This work reports the identification of a new regulator of axillary meristem formation in maize, showing that BARREN STALK FASTIGIATE1 likely functions as a boundary determinant for axillary meristem primordia. In addition, it reveals that BARREN STALK FASTIGIATE1 is involved in the regulation of Barren stalk1, a major player in the initiation of axillary meristems.
Abstract
Ears are the seed-bearing inflorescences of maize (Zea mays) plants and represent a crucial component of maize yield. The first step in the formation of ears is the initiation of axillary meristems in the axils of developing leaves. In the classic maize mutant barren stalk fastigiate1 (baf1), first discovered in the 1950s, ears either do not form or, if they do, are partially fused to the main stalk. We positionally cloned Baf1 and found that it encodes a transcriptional regulator containing an AT-hook DNA binding motif. Single coorthologs of Baf1 are found in syntenic regions of brachypodium (Brachypodium distachyon), rice (Oryza sativa), and sorghum (Sorghum bicolor), suggesting that the gene is likely present in all cereal species. Protein–protein interaction assays suggest that BAF1 is capable of forming homodimers and heterodimers with other members of the AT-hook family. Another transcriptional regulator required for ear initiation is the basic helix-loop-helix protein BARREN STALK1 (BA1). Genetic and expression analyses suggest that Baf1 is required to reach a threshold level of Ba1 expression for the initiation of maize ears. We propose that Baf1 functions in the demarcation of a boundary region essential for the specification of a stem cell niche.
INTRODUCTION
The developmental plasticity of plants, that is, the ability of plants to modify their morphology according to different environmental stimuli, relies on the production and activity of axillary meristems. Axillary meristems form in the axils of leaf or bract primordia on the adaxial (upper) side of the region where the primordium and the main stem join (Bennett and Leyser, 2006). Formed postembryonically during both vegetative and reproductive development, axillary meristems are eventually responsible for the establishment of secondary axes of growth, such as lateral branches and flowers. The production and activity of axillary meristems determine the overall elaboration of different plant architectures and, during reproductive development, of different inflorescence morphologies in all flowering plants.
In the case of maize (Zea mays), two distinct inflorescences, the tassel and the ear, are borne on separate parts of the plant (Kiesselbach, 1949). Whereas the apical male inflorescence, the tassel (Figure 1A), derives directly from the shoot apical meristem formed during embryogenesis, the female inflorescences, the ears (Figure 1D), derive from axillary meristems that are formed soon after germination in the axils of leaves (Figures 1H and 1I). During vegetative development, several basal axillary meristems may originate lateral branches, also known as tillers. Other axillary meristems form buds that remain quiescent until the plant switches to reproductive development and are enclosed in the prophyll (Kiesselbach, 1949). During reproductive development, the highest axillary buds transition to ear formation. Therefore, a prerequisite for the formation of maize ears is the initiation of a series of axillary meristems in the axils of leaves that occurs very early in development. In both ear and tassel development, the primary apical meristems (also called inflorescence meristems) produce additional axillary meristems that are eventually responsible for the formation of all reproductive structures. In maize, these axillary meristems are named for the structures that they form: branch meristems form tassel branches, spikelet-pair meristems form spikelet pairs, spikelet meristems form spikelets, and floral meristems form florets (McSteen et al., 2000). Spikelets are grass-specific structures bearing florets, and in maize they are paired, with one spikelet being pedicellate and the other sessile.
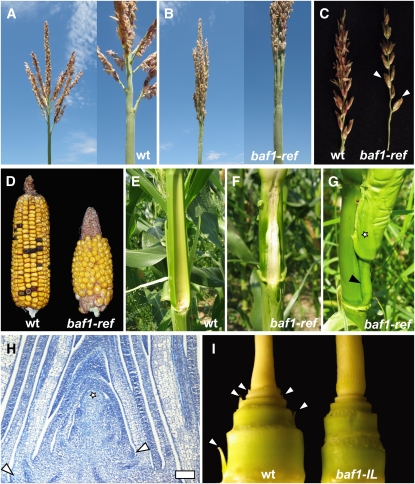
Figure 1.
The Phenotype of the baf1 Mutant.
(A) Wild-type (wt) tassel.
(B) baf1-ref mutant tassel.
(C) Primary branches from wild-type and baf1-ref tassels. Arrowheads point to single spikelets in the baf1-ref mutant.
(D) Wild-type and baf1-ref mutant ears.
(E) and (F) Internodes subtending mature ears form an indentation in wild-type plants (E) that is absent in baf1-ref mutants (F) (ears have been removed).
(G) In baf1-ref mutants, the ear is borne on a slightly elongated internode (arrowhead). The star marks the prophyll.
(H) Longitudinal section stained with Toluidine Blue of a wild-type seedling, showing the shoot apical meristem (star) and axillary meristems forming at the axils of developing leaves in an alternate arrangement (arrowheads). Bar = 100 μm.
(I) Seedlings of both wild-type and baf1-ref mutant plants were stripped of most leaves, revealing axillary buds (arrowheads) in normal but not in baf1-IL mutant plants.
Axillary meristem initiation involves the correct positioning of the meristem anlagen, followed by the delineation of the boundaries for proper primordium separation, and then the establishment of the meristem itself. In Arabidopsis thaliana, the transport of the hormone auxin in and out of the emerging primordium by the activity of polar auxin transporters, an essential step in primordium positioning, is followed by upregulation of boundary genes and the concomitant expression of meristem identity and maintenance genes (Heisler et al., 2005). The establishment of primordium boundaries and the formation of new meristems are therefore deeply interconnected processes, and the boundary region is thought to be important for communication between the axillary meristem and the leaf primordium. Genes affecting boundary formation are generally involved in promoting meristem initiation as well. A number of transcription factors have been described as having a role in axillary meristem formation and lateral organ separation in Arabidopsis, such as CUP SHAPED COTYLEDONS1,2,3 (CUC1,2,3) (Hibara et al., 2006), LATERAL SUPPRESSOR (LAS) (Greb et al., 2003), LATERAL ORGAN BOUNDARY (Shuai et al., 2002), LATERAL ORGAN FUSION1,2 (LOF1,2) (Lee et al., 2009), and REGULATOR OF AXILLARY MERISTEMS1 (RAX1) (Keller et al., 2006), all showing a boundary-specific mRNA expression pattern. Functional orthologs of some of these genes have been also found in both maize and rice (Oryza sativa), and the role of auxin transport seems conserved (Li et al., 2003; McSteen et al., 2007; Gallavotti et al., 2008b). In Arabidopsis, mutations affecting axillary meristem formation during vegetative development do not necessarily affect reproductive development. In las mutants, for example, despite defects in the initiation of vegetative axillary meristems, floral meristems form normally (Greb et al., 2003).
In maize, many aspects of the regulation of axillary meristem initiation are common to both vegetative and reproductive development, and mutations that affect axillary meristem development usually impair the formation of both vegetative and reproductive axillary structures (tillers, spikelets, and florets). The barren stalk1 (ba1), Barren inflorescence1 (Bif1), bif2, and sparse inflorescence1 (spi1) mutants of maize all have inflorescences devoid or depleted of branches, spikelets, and flowers. With the exception of the semidominant Bif1, these mutants are also impaired in tiller formation, as seen in double mutant combinations with teosinte branched1 (tb1), a highly tillered mutant (Doebley et al., 1997; Ritter et al., 2002; McSteen et al., 2007; Barazesh and McSteen, 2008; Gallavotti et al., 2008a). One of the most striking phenotypes is observed in the ba1 mutants, in which plants with the most severe alleles completely lack vegetative and reproductive branches (Ritter et al., 2002). Ba1 encodes a basic helix-loop-helix (bHLH) transcription factor (Gallavotti et al., 2004) that is phosphorylated by the Ser-Thr kinase BIF2 (McSteen et al., 2007; Skirpan et al., 2008), which is known to be involved in polar auxin transport (Skirpan et al., 2009). Whereas the identity of the Bif1 gene is still unknown, Spi1 was shown to encode an enzyme involved in auxin biosynthesis (Gallavotti et al., 2008a). Ba1 and its rice ortholog LAX1 are also expressed in a specific boundary region adaxial to the initiating axillary meristems (Komatsu et al., 2003; Gallavotti et al., 2004), they are both required for the formation of these meristems, and the LAX1 protein was also reported to move from the site of expression into the newly forming meristem (Oikawa and Kyozuka, 2009). The role of Ba1 in Arabidopsis has yet to be determined, although a putative ortholog has been recently identified by phylogenetic analysis (Woods et al., 2011).
The mechanisms regulating axillary meristem initiation in vascular plants are unclear. Traditionally, it was considered that axillary meristems develop either from cells that originate in the shoot apical meristem (detached-meristem theory) or from cells that acquire a meristematic identity (de novo theory). However, analyses in Arabidopsis and tomato (Solanum lycopersicum) suggest that both forms of axillary meristems exist in the same plant and might reflect opposite extremes of the same process regulated by the same genetic mechanisms (Long and Barton, 2000; Greb et al., 2003; Leyser, 2003; Bennett and Leyser, 2006). The number of taxa possessing both forms of axillary meristem initiation has yet to be investigated, but in rice, a careful expression analysis of a series of genes regulating axillary meristem formation supports the de novo origin of axillary meristems (Oikawa and Kyozuka, 2009). Identifying genes that regulate axillary meristem initiation and are restricted to grasses will provide support to the hypothesis that monocots (or at least grasses) and eudicots have lineage-specific mechanisms (Oikawa and Kyozuka, 2009).
Here, we report the identification of a new regulator of axillary meristem formation in maize, the Barren stalk fastigiate1 (Baf1) gene. Baf1 encodes a putative transcriptional regulator containing an AT-hook DNA binding motif and a conserved domain of unknown function, called the Plant and Prokaryote Conserved (PPC) domain. Baf1 coorthologs are present in different grasses, and four potential coorthologs were identified in Arabidopsis, albeit without strong support. We show that BAF1 can form homo- and heterodimers with other putative AT-hook DNA binding proteins, revealing a previously unrecognized aspect of this protein family. Mutations in Baf1 give rise to plants that either lack ears or produce fusion defects, suggesting that Baf1 is involved in demarcating the boundary region where the axillary meristem is forming and plays a role in axillary meristem initiation. We show by genetic and expression analysis that Baf1 functions upstream of Ba1 in this pathway.
RESULTS
The baf1 Mutant Has Defects in Tassel and Ear Development
baf1-ref is a recessive mutant (Coe and Beckett, 1987) originally identified by Charles R. Burnham in the 1950s. baf1-ref mutant plants are characterized by a more compact tassel (Figures 1A and 1B). Normal tassels carry several basal long branches, derived from the activity of branch meristems, emerging from the central spike (Figure 1A). In baf1 mutant tassels, the primary branches are borne in a more upright orientation than in normal tassels, hence the name fastigiate, indicating a cluster of erect branches. A slight increase (of 3.73 branches on average) in the number of tassel branches was also observed in the baf1-ref mutant compared with normal plants when introgressed in the maize inbred line B73 (Table 1). Furthermore, these branches appeared to be slightly shorter compared with normal (Table 1). The barren stalk denomination refers to the tendency of baf1 mutant plants to be earless. In baf1-ref mutants, ~55% of the plants failed to form ears (n = 69).
Table 1.
Tassel Measurements
| Phenotype | n | Tassel Length (cm) | No. of Primary Branches | Branch Length (cm) | % Single Spikelets on Branches (n > 600) | Branch Radial Size (mm; n > 50) |
| Wild type | 27 | 27.94 (2.98) | 10.67 (2.47) | 15.65 (3.12) | 1.9% | 1.96 (0.36) |
| baf1-ref | 10 | 23.85 (4.97)* | 14.40 (3.47)*** | 13.55 (3.85)**** | 14.3%** | 1.60 (0.46)** |
| Wild type | 10 | 32.25 (1.78) | 6.10 (2.13) | 16.79 (2.29) | 5.1% | n.d. |
| baf1-IL | 11 | 28.32 (1.90)** | 4.64 (1.03) | 11.97 (4.77)** | 37%** | n.d. |
Tassel measurements on the wild type and baf1-ref mutants in the B73 background (BC2) and the wild-type and baf1-IL in the A619 background (BC1). Average numbers (sd in parentheses). All compared values are in the same genetic background. *t test, P = 0.041; **t test, P ≤ 0.0001; ***t test, P = 0.0008; ****t test, P = 0.0009. n.d., not determined.
A leaf, its axillary meristem, and the associated internode together make up a developmental unit referred to as a phytomer (Galinat, 1959). In wild-type plants, the internode of an ear-bearing phytomer typically develops an indentation to accommodate the mature inflorescence (Figure 1E). When ears form in baf1-ref mutants, the proximal region of the ear branch develops fused to the internode, and no indentation is observed (Figure 1F). The fusion occurs mainly between the stem internode and the first internode of the lateral branch bearing the ear that is sometimes unusually elongated (Figure 1G; see Supplemental Figure1 online). Occasionally, the fusion also includes the basal part of the prophyll. Often a notch is formed in the stem, likely caused by the mechanical stress posed by the elongating internode fused to the axillary bud (Figure 2B). When ears form, they are generally shorter than normal and show unorganized rows of kernels (Figure 1D).
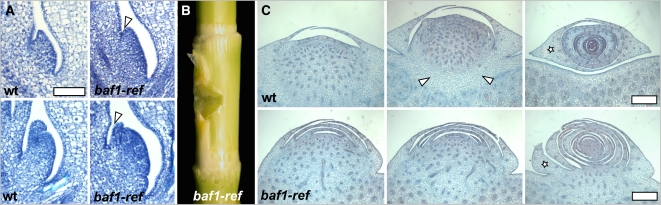
Figure 2.
The Fusion Defects in the Development of baf1 Mutant Ears.
(A) Toluidine Blue staining of sections of young axillary meristems during vegetative development in wild-type (wt) and baf1-ref plants. Arrowheads point to the groove between the axillary meristem and the upper leaves. Bar = 50 μm.
(B) In baf1-ref internodes, axillary meristems fused to the culm cause the formation of a notch.
(C) Series of cross sections of developing ears, stained with Safranin O-Alcian Blue, reveal the lack of a separation layer in baf1-ref mutants between the stem and the developing axillary inflorescence. Arrowheads demarcate the separation zone normally observed in wild-type plants. The star marks the prophyll.
Bars = 500 μm.
baf1-ref mutants are also defective in the formation of paired spikelets. Spikelets are formed by spikelet-pair meristems, very short branches that produce one terminal and one lateral spikelet (McSteen et al., 2000). In the baf1 mutant, there is a tendency to form more single spikelets in tassel branches (Table 1, Figure 1C) but not on the central spike of the tassel. We found an increase of more than sevenfold in the percentage of single spikelets borne on tassel branches (Table 1). This may result from the smaller size of branch meristems and the consequent smaller than normal group of meristematic cells available for the formation of spikelet-pair meristems, which initiate on the flanks of branch meristems. We therefore measured the radial size of the base of tassel branches in both mutant and wild-type plants and found a significant decrease in baf1-ref mutants (Table 1).
An additional mutant allele of Baf1 was identified segregating in an M2 family generated by ethyl methanesulfonate (EMS) mutagenesis in the inbred line A619 (baf1-IL). The tassel defects observed in baf1-IL mutant plants were similar to those described above for baf1-ref mutants, although there was no increase in the number of primary tassel branches (Table 1). baf1-IL plants showed a completely earless phenotype and failed to complement the baf1-ref mutant. In baf1-IL seedlings, the axillary buds that would normally give rise to ears fail to initiate (Figure 1I).
To confirm that the baf1 mutant is defective in the initiation of axillary meristems, we crossed both baf1 alleles to the tb1 mutant of maize, defective in the repression of axillary meristem outgrowth (Doebley et al., 1997). tb1 mutant plants are characterized by bushy growth, with a large number of tillers. In addition, ears are replaced by masculinized inflorescences borne on lateral branches with elongated internodes (Doebley et al., 1997). In wild-type maize, ears normally form at the apex of a lateral branch atop a series of suppressed internodes (Kiesselbach, 1949). tb1 baf1-IL double mutant plants had significantly fewer tillers than did tb1 mutants (Table 2), indicating that the baf1 mutation affects axillary meristem initiation, but when an axillary meristem is formed, baf1 does not seem to influence its subsequent outgrowth. The analysis of these crosses also showed that the earless phenotype, rather than depending on a particular mutant allele, is influenced by the genetic background because baf1-IL mutant plants segregating in this cross were not always earless (Table 2). Furthermore, a closer look at the base of the lateral inflorescences in these double mutants showed complete fusion of the first elongated internode of the branch to that of the main stem (see Supplemental Figure 1 online).
Table 2.
The tb1 baf1 Double Mutant
| Genotype | n | No. of Tillers | % Earless |
| tb1 | 27 | 7.30 (2.41) | 0% |
| baf1-IL | 20 | 0.65 (0.81)* | 15% |
| tb1 baf1-IL | 7 | 2.14 (1.95)* | 42% |
Quantification of the average number of tillers in tb1 baf1 double mutants (sd in parentheses). All values were compared to the tb1 single mutant. *t test, P < 0.0001.
To understand better the organ fusion phenotype of the developing baf1-ref ears, we analyzed sections of wild-type and mutant ears (Figure 2). Longitudinal sections of axillary meristems during vegetative development reveal subtle differences, such as a less pronounced groove between the upper leaf and the prophyll in baf1 mutants (Figure 2A, arrowheads). Transverse sections of wild-type ears have an obvious cellular separation layer between the stem and the base of the ear prophyll (Figures 2B and 2C, arrowheads). This layer is eventually resolved into two distinct structures, the axillary bud and the indentation of the internode. In baf1-ref plants, this separation layer is clearly missing throughout the sections and vascular strands are also formed in this area. These defects eventually result in the organ fusion observed in mature ears (Figure 1F).
We also sectioned the base of the tassel branches prior to anthesis (see Supplemental Figure 2 online). The erect tassel branch phenotype has also been observed in other mutants, such as the ramosa2 (ra2) (Bortiri et al., 2006) and the ramosa1 enhancer locus2 (rel2) (Gallavotti et al., 2010) mutants, both involved in regulation of spikelet-pair meristem determinacy. The axils of the tassel branches bear a specific structure, the pulvinus (Galinat, 1959), whose cells are responsible for the opening of the tassel branch angle at anthesis. We sectioned and stained the area at the base of the tassel branches and found that baf1-ref mutant branches are significantly thinner, and their pulvinal cells, stained blue, are formed but reduced in numbers (see Supplemental Figure 2 online). These defects appear different from what was previously reported for both ra2 and rel2 (Gallavotti et al., 2010), in which cellular and structural organization is affected.
Baf1 Encodes a Putative AT-Hook DNA Binding Protein
The Baf1 locus was originally mapped to the short arm of chromosome 9 (Coe and Beckett, 1987; Neuffer et al., 1997). To clone Baf1, we crossed the baf1-ref allele, in an unknown genetic background, into the inbred line B73 to generate an F2 mapping population. We first identified two simple sequence repeat markers (umc1893 and umc1586) in proximal and distal linkage to the Baf1 locus. Subsequently, a small population of 141 homozygous mutant plants was used to narrow the region containing the Baf1 locus to between newly developed molecular markers MAGIv4_59303 and MAGIv4_46990 (see Methods) (Figure 3A). Syntenic regions of both sorghum (Sorghum bicolor) and rice were analyzed for predicted genes in the corresponding interval, and a few potential candidate genes were sequenced in the baf1-ref mutant (Figure 3A). One of these gene models, GRMZM2G072274, carried a 151-bp deletion starting at position +654 in the predicted coding sequence and a small insertion (GGTGGA), presumably a footprint of a transposition event, that caused a frame shift of the corresponding protein sequence. We therefore sequenced the baf1-IL mutant at the same locus and identified at position +121 a single nucleotide change that created a premature stop codon (CAG changed into TAG). The occurrence in both baf1-ref and baf1-IL mutants of two independent mutations in the coding sequence of the same gene confirmed that GRMZM2G072274 corresponded to the baf1 gene.
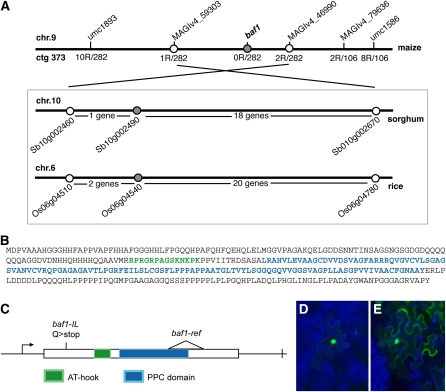
Figure 3.
Baf1 Encodes an AT-Hook Transcriptional Regulator.
(A) Schematic representation of the map-based cloning of the Baf1 locus. Boxed are the syntenic regions analyzed for candidate genes in both sorghum and rice.
(B) Amino acid sequence of the BAF1 protein. In green is the amino acid sequence of the AT-hook DNA binding motif, whereas in blue is the PPC domain (DUF296).
(C) Schematic representation of the Baf1 gene and of the lesions identified in the baf1-ref and baf1-IL mutants. The white triangle represents a deletion.
(D) and (E) Transient expression of the BAF1-YFP fusion protein (D) and YFP (E) in tobacco leaves.
Baf1 is an intronless gene (Figure 3C) with a coding sequence of 1026 bp, highly enriched in GC (75%). The predicted protein sequence of 341 amino acids carries an AT-hook DNA binding motif at the N terminus, centered around a Gly-Arg-Pro tripeptide (Aravind and Landsman, 1998), and a PPC domain, also identified as Domain of Unknown Function 296 (DUF296), in the middle portion of the protein sequence (Figures 3B and 3C). The combination of a single AT-hook motif and the PPC domain is a feature observed only in plants (Fujimoto et al., 2004). AT-hook motif-containing proteins are generally considered a class of transcriptional regulators that bind to AT-rich regions and make chromatin more accessible to other transcription factors, possibly through changes in the structure of DNA (Aravind and Landsman, 1998). An obvious prerequisite for transcriptional regulator activity is the nuclear localization of the encoded protein. To test for nuclear localization, we transiently expressed a BAF1-yellow fluorescent protein (YFP) fusion using a cauliflower mosaic virus 35S promoter in tobacco (Nicotiana tabacum) cells, and, as expected, we observed a specific nuclear localization for the BAF1-YFP fusion protein but not for YFP alone (Figures 3D and 3E), consistent with BAF1’s predicted role as a transcriptional regulator.
Baf1 Belongs to a Monocot-Specific Clade
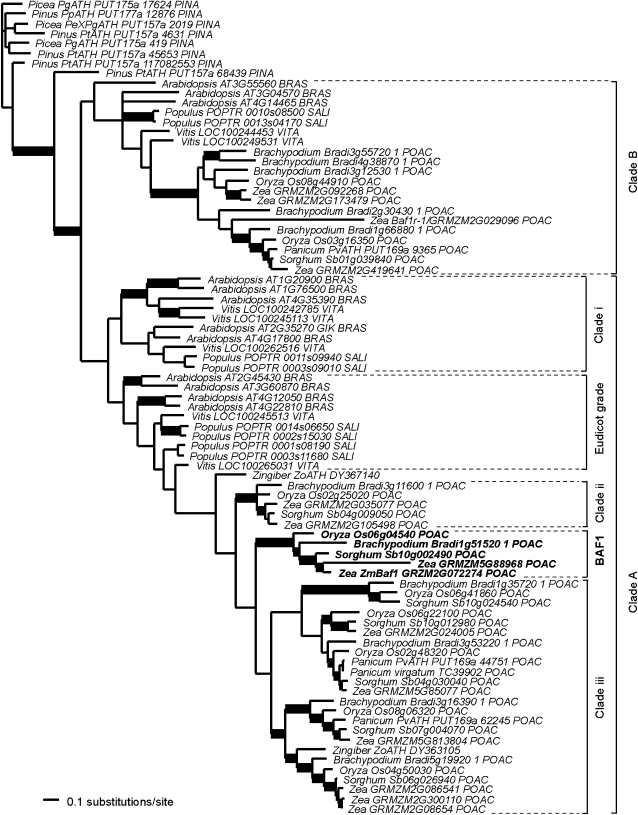
AT-hook motif-containing proteins are found in all sampled land plants, with several sequences each in Physcomitrella patens (a moss) and Selaginella moellendorffii (a lycophyte), as well as multiple gymnosperm and angiosperm sequences. Using only the conserved domain of the protein, and rooting the trees at P. patens, all the moss sequences formed a clade, as did the S. moellendorffii sequences. All remaining seed plant sequences were a single clade (see Supplemental Figure 3 online). The phylogeny of the seed plant sequences (Figure 4) shows several well-supported clades. Gymnosperms form a grade with the conifer sequence Pinus taeda Pt ATH PUT-157a 68439, estimated as the well-supported (1.00 posterior probability) sister to the angiosperm clade. The flowering plant clades A and B include both monocot and eudicot sequences, indicating that the AT-hook proteins had diversified before the common ancestor of the extant angiosperms. BAF1 falls in a strongly supported (1.00 posterior probability) clade (BAF1) with another maize locus (Zm GRMZM5G88968), plus homologs from sorghum, Brachypodium, and rice. In the Bayesian consensus tree, the eudicot grade, plus the monocot BAF1 clade, clades i, ii, and iii, and the ginger sequence (Zingiber officinale DY367140) comprise clade A, but without strong support (0.63 posterior probability).
Figure 4.
Phylogenetic Analysis.
Bayesian consensus phylogram of 87 BAF1-like AT-hook DNA binding proteins from phylogenetic analyses comprising 50 million generations and the General Time Reversible model of evolution with invariant sites and gamma distributed rates (GTR+I+G) partitioned according to codon position. Bold branches supported by ≥0.95 posterior probability. Gymnosperms: PINA, Pinaceae; Eudicots: BRAS, Brassicaceae; SALI, Salicaceae; VITA, Vitaceae; Monocots: POAC, Poaceae; ZING, Zingiberaceae.
The AT-hook genes differ considerably in GC content. The eudicot sequences are 47.9 to 61.2% GC, whereas the monocot sequences (mostly grasses) are 68.4 to 76.7% GC. In the conserved AT-hook motif, the bias is even more striking, with the eudicots being 48.1 to 62.7% GC, whereas the monocots are 69.9 to 78.8% GC. Baf1 itself is near the higher end of the monocot range, with 75.7% GC over the entire gene and 78.2% in the AT-hook motif. The effect of this bias on gene function is unknown, but it does affect the ability to recover an accurate phylogeny. In analyses of the AT-hook motif alone, the eudicot and grass sequences formed separate clades, corresponding to low and high GC content respectively. Broader taxonomic sampling, in particular the addition of more nongrass monocots, will likely make it easier to recover the phylogenetic signal.
The Baf1 (Zm Baf1A) genomic region shows good synteny with rice, sorghum, and Brachypodium (see Supplemental Figure 4 online), consistent with the results from the phylogenetic analysis. Immediately downstream of Baf1 is an endopeptidase (A) followed by a phosphopyruvate hydratase (B), and immediately upstream of Baf1 is a microtubule motor activity protein (C) whose positions are conserved on rice chromosome 6, Brachypodium chromosome 1, sorghum chromosome 10, and maize chromosome 9, but are lacking from maize chromosome 6 (see Supplemental Figure 4 online). It therefore seems that GRMZM5G88968 (Zm Baf1B), the duplicate of Zm Baf1A, is the only remaining open reading frame from the corresponding region of chromosome 6 following the maize-tripsacum duplication event (Gaut and Doebley, 1997; Bomblies and Doebley, 2005).
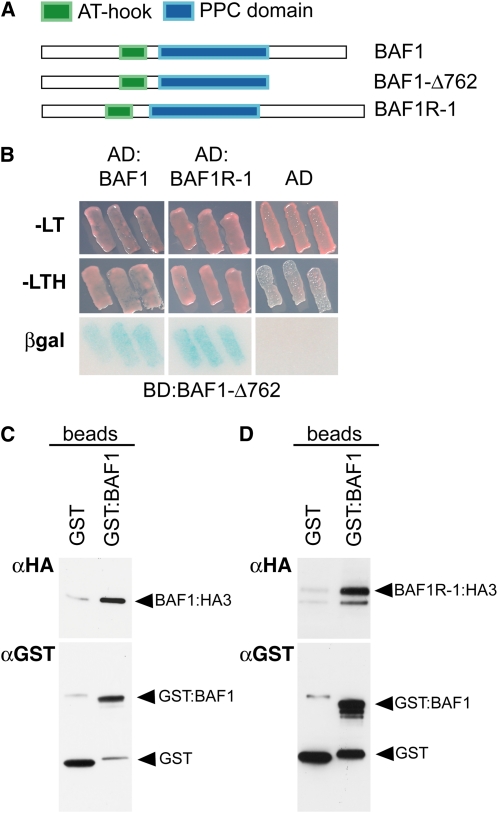
BAF1 Can Form Homodimers and Can Heterodimerize with Different AT-Hook Motif-Containing Proteins
To identify potential interacting partners of the BAF1 protein and understand its mode of action further, we used a yeast two-hybrid system. We screened an immature ear (2 mm) cDNA library in a pAD-GAL4 vector using a BD:Baf1 fusion construct truncated at the C terminus but containing the full AT-hook and PPC domains (Baf1Δ762). This was necessary to prevent the self-activation of the reporter genes observed with the full-length Baf1 construct, attributable to the C terminus of the BAF1 protein. We assayed the interactions using two reporter gene systems, growth on His-lacking media and β-galactosidase activity. This screen identified a short list of potential interactors (Table 3). Notably, two of the recovered clones encoded other members of the AT-hook protein family. Both clones encoded truncated fusion proteins that were missing the N terminus. We focused on one of these (GRMZM2G029096), also expressed in developing inflorescences, and cloned the corresponding full-length coding sequence; we named it Baf1r-1 (Baf1related-1). Baf1r-1 is also an intronless gene and encodes a protein of 351 amino acids with a domain organization similar to that of Baf1 (Figure 5A), although the overall sequence identity of BAF1 and BAF1R-1 proteins is only 52%. This clone was used to confirm the interaction with BAF1 in yeast (Figure 5B). We also confirmed the interaction of BAF1 and BAF1R-1 with an in vitro pull-down assay, using a glutathione S-transferase (GST)-BAF1 fusion protein bound to glutathione-coupled beads (Figure 5C). Furthermore, we detected interaction of BAF1 with itself in both yeast (BD:BAF1Δ762/AD:BAF1; Figure 5B) and in vitro pull-down assays (Figure 5D). These data suggest that AT-hook and PPC domain-containing proteins can form homo- and heterodimers in vitro and in vivo, a previously unrecognized property of this family of transcriptional regulators.
Table 3.
Yeast Two-Hybrid Results
| Clone No. | n | Homology | Gene Model |
| 1 | 4 | AT-hook DNA binding transcriptional regulator | GRMZM2G029096 |
| 2 | 1 | AT-hook DNA binding transcriptional regulator | GRMZM2G024005 |
| 3 | 1 | Zinc-finger transcription factor | GRMZM2G055116 |
| 4 | 1 | Hypothetical protein | GRMZM2G097249 |
List of BAF1Δ762 interactors identified in a yeast two-hybrid library screen.
Figure 5.
BAF1 Proteins Form Homo- and Heterodimers.
(A) Schematic representation of the constructs used in yeast two-hybrid and in vitro pull-down assays.
(B) Targeted yeast two-hybrid assay. Yeast transformants were grown on selective medium for both bait (BD:BAF1-Δ762) and prey (AD:BAF1, AD:BAF1R-1, and AD) constructs (-LT) and on medium lacking His (-LTH) for detecting interactions. Interactions are also verified by β-galactosidase activity (βgal). AD, GAL4 activation domain; BD, GAL4 DNA binding domain.
(C) and (D) In vitro pull-down assays. BAF1 (C) and BAF1R-1 (D) fused to a 3XHA tag were obtained by in vitro transcription/translation and were incubated with GST-only (negative control) or GST-BAF1 bound beads. Anti-GST antibody shows even loading of GST-only and GST-BAF1 proteins, whereas anti-HA antibody shows preferential retention of the input only on GST-BAF1 beads.
Baf1 Expression Is Detected in a Narrow Pattern in Immature Inflorescences
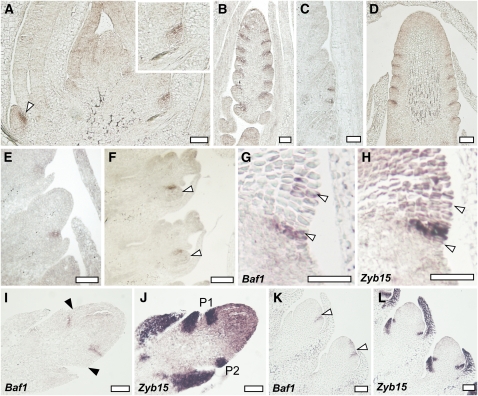
We investigated the domain of Baf1 expression using mRNA in situ hybridizations during maize vegetative and reproductive development. At the seedling stage, Baf1 expression was observed in a narrow domain adaxial to the developing axillary meristems (Figure 6A, arrowhead). After the switch to reproductive development, we observed a similar narrow domain of expression in both immature tassels and ears. The expression domain seemed to mark a boundary between the newly forming spikelet-pair meristems and the inflorescence meristems (Figures 6B and 6D) and in spikelet-pair meristems borne on tassel branch meristems (Figure 6C). In later stages, a similar expression pattern was observed in developing spikelets in positions where new floral meristems were forming (Figure 6E), and it remained adaxial to the lower floral meristems during floral organ development (Figure 6F, arrowheads).
Figure 6.
The Expression Pattern of Baf1.
(A) to (F) In situ hybridizations of Baf1 during vegetative and reproductive development in the wild type.
(A) Longitudinal section of a seedling showing Baf1 expression adaxial to the newly developing axillary meristems (arrowhead and close-up).
(B) and (C) Localization of Baf1 in an immature tassel (B) and a tassel branch (C) in a narrow domain where the axillary meristems are forming.
(D) A similar localization is observed in immature ears.
(E) and (F) In developing spikelet meristems, Baf1 is localized where a new floral meristem is forming (arrowheads).
(G) to (L) Consecutive sections hybridized with probes for either Baf1 or Zyb15, a marker for suppressed bract primordia.
(G) and (H) flank of an inflorescence meristem. Arrowheads point to a few Baf1-expressing cells in (G) and the corresponding region in (H).
(I) and (J) Immature developing ear.
(K) and (L) Developing spikelet meristems.
Black arrowheads in (I) mark the corresponding P1 and P2 sites of Zyb15 expression in (J). White arrowheads in (K) mark the Baf1 expression domain. P1 and P2, suppressed bract primordia. Bars = 50 μm.
To determine if the expression of Baf1 coincides with the suppressed bract domain, we used as a marker the maize FILAMENTOUS FLOWER ortholog Zyb15 (Juarez et al., 2004), whose expression is observed in initiating suppressed bracts (Whipple et al., 2010). Consecutive sections of the flanks of inflorescence meristems hybridized with both Baf1 and Zyb15 (Figures 6G to 6L) show that the expression domains of the two genes do not overlap, but rather they seem expressed initially in adjacent regions, with Baf1 expressed above the suppressed bract (Figures 6G and 6H). Since maize inflorescence development is acropetal, Baf1 expression is also observed below the suppressed bract (Figures 6G and 6H and P1 in Figures 6I and 6J), as an axillary meristem primordium starts to develop (P2 in Figures 6I and 6J). The distinct domains of expression for Baf1 and Zyb15 are clearly observable in later stages of meristem development, such as in developing spikelet meristems (Figures 6K and 6L). Therefore, we conclude that initial Baf1 expression marks a specific boundary domain flanking the apical meristem and that this domain remains adaxial to both the suppressed bract and the meristem primordium as it continues to develop.
Baf1 Is Required for Ba1 Expression during Axillary Meristem Formation
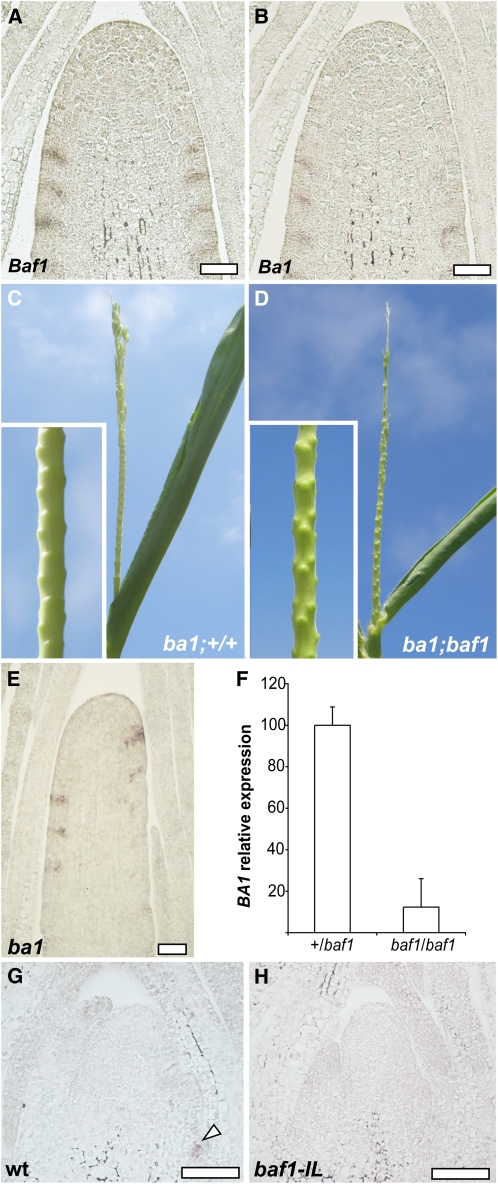
The very specific domain of expression of Baf1 was remarkably similar to the expression pattern of the bHLH transcription factor Ba1 (Gallavotti et al., 2004). Consecutive sections of wild-type ear inflorescences hybridized with either Baf1 or Ba1 probes showed that the two genes are indeed expressed in overlapping domains (Figures 7A and 7B). We next evaluated ba1-ref baf1-ref double mutant plants and found an extremely severe phenotype: barren tassels with neither branches nor spikelets and no ears. These phenotypes result from the lack of axillary meristem initiation at every stage of development (Ritter et al., 2002; Gallavotti et al., 2004). In ba1 mutant tassels, only the suppressed bracts, in whose axils meristems usually form, are observed (Figure 7C) (Ritter et al., 2002). In ba1 baf1 double mutants, the suppressed bracts still form, and we could not detect any phenotypic difference when compared with ba1 single mutants (Figures 7C and 7D), suggesting that ba1 and baf1 work in the same pathway. We then checked whether the Baf1 gene was expressed in ba1 mutant tassels and observed a similar pattern of Baf1 expression to the one observed in wild-type tassels, suggesting that Baf1 either functions upstream of Ba1 or in a parallel pathway in the formation of axillary meristems and that Baf1 marks the domain where axillary meristems will form (Figure 7E).
Figure 7.
The Relationship between Baf1 and Ba1.
(A) and (B) Consecutive sections of an inflorescence meristem of a developing ear hybridized with Baf1 (A) or Ba1 (B) probes show overlapping expression patterns.
(C) and (D) ba1 baf1 double mutant tassels show no noticeable phenotypic differences from single ba1 mutants. Suppressed bracts are still evident in both single and double mutants (close-ups).
(E) In situ localization of Baf1 in a ba1 mutant tassel shows a regular pattern of expression of Baf1.
(F) Quantitative RT-PCR of Ba1 in pools of seedlings of heterozygous or homozygous baf1-IL mutant plants. The expression levels are relative to actin, and the expression in the heterozygous sample is set to 100. Error bars represent sd.
(G) and (H) In situ localization of Ba1 in wild-type (wt) (G) and baf1-IL mutant (H) seedlings. Arrowhead points to Ba1 expression in a developing axillary meristem.
Bars = 50 μm.
To verify if Baf1 could regulate the expression of Ba1, we performed quantitative RT-PCR of Ba1 on pools of homozygous and heterozygous baf1-IL seedlings in the original A619 background. In these samples, although Ba1 expression was not completely lost, it was severely reduced (approximately eightfold) compared with wild-type seedlings (Figure 7F). To assess the extent of meristem development in these seedlings, we used in situ hybridizations of the Knotted1 (Kn1) gene, which encodes a homeobox transcription factor expressed in meristematic cells (Jackson et al., 1994). It is clear from these in situ hybridizations that axillary meristems are not initiated in mutant seedlings but are visible in wild-type samples (see Supplemental Figure 5 online). We also checked Ba1 expression by in situ hybridizations and failed to detect a clear signal in baf1-IL mutant seedlings (Figures 7G and 7H). Finally, we tested whether BAF1 and BA1 proteins could interact in a targeted yeast two-hybrid assay, but no interaction was detected (see Supplemental Figure 6 online). Altogether, these results suggest that Baf1 is required for normal levels of Ba1 expression.
DISCUSSION
Here, we report the isolation of BAF1, a putative transcriptional regulator required for the formation of maize ears. Phylogenetic and genomic analyses estimate that clear coorthologs of Baf1 are restricted to grasses, although four Arabidopsis genes are possibly close relatives, despite lack of strong Bayesian support. We show that Baf1 functions in the early steps of axillary meristem initiation as a boundary determinant for the axillary meristem primordium, and, accordingly, baf1 mutants show fusion defects between the newly developing axillary meristems and the leaf or bract axils. Genetic and expression analyses suggest that Baf1 acts upstream of the bHLH transcription factor Ba1, a key gene required for the initiation of new axillary meristems whose function is also required for the formation of maize ears.
Baf1 Reveals a New Function and Properties of AT-Hook DNA Binding Proteins in Plants
Baf1 encodes a putative transcriptional regulator containing an AT-hook DNA binding motif and a PPC domain of unknown function (DUF296). In Arabidopsis, these combined features are found in 29 members of the AT-hook type DNA binding family (Matsushita et al., 2007), whereas AT-hook motifs alone, in single or multiple copies, can be found in proteins from a wide range of organisms (Aravind and Landsman, 1998). AT-hook transcriptional regulators are generally considered architectural transcriptional regulators, making chromatin more accessible for DNA binding, possibly through structural changes. They are predicted to bind AT-rich DNA regions, such as the matrix attachment regions, putative structural components of nuclei (Morisawa et al., 2000; Fujimoto et al., 2004). Several AT-hook DNA binding proteins with a similar domain organization to BAF1 have been described in plants. They were mainly identified by yeast one-hybrid or activation-tagging screens and have been implicated in jasmonate signaling, gibberellin biosynthesis, senescence, and hypocotyl elongation (Vom Endt et al., 2007; Lim et al., 2007; Matsushita et al., 2007; Street et al., 2008). Recently GIANT KILLER (GIK), another AT-hook transcriptional regulator, was identified as a target of the floral homeotic protein AGAMOUS. GIK has been proposed to regulate the expression of multiple genes involved in patterning and differentiation of floral organs, and its activity is associated with epigenetic modifications at the targeted genomic regions (Ng et al., 2009).
The baf1 mutants are distinct from other barren mutants in that they are still capable of forming tassels with most branches, spikelets, and florets, whereas they often fail to produce ears. Another mutation that also affects ear formation without affecting tassel fertility is ba2, first reported, together with ba1, in the 1920s (Hofmeyr, 1931; Neuffer et al., 1997). No Ba2 gene has been isolated yet, and the original mutation might have been lost from mutant collections (see Methods). baf1 loss-of-function mutants show defects in the formation of axillary meristems, a previously unknown role for AT-hook transcriptional regulators. The main defects of baf1 mutants are observed in the first axillary meristems formed, during both vegetative and reproductive development (i.e., the axillary meristems formed at the axils of leaves after germination [from which ears eventually develop] and the branch meristems in the tassel, which in baf1 give rise to smaller branches fused at the base to the central spike and borne in an upright position). However, we also observed that the tassel branches of baf1 mutants have a higher percentage of solitary versus paired spikelets (Table 1), suggesting that some spikelet-pair meristems fail to initiate one of the spikelet meristems. The formation of single spikelets correlates with a reduction in tassel branch size observed in baf1 mutants, whereas in the central spike, spikelet meristems form normal paired spikelets. The formation of floral meristems is not visibly affected in tassels and ears, suggesting that other factors may act redundantly with Baf1 or that the role of Baf1 is restricted mainly to vegetative axillary meristems and branch meristems. Baf1 is expressed in developing spikelet and floral meristems as well (Figure 6), suggesting that redundancy with similar AT-hook transcriptional regulators may exist. At least one other gene belongs to the BAF1 clade and several others are found in clades ii and iii (Figure 4). We also showed that the Baf1r-1, which belongs to a different clade (Clade B), encodes a protein capable of interaction with BAF1 in yeast and in vitro assays. Together with the ability of BAF1 proteins to homodimerize, these data suggest that homo- and heterodimerization of AT-hook/PPC domain-containing proteins, a previously uncharacterized feature of this family of transcriptional regulators, may play a role in their mechanism of action and that other genes related to Baf1 may work in the same pathway.
Baf1 Is Conserved in Grasses
Zm BAF1 appears to be a protein without much similarity to other known proteins in the gene family. Orthologous genes were identified in sorghum, rice, and Brachypodium, but the BAF1 clade might have been produced by a duplication event prior to the divergence of Commelinid monocots based on the position of the ginger sequence DY363105 in Clade A-iii, and BAF1 has no immediate relatives in Arabidopsis. We find weak evidence, with little support (0.63 posterior probability), for clade A that includes the BAF1 clade and the groups that we have called eudicot grade, and clades ii and iii (Figure 4). Based on the phylogenetic analysis, it is therefore unclear if any of the four Arabidopsis genes belonging to the eudicot grade is a possible ortholog of Baf1. Based on the lack of a Baf1 duplicate in sorghum and rice and the partial synteny among noncoding regions of maize chromosomes 6 and 9, the Baf1 paralog in maize (GRMZM5G88968/Baf1B) appears to have been produced by the maize tetraploidy event (Gaut and Doebley, 1997).
The AT-hook proteins are at least as old as the land plants and have radiated extensively among the seed plants. It appears that the ancestral land plant may have had a single gene but that duplications occurred independently in the bryophytes, lycophytes, gymnosperms, and angiosperms. All studied angiosperms have multiple members of this gene family; the phylogeny indicates that at least four loci were present in the ancestor of the flowering plants. Additional duplications in the grasses are consistent with the well-documented whole-genome duplication at the origin of the grasses (Paterson et al., 2004) and the additional duplication in the history of Zea (Gaut and Doebley, 1997; Bomblies and Doebley, 2005). As additional monocot sequences become available in the databases, it is likely that the details of gene duplications in this gene family will become clearer and many of the relationships will become better supported.
A microarray analysis of the rice mutant lax1 uncovered an AT-hook DNA binding protein called b-2 whose expression was downregulated in the lax1 mutant compared with wild-type rice plants (Furutani et al., 2006). LAX1 encodes a bHLH transcription factor (Komatsu et al., 2003), the ortholog of the maize gene Ba1 (Gallavotti et al., 2004). In situ hybridizations of b-2 in developing panicles showed that b-2 had a similar expression pattern to LAX1. Furthermore, b-2 was expressed in lax mutant panicles, therefore suggesting that b-2 did not function downstream of LAX1 and that the observed downregulation of b-2 expression was likely a secondary effect of the inherent lack of axillary meristems in lax1 mutants. b-2 maps in the region syntenic to where the Baf1 locus is located (see Supplemental Figure 4 online), corresponds to Os06g04540, and is likely the rice ortholog of Baf1. Therefore, we can infer that, in accordance with the phylogenetic analysis, the function of Baf1 may be conserved between rice and maize and likely in all grasses.
The Role of Baf1 in Axillary Meristem Initiation
During reproductive development in maize, a major role in phytomers is played by axillary meristems, at the expense of both leaf development and internode elongation. In situ hybridizations of Baf1 and Zyb15, a marker for suppressed bract primordia, showed that Baf1 expression marks a domain adaxial to the Zyb15 expression domain (Figures 6G and 6H). Based on our analysis, Zyb15 expression seems to precede that of Baf1 during reproductive development (P1 in Figures 6I and 6J), and during vegetative development, leaf primordia are clearly visible before Baf1 is expressed (Figure 6A). The specification of a boundary domain at the flanks of the inflorescence meristem represents the first discernible step in the pathway for axillary meristem initiation, marked by the expression of both Baf1 and Ba1, whose expression patterns overlap (Figures 7A and 7B). This boundary domain remains distinct from the domain of the suppressed bract primordium and remains adaxial to axillary meristems throughout development (Figures 6K and 6L). The fusion defects observed in early axillary meristem development of baf1 mutants also indicate that organ fusion occurs between the primordia formed by axillary meristems (the prophyll and the first basal internode) and the internode above it. Therefore, Baf1 seems to function in establishing a boundary region required for axillary meristem initiation and separating the emerging axillary buds from the stem internode. In baf1 mutants, this boundary is partially or totally lost, and, as a consequence, axillary meristems either do not form, as observed during vegetative development, or remain smaller if they do form, as noticed in tassel branch meristems.
Several mutations are known to make maize plants earless, such as ba1 (Ritter et al., 2002) and loss-of-function alleles of Kn1, a homeobox gene whose product specifies meristematic cell identity (Kerstetter et al., 1997). In both mutants though, the tassel is also affected, making it an almost or completely sterile inflorescence. Ba1 and Baf1 are expressed in an overlapping narrow domain, whereas Kn1 is expressed in the meristematic core (Jackson et al., 1994). ba1 baf1 double mutant plants fail to form axillary meristems, and the suppressed bracts are as visible as they are in ba1 single mutant tassels (Figures 7A and 7B), suggesting that Baf1 functions upstream of Ba1 in the initiation of axillary meristems. In situ hybridizations revealing normal Baf1 expression in ba1 mutant tassels and quantitative RT-PCR and in situ hybridizations showing a dramatic decrease in Ba1 expression in baf1 mutant seedlings support the hypothesis that Baf1 function precedes Ba1 in the establishment of new meristems or that it is required for maintaining Ba1 expression. Ba1 expression is in turn needed for the development of a new meristem primordium that will subsequently express Kn1 (Ritter et al., 2002) (Figure 8). In baf1 mutant tassels, Ba1 expression levels are not affected (see Supplemental Figure 6 online). As mentioned above, it is possible that in the tassel other AT-hook transcriptional regulators function redundantly with BAF1, and this could explain why baf1 tassels are not similar to ba1 mutant tassels. It remains unclear if Baf1 directly regulates Ba1 expression in vegetative axillary meristems or alternatively if it is required for the correct establishment of the domain where Ba1 is subsequently expressed. In baf1-IL seedlings, which lack visible axillary meristems (see Supplemental Figure 5 online), Ba1 is still expressed, though at significantly lower levels (Figure 7F). This implicates other unknown factors in the regulation of Ba1 expression and also suggests that a threshold level of Ba1 expression is necessary for the initiation and development of these axillary meristems. The correct timing of expression, as well as stochastic changes in levels of Ba1 influenced by the environment, may determine whether or not baf1 mutants initiate axillary meristems in the axils of leaves during vegetative development and, hence, whether ears subsequently develop. Furthermore, the suppression of the earless phenotype in outcrosses of the baf1-IL allele with tb1 also suggests the presence of genetic modifiers of the baf1 phenotype that may influence the ability to form ears, a common occurrence also in kn1 mutants (Kerstetter et al., 1997; Vollbrecht et al., 2000).
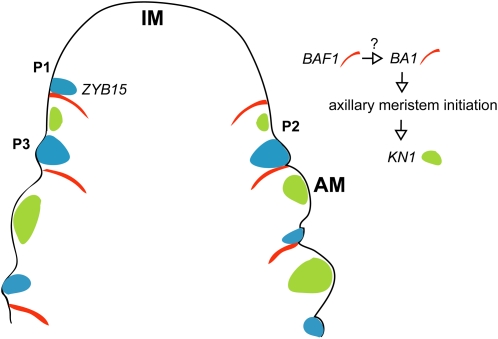
Figure 8.
Model for Baf1 Function in Axillary Meristem Initiation.
In developing inflorescences, the expression of the suppressed bract marker Zyb15 (P1, in blue) is followed by the expression of Baf1 in a boundary domain (P2, in red). Baf1 is required, together with other unknown factors, for proper expression of Ba1. Ba1 triggers axillary meristem initiation at the axils of the suppressed bracts (P2) and the establishment of a population of meristematic cells, expressing Kn1 (in green). IM, inflorescence meristem; AM, axillary meristem. P1, P2, and P3 mark suppressed bract primordia, with P1 marking the youngest primordium.
Our analysis shows that Baf1 likely functions in specifying a boundary domain where a new population of stem cells is established that will give rise to an axillary meristem. In Arabidopsis, genes with a similar function have been previously reported, such as RAX1, CUC1,2,3, LOF1,2, and LAS (Greb et al., 2003; Hibara et al., 2006; Keller et al., 2006; Lee et al., 2009). Our phylogenetic analysis did not rule out the possibility that a functional ortholog may exist in Arabidopsis, whereas it clearly identified orthologous genes in the grass family. Therefore, it remains to be determined if other transcriptional regulators related to Baf1 play the same role in dicotyledonous species or if in grasses AT-hook and bHLH transcriptional regulators, such as Ba1, have acquired a predominant role in the establishment of new axillary meristems throughout development.
METHODS
Plant Materials and Phenotyping
The baf1-ref allele was obtained from the Maize Genetics Cooperation Stock Center (913E baf1). The baf1-IL allele was generated by EMS mutagenesis by the Maize Inflorescence Architecture Project and corresponds to 03IL-A619TR-1051 (http://gremlin1.gdcb.iastate.edu/MIP/EMSphenotypeDB/index.php). In the course of this project, we realized that the ba2 mutant seeds deposited at the Maize Genetics Cooperation Stock Center (218E ba2 and 217H ba2 v4) carry the same mutation as the baf1-ref allele. Marty Sachs and Phil Stinard at the Maize Genetics Cooperation Stock Center investigated the pedigree records dating back to the 1950s and provided a hypothesis on what it might have happened. When the Maize Genetics Cooperation Stock Center moved to Illinois in 1953, it received a stock called ba2 from Cornell University that had been part of the COOP's collection since 1937. The same year it also received a baf1 stock from Charles R. Burnham. They were grown next to each other in the first crop in Illinois in 1953 and crossed together. When grown in 1954, none of the progeny from the cross were barren, suggesting that the two mutant loci were different. The baf1 stock that Burnham provided was originally called ba-s. At that time, Burnham's handwriting was difficult to read, and it is possible therefore that Burnham actually sent his ba-s mutant in 1953 and it was misread as ba2. From then on, only the Burnham ba2 was propagated and the original ba2 allele, which maps on chromosome 2 (Hofmeyr, 1931), was therefore lost. Burnham separately sent his ba-s to a maize geneticist and this was subsequently called baf1 and listed separately at the Maize COOP from the ba2 stock.
For double mutant analysis, the tb1 (117D) mutant was obtained from the Maize Genetics Cooperation Stock Center. The ba1-ref allele used for ba1 baf1 double mutant analysis is in a mixed genetic background (A188/W23) (Ritter et al., 2002). For tb1 baf1-IL double mutants, the baf1-IL mutation was genotyped by sequencing. For ba1 baf1-ref double mutants, baf1-ref was genotyped using the following primers: forward, 5′-TCGTCGATCGGGACGCACCAGCTC-3′; and reverse 5′-AAGGAGGAGGGATTGAGTCTTAGC-3′.
For tassel measurements, we considered tassel length as the distance from the base of the first tassel branch to the tip of the tassel and branch number as the number of branches with more than two spikelets. Branch length and size were calculated on the four longest branches of any tassel. Branch size was calculated by hand-sectioning the base of the tassel branches, putting the sections on slides, and taking pictures under a light microscope. Measurements on these sections were taken using ImageJ software. Given that the branch sections are not perfectly round, but more often elliptical in shape, we measured the longest distance between two opposite margins. Student’s t test was used to calculate statistical significance.
Positional Cloning
For map-based cloning of the Baf1 locus, we proceeded by developing new molecular markers using genome sequence information available at the Maize Assembled Genomic Island and maizesequence.org databases (Fu et al., 2005; Schnable et al., 2009). These molecular markers are listed in Supplemental Table 1 online. After narrowing down the genomic region surrounding the Baf1 locus, we analyzed syntenic regions in sorghum (Sorghum bicolor) and rice (Oryza sativa) using sequence information available at phytozome.net.
Expression Analysis and Histology
For in situ hybridizations, we used an antisense mRNA probe including the 3′-untranslated region of the Baf1 gene (primers: forward, 5′-GCCCCGTACTGATCGATCGAATC-3′; reverse, 5′-AGACTGACATGCAGTTCCAAGC-3′). The Ba1, Zyb15, and Kn1 in situ probes and the protocol used for hybridization have been previously described (Gallavotti et al., 2004, 2008b; Whipple et al., 2010).
Quantitative RT-PCR was performed in a Roche LightCycler apparatus using the QuantiFast SYBR Green RT-PCR kit (Qiagen). The reactions were run on two pools of baf1/baf1 and on two pools of +/baf1 seedlings (three plants per pool). The seedling samples, without roots, were stripped of most leaves to minimize the dilution of mRNAs from meristematic cells. Ba1 expression levels represent the average of seven technical replicates of two distinct biological pools relative to actin. Primers for RT-PCR of Rel2 were previously described (Gallavotti et al., 2010). For quantitative RT-PCR of both Ba1 and Actin, we used the following primers: Ba1 forward, 5′-GTGGTTGGTGACAACGAGGT-3′, and Ba1 reverse, 5′-CGAGGAAGATGCAAGAAGCAG-3′; Actin forward, 5′-CTCATGCTATTCTCCGTTTGG-3′, and Actin reverse, 5′-TCAGGCATCTCGTAGCTCTTC-3′.
For transient expression in tobacco (Nicotiana benthamiana), Baf1 and YFP were amplified using the following primers: Baf1 forward, 5′-ATTCTAGAATGGATCCCGTGGCGGCGGCGCAC-3′, and Baf1 reverse, 5′-ATGGATCCAGCAGCTTCGTACGGGGCGACGCGGC-3′; YFP forward, 5′-GAAGCTGCTGGATCCATGGTGAGCAAGGGCGAGGAG-3′, and YFP reverse 5′-ATGAATTCCTAGGCCCCAGCGGCCGCAGCA-3′. A Baf1-YFP PCR fusion product of the two fragments was subsequently obtained using Baf1 forward and YFP reverse primers. This product was cloned in the pGREEN0229 vector, carrying a 35S promoter, using XbaI and EcoRI restriction sites. YFP was amplified (primers are listed in Supplemental Table 2 online) and cloned in the same vector using SmaI and EcoRI restriction enzyme sites. Agrobacterium tumefaciens transformed with either BAF1-YFP or YFP constructs was washed in 10 mM MgCl2 and infiltrated in tobacco leaves using a 3-mL syringe (without needle). Images were acquired 3 to 4 d following infiltration with a Nikon Eclipse TE2000-U fluorescent microscope and processed using the ImageJ software.
The procedure used for fixing tissue for staining of both tassel and ear sections has also been previously described (Gallavotti et al., 2010). Toulidine Blue staining was performed by immersing the slides in a 0.1% solution (in 0.6% boric acid) for 2 min, followed by rinsing in water, whereas Safranin O-Alcian Blue (0.01 to 0.05% in 0.1 M Na acetate, pH 5.0) staining was carried out for 20 min, followed by rinsing in water. After drying, stained slides were mounted using Permount (Fisher).
Protein–Protein Interaction Assays
A cDNA yeast two-hybrid library of 2-mm ears was created in the GAL4 Two-Hybrid system (Stratagene). This library was screened using a construct carrying Baf1Δ762 (deleted at position 763 of the coding sequence) in the pBD:GAL4 CAM vector as bait, obtained using the primers and restriction sites listed in Supplemental Table 2 online. The library screen was performed following the manufacturer’s instructions. For targeted yeast two-hybrid assays, the prey genes were cloned in the pAD-GAL4 2.1 vector (Stratagene; primers and restriction sites are listed in Supplemental Table 2 online).
For pull-down assays, we used the MagneGST pull-down system from Promega, and we followed the manufacturer’s instructions. Incubations of in vitro–transcribed/translated products with GST fusion proteins bound to magnetic beads were performed for 2 to 4 h rotating at 4°C. For immunoblots, we used monoclonal anti-HA (Covance) and anti-GST (Cell Signaling Technology) antibodies as previously described (Gallavotti et al., 2010). Baf1 was cloned in pGEX-4TK (GE Healthcare) for expressing GST-BAF1 fusion protein using the primers and restriction sites listed in Supplemental Table 2 online. For cloning Baf1-HA3, PCR products were obtained by amplifying Baf1 and a 3XHA tag using the following primers: Baf1 forward, 5′-ATCCATGGATCCCGTGGCGGCGGCGCAC-3′, and Baf1 reverse 5′-TGGGTAACCTGCCATGTACGGGGCGACGCGGC-3′; HA3 forward, 5′-CGCGTCGCCCCGTACATGGCAGGTTACCCATACG-3′, and HA3 reverse, 5′-ATGAATTCTCAAGAGGAACGTAGAGAAGCGT-3′. The Baf1 forward and HA3 reverse primers were subsequently used for obtaining a fusion of the amplified products and cloned in pCITE-2a (Novagen) digested with NcoI and EcoRI. A pCITE-2a clone containing a 3XHA tag inserted between EcoRI and NotI sites was used to clone Baf1r-1 amplified with the primers listed in Supplemental Table 2 online.
Phylogenetic Analysis
Using ZmBAF1 as a query sequence, other AT-hook proteins were downloaded from a variety of gene and genome databases, to give a data set of 90 sequences with the conserved AT-hook motif (see Supplemental Data Set 1 online). The domain consists of 184 aligned amino acids, or 552 nucleotides, with only a couple of one-codon gaps inserted to improve alignments. Alignments were done on translations of the nucleotide sequences using MUSCLE (Edgar, 2004) and then were converted back to nucleotides preserving the reading frame. Preliminary phylogenetic trees were constructed using the neighbor-joining algorithm as implemented in MEGA (Tamura et al., 2007) and a Bayesian analysis implemented in MrBayes (Huelsenbeck and Ronquist, 2001), consisting of two separate runs of 10 million generations and the General Time Reversible model of evolution with invariant sites and gamma distributed rates partitioned according to codon position run on the Grethor Parallel processing cluster at the University of Missouri, St. Louis. To improve the quality of alignments to explore the evolution of the sequences in more detail, the data set was pruned successively based on the results of the preliminary phylogenetic analyses. All sequences from seed plants formed a monophyletic group (see Supplemental Figure 3 online) so the outgroup sequences from Physcomitrella patens and Selaginella moellendorffii were removed. The preliminary analysis also identified a clade consisting of divergent monocot and dicot sequences, and these were also omitted from subsequent analyses. The remaining seed plant sequences were aligned based on amino acids in MacClade 4.08 for OSX (Maddison and Maddison, 2003) and then returned to nucleotide sequences for analysis (see Supplemental Data Set 2 online). The final Bayesian phylogenetic analysis consisted of two separate searches of 30 million generations using flat priors and the General Time Reversible model of sequence evolution with invariant sites and gamma distributed rates partitioned according to codon position with trees being sampled every 1000 generations. At the end of the run, the first 25% of trees were removed as burn-in and clade credibility values estimated using MrBayes.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Baf1 (JF309450) and Baf1r-1 (JF309451). Accession numbers for the sequences used in the phylogenetic analysis are labeled on Figure 4 and provided in Supplemental Data Sets 1 and 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of the Ear Insertion Site in Different Mutant Combinations.
Supplemental Figure 2. The Tassel Phenotype of baf1 Mutants.
Supplemental Figure 3. Bayesian Consensus Phylogram of Land Plants.
Supplemental Figure 4. Comparison of the Genomic Regions Encompassing the Baf1 Locus in Maize, Rice, Brachypodium, and Sorghum.
Supplemental Figure 5. In Situ Localization of Kn1.
Supplemental Figure 6. Protein–Protein Interaction of BAF1 and BA1, and RT-PCR of Ba1 in baf1 Tassels.
Supplemental Table 1. List of New Molecular Markers Developed for the Positional Cloning of Baf1.
Supplemental Table 2. List of Primers and Vectors Used for Cloning.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 3 Online.
Supplemental Data Set 2. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 4.
Acknowledgments
We thank Darren Hall, Zara Tabi, Amy Buck, and Fausto Bustos for help with genotyping, Jinshun Zhong for help with initial database searches and alignments, and Marty Sachs and Phil Stinard at the Maize Genetics Cooperation Stock Center for clarifying the origin of the baf1 and ba2 mutant seed stocks. This work was supported by grants from the National Science Foundation to R.J.S. and E.K. (NSF DBI-0604923) and S.M. and A.G. (NSF IOS-0820729).
References
- Aravind L., Landsman D. (1998). AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26: 4413–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazesh S., McSteen P. (2008). Barren inflorescence1 functions in organogenesis during vegetative and inflorescence development in maize. Genetics 179: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T., Leyser O. (2006). Something on the side: Axillary meristems and plant development. Plant Mol. Biol. 60: 843–854 [DOI] [PubMed] [Google Scholar]
- Bomblies K., Doebley J.F. (2005). Molecular evolution of FLORICAULA/LEAFY orthologs in the Andropogoneae (Poaceae). Mol. Biol. Evol. 22: 1082–1094 [DOI] [PubMed] [Google Scholar]
- Bortiri E., Chuck G., Vollbrecht E., Rocheford T., Martienssen R., Hake S. (2006). ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe E., Beckett J. (1987). Barren-stalk-fastigate, baf, chromosome 9S. Maize Genet. Coop. News Lett. 61: 46–47 [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Emrich S.J., Guo L., Wen T.J., Ashlock D.A., Aluru S., Schnable P.S. (2005). Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc. Natl. Acad. Sci. USA 102: 12282–12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S., Matsunaga S., Yonemura M., Uchiyama S., Azuma T., Fukui K. (2004). Identification of a novel plant MAR DNA binding protein localized on chromosomal surfaces. Plant Mol. Biol. 56: 225–239 [DOI] [PubMed] [Google Scholar]
- Furutani I., Sukegawa S., Kyozuka J. (2006). Genome-wide analysis of spatial and temporal gene expression in rice panicle development. Plant J. 46: 503–511 [DOI] [PubMed] [Google Scholar]
- Galinat W. (1959). The phytomer in relation to the floral homologies in the American Maydea. Bot. Mus. Lealf. Harv. Univ. 19: 1–32 [Google Scholar]
- Gallavotti A., Barazesh S., Malcomber S., Hall D., Jackson D., Schmidt R.J., McSteen P. (2008a). sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA 105: 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Long J.A., Stanfield S., Yang X., Jackson D., Vollbrecht E., Schmidt R.J. (2010). The control of axillary meristem fate in the maize ramosa pathway. Development 137: 2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Yang Y., Schmidt R.J., Jackson D. (2008b). The relationship between auxin transport and maize branching. Plant Physiol. 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Zhao Q., Kyozuka J., Meeley R.B., Ritter M.K., Doebley J.F., Pè M.E., Schmidt R.J. (2004). The role of barren stalk1 in the architecture of maize. Nature 432: 630–635 [DOI] [PubMed] [Google Scholar]
- Gaut B.S., Doebley J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94: 6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schafer E., Muller D., Herrero R., Schmitz G., Theres K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17: 1175–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hibara K., Karim M.R., Takada S., Taoka K., Furutani M., Aida M., Tasaka M. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeyr J.D.J. (1931). The Inheritance and Linkage Relationships of barren stalk-1 and barren stalk-2, Two Mature-Plant Characters of Maize. (Ithaca, NY: Cornell University; ). [Google Scholar]
- Huelsenbeck J.P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Jackson D., Veit B., Hake S. (1994). Expression of Maize Knotted1 Related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Juarez M.T., Twigg R.W., Timmermans M.C. (2004). Specification of adaxial cell fate during maize leaf development. Development 131: 4533–4544 [DOI] [PubMed] [Google Scholar]
- Keller T., Abbott J., Moritz T., Doerner P. (2006). Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18: 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R.A., Laudencia-Chingcuanco D., Smith L.G., Hake S. (1997). Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054 [DOI] [PubMed] [Google Scholar]
- Kiesselbach T. (1949). The Structure and Reproduction of Corn. (Lincoln, NE: University of Nebraska College of Agriculture; ). [Google Scholar]
- Komatsu K., Maekawa M., Ujiie S., Satake Y., Furutani I., Okamoto H., Shimamoto K., Kyozuka J. (2003). LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 100: 11765–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.K., Geisler M., Springer P.S. (2009). LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 136: 2423–2432 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2003). Regulation of shoot branching by auxin. Trends Plant Sci. 8: 541–545 [DOI] [PubMed] [Google Scholar]
- Li X., et al. (2003). Control of tillering in rice. Nature 422: 618–621 [DOI] [PubMed] [Google Scholar]
- Lim P.O., Kim Y., Breeze E., Koo J.C., Woo H.R., Ryu J.S., Park D.H., Beynon J., Tabrett A., Buchanan-Wollaston V., Nam H.G. (2007). Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 52: 1140–1153 [DOI] [PubMed] [Google Scholar]
- Long J., Barton M.K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218: 341–353 [DOI] [PubMed] [Google Scholar]
- Maddison D., Maddison W. (2003). MacClade 4: Analysis of Phylogeny and Character Evolution, Version 4.06. (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Matsushita A., Furumoto T., Ishida S., Takahashi Y. (2007). AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol. 143: 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P., Laudencia-Chingcuanco D., Colasanti J. (2000). A floret by any other name: Control of meristem identity in maize. Trends Plant Sci. 5: 61–66 [DOI] [PubMed] [Google Scholar]
- McSteen P., Malcomber S., Skirpan A., Lunde C., Wu X., Kellogg E., Hake S. (2007). barren inflorescence2 Encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol. 144: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisawa G., Han-Yama A., Moda I., Tamai A., Iwabuchi M., Meshi T. (2000). AHM1, a novel type of nuclear matrix-localized, MAR binding protein with a single AT hook and a J domain-homologous region. Plant Cell 12: 1903–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer M., Coe E., Wessler S. (1997). Mutants of Maize. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Ng K.H., Yu H., Ito T. (2009). AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol. 7: e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T., Kyozuka J. (2009). Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21: 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., Bowers J.E., Chapman B.A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101: 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter M.K., Padilla C.M., Schmidt R.J. (2002). The maize mutant barren stalk1 is defective in axillary meristem development. Am. J. Bot. 89: 203–210 [DOI] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Shuai B., Reynaga-Peña C.G., Springer P.S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirpan A., Culler A.H., Gallavotti A., Jackson D., Cohen J.D., McSteen P. (2009). BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development. Plant Cell Physiol. 50: 652–657 [DOI] [PubMed] [Google Scholar]
- Skirpan A., Wu X., McSteen P. (2008). Genetic and physical interaction suggest that BARREN STALK 1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J. 55: 787–797 [DOI] [PubMed] [Google Scholar]
- Street I.H., Shah P.K., Smith A.M., Avery N., Neff M.M. (2008). The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J. 54: 1–14 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Reiser L., Hake S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172 [DOI] [PubMed] [Google Scholar]
- Vom Endt D., Soares e Silva M., Kijne J.W., Pasquali G., Memelink J. (2007). Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol. 144: 1680–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple C.J., Hall D.H., DeBlasio S., Taguchi-Shiobara F., Schmidt R.J., Jackson D.P. (2010). A conserved mechanism of bract suppression in the grass family. Plant Cell 22: 565–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D.P., Hope C.L., Malcomber S.T. (2011). Phylogenomic analyses of the BARREN STALK1/LAX PANICLE1 (BA1/LAX1) genes and evidence for their roles during axillary meristem development. Mol. Biol. Evol., in press. [DOI] [PubMed] [Google Scholar]