden CD, Jenner P, Halliwell B. Oxidative DNA damage in the parkinsonian brain: an apparent selective increase in 8-hydroxyguanine levels in substantia nigra. J Neurochem. 1997;69:1196–203. doi: 10.1046/j.1471-4159.1997.69031196.x. [DOI] [PubMed] [Google Scholar]
- Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci U S A. 1993;90:7915–22. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, Ren K. Fanconi anemia proteins, DNA interstrand crosslink repair pathways, and cancer therapy. Curr Cancer Drug Targets. 2009;9:101–17. doi: 10.2174/156800909787314011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews AD, Barrett SF, Robbins JH. Xeroderma pigmentosum neurological abnormalities correlate with colony-forming ability after ultraviolet radiation. Proc Natl Acad Sci U S A. 1978;75:1984–8. doi: 10.1073/pnas.75.4.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angele S, Romestaing P, Moullan N, Vuillaume M, Chapot B, Friesen M, Jongmans W, Cox DG, Pisani P, Gerard JP, Hall J. ATM haplotypes and cellular response to DNA damage: association with breast cancer risk and clinical radiosensitivity. Cancer Res. 2003;63:8717–25. [PubMed] [Google Scholar]
- Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem. 2001;276:18665–72. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- Baird DM, Davis T, Rowson J, Jones CJ, Kipling D. Normal telomere erosion rates at the single cell level in Werner syndrome fibroblast cells. Hum Mol Genet. 2004;13:1515–24. doi: 10.1093/hmg/ddh159. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–95. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Balajee AS, May A, Dianov GL, Friedberg EC, Bohr VA. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci U S A. 1997;94:4306–11. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barja G, Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J. 2000;14:312–8. doi: 10.1096/fasebj.14.2.312. [DOI] [PubMed] [Google Scholar]
- Barja G. Aging in vertebrates, and the effect of caloric restriction: a mitochondrial free radical production-DNA damage mechanism? Biol Rev Camb Philos Soc. 2004a;79:235–51. doi: 10.1017/s1464793103006213. [DOI] [PubMed] [Google Scholar]
- Barja G. Free radicals and aging. Trends Neurosci. 2004b;27:595–600. doi: 10.1016/j.tins.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Barzilai A. The contribution of the DNA damage response to neuronal viability. Antioxid Redox Signal. 2007;9:211–8. doi: 10.1089/ars.2007.9.211. [DOI] [PubMed] [Google Scholar]
- Baynton K, Otterlei M, Bjoras M, von Kobbe C, Bohr VA, Seeberg E. WRN interacts physically and functionally with the recombination mediator protein RAD52. J Biol Chem. 2003;278:36476–86. doi: 10.1074/jbc.M303885200. [DOI] [PubMed] [Google Scholar]
- Beal MF. Mitochondria take center stage in aging and neurodegeneration. Ann Neurol. 2005;58:495–505. doi: 10.1002/ana.20624. [DOI] [PubMed] [Google Scholar]
- Becherel OJ, Jakob B, Cherry AL, Gueven N, Fusser M, Kijas AW, Peng C, Katyal S, McKinnon PJ, Chen J, Epe B, Smerdon SJ, Taucher-Scholz G, Lavin MF. CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response. Nucleic Acids Res. 2010;38:1489–503. doi: 10.1093/nar/gkp1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherel OJ, Gueven N, Birrell GW, Schreiber V, Suraweera A, Jakob B, Taucher-Scholz G, Lavin MF. Nucleolar localization of aprataxin is dependent on interaction with nucleolin and on active ribosomal DNA transcription. Hum Mol Genet. 2006;15:2239–49. doi: 10.1093/hmg/ddl149. [DOI] [PubMed] [Google Scholar]
- Bell GI, Burant CF, Takeda J, Gould GW. Structure and function of mammalian facilitative sugar transporters. J Biol Chem. 1993;268:19161–4. [PubMed] [Google Scholar]
- Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, Taylor RW, Turnbull DM. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–7. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- Bennett RA, Wilson DM, 3rd, Wong D, Demple B. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc Natl Acad Sci U S A. 1997a;94:7166–9. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SE, Umar A, Oshima J, Monnat RJ, Jr, Kunkel TA. Mismatch repair in extracts of Werner syndrome cell lines. Cancer Res. 1997b;57:2956–60. [PubMed] [Google Scholar]
- Biton S, Barzilai A, Shiloh Y. The neurological phenotype of ataxia-telangiectasia: solving a persistent puzzle. DNA Repair (Amst) 2008;7:1028–38. doi: 10.1016/j.dnarep.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Blandini F, Porter RH, Greenamyre JT. Glutamate and Parkinson’s disease. Mol Neurobiol. 1996;12:73–94. doi: 10.1007/BF02740748. [DOI] [PubMed] [Google Scholar]
- Blank A, Bobola MS, Gold B, Varadarajan S, DDK, Meade EH, Rabinovitch PS, Loeb LA, Silber JR. The Werner syndrome protein confers resistance to the DNA lesions N3-methyladenine and O6-methylguanine: implications for WRN function. DNA Repair (Amst) 2004;3:629–38. doi: 10.1016/j.dnarep.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Bohr VA, Cooper M, Orren D, Machwe A, Piotrowski J, Sommers J, Karmakar P, Brosh R. Werner syndrome protein: biochemical properties and functional interactions. Exp Gerontol. 2000;35:695–702. doi: 10.1016/s0531-5565(00)00145-5. [DOI] [PubMed] [Google Scholar]
- Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–20. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillee S, Vande Velde C, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Bokov A, Chaudhuri A, Richardson A. The role of oxidative damage and stress in aging. Mech Ageing Dev. 2004;125:811–26. doi: 10.1016/j.mad.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Brazel CY, Rao MS. Aging and neuronal replacement. Ageing Res Rev. 2004;3:465–83. doi: 10.1016/j.arr.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Halaban R, van Gool AJ, Henning KA, Friedberg EC, Warren SL. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci U S A. 1996;93:11586–90. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C. Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry. 2002;41:9003–14. doi: 10.1021/bi025942q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K, Tarone RE, Robbins JH. The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem. 2000;275:22355–62. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- Brooks PJ. The case for 8,5′-cyclopurine-2′-deoxynucleosides as endogenous DNA lesions that cause neurodegeneration in xeroderma pigmentosum. Neuroscience. 2007;145:1407–17. doi: 10.1016/j.neuroscience.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–44. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–28. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce-Keller AJ, Begley JG, Fu W, Butterfield DA, Bredesen DE, Hutchins JB, Hensley K, Mattson MP. Bcl-2 protects isolated plasma and mitochondrial membranes against lipid peroxidation induced by hydrogen peroxide and amyloid beta-peptide. J Neurochem. 1998;70:31–9. doi: 10.1046/j.1471-4159.1998.70010031.x. [DOI] [PubMed] [Google Scholar]
- Bruckner BA, Ammini CV, Otal MP, Raizada MK, Stacpoole PW. Regulation of brain glucose transporters by glucose and oxygen deprivation. Metabolism. 1999;48:422–31. doi: 10.1016/s0026-0495(99)90098-7. [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–49. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- Bukowy Z, Harrigan JA, Ramsden DA, Tudek B, Bohr VA, Stevnsner T. WRN Exonuclease activity is blocked by specific oxidatively induced base lesions positioned in either DNA strand. Nucleic Acids Res. 2008;36:4975–87. doi: 10.1093/nar/gkn468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcham PC. Genotoxic lipid peroxidation products: their DNA damaging properties and role in formation of endogenous DNA adducts. Mutagenesis. 1998;13:287–305. doi: 10.1093/mutage/13.3.287. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Hensley K, Harris M, Mattson M, Carney J. beta-Amyloid peptide free radical fragments initiate synaptosomal lipoperoxidation in a sequence-specific fashion: implications to Alzheimer’s disease. Biochem Biophys Res Commun. 1994;200:710–5. doi: 10.1006/bbrc.1994.1508. [DOI] [PubMed] [Google Scholar]
- Cabelof DC, Raffoul JJ, Yanamadala S, Ganir C, Guo Z, Heydari AR. Attenuation of DNA polymerase beta-dependent base excision repair and increased DMS-induced mutagenicity in aged mice. Mutat Res. 2002;500:135–45. doi: 10.1016/s0027-5107(02)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW, Tucker JD, Stanker LH, Thompson LH. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–43. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–94. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays. 2001;23:447–55. doi: 10.1002/bies.1063. [DOI] [PubMed] [Google Scholar]
- Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003;2:955–69. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- Canevari L, Abramov AY, Duchen MR. Toxicity of amyloid beta peptide: tales of calcium, mitochondria, and oxidative stress. Neurochem Res. 2004;29:637–50. doi: 10.1023/b:nere.0000014834.06405.af. [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri I, Lin MT, Zheng K, Keller-McGandy CE, Betensky RA, Johns DR, Beal MF, Standaert DG, Simon DK. Somatic mitochondrial DNA mutations in single neurons and glia. Neurobiol Aging. 2005;26:1343–55. doi: 10.1016/j.neurobiolaging.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Canugovi C, Maynard S, Bayne AC, Sykora P, Tian J, de Souza-Pinto NC, Croteau DL, Bohr VA. The mitochondrial transcription factor A functions in mitochondrial base excision repair. DNA Repair (Amst) 2010;9:1080–9. doi: 10.1016/j.dnarep.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J Biol Chem. 1997;272:23970–5. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–86. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Caspersen C, Wang N, Yao J, Sosunov A, Chen X, Lustbader JW, Xu HW, Stern D, McKhann G, Yan SD. Mitochondrial Abeta: a potential focal point for neuronal metabolic dysfunction in Alzheimer’s disease. FASEB J. 2005;19:2040–1. doi: 10.1096/fj.05-3735fje. [DOI] [PubMed] [Google Scholar]
- Chalmers AJ. Poly(ADP-ribose) polymerase-1 and ionizing radiation: sensor, signaller and therapeutic target. Clin Oncol (R Coll Radiol) 2004;16:29–39. doi: 10.1016/s0936-6555(03)00223-1. [DOI] [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–15. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- Cheng WH, von Kobbe C, Opresko PL, Arthur LM, Komatsu K, Seidman MM, Carney JP, Bohr VA. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J Biol Chem. 2004;279:21169–76. doi: 10.1074/jbc.M312770200. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Kusumoto R, Opresko PL, Sui X, Huang S, Nicolette ML, Paull TT, Campisi J, Seidman M, Bohr VA. Collaboration of Werner syndrome protein and BRCA1 in cellular responses to DNA interstrand cross-links. Nucleic Acids Res. 2006;34:2751–60. doi: 10.1093/nar/gkl362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Muftic D, Muftuoglu M, Dawut L, Morris C, Helleday T, Shiloh Y, Bohr VA. WRN is required for ATM activation and the S-phase checkpoint in response to interstrand cross-link-induced DNA double-strand breaks. Mol Biol Cell. 2008;19:3923–33. doi: 10.1091/mbc.E07-07-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW. Glutamate neurotoxicity and diseases of the nervous system. Neuron. 1988;1:623–34. doi: 10.1016/0896-6273(88)90162-6. [DOI] [PubMed] [Google Scholar]
- Choi J, Shendrik I, Peacocke M, Peehl D, Buttyan R, Ikeguchi EF, Katz AE, Benson MC. Expression of senescence-associated beta-galactosidase in enlarged prostates from men with benign prostatic hyperplasia. Urology. 2000;56:160–6. doi: 10.1016/s0090-4295(00)00538-0. [DOI] [PubMed] [Google Scholar]
- Chun HH, Gatti RA. Ataxia-telangiectasia, an evolving phenotype. DNA Repair (Amst) 2004;3:1187–96. doi: 10.1016/j.dnarep.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Chung FL, Pan J, Choudhury S, Roy R, Hu W, Tang MS. Formation of trans-4-hydroxy-2-nonenal- and other enal-derived cyclic DNA adducts from omega-3 and omega-6 polyunsaturated fatty acids and their roles in DNA repair and human p53 gene mutation. Mutat Res. 2003;531:25–36. doi: 10.1016/j.mrfmmm.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Clayton DA, Doda JN, Friedberg EC. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc Natl Acad Sci U S A. 1974;71:2777–81. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218:652–6. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, Brenner C, Moreira MC, Taylor AM, Caldecott KW. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 2004;3:1493–502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Cline SD, Riggins JN, Tornaletti S, Marnett LJ, Hanawalt PC. Malondialdehyde adducts in DNA arrest transcription by T7 RNA polymerase and mammalian RNA polymerase II. Proc Natl Acad Sci U S A. 2004;101:7275–80. doi: 10.1073/pnas.0402252101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L. RecQ helicases: lessons from model organisms. Nucleic Acids Res. 2006;34:4106–14. doi: 10.1093/nar/gkl557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey G, Lakshmipathy U, Campbell C. Mammalian mitochondrial extracts possess DNA end-binding activity. Nucleic Acids Res. 1999;27:3348–54. doi: 10.1093/nar/27.16.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, Leach DR. Repair of DNA covalently linked to protein. Mol Cell. 2004;13:307–16. doi: 10.1016/s1097-2765(04)00056-5. [DOI] [PubMed] [Google Scholar]
- Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–12. [PMC free article] [PubMed] [Google Scholar]
- Cullinane C, Bohr VA. DNA interstrand cross-links induced by psoralen are not repaired in mammalian mitochondria. Cancer Res. 1998;58:1400–4. [PubMed] [Google Scholar]
- Cullinane C, Cutts SM, Panousis C, Phillips DR. Interstrand cross-linking by adriamycin in nuclear and mitochondrial DNA of MCF-7 cells. Nucleic Acids Res. 2000;28:1019–25. doi: 10.1093/nar/28.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Errico M, Parlanti E, Teson M, de Jesus BM, Degan P, Calcagnile A, Jaruga P, Bjoras M, Crescenzi M, Pedrini AM, Egly JM, Zambruno G, Stefanini M, Dizdaroglu M, Dogliotti E. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006;25:4305–15. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza Pinto N, Ramos W, Greenberg MM, Hazra TK, Mitra S, Bohr VA. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J Biol Chem. 2007;282:26591–602. doi: 10.1074/jbc.M703343200. [DOI] [PubMed] [Google Scholar]
- Das BB, Dexheimer TS, Maddali K, Pommier Y. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci U S A. 2010;107:19790–5. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, Sakai T, Takahashi T, Nagatomo H, Sekijima Y, Kawachi I, Takiyama Y, Nishizawa M, Fukuhara N, Saito K, Sugano S, Tsuji S. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29:184–8. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- Davis T, Singhrao SK, Wyllie FS, Haughton MF, Smith PJ, Wiltshire M, Wynford-Thomas D, Jones CJ, Faragher RG, Kipling D. Telomere-based proliferative lifespan barriers in Werner-syndrome fibroblasts involve both p53-dependent and p53-independent mechanisms. J Cell Sci. 2003;116:1349–57. doi: 10.1242/jcs.00331. [DOI] [PubMed] [Google Scholar]
- Davydov V, Hansen LA, Shackelford DA. Is DNA repair compromised in Alzheimer’s disease? Neurobiol Aging. 2003;24:953–68. doi: 10.1016/s0197-4580(02)00229-4. [DOI] [PubMed] [Google Scholar]
- de Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21:453–60. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- de la Monte SM, Luong T, Neely TR, Robinson D, Wands JR. Mitochondrial DNA damage as a mechanism of cell loss in Alzheimer’s disease. Lab Invest. 2000;80:1323–35. doi: 10.1038/labinvest.3780140. [DOI] [PubMed] [Google Scholar]
- de Rijk MC, Rocca WA, Anderson DW, Melcon MO, Breteler MM, Maraganore DM. A population perspective on diagnostic criteria for Parkinson’s disease. Neurology. 1997;48:1277–81. doi: 10.1212/wnl.48.5.1277. [DOI] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, Lebel M, Stevnsner TV, Rasmussen LJ, Bohr VA. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst) 2009;8:704–19. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat Res. 2009;668:11–9. doi: 10.1016/j.mrfmmm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- de Waard H, de Wit J, Gorgels TG, van den Aardweg G, Andressoo JO, Vermeij M, van Steeg H, Hoeijmakers JH, van der Horst GT. Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair (Amst) 2003;2:13–25. doi: 10.1016/s1568-7864(02)00188-x. [DOI] [PubMed] [Google Scholar]
- Demple B, Sung JS. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair (Amst) 2005;4:1442–9. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- den Dunnen WF, Brouwer WH, Bijlard E, Kamphuis J, van Linschoten K, Eggens-Meijer E, Holstege G. No disease in the brain of a 115-year-old woman. Neurobiol Aging. 2008;29:1127–32. doi: 10.1016/j.neurobiolaging.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Dianov G, Bischoff C, Sunesen M, Bohr VA. Repair of 8-oxoguanine in DNA is deficient in Cockayne syndrome group B cells. Nucleic Acids Res. 1999;27:1365–8. doi: 10.1093/nar/27.5.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov GL, Houle JF, Iyer N, Bohr VA, Friedberg EC. Reduced RNA polymerase II transcription in extracts of cockayne syndrome and xeroderma pigmentosum/Cockayne syndrome cells. Nucleic Acids Res. 1997;25:3636–42. doi: 10.1093/nar/25.18.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digweed M, Sperling K. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks. DNA Repair (Amst) 2004;3:1207–17. doi: 10.1016/j.dnarep.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Ding G, Franki N, Kapasi AA, Reddy K, Gibbons N, Singhal PC. Tubular cell senescence and expression of TGF-beta1 and p21(WAF1/CIP1) in tubulointerstitial fibrosis of aging rats. Exp Mol Pathol. 2001;70:43–53. doi: 10.1006/exmp.2000.2346. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M, Karakaya A, Jaruga P, Slupphaug G, Krokan HE. Novel activities of human uracil DNA N-glycosylase for cytosine-derived products of oxidative DNA damage. Nucleic Acids Res. 1996;24:418–22. doi: 10.1093/nar/24.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M, Jaruga P, Rodriguez H. Identification and quantification of 8,5′-cyclo-2′-deoxy-adenosine in DNA by liquid chromatography/mass spectrometry. Free Radic Biol Med. 2001;30:774–84. doi: 10.1016/s0891-5849(01)00464-6. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat Res. 2005;591:45–59. doi: 10.1016/j.mrfmmm.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Dumont P, Burton M, Chen QM, Gonos ES, Frippiat C, Mazarati JB, Eliaers F, Remacle J, Toussaint O. Induction of replicative senescence biomarkers by sublethal oxidative stresses in normal human fibroblast. Free Radic Biol Med. 2000;28:361–73. doi: 10.1016/s0891-5849(99)00249-x. [DOI] [PubMed] [Google Scholar]
- El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–13. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- El-Khamisy SF, Hartsuiker E, Caldecott KW. TDP1 facilitates repair of ionizing radiation-induced DNA single-strand breaks. DNA Repair (Amst) 2007;6:1485–95. doi: 10.1016/j.dnarep.2007.04.015. [DOI] [PubMed] [Google Scholar]
- El-Khamisy SF, Katyal S, Patel P, Ju L, McKinnon PJ, Caldecott KW. Synergistic decrease of DNA single-strand break repair rates in mouse neural cells lacking both Tdp1 and aprataxin. DNA Repair (Amst) 2009;8:760–6. doi: 10.1016/j.dnarep.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres M, Biniszkiewicz D, Sobol RW, Harms C, Ahmadi M, Lipski A, Katchanov J, Mergenthaler P, Dirnagl U, Wilson SH, Meisel A, Jaenisch R. Increased postischemic brain injury in mice deficient in uracil-DNA glycosylase. J Clin Invest. 2004;113:1711–21. doi: 10.1172/JCI20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner’s syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore) 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Fan J, Wilson DM., 3rd Protein-protein interactions and posttranslational modifications in mammalian base excision repair. Free Radic Biol Med. 2005;38:1121–38. doi: 10.1016/j.freeradbiomed.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Fang-Kircher SG, Labudova O, Kitzmueller E, Rink H, Cairns N, Lubec G. Increased steady state mRNA levels of DNA-repair genes XRCC1, ERCC2 and ERCC3 in brain of patients with Down syndrome. Life Sci. 1999;64:1689–99. doi: 10.1016/s0024-3205(99)00107-1. [DOI] [PubMed] [Google Scholar]
- Flangas AL, Bowman RE. Differential metabolism of RNA in neuronal-enriched and glial-enriched fractions of rat cerebrum. J Neurochem. 1970;17:1237–45. doi: 10.1111/j.1471-4159.1970.tb03373.x. [DOI] [PubMed] [Google Scholar]
- Flohr C, Burkle A, Radicella JP, Epe B. Poly(ADP-ribosyl)ation accelerates DNA repair in a pathway dependent on Cockayne syndrome B protein. Nucleic Acids Res. 2003;31:5332–7. doi: 10.1093/nar/gkg715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini P, Pascucci B, Parlanti E, D’Errico M, Simonelli V, Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85:1053–71. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–82. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Meira LB. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage Version 7. DNA Repair (Amst) 2006;5:189–209. doi: 10.1016/j.dnarep.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Frouin I, Maga G, Denegri M, Riva F, Savio M, Spadari S, Prosperi E, Scovassi AI. Human proliferating cell nuclear antigen, poly(ADP-ribose) polymerase-1, and p21waf1/cip1. A dynamic exchange of partners. J Biol Chem. 2003;278:39265–8. doi: 10.1074/jbc.C300098200. [DOI] [PubMed] [Google Scholar]
- Fujimura M, Morita-Fujimura Y, Sugawara T, Chan PH. Early decrease of XRCC1, a DNA base excision repair protein, may contribute to DNA fragmentation after transient focal cerebral ischemia in mice. Stroke. 1999;30:2456–62. doi: 10.1161/01.str.30.11.2456. discussion 2463. [DOI] [PubMed] [Google Scholar]
- Fujimura M, Morita-Fujimura Y, Noshita N, Yoshimoto T, Chan PH. Reduction of the DNA base excision repair protein, XRCC1, may contribute to DNA fragmentation after cold injury-induced brain trauma in mice. Brain Res. 2000;869:105–11. doi: 10.1016/s0006-8993(00)02375-1. [DOI] [PubMed] [Google Scholar]
- Fukae J, Takanashi M, Kubo S, Nishioka K, Nakabeppu Y, Mori H, Mizuno Y, Hattori N. Expression of 8-oxoguanine DNA glycosylase (OGG1) in Parkinson’s disease and related neurodegenerative disorders. Acta Neuropathol. 2005;109:256–62. doi: 10.1007/s00401-004-0937-9. [DOI] [PubMed] [Google Scholar]
- Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–40. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J Biol Chem. 1997;272:24522–9. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- Gary R, Kim K, Cornelius HL, Park MS, Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J Biol Chem. 1999;274:4354–63. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
- Gatti RA, Becker-Catania S, Chun HH, Sun X, Mitui M, Lai CH, Khanlou N, Babaei M, Cheng R, Clark C, Huo Y, Udar NC, Iyer RK. The pathogenesis of ataxia-telangiectasia. Learning from a Rosetta Stone. Clin Rev Allergy Immunol. 2001;20:87–108. doi: 10.1385/CRIAI:20:1:87. [DOI] [PubMed] [Google Scholar]
- Golden TR, Hinerfeld DA, Melov S. Oxidative stress and aging: beyond correlation. Aging Cell. 2002;1:117–23. doi: 10.1046/j.1474-9728.2002.00015.x. [DOI] [PubMed] [Google Scholar]
- Gorodetsky E, Calkins S, Ahn J, Brooks PJ. ATM, the Mre11/Rad50/Nbs1 complex, and topoisomerase I are concentrated in the nucleus of Purkinje neurons in the juvenile human brain. DNA Repair (Amst) 2007;6:1698–707. doi: 10.1016/j.dnarep.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M. Hierarchical deterioration of body systems in Werner’s syndrome: implications for normal ageing. Mech Ageing Dev. 1997;98:239–54. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- Graziewicz MA, Sayer JM, Jerina DM, Copeland WC. Nucleotide incorporation by human DNA polymerase gamma opposite benzo[a]pyrene and benzo[c]phenanthrene diol epoxide adducts of deoxyguanosine and deoxyadenosine. Nucleic Acids Res. 2004;32:397–405. doi: 10.1093/nar/gkh213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredilla R, Weissman L, Yang JL, Bohr VA, Stevnsner T. Mitochondrial base excision repair in mouse synaptosomes during normal aging and in a model of Alzheimer’s disease. Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–67. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, Kisselev AF, Harel-Bellan A, Nakatani Y. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 2006;20:1429–34. doi: 10.1101/gad.378206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryk MR, Marintchev A, Maciejewski MW, Robertson A, Wilson SH, Mullen GP. Mapping of the interaction interface of DNA polymerase beta with XRCC1. Structure. 2002;10:1709–20. doi: 10.1016/s0969-2126(02)00908-5. [DOI] [PubMed] [Google Scholar]
- Gu G, Reyes PE, Golden GT, Woltjer RL, Hulette C, Montine TJ, Zhang J. Mitochondrial DNA deletions/rearrangements in parkinson disease and related neurodegenerative disorders. J Neuropathol Exp Neurol. 2002;61:634–9. doi: 10.1093/jnen/61.7.634. [DOI] [PubMed] [Google Scholar]
- Gueven N, Becherel OJ, Kijas AW, Chen P, Howe O, Rudolph JH, Gatti R, Date H, Onodera O, Taucher-Scholz G, Lavin MF. Aprataxin, a novel protein that protects against genotoxic stress. Hum Mol Genet. 2004;13:1081–93. doi: 10.1093/hmg/ddh122. [DOI] [PubMed] [Google Scholar]
- Gumy-Pause F, Wacker P, Sappino AP. ATM gene and lymphoid malignancies. Leukemia. 2004;18:238–42. doi: 10.1038/sj.leu.2403221. [DOI] [PubMed] [Google Scholar]
- Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–21. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH, Jr, Scherer SW, Rouleau GA, Hayden MR, Ikeda JE. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet. 2001;29:166–73. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- Hadano S, Kunita R, Otomo A, Suzuki-Utsunomiya K, Ikeda JE. Molecular and cellular function of ALS2/alsin: implication of membrane dynamics in neuronal development and degeneration. Neurochem Int. 2007;51:74–84. doi: 10.1016/j.neuint.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Hagelstrom RT, Blagoev KB, Niedernhofer LJ, Goodwin EH, Bailey SM. Hyper telomere recombination accelerates replicative senescence and may promote premature aging. Proc Natl Acad Sci U S A. 2010;107:15768–73. doi: 10.1073/pnas.1006338107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, Kewitt K, Walter CA, Richardson A. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci U S A. 2001;98:10469–74. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007;64:2306–22. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28:425–33. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–7. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- Harman D. The aging process. Proc Natl Acad Sci U S A. 1981;78:7124–8. doi: 10.1073/pnas.78.11.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrigan JA, Opresko PL, von Kobbe C, Kedar PS, Prasad R, Wilson SH, Bohr VA. The Werner syndrome protein stimulates DNA polymerase beta strand displacement synthesis via its helicase activity. J Biol Chem. 2003;278:22686–95. doi: 10.1074/jbc.M213103200. [DOI] [PubMed] [Google Scholar]
- Harrigan JA, Wilson DM, 3rd, Prasad R, Opresko PL, Beck G, May A, Wilson SH, Bohr VA. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase beta. Nucleic Acids Res. 2006;34:745–54. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrigan JA, Fan J, Momand J, Perrino FW, Bohr VA, Wilson DM., 3rd WRN exonuclease activity is blocked by DNA termini harboring 3′ obstructive groups. Mech Ageing Dev. 2007;128:259–66. doi: 10.1016/j.mad.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JL, Jakob B, Taucher-Scholz G, Dianov GL, Becherel OJ, Lavin MF. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum Mol Genet. 2009;18:4102–17. doi: 10.1093/hmg/ddp359. [DOI] [PubMed] [Google Scholar]
- Hashiguchi K, Stuart JA, de Souza-Pinto NC, Bohr VA. The C-terminal alphaO helix of human Ogg1 is essential for 8-oxoguanine DNA glycosylase activity: the mitochondrial beta-Ogg1 lacks this domain and does not have glycosylase activity. Nucleic Acids Res. 2004;32:5596–608. doi: 10.1093/nar/gkh863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins AJ, Subler MA, Akopiants K, Wiley JL, Taylor SM, Rice AC, Windle JJ, Valerie K, Povirk LF. In vitro complementation of Tdp1 deficiency indicates a stabilized enzyme-DNA adduct from tyrosyl but not glycolate lesions as a consequence of the SCAN1 mutation. DNA Repair (Amst) 2009;8:654–63. doi: 10.1016/j.dnarep.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa M, Hattori K, Sugiyama S, Ozawa T. Age-associated oxygen damage and mutations in mitochondrial DNA in human hearts. Biochem Biophys Res Commun. 1992;189:979–85. doi: 10.1016/0006-291x(92)92300-m. [DOI] [PubMed] [Google Scholar]
- He X, van Waardenburg RC, Babaoglu K, Price AC, Nitiss KC, Nitiss JL, Bjornsti MA, White SW. Mutation of a conserved active site residue converts tyrosyl-DNA phosphodiesterase I into a DNA topoisomerase I-dependent poison. J Mol Biol. 2007;372:1070–81. doi: 10.1016/j.jmb.2007.07.055. [DOI] [PubMed] [Google Scholar]
- Heeres JT, Hergenrother PJ. Poly(ADP-ribose) makes a date with death. Curr Opin Chem Biol. 2007;11:644–53. doi: 10.1016/j.cbpa.2007.08.038. [DOI] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst) 2007;6:923–35. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Hentati F, Ben Hamida C, Zeghal M, Kamoun M, Fezaa B, Ben Hamida M. Age-dependent axonal loss in nerve biopsy of patients with xeroderma pigmentosum. Neuromuscul Disord. 1992;2:361–9. doi: 10.1016/s0960-8966(06)80007-6. [DOI] [PubMed] [Google Scholar]
- Hinerfeld D, Traini MD, Weinberger RP, Cochran B, Doctrow SR, Harry J, Melov S. Endogenous mitochondrial oxidative stress: neurodegeneration, proteomic analysis, specific respiratory chain defects, and efficacious antioxidant therapy in superoxide dismutase 2 null mice. J Neurochem. 2004;88:657–67. doi: 10.1046/j.1471-4159.2003.02195.x. [DOI] [PubMed] [Google Scholar]
- Hinz JM. Role of homologous recombination in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51:582–603. doi: 10.1002/em.20577. [DOI] [PubMed] [Google Scholar]
- Hirano M, Yamamoto A, Mori T, Lan L, Iwamoto TA, Aoki M, Shimada K, Furiya Y, Kariya S, Asai H, Yasui A, Nishiwaki T, Imoto K, Kobayashi N, Kiriyama T, Nagata T, Konishi N, Itoyama Y, Ueno S. DNA single-strand break repair is impaired in aprataxin-related ataxia. Ann Neurol. 2007a;61:162–74. doi: 10.1002/ana.21078. [DOI] [PubMed] [Google Scholar]
- Hirano R, Interthal H, Huang C, Nakamura T, Deguchi K, Choi K, Bhattacharjee MB, Arimura K, Umehara F, Izumo S, Northrop JL, Salih MA, Inoue K, Armstrong DL, Champoux JJ, Takashima H, Boerkoel CF. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007b;26:4732–43. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Yamaguchi R, Asami S, Iwamoto N, Kasai H. 8-hydroxyguanine levels in nuclear DNA and its repair activity in rat organs associated with age. J Gerontol A Biol Sci Med Sci. 1996;51:B303–7. doi: 10.1093/gerona/51a.5.b303. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Holden JA, Rahn MP, Jolles CJ, Vorobyev SV, Bronstein IB. Immunohistochemical detection of DNA topoisomerase I in formalin fixed, paraffin wax embedded normal tissues and in ovarian carcinomas. Mol Pathol. 1997;50:247–53. doi: 10.1136/mp.50.5.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JK, Prasad R, Hou E, Wilson SH. Protection against methylation-induced cytotoxicity by DNA polymerase beta-dependent long patch base excision repair. J Biol Chem. 2000;275:2211–8. doi: 10.1074/jbc.275.3.2211. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Scuric Z, D’Andrea AD, Schiestl RH. Impaired DNA double strand break repair in cells from Nijmegen breakage syndrome patients. DNA Repair (Amst) 2006;5:251–7. doi: 10.1016/j.dnarep.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′-->5′ exonuclease. Nat Genet. 1998;20:114–6. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Lee L, Hanson NB, Lenaerts C, Hoehn H, Poot M, Rubin CD, Chen DF, Yang CC, Juch H, Dorn T, Spiegel R, Oral EA, Abid M, Battisti C, Lucci-Cordisco E, Neri G, Steed EH, Kidd A, Isley W, Showalter D, Vittone JL, Konstantinow A, Ring J, Meyer P, Wenger SL, von Herbay A, Wollina U, Schuelke M, Huizenga CR, Leistritz DF, Martin GM, Mian IS, Oshima J. The spectrum of WRN mutations in Werner syndrome patients. Hum Mutat. 2006;27:558–67. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic Res. 1998;29:573–9. doi: 10.1080/10715769800300611. [DOI] [PubMed] [Google Scholar]
- Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Iida T, Furuta A, Nishioka K, Nakabeppu Y, Iwaki T. Expression of 8-oxoguanine DNA glycosylase is reduced and associated with neurofibrillary tangles in Alzheimer’s disease brain. Acta Neuropathol. 2002;103:20–5. doi: 10.1007/s004010100418. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Kohmoto T, Tabata R, Seki Y. Differential intracellular localization of the human and mouse endonuclease III homologs and analysis of the sorting signals. DNA Repair (Amst) 2002;1:847–54. doi: 10.1016/s1568-7864(02)00145-3. [DOI] [PubMed] [Google Scholar]
- Intano GW, Cho EJ, McMahan CA, Walter CA. Age-related base excision repair activity in mouse brain and liver nuclear extracts. J Gerontol A Biol Sci Med Sci. 2003;58:205–11. doi: 10.1093/gerona/58.3.b205. [DOI] [PubMed] [Google Scholar]
- Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc Natl Acad Sci U S A. 2001;98:12009–14. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Interthal H, Chen HJ, Kehl-Fie TE, Zotzmann J, Leppard JB, Champoux JJ. SCAN1 mutant Tdp1 accumulates the enzyme--DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 2005;24:2224–33. doi: 10.1038/sj.emboj.7600694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Yurov YB. Increased chromosome instability dramatically disrupts neural genome integrity and mediates cerebellar degeneration in the ataxia-telangiectasia brain. Hum Mol Genet. 2009;18:2656–69. doi: 10.1093/hmg/ddp207. [DOI] [PubMed] [Google Scholar]
- Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26:231–43. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Sawada H, Yamamoto N, Kume T, Katsuki H, Shimohama S, Akaike A. Iron accelerates the conversion of dopamine-oxidized intermediates into melanin and provides protection in SH-SY5Y cells. J Neurosci Res. 2005;82:126–37. doi: 10.1002/jnr.20595. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Yamamoto N, Matsuo T, Wakita S, Takeuchi H, Kume T, Katsuki H, Sawada H, Akaike A. Vulnerability to glutamate toxicity of dopaminergic neurons is dependent on endogenous dopamine and MAPK activation. J Neurochem. 2009;110:745–55. doi: 10.1111/j.1471-4159.2009.06178.x. [DOI] [PubMed] [Google Scholar]
- Jaiswal MK, Keller BU. Cu/Zn superoxide dismutase typical for familial amyotrophic lateral sclerosis increases the vulnerability of mitochondria and perturbs Ca2+ homeostasis in SOD1G93A mice. Mol Pharmacol. 2009;75:478–89. doi: 10.1124/mol.108.050831. [DOI] [PubMed] [Google Scholar]
- Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–14. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–46. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Fink SP, Marnett LJ. Repair of propanodeoxyguanosine by nucleotide excision repair in vivo and in vitro. J Biol Chem. 1997;272:11434–8. doi: 10.1074/jbc.272.17.11434. [DOI] [PubMed] [Google Scholar]
- Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- Kajander OA, Karhunen PJ, Holt IJ, Jacobs HT. Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep. 2001;2:1007–12. doi: 10.1093/embo-reports/kve233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb AS, Shen JC, Loeb LA, Fry M. Werner syndrome protein. II. Characterization of the integral 3′--> 5′ DNA exonuclease. J Biol Chem. 1998;273:34145–50. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- Kamenisch Y, Fousteri M, Knoch J, von Thaler AK, Fehrenbacher B, Kato H, Becker T, Dolle ME, Kuiper R, Majora M, Schaller M, van der Horst GT, van Steeg H, Rocken M, Rapaport D, Krutmann J, Mullenders LH, Berneburg M. Proteins of nucleotide and base excision repair pathways interact in mitochondria to protect from loss of subcutaneous fat, a hallmark of aging. J Exp Med. 2010;207:379–90. doi: 10.1084/jem.20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Tahara S, Matsuo M. Non-linear accumulation of 8-hydroxy-2′-deoxyguanosine, a marker of oxidized DNA damage, during aging. Mutat Res. 1996;316:277–85. doi: 10.1016/s0921-8734(96)90010-7. [DOI] [PubMed] [Google Scholar]
- Karanjawala ZE, Murphy N, Hinton DR, Hsieh CL, Lieber MR. Oxygen metabolism causes chromosome breaks and is associated with the neuronal apoptosis observed in DNA double-strand break repair mutants. Curr Biol. 2002;12:397–402. doi: 10.1016/s0960-9822(02)00684-x. [DOI] [PubMed] [Google Scholar]
- Karmakar P, Piotrowski J, Brosh RM, Jr, Sommers JA, Miller SP, Cheng WH, Snowden CM, Ramsden DA, Bohr VA. Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J Biol Chem. 2002;277:18291–302. doi: 10.1074/jbc.M111523200. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Katyal S, el-Khamisy SF, Russell HR, Li Y, Ju L, Caldecott KW, McKinnon PJ. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–31. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JN, Schmitt FA, Scheff SW, Ding Q, Chen Q, Butterfield DA, Markesbery WR. Evidence of increased oxidative damage in subjects with mild cognitive impairment. Neurology. 2005;64:1152–6. doi: 10.1212/01.WNL.0000156156.13641.BA. [DOI] [PubMed] [Google Scholar]
- Khobta A, Kitsera N, Speckmann B, Epe B. 8-Oxoguanine DNA glycosylase (Ogg1) causes a transcriptional inactivation of damaged DNA in the absence of functional Cockayne syndrome B (Csb) protein. DNA Repair (Amst) 2009;8:309–17. doi: 10.1016/j.dnarep.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Kikuchi H, Furuta A, Nishioka K, Suzuki SO, Nakabeppu Y, Iwaki T. Impairment of mitochondrial DNA repair enzymes against accumulation of 8-oxo-guanine in the spinal motor neurons of amyotrophic lateral sclerosis. Acta Neuropathol. 2002;103:408–14. doi: 10.1007/s00401-001-0480-x. [DOI] [PubMed] [Google Scholar]
- Kirkali G, de Souza-Pinto NC, Jaruga P, Bohr VA, Dizdaroglu M. Accumulation of (5′S)-8,5′-cyclo-2′-deoxyadenosine in organs of Cockayne syndrome complementation group B gene knockout mice. DNA Repair (Amst) 2009;8:274–8. doi: 10.1016/j.dnarep.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisby GE, Kohama SG, Olivas A, Churchwell M, Doerge D, Spangler E, de Cabo R, Ingram DK, Imhof B, Bao G, Kow YW. Effect of caloric restriction on base-excision repair (BER) in the aging rat brain. Exp Gerontol. 2010;45:208–16. doi: 10.1016/j.exger.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisby GE, Milne J, Sweatt C. Evidence of reduced DNA repair in amyotrophic lateral sclerosis brain tissue. Neuroreport. 1997;8:1337–40. doi: 10.1097/00001756-199704140-00004. [DOI] [PubMed] [Google Scholar]
- Kisby GE, Lesselroth H, Olivas A, Samson L, Gold B, Tanaka K, Turker MS. Role of nucleotide- and base-excision repair in genotoxin-induced neuronal cell death. DNA Repair (Amst) 2004;3:617–27. doi: 10.1016/j.dnarep.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes Dev. 2004;18:1423–38. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44:239–67. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- Kobayashi J, Okui M, Asaithamby A, Burma S, Chen BP, Tanimoto K, Matsuura S, Komatsu K, Chen DJ. WRN participates in translesion synthesis pathway through interaction with NBS1. Mech Ageing Dev. 2010;131:436–44. doi: 10.1016/j.mad.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol. 2002;12:1846–51. doi: 10.1016/s0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- Kobayashi J. Molecular mechanism of the recruitment of NBS1/hMRE11/hRAD50 complex to DNA double-strand breaks: NBS1 binds to gamma-H2AX through FHA/BRCT domain. J Radiat Res (Tokyo) 2004;45:473–8. doi: 10.1269/jrr.45.473. [DOI] [PubMed] [Google Scholar]
- Kohji T, Hayashi M, Shioda K, Minagawa M, Morimatsu Y, Tamagawa K, Oda M. Cerebellar neurodegeneration in human hereditary DNA repair disorders. Neurosci Lett. 1998;243:133–6. doi: 10.1016/s0304-3940(98)00109-8. [DOI] [PubMed] [Google Scholar]
- Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–11. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–52. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987;123:241–50. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, DiGiovanna JJ. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–96. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–20. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476:73–7. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- Kruman, Schwartz E, Kruman Y, Cutler RG, Zhu X, Greig NH, Mattson MP. Suppression of uracil-DNA glycosylase induces neuronal apoptosis. J Biol Chem. 2004a;279:43952–60. doi: 10.1074/jbc.M408025200. [DOI] [PubMed] [Google Scholar]
- Kruman, Wersto RP, Cardozo-Pelaez F, Smilenov L, Chan SL, Chrest FJ, Emokpae R, Jr, Gorospe M, Mattson MP. Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron. 2004b;41:549–61. doi: 10.1016/s0896-6273(04)00017-0. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–70. [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. Nucleotide repeats. Slippery DNA and diseases. Nature. 1993;365:207–8. doi: 10.1038/365207a0. [DOI] [PubMed] [Google Scholar]
- Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci U S A. 2000;97:3832–7. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto R, Muftuoglu M, Bohr VA. The role of WRN in DNA repair is affected by post-translational modifications. Mech Ageing Dev. 2007;128:50–7. doi: 10.1016/j.mad.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Kusumoto R, Dawut L, Marchetti C, Wan Lee J, Vindigni A, Ramsden D, Bohr VA. Werner protein cooperates with the XRCC4-DNA ligase IV complex in end-processing. Biochemistry. 2008;47:7548–56. doi: 10.1021/bi702325t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyng KJ, May A, Stevnsner T, Becker KG, Kolvra S, Bohr VA. Gene expression responses to DNA damage are altered in human aging and in Werner Syndrome. Oncogene. 2005;24:5026–42. doi: 10.1038/sj.onc.1208692. [DOI] [PubMed] [Google Scholar]
- Laine JP, Egly JM. When transcription and repair meet: a complex system. Trends Genet. 2006;22:430–6. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C. Double strand break rejoining by mammalian mitochondrial extracts. Nucleic Acids Res. 1999;27:1198–204. doi: 10.1093/nar/27.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmipathy U, Campbell C. Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res. 2000;28:3880–6. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugel V, Dalloz C, Durand M, Sauvanaud F, Kristensen U, Vincent MC, Pasquier L, Odent S, Cormier-Daire V, Gener B, Tobias ES, Tolmie JL, Martin-Coignard D, Drouin-Garraud V, Heron D, Journel H, Raffo E, Vigneron J, Lyonnet S, Murday V, Gubser-Mercati D, Funalot B, Brueton L, Sanchez Del Pozo J, Munoz E, Gennery AR, Salih M, Noruzinia M, Prescott K, Ramos L, Stark Z, Fieggen K, Chabrol B, Sarda P, Edery P, Bloch-Zupan A, Fawcett H, Pham D, Egly JM, Lehmann AR, Sarasin A, Dollfus H. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat. 2010;31:113–26. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- Le Ber I, Moreira MC, Rivaud-Pechoux S, Chamayou C, Ochsner F, Kuntzer T, Tardieu M, Said G, Habert MO, Demarquay G, Tannier C, Beis JM, Brice A, Koenig M, Durr A. Cerebellar ataxia with oculomotor apraxia type 1: clinical and genetic studies. Brain. 2003;126:2761–72. doi: 10.1093/brain/awg283. [DOI] [PubMed] [Google Scholar]
- LeDoux SP, Wilson GL, Beecham EJ, Stevnsner T, Wassermann K, Bohr VA. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–73. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- LeDoux SP, Patton NJ, Avery LJ, Wilson GL. Repair of N-methylpurines in the mitochondrial DNA of xeroderma pigmentosum complementation group D cells. Carcinogenesis. 1993;14:913–7. doi: 10.1093/carcin/14.5.913. [DOI] [PubMed] [Google Scholar]
- Lee Y, Barnes DE, Lindahl T, McKinnon PJ. Defective neurogenesis resulting from DNA ligase IV deficiency requires Atm. Genes Dev. 2000;14:2576–80. doi: 10.1101/gad.837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Chong MJ, McKinnon PJ. Ataxia telangiectasia mutated-dependent apoptosis after genotoxic stress in the developing nervous system is determined by cellular differentiation status. J Neurosci. 2001;21:6687–93. doi: 10.1523/JNEUROSCI.21-17-06687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, McKinnon PJ. Responding to DNA double strand breaks in the nervous system. Neuroscience. 2007;145:1365–74. doi: 10.1016/j.neuroscience.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–73. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Lehmann AR. New functions for Y family polymerases. Mol Cell. 2006;24:493–5. doi: 10.1016/j.molcel.2006.10.021. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Leppard JB, Dong Z, Mackey ZB, Tomkinson AE. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol Cell Biol. 2003;23:5919–27. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesher DT, Pommier Y, Stewart L, Redinbo MR. 8-Oxoguanine rearranges the active site of human topoisomerase I. Proc Natl Acad Sci U S A. 2002;99:12102–7. doi: 10.1073/pnas.192282699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr Biol. 2000;10:919–22. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- Levin DS, Vijayakumar S, Liu X, Bermudez VP, Hurwitz J, Tomkinson AE. A conserved interaction between the replicative clamp loader and DNA ligase in eukaryotes: implications for Okazaki fragment joining. J Biol Chem. 2004;279:55196–201. doi: 10.1074/jbc.M409250200. [DOI] [PubMed] [Google Scholar]
- Li B, Comai L. Functional interaction between Ku and the werner syndrome protein in DNA end processing. J Biol Chem. 2000;275:39800. doi: 10.1074/jbc.C000289200. [DOI] [PubMed] [Google Scholar]
- Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- Li H, Vogel H, Holcomb VB, Gu Y, Hasty P. Deletion of Ku70, Ku80, or both causes early aging without substantially increased cancer. Mol Cell Biol. 2007a;27:8205–14. doi: 10.1128/MCB.00785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wu H, Yang S, Chen D. Ischemic preconditioning induces XRCC1, DNA polymerase-beta, and DNA ligase III and correlates with enhanced base excision repair. DNA Repair (Amst) 2007b;6:1297–306. doi: 10.1016/j.dnarep.2007.02.027. [DOI] [PubMed] [Google Scholar]
- Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington’s disease. Trends Genet. 2004;20:146–54. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Licht CL, Stevnsner T, Bohr VA. Cockayne syndrome group B cellular and biochemical functions. Am J Hum Genet. 2003;73:1217–39. doi: 10.1086/380399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–20. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–95. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Linnane AW, Marzuki S, Ozawa T, Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–5. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- Liu FJ, Barchowsky A, Opresko PL. The Werner syndrome protein functions in repair of Cr(VI)-induced replication-associated DNA damage. Toxicol Sci. 2009;110:307–18. doi: 10.1093/toxsci/kfp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–87. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman M, Lane DP. Transcription - guarding the genome by sensing DNA damage. Nat Rev Cancer. 2004;4:727–37. doi: 10.1038/nrc1435. [DOI] [PubMed] [Google Scholar]
- Lovell MA, Xie C, Markesbery WR. Decreased base excision repair and increased helicase activity in Alzheimer’s disease brain. Brain Res. 2000;855:116–23. doi: 10.1016/s0006-8993(99)02335-5. [DOI] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–91. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol. 2003;5:255–60. doi: 10.1038/ncb945. [DOI] [PubMed] [Google Scholar]
- Ly CV, Verstreken P. Mitochondria at the synapse. Neuroscientist. 2006;12:291–9. doi: 10.1177/1073858406287661. [DOI] [PubMed] [Google Scholar]
- Machwe A, Ganunis R, Bohr VA, Orren DK. Selective blockage of the 3′-->5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA. Nucleic Acids Res. 2000;28:2762–70. doi: 10.1093/nar/28.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Orren DK. TRF2 recruits the Werner syndrome (WRN) exonuclease for processing of telomeric DNA. Oncogene. 2004;23:149–56. doi: 10.1038/sj.onc.1206906. [DOI] [PubMed] [Google Scholar]
- Maddukuri L, Speina E, Christiansen M, Dudzinska D, Zaim J, Obtulowicz T, Kabaczyk S, Komisarski M, Bukowy Z, Szczegielniak J, Wojcik A, Kusmierek JT, Stevnsner T, Bohr VA, Tudek B. Cockayne syndrome group B protein is engaged in processing of DNA adducts of lipid peroxidation product trans-4-hydroxy-2-nonenal. Mutat Res. 2009;666:23–31. doi: 10.1016/j.mrfmmm.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistretti PJ, Pellerin L. Cellular bases of brain energy metabolism and their relevance to functional brain imaging: evidence for a prominent role of astrocytes. Cereb Cortex. 1996;6:50–61. doi: 10.1093/cercor/6.1.50. [DOI] [PubMed] [Google Scholar]
- Manczak M, Anekonda TS, Henson E, Park BS, Quinn J, Reddy PH. Mitochondria are a direct site of A beta accumulation in Alzheimer’s disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum Mol Genet. 2006;15:1437–49. doi: 10.1093/hmg/ddl066. [DOI] [PubMed] [Google Scholar]
- Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–3. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- Marnett LJ. Lipid peroxidation-DNA damage by malondialdehyde. Mutat Res. 1999;424:83–95. doi: 10.1016/s0027-5107(99)00010-x. [DOI] [PubMed] [Google Scholar]
- Martin LJ, Liu Z, Chen K, Price AC, Pan Y, Swaby JA, Golden WC. Motor neuron degeneration in amyotrophic lateral sclerosis mutant superoxide dismutase-1 transgenic mice: mechanisms of mitochondriopathy and cell death. J Comp Neurol. 2007;500:20–46. doi: 10.1002/cne.21160. [DOI] [PubMed] [Google Scholar]
- Maser RS, Zinkel R, Petrini JH. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat Genet. 2001;27:417–21. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- Mason PA, Matheson EC, Hall AG, Lightowlers RN. Mismatch repair activity in mammalian mitochondria. Nucleic Acids Res. 2003;31:1052–8. doi: 10.1093/nar/gkg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–4. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–9. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh PJ, Sarkar S. DNA interstrand cross-link repair in the cell cycle: a critical role for polymerase zeta in G1 phase. Cell Cycle. 2006;5:1044–7. doi: 10.4161/cc.5.10.2763. [DOI] [PubMed] [Google Scholar]
- McKinnon PJ. ATM and ataxia telangiectasia. EMBO Rep. 2004;5:772–6. doi: 10.1038/sj.embor.7400210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, Pommier Y. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair (Amst) 2006;5:1489–94. doi: 10.1016/j.dnarep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–40. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- Missura M, Buterin T, Hindges R, Hubscher U, Kasparkova J, Brabec V, Naegeli H. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 2001;20:3554–64. doi: 10.1093/emboj/20.13.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Hoeijmakers JH, Niedernhofer LJ. Divide and conquer: nucleotide excision repair battles cancer and ageing. Curr Opin Cell Biol. 2003;15:232–40. doi: 10.1016/s0955-0674(03)00018-8. [DOI] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403:451–6. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, Tomkinson AE. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998;17:3786–95. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore H, Greenwell PW, Liu CP, Arnheim N, Petes TD. Triplet repeats form secondary structures that escape DNA repair in yeast. Proc Natl Acad Sci U S A. 1999;96:1504–9. doi: 10.1073/pnas.96.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–93. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Geller HM. Induction of neuronal apoptosis by camptothecin, an inhibitor of DNA topoisomerase-I: evidence for cell cycle-independent toxicity. J Cell Biol. 1996;134:757–70. doi: 10.1083/jcb.134.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J. 2000;19:463–71. doi: 10.1093/emboj/19.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LH, Fousteri MI. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol Cell. 2007;27:311–23. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Mouradian MM. Recent advances in the genetics and pathogenesis of Parkinson disease. Neurology. 2002;58:179–85. doi: 10.1212/wnl.58.2.179. [DOI] [PubMed] [Google Scholar]
- Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M, Bohr VA. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J Biol Chem. 2009;284:9270–9. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multani AS, Chang S. WRN at telomeres: implications for aging and cancer. J Cell Sci. 2007;120:713–21. doi: 10.1242/jcs.03397. [DOI] [PubMed] [Google Scholar]
- Murakami T, Nagai M, Miyazaki K, Morimoto N, Ohta Y, Kurata T, Takehisa Y, Kamiya T, Abe K. Early decrease of mitochondrial DNA repair enzymes in spinal motor neurons of presymptomatic transgenic mice carrying a mutant SOD1 gene. Brain Res. 2007;1150:182–9. doi: 10.1016/j.brainres.2007.02.057. [DOI] [PubMed] [Google Scholar]
- Myers KA, Saffhill R, O’Connor PJ. Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat. Carcinogenesis. 1988;9:285–92. doi: 10.1093/carcin/9.2.285. [DOI] [PubMed] [Google Scholar]
- Nakae D, Akai H, Kishida H, Kusuoka O, Tsutsumi M, Konishi Y. Age and organ dependent spontaneous generation of nuclear 8-hydroxydeoxyguanosine in male Fischer 344 rats. Lab Invest. 2000;80:249–61. doi: 10.1038/labinvest.3780028. [DOI] [PubMed] [Google Scholar]
- Narciso L, Fortini P, Pajalunga D, Franchitto A, Liu P, Degan P, Frechet M, Demple B, Crescenzi M, Dogliotti E. Terminally differentiated muscle cells are defective in base excision DNA repair and hypersensitive to oxygen injury. Proc Natl Acad Sci U S A. 2007;104:17010–5. doi: 10.1073/pnas.0701743104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–87. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–5. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouspikel T, Hanawalt PC. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol Cell Biol. 2000;20:1562–70. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouspikel T, Hanawalt PC. DNA repair in terminally differentiated cells. DNA Repair (Amst) 2002;1:59–75. doi: 10.1016/s1568-7864(01)00005-2. [DOI] [PubMed] [Google Scholar]
- Nouspikel T. Nucleotide excision repair and neurological diseases. DNA Repair (Amst) 2008;7:1155–67. doi: 10.1016/j.dnarep.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Nowak-Wegrzyn A, Crawford TO, Winkelstein JA, Carson KA, Lederman HM. Immunodeficiency and infections in ataxia-telangiectasia. J Pediatr. 2004;144:505–11. doi: 10.1016/j.jpeds.2003.12.046. [DOI] [PubMed] [Google Scholar]
- O’Driscoll M, Jeggo PA. The role of double-strand break repair - insights from human genetics. Nat Rev Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- O’Driscoll M, Jeggo PA. The role of the DNA damage response pathways in brain development and microcephaly: insight from human disorders. DNA Repair (Amst) 2008;7:1039–50. doi: 10.1016/j.dnarep.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Oba D, Hayashi M, Minamitani M, Hamano S, Uchisaka N, Kikuchi A, Kishimoto H, Takagi M, Morio T, Mizutani S. Autopsy study of cerebellar degeneration in siblings with ataxia-telangiectasia-like disorder. Acta Neuropathol. 2010;119:513–20. doi: 10.1007/s00401-010-0639-4. [DOI] [PubMed] [Google Scholar]
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, Mullenders LH, Yamashita S, Fousteri MI, Lehmann AR. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37:714–27. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8:640–2. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- Ohtsubo T, Nishioka K, Imaiso Y, Iwai S, Shimokawa H, Oda H, Fujiwara T, Nakabeppu Y. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–64. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277:41110–9. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc Natl Acad Sci U S A. 2006;103:10017–22. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenbroch PO, Auk-Emblem P, Halsne R, Strand J, Forstrom RJ, van der Pluijm I, Eide L. Accumulation of mitochondrial DNA damage and bioenergetic dysfunction in CSB defective cells. FEBS J. 2009;276:2811–21. doi: 10.1111/j.1742-4658.2009.07004.x. [DOI] [PubMed] [Google Scholar]
- Otterlei M, Bruheim P, Ahn B, Bussen W, Karmakar P, Baynton K, Bohr VA. Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J Cell Sci. 2006;119:5137–46. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- Ouyang KJ, Woo LL, Ellis NA. Homologous recombination and maintenance of genome integrity: cancer and aging through the prism of human RecQ helicases. Mech Ageing Dev. 2008;129:425–40. doi: 10.1016/j.mad.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Owen BA, Yang Z, Lai M, Gajec M, Badger JD, 2nd, Hayes JJ, Edelmann W, Kucherlapati R, Wilson TM, McMurray CT. (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat Struct Mol Biol. 2005;12:663–70. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–4. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- Parsons JL, Dianova, Dianov GL. APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004;32:3531–6. doi: 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascucci B, Versteegh A, van Hoffen A, van Zeeland AA, Mullenders LH, Dogliotti E. DNA repair of UV photoproducts and mutagenesis in human mitochondrial DNA. J Mol Biol. 1997;273:417–27. doi: 10.1006/jmbi.1997.1268. [DOI] [PubMed] [Google Scholar]
- Patrick SM, Turchi JJ. Xeroderma pigmentosum complementation group A protein (XPA) modulates RPA-DNA interactions via enhanced complex stability and inhibition of strand separation activity. J Biol Chem. 2002;277:16096–101. doi: 10.1074/jbc.M200816200. [DOI] [PubMed] [Google Scholar]
- Pettepher CC, LeDoux SP, Bohr VA, Wilson GL. Repair of alkali-labile sites within the mitochondrial DNA of RINr 38 cells after exposure to the nitrosourea streptozotocin. J Biol Chem. 1991;266:3113–7. [PubMed] [Google Scholar]
- Pitsikas P, Lee D, Rainbow AJ. Reduced host cell reactivation of oxidative DNA damage in human cells deficient in the mismatch repair gene hMSH2. Mutagenesis. 2007;22:235–43. doi: 10.1093/mutage/gem008. [DOI] [PubMed] [Google Scholar]
- Plo I, Liao ZY, Barcelo JM, Kohlhagen G, Caldecott KW, Weinfeld M, Pommier Y. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair (Amst) 2003;2:1087–100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- Polidori MC, Mecocci P, Browne SE, Senin U, Beal MF. Oxidative damage to mitochondrial DNA in Huntington’s disease parietal cortex. Neurosci Lett. 1999;272:53–6. doi: 10.1016/s0304-3940(99)00578-9. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, Zhang H, Kohn KW. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Poot M, Yom JS, Whang SH, Kato JT, Gollahon KA, Rabinovitch PS. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15:1224–6. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- Poot M, Gollahon KA, Emond MJ, Silber JR, Rabinovitch PS. Werner syndrome diploid fibroblasts are sensitive to 4-nitroquinoline-N-oxide and 8-methoxypsoralen: implications for the disease phenotype. FASEB J. 2002;16:757–8. doi: 10.1096/fj.01-0906fje. [DOI] [PubMed] [Google Scholar]
- Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science. 1999;286:552–5. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- Povirk LF, Zhou T, Zhou R, Cowan MJ, Yannone SM. Processing of 3′-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J Biol Chem. 2007;282:3547–58. doi: 10.1074/jbc.M607745200. [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–83. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- Prasad R, Lavrik OI, Kim SJ, Kedar P, Yang XP, Vande Berg BJ, Wilson SH. DNA polymerase beta -mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J Biol Chem. 2001;276:32411–4. doi: 10.1074/jbc.C100292200. [DOI] [PubMed] [Google Scholar]
- Ranalli TA, Tom S, Bambara RA. AP endonuclease 1 coordinates flap endonuclease 1 and DNA ligase I activity in long patch base excision repair. J Biol Chem. 2002;277:41715–24. doi: 10.1074/jbc.M207207200. [DOI] [PubMed] [Google Scholar]
- Randerath K, Zhou GD, Somers RL, Robbins JH, Brooks PJ. A 32P-postlabeling assay for the oxidative DNA lesion 8,5′-cyclo-2′-deoxyadenosine in mammalian tissues: evidence that four type II I-compounds are dinucleotides containing the lesion in the 3′ nucleotide. J Biol Chem. 2001;276:36051–7. doi: 10.1074/jbc.M105472200. [DOI] [PubMed] [Google Scholar]
- Rapic-Otrin V, Navazza V, Nardo T, Botta E, McLenigan M, Bisi DC, Levine AS, Stefanini M. True XP group E patients have a defective UV-damaged DNA binding protein complex and mutations in DDB2 which reveal the functional domains of its p48 product. Hum Mol Genet. 2003;12:1507–22. doi: 10.1093/hmg/ddg174. [DOI] [PubMed] [Google Scholar]
- Rapin I, Lindenbaum Y, Dickson DW, Kraemer KH, Robbins JH. Cockayne syndrome and xeroderma pigmentosum. Neurology. 2000;55:1442–9. doi: 10.1212/wnl.55.10.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasouli-Nia A, Karimi-Busheri F, Weinfeld M. Stable down-regulation of human polynucleotide kinase enhances spontaneous mutation frequency and sensitizes cells to genotoxic agents. Proc Natl Acad Sci U S A. 2004;101:6905–10. doi: 10.1073/pnas.0400099101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Actions of aprataxin in multiple DNA repair pathways. J Biol Chem. 2007a;282:9469–74. doi: 10.1074/jbc.M611489200. [DOI] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007b;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Molecular mechanism of DNA deadenylation by the neurological disease protein aprataxin. J Biol Chem. 2008;283:33994–4001. doi: 10.1074/jbc.M807124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond AC, Staker BL, Burgin AB., Jr Substrate specificity of tyrosyl-DNA phosphodiesterase I (Tdp1) J Biol Chem. 2005;280:22029–35. doi: 10.1074/jbc.M502148200. [DOI] [PubMed] [Google Scholar]
- Reddy YV, Ding Q, Lees-Miller SP, Meek K, Ramsden DA. Non-homologous end joining requires that the DNA-PK complex undergo an autophosphorylation-dependent rearrangement at DNA ends. J Biol Chem. 2004;279:39408–13. doi: 10.1074/jbc.M406432200. [DOI] [PubMed] [Google Scholar]
- Reynolds JJ, El-Khamisy SF, Katyal S, Clements P, McKinnon PJ, Caldecott KW. Defective DNA ligation during short-patch single-strand break repair in ataxia oculomotor apraxia 1. Mol Cell Biol. 2009;29:1354–62. doi: 10.1128/MCB.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee DB, Ghosh A, Lu J, Bohr VA, Liu Y. Factors that influence telomeric oxidative base damage and repair by DNA glycosylase OGG1. DNA Repair (Amst) 2011;10:34–44. doi: 10.1016/j.dnarep.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–24. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Richardson C, Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol. 2000;20:9068–75. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci U S A. 1988;85:6465–7. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JH, Brumback RA, Mendiones M, Barrett SF, Carl JR, Cho S, Denckla MB, Ganges MB, Gerber LH, Guthrie RA, et al. Neurological disease in xeroderma pigmentosum. Documentation of a late onset type of the juvenile onset form. Brain. 1991;114(Pt 3):1335–61. doi: 10.1093/brain/114.3.1335. [DOI] [PubMed] [Google Scholar]
- Rodriguez MC, Obeso JA, Olanow CW. Subthalamic nucleus-mediated excitotoxicity in Parkinson’s disease: a target for neuroprotection. Ann Neurol. 1998;44:S175–88. doi: 10.1002/ana.410440726. [DOI] [PubMed] [Google Scholar]
- Rossi MN, Carbone M, Mostocotto C, Mancone C, Tripodi M, Maione R, Amati P. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J Biol Chem. 2009;284:31616–24. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–15. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo I, Cosentino C, Del Giudice E, Broccoletti T, Amorosi S, Cirillo E, Aloj G, Fusco A, Costanzo V, Pignata C. In ataxia-teleangiectasia betamethasone response is inversely correlated to cerebellar atrophy and directly to antioxidative capacity. Eur J Neurol. 2009;16:755–9. doi: 10.1111/j.1468-1331.2009.02600.x. [DOI] [PubMed] [Google Scholar]
- Rutten BP, Schmitz C, Gerlach OH, Oyen HM, de Mesquita EB, Steinbusch HW, Korr H. The aging brain: accumulation of DNA damage or neuron loss? Neurobiol Aging. 2007;28:91–8. doi: 10.1016/j.neurobiolaging.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, Maser RS, Tonon G, Foerster F, Xiong R, Wang YA, Shukla SA, Jaskelioff M, Martin ES, Heffernan TP, Protopopov A, Ivanova E, Mahoney JE, Kost-Alimova M, Perry SR, Bronson R, Liao R, Mulligan R, Shirihai OS, Chin L, DePinho RA. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–65. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai K, Takagi A, Umemura T, Hasegawa R, Kurokawa Y. Changes of 8-hydroxydeoxyguanosine levels in rat organ DNA during the aging process. J Environ Pathol Toxicol Oncol. 1992;11:139–43. [PubMed] [Google Scholar]
- Sanz A, Caro P, Gomez J, Barja G. Testing the vicious cycle theory of mitochondrial ROS production: effects of H2O2 and cumene hydroperoxide treatment on heart mitochondria. J Bioenerg Biomembr. 2006;38:121–7. doi: 10.1007/s10863-006-9011-8. [DOI] [PubMed] [Google Scholar]
- Sarkander HI, Uthoff CG. Comparison of the number of RNA initiation sites in rat brain fractions enriched in neuronal or glial nuclei. FEBS Lett. 1976;72:53–6. doi: 10.1016/0014-5793(76)80897-6. [DOI] [PubMed] [Google Scholar]
- Sarkander HI, Dulce HJ. Studies on the regulation of RNA synthesis in neuronal and glial nuclei isolated from rat brain. Exp Brain Res. 1978;31:317–27. doi: 10.1007/BF00237292. [DOI] [PubMed] [Google Scholar]
- Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, Cooper PK. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol Cell. 2005;20:187–98. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Satoh MS, Huh N, Rajewsky MF, Kuroki T. Enzymatic removal of O6-ethylguanine from mitochondrial DNA in rat tissues exposed to N-ethyl-N-nitrosourea in vivo. J Biol Chem. 1988;263:6854–6. [PubMed] [Google Scholar]
- Satoh MS, Jones CJ, Wood RD, Lindahl T. DNA excision-repair defect of xeroderma pigmentosum prevents removal of a class of oxygen free radical-induced base lesions. Proc Natl Acad Sci U S A. 1993;90:6335–9. doi: 10.1073/pnas.90.13.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saydam N, Kanagaraj R, Dietschy T, Garcia PL, Pena-Diaz J, Shevelev I, Stagljar I, Janscak P. Physical and functional interactions between Werner syndrome helicase and mismatch-repair initiation factors. Nucleic Acids Res. 2007;35:5706–16. doi: 10.1093/nar/gkm500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg GD, Miki T, Yu C-E, Nakura J. The metabolic and molecular basis of inherited disease. McGraw-Hill; New York, N.Y: 2001. [Google Scholar]
- Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277:23028–36. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA, Ogburn CE, McKay J, Jarzebowicz AA, Edland SD, Martin GM. Accelerated loss of telomeric repeats may not explain accelerated replicative decline of Werner syndrome cells. Hum Genet. 1996;97:750–4. doi: 10.1007/BF02346184. [DOI] [PubMed] [Google Scholar]
- Seidle HF, Bieganowski P, Brenner C. Disease-associated mutations inactivate AMP-lysine hydrolase activity of Aprataxin. J Biol Chem. 2005;280:20927–31. doi: 10.1074/jbc.M502889200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J, Ferguson DO, Chen HT, Yang EM, Earle J, Frank K, Whitlow S, Gu Y, Xu Y, Nussenzweig A, Alt FW. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proc Natl Acad Sci U S A. 2001;98:3243–8. doi: 10.1073/pnas.051632098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci U S A. 1997;94:11205–9. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci U S A. 2004;101:7624–9. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow RB, Regan JD, German J, Carrier WL. Evidence that xeroderma pigmentosum cells do not perform the first step in the repair of ultraviolet damage to their DNA. Proc Natl Acad Sci U S A. 1969;64:1035–41. doi: 10.1073/pnas.64.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh AY, Martin LJ. DNA base-excision repair enzyme apurinic/apyrimidinic endonuclease/redox factor-1 is increased and competent in the brain and spinal cord of individuals with amyotrophic lateral sclerosis. Neuromolecular Med. 2002;2:47–60. doi: 10.1007/s12017-002-0038-7. [DOI] [PubMed] [Google Scholar]
- Shao C, Xiong S, Li GM, Gu L, Mao G, Markesbery WR, Lovell MA. Altered 8-oxoguanine glycosylase in mild cognitive impairment and late-stage Alzheimer’s disease brain. Free Radic Biol Med. 2008;45:813–9. doi: 10.1016/j.freeradbiomed.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–37. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Gray MD, Oshima J, Kamath-Loeb AS, Fry M, Loeb LA. Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. J Biol Chem. 1998;273:34139–44. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- Shen S, Cooley DM, Glickman LT, Glickman N, Waters DJ. Reduction in DNA damage in brain and peripheral blood lymphocytes of elderly dogs after treatment with dehydroepiandrosterone (DHEA) Mutat Res. 2001;480–481:153–62. doi: 10.1016/s0027-5107(01)00179-8. [DOI] [PubMed] [Google Scholar]
- Sherer TB, Betarbet R, Stout AK, Lund S, Baptista M, Panov AV, Cookson MR, Greenamyre JT. An in vitro model of Parkinson’s disease: linking mitochondrial impairment to altered alpha-synuclein metabolism and oxidative damage. J Neurosci. 2002;22:7006–15. doi: 10.1523/JNEUROSCI.22-16-07006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Shimura-Miura H, Hattori N, Kang D, Miyako K, Nakabeppu Y, Mizuno Y. Increased 8-oxo-dGTPase in the mitochondria of substantia nigral neurons in Parkinson’s disease. Ann Neurol. 1999;46:920–4. [PubMed] [Google Scholar]
- Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res. 2008;18:64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull ER, Lee Y, Nakane H, Stracker TH, Zhao J, Russell HR, Petrini JH, McKinnon PJ. Differential DNA damage signaling accounts for distinct neural apoptotic responses in ATLD and NBS. Genes Dev. 2009;23:171–80. doi: 10.1101/gad.1746609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijbers AM, van Voorst Vader PC, Snoek JW, Raams A, Jaspers NG, Kleijer WJ. Homozygous R788W point mutation in the XPF gene of a patient with xeroderma pigmentosum and late-onset neurologic disease. J Invest Dermatol. 1998;110:832–6. doi: 10.1046/j.1523-1747.1998.00171.x. [DOI] [PubMed] [Google Scholar]
- Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Singh NP, Ogburn CE, Wolf NS, van Belle G, Martin GM. DNA double-strand breaks in mouse kidney cells with age. Biogerontology. 2001;2:261–70. doi: 10.1023/a:1013262327193. [DOI] [PubMed] [Google Scholar]
- Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–6. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- Spiro C, Pelletier R, Rolfsmeier ML, Dixon MJ, Lahue RS, Gupta G, Park MS, Chen X, Mariappan SV, McMurray CT. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol Cell. 1999;4:1079–85. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Scharer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–20. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevnsner T, Nyaga S, de Souza-Pinto NC, van der Horst GT, Gorgels TG, Hogue BA, Thorslund T, Bohr VA. Mitochondrial repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Oncogene. 2002;21:8675–82. doi: 10.1038/sj.onc.1205994. [DOI] [PubMed] [Google Scholar]
- Stevnsner T, Muftuoglu M, Aamann MD, Bohr VA. The role of Cockayne Syndrome group B (CSB) protein in base excision repair and aging. Mech Ageing Dev. 2008;129:441–8. doi: 10.1016/j.mad.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–87. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103:2729–59. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- Stuart JA, Mayard S, Hashiguchi K, Souza-Pinto NC, Bohr VA. Localization of mitochondrial DNA base excision repair to an inner membrane-associated particulate fraction. Nucleic Acids Res. 2005;33:3722–32. doi: 10.1093/nar/gki683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Ng JM, Masutani C, Maekawa T, Uchida A, van der Spek PJ, Eker AP, Rademakers S, Visser C, Aboussekhra A, Wood RD, Hanaoka F, Bootsma D, Hoeijmakers JH. Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol Cell Biol. 1997;17:6924–31. doi: 10.1128/mcb.17.12.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K. UV-induced ubiquitylation of XPC complex, the UV-DDB-ubiquitin ligase complex, and DNA repair. J Mol Histol. 2006;37:189–202. doi: 10.1007/s10735-006-9044-7. [DOI] [PubMed] [Google Scholar]
- Sugawara M, Wada C, Okawa S, Kobayashi M, Sageshima M, Imota T, Toyoshima I. Purkinje cell loss in the cerebellar flocculus in patients with ataxia with ocular motor apraxia type 1/early-onset ataxia with ocular motor apraxia and hypoalbuminemia. Eur Neurol. 2008;59:18–23. doi: 10.1159/000109256. [DOI] [PubMed] [Google Scholar]
- Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J. 2000;19:1397–404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup JQ. Mechanisms of transcription-coupled DNA repair. Nat Rev Mol Cell Biol. 2002;3:21–9. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- Sweasy JB, Lang T, DiMaio D. Is base excision repair a tumor suppressor mechanism? Cell Cycle. 2006;5:250–9. doi: 10.4161/cc.5.3.2414. [DOI] [PubMed] [Google Scholar]
- Swerdlow RH, Parks JK, Miller SW, Tuttle JB, Trimmer PA, Sheehan JP, Bennett JP, Jr, Davis RE, Parker WD., Jr Origin and functional consequences of the complex I defect in Parkinson’s disease. Ann Neurol. 1996;40:663–71. doi: 10.1002/ana.410400417. [DOI] [PubMed] [Google Scholar]
- Swerdlow RH, Parks JK, Cassarino DS, Maguire DJ, Maguire RS, Bennett JP, Jr, Davis RE, Parker WD., Jr Cybrids in Alzheimer’s disease: a cellular model of the disease? Neurology. 1997;49:918–25. doi: 10.1212/wnl.49.4.918. [DOI] [PubMed] [Google Scholar]
- Szekely AM, Bleichert F, Numann A, Van Komen S, Manasanch E, Ben Nasr A, Canaan A, Weissman SM. Werner protein protects nonproliferating cells from oxidative DNA damage. Mol Cell Biol. 2005;25:10492–506. doi: 10.1128/MCB.25.23.10492-10506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Tada M, Igarashi S, Koyama A, Date H, Yokoseki A, Shiga A, Yoshida Y, Tsuji S, Nishizawa M, Onodera O. Aprataxin, causative gene product for EAOH/AOA1, repairs DNA single-strand breaks with damaged 3′-phosphate and 3′-phosphoglycolate ends. Nucleic Acids Res. 2007;35:3797–809. doi: 10.1093/nar/gkm158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–72. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- Tan Z, Sun N, Schreiber SS. Immunohistochemical localization of redox factor-1 (Ref-1) in Alzheimer’s hippocampus. Neuroreport. 1998;9:2749–52. doi: 10.1097/00001756-199808240-00012. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Engelender S, Igarashi S, Rao RK, Wanner T, Tanzi RE, Sawa A, VLD, Dawson TM, Ross CA. Inducible expression of mutant alpha-synuclein decreases proteasome activity and increases sensitivity to mitochondria-dependent apoptosis. Hum Mol Genet. 2001;10:919–26. doi: 10.1093/hmg/10.9.919. [DOI] [PubMed] [Google Scholar]
- Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, van Heems D, Ito E, Nakamura A, Sonoda E, Takata M, Takeda S, Matsuura S, Komatsu K. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–8. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Groom A, Byrd PJ. Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA Repair (Amst) 2004;3:1219–25. doi: 10.1016/j.dnarep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Thomas B, Beal MF. Parkinson’s disease. Hum Mol Genet. 2007;16(Spec No 2):R183–94. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- Thompson LH, West MG. XRCC1 keeps DNA from getting stranded. Mutat Res. 2000;459:1–18. doi: 10.1016/s0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- Thorslund T, von Kobbe C, Harrigan JA, Indig FE, Christiansen M, Stevnsner T, Bohr VA. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol Cell Biol. 2005;25:7625–36. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorstenson YR, Roxas A, Kroiss R, Jenkins MA, Yu KM, Bachrich T, Muhr D, Wayne TL, Chu G, Davis RW, Wagner TM, Oefner PJ. Contributions of ATM mutations to familial breast and ovarian cancer. Cancer Res. 2003;63:3325–33. [PubMed] [Google Scholar]
- Thyagarajan B, Padua RA, Campbell C. Mammalian mitochondria possess homologous DNA recombination activity. J Biol Chem. 1996;271:27536–43. doi: 10.1074/jbc.271.44.27536. [DOI] [PubMed] [Google Scholar]
- Tijsterman M, Brouwer J. Rad26, the yeast homolog of the cockayne syndrome B gene product, counteracts inhibition of DNA repair due to RNA polymerase II transcription. J Biol Chem. 1999;274:1199–202. doi: 10.1074/jbc.274.3.1199. [DOI] [PubMed] [Google Scholar]
- Tomkinson AE, Bonk RT, Linn S. Mitochondrial endonuclease activities specific for apurinic/apyrimidinic sites in DNA from mouse cells. J Biol Chem. 1988;263:12532–7. [PubMed] [Google Scholar]
- Tornaletti S, Reines D, Hanawalt PC. Structural characterization of RNA polymerase II complexes arrested by a cyclobutane pyrimidine dimer in the transcribed strand of template DNA. J Biol Chem. 1999;274:24124–30. doi: 10.1074/jbc.274.34.24124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Cooper PK. Transcription-coupled repair of oxidative DNA damage in human cells: mechanisms and consequences. Cold Spring Harb Symp Quant Biol. 2000;65:201–15. doi: 10.1101/sqb.2000.65.201. [DOI] [PubMed] [Google Scholar]
- Tuo J, Jaruga P, Rodriguez H, Dizdaroglu M, Bohr VA. The cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J Biol Chem. 2002;277:30832–7. doi: 10.1074/jbc.M204814200. [DOI] [PubMed] [Google Scholar]
- Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 2003;17:668–74. doi: 10.1096/fj.02-0851com. [DOI] [PubMed] [Google Scholar]
- Valerie K, Povirk LF. Regulation and mechanisms of mammalian double-strand break repair. Oncogene. 2003;22:5792–812. doi: 10.1038/sj.onc.1206679. [DOI] [PubMed] [Google Scholar]
- van Brabant AJ, Stan R, Ellis NA. DNA helicases, genomic instability, and human genetic disease. Annu Rev Genomics Hum Genet. 2000;1:409–59. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- van Dellen A, Grote HE, Hannan AJ. Gene-environment interactions, neuronal dysfunction and pathological plasticity in Huntington’s disease. Clin Exp Pharmacol Physiol. 2005;32:1007–19. doi: 10.1111/j.1440-1681.2005.04313.x. [DOI] [PubMed] [Google Scholar]
- van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D, Hoeijmakers JH. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–65. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, Stumm M, Weemaes CM, Gatti RA, Wilson RK, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–76. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci U S A. 2006;103:1864–9. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasko MR, Guo C, Kelley MR. The multifunctional DNA repair/redox enzyme Ape1/Ref-1 promotes survival of neurons after oxidative stress. DNA Repair (Amst) 2005;4:367–79. doi: 10.1016/j.dnarep.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV. The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci U S A. 1990;87:4707–11. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierck J, O’Reilly B, Hossner K, Antonio J, Byrne K, Bucci L, Dodson M. Satellite cell regulation following myotrauma caused by resistance exercise. Cell Biol Int. 2000;24:263–72. doi: 10.1006/cbir.2000.0499. [DOI] [PubMed] [Google Scholar]
- Viswanathan A, You HJ, Doetsch PW. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science. 1999;284:159–62. doi: 10.1126/science.284.5411.159. [DOI] [PubMed] [Google Scholar]
- Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci U S A. 1999;96:10770–5. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kobbe C, Harrigan JA, May A, Opresko PL, Dawut L, Cheng WH, Bohr VA. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol Cell Biol. 2003;23:8601–13. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kobbe C, Harrigan JA, Schreiber V, Stiegler P, Piotrowski J, Dawut L, Bohr VA. Poly(ADP-ribose) polymerase 1 regulates both the exonuclease and helicase activities of the Werner syndrome protein. Nucleic Acids Res. 2004a;32:4003–14. doi: 10.1093/nar/gkh721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Kobbe C, May A, Grandori C, Bohr VA. Werner syndrome cells escape hydrogen peroxide-induced cell proliferation arrest. FASEB J. 2004b;18:1970–2. doi: 10.1096/fj.04-1895fje. [DOI] [PubMed] [Google Scholar]
- Wang H, Powell SN, Iliakis G, Wang Y. ATR affecting cell radiosensitivity is dependent on homologous recombination repair but independent of nonhomologous end joining. Cancer Res. 2004;64:7139–43. doi: 10.1158/0008-5472.CAN-04-1289. [DOI] [PubMed] [Google Scholar]
- Wang J, Xiong S, Xie C, Markesbery WR, Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer’s disease. J Neurochem. 2005;93:953–62. doi: 10.1111/j.1471-4159.2005.03053.x. [DOI] [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- Warita H, Hayashi T, Murakami T, Manabe Y, Abe K. Oxidative damage to mitochondrial DNA in spinal motoneurons of transgenic ALS mice. Brain Res Mol Brain Res. 2001;89:147–52. doi: 10.1016/s0169-328x(01)00029-8. [DOI] [PubMed] [Google Scholar]
- Weidenheim KM, Dickson DW, Rapin I. Neuropathology of Cockayne syndrome: Evidence for impaired development, premature aging, and neurodegeneration. Mech Ageing Dev. 2009 doi: 10.1016/j.mad.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Weissman L, de Souza-Pinto NC, Stevnsner T, Bohr VA. DNA repair, mitochondria, and neurodegeneration. Neuroscience. 2007a;145:1318–29. doi: 10.1016/j.neuroscience.2006.08.061. [DOI] [PubMed] [Google Scholar]
- Weissman L, Jo DG, Sorensen MM, de Souza-Pinto NC, Markesbery WR, Mattson MP, Bohr VA. Defective DNA base excision repair in brain from individuals with Alzheimer’s disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007b;35:5545–55. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman L, de Souza-Pinto NC, Mattson MP, Bohr VA. DNA base excision repair activities in mouse models of Alzheimer’s disease. Neurobiol Aging. 2009;30:2080–1. doi: 10.1016/j.neurobiolaging.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–45. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–17. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- Wiedemann FR, Manfredi G, Mawrin C, Beal MF, Schon EA. Mitochondrial DNA and respiratory chain function in spinal cords of ALS patients. J Neurochem. 2002;80:616–25. doi: 10.1046/j.0022-3042.2001.00731.x. [DOI] [PubMed] [Google Scholar]
- Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–20. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- Wilson DM., 3rd Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J Mol Biol. 2003;330:1027–37. doi: 10.1016/s0022-2836(03)00712-5. [DOI] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–59. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Mattson MP. Neurodegeneration: nicked to death. Curr Biol. 2007;17:R55–8. doi: 10.1016/j.cub.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Wilson SH. Mammalian base excision repair and DNA polymerase beta. Mutat Res. 1998;407:203–15. doi: 10.1016/s0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- Winkler GS, Araujo SJ, Fiedler U, Vermeulen W, Coin F, Egly JM, Hoeijmakers JH, Wood RD, Timmers HT, Weeda G. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem. 2000;275:4258–66. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
- Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., 3rd Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–13. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- Wu X, Li J, Li X, Hsieh CL, Burgers PM, Lieber MR. Processing of branched DNA intermediates by a complex of human FEN-1 and PCNA. Nucleic Acids Res. 1996;24:2036–43. doi: 10.1093/nar/24.11.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman C, Ristic D, Kanaar R. Homologous recombination-mediated double-strand break repair. DNA Repair (Amst) 2004;3:827–33. doi: 10.1016/j.dnarep.2004.03.037. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–91. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Herzig M, Rotrekl V, Walter CA. Base excision repair, aging and health span. Mech Ageing Dev. 2008;129:366–82. doi: 10.1016/j.mad.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–9. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Nakamura Y, Kobayashi N, Iwamoto T, Yoshioka A, Kuniyasu H, Kishimoto T, Mori T. Neurons and astrocytes exhibit lower activities of global genome nucleotide excision repair than do fibroblasts. DNA Repair (Amst) 2007;6:649–57. doi: 10.1016/j.dnarep.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Yan Z. Skeletal muscle adaptation and cell cycle regulation. Exerc Sport Sci Rev. 2000;28:24–6. [PubMed] [Google Scholar]
- Yang JL, Tadokoro T, Keijzers G, Mattson MP, Bohr VA. Neurons efficiently repair glutamate-induced oxidative DNA damage by a process involving CREB-mediated up-regulation of apurinic endonuclease 1. J Biol Chem. 2010;285:28191–9. doi: 10.1074/jbc.M109.082883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, Weissman L, Bohr VA, Mattson MP. Mitochondrial DNA damage and repair in neurodegenerative disorders. DNA Repair (Amst) 2008;7:1110–20. doi: 10.1016/j.dnarep.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Burgin AB, Jr, Huizenga BN, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci U S A. 1996;93:11534–9. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkovic K. 4-hydroxynonenal and neurodegenerative diseases. Mol Aspects Med. 2003;24:293–303. doi: 10.1016/s0098-2997(03)00024-4. [DOI] [PubMed] [Google Scholar]
- Zhang J, Perry G, Smith MA, Robertson D, Olson SJ, Graham DG, Montine TJ. Parkinson’s disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am J Pathol. 1999;154:1423–9. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Yu JT, Wang P, Chen W, Wu ZC, Jiang H, Tan L. Mitochondrial transcription factor A (TFAM) polymorphisms and risk of late-onset Alzheimer’s disease in Han Chinese. Brain Res. 2011;1368:355–60. doi: 10.1016/j.brainres.2010.10.074. [DOI] [PubMed] [Google Scholar]
- Zhou T, Lee JW, Tatavarthi H, Lupski JR, Valerie K, Povirk LF. Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1) Nucleic Acids Res. 2005;33:289–97. doi: 10.1093/nar/gki170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Akopiants K, Mohapatra S, Lin PS, Valerie K, Ramsden DA, Lees-Miller SP, Povirk LF. Tyrosyl-DNA phosphodiesterase and the repair of 3′-phosphoglycolate-terminated DNA double-strand breaks. DNA Repair (Amst) 2009;8:901–11. doi: 10.1016/j.dnarep.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Doetsch PW. Transcription bypass or blockage at single-strand breaks on the DNA template strand: effect of different 3′ and 5′ flanking groups on the T7 RNA polymerase elongation complex. Biochemistry. 1994;33:14926–34. doi: 10.1021/bi00253a032. [DOI] [PubMed] [Google Scholar]
- Ziegler M, Oei SL. A cellular survival switch: poly(ADP-ribosyl)ation stimulates DNA repair and silences transcription. Bioessays. 2001;23:543–8. doi: 10.1002/bies.1074. [DOI] [PubMed] [Google Scholar]
- Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson Iii DM, Stevnsner T, Bohr VA. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010 doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]