Abstract
In the present work, we have measured the messenger RNA expression of specific genes both from total RNA and cells encapsulated in droplets. The microfluidic chip introduced includes the following functionalities: RNA∕cell encapsulation, lysis, reverse transcription and real-time polymerase chain reaction. We have shown that simplex and duplex gene expression measurements can be carried out over a population of 100 purified RNA samples encapsulated simultaneously in 2 nl droplets in less than 2 h. An analysis of 100 samples containing one to three cells has shown excellent consistency with standard techniques regarding average values. The cell-to-cell distributions of the E-cadherin expression suggest fluctuations on the order of 80% in the number of transcripts, which is highly consistent with the general findings from the literature. A mathematical model has also been introduced to strengthen the interpretation of our results. The present work paves the way for the systematic acquisition of such information in biological and biomedical studies.
INTRODUCTION
In functional biology, the widespread existence of cell-to-cell variations of gene expressions in genetically homogeneous populations, both in a rest state and when exposed to stimuli, raises the challenge of quantifying expressions at the single-cell level and, preferably, for large cell populations.
This challenge was being met by the biological community once appropriate sensitive tools for molecular biology became available. Most experiments conducted over the past 20 years have focused on determining protein expression profiles of single cells using fluorescent reporters, immunoassays, or cell sorters. A wealth of information has been acquired with these techniques, including estimates of noise level in the expression,1 an evaluation of gene regulation functions,2 and a demonstration of the bursty nature of translation.3 Some limitations inherent to these approaches can be found, with the most important one being an inability to investigate weakly expressed proteins, due to the moderate sensitivity built into the technique. To overcome such limitations, another approach consists of measuring the number of transcripts in each cell, through the use of the reverse transcription real-time polymerase chain reaction (RT-qPCR) technique. The extreme sensitivity associated with the PCR-based approach allows quantifying the gene expression of cells down to a few transcripts. This approach requires inputting the number of biological reactions to be performed (i.e., cell lysis, RT, and real-time qPCR), which, in turn, needs the cells to be isolated in capillaries or chambers and exposed to lytic agents, primers, fluorescence quenchers, etc. In practice, this step necessitates being able to handle volumes on the order of 1 μl or less, in which a single cell may be statistically isolated and where lysis and RT-qPCR reactions may be processed. Although recent contributions have demonstrated that this step can be achieved by manual manipulation of capillaries,4, 5, 6 microfluidic technology obviously appears to be well adapted to perform this task, as a result of the small reaction volumes typically handled and a natural capacity for automation and parallelization.7
The power of microfluidics for single-cell measurements has already been demonstrated: cells were isolated in microfluidic chambers in order to perform mRNA analysis8 or collect data on cell expression levels once the genes had been reverse-transcribed and externally preamplified.9, 10 Information regarding cell-to-cell expression variability at various ages of development11 as well as RT performance in nanoliter volumes12 could be obtained. In this domain, droplet-based microfluidic technology seems to be particularly attractive as a result of naturally generating reservoirs capable of isolating individual cells and having the capability to handle an extremely large number of droplets, as recently found,13, 14, 15 thus creating a path to the high-throughput analysis of large populations of cells or highly resolved statistics. A droplet-based microfluidic approach applied to the measurements of single RNA virions was recently demonstrated.16 As part of the present work, the content of RNA virions, at the single-virus level and in a simplex mode, was analyzed for a population of 100 virions. Although the microfluidic approach offers considerable potential for single-cell studies, the demonstration that this technology is capable of quantitatively analyzing the single-cell expression, at the single-cell level and under high-throughput conditions, still lies ahead. In this context, it is important to operate in a duplex mode so as to compare the target gene with a reference gene. This step has yet to be performed. We must also concentrate on cell samples rather than purified RNA samples in order to determine whether the RT-qPCR amplification process can be affected by interactions with cell constituents within the small droplet. We have taken a step forward on these issues by measuring the expression of two genes simultaneously in an RNA sample and then analyzing the outcome on the chip of RT-qPCR amplification performed directly in the droplets. In this paper, we will thus provide averaged measurements of the threshold cycle and data on the cell-to-cell variability for 100 cells in a single run. This work paves the way for characterizing biological samples, at the single-cell level, under high-throughput conditions.
RESULTS AND DISCUSSION
Performance of the mRNA reverse transcription followed by real-time polymerase chain reaction in microdroplets
Our microfluidic device consisted of a flow-focusing junction (see Fig. 1), where 2 nl droplets (containing RT-PCR reagents and the RNA or cell solution) were formed in oil and stopped in the device. After immobilizing the droplets, the RT-PCR thermal cycles were applied to the device and fluorescence images were captured at the end of each PCR cycle (see the discussion in Sec. 3E). The different biochemical steps occurring in droplets are depicted in Fig. 1c.
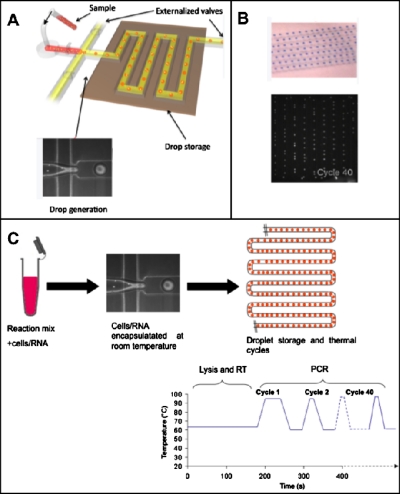
Figure 1.
(a) Schematic view of the droplet-making device. Droplets were formed at a flow-focusing junction and subsequently stored in the array of channels. (b) Brightfield image of aqueous droplets stained with a food dye (top) stored in the channels and fluorescent image of droplets containing an RT-PCR mixture and RNA, stored in the channels, after 40 PCR cycles (bottom). (c) Schematic representation of the different steps occurring in droplets.
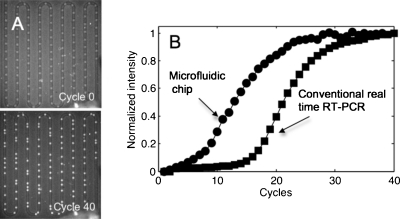
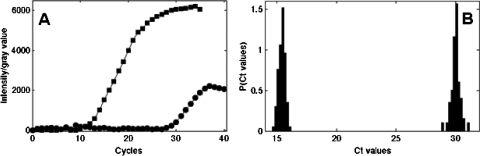
We studied the efficiency and reproducibility of the microfluidic RT-qPCR chip by considering the gene β-actin using total RNA samples at concentrations ranging from 0.04 to 20 pg∕droplet. The intensity curves obtained in a 10 μl tube inside the thermocycler Lightcycler 480 (Roche Applied Science), in addition to the microdroplets on a chip at the same concentration (10 pg∕nl), are shown in Fig. 2b. Figure 2a displays the corresponding fluorescence images of the droplets, from which the round symbols in Fig. 2b are derived. The various curve amplification phases are identical according to both approaches; however, the threshold cycle (Ct) is shorter with the microfluidic chip than with the bulk technique. Two factors explain this behavior: first, the detection device used on the chip is more sensitive than the thermocycler and, second, miniaturization favors higher efficiency in the RT reaction.12
Figure 2.
(a) Fluorescence images of the on-chip droplet parking at the initial time (cycle 0, top) and at cycle 40 (bottom). (b) Comparison between an amplification curve of reverse-transcribed tRNA obtained for the same concentration (10 pg∕nl) in a conventional real-time PCR device operating with 10 μl of sample (square symbols) and a curve derived using microfluidic droplet-based technology (round symbols, corresponding to a 2 nl droplet).
Let us now analyze the RNA sample reaction performance in greater detail. Observations indicating the evolution in real-time PCR signal as a function of RNA concentration are presented in Fig. 3a. The negative control (PCR mix emulsified without a template) shows that the fluorescence signal remains negligible within the range located below 35 cycles; beyond this value, the intensity increases, thus suggesting that a partial hydrolysis of the FRET qPCR probes has occurred. As would be expected, Ct increases as concentration decreases. The corresponding evolution in the threshold cycle versus RNA concentration is depicted in Fig. 3b. A linear dependence of Ct on the log of RNA concentration is obtained. For real-time PCR, the following classical formula can be written:
| (1) |
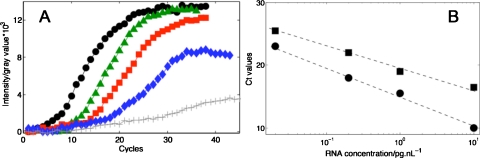
where E denotes PCR efficiency, C is the total RNA concentration, and CB is the number reflecting experimental noise (quantitatively speaking, CB is the ratio of the concentration of synthesized cDNA when the number of cycles reaches Ct to the RT efficiency). This expression fits the measurements well. In our system, we have estimated CB=2.3×104 pg∕nl and E=1.8, i.e., for a 90% efficiency rating. Interestingly, the microfluidic data lie substantially above the findings from the tube technique [see Fig. 2b]. The microfluidic method is therefore more sensitive than the benchtop real-time PCR instrument. The difference between these two methods is significant: we reached an 8-cycle shift between the two methods, which represents two orders of magnitude in terms of RNA concentration. The origin of this remarkable improvement is likely due to the highly sensitive camera we installed, offering reduced noise compared to the CCD camera fitted on the Lightcycler 480. RT efficiency also makes a contribution perhaps, given that efficiency tends to increase as volume shrinks.12 Upon closer inspection of the curve in Fig. 3a, the detection limit of our system can be estimated for the case of total purified RNA and β-actin amplification: 0.02 pg∕nl total RNA, which means that our detection limit for purified RNA is far less than the equivalent tRNA content of a cell (10 pg).
Figure 3.
(a) Real time amplification curve of β-actin transcripts from total universal RNA at various concentrations. The curves from left to right correspond to 0, 0.02, 0.2, 1, and 10 pg/nl of RNA. (b) Evolution in Ct values vs the RNA concentration obtained in microdroplets (round symbols) and, in a conventional manner, in the Lightcycler 480.
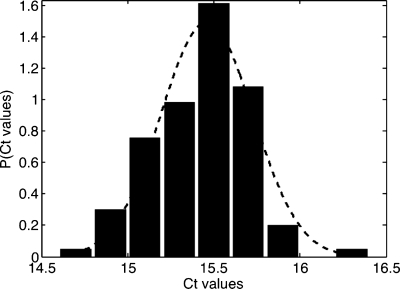
A distribution of the threshold cycles for β-actin at a concentration of 1 pg∕nl is shown in Fig. 4. It is apparent that the Ct is peaked around a well-defined value, equal to 15.4±0.3. By assuming that the number of copies is proportional to the total RNA concentration and by using relation 1, we can estimate the corresponding coefficient of variation for the number of mRNA copies through applying the formula: CV=δCT log E, where δCT is the standard deviation of the threshold cycle distribution. A CV value on the order of 20% is derived for the number of copies; this value provides an estimate of mRNA copy measurement accuracy using the microfluidic technique. This uncertainty has many possible sources, such as variations due to sampling statistics, variability in droplet sizes, noise inherent to the image analysis process, and errors generated during RT and PCR reactions.1, 17 A detailed analysis of the various contributions appears to be a challenging task at present.
Figure 4.
Ct distribution obtained from 50 droplets for a concentration of 1 pg∕nl of tRNA.
Duplex gene expression measurements
In real-time RT-qPCR, renormalization is an important step in quantifying the expression of a targeted gene since it removes nonbiological variation from the measurements. The preferred normalization procedure currently consists of measuring the expression of reference genes.18 Using an image splitter, it is possible to simultaneously record images at two different optical wavelengths; duplex experiments have been performed on β-actin and TATA-binding protein (TBP) genes with an initial tRNA concentration of 1 pg∕nl [Fig. 5a]. Even though β-actin is shown to be differentially expressed in certain tumor cells,19 it is still a common and historically used reference gene. In this case, the RNA was extracted from a mixture containing different cells; the working concentration ensures the presence of a large number of RNA molecules along with a homogeneous representation per droplet. We have simultaneously measured the distribution of β-actin and TBP for 50 droplets; the corresponding distributions are presented in Fig. 5b. The standard deviations of the Ct distributions for both genes are on the order of one cycle and are similar to those obtained in simplex. The difference between the Ct of genes in the device, 15, is consistent with the typical value obtained in the Lightcycler.
Figure 5.
(a) Typical real-time duplex PCR curves of β-actin (square symbols) and TBP (round symbols) obtained for a concentration of 1 pg∕nl of tRNA. (b) The corresponding Ct distribution obtained from 50 droplets.
Analysis of cell-to-cell variability
Let us now consider the case where cells are introduced in the droplets; the encapsulation process follows the Poisson statistics:20, 21
| (2) |
where k denotes the number of cells in the drop and λ is the average number of cells per drop.
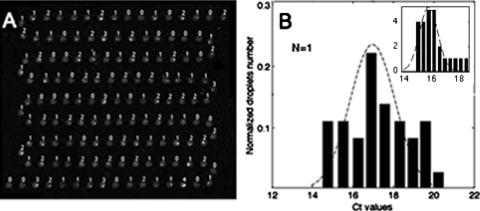
The distribution of the number of cells per drop (k) was found in all cases to be in good agreement with a Poisson distribution.22 Since the cell number distribution per droplet is broad,22 it is important to determine the number of cells for each droplet in order to establish quantitative estimates of expression variability. The gene studied herein is E-cadherin, and the MDCK cells used express the histone 2B-red fluorescent protein (H2B-RFP) fusion gene. Consequently, cells can be visualized by fluorescence and counted in each droplet. A reconstructed fluorescence image of the entire device before the RT is shown in Fig. 6a; these images were shot with a 20× lens at different locations in the system and then assembled into a single image. With 130 droplets containing random numbers of MDCK cells, lying between 0 and 3, we selected the 30 droplets encapsulating single cells as being relevant for the study of cell-to-cell variability. The distribution of threshold cycles obtained with this subpopulation of droplets is depicted in Fig. 6b. The distribution is clearly broader than with RNA samples. By analyzing the distributions in Fig. 6, it is found that the standard deviation of Ct distributions is on the order of 1.7 cycles. This estimate, when converted into transcript numbers, leads to CVs on the order of 80%, i.e., a factor four times higher than the fluctuation level measured for RNA samples.
Figure 6.
(a) Fluorescence image of the droplets, with the number of cells being shown for each droplet. (b) Ct distributions for droplets containing a single cell: the experiment corresponds to bars and the dashed line is a Gaussian fit. (Inset) Ct distributions of single cells obtained with the numerical model (1) (see supplementary materials) for a cell distribution in the droplets identical to the experiment and a total of 100 droplets. The parameters used in the model are D=0.3 and CR=1; in the model, CmRNA(x) is a random number whose standard type deviation is 80%. Assuming a broad distribution of CmRNA(x) is the only way to match the broad distributions obtained experimentally. The dashed line is the Gaussian fit.
In order to interpret these observations on a quantitative basis and assess the experimental uncertainties, we have developed a model assuming that droplets contain, in addition to the cells, a homogeneous concentration of RNA resulting from cell debris appearing during sample preparation.22 Moreover, we have taken into account potential entity transfers (fluorescent probes or amplicons) between the cells and their neighbors (droplet-droplet diffusive transfers may induce intensity changes over time, which, in turn, favor a broadening of threshold cycle distributions23, 24). We have made further assumption that PCR efficiency is homogeneous. This model is described in the supplementary material (see “Theoretical model”).22 Specifically, the model is based on a one-dimensional diffusion equation with a time diffusion constant on the order of one cycle. This time constant, which is several orders of magnitude less than a diffusive time based on the interdroplet distance, should provide an upper estimate for possible interdiffusion exchanges between droplets. The model also includes a source term that represents the amplified cDNA present in droplets originating from the randomly distributed cells in the droplets along with (homogeneously distributed) debris from cells lysed during sample preparation prior to their encapsulation [see Eq. (1) of the supplementary material22]. With this model, we are able to quantify the effects of the debris and interdiffusion; a typical distribution of Ct, obtained for 30 isolated cells, taken among 100 droplets, is shown in the inset of Fig. 6b. We obtain a qualitative agreement between the experiment and the model. The model allows one to analyze the various contributions to the Ct distribution separately: concerning interdiffusion, we found that the resulting variability in threshold cycles does not exceed ±0.5 cycles, i.e., a CV of 30% of the number of transcripts. The reason originates from the fact that diffusion is much slower than the PCR amplification process (and therefore its contribution on the Ct variability is weak). On the other hand, debris are homogeneously spread in the droplets and therefore do not contribute to the broadening of the Ct distributions. All this suggests that the main origin of the broad threshold cycle distribution is likely to be due to the intrinsic cell-to-cell variability in the number of transcripts. Using this model, we are able to suggest that the fluctuation in number of transcripts per cell for E-cadherin has a CV on the order of 80%, the other contributions playing a neglecting role, provided that PCR efficiency is homogeneous throughout the droplet population. Nonetheless, owing to the oversimplified nature of the model, this information should be taken as semiquantitative. The estimate of the fluctuation in number of transcripts we obtain is nonetheless consistent with the general estimate of cell-to-cell fluctuations reported in literature, albeit for other genes.1, 25, 26, 27
EXPERIMENTAL PROCEDURE
Device fabrication
Microfluidic chips were produced by means of soft imprint technology on a PDMS mold using UV-polymerizable materials.28 A PDMS stamp was made by replica molding an SU8-mold patterned by UV-lithography. A UV-curable glue (NOA 81, Norland) was applied between the stamp and a PDMS flat and then cured by a uniform 365-nm illumination (LC8 Lamp Hamamatsu). After curing, the PDMS stamp was carefully removed and replaced by a glass slide with holes drilled by a sandblaster (Texas Airsonics); the presence of O2 between the glue and the PDMS, due to the permeability of PDMS to gas, ensures the presence of a thin layer of uncured polymer. As a result, a second curing bonds the micropatterned glue to the glass slide.
In order to interface the systems with external fluids, we introduced PDMS-based microvalve29 connectors bonded to the glass slide with an O2 plasma cleaner; these connectors were aligned with the holes. The microvalves were generated using multilayer soft lithography.30 The typical valve dimensions were 40 μm high and 500 μm wide for the fluidic, parabolic-shaped channel, whereas the rectangular control channel featured a height of 80 μm and a width of 500 μm.
Sample preparation for simplex experiments
PCR mixtures were prepared in 10 μl volumes using a one-step RT-PCR kit (Lightcycler 480, RNA Master Hydrolysis Probes, Roche Applied Science), containing: 3.2 μl 2.7X conc. Lightcycler 480, RNA Master Hydrolysis Probes, 3.7 μl activator, 0.5 μl enhancer, PCR grade water, a solution of bovine sulfate albumin (BSA) at a final concentration of 625 ng∕ml (Sigma), 0.4 μM each of forward and reverse primer (Eurogentec), and a 0.4 μM LNA-based probe (Eurogentec). The reporters∕quenchers of probes were Texas Red∕BHQ-2 and FAM∕BHQ-1 for β-actin and TBP, respectively. BSA prevents the enzyme from being denatured at the oil∕water interface.31 We verified that an absence of BSA leads to low amplification efficiency.
The carrier fluid forming the continuous phase was mineral oil (Sigma), to which 2% (w∕w) of the nonionic surfactant Abil EM 90 (Evonik) was added. This biocompatible surfactant31 allows obtaining a full wetting of the continuous phase with respect to the walls,32 along with emulsion stability during the PCR cycles. Droplets were formed at a flow rate of 900 μl∕h for the dispersed phase and 1800 μl∕h for the continuous phase.
Sample preparation for duplex experiments
For duplex experiments (β-actin and TPB), we employed the same kit and same reporters∕quenchers as for the simplex experiments; however, we increased the enzyme concentration. The reason for this modification is that in the duplex case, abundant mRNAs may consume PCR reagents during the early PCR cycles, thereby jeopardizing the possibility of observing weakly expressed genes. We thus adapted the protocol in order to obtain equivalent amplification of β-actin and TBP in both the simplex and the duplex experiments. For this purpose, we conducted a set of experiments on total RNA at 18.4 pg∕nl using conventional qPCR. β-actin and TBP were amplified in duplex using the same concentration of reagents as in the simplex and duplex experiments, with the addition of an enzyme (2.5 U of Tth DNA polymerase) that amplifies the PCR signal without altering the underlying amplification process. The corresponding calibration measurements are shown in the supporting information section.22 As would be expected, adding the enzyme increases the fluorescence level of TBP, i.e., the weakly expressed gene (as compared to the duplex experiment without an additional enzyme), which reaches a level comparable to the simplex experiment while exerting no significant influence on the PCR curve of the β-actin gene.
Culture and preparation of the cells
The H2B-RFP MDCK cells were cultured in MEM (Gibco) supplemented with 10% fetal calf serum (Fisher Scientific) and 1% penicillin∕streptomycin (Gibco). The cells were washed with PBS, incubated 3 min at 37 °C with a solution of trypsin at 100 μg∕ml in PBS, in order to detach them from the flask. The trypsin was then blocked by adding a culture medium. The cells were counted, centrifuged three times (8 min at 800 rpm), and washed twice in PBS. After the second washing, they were resuspended in the correct amount of PBS to adjust concentration to the desired value.
Device operations
Aqueous droplets in oil were formed at a flow-focusing junction (see Fig. 1). The height and width of the channels were 120 and 150 μm, respectively. Both aqueous and oil phases were controlled by a commercially available pressure controller (MFCS 8C, Fluigent). In order to reduce reagent consumption, the sample (i.e., the RT-PCR reagents and cell solution) was first loaded into a 10 μl pipette tip before being injected in the chip. Externalized valves were used to stop the flow and maintain the droplets immobile. After loading droplets into the channel, the valves were closed and the aqueous sample was replaced by photocurable glue (NOA 89, Norland). The valves were then opened again to allow a small amount of glue to enter the flow-focusing junction, which was cured by short UV exposure so as to definitively close the system and limit droplet movement. This same procedure was repeated at the device outlet. Due to material expansion during thermal cycles, however, the droplets moved over a length on the order of one interdroplet distance from one cycle to the next, yet movement amplitude was small enough to easily track the droplets.
The PCR cycles were controlled for temperature by means of a commercially available thermocycler (PTC 200 DNA Engine, MJ Research). The programed RT-PCR profile was 3 min at 63 °C for both lysis of cells and the RT step, followed by 30 s at 95 °C and 40 cycles of 95 °C for 10 s and then 60 °C for 30 s. We implemented real-time fluorescence detection of each droplet in parallel by placing the chip on the thermocycler under an epifluorescence microscope (AZ100, Nikon), equipped with a sensitive EM-CCD camera (C9100, Hamamatsu) and a mercury lamp (Intensilight, Nikon) with adapted filter cubes (Semrock). The fluorescence emission of droplets was imaged at each cycle, at the end of the elongation step.
In the duplex experiments, we added an image splitter (Optosplit, Cairn Research) to enable the use of a single camera for recording images simultaneously at two different optical wavelengths. This optical system was composed of a dichroic beamsplitter and interference filters placed along two pathways.
Data analysis
Images were analyzed by a program encoded in MATLAB. Under conventional conditions, the threshold cycle (Ct) was determined as a parameter of the quantification of each PCR assay. Moreover, it was defined as the cycle where background intensity intersects the amplification part of the curve. We calculated Ct with a method that uses the maximum second derivative of the curve.
CONCLUSION
In the present paper, we have applied microfluidic technology in order to study the expression of genes from purified RNA samples, in a duplex mode, and from eukaryotic cell solutions. These steps have confirmed our ability to analyze tens of isolated cells encapsulated in 2 nl droplets, within a single run. Neat measurements of averaged values, consistent with the current technology, have been obtained on both RNA and the cell samples. Cell-to-cell variability measurements proved more difficult, and our results must be taken as semiquantitative. For any discussion on this topic, the noise∕interference must certainly be assessed with the cell material before reaching full quantitative conclusions. Along these lines, the neat results obtained with RNA samples provide an encouraging reference point. In any event, the present work constitutes a step forward in microfluidic single-cell studies by demonstrating that high-throughput measurements of cell-to-cell expressions are indeed feasible using microfluidic technology. This study has paved the way for the systematic acquisition of such information in biological and biomedical studies.
ACKNOWLEDGMENTS
The authors would like to thank Pascal Poncet, Fabrice Monti, Jean-Christophe Galas, Avin Babateheri, Massimo Vergassola, Alberto Puliafito, and Jean-Baptiste Sibarita for their assistance with the image analysis. This work was financed by France’s Ministry of Higher Education and Research, as well as the ANR∕PNANO and CNRS organizations.
References
- Bengtsson M., Hemberg M., Rorsman P., and Stahlberg A., BMC Mol. Biol. 63, 9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Ding J. Q., Qu Y., Hu H. L., Lin M. H., Datta A., Larson A., Liu G. E., and Li B. R., Immunology 127, 83 (2009). 10.1111/j.1365-2567.2008.02926.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Friedman N., and Xie X. S., Nature (London) 440, 358 (2006). 10.1038/nature04599 [DOI] [PubMed] [Google Scholar]
- Fink L., Seeger W., Ermert L., Hanze J., Stahl U., Grimminger F., Kummer W., and Bohle R. M., Nat. Med. 4, 1329 (1998). 10.1038/3327 [DOI] [PubMed] [Google Scholar]
- Hinkle D. A. and Eberwine J. H., Biol. Psychiatry 54, 413 (2003). 10.1016/S0006-3223(03)00322-6 [DOI] [PubMed] [Google Scholar]
- Esumi S., Kaneko R., Kawamura Y., and Yagi T., Nat. Protoc. 1, 2143 (2006). 10.1038/nprot.2006.343 [DOI] [PubMed] [Google Scholar]
- Tabeling P., Introduction to Microfluidics (Oxford University Press, Oxford, 2005). [Google Scholar]
- Marcus J. S., Anderson W. F., and Quake S. R., Anal. Chem. 78, 3084 (2006). 10.1021/ac0519460 [DOI] [PubMed] [Google Scholar]
- Warren L., Bryder D., Weissman I. L., and Quake S. R., Proc. Natl. Acad. Sci. U.S.A. 103, 17807 (2006). 10.1073/pnas.0608512103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong D., Hahn-Windgassen A., Foygel K., Jun S., Behr B., and Yao M., Nat. Protoc. 2009 (2009).
- Warren L. A., Rossi D. J., Schiebinger G. R., Weissman I. L., Kim S. K., and Quake S. R., Aging Cell 6, 775 (2007). 10.1111/j.1474-9726.2007.00337.x [DOI] [PubMed] [Google Scholar]
- Bontoux N., Dauphinot L., Vitalis T., Studer V., Chen Y., Rossier J., and Potier M. C., Lab Chip 8, 443 (2008). 10.1039/b716543a [DOI] [PubMed] [Google Scholar]
- Kiss M. M., Ortoleva-Donnelly L., Beer N. R., Warner J., Bailey C. G., Colston B. W., Rothberg J. M., Link D. R., and Leamon J. H., Anal. Chem. 80, 8975 (2008). 10.1021/ac801276c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R., Warner J. B., Nakano M., Libby B., Medkova M., David P. H., Kotsopoulos S. K., Samuels M. L., Hutchison J. B., Larson J. W., Topol E. J., Weiner M. P., Harismendy O., Olson J., Link D. R., and Frazer K. A., Nat. Biotechnol. 27, 1025 (2009). 10.1038/nbt.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agresti J. J., Antipov E., Abate A. R., Ahn K., Rowat A. C., Baret J. C., Marquez M., Klibanov A. M., Griffiths A. D., and Weitz D. A., Proc. Natl. Acad. Sci. U.S.A. 107, 4004 (2010). 10.1073/pnas.0910781107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer N. R., Wheeler E. K., Lee-Houghton L., Watkins N., Nasarabadi S., Hebert N., Leung P., Arnold D. W., Bailey C. G., and Colston B. W., Anal. Chem. 80, 1854 (2008). 10.1021/ac800048k [DOI] [PubMed] [Google Scholar]
- Tichopad A., Kitchen R., Riedmaier I., Becker C., Stahlberg A., and Kubista M., Clin. Chem. 55, 1816 (2009). 10.1373/clinchem.2009.126201 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., Kubista M., and Pfaffl M. W., in Real-Time PCR: Current Technology and Applications, edited by Logan J., Edwards K., and Saunders N. (Caister Academic, Norwich, U.K., 2009). [Google Scholar]
- Blomberg J., Andersson M., and Faldt R., Br. J. Haematol. 65, 83 (1987). 10.1111/j.1365-2141.1987.tb06139.x [DOI] [PubMed] [Google Scholar]
- Clausell-Tormos J., Lieber D., Baret J. C., El-Harrak A., Miller O. J., Frenz L., Blouwolff J., Humphry K. J., Koster S., Duan H., Holtze C., Weitz D. A., Griffiths A. D., and Merten C. A., Chem. Biol. 15, 427 (2008). 10.1016/j.chembiol.2008.04.004 [DOI] [PubMed] [Google Scholar]
- Köster S., Angilè F. E., Duan H., Agresti J. J., Wintner A., Schmitz C., Rowat A. C., Merten C. A., Pisignano D., Griffiths A. D., and Weitz D. A., Lab Chip 8, 1110 (2008). 10.1039/b802941e [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.3596394 for Fig. S1 for a comparison between the experimental distributions of cells per droplet and the Poisson statistics, a description of the model and the optimization of the duplex PCR in tube.
- Courtois F., Olguin L. F., Whyte G., Theberge A. B., Huck W. T. S., Hollfelder F., and Abell C., Anal. Chem. 81, 3008 (2009). 10.1021/ac802658n [DOI] [PubMed] [Google Scholar]
- Dorfman K. D., Chabert M., Codarbox J. H., Rousseau G., de Cremoux P., and Viovy J. L., Anal. Chem. 77, 3700 (2005). 10.1021/ac050031i [DOI] [PubMed] [Google Scholar]
- Bengtsson M., Stahlberg A., Rorsman P., and Kubista M., Genome Res. 15, 1388 (2005). 10.1101/gr.3820805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K., Kajiyama T., and Kambara H., Nat. Methods 6, 503 (2009). 10.1038/nmeth.1338 [DOI] [PubMed] [Google Scholar]
- Elowitz M. B., Levine A. J., Siggia E. D., and Swain P. S., Science 297, 1183 (2002). 10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- Bartolo D., Degre G., Nghe P., and Studer V., Lab Chip 8, 274 (2008). 10.1039/b712368j [DOI] [PubMed] [Google Scholar]
- Unger M. A., Chou H. P., Thorsen T., Scherer A., and Quake S. R., Science 288, 113 (2000). 10.1126/science.288.5463.113 [DOI] [PubMed] [Google Scholar]
- Galas J. C., Bartolo D., and Studer V., New J. Phys. 11, 075027 (2009). [Google Scholar]
- Williams R., Peisajovich S. G., Miller O. J., Magdassi S., Tawfik D. S., and Griffiths A. D., Nat. Methods 3, 545 (2006). 10.1038/nmeth896 [DOI] [PubMed] [Google Scholar]
- Dreyfus R., Tabeling P., and Willaime H., Phys. Rev. Lett. 90, 144505 (2003). 10.1103/PhysRevLett.90.144505 [DOI] [PubMed] [Google Scholar]