This research assesses how plant leaf growth is regulated under water-limiting conditions at the cellular and molecular level. It demonstrates that growth and, more specifically, cell division responds to stress in a highly dynamic manner. Growth inhibition is mediated by ethylene signaling followed by adaptation and recovery.
Abstract
Despite its relevance for agricultural production, environmental stress-induced growth inhibition, which is responsible for significant yield reductions, is only poorly understood. Here, we investigated the molecular mechanisms underlying cell cycle inhibition in young proliferating leaves of the model plant Arabidopsis thaliana when subjected to mild osmotic stress. A detailed cellular analysis demonstrated that as soon as osmotic stress is sensed, cell cycle progression rapidly arrests, but cells are kept in a latent ambivalent state allowing a quick recovery (pause). Remarkably, cell cycle arrest coincides with an increase in 1-aminocyclopropane-1-carboxylate levels and the activation of ethylene signaling. Our work showed that ethylene acts on cell cycle progression via inhibition of cyclin-dependent kinase A activity independently of EIN3 transcriptional control. When the stress persists, cells exit the mitotic cell cycle and initiate the differentiation process (stop). This stop is reflected by early endoreduplication onset, in a process independent of ethylene. Nonetheless, the potential to partially recover the decreased cell numbers remains due to the activity of meristemoids. Together, these data present a conceptual framework to understand how environmental stress reduces plant growth.
INTRODUCTION
When subjected to environmental stress, plants actively reduce their vegetative growth to conserve and redistribute resources and thus increase their chance of survival if the stress becomes severe (Skirycz and Inzé, 2010). However, when the stress does not threaten survival, growth inhibition is counterproductive because it leads to an unnecessary drop in productivity and substantial yield penalties. Bolder plants that are able to grow during mild stress episodes might prove an efficient way to boost productivity in regions that do not experience severe weather conditions (Tardieu, 2003). Therefore, understanding the mechanisms underlying growth inhibition in response to stress is essential not only from an academic but also from a socioeconomic point of view.
In plants, organ growth is driven by two tightly controlled and dynamic processes: cell proliferation and subsequent cell expansion. The coordination of these two processes during leaf growth ultimately determines leaf size and shape. In dicots, such as the model species Arabidopsis thaliana, leaves initiate at the flank of the meristem, and, in the initial phase, their growth is driven exclusively by cell proliferation (Donnelly et al., 1999). In somewhat older leaves, cells will exit the mitotic cell cycle and begin to expand starting from the tip onward. This transition is marked by the onset of endoreduplication that is a modified cell cycle in which DNA replication proceeds without mitosis, resulting in higher ploidy levels such as 4C, 8C, etc. (Beemster et al., 2005). In water-limited environments, plants respond to this drought stress by a rapid initial growth reduction followed by growth adaptation, giving rise to leaves with fewer and smaller cells (Schuppler et al., 1998; Granier and Tardieu, 1999; Aguirrezabal et al., 2006; Skirycz et al., 2010). Whereas previously we investigated processes involved in growth adaptation to long-term exposure to stress (Skirycz et al., 2010), the aim of this research was to learn more about the mechanisms underlying acute stress-mediated growth inhibition. Although reduction of cell proliferation upon stress onset is a well-known phenomenon, how changes in the environment translate into reduced proliferation rates is only poorly understood.
At the level of the cell cycle machinery, the most often proposed scenario that mediates stress-induced cell cycle inhibition assumes transcriptional upregulation of cell cycle inhibitors that belong to the cyclin-dependent kinase (CDK) inhibitor (ICK)/KIP-related protein (KRP) and/or the SIAMESE family. These inhibitors are thought to transiently arrest cell proliferation by inhibiting CYCLIN-DEPENDENT KINASE A (CDKA)/cyclin complexes (De Veylder et al., 2001; Churchman et al., 2006; Peres et al., 2007; Rymen et al., 2007). CDKA activity, which is a main driver of cell cycle progression, can also be reduced via targeted degradation of cyclins and/or inhibitory phosphorylation, as shown for wheat (Triticum aestivum) plants subjected to drought stress (Schuppler et al., 1998). Upstream of the cell cycle machinery, the plant hormone abscisic acid (ABA) has been demonstrated to affect the expression of ICK/KRP and/or SIAMESE (Wang et al., 1998; Pettkó-Szandtner et al., 2006). Another classical stress hormone is ethylene, which was shown to accumulate upon drought (Kalantari et al., 2000; Sobeih et al., 2004), and similarly to ABA, the ethylene precursor 1-aminocyclopropane-1-carboxylate (ACC) is known to be transported from root to shoot (reviewed in Sobeih et al., 2004). As such, ABA and ethylene are considered good candidates to communicate changes in the soil water status to the meristems. Examples of positive and negative effects of ethylene and ABA on growth can be found in the literature (reviewed in Sharp and LeNoble, 2002; Pierik et al., 2006), but their exact role in cell cycle regulation remains largely unknown.
Here, we examined how mild drought stress affects cell proliferation during early leaf development. In contrast with expanding leaves, Arabidopsis leaves in the very early stage in which cells are proliferating only and not yet expanding are extremely small (<0.1 mm2 in size). Thus, it is a technical challenge to analyze the molecular basis of stress-induced cell cycle arrest with sufficient developmental and temporal resolution. To this end, we devised an experimental setup to enable the simultaneous analysis of growth-related parameters and molecular mechanisms specifically in the proliferating leaves upon short-term exposure to stress. Unlike many previous studies focusing on very severe stress in mature leaves or complete seedlings (e.g., Fujita et al., 2007; Kant et al., 2007; Papdi et al., 2008), our mild stress setup slowed growth without affecting plant survival. Our data clearly demonstrated that cell cycle arrest is a very rapid response to stress mediated by posttranscriptional mechanisms rather than by a transcriptional cascade, with the plant hormone ethylene upstream of the reversible cell cycle arrest. Whereas ethylene is a primary signal for growth arrest, the subsequent ethylene-independent cell cycle exit occurs relatively late and only when the stress persists. Such highly temporal regulation allows plants to fine-tune their growth response according to the stress duration.
RESULTS
Osmotic Stress Affects Cell Proliferation and the Onset of Endoreduplication
To decipher the mechanisms by which water deficit inhibits cell proliferation, an experimental setup was developed that reproducibly reduced the leaf area by ~50%. The best results were obtained by addition of mannitol to the growth medium at a low concentration (25 mM), thereby decreasing the water potential of the medium and, hence, water uptake of the exposed roots. Arabidopsis seedlings were germinated and grown on nylon meshes overlaying control medium (without mannitol) until 9 d after stratification (DAS). At this point, the third true leaf is fully proliferating (Skirycz et al., 2010), and seedlings were subsequently transferred to control or mannitol-containing medium (Figures 1A to 1C). Kinematic analysis was performed, whereby leaves were harvested daily throughout development of leaf 3 (9 to 20 DAS) and, based on drawings of the abaxial epidermis, cell number, cell area, the number of guard cells, and the rates of cell division and cell expansion were calculated (De Veylder et al., 2001).
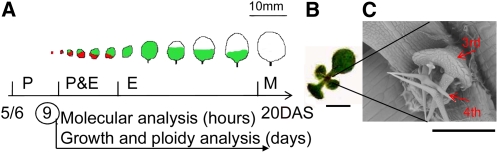
Figure 1.
Experimental Setup.
(A) Schematic representation of leaf 3 development with proliferating (P; red), expanding (E; green), and mature (M; white) cells. At 9 DAS, plants were transferred to mannitol, and leaf 3 was dissected for growth, ploidy, and molecular analysis.
(B) Nine-day-old seedling.
(C) Electron micrograph of the 3rd and 4th leaves at 9 DAS.
Bars = 2 mm in (B) and 200 μm in (C).
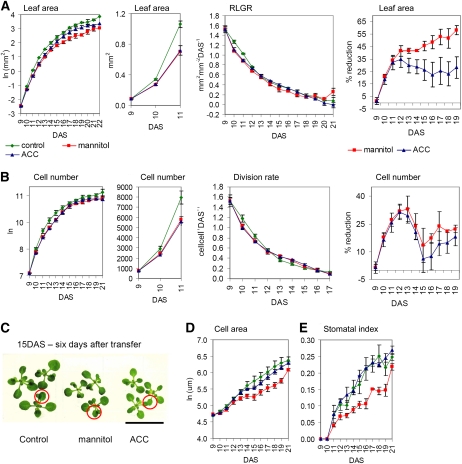
A decrease in leaf area was already apparent 24 h after the transfer (Figure 2A; t test, P value = 0.003) and resulted from reduced proliferation rates, as demonstrated by cellular measurements (Figure 2B). However, this reduction was short term: cell division rates of stressed plants were indistinguishable from controls within 72 h of transfer and afterward even slightly increased as a compensation for the initial decrease (Figure 2B). Importantly, transfer itself did not inhibit leaf growth, and the reduced cell numbers could be fully attributed to osmotic stress (see Supplemental Figure 1A online). Leaf and plant morphology were not altered by mannitol (Figure 2C). Cell expansion was also affected by stress, and the reduced final leaf area was a combination of fewer and smaller cells (Figure 2D). The stomatal index, which represents the number of stomata as a fraction of the total number of cells, was reduced as well (Figure 2E).
Figure 2.
Kinematic Analysis of Leaf 3 Dissected from Plants Transferred to Control, Mannitol-, or ACC-Containing Media at 9 DAS, When the Third Leaf Is Fully Proliferating.
(A) Leaf area, relative leaf growth rate (RLGR), and percentage of reduction of leaf area caused by mannitol or ACC. RLGR is expressed as increase in leaf area (mm2) relative to the initial leaf area per unit of time (day).
(B) Cell number, relative cell division rates, and percentage of reduction of cell number caused by mannitol or ACC. Relative cell division rates is expressed as increase in cell numbers relative to the initial cell numbers per unit of time (day).
(C) Plants 6 d after transfer to control, mannitol-, or ACC-containing media. The red circle marks the 3rd leaf. Bar = 2 cm.
(D) Cell area.
(E) Stomatal index that represents the number of stomata as a fraction of the total number of cells
Data for (A) to (E) are means ± se from three independent experiments. Leaf area was measured for 8 to 10 leaves in each experiment. Cellular data are from four leaves in each experiment.
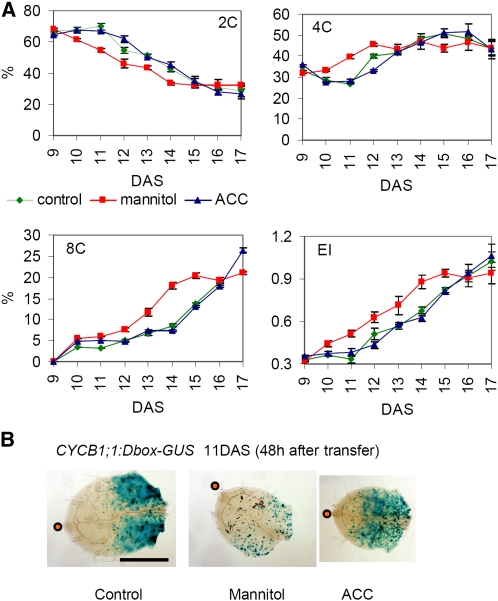
To learn more about the mechanisms underlying the rapid reduction of cell division upon stress onset, we harvested leaf samples daily after transfer. As a measure of cellular differentiation, the ploidy distribution was examined by flow cytometry. This revealed significant differences starting from 48 h after stress imposition (Figure 3A). In stressed leaves, 4C nuclei started to accumulate approximately 1 d earlier than in controls at the expense of 2C nuclei (Figure 3A). Analogously, the number of 8C nuclei increased sharply at 14 DAS in stressed leaves but only at 15 DAS in control samples (Figure 3A). Again, transfer itself had no effect on ploidy levels (see Supplemental Figure 1B online). Faster onset of endoreduplication implied that stress induced mitotic exit.
Figure 3.
Osmotic Stress Arrests Cell Cycle and Subsequently Triggers Cell Cycle Exit.
(A) Ploidy analysis of leaf 3 dissected from plants transferred to control, mannitol-, or ACC-containing media at 9 DAS when the 3rd leaf is fully proliferating. Percentage of 2C, 4C, and 8C nuclei is presented. EI stands for endoreduplication index and represents the average number of endocycles undergone by a typical nucleus (EI = 1*4C+2*8C+3*16C). Data show means ± se from three independent experiments with multiple leaves pooled in each experiment.
(B) Leaf 3 of CYCB1;1:DBox-GUS plants 48 h (11 DAS) after transfer to control, mannitol-, or ACC-containing medium. Blue staining indicates mitotic activity. Orange dot indicates leaf tip. Bar = 0.5 mm.
This observation was further investigated with a CYCB1;1:Dbox-GUS reporter line. Staining for CYCB1;1:Dbox-GUS activity visualizes cells at the G2-to-M transition, reflecting mitotic activity (Colón-Carmona et al., 1999). The most apparent differences were observed 48 h after transfer. Whereas the developmental differentiation manifested by strong staining at the leaf base and lack of staining at the leaf tip could be clearly seen in both control and stressed leaves, the overall β-glucuronidase (GUS) activity was much weaker in the stressed leaves corresponding to a reduced number of mitotic cells (Figure 3B). Although the relative size of the cell proliferation zone was similar in mannitol-treated leaves, the remaining proliferating cells were found in a dispersed pattern throughout this zone. In conclusion, exposure of proliferating leaves to mild osmotic stress leads to a rapid decrease in cell division rates and a faster onset of endoreduplication, indicative of an early mitotic exit as also observed by the reduced expression of CYCB1;1:Dbox-GUS upon stress treatment. Within a few days after transfer, division rates adapted to the new conditions and compensation effects were observed.
Cell Cycle Arrest and Exit Depend on Stress Duration
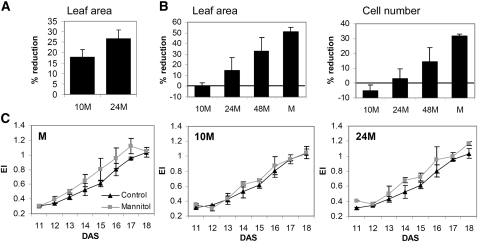
To investigate the dynamics of stress signaling, we transferred 9-DAS-old seedlings to mannitol for 10, 24, or 48 h and subsequently transferred them back to control medium. Whereas in all cases osmotic stress resulted in a cell cycle arrest, illustrated by a reduced leaf area measured at 10 DAS (24 h after the initial transfer) (Figure 4A), 10 h of stress imposition was too short to trigger mitotic exit, but after 24 h, some early differentiation was visible (Figure 4C). Moreover, the initial reduction in cell number measured after exposure for 10 and 24 h was completely compensated for by additional cell division, and no changes in cell number could be detected at 14 DAS, while this recovery was only partial for plants exposed to stress for 48 h (Figure 4B). Along with cell numbers, the stomatal index recovered as well in these plants (see Supplemental Figure 2 online). In conclusion, a short stress exposure reversibly arrests cell cycle, and only when stress persists are mitotic exit and differentiation observed.
Figure 4.
Effects of Varying Duration of Stress on Proliferating Leaves.
Plants were transferred to mannitol-containing plates at 9 DAS and then transferred back to control plates after 10 h (10M), 24 h (24M), or 48 h (48M) of mannitol treatment or kept on mannitol plates (M). Leaf 3 was dissected for further analysis.
(A) Reduction of leaf area at 10 DAS (24 h after first transfer).
(B) Reduction of leaf area and cell number at 14 DAS.
(C) Ploidy analysis. EI stands for endoreduplication index and represents the average number of endocycles undergone by a typical nucleus.
Data in (A) to (C) show means ± se from three independent experiments with multiple leaves measured in each experiment.
Increased Meristemoid Division Activity Aids in Cell Number Recovery
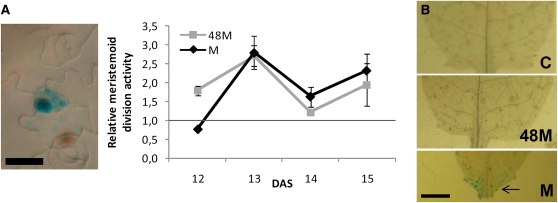
After the initial arrest of cell division in the cell proliferation zone and subsequent mitotic cell cycle exit, cell division rates recovered and became slightly higher in mannitol-treated leaves (Figure 2B) starting at 13 DAS, a time point at which the defined cell division zone is reduced to a very narrow zone near the base of the leaf. Furthermore, when plants were stressed for 48 h, at which time differentiation had occurred, and were then transferred back to control medium, their cell numbers partially recovered and, simultaneously, stomatal index fully recovered as well (Figure 4C; see Supplemental Figure 2 online). The divisions associated with the formation of stomata might account for this recovery. Using the CYCB1;1:DBox-GUS line, we observed that at the time of cell number recovery (13 to 14 DAS), meristemoid division activity was higher in mannitol-treated samples and in samples recovering from mannitol treatment than in control samples (Figure 5A). As the meristemoid lineage is restricted to the epidermis, we also checked for division activity in the internal tissues of the leaf. At 14 DAS there was still some mitotic activity at the base of the leaf of mannitol-grown plants, while in control leaves the cell proliferation zone had completely disappeared (Figure 5B).
Figure 5.
Effects of Osmotic Stress on Meristemoid Division Activity and Proliferation Zone.
Plants were transferred to mannitol-containing plates at 9 DAS and afterward transferred back to control (C) plates after 48 h of mannitol treatment (48M) or kept on mannitol plates (M). Leaf 3 was dissected for further analysis.
(A) Meristemoid division activity as determined by CYCB1;1:DBox-GUS staining, expressed relative to control. Data on right are means ± se from three independent experiments with 6 to 12 leaves measured in each experiment. Photo on left shows a representative active meristemoid. Bar = 10 μm.
(B) Leaf base of leaf 3 stained for CYCB1;1:DBox-GUS expression at 14 DAS. Some mitotic activity can still be seen in the proliferation zone of mannitol-treated leaves (indicated with an arrow). Bar = 0.5 mm.
Molecular Insight into Growth Inhibition Revealed by Transcript Profiling
To obtain a better understanding of the molecular mechanisms inhibiting cell division during stress, we subjected proliferating leaf primordia to whole-genome transcript profiling. Statistical analysis identified 27, 189, 351, and 886 upregulated and 31, 145, 84, and 622 downregulated transcripts at 1.5, 3, 12, and 24 h after transfer to 25 mM mannitol, respectively (see Supplemental Data Set 1 online). Selected microarray data could be validated with an nCounter platform containing probes for 100 genes involved in growth, stress, and hormonal regulation (see Supplemental Figure 3 online). Differentially expressed transcripts were used to construct Venn diagrams and were subjected to k-means clustering (MacQueen, 1967) (see Supplemental Figure 4 online). The number of differentially expressed genes was proportional to the stress exposure time, and the majority of the genes that were up- or downregulated at earlier time points remained high or low at 24 h, respectively.
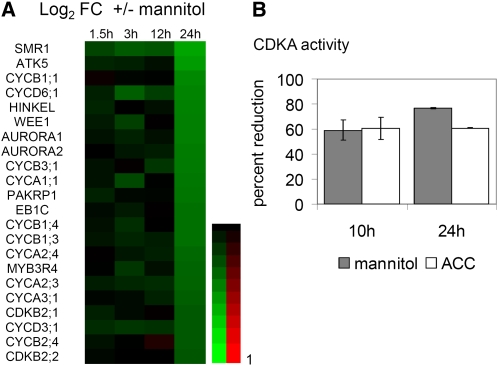
Subsequently, the differential transcripts were examined with PageMan to calculate the functional overrepresentation of MapMan categories (Thimm et al., 2004; Usadel et al., 2006) (see Supplemental Figure 5 online) and were compared with selected publicly available microarray experiments (see Methods; see Supplemental Table 1 online). To provide a possible explanation for the reduced cell proliferation rates, cell cycle genes were among the prime suspects. Indeed, a number of transcripts encoding A-, B-, and D-type cyclins, CDKB, SIAMESE-related proteins, and a MYB3R4 transcription factor were significantly downregulated (Figure 6A). Comparison with expression data obtained from synchronized cell cultures (Menges et al., 2003) revealed a significant overrepresentation of mitotic genes among the downregulated transcripts, such as the AURORA kinases and the kinesin HINKEL that are involved in cytokinesis (see Supplemental Table 1 online; Figure 6A). Strikingly, the magnitude of change for all of the above-mentioned cell cycle genes was similar (~30%) and occurred only at 24 h (Figure 6A).
Figure 6.
Osmotic Stress Effects on Cell Cycle.
(A) Heat map of selected cell cycle genes differentially regulated by osmotic stress in the 3rd fully proliferating leaf 1.5, 3, 12, and 24 h after stress imposition. Data are from Affymetrix ATH1 arrays and are expressed as the log2 of fold change (mannitol, control). Red and green indicate upregulation and downregulation, respectively. A key to the gene names is provided in the Supplemental Table 3 online.
(B) Relative CDKA activity measured in the 3rd proliferating leaf, microdissected from plants transferred to control, mannitol-, or ACC-containing medium 10 and 24 h after transfer. Data show means ± se from two (24 h) or three (10 h) independent experiments with ± 50 leaves pooled in each experiment.
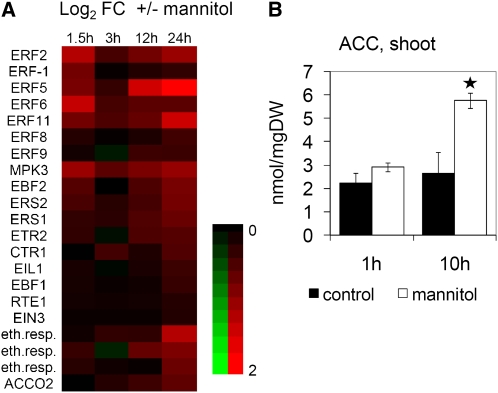
Besides the cell cycle–related transcripts, we were particularly interested in genes related to hormonal signaling. Both the comparison to publicly available hormone addition experiments (Goda et al., 2008) and the PageMan analysis revealed changes in ethylene signaling (see Supplemental Table 1 and Supplemental Figure 5 online). ACC-responsive genes were enriched among the transcripts upregulated in proliferating leaves as early as 1.5 and 3 h after stress onset (see Supplemental Table 1 online). The expression of genes directly involved in ethylene signaling was also induced, namely, the ethylene receptors (ETR2 and ESR1), CTR1, and MPK3 mitogen-activated protein kinases (MAPKs), EIN3 and EIL1 transcription factors, EBF1 and EBF2 involved in EIN3 protein degradation, and a number of ethylene-responsive transcription factors (ERF1, ERF2, ERF5, ERF6, and ERF11) (Figure 7A). While transcripts encoding ACC biosynthetic enzymes were not affected, ACC oxidase (ACO2), which converts ACC to ethylene, was upregulated (Figure 7A). Significantly, no activation of neither ABA nor jasmonate signaling, two other classical stress hormones, was apparent from the transcriptome analysis. Importantly, transfer itself did not affect the expression of selected cell cycle and stress genes as measured by quantitative RT-PCR (qRT-PCR), further showing the lack of basal stress response in controls due to the transfer that could mask stress responses, such as an ABA response, in plants exposed to mannitol (see Supplemental Figure 1C online). In summary, short-term stress exposure resulted in rapid induction of genes involved in ethylene signaling. Cell cycle–related genes were concomitantly downregulated, but only 24 h after the onset of stress.
Figure 7.
Rapid Increase in ACC Levels after Stress Imposition.
(A) Heat map of selected ethylene signaling genes differentially regulated by osmotic stress in the 3rd fully proliferating leaf 1.5, 3, 12, and 24 h after stress imposition. Data are from Affymetrix ATH1 arrays and are expressed as the log2 of fold change (mannitol, control). Red and green indicate upregulation and downregulation, respectively. A key to the gene names is provided in the Supplemental Table 3 online.
(B) ACC levels determined in shoots of 9-DAS seedlings 1 and 10 h after transfer to mannitol. Data show means ± se of ~100 to 150 plants from three independent experiments. Asterisk indicates significance (t test, P value < 0.05).
ACC Accumulates in Shoots of Stressed Plants
To find out whether differential expression of transcripts reflects changes in hormone levels, we determined concentrations of the ethylene precursor ACC in complete shoots of 9-d-old plants transferred to mannitol. As soon as 1 h after stress onset, ACC levels were 30% higher in stressed than in control samples, although this was not statistically significant, and by 10 h, the increase was more than twofold and significant (Figure 7B). At the time of analysis, the shoot samples were mainly composed of expanding cells, reflecting the overall importance of ethylene signaling for the response of growing tissues to stress.
CDKA Activity Is Reduced within Hours of Stress Onset
As transcripts of the cell cycle genes were downregulated by stress relatively late, it is unlikely that transcriptional mechanisms contribute to the rapid cell cycle arrest. To study the involvement of posttranscriptional mechanisms, we investigated the activity and protein abundance of CDKA. CDKA is a nonredundant protein central to cell cycle regulation and promotes both G1-to-S and G2-to-M transitions (Inzé and De Veylder, 2006). Although transcript and protein levels of CDKA were stable throughout the stress treatment, CDKA activity decreased as early as 10 h after stress onset and remained low at 24 h (Figure 6B; see Supplemental Figure 6A online). The rapid decrease in CDKA activity upon stress coincides with the cell cycle arrest.
Ethylene Arrests the Mitotic Cell Cycle
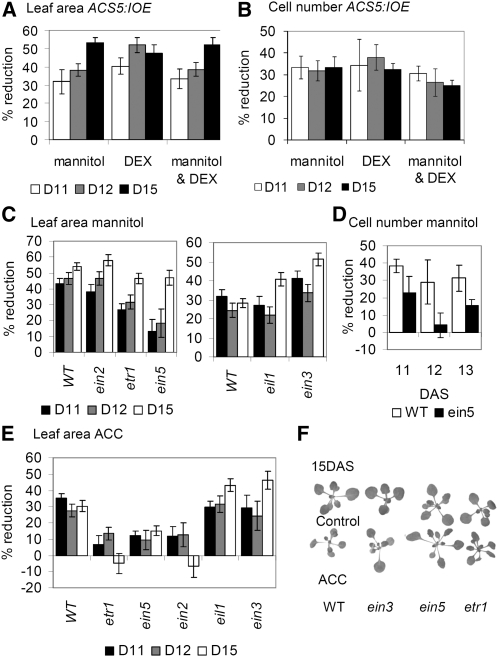
The transcriptome analysis revealed an early stress-dependent activation of ethylene-responsive genes in leaf primordia. To assess the role of ethylene in cell cycle regulation, we analyzed the effect of ACC. To this end, seedlings were transferred to medium containing 5 μM ACC (Goda et al., 2008) with the same experimental setup as that used for the mannitol treatments. Similarly to the effect of osmotic stress, transfer of seedlings to ACC resulted in a rapid reduction of cell proliferation rates (Figure 2B), and CDKA activity decreased as early as 10 h after transfer (Figure 6B). However, in contrast with mannitol treatment, the ploidy analysis revealed no changes in endoreduplication onset (Figure 3A) and no difference in the expression of the mitotic cell cycle genes CDKB2;1 and CYCB1;1 (see Supplemental Figure 7A online). Consistently, staining for CYCB1;1:DBox-GUS activity revealed no changes in the number of mitotic cells (Figure 3B). Stomatal index was also not affected (Figure 2E). To complement the ACC addition experiments, an inducible ACC synthase 5 (ACS5)-overexpressing line (ACS5:IOE) in which ethylene production can be triggered by dexamethasone (DEX) was analyzed. As with ACC treatment, transfer to DEX resulted in a decrease in cell numbers and leaf size (Figures 8A and 8B) but, as for the ACC treatment, did not affect the endoreduplication onset (see Supplemental Figure 7B online). Additionally, to test whether mannitol and ACC treatments have an additive effect on leaf size, ACS5:IOE plants were transferred to a combination of DEX and mannitol, but the results were comparable to those for either mannitol or DEX alone (Figures 8A and 8B).
Figure 8.
Involvement of Ethylene Signaling in Cell Cycle Arrest.
(A) and (B) Percentage of reduction of leaf area (A) and cell number (B) in the 3rd leaf of plants harboring the inducible ACS5 construct (ACS5:IOE5) transferred to media containing DEX, mannitol, or DEX and mannitol at 9 DAS, compared with transfer to control medium.
(C) to (F) Phenotypes of ethylene-insensitive mutants. Percentage of reduction in leaf area (C) and in cell number (D) of the 3rd leaf of mannitol-treated ethylene-insensitive mutants versus wild-type (WT) plants. Percentage of reduction in leaf area of the 3rd leaf of ACC-treated ethylene-insensitive mutants versus wild-type plants (E). Photographs of wild-type, ein3, etr1, and ein5 seedlings 6 d after transfer to ACC (F).
Data in (A) to (E) show means ± se for two or three independent experiments. Leaf area was measured for minimum 8 to 10 leaves in each experiment. Cellular data were from four leaves in each experiment. D, DAS.
The possible involvement of ethylene in the stress-induced inhibition of cell division predicts that ethylene-insensitive mutants would be less affected by transfer to mannitol. To test this prediction, we selected mutants with no or little effect on growth under normal conditions. This hypothesis proved true for the ethylene receptor mutant etr1.3 and for ein5.1, which is defective in the activity of the XRN4 exoribonuclease upstream of the EBF1 and EBF2 F-Box proteins (Figures 8C and 8D). A particularly pronounced difference was measured 72 h after transfer (12 DAS); whereas the reduction in leaf area of the wild type was ~45%, it was only ~20 and ~30% for ein5.1 and etr1.3, respectively (Figure 8C). A difference, albeit not significant, was also measured for the mkk9 mutant (see Supplemental Figure 8 online). However, neither ein2.5, eil1, nor ein3.1 (Figures 8C and 8D) showed this partial relief of inhibition.
Importantly, early endoreduplication onset measured in stress-treated wild-type leaves could also be detected in the ein5.1 mutant (see Supplemental Figure 7C online) exposed to mannitol. As reported before, when left on mannitol-containing medium until 22 DAS, all of the mutants, and particularly ein2.5, developed severe phenotypes and were overall much more affected by stress than the wild-type plants (Skirycz et al., 2010). In addition to mannitol, leaf growth of ethylene-insensitive mutants was also tested after transfer to ACC. Whereas ACC-related decrease in leaf area in ein3.1 and eil1 mutants was comparable to that of the wild type, it was significantly less in etr1.3, ein5.1, mkk9, and ein2.5 mutants (Figures 8E and 8F; see Supplemental Figure 8 online). Consistently, CDKA activity was reduced by ACC treatment in the wild type and ein3.1 but not etr1.3 and ein5.1 mutants (see Supplemental Figure 6B online). In conclusion, exogenous ACC addition or activation of ethylene production reduces cell proliferation without significantly affecting the onset of endoreduplication and subsequent cellular differentiation in an EIN3-independent manner.
DISCUSSION
Growth Responds to Stress in a Dynamic Fashion
One of the main characteristics of growth is its highly dynamic nature, as nicely illustrated by daily expansion rhythms measured in Arabidopsis leaves (Poiré et al., 2010). Similarly, plant growth responds dynamically to osmotic stress. First, stress exposure resulted in a rapid decrease in cell division rates, referred to as acute growth response, and by 24 h after stress onset, cell numbers were markedly reduced. Nevertheless, within 72 h, cell proliferation rates of stressed and control plants were again identical, illustrating the stable and relatively mild character of the treatment; later, division rates became even slightly higher in stressed leaves than in controls. This suggests that leaves adapted to the restrictive environment and established a new steady state, referred to as adaptive growth response (Skirycz and Inzé, 2010). Similar acute and adaptive growth responses to stress have been reported previously for roots (Burssens et al., 2000; Hsiao and Xu, 2000; West et al., 2004), monocot leaves (Hsiao and Xu, 2000; Veselov et al., 2002; Fricke et al., 2006), and sunflower (Helianthus annuus) leaves (Granier and Tardieu, 1999), but in all cases, organ elongation rates were measured, revealing mainly effects on expansion. We show that cell proliferation is subject to acute and adaptive growth responses upon exposure to mild stress as well. These findings demonstrate that molecular data on stress responses have to be interpreted with care and, importantly, have to be accompanied by a detailed growth analysis. As illustrated, samples taken 72 h, instead of 24 h, after stress onset would reflect mechanisms that allow adapted cells to proliferate under stress conditions but would not disclose much information on acute cell cycle inhibition. For technical reasons, these measurements were performed on the epidermis, although the leaf is composed of many cell types, but, in a similar setup, development of epidermal cells has been demonstrated to reflect that of the majority of the cells in the leaf (Beemster et al., 2005). Moreover, the epidermis has been shown to be the tissue driving organ growth (Savaldi-Goldstein et al., 2007; Marcotrigiano, 2010; Hacham et al., 2011).
Stress Inhibits Growth by Reducing the Number of Proliferating Cells
Expression data revealed a downregulation of cell cycle–related transcripts only 24 h after stress imposition, and, intriguingly, negative and positive cell cycle regulators were downregulated to the same extent. Furthermore, ploidy analysis did not provide evidence for an arrest at a specific point in the cell cycle, such as the G2/M block found in salt-treated Arabidopsis roots (West et al., 2004). Hence, adaptation of growth to stress might not affect the rate of individual cell division but rather reduce the number of dividing cells. Water deficit was previously shown to shorten the cell division zone in wheat and maize (Zea mays) leaves (Schuppler et al., 1998; Tardieu et al., 2000), while salt stress treatment was found to reduce the Arabidopsis root meristem size (West et al., 2004), although effects on cell cycle duration in wheat (Schuppler et al., 1998) and sunflower leaves (Granier and Tardieu, 1999) have been reported as well. The early onset of endoreduplication and differentiation observed in mannitol-stressed leaves also implies that a fraction of cells exit their mitotic cycle in favor of endoreduplication. Further confirmation was obtained with plants expressing a CYCB1;1:Dbox-GUS construct that clearly showed that the mannitol treatment reduced the number of mitotic cells. However, the cell division zone was not shortened, but the dividing cells were found in a more dispersed pattern throughout the proliferation zone in stress-treated leaves. In other words, stress-induced exit from the mitotic cell cycle does not proceed in the same organized manner as developmental differentiation that starts at the leaf tip and progresses to the leaf base (Donnelly et al., 1999). Therefore, we postulate that developmental and stress-induced differentiation are regulated by different mechanisms.
Enhanced Meristemoid Division Activity Is Responsible for Partial Cell Number Recovery
When stress was relieved after the occurrence of the mitotic exit, and even when stable stress conditions persisted, cell numbers partially recovered at the later stages of leaf development. At this stage, the cell proliferation zone had almost completely disappeared and divisions were largely restricted to dispersed meristemoids forming stomata and generating pavement cells in the process (Bergmann and Sack, 2007). Meristemoid division activity was higher in both conditions, indicating that meristemoid divisions, at least partially, account for this recovery. Intriguingly though, when mannitol-treated plants were examined, this higher meristemoid division activity did not appear to be reflected in stomatal indices, which are greatly reduced. This could be explained by a modulation of the number of amplifying divisions, allowing meristemoids to generate many pavement cells through repeated asymmetric amplification divisions, before finally differentiating into guard cells. Meristemoid longevity was shown to be under genetic control of MUTE (Pillitteri et al., 2007), and preliminary data suggest that MUTE expression may be reduced in mannitol-treated leaves. To assess whether this growth recovery in the epidermis does not create a growth imbalance between different leaf tissues, the mesophyll was probed for cell division activity, revealing extended cell division in the mesophyll at the leaf base. Furthermore, in agreement with the model in which the epidermis is the tissue driving leaf growth (Savaldi-Goldstein et al., 2007; Marcotrigiano, 2010), we propose that extra divisions in the epidermis also allow for increased cell expansion in the internal tissues, thereby ensuring a balance in growth between the different cell layers of the leaf.
Ethylene Arrests Cell Proliferation Posttranscriptionally
Transcriptome analysis revealed a rapid upregulation of ethylene biosynthesis and signaling genes in fully proliferating leaves upon osmotic stress perception, consistent with increased ACC levels measured in complete shoots. These findings make ethylene a good candidate to mediate cell cycle arrest. In support of this role, exogenous ACC or inducible activation of ethylene production in ACS5:IOE plants reduced cell numbers but remarkably did not affect cellular differentiation. Also in support of a role for ACC in mediating stress-related growth inhibition, the combined ACC and mannitol treatment had no additive effect when compared with either ACC or mannitol alone. Furthermore, ein5.1 and etr1.3, two ethylene-insensitive mutants, were partially resistant to leaf growth inhibition by osmotic stress. These findings are in agreement with the work of Sobeih et al. (2004) demonstrating that mild drought increases leaf ethylene evolution and thereby inhibits leaf growth in tomato (Solanum lycopersicum), although expanding leaves were studied. The authors suggested ACC as the primary root-borne signal responsible for inhibiting leaf growth, whereas ABA would be responsible for drought responses in mature leaves. We also found no evidence for ABA signaling in the stress response of proliferating leaves. Moreover, only ACO2, encoding an ACC oxidase, but none of the genes encoding ACC synthases were upregulated in proliferating leaves, leaving the possibility that also in Arabidopsis ACC might be a mobile root-borne signal mediating reduction in cell proliferation activity in response to stress; nevertheless, we cannot rule out that it might be synthesized in other parts of the shoot and then transported to proliferating leaves.
At the level of the cell cycle machinery, both osmotic stress and ACC reduced CDKA activity already 10 h after stress onset. A decrease in CDKA activity upon stress had been reported previously for leaves of wheat (Schuppler et al., 1998) and maize (Granier et al., 2000) and for Arabidopsis roots (West et al., 2004), making it a primary target of stress-mediated cell cycle arrest. This would affect both the G1-to-S and the G2-to-M transitions and could explain why no arrest at a specific stage was observed. Importantly, the absence of specific upregulation of transcripts encoding cell cycle inhibitors belonging to the ICK/KRP and/or SIAMESE family and the finding that changes in cell division precede effects on cell cycle–related transcripts strongly suggest that ethylene arrests cell proliferation and CDKA activity by a posttranscriptional mechanism. Importantly, the observation that the leaf growth reduction caused by both mannitol and ACC was not relieved in the ein3 mutant demonstrated that cell cycle arrest can be mediated through a branch of ethylene signaling independently of the EIN3 transcriptional control, similarly to the rapid inhibition of hypocotyl elongation in seedlings exposed to ethylene (Binder et al., 2004).
Moreover, the lack of a growth phenotype in ein3 but clear relief of cell cycle arrest in the ein5.1 mutant, situated upstream of EIN3 and controlling its stability (Olmedo et al., 2006; Potuschak et al., 2006), hint at still unknown roles of the EIN5/XRN4 endoribonuclease in the ethylene signaling pathway. Besides EIN5, other good candidates to regulate cell cycle arrest independently of EIN3 are the MKK9-MPK3/MPK6 MAPK cascade or still unidentified MAPKs that might be implicated in ethylene signaling, although their precise role is still under debate (Xu et al., 2008; Yoo et al., 2008; Hahn and Harter, 2009). In support of this possibility, the mkk9 mutant was partially resistant to growth inhibition by ACC and mannitol, but the latter was not significant possibly due to redundancy with other MAPK kinases.
In summary, we present evidence for the involvement of ethylene in cell cycle arrest upon stress sensing in an EIN3-independent manner and through a posttranscriptional cascade that regulates CDKA activity. While the role of ethylene in growth regulation is well established, the link to cell cycle arrest upon stress is, to our knowledge, novel.
Multiple Signals Act together to Pause-or-Stop Cell Cycle under Osmotic Stress
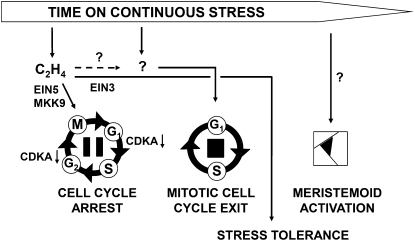
The finding that cell cycle arrest and onset of differentiation following osmotic stress are regulated by different mechanisms, together with the fact that the early cell cycle arrest is brought about entirely by posttranscriptional means, led us to hypothesize that this arrest is reversible and becomes irreversible only once cells start to differentiate. To test this hypothesis, we exposed plants to stress for a limited time after which it was relieved, showing that cell cycle arrest caused by stress that persists for 24 h or less could be fully overcome and stress that persists for 48 h only partially. We propose a model (Figure 9) in which stress causes ethylene to very quickly and reversibly pause cell division in a fraction of cells, but these cells are kept in an uncommitted, quiescent state allowing them to quickly recover when the environmental conditions improve. However, when the stress persists, cells are irreversibly pushed into differentiation, thereby removing their potential for proliferation, in a process probably mediated by DELLA proteins (H. Claeys, A. Skirycz, and D. Inzé, personal communication). Nonetheless, even when the cell division zone has completely disappeared, there is still some capacity to compensate for the reduced cell numbers even in the leaves that experience continuous stress, most likely through meristemoid division activity that might account for an important portion of cell division especially at the later stages of leaf development (M. Andriankaja, S. Dhondt, and D. Inzé, personal communication). This leads to more epidermal cells sustaining larger leaves, while the number of stomata and thereby transpiration losses are not increased. In a rapidly changing environment, such a mechanism of fast, but reversible, modulation of growth is without doubt essential for plants to maintain a balance between growth and survival.
Figure 9.
Simplified Scheme Depicting the Regulation of Cell Cycle Inhibition and Cell Differentiation in Response to Osmotic Stress.
Very rapidly (within hours) after stress imposition, ethylene (C2H4) production is triggered, inhibiting CDKA activity through a posttranscriptional mechanism that reversibly inhibits the cell cycle by G1/S and G2/M arrest. Cell cycle arrest is independent of EIN3 transcriptional control and possibly mediated by a MAPK signaling pathway or the ribonuclease EIN5. In a later phase, a different signal leads to permanent inhibition and exit from the mitotic cell cycle in favor of the endocycle and cell differentiation. Later in leaf development, meristemoid division activity becomes higher in stressed leaves and the enhanced meristemoid division results in a small increase in cell numbers.
METHODS
Plant Growth
Seedlings of Arabidopsis thaliana ecotype Columbia-0 were grown in vitro in half-strength Murashige and Skoog medium (Murashige and Skoog, 1962), supplemented with 1% Suc under a 16-h-day (110 μmol m−2 s−1) and 8-h-night regime. Plates were overlaid with nylon mesh (Prosep) of 20-μm pore size to prevent roots from growing into the medium. Depending on the experiment, 32 or 64 seeds were equally distributed on a 150-mm-diameter plate. Mutant plants were grown together with their corresponding wild-type controls on the same plate.
Stress and Chemical Treatments
At 9 DAS, when the third leaf is fully proliferating, seedlings were transferred to plates containing control medium (half-strength Murashige and Skoog) or medium supplemented with either 25 mM mannitol (Sigma-Aldrich) or 5 μM ACC (Sigma-Aldrich) by gently lifting the nylon mesh with forceps. For the CDKA activity assay, the transfer was done at 11 DAS; at this stage, the third leaf is still dividing and can be quickly harvested without the need for RNAlater solution (see below) that would inhibit CDKA activity. All transfers were performed 2 to 3 h after the beginning of the 16-h-day period.
Growth Analysis
Growth was analyzed on the third true leaf harvested at different time points after transfer. After clearing with 70% ethanol, leaves were mounted in 100% lactic acid on microscopy slides. For each experiment, 8 to 12 leaves were photographed with a binocular microscope, and epidermal cells (40 to 300) were drawn for four representative leaves with a DMLB microscope (Leica) fitted with a drawing tubus and a differential interference contrast objective. Photographs of leaves and drawings were used to measure leaf and cell area, respectively, with ImageJ v1.41o (NIH; http://rsb.info.nih.gov/ij/), from which the cell numbers were calculated. The stomatal index was defined as the percentage of stomata among all cells. For the kinematic analysis, ln-transformed means of leaf area, cell size, and cell number were locally fitted to a quadratic function of which the first derivative was taken as the relative growth, expansion, and division rate, respectively (De Veylder et al., 2001).
Sampling for Expression Analysis
Leaf 3 was harvested from plants at 1.5, 3, 12, and 24 h after transfer to mannitol. Briefly, whole seedlings were harvested in an excess of RNAlater solution (Ambion) and, after overnight storage at 4°C, dissected under a binocular microscope on a cooling plate with precision microscissors. Dissected leaves were transferred to a new test tube, frozen in liquid nitrogen, and ground with a Retsch machine (Retsch) and 3-mm metal balls. Samples were obtained from three independent biological experiments and from multiple plates within the experiment.
RNA Extraction
RNA was extracted with TriZol (Invitrogen) according to the manufacturer’s instructions with 4 μg of glycogen as carrier during the precipitation step. RNA samples were subjected to DNA digestion with RNase-free DNase I (Roche), and subsequently impurities were removed with the RNeasy mini kit (Qiagen).
ATH1 Expression Profiling and Data Analysis
RNA samples were hybridized to single Affymetrix ATH1 Genome arrays at the VIB Microarray Facility (Leuven, Belgium). Expression data were processed with robust multichip average (RMA) (background correction, normalization, and summarization) as implemented in BioConductor (Irizarry et al., 2003a, 2003b; Gentleman et al., 2004). An alternative chip definition file (cdf; named tinesath1cdf) was used, in which each probe is uniquely assigned to one transcript (Casneuf et al., 2007) (http://www.bioconductor.org/packages/release/data/experiment/html/tinesath1cdf.html). The BioConductor package Limma was used to identify differentially expressed genes (Smyth, 2004). For comparisons of interest, moderated t statistics were calculated using the eBayes function, and P values were corrected for multiple testing for each contrast separately using topTable (Smyth, 2004). False discovery rate (FDR)–corrected P value < 0.05 was used as a cutoff (Benjamini and Hochberg, 1995).
Flow Cytometry
For flow cytometry analysis, nuclei were extracted by chopping 4 to 32 leaves with a razor blade in 1 mL of 45 mM MgCl2, 30 mM sodium citrate, 20 mM 3-(N-morpholino)propanesulfonic acid, pH 7, and 1% Triton X-100 (Galbraith et al., 1983). From a stock of 1 mg/mL 4',6-diamidino-2-phenylindole, 1 μL was added to the filtered supernatant. The nuclei were analyzed with a CyFlow flow cytometer with the FloMax Software (Partec).
CDKA Activity Assay
Total soluble protein was extracted from 50 leaves by adding extraction buffer (Van Leene et al., 2007) to ground samples, followed by two freeze-thaw steps and two centrifugation steps (20,817g, 10 min, 4°C), with the supernatant being collected each time. Equal amounts of total protein were incubated with p9CKS1Hs-sepharose beads (De Veylder et al., 1997), and kinase assays were performed as described (De Veylder et al., 1997) with histone H1 (Millipore) as CDK substrate. To correct for the amount of CDKA protein purified by p9CKS1Hs-sepharose beads, an aliquot of each sample was used for protein gel blot analysis with primary rabbit anti-PSTAIRE antibodies (Santa Cruz) (diluted 1:5000) and secondary horseradish peroxidase–conjugated donkey anti-rabbit antibodies (GE-Healthcare) (diluted 1:10,000). Proteins were detected by chemiluminescence (Western Lightning Plus ECL; Perkin-Elmer Life Sciences). CDK activity and CDKA amount were quantified with ImageJ v1.41o. Control samples were arbitrarily set at 100%.
Comparison to Publicly Available Microarray Data
Selected publicly available microarray data were grouped according to experiment type (such as abiotic stress and hormone treatment) (see Supplemental Table 1 online). Groups of experiments were RMA processed and subjected to Limma analysis, as described above. Sets of responsive genes were delineated always with a twofold expression change and FDR-corrected P value < 0.05 cutoffs. Although these cutoffs were chosen somewhat arbitrarily, we assessed the robustness of the results by testing more and less stringent cutoffs. All tests gave very similar results. The lists of responsive genes were compared with those identified in our microarray experiment to identify global trends in the functional repertoire of the affected genes that were used as hints to explore the results in more detail. Overrepresentation was tested by means of Fisher exact tests (fisher.test function in R) followed by Bonferroni P value correction (Hochberg, 1988).
qRT-PCR
For cDNA synthesis, 100 ng to 2 μg of RNA was used with the SuperScript Reverse III reagent (Invitrogen) according to the manufacturer's instructions. Primers were designed with the QuantPrime website (Arvidsson et al., 2008; Skirycz et al., 2010). qRT-PCR was done on a LightCycler 480 (Roche Diagnostics) in 384-well plates with LightCycler 480 SYBR Green I Master (Roche) according to the manufacturer's instructions. Melting curves were analyzed to check primer specificity. Normalization was done against the average of housekeeping genes UBQ10, GAPDH, and CBP20; Δcycle threshold (Ct) = Ct (gene) – Ct (mean [housekeeping genes]) and ΔΔCt = ΔCt (control) − ΔCt (mannitol). ΔCt values for the three biological replicates were used for statistical analysis. Ct refers to the number of cycles at which SYBR Green fluorescence reaches an arbitrary value during the exponential phase of the cDNA amplification.
Analysis of mRNA Expression
mRNA expression levels were measured using an nCounter Analysis System (NanoString Technologies) by the VIB MicroArray Facility (www.microarrays.be) as described (Geiss et al., 2008). Total RNA extract (100 ng) was hybridized. Gene expression was measured simultaneously for all genes in multiplexed reactions. The nCounter code set contained probe pairs for 100 Arabidopsis genes (for the full list, see Supplemental Table 2 online). The data were normalized by a two-step procedure with internal spike-in controls and the three most stable reference genes included in the probe set (CDKA_1, UBC, and CBP20).
Quantification of ACC
Freeze-dried samples were dissolved in 500 μL of methanol, including two internal standard compounds (4 μM Met sulfone used for compensation of the peak area after capillary electrophoresis–mass spectrometry [CE-MS] analysis and 0.2 μM D4-ACC for quantification of ACC). After addition of 500 μL of chloroform and 200 μL of water, the mixture was vortexed for 3 min and centrifuged at 20,400g for 3 min at 4°C. The upper layer was evaporated for 30 min at 45°C by a centrifugal concentrator and then separated into two layers. The upper layer (100 to 200 μL) was centrifugally filtered through a Millipore 5-kD cutoff filter at 9100g for 90 min. The filtrate was dried for 120 min by a centrifugal concentrator at room temperature. The residue was dissolved in 10 μL of water containing a reference compound (3-aminopyrrolidine). The final solution (10 μL) was used to quantify the contents of ACC by cation analysis using CE-MS. The CE-MS system and conditions were as described (Watanabe et al., 2008).
GUS Staining
Whole plantlets were incubated in heptane for 10 min, washed in 100 mM Tris-HCl/50 mM NaCl, pH 7.0, and subsequently incubated in 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) buffer (100 mM Tris-HCl/50 mM NaCl buffer, pH 7.0, 2 mM K3[Fe(CN)6] and 4 mM X-Gluc) at 37°C for 24 h. Seedlings were washed in 100 mM Tris-HCl/50 mM NaCl, pH 7.0, and cleared overnight in 90% lactic acid. Samples were photographed with a differential interference contrast microscope (Leica).
Meristemoid Division Activity Measurements
Leaf 3 was cut from CYCB1;1:DBox-GUS plants at several time points in three independent biological replicates and stained as described above. Using a differential interference contrast microscope (Leica), stained meristemoids were counted in a fixed area near the leaf tip, where all normal cell proliferation had ceased on 6 to 12 leaves per experiment. Relative values (compared with control samples) were calculated for each experiment separately and averaged over the replicates.
Transgenic Lines and Mutants
Seeds of ACS5-inducible overexpressing lines were kindly provided by J. Ecker (SALK Institute, La Jolla, CA). EBF1-overexpressing plants were kind gifts of T. Potuschak (Centre National de la Recherche Scientifique, Strasbourg, France). Ethylene-insensitive mutants were obtained from the Arabidopsis Seed Stock Center (ein2.5 [N8844], ein3.1 [N8052], etr1.3 [N3070], and ein5.1 [N8053], previously annotated as ein4). All transgenic lines and mutants were in Columbia-0 background.
Accession Numbers
Microarray data were deposited in the Gene Expression Omnibus database (GSE22107). Accession numbers for the genes analyzed in Figures 6 and 7 are provided in Supplemental Table 3 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of Leaf 3 Dissected from Plants Left to Grow (No Transfer) or Transferred to Control Media at 9 DAS.
Supplemental Figure 2. Effects of Stress during a Limited Period on Proliferating Leaves.
Supplemental Figure 3. Expression of Selected Genes from Microarray Analysis Validated with nCounter Technology in Three Independent Experiments (see Supplemental Table 2 online).
Supplemental Figure 4. Differential Transcripts (FDR < 0.05) Identified by ATH1 Microarray Analysis Used to Generate Venn Diagrams (Separate for Up- and Downregulated Genes).
Supplemental Figure 5. PageMan Analysis of the Biological Processes.
Supplemental Figure 6. Representative Blot Photos Used to Quantify CDKA Activity.
Supplemental Figure 7. Ethylene Effects on Leaf Growth.
Supplemental Figure 8. Percentage of Reduction in Leaf Area of the Third Leaf of Mannitol- and ACC-Treated mkk9 Mutants versus Wild-Type Plants.
Supplemental Table 1. Comparison of Transcripts Differentially Regulated by Osmotic Stress using Publicly Available Expression Data Sets.
Supplemental Table 2. List of nCounter Probes.
Supplemental Table 3. Accession Numbers of Genes Whose Expression Was Analyzed in Figures 6 and 7.
Supplemental Data Set 1. Expression Data for the Mannitol Treatment.
Acknowledgments
We thank the Systems Biology of Yield group for the stimulating scientific atmosphere, Patrick Achard and Pascal Genschik (Institut de Biologie Moléculaire des Plantes, Strasbourg, France) for helpful discussions, Joe Kieber (University of North Carolina, Chapel Hill, NC) for kindly providing the ACS5:IOE line, Thomas Potuschak (Institut de Biologie Moléculaire des Plantes, Strasbourg, France) for kindly providing EBF1:OE seeds, Yuji Kamiya and Yusuke Jikumaru (Plant Science Center, RIKEN, Yokohama, Japan) for help with the ACC measurements, and Martine De Cock for help in preparing the manuscript. This work was supported by grants from Ghent University (“Bijzonder Onderzoeksfonds Methusalem project” No. BOF08/01M00408 and Multidisciplinary Research Partnership “Biotechnology for a Sustainable Economy” No. 01MRB510W), the Interuniversity Attraction Poles Programme (IUAP VI/33), initiated by the Belgian State, Science Policy Office, the European Community 6th Framework Programme (AGRON-OMICS; LSHG-CT-2006-037704), and the National Research Foundation of Korea (2010-0016509). H.C. is a predoctoral and S.D.B. a postdoctoral fellow of the Research Foundation-Flanders.
References
- Aguirrezabal L., Bouchier-Combaud S., Radziejwoski A., Dauzat M., Cookson S.J., Granier C. (2006). Plasticity to soil water deficit in Arabidopsis thaliana: Dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ. 29: 2216–2227 [DOI] [PubMed] [Google Scholar]
- Arvidsson S., Kwasniewski M., Riaño-Pachón D.M., Mueller-Roeber B. (2008). QuantPrime–A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster G.T.S., De Veylder L., Vercruysse S., West G., Rombaut D., Van Hummelen P., Galichet A., Gruissem W., Inzé D., Vuylsteke M. (2005). Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Methodol. 57: 289–300 [Google Scholar]
- Bergmann D.C., Sack F.D. (2007). Stomatal development. Annu. Rev. Plant Biol. 58: 163–181 [DOI] [PubMed] [Google Scholar]
- Binder B.M., Mortimore L.A., Stepanova A.N., Ecker J.R., Bleecker A.B. (2004). Short-term growth responses to ethylene in Arabidopsis seedlings are EIN3/EIL1 independent. Plant Physiol. 136: 2921–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burssens S., Himanen K., van de Cotte B., Beeckman T., Van Montagu M., Inzé D., Verbruggen N. (2000). Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta 211: 632–640 [DOI] [PubMed] [Google Scholar]
- Casneuf T., Van de Peer Y., Huber W. (2007). In situ analysis of cross-hybridisation on microarrays and the inference of expression correlation. BMC Bioinformatics 8: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman M.L., et al. (2006). SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18: 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508 [DOI] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G.T.S., Krols L., Terras F., Landrieu I., van der Schueren E., Maes S., Naudts M., Inzé D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Segers G., Glab N., Casteels P., Van Montagu M., Inzé D. (1997). The Arabidopsis Cks1At protein binds the cyclin-dependent kinases Cdc2aAt and Cdc2bAt. FEBS Lett. 412: 446–452 [DOI] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Fricke W., Akhiyarova G., Wei W., Alexandersson E., Miller A., Kjellbom P.O., Richardson A., Wojciechowski T., Schreiber L., Veselov D., Kudoyarova G., Volkov V. (2006). The short-term growth response to salt of the developing barley leaf. J. Exp. Bot. 57: 1079–1095 [DOI] [PubMed] [Google Scholar]
- Fujita M., Mizukado S., Fujita Y., Ichikawa T., Nakazawa M., Seki M., Matsui M., Yamaguchi-Shinozaki K., Shinozaki K. (2007). Identification of stress-tolerance-related transcription-factor genes via mini-scale Full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem. Biophys. Res. Commun. 364: 250–257 [DOI] [PubMed] [Google Scholar]
- Galbraith D.W., Harkins K.R., Maddox J.M., Ayres N.M., Sharma D.P., Firoozabady E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 [DOI] [PubMed] [Google Scholar]
- Geiss G.K., et al. (2008). Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26: 317–325 Erratum. Nat. Biotechnol. 26: 709 [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H., et al. (2008). The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Granier C., Inzé D., Tardieu F. (2000). Spatial distribution of cell division rate can be deduced from that of p34cdc2 kinase activity in maize leaves grown at contrasting temperatures and soil water conditions. Plant Physiol. 124: 1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C., Tardieu F. (1999). Water deficit and spatial pattern of leaf development. Variability in responses can be simulated using a simple model of leaf development. Plant Physiol. 119: 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y., Holland N., Butterfield C., Ubeda-Tomas S., Bennett M.J., Chory J., Savaldi-Goldstein S. (2011). Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A., Harter K. (2009). Mitogen-activated protein kinase cascades and ethylene: Signaling, biosynthesis, or both? Plant Physiol. 149: 1207–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y. (1988). A sharper Bonferroni procedure for multiple tests of significance. Biometrika 75: 800–802 [Google Scholar]
- Hsiao T.C., Xu L.-K. (2000). Sensitivity of growth of roots versus leaves to water stress: Biophysical analysis and relation to water transport. J. Exp. Bot. 51: 1595–1616 [DOI] [PubMed] [Google Scholar]
- Inzé D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. (2003a). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003b). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Kalantari K.M., Smith A.R., Hall M.A. (2000). The effect of water stress on 1-(malonylamino)cyclopropane-1-carboxylic acid concentration in plant tissues. Plant Growth Regul. 31: 183–193 [Google Scholar]
- Kant P., Kant S., Gordon M., Shaked R., Barak S. (2007). STRESS RESPONSE SUPPRESSOR1 and STRESS RESPONSE SUPPRESSOR2, two DEAD-box RNA helicases that attenuate Arabidopsis responses to multiple abiotic stresses. Plant Physiol. 145: 814–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen J.B. (1967). Some methods for classification and analysis of multivariate observations. In Proceedings of the 5th Berkeley Symposium on Mathematical Statistics and Probability, Le Cam L.M., Neyman J., (Berkeley, CA: University of California Press; ), pp. 281–297 [Google Scholar]
- Marcotrigiano M. (2010). A role for leaf epidermis in the control of leaf size and the rate and extent of mesophyll cell division. Am. J. Bot. 97: 224–233 [DOI] [PubMed] [Google Scholar]
- Menges M., Hennig L., Gruissem W., Murray J.A.H. (2003). Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53: 423–442 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Olmedo G., Guo H., Gregory B.D., Nourizadeh S.D., Aguilar-Henonin L., Li H., An F., Guzman P., Ecker J.R. (2006). ETHYLENE-INSENSITIVE5 encodes a 5′→3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl. Acad. Sci. USA 103: 13286–13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C., Abrahám E., Joseph M.P., Popescu C., Koncz C., Szabados L. (2008). Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol. 147: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres A., et al. (2007). Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. J. Biol. Chem. 282: 25588–25596 [DOI] [PubMed] [Google Scholar]
- Pettkó-Szandtner A., Mészáros T., Horváth G.V., Bakó L., Csordás-Tóth É., Blastyák A., Zhiponova M., Miskolczi P., Dudits D. (2006). Activation of an alfalfa cyclin-dependent kinase inhibitor by calmodulin-like domain protein kinase. Plant J. 46: 111–123 [DOI] [PubMed] [Google Scholar]
- Pierik R., Tholen D., Poorter H., Visser E.J.W., Voesenek L.A.C.J. (2006). The Janus face of ethylene: Growth inhibition and stimulation. Trends Plant Sci. 11: 176–183 [DOI] [PubMed] [Google Scholar]
- Pillitteri L.J., Sloan D.B., Bogenschutz N.L., Torii K.U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445: 501–505 [DOI] [PubMed] [Google Scholar]
- Poiré R., Wiese-Klinkenberg A., Parent B., Mielewczik M., Schurr U., Tardieu F., Walter A. (2010). Diel time-courses of leaf growth in monocot and dicot species: Endogenous rhythms and temperature effects. J. Exp. Bot. 61: 1751–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T., Vansiri A., Binder B.M., Lechner E., Vierstra R.D., Genschik P. (2006). The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell 18: 3047–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymen B., Fiorani F., Kartal F., Vandepoele K., Inzé D., Beemster G.T.S. (2007). Cold nights impair leaf growth and cell cycle progression in maize through transcriptional changes of cell cycle genes. Plant Physiol. 143: 1429–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Peto C., Chory J. (2007). The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202 [DOI] [PubMed] [Google Scholar]
- Schuppler U., He P.-H., John P.C.L., Munns R. (1998). Effect of water stress on Cdc2-like cell-cycle kinase activity in wheat leaves. Plant Physiol. 117: 667–678 Erratum. Plant Physiol. 117: 1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp R.E., LeNoble M.E. (2002). ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 53: 33–37 [PubMed] [Google Scholar]
- Skirycz A., De Bodt S., Obata T., De Clercq I., Claeys H., De Rycke R., Andriankaja M., Van Aken O., Van Breusegem F., Fernie A.R., Inzé D. (2010). Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol. 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A., Inzé D. (2010). More from less: Plant growth under limited water. Curr. Opin. Biotechnol. 21: 197–203 [DOI] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article 3. [DOI] [PubMed] [Google Scholar]
- Sobeih W.Y., Dodd I.C., Bacon M.A., Grierson D., Davies W.J. (2004). Long-distance signals regulating stomatal conductance and leaf growth in tomato (Lycopersicon esculentum) plants subjected to partial root-zone drying. J. Exp. Bot. 55: 2353–2363 [DOI] [PubMed] [Google Scholar]
- Tardieu F. (2003). Virtual plants: Modelling as a tool for the genomics of tolerance to water deficit. Trends Plant Sci. 8: 9–14 [DOI] [PubMed] [Google Scholar]
- Tardieu F., Reymond M., Hamard P., Granier C., Muller B. (2000). Spatial distributions of expansion rate, cell division rate and cell size in maize leaves: A synthesis of the effects of soil water status, evaporative demand and temperature. J. Exp. Bot. 51: 1505–1514 [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Steinhauser D., Gibon Y., Bläsing O.E., Redestig H., Sreenivasulu N., Krall L., Hannah M.A., Poree F., Fernie A.R., Stitt M. (2006). PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Veselov D.S., Mustafina A.R., Sabirjanova I.B., Akhiyarova G.R., Dedov A.V., Veselov S.U., Kudoyarova G.R. (2002). Effect of PEG-treatment on the leaf growth response and auxin content in shoots of wheat seedlings. Plant Growth Regul. 38: 191–194 [Google Scholar]
- Wang H., Qi Q., Schorr P., Cutler A.J., Crosby W.L., Fowke L.C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15: 501–510 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Kusano M., Oikawa A., Fukushima A., Noji M., Saito K. (2008). Physiological roles of the β-substituted alanine synthase gene family in Arabidopsis. Plant Physiol. 146: 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West G., Inzé D., Beemster G.T.S. (2004). Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 135: 1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li Y., Wang Y., Liu H., Lei L., Yang H., Liu G., Ren D. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283: 26996–27006 [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]