Summary
Background
Previous genetic evidence suggested that the C. elegans TGF-β Dauer pathway is responsible solely for regulation of dauer formation, with no role in longevity regulation, while the Insulin/IGF-1 signaling (IIS) pathway regulates both dauer formation and longevity.
Results
We have uncovered a significant longevity-regulating activity by the TGF-β Dauer pathway that is masked by an egg-laying (Egl) phenotype; mutants in the pathway display up to two-fold increases in lifespan. The expression profiles of adult TGF-β mutants overlap significantly with IIS pathway profiles: adult TGF-β mutants regulate the transcription of many DAF-16-regulated genes, including genes that regulate lifespan; the two pathways share enriched Gene Ontology categories; and a motif previously associated with DAF-16-regulated transcription (the DAE, or DAF-16-Associated Element) is highly overrepresented in the promoters of TGF-β regulated genes. The TGF-β Dauer pathway’s regulation of longevity appears to be mediated at least in part through insulin interactions with the IIS pathway and the regulation of DAF-16 localization.
Conclusions
Together, our results suggest there are TGF-β-specific downstream targets and functions, but that the TGF-β and IIS pathways may be more tightly linked in the regulation of longevity than has been previously appreciated.
Introduction
In times of environmental stress, crowded conditions, and limited food, juvenile C. elegans develop into an alternative larval state called dauer that is highly stress-resistant and long-lived [1]. The decision to enter the dauer state is made during the first larval stage, and is regulated by a complex branched pathway of more than 30 daf (dauer formation) genes [1]. Genetic epistasis tests suggested that components of one of the two canonical TGF-β signaling pathways (the TGF-β-like ligand DAF-7, the Type 1 and 2 receptors DAF-1 and DAF-4, and the downstream DAF-3 Smad and DAF-5 Sno/Ski) make up one branch of the dauer regulation pathway, while Insulin/IGF-1 Signaling (IIS) pathway genes, including the Insulin receptor DAF-2 and the FOXO transcription factor DAF-16, occupy a separate downstream branch regulating both dauer formation and longevity [1–3]. The TGF-β dauer pathway has not been previously implicated in the regulation of longevity.
Transcriptional analyses have been carried out on wild-type dauer larvae [4], TGF-β mutant dauer larvae [5], and adult IIS pathway mutants [6–8]. However, the transcriptional profiles of TGF-β Dauer pathway mutant adults have not yet been examined; this is important in order to temporally separate developmental and adult targets. We hypothesized that the transcriptional targets of these pathways likely share dauer-specific targets [5, 8], but should diverge significantly in adults if longevity determination is a primary output solely of the IIS pathway.
Here, we have examined the transcriptional output from the adult TGF-β pathway and compared it to the profiles of TGF-β dauer animals and adult IIS pathway mutants. Surprisingly, the adult TGF-β profiles correlated well with the IIS pathway profile, suggesting that we should re-examine the longevity phenotype of the TGF-β mutants and the relationship between the TGF-β and Insulin/IGF-1 Signaling pathways.
Results
The Adult TGF-β Dauer pathway shares transcriptional outputs and Gene Ontology categories with the Insulin-IGF-1 Receptor/FOXO pathway
To identify the downstream targets of the TGF-β pathway in adulthood, we compared the dauer-constitutive mutants daf-7(e1372), daf-7(m62), and daf-1(m40) with dauer-defective mutants daf-3(mgDf90), daf-5(e1386), and daf-7(e1372);daf-3(mgDf90) double mutants at the permissive temperature, 20°C, on the first day of adulthood (see Fig. 1 and Exp. Proc. for hybridization details). Because daf-3 and daf-5 are epistatic to daf-7 and daf-1, these comparisons should identify targets that act downstream of this linear pathway. We compared these profiles with expression data from Liu et al [5] of dauer-stage daf-7(e1372), daf-8(e1393), and daf-14(m77) mutants vs wild type L2/L3. Surprisingly, the Pearson correlation between the transcriptional outputs of the TGF-β dauer and adult pathways is low (0.004; Fig. 1a), suggesting that the downstream targets of this pathway vary significantly from dauers to adults.
Fig. 1. Hierarchical clustering and Correlations between the TGF-β dauer and Insulin/IGF-1 Signaling pathways.
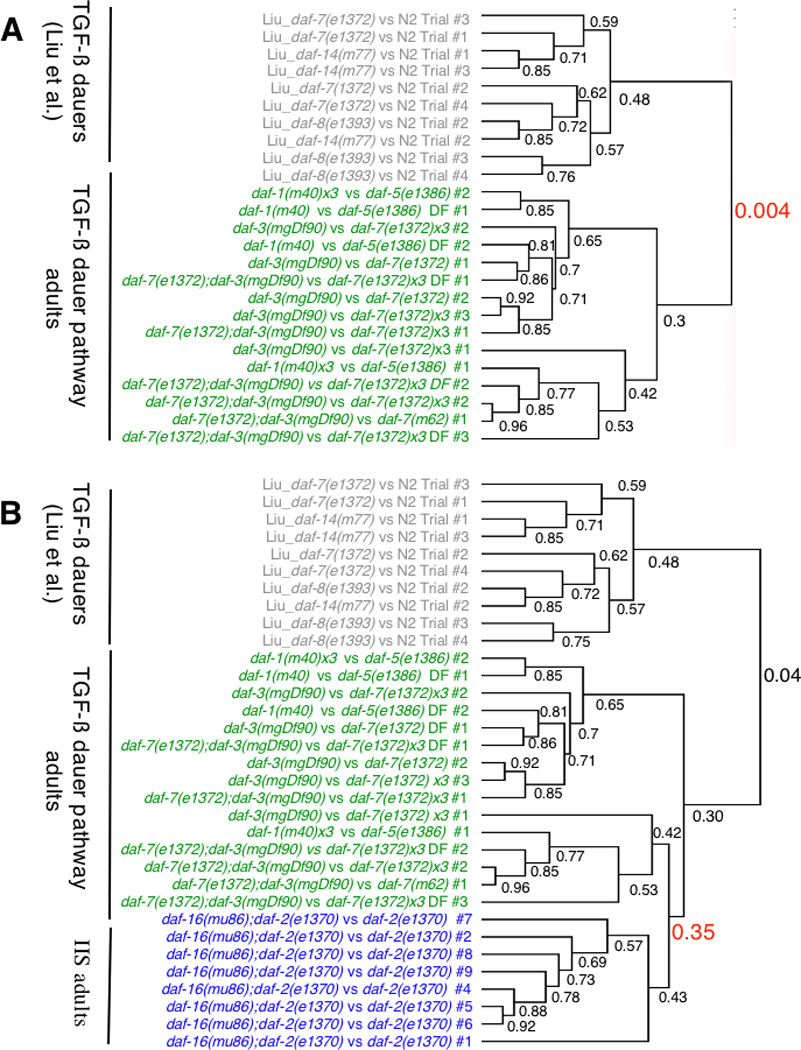
(A) TGF-β dauer stage (Liu, et al.) and TGF-β adult profiles (Pearson correlation = 0.004); (B) TGF-β adults and IIS adults (8 IIS arrays), Pearson corr. = 0.35. Note: TGF-β adults and IIS adults Pearson correlations remained high with six IIS arrays (0.42; as in Supp. Fig. 1), when only the adult profiles for the two pathways were compared (0.348), and when all of the TGF-β arrays are forced into a single clade and compared with the IIS clade (0.32).
We used SAM [9], which provides an estimate of false-discovery rate for multiple testing, to identify genes consistently and significantly changed in TGF-β adults (Supp. Table 2, Supp. Fig. 2) and compared them with the Liu, et al. dauer data [5]. As the Pearson correlation suggested, very few genes are commonly regulated (Supp. Table 1); in fact, only 14% of the upregulated and 37% of the down-regulated genes are shared between dauer and adult TGF-β pathway mutants. By contrast, the IIS pathway and the adult TGF-β pathway share significant similarity in transcriptional output, as demonstrated by a Pearson correlation of 0.35, almost 90-fold higher than the adult/dauer correlation (Fig. 1b, Supp. Fig. 1). We compared the SAM-determined significantly-regulated genes in the Insulin-IGF-1 Signaling (IIS) pathway [6], and found that 55% of the genes significantly up-regulated by the IIS pathway and 66% of the down-regulated genes are also regulated by the adult TGF-β pathway in the same direction. The transcriptional profiles of adult TGF-β and IIS pathway mutants show that these pathways share many targets in adulthood (Supp. Fig. 3).
To compare the biological roles that the genes regulated by each of these pathways might play, we submitted the lists of significantly up- and down-regulated genes from both the adult TGF-β SAM analysis and from the TGF-β dauer arrays [5] to DAVID [10] for Gene Ontology (GO) analysis. The TGF-β adults and TGF-β dauers’ most-enriched GO categories were then compared (Fig. 2; note that because GO terms are not independent, a statistical assessment of the degree of overlap is not possible, thus, we have graphically compared sets of enriched GO terms). Some categories are shared between the dauer and adult upregulated (Fig. 2a, 9 of 37) and downregulated (Fig. 2b, 14 of 47) sets. However, the majority of the top-enriched categories from the up-(28/37) and downregulated (33/47) gene sets are specific for either the adult or the dauer-stage TGF-β worms.
Fig. 2. Gene Ontology analysis of TGF-β targets in dauer and adults.
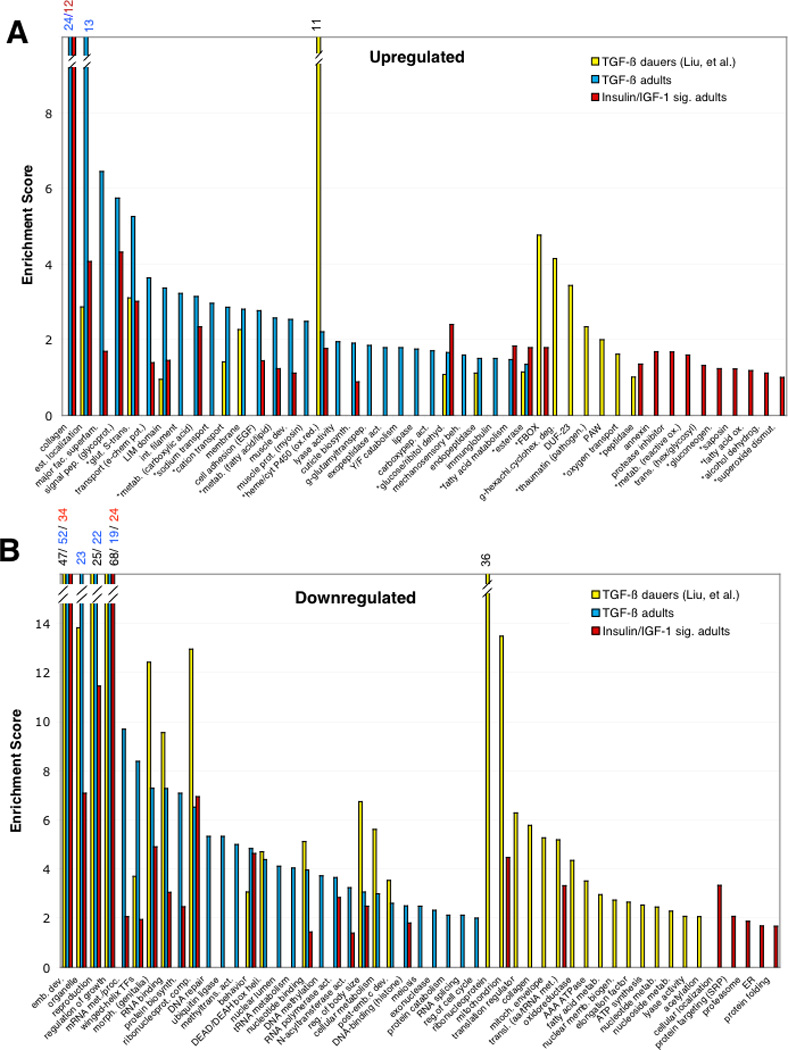
SAM-determined significantly-changed genes in TGF-β dauers (Liu, et al.), TGF-β dauer pathway adults (Supp. Table. 2), and IIS mutants were submitted for GO analysis. (A) GO categories of genes upregulated by TGF-β dauers (1381), TGF-β adults (2181 genes), and IIS mutants (1390); (B) GO categories of genes downregulated by TGF-β dauers (2725), TGF-β adults (top 3000 genes), and IIS mutants (1054). Asterisks indicate GO categories that are known to function in longevity regulation.
Comparison of the GO categories most highly enriched by the IIS and adult TGF-β pathways reveals that several of the highest-scoring GO categories in both the up- and down-regulated sets overlap: 16 of the 41 upregulated categories (Fig. 2a) and 16 of the 38 downregulated categories (Fig. 2b) are shared between the TGF-β adult and IIS profiles, which exceeds the overlap between TGF-β adult and dauer profiles. Thus, by Pearson correlation of whole transcriptome, percentage of significantly-regulated genes, and GO categories, the adult TGF-β pathway appears to have much more in common with the adult IIS pathway than might have been expected. Many of the GO categories that are known to function in longevity regulation (Fig. 2a, asterisks) [6] were enriched in the TGF-β mutant adults.
Strikingly, many of the “dod” genes (downstream of DAF-16) that are responsible for daf-2 insulin receptor mutants’ long life [6] are highly regulated by the TGF-β pathway, in the same direction as in the IIS pathway [6]. Specifically, sod-3, mtl-1, dod-3, acdh-1/dod-12, sodh-1/dod-11, lys-7, dao-3, gei-7, dod-4/aqp-1, dod-9/acs-17, ges-1, dod-7/asah-1, gcp-1, dod-6, ins-18, fat-7, mdl-1, spp-1, dod-24, dod-22, and dod-18, which had all been previously identified as lifespan-regulating dod genes [6], were notable because of their strong expression in TGF-β adults (Supp. Fig. 3b).
The DAF-16 Associated Element (DAE) is enriched in the promoters of Adult TGF-β Dauer pathway targets
We then looked for clusters [11] of genes that are up- or down-regulated and shared or distinct across the strains (Fig. 3, Supp. Table 1), similar to the approach previously used to find longevity genes regulated by daf-2 and daf-16 [6]. These comparisons allowed us to identify TGF-β-regulated genes that are specific to and shared in dauer and adulthood. We submitted the promoter sequences from the cluster gene lists to two complementary motif-finding algorithms, BioProspector [12] and Weeder [13], to identify overrepresented promoter sequences (Fig. 3). The yeast-one-hybrid-identified DAF-3 binding sequence, GTCTG, which may direct pharyngeal gene expression during larval development [14] was not apparent in any of the high-scoring motifs. By contrast, the DAE (DAF-16 associated element, CTTATCA), a GATA-like motif that was first identified as an overrepresented sequence in the promoters of genes regulated by DAF-16 [6], and variations of the DAE were the most common sequences in the promoters of these genes. While the presence of the DAE in the promoters of genes regulated by both the IIS and adult TGF-β pathway could be explained by the presence of the IIS genes, the DAE was also strongly associated with Cluster 1 (Fig. 3), which is regulated specifically by the TGF-β pathway. Several novel motifs were also associated with specific clusters.
Fig. 3. Transcriptional targets of TGF-β dauer stage, TGF-β adults, and IIS adults with WebLogos of associated motifs.

1) TGF-β adult-specific up/dauer downregulated targets; 2) TGF-β/IIS shared downregulated targets; 3) TGF-β adult/IIS shared upregulated; 4) TGF-β adult-specific down/IIS upregulated; 5) TGF-β/IIS shared upregulated; and 6) TGF-β dauer stage upregulated targets. Arrays deposited into Princeton University MicroArray database (PUMA, puma.princeton.edu). Arrays are log2 expression ratios as indicated on scale bar. 1.5 kb of promoter sequences from genes in each cluster were submitted to BioProspector [12] and Weeder [12] to identify overrepresented motifs; high-scoring motifs are depicted by WebLogo [31].
TGF-β Dauer Pathway Mutants Regulate Longevity
The striking regulation of insulin transcriptional targets previously demonstrated to regulate lifespan [6] by adult TGF-β mutants (Supp. Fig. 3b) suggested that the TGF-β pathway might also regulate lifespan, despite previously-published longevity measurements [2, 3]. To test this hypothesis, we measured the lifespans of ten different TGF-β Dauer pathway mutant alleles. Because these mutants are known to have severe egg-laying (Egl) defects [3, 15], we were concerned that matricide (progeny hatching within the mother, or “bagging”), might cause premature death. In fact, we found that daf-7 animals display high rates of bagging very early in adulthood compared with wild type (Fig. 4a). Therefore, we used the chemical 5-fluorodeoxyuridine (FUdR), an inhibitor of DNA synthesis, to prevent progeny development [16]. FUdR treatment itself has minimal effects on wild type lifespan [16] (Table 1, Supp. Tables 4–6).
Fig. 4. Longevity and thermotolerance are regulated by TGF-β Dauer pathway signaling in a DAF-3- and DAF-16-dependent manner (see Supp. Data for expanded legend including lifespan data).
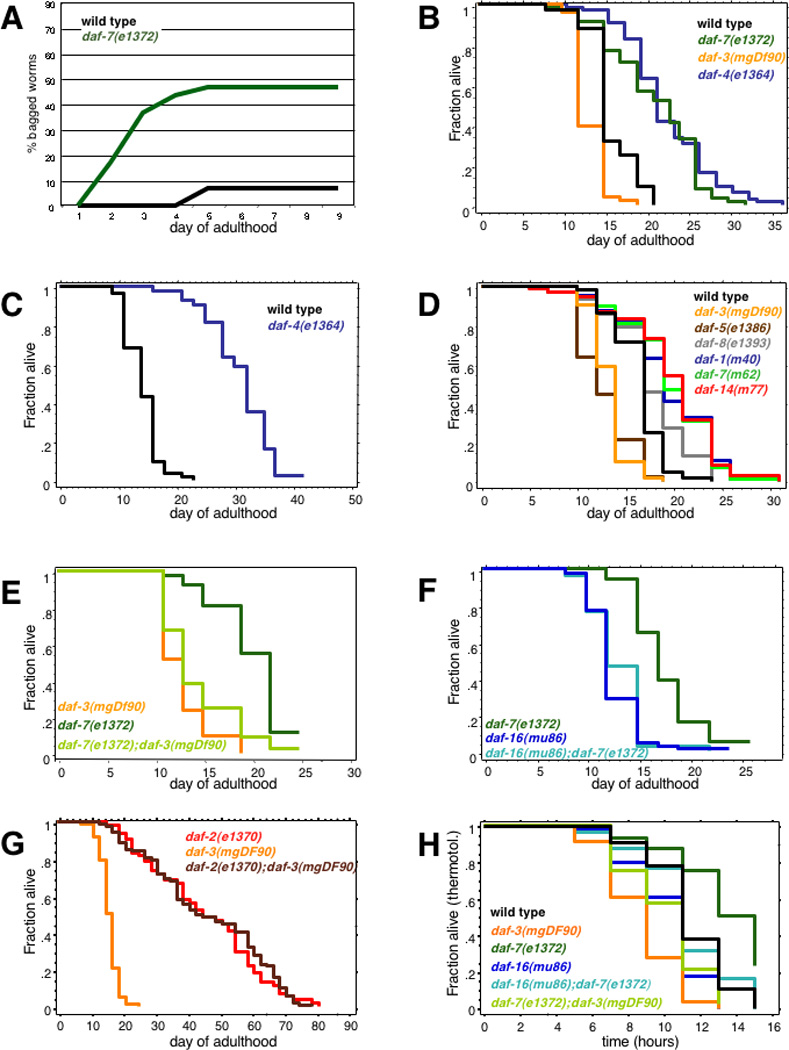
(A) daf-7(e1372) matricide rates (20°C, no FUdR). (B–G) Lifespans of hermaphrodites treated with 50 µM FUdR during early adulthood to prevent progeny development. (B, C, D) Daf-c mutants (daf-7, daf-4, daf-8, daf-1, and daf-14) are long-lived, while Daf-d mutants (daf-3 and daf-5) are short-lived. (E, F) daf-7(e1372) mutant lifespan extension is dependent on both daf-3 (E, 8 replicates, Supp. Table 4) and daf-16 activity (F, 6 replicates, Supp. Table 5). (G) daf-2 longevity is independent of daf-3 (7 replicates, Supp Table 6). (H) TGF-β mutants are thermotolerant (35°C) in a daf-3-, daf-16-dependent manner. (daf-2(e1370) was very thermotolerant in this assay, with a mean of 21.2 hours.)
Table 1. Lifespans of TGF-β Dauer pathway mutants.
Lifespan experiments were carried out at 20°C on 50 µM FUdR; n > 60 for each sample.
| Genotype | Mean LS |
Std error |
P-value | % change |
|---|---|---|---|---|
| wild type | 15.8 | 0.4 | -- | -- |
| daf-7(e1372) | 21.1 | 0.7 | <0.0001 | +33% |
| daf-4(e1364) | 22.1 | 0.6 | <0.0001 | +40% |
| daf-3(mgDf90) | 13.2 | 0.3 | <0.0001 | −16% |
| wild type | 15.7 | 0.4 | -- | - |
| daf-7(e1372) | 17.8 | 0.8 | 0.15 | +13% |
| daf-3(mgDf90) | 13.2 | 0.3 | <0.0001 | −16% |
| wild-type | 14.4 | 0.3 | -- | -- |
| daf-1(m40) | 21.0 | 0.6 | <0.0001 | +46% |
| bra-1(nk1) | 10.8 | 0.2 | <0.0001 | −25% |
| daf-3(e1376) | 14.4 | 0.2 | 0.76 | 0% |
| wild type | 13.5 | 0.3 | -- | -- |
| daf-7(e1372) | 18.8 | 0.8 | <0.0001 | +39% |
| daf-7(m62) | 16.3 | 0.5 | <0.0001 | +21% |
| daf-8(e1393) | 17.7 | 0.5 | <0.0001 | +31% |
| daf-14(m77) | 19.4 | 0.5 | <0.0001 | +44% |
| daf-3(mgDf90) | 12.0 | 0.2 | <0.0001 | −11% |
| wild type | 14.2 | 0.4 | -- | -- |
| daf-7(e1372) | 18.9 | 0.7 | <0.0001 | +33% |
| daf-7(m62) | 18.0 | 0.5 | <0.0001 | +27% |
| daf-4(e1364) | 31.0 | 0.8 | <0.0001 | +120% |
| wild type | 16.4 | 0.2 | -- | -- |
| daf-7(e1372) | 16.5 | 0.4 | 0.45 | 0% |
| daf-7(m62) | 19.7 | 0.5 | <0.0001 | +20% |
| daf-1(m40) | 19.5 | 0.5 | <0.0001 | +19% |
| daf-8(e1393) | 17.8 | 0.4 | 0.3 | +9% |
| daf-14(m77) | 19.9 | 0.6 | <0.0001 | +21% |
| bra-1(nk1) | 15.4 | 0.3 | 0.2 | −6% |
| daf-5(e1386) | 13.3 | 0.4 | <0.0001 | −19% |
| daf-3(e1376) | 15.5 | 0.4 | 0.03 | −6% |
| daf-3(mgDf90) | 13.3 | 0.2 | <0.0001 | −19% |
| wild type | 17 | 0.4 | -- | -- |
| daf-7(e1372) × 3 | 21 | 1.7 | <0.0001 | +24% |
| daf-7(m62) × 3 | 22 | 0.6 | <0.0001 | +29% |
| daf-14(m77) × 3 | 21 | 0.4 | <0.0001 | +24% |
| daf-1(m40) × 3 | 20 | 0.40 | <0.0001 | +18% |
Surprisingly, most mutants in the Dauer pathway extended lifespan substantially and consistently: we observed a significant increase in lifespan in daf-7, daf-1, daf-4, daf-8, and daf-14 mutants relative to wild type worms under the same conditions (Fig. 4b–d; Table 1). In fact, daf-4(e1364) mutants lived more than twice as long as wild type in one trial (Fig. 4c, Table 1). Meanwhile, negative regulators of the dauer pathway (bra-1, daf-5, and daf-3) displayed significant shortening of lifespan relative to wild type (Fig. 4d; Table 1). The long lifespan of daf-7 mutants was suppressed by daf-3 mutations (Fig. 4e, Supp. Table 4), similar to daf-3’s suppression of daf-7-mediated dauer formation.
Regulation of Lifespan by the TGF-β Dauer pathway requires DAF-16 activity
Previous studies suggested that the TGF-β and IIS pathways function independently [2, 3], although daf-16 mutations have been reported to partially suppress dauer formation of TGF-β mutants [3]. To investigate the interaction between the two pathways in longevity regulation, we measured the lifespan of the daf-16(mu86);daf-7(e1372) double mutant. We found that loss of daf-16 activity completely suppressed the longevity phenotype of daf-7(−) mutants or RNAi (Fig. 4f, Supp. Table 5). Furthermore, loss of daf-16 suppressed the slight thermotolerance displayed by daf-7 (Fig. 4h). By contrast, loss of daf-3 was not able to suppress the lifespan extension caused by daf-2 mutations or RNAi (Fig. 4g, Supp. Table 6), suggesting unidirectional communication from the TGF-β Dauer pathway to the IIS/FOXO pathway in longevity regulation.
The TGF-β Dauer pathway acts in adulthood to regulate longevity
Reduction of Insulin/IGF-1 (IIS) pathway signaling during adulthood is sufficient to increase longevity [17], but the dauer decision is made in the L1 larval stage [1]. To determine when TGF-β pathway activity affects lifespan, we carried out a series of temporal temperature-shift and double-stranded RNA interference experiments. When we raised daf-7(e1372)ts or daf-7(m62)ts animals at 20°C and then shifted them to 15°C as young adults, no lifespan extension was observed (Fig. 5b). By contrast, when these worms were raised at 15°C and then shifted to 20°C as L4/ young adults, lifespan was increased significantly (p < 0.0001) (Fig. 5c). Furthermore, when wild type worms were treated with daf-7 RNAi only in adulthood, lifespan was also increased (p < 0.0001) (Fig. 5d). Thus, like the insulin pathway, the TGF-β pathway acts during adulthood rather than in larval stages to regulate longevity, suggesting that this activity is separable from the dauer decision- and formation-requiring daf-7 activity in larval stages.
Fig. 5. Temporal Analysis of daf-7 Longevity Regulation.
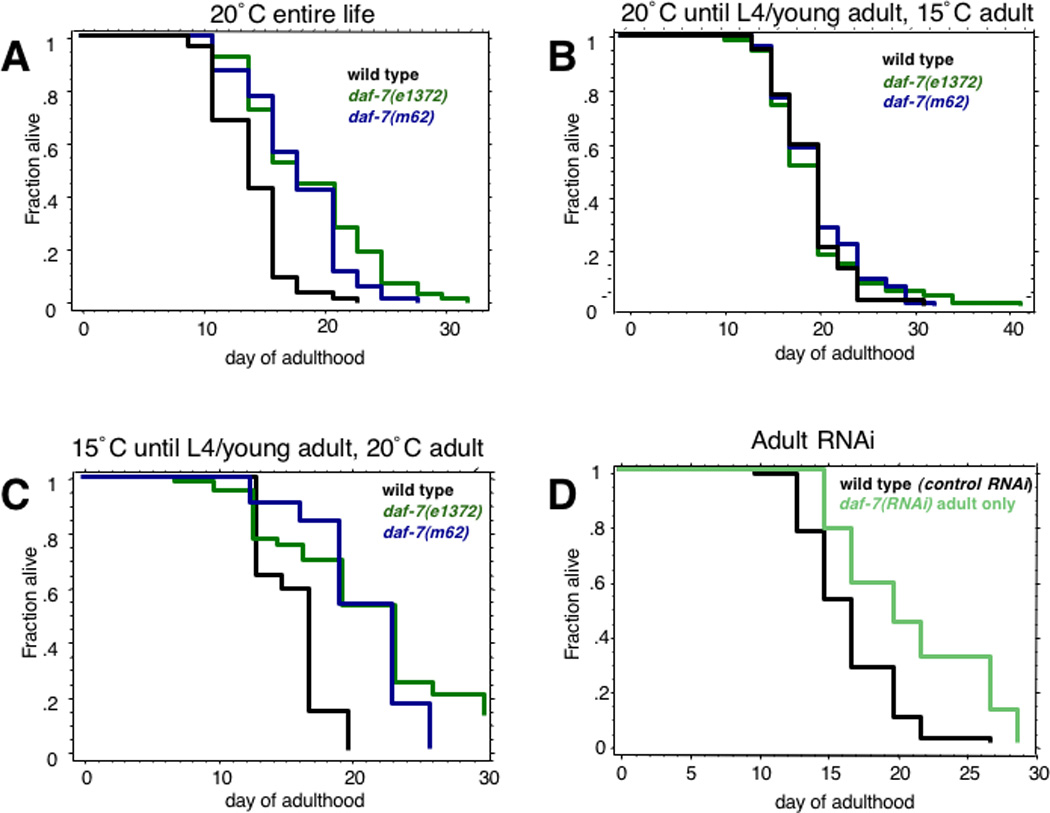
(A) Lifespan of wild type (14.2 ± 0.4), daf-7(e1372) (18.9 ± 0.7, p<0.0001), and daf-7(m62) (18 ± 0.5, p<0.0001) worms at 20°C their entire life. (B) Lifespan of wild type (20.1 ± 0.5), daf-7(e1372) (20.3 ± 0.6, p=0.9), and daf-7(m62) (20.8±0.5, p=0.9) worms raised at 20°C until L4/young adulthood then shifted to 15°C. (C) Lifespan of wild type (15.9 ± 0.5), daf-7(e1372) (21.6 ± 0.9, p<0.0001), and daf-7(m62) (20.8 ± 0.5, p<0.0001) worms raised at 15°C until L4/young adulthood, then shifted to 20°C. (D) Lifespan of wild type worms treated with vector control (16.7 ± 0.4), or daf-7 RNAi during adulthood only (21.1 ± 1.1, p<0.0001).
The TGF-β Dauer pathway regulates DAF-16 localization and DAF-16 target gene transcription
The suppression of daf-7 mutants’ extended lifespan by daf-16 mutations suggests that the TGF-β Dauer pathway regulates IIS pathway activity in adults. When daf-2 insulin receptor signaling is abrogated, DAF-16/FOXO becomes nuclearly localized [18, 19], and mutations in daf-18, the PTEN phosphatase that opposes insulin/PI-3-kinase signaling, promote cytoplasmic retention of DAF-16 [18]. daf-7(m62) dauer animals were previously shown to nuclearly localize DAF-16 during the L2d pre-dauer state [20]. We found that DAF-16::GFP was excluded from nuclei in many adult daf-3 mutants (Fig. 6a–c) and with daf-3 RNAi, similar to daf-18 mutants [18]. daf-7(e1372);daf-16::gfp adults were also heterogeneous, but generally shifted DAF-16::GFP localization from diffuse to nuclearly localized (Fig. 6a–c). These results suggest that the TGF-β Dauer pathway may act through an insulin-signaling-like regulation of DAF-16 localization.
Fig. 6. The TGF-β Dauer pathway regulates DAF-16 localization and sod-3 transcription.
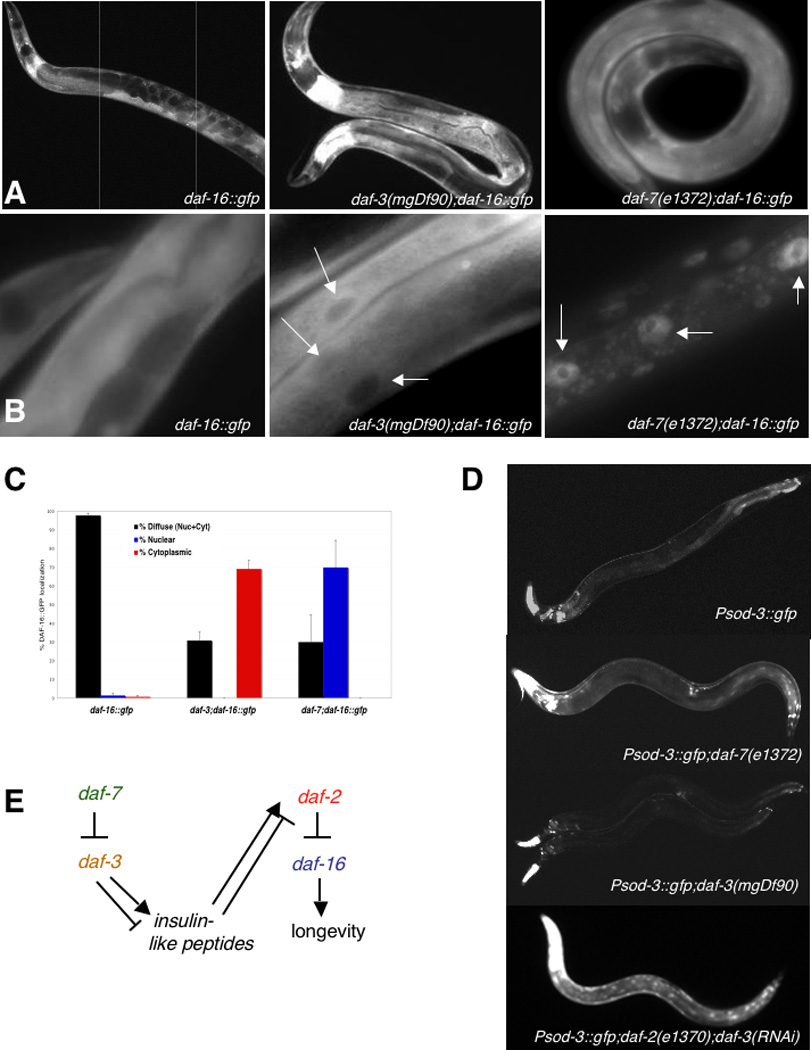
(A) 100× and (B) 400× images of DAF-16::GFP animals. DAF-16::GFP remains diffuse in a wild-type background, but is excluded from nuclei in many daf-3 animals, and is partially nuclearly localized in many daf-7 mutants. (C) Ratios of animals with diffuse (black), cytoplasmic (red), and nuclearly-localized (blue) DAF-16::GFP in wild type and daf-7(e1372) and daf-3(mgDf90) mutant backgrounds. (D) Psod-3::gfp, Psod-3::gfp;daf-7(e1372), Psod-3::gfp;daf-3(mgDf90), and daf-2(e1370);Psod-3::gfp animal treated with daf-3 RNAi. (E) Model of TGF-β Dauer and IIS pathway interactions regulating longevity.
sod-3 (superoxide dismutase) is a direct transcriptional target of DAF-16 [21, 22], and the expression of a Psod-3::gfp reporter is increased broadly in daf-2 mutants [23]. Our microarray results suggested that like the IIS pathway, TGF-β Dauer mutants regulate the transcription of sod-3 (Fig. 3c). We examined daf-7(e1372);Psod-3::gfp mutants and found that fluorescence was increased in intestines, hypodermis, and cuticle, although this pattern was weaker and more heterogeneous than in daf-2 mutants. Psod-3::gfp expression was not noticeably altered below wild type levels in daf-3(mgDf90) mutants (Fig. 6d), where expression is already low and restricted to the pharynx and tail. daf-3 RNAi did not have a notable effect on the high Psod-3::gfp expression in daf-2 mutants, correlating with its lack of daf-2 longevity suppression. Together these results suggest that the daf-2/daf-16 pathway acts downstream of daf-7 and daf-3.
Discussion
The TGF-β pathway regulates longevity through Insulin/IGF-1 Signaling
Our transcriptional analyses of adult TGF-β mutants have identified a number of adult transcriptional targets, functional categories, and regulatory motifs [5][6] that are shared between the TGF-β and IIS pathways, including many daf-2/daf-16 target genes demonstrated to regulate longevity [6]. These transcriptional results directly suggested the possibility that TGF-β mutants might live long. Supportive of this, we found that TGF-β mutants are indeed long-lived, that the TGF-β pathway acts during adulthood to regulate lifespan, and that this regulation depends on the FOXO transcription factor DAF-16. Together, our results suggest that the TGF-β and Insulin/IGF-1 pathways use similar transcriptionally-regulated mechanisms to survive, and that these pathways are more closely linked than previously appreciated.
One interesting difference between dauer and longevity regulation is that loss of daf-16 is only able to partially suppress daf-7 dauer formation [3], but completely suppresses the lifespan extension of daf-7, which is weaker than daf-2’s longevity effect. This may indicate a greater role by DAF-3 in dauer formation and a more specific role for DAF-16 in longevity regulation. Nevertheless, DAF-16 may play at least a partial role in the regulation of daf-7 dauer transcriptional targets and the partial suppression of dauer formation.
Lee, et al. also noted daf-7 regulation of DAF-16::GFP in L2d animals [20], and suggested that an insulin might mediate the interaction between the TGF-β and IIS/FOXO pathways, while Liu, et al. found several insulins regulated by TGF-β mutants in dauer [5]. The regulation of an insulin-like peptide gene or genes by DAF-3 in adulthood (Fig. 6e, Supp. Fig. 3) would provide a powerful mechanism for the activation of the IIS pathway and the coordination of the two pathways, similar to the proposed coordination of the IIS pathway in the whole animal through ins-7 signaling [6]. An insulin-based mechanism might allow the coordination of the TGF-β and IIS pathways throughout the animal, reinforcing the dauer decision during larval stages and longevity regulation in adults.
Matricide may significantly mask longevity phenotypes of TGF-β Dauer mutants
Our finding that the TGF-β dauer pathway can regulate lifespan through its interaction with the Insulin/IGF-1 pathway suggests that dauer survival and longevity regulation may be even more closely linked than earlier genetic studies had indicated. While it is surprising that such significant lifespan increases had not been previously reported for any of the mutants in this pathway, it should be noted that some studies had not prevented progeny production to reduce matricide. In other studies, fer mutations were used to confer partial sterility (due to the loss of sperm progeny production); however, we have observed that fer mutations may also have a slightly deleterious effect on lifespan in TGF-β mutant backgrounds (Supp. Fig. 4). Previous studies also measured lifespan at 25.5°C, which may have caused more severe deleterious effects, either from TGF-β dauer mutant or fer pleiotropies, than the temperature we have used here (20°C). Finally, complete deletion of bagged animals from the entire lifespan, rather than standard survival analysis censoring (in which the animal contributes to the “live” and total population until the point at which it is removed), was used in the calculation of lifespan in one study [2], and the second did not censor for bagging events after day 7 [3], when we continue to observe matricide (Fig. 4a). It is also not surprising that RNAi longevity screens did not pick up these genes, despite the use of sterile strains [24–27], because the daf-7 RNAi clone appears to act more weakly than does the mutant allele and might not have met the maximum longevity requirement of these screens (W. Shaw and C. T. Murphy, unpublished). Interestingly, another study did identify daf-7 and daf-1 in tests of L1 starvation-induced increased longevity [28], and noted their increased thermotolerance as well, but discounted them for further adult longevity tests based on previously-published lifespan reports. In general, it is possible that matricide contributes to deaths more frequently than is currently appreciated, and the suppression of matricide may reveal additional longevity regulators.
Implications for higher organisms
The conservation of TGF-β and insulin signaling pathways between C. elegans and mammals is significant. It is possible that more interaction between these pathways than has been previously suspected may also exist in higher organisms, and may affect the survival of specific tissues and ultimately, longevity of these animals.
Experimental Procedures
C. elegans genetics
All strains were cultured at 20°C unless otherwise noted using standard methods [29]. Strain and allele information listed in Supp. Data.
RNA preparation and Microarray hybridization
Standard RNA purification, cRNA generation, labeling, and hybridization on Agilent 4 × 44K C. elegans arrays were performed (Supp. Data). 15 replicates of TGF-β adults were hybridized (4 × daf-1 v daf-5 (2 dye-flipped), 10 × daf-7(e1372) v daf-3(mgDf90) or daf-7(e1372);daf-3(mgDf90) (4 dye-flipped), 1 × daf-7(m62) v daf-7(e1372);daf-39(mgDf90). Nine replicates of daf-2(e1370) v daf-16(mu86);daf-2(e1370) (4 dye-flips) were hybridized.
Microarray Analysis
Data was loaded onto the Princeton University MicroArray database (PUMA, puma.princeton.edu), filtered for array and spot quality, and replicate spots were collapsed to an average value. One-class analysis in SAM [9] was performed with no fold-expression cutoff. Hierarchical clustering by gene and arrays of log2 expression rations was done after filtering for genes that were present in 80% of the arrays (uncentered correlation, average linking) in Cluster and displayed in TreeView [11].
Gene Ontology analysis
Up-and down-regulated genes identified in the SAM analyses were converted to SwissProt ID and TrEMBLE ID’s in wormbase and submitted to DAVID for conversion to DAVID ID’s for functional annotation clustering (>70% converted). Molecular/biological functional annotation was assigned to the upregulated and downregulated genes using the default C. elegans Gene Ontology criteria in DAVID (http://david.abcc.ncifcrf.gov/), and Enrichment Score (the −log of the Fisher Exact test p-value) was determined [10]. All categories with an enrichment score >1 or the top 30 GO categories are shown (Fig. 2).
Motif analysis
1.5 kb upstream sequences (WormBase Release WS170) of were submitted to two complementary algorithms, BioProspector [12] (a Gibbs sampling-based algorithm) and Weeder [13] (a consensus-based algorithm), to identify overrepresented sequences. p-values for BioProspector motifs were generated by performing thirty-five Monte Carlo simulations to estimate the null motif score distribution [12]; Weeder’s p-value calculator was used for motif significance. Motifs were displayed in WebLogo [30].
Survival analysis
Standard Kaplan-Meier survival analysis (1st day of adulthood as t=0, 20°C unless otherwise indicated, n>60, 50 µM FUdR) was performed (Supp Data).
RNAi
Bacterial feeding RNAi experiments were carried out as described previously [32][17] after verification by PCR and sequencing, on 0.1 M IPTG.
Thermotolerance assay
n> 60 Day 1 adult worms (treated with FUdR from L4) were shifted from 20°C to 35°C at t=0, and time points were taken every 2 hours after 3 hours of incubation at 35°C; standard Kaplan-Meier survival analysis was performed.
DAF-16::GFP localization assay
Day 1 adult worms were scored (3 assays) for nuclear, cytoplasmic, or diffuse GFP localization, as in [33], and plotted as a percentage of the total, with SEP.
Supplementary Material
Acknowledgments
We thank the Center for C. elegans Genetics for strains, John Matese and Laurie Kramer for assistance with PUMA, Hani Goodarzi, Max Jan and Kieran Dilks for technical assistance, Zemer Gitai for comments on the manuscript, and Chad Myers for discussion. WMS and CTM designed the experiments, WMS, SL, JA, and CTM carried out the experiments, JL performed motif analyses, and WMS, SL, JL, and CTM wrote the manuscript. WMS is a fellow of the New Jersey Commission on Cancer Research. CTM is an Alfred P. Sloan Fellow and a Pew Scholar in the Biomedical Sciences.
References
- 1.Riddle DL, Albert PS. Genetic and Environmental Regulation of Dauer Larva Development. In: Riddle DL II, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans. Cold Spring Harbor Laboratory Press; 1997. pp. 739–768. [PubMed] [Google Scholar]
- 2.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 3.Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development (Cambridge, England) 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- 5.Liu T, Zimmerman KK, Patterson GI. Regulation of signaling genes by TGFbeta during entry into dauer diapause in C. elegans. BMC Dev Biol. 2004;4:11. doi: 10.1186/1471-213X-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 7.McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 8.McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- 9.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- 11.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Brutlag DL, Liu JS. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pacific Symposium on Biocomputing. 2001:127–138. [PubMed] [Google Scholar]
- 13.Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic acids research. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thatcher JD, Haun C, Okkema PG. The DAF-3 Smad binds DNA and represses gene expression in the Caenorhabditis elegans pharynx. Development (Cambridge, England) 1999;126:97–107. doi: 10.1242/dev.126.1.97. [DOI] [PubMed] [Google Scholar]
- 15.Trent C, Tsuing N, Horvitz HR. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics. 1983;104:619–647. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gandhi S, Santelli J, Mitchell DH, Stiles JW, Sanadi DR. A simple method for maintaining large, aging populations of Caenorhabditis elegans. Mech Ageing Dev. 1980;12:137–150. doi: 10.1016/0047-6374(80)90090-1. [DOI] [PubMed] [Google Scholar]
- 17.Dillin A, Crawford DK, Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- 18.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nature genetics. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 19.Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 20.Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 21.Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- 22.Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nature genetics. 2005 doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 23.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 24.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes & development. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen M, Hsu AL, Dillin A, Kenyon C. New Genes Tied to Endocrine, Metabolic, and Dietary Regulation of Lifespan from a Caenorhabditis elegans Genomic RNAi Screen. PLoS Genet. 2005;1:e17. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SS, Lee RYN, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature. 2002 doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 28.Munoz MJ, Riddle DL. Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics. 2003;163:171–180. doi: 10.1093/genetics/163.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawless JF. Models and Methods for Lifetime Data. New York: Wiley; 1982. [Google Scholar]
- 32.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 33.Hertweck M, Gobel C, Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


