Summary
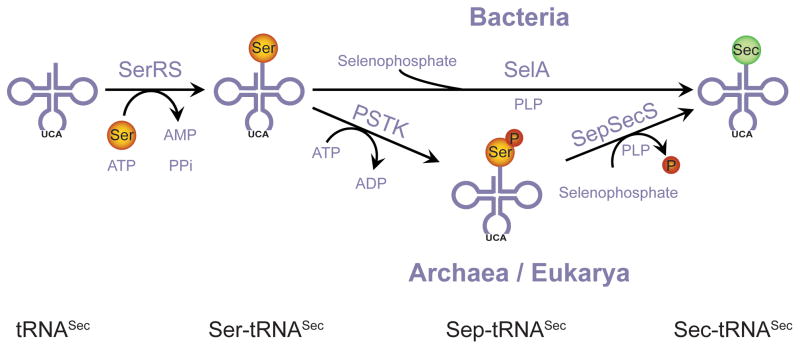
In Archaea selenocysteine (Sec) is synthesized in three steps. First seryl-tRNA synthetase acylates tRNASec with serine to generate Ser-tRNASec. Then phosphoseryl-tRNASec kinase (PSTK) forms Sep-tRNASec, which is converted to Sec-tRNASec by Sep-tRNA:Sec-tRNA synthase (SepSecS) in the presence of selenophosphate produced by selenophosphate synthetase (SelD). A complete in vivo analysis of the archaeal Sec biosynthesis pathway is still unavailable, and the existence of a redundant pathway or of a rescue mechanism based on the conversion of Sep-tRNASec to Cys-tRNASec during selenium starvation, cannot be excluded.
Here we present a mutational analysis of Sec biosynthesis in Methanococcus maripaludis strain Mm900. Sec formation is abolished upon individually deleting the genes encoding SelD, PSTK, or SepSecS; the resulting mutant strains could no longer grow on formate while growth with H2 + CO2 remained unaffected. However, deletion of the PSTK and SepSecS genes was not possible unless the selenium-free [NiFe]-hydrogenases Frc and Vhc were expressed. This required the prior deletion of either the gene encoding SelD or that of HrsM, a LysR-type regulator suppressing transcription of the frc and vhc operons in the presence of selenium. These results show that M. maripaludis Mm900 is facultatively selenium-dependent with a single pathway of Sec-tRNASec formation.
Keywords: Selenocysteine, Selenophosphate synthetase, O-phosphoseryl-tRNASec kinase, Sep-tRNA:Sec-tRNA synthase
Introduction
Selenium is an important micronutrient for a subset of organisms from all three lines of descent (Bacteria, Archaea and Eukarya). Its most relevant biological form is the rare amino acid selenocysteine which is made in a tRNA-dependent manner. First tRNASec is acylated with serine (Ser) by seryl-tRNA synthetase (SerRS), followed by conversion of Ser-tRNASec to Sec-tRNASec. Two pathways for Sec formation have been found in nature (Fig. 1). In bacteria Ser-tRNASec to Sec-tRNASec conversion occurs in a single step, catalyzed by the PLP-dependent enzyme selenocysteine synthase (SelA) (Forchhammer and Böck, 1991; Böck et al., 2005). In eukaryotes and archaea, an additional step is required, in which Ser-tRNASec is first converted to O-phosphoseryl-tRNASec (Sep-tRNASec) by O-phosphoseryl-tRNA kinase (PSTK) (Carlson et al., 2004; Sherrer et al., 2008a). The transformation from Sep-tRNASec to Sec-tRNASec is then catalyzed by a distant homolog of SelA, Sep-tRNA:Sec-tRNA synthase (SepSecS), in a PLP-dependent manner (Yuan et al., 2006; Xu et al., 2007). Both SelA and SepSecS use as selenium donor selenophosphate, which is produced by selenophosphate synthetase (SelD) from selenide and ATP (Leinfelder et al., 1990). Besides its role in Sec formation, selenophosphate has also been described as a selenium donor for the synthesis of 2-selenouridine, a modified nucleoside found in certain tRNAs (Wittwer, 1983; Ching, 1984; Ching et al., 1984; Ching et al., 1985).
Fig. 1.
tRNA-dependent pathways leading to Sec-formation. In bacteria, Ser-tRNASec to Sec-tRNASec transformation is catalyzed by SelA. The archaeal/eukaryal route is dependent on PSTK and SepSecS.
Sec is co-translationally inserted into proteins by tRNASec at certain UGA codons, that would otherwise signal stop (Flohe et al., 1973; Rotruck et al., 1973). Recoding of UGA into a Sec-codon requires two components. The first is the SECIS (Sec insertion sequence) element, a distinct stem-loop structure in the selenoprotein mRNA. The second is a Sec-specific elongation factor (SelB or EF Sec) that binds GTP and the SECIS element and delivers Sec-tRNASec to the ribosome (Forchhammer et al., 1989; Berry et al., 1991; Fagegaltier et al., 2000).
Only a small number of archaea encode selenoproteins; this includes all representatives of the orders Methanopyrales and Methanococcales with known genome sequences (Rother et al., 2001; Kryukov and Gladyshev, 2004). Members of these orders are obligate anaerobes which gain energy exclusively by the reduction of CO2 (with H2) and/or formate to methane. Methanococcus maripaludis S2 encodes ten Sec containing proteins in its genome (Hendrickson et al., 2004; Kryukov and Gladyshev, 2004), of which eight are involved in hydrogenotrophic energy metabolism. This includes subunits of F420-dependent (FruA; MMP1382) and F420-independent hydrogenase (VhuD; MMP1696 and VhuU; MMP1693), tungsten-containing formyl-methanofuran dehydrogenase (FwdB; MMP1691), molybdenum containing formylmethanofuran dehydrogenase (FmdB; MMP0511), heterodisulfide reductase (HdrA; MMP1697) and formate dehydrogenase (FdhA1; MMP1298 and FdhA2; MMP0138). In addition, a backup system consisting of selenium-free, cysteine-containing homologs for most of these selenoenzymes is encoded in the genome, with the exception of formate dehydrogenase. Consequently, under selenium limitation M. maripaludis should be able to grow with H2 + CO2 but not with formate. Indeed, this has been shown for selB and selD deletion strains of M. maripaludis JJ (Rother et al., 2003; Stock et al., 2010).
Apart from these genetic studies demonstrating the physiological functions of SelB (Rother et al., 2003) and SelD (Stock et al., 2010), most of our knowledge on Sec biosynthesis in archaea is based on in vitro analyses of PSTK from Methanocaldococcus jannaschii (Kaiser et al., 2005; Sherrer et al., 2008b) and SepSecS from M. maripaludis (Araiso et al., 2008), as well as on in vivo complementation studies using M. jannaschii PSTK and SepSecS in a heterologous bacterial system (Yuan et al., 2006; Araiso et al., 2008; Sherrer et al., 2008b). Although these experiments clearly indicate that Sec formation in archaea and eukaryotes is of a common origin, a number of questions remain unresolved. We recently demonstrated that Sep-tRNA:Cys-tRNA synthase (SepCysS), an enzyme catalyzing the conversion of Sep-tRNACys to Cys-tRNACys in a number of archaea, including M. maripaludis (Sauerwald et al., 2005), can also recognize Sep-tRNASec as a substrate, and thus can replace SepSecS in the heterologous Escherichia coli system (Yuan et al., 2010). This raises the intriguing possibility that SepCysS could rescue a SepSecS deletion in M. maripaludis. Moreover, it remains unknown why bacteria can convert Ser to Sec in a single step via the action of SelA, while archaea and eukaryotes require two enzymes, PSTK and SepSecS, which are phylogenetically remarkably distinct from SelA (Araiso et al., 2008). Yet, it raises the question whether PSTK and/or SepSecS perform other physiological functions that are essential for these organisms.
In the present study, we took advantage of the genetic tractability of the M. maripaludis Mm900 strain (Moore and Leigh, 2005) to examine the archaeal mechanism of Sec biosynthesis in vivo, and to investigate the key roles of SelD, PSTK and SepSecS.
Results
Construction of marked and unmarked gene deletions in M. maripaludis
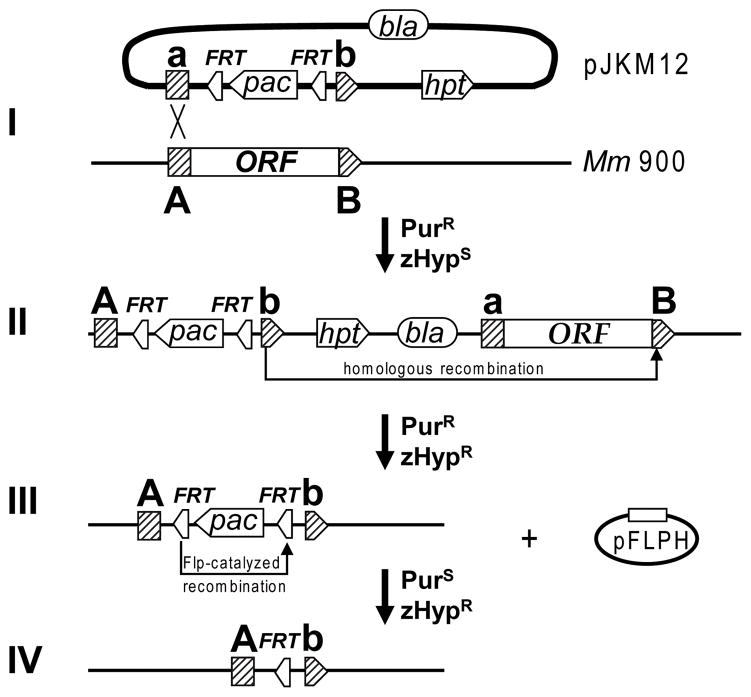
To analyze the Sec-tRNASec formation pathway in M. maripaludis we constructed deletion mutants of the genes encoding SelD (MMP0904), PSTK (MMP1490; encoded by pstK) and SepSecS (MMP0595; encoded by spcS). We chose a “knock-in-knock-out” approach (Fig. 2) which allows generation of both marked and markerless gene deletions using the same suicide-plasmid. To construct marked gene deletions (Fig. 2, I–III), approximately 0.5 to 1.0 kb long DNA segments flanking the desired gene were first cloned into the suicide plasmid pJKM12, upstream and downstream of the puromycin resistance cassette (pac) which is flanked by two flp recombinase recognition sites (FRT). The resulting plasmid was then transformed into M. maripaludis Mm900 (M. maripaludis S2 Δhpt mutant) with selection for puromycin resistance (Fig. 2, I). Since the plasmid cannot replicate in Methanococcus, recombinants arise by chromosomal integration of the vector into the flanking region of the target gene, mediated by a single homologous recombination event (Fig. 2, II). The resulting merodiploids were resistant to puromycin and were sensitive to the toxic purine analog 8-azahypoxanthine (zHyp) due to the presence of the vector-encoded hpt. Under continuous selection with puromycin, a second homologous recombination can take place, removing the wild-type allele together with the vector backbone from the chromosome, thus resolving the unstable merodiploid state. These recombinants were selected as puromycin and zHyp resistant clones (Fig. 2, III). To allow reuse of puromycin as a marker, the pac-cassette could be removed by intrachromosomal recombination (Fig. 2, IV) mediated by Flp-recombinase, which was expressed from plasmid pFLPH. By this method we readily generated marked deletion mutants of selD. Plating of ca. 103 cells that had undergone the second homologous recombination event gave rise to six puromycin and zHyp resistant colonies. Southern blot analysis revealed that in all six clones selD had been deleted (Fig S1 B). From one strain, designated FSD6, the pac-cassette was subsequently removed by Flp-recombinase mediated recombination to yield the markerless selD mutant FSD10 (Fig. S1). Surprisingly however, we were not able to delete the genes encoding PSTK and SepSecS from the M. maripaludis Mm900 chromosome, neither by using the “knock-in-knock-out” gene deletion strategy described above, nor by applying previously established methods (Stathopoulos et al., 2001; Moore and Leigh, 2005).
Fig. 2.
Generation of marked and markerless chromosomal deletions in M. maripaludis. (I) Plasmid pJKM12 which encodes M. maripaludis hpt, as well as a puromycin resistance cassette (pac) flanked by two flp recombinase recognition sites (FRT) and regions homologous to the target gene (a/A and b/B) is transformed into M. maripaludis Mm900 (Δhpt). (II) Integration of pJKM12 into the chromosome creates an unstable merodiploid state; a second homologous recombination event removes the vector backbone together with the wild-type allele of the target gene from the chromosome. (III) The target gene is replaced by pac bracketed by two FRT sites; cells are resistant to puromycin and zHyp. (IV) Expression of Flp recombinase from plasmid pFLPH results in removal of the puromycin cassette.
Characterization of the SelD mutant
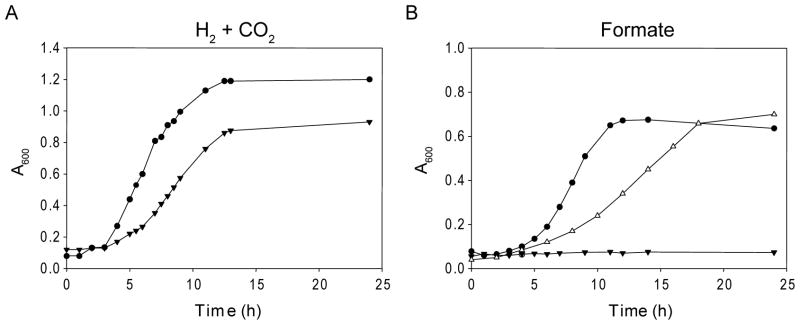
To determine how the deletion of SelD affects the growth phenotype compared to the wild-type, the knockout mutant FSD10 was subjected to cultivation experiments. Grown in the presence of H2 + CO2, the ΔselD strain had a slightly longer doubling time than the wild-type, and it was not able to grow on formate alone (Fig. 3). Given that both formate dehydrogenases in M. maripaludis are selenoenzymes (Wood et al., 2003), this phenotype indicated that the Sec forming pathway has been impaired in this mutant. To directly follow selenium incorporation into protein and tRNAs, wild-type and deletion mutants were grown in the presence of [75Se]selenite, followed by separation of cell extracts on a polyacrylamide gel and visualization by autoradiography. While in wild-type cell extract selenium incorporation occurred in both proteins and tRNAs (Fig. 4, lane 1), in cell extracts from the ΔselD mutant neither selenoproteins nor selenium-modified tRNAs were detectable (Fig. 4, lane 2). This confirms the previous finding that SelD is the predicted archaeal selenophosphate synthetase which provides the selenium donor for both Sec and selenouridine synthesis (Stock et al., 2010). In an attempt to rescue the selD deletion, FSD10 was transformed with plasmid pECSD from which E. coli SelD was expressed under the control of a constitutive Methanococcus promoter. In vivo labeling experiments with [75Se]selenite show that both selenoprotein synthesis and tRNA modification were reestablished in this strain (Fig. 4, lane 3); thus, archaeal and bacterial SelD are interchangeable (Stock et al., 2010). Consequently, the E. coli selD complemented strain is able to use formate as sole substrate, demonstrating the expression of a functional formate dehydrogenase (Fig. 3B).
Fig. 3.
Effects of selenium on growth. (A) Growth of M. maripaludis Mm900 wild-type (solid circles) and of FSD10 (selD deletion mutant; solid triangles) cultivated on H2 + CO2. (B) Cultivation on formate. Same symbols as in A. Additionally: FSD10 was complemented with E. coli selD (open triangles).
Fig. 4.
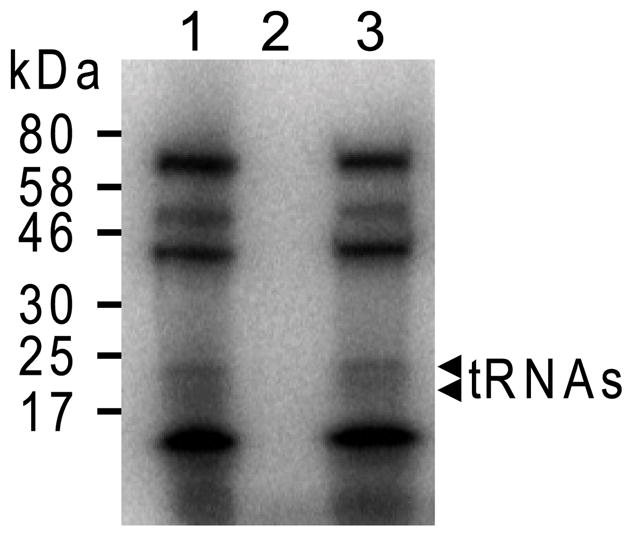
75Se incorporation into protein and tRNA. Cell extracts of wild-type Mm900 (lane 1), FSD10 (lane 2), and FSD10 containing the E. coli selD expression vector (lane 3), were prepared following growth on McCas medium in the presence of [75Se]selenite. Extracts were separated on a SDS/PAGE gel (4–20%), and the insertion of 75Se into macromolecules was followed by autoradiography. Migration position of 75Se-labeled RNA is indicated on the right.
Deletion of PSTK or SepSecS genes from the SelD mutant
Given that selenoprotein formation is not essential for the cultivation of M. maripaludis Mm900 on H2 + CO2, the finding that PSTK and SepSecS could not be deleted from this strain indicates that PSTK and/or SepSecS might have essential cellular functions in addition to Sec synthesis. If this were the case, the deletion of their corresponding genes should generally not be possible. Alternatively, an as-yet-unidentified regulatory process could make the presence of PSTK and/or SepSecS necessary as long as selenium is available to the organism. In that case, PSTK and SepSecS should become dispensable when M. maripaludis is grown on H2 + CO2 in a selenium-free medium or generally speaking, under conditions where no selenium donor for Sec formation is available. Due to the difficulty of preparing a medium free of any trace contamination with selenium, we decided to use a selD mutant (which is unable to form selenophosphate) as background to delete pstK and spcS. Transformation of FSD10 with suicide plasmids pJKM12ΔpstK or pJKM12ΔspcS yielded both knockouts of pstK (Fig. S2) and spcS (Fig. S3), respectively. Thus, neither PSTK nor SepSecS has further essential functions besides Sec formation.
SepSecS is required for Sec formation in M. maripaludis
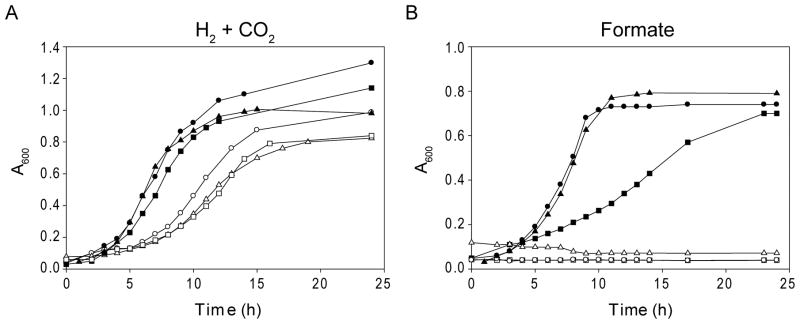
In order to assess the phenotype of the SepSecS deletion, we complemented the selD/spcS double-deletion mutant (designated FSD103) with the E. coli selD-encoding plasmid. Cultivated on H2 + CO2, the resulting strain had a longer doubling time than the wild-type comparable to the selD deletion mutant, and growth on formate was still impaired (Fig. 5). Furthermore, no selenium containing proteins were detectable after in vivo labeling with [75Se]selenite (Fig. 6, lane 3). The presence of active SelD, on the other hand, was evidenced by the formation of selenium-modified tRNAs. This result demonstrates that SepSecS is the only Sec synthase present in M. maripaludis. Also, the inability to grow on formate revealed that SepCysS cannot complement the deletion of SepSecS in vivo.
Fig. 5.
Effects of selenium on growth. (A) Growth of M. maripaludis wild-type Mm900 (solid circles), FSD10 (open circles), FSD103 (selD spcS double-deletion) containing the E. coli selD expression vector (open triangles), Mm900 pstK deletion mutant, pre-transformed with the E. coli selA expression vector (solid triangles), FHM1 (hrsM deletion; solid squares), and FHM11 (hrsM pstK double-deletion; open squares) cultivated on H2 + CO2. (B) Cultivation on formate. Same symbols as in A.
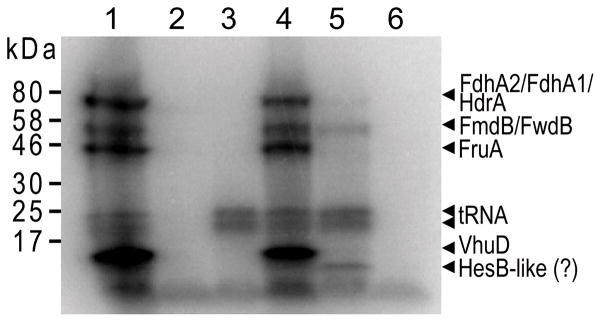
Fig. 6.
Selenoprotein synthesis by M. maripaludis. Autoradiograph of a SDS/PAGE gel after electrophoresis of cell lysates from 75Se-labeled M. maripaludis Mm900 (lane 1), FSD103 (selD spcS double-deletion; lane 2), FSD103 containing the E. coli selD expression vector (lane 3), Mm900 ΔpstK mutant pre-transformed with the E. coli selA expression vector (lane 4), FHM1 (hrsM deletion; lane 5), and FHM11 (hrsM pstK double-deletion; lane 6), respectively. The migration positions of the standard proteins are indicated on the left; the positions predicted for the putative selenoproteins in M. maripaludis and of Se-containing tRNAs are indicated on the right side.
The archaeal and bacterial Sec formation pathways are compatible
In order to address the in vivo role of PSTK, we attempted to complement the selD/pstK double deletion mutant (designated FSD102) with E. coli selD in the same manner as described above for FSD103. The SelD expression plasmid, however, was unstable in this strain despite neomycin selection. In an alternative approach to construct a pstK deletion mutant, we transformed M. maripaludis Mm900 with plasmid pECSA, encoding E. coli Sec synthase (SelA), a neomycin marker, and the Hpt gene under control of constitutive archaeal promoters. From this transformant the PSTK gene could subsequently be deleted, resulting in a strain able to grow with H2 + CO2 as well as with formate (Fig 5). Selenoprotein formation was not impaired in this strain (Fig. 6, lane 4), demonstrating that the bacterial Sec formation pathway can replace its archaeal counterpart. Nevertheless, successive attempts to remove the selA-expression plasmid by counter selection with 8-azahypoxanthine failed. This observation is adding further support to our previous speculation that a regulatory mechanism may be present in M. maripaludis that renders the Sec biosynthesis pathway indispensable as long as selenium is available to the organism.
Deletion of PSTK requires expression of selenium-free [NiFe]-hydrogenases
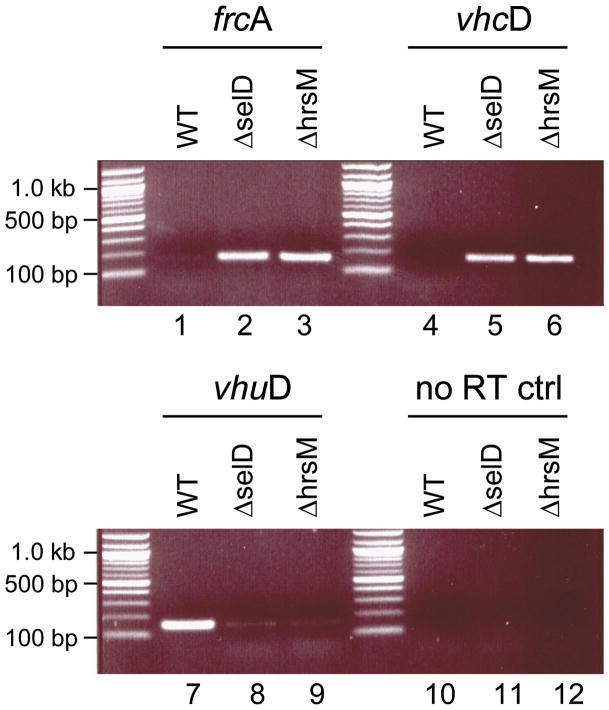
Selenium-dependent gene regulatory mechanisms were observed previously in the closely related Methanococcus voltae. It has been shown that the two selenium-free [NiFe]-hydrogenases Frc and Vhc are only expressed under selenium starvation, whereas in the presence of selenium only the two Sec containing homologs Fru and Vhu are produced (Berghöfer et al., 1994). In M. voltae, the LysR-family regulator HrsM was shown to repress the transcription of the Frc and Vhc operons when selenium is available (Sun and Klein, 2004). The presence of an HrsM homolog (MMP1712), together with a conserved organization for the Frc and Vhc operons, indicate that the same regulatory mechanism is likely to be present in M. maripaludis S2. To verify this, we constructed an hrsM deletion mutant (designated FHM1). RT-PCR experiments were used to evaluate transcription of the FrcA and VhcD encoding genes in M. maripaludis Mm900, FSD10 and FHM1 strains grown in selenite containing medium under an H2 + CO2 atmosphere. While transcription of the frcA and vhcD genes was repressed in the wild-type under these conditions, their expression was detected in the ΔhrsM and in the ΔselD strains (Fig. 7, lanes 1–6). On the other hand, transcription of the vhuD gene, encoding a subunit of the Sec-containing [NiFe]-hydrogenase Vhu, occurred at a significantly lower level in the ΔhrsM and in the ΔselD strains compared to wild-type (Fig. 7, lanes 7–9). Using suicide plasmid pJKM12NΔpstK, we were able to delete pstK in the ΔhrsM strain (resulting in strain FHM11; Fig. S4), indicating that the tight repression of the selenium-free [NiFe]-hydrogenases (Frc and Vhc) was affecting the dispensability of PSTK.
Fig. 7.
Transcription of genes encoding hydrogenase subunits in M. maripaludis Mm900, FHM1, and FSD10 strains, analyzed by RT-PCR. DNA-free total RNA from Mm900 wild-type (lanes 1, 4, 7, 10), or from deletion mutants carrying an insertion in the hrsM gene (lanes 2, 5, 8, 11) or the selD gene (lanes 3, 6, 9, 12) was subjected to RT-PCR. Primer pairs targeting fractions of the indicated genes encoding of the Se-free hydrogenases (frcA and vhcA), or on of the Se-containing hydrogenase (vhuD) were used. The primer pair directed against vhuD was also used in a control experiment where reverse-transcriptase has been omitted to exclude contamination with genomic DNA (lanes 10–12). The expected PCR product sizes were: frcA, 167 bp; vhcD, 165 bp; vhuD, 160 bp.
Sep-tRNASec is an essential intermediate in the archaeal Sec formation pathway
The hrsM deletion and hrsM/pstK double-deletion strains were characterized in cultivation experiments and by in vivo [75Se] incorporation. The ΔhrsM mutant was able to synthesize [75Se]-labeled proteins and seleno-modified tRNAs (Fig. 6, lane 5). Protein bands corresponding to FruA and VhuD, subunits of the selenium-containing hydrogenases, were not detectable. This did not significantly affect the growth rate of this strain on H2 + CO2 (Fig. 5), presumably due to the expression of Frc and Vhc. On formate medium, the ΔhrsM mutant exhibited a slightly slower growth rate compared to the wild-type (Fig. 5). This might be explained by a weaker expression of either one or both of the two formate dehydrogenases. In agreement with this, the labeled protein band corresponding to formate dehydrogenase subunits FdhA1 and FdhA2 appeared to be of significantly lesser intensity (Fig. 6, lane 5). On the other hand, the hrsM/pstK double-deletion mutant exhibited a slower growth rate on H2 + CO2 and completely lost the capacity to grow on formate (Fig. 5). Also, [75Se] incorporation into both proteins and tRNAs was undetectable (Fig. 6, lane 6). The absence of [75Se]-labeled tRNAs was anticipated, since M. maripaludis SelD, which provides the activated Se-donor for selenouridine formation, is a selenoenzyme itself (Moore and Leigh, 2005; Stock et al., 2010). Our results confirm previous in vitro experiments showing that Sep-tRNASec formed by PSTK is a crucial intermediate in the archaeal Sec formation pathway (Yuan et al., 2006).
DISCUSSION
A number of powerful genetic tools have been developed for M. maripaludis, making this organism an important model for molecular studies within the archaeal domain (Sandbeck and Leigh, 1991; Blank et al., 1995; Argyle et al., 1996; Whitman et al., 1997; Moore and Leigh, 2005; Rother and Metcalf, 2005; Chaban et al., 2007; VanDyke et al., 2009). A recent report that M. maripaludis is polyploid and that single cells may contain more than 50 genome copies during exponential growth (Hildenbrand et al., 2011) makes clear, however, that genetic manipulation of this organism is not trivial. Attempts to construct knockout mutants may result in strains carrying both the wild-type and mutant allele even when the targeted gene is not essential, and careful evaluation of the resulting genotype is therefore crucial. A further limitation is the availability of only two antibiotic resistance markers (providing resistance against puromycin and neomycin) for selection in Methanococcus. Thus, it is challenging to construct strains carrying several mutations, an essential requirement for investigating interactions among various genes or to examine functional genetic redundancies. A previously reported mutagenesis method (Moore and Leigh, 2005) overcame this limitation by applying two consecutive allelic exchange reactions; they yield either a markerless deletion of the targeted gene, or restore the wild-type. Screening for the desired mutant is necessary, and is hampered when the mutant has a slower growth rate than the wild-type (Rother and Metcalf, 2005). A more straightforward strategy, the Saccharomyces cerevisiae Flp-FRT system (Sadowski, 1995), which allows flp recombinase-mediated removal of the antibiotic marker by an intra-chromosomal sequence-specific recombination, was successfully used in bacterial (Cherepanov and Wackernagel, 1995; Schweizer, 2003), archaeal (Welander and Metcalf, 2008) and eukaryotic (Dymecki, 1996; Lyznik et al., 1996) systems. Our adaptation of the Flp-FRT recombination system for use in M. maripaludis to generate single and consecutive gene deletions was the basis for successfully obtaining deletions of the three genes involved in Sec biosynthesis and the demonstration that this biosynthetic role is their only cellular function.
Selenophosphate has previously been shown to serve as the activated selenium donor for both Sec biosynthesis and for the formation of selenium-modified tRNAs (Leinfelder et al., 1990). In agreement with this, deletion of the selenophosphate synthetase encoding selD from the M. maripaludis JJ genome eradicated this organism’s ability of selenium incorporation into proteins and tRNA (Stock et al., 2010). However, attempts to delete selD (encoded by ORF MMP0904) in M. maripaludis S2 failed, leading the authors to conclude that viability of this strain is strictly selenium dependent (Stock et al., 2010). In the present study MMP0904 was readily deleted from M. maripaludis S2 derived strain Mm900. Selenoprotein formation, and consequently the ability to grow on formate, was abolished in this mutant. Growth on H2 + CO2 remained unaffected demonstrating that, as in M. maripaludis JJ, the presence of SelD is not required for hydrogenotrophic growth. Complementation of the ΔselD strain FSD10 with E. coli SelD restored selenoprotein formation. The observed slower growth rate on formate for this complemented strain might be due to a lower efficiency of E. coli SelD in M. maripaludis or might result from a low expression level of the heterologous enzyme.
While earlier in vitro experiments and genetic studies with the heterologous E. coli in vivo system established an archaeal Sec biosynthesis pathway dependent on PSTK and SepSecS (Yuan et al., 2006), the presence of an additional SepSecS-independent route could not be fully excluded. Indeed, Methanocaldococcus and Methanococcus species encode a bacterial SelA homolog; but the corresponding recombinant M. jannaschii enzyme (MJ0158) could not form Sec-tRNASec from either Ser-tRNASec or Sep-tRNASec in vitro (Kaiser et al., 2005). While this led to the notion that this hypothetical SelA enzyme might require further proteins for activity, the experiments reported here reveal that in vivo M. maripaludis uses only the essential enzymes PSTK and SepSecS for Sec formation. Recent in vivo studies in Trypanosoma brucei indicate that this is the only pathway present in eukaryotes (Aeby et al., 2009). Our experiments also showed that E. coli SelA complements the ΔpstK strain. This is was expected as E. coli Sec synthase converted archaeal Ser-tRNASec to Sec-tRNASec in vitro (Kaiser et al., 2005). This leaves us with the intriguing question as to why during the eukaryotic and archaeal evolution these lineages maintained the two-step pathway for Sec-tRNA formation without any organism adopting the single-step bacterial mechanism via lateral gene transfer.
The deletion of pstK or spcS required the prior disruption of Sec-formation by deleting the gene encoding either SelD or the repressor HrsM. The deletion of selD or hrsM led to transcription of the genes encoding the selenium-free [NiFe]-hydrogenases Frc and Vhc, necessary for energy generation under selenium deprivation. LysR-type gene regulators (e.g., HrsM) usually require activation by an effector molecule. The effector for HrsM is unknown, but a likely candidate may be selenophosphate.
In this context, our finding that the selD/pstK double-mutant could be complemented with E. coli selD is surprising, as one would expect that the presence of selenophosphate (which is evidenced by the formation of selenium-modiefied tRNAs in this strain) would lead to repression of the Frc/Vhc operon. Possibly, the observed lower efficiency of E. coli SelD in the heterologous background results in cellular selenophosphate levels low enough to allow expression of the selenium-free hydrogenases.
Overall, our results demonstrate that in M. maripaludis Sec biosynthesis and the expression of certain selenoproteins (e.g., Fru and Vhu) are linked by a complex regulatory feedback mechanism.
The pathway catalyzed by PSTK and SepSecS is reminiscent of the indirect Cys-tRNACys formation in methanogens where SepCysS converts Sep-tRNACys to Cys-tRNACys (Sauerwald et al., 2005). We showed recently that an E. coli ΔselA strain expresses active formate dehydrogenase (normally a selenoenzyme in E. coli) when complemented with M. jannaschii PSTK and SepCysS (Yuan et al., 2010). Since no selenoproteins were formed in this strain, SepCysS might convert Sep-tRNASec to Cys-tRNASec, probably leading to a Cys homolog of formate dehydrogenase H. A similar SepCysS induced read-through of Sec-codons might be imaginable in M. maripaludis mutants with a disrupted Sec biosynthesis pathway. However, neither the SelD, PSTK, nor SepSecS deletion mutant was able to grow with formate as the only carbon and energy source, indicating the absence of a sufficient amount of active formate dehydrogenase in these strains. The mutation of the Sec residue in the catalytic active site of E. coli formate dehydrogenase H to cysteine results in a 110-fold reduction of catalytic activity compared to the wild-type enzyme (Axley et al., 1991). The effect of a Sec to Cys mutation in the M. maripaludis paralog formate dehydrogenase A has not been studied so far, but might render this enzyme inactive. Alternatively, control mechanisms might be present in M. maripaludis to prevent Cys-tRNASec from entering protein biosynthesis. Further biochemical studies and genetic experiments using suitable reporter systems will be necessary to clarify these issues.
Experimental procedures
Microbial strains and growth conditions
M. maripaludis strains (Table S1) were grown at 37 °C on solid or liquid McCas medium (Moore and Leigh, 2005) under strictly anaerobic conditions with 276 kPa H2/CO2 gas (80:20 [vol/vol]) as described before (Balch et al., 1979). Puromycin (2.5 μg/ml), neomycin (1.0 mg/ml), and 8-azahypoxanthine (0.25 mg/ml) were added to the medium as needed. For growth on formate, the medium contained 2% sodium formate and 80 mM morpholinepropanesulfonic acid, and cultures were pressurized with 100 kPa N2/CO2. Escherichia coli Dh5α, grown at 37 °C on Luria-Bertani medium supplemented with ampicillin (100 μg/ml) was used for plasmid cloning.
DNA methods, plasmid construction, and transformation
Standard methods were used for isolation of genomic DNA from M. maripaludis and of plasmid and genomic DNA from E. coli. All plasmids and primers used in this study are described in Tables S2 and S3. Southern blot analyses were performed using the DIG system (Roche, Indianapolis, IN) according to manufacturer guidelines. M. maripaludis strains were transformed as described previously (Tumbula et al., 1994).
Generation of M. maripaludis deletion strains
M. maripaludis gene deletions were generated in the Δhpt strain Mm900 (Moore and Leigh, 2005) by homologous-recombination mediated gene replacement. Approximately 0.5 to 1.0 kb long DNA segments flanking the 5′ region and the 3′ region of the gene of interest were PCR amplified using M. maripaludis Mm900 genomic DNA as template. Each fragment was then cloned into plasmid pJKM12, upstream and downstream of the pac-cassette which is flanked by two Frt sites in the same orientation. 5 μg of the resulting plasmid were transformed into M. maripaludis, and transformants were incubated without antibiotic selection for 6 h. Cultures were then transferred into 5 ml McCas medium containing puromycin, grown to an A600 ≈0.8 and, after adequate dilution, plated on McCas plates supplemented with puromycin and 8-azahypoxanthine. Single clones were PCR-screened, and replacement of the wild-type locus with the Frt-pac-Frt cassette was confirmed by Southern blotting.
When additional gene deletions were required, the puromycin marker was removed by transforming the knock-out strains with the replicative plasmid pFLPH, which encodes Flp recombinase under control of the hmvA promoter. Transformants were grown for 5 to 10 consecutive passages in McCas medium under neomycin selection, single colony purified, and screened by PCR for removal of the pac cassette. Markerless deletion of the wild-type locus was confirmed by Southern analysis. Strains were cured from the Flp recombinase encoding plasmid by 5 consecutive transfers on McCas medium without antibiotic selection. After plating, clones which had lost the plasmid were identified by PCR and by their sensitivity to neomycin.
Reverse transcriptase PCR
Total cellular RNA from 5 ml M. maripaludis cultures was purified using the RNeasy Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Two mg RNA in H2O was treated with RNase-free DNase I (20 units mg−1 RNA) for 2 h at 37 °C. The DNase was inactivated by heating at 75 °C for 10 min. The RNA was then used as template for cDNA synthesis using random hexamer primers and the Superscript II RT system (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol.
Primer combinations FRCAF-FRCAR, VHCDF-VHCDR, and VHUF-VHUR (Table S3) were used to amplify fragments of frcA, vhcD, and vhuD, respectively.
Metabolic labeling with [75Se]selenite
For labeling, M. maripaludis cultures grown to an A600 of 0.8 were diluted 1:50 in 5 ml McCas medium supplemented with 7.5 μCi [75Se]selenite, and incubated at 37 °C for 48 h. Tubes were re-pressurized with H2/CO2 after 24 h. Cells were harvested by centrifugation, washed in McCas medium, and lysed by resuspending in H2Obidest. Cell lysates were subjected to SDS/PAGE, followed by autoradiography.
Supplementary Material
Acknowledgments
We are indebted to Iris Porat for gifts of plasmids and strains and for experimental advice. We thank R. Lynn Sherrer, Yuchen Liu, and Patrick O’Donoghue for helpful discussions and critical reading of the manuscript. M.J.H. was a Feodor Lynen Fellow of the Alexander von Humboldt Foundation (Bonn, Germany). This work was supported by grants from Department of Energy Office of Basic Energy Sciences and the National Institutes of General Medical Sciences (to D.S.).
References
- Aeby E, Palioura S, Pusnik M, Marazzi J, Lieberman A, Ullu E, et al. The canonical pathway for selenocysteine insertion is dispensable in Trypanosomes. Proc Natl Acad Sci USA. 2009;106:5088–5092. doi: 10.1073/pnas.0901575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araiso Y, Palioura S, Ishitani R, Sherrer RL, O’Donoghue P, Yuan J, et al. Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formation. Nucleic Acids Res. 2008;36:1187–1199. doi: 10.1093/nar/gkm1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyle JL, Tumbula DL, Leigh JA. Neomycin resistance as a selectable marker in Methanococcus maripaludis. Appl Env Microbiol. 1996;62:4233–4237. doi: 10.1128/aem.62.11.4233-4237.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axley MJ, Böck A, Stadtman TC. Catalytic properties of an Escherichia coli formate dehydrogenase mutant in which sulfur replaces selenium. Proc Natl Acad Sci USA. 1991;88:8450–8454. doi: 10.1073/pnas.88.19.8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghöfer Y, Agha-Amiri K, Klein A. Selenium is involved in the negative regulation of the expression of selenium-free [NiFe] hydrogenases in Methanococcus voltae. Mol Gen Genet. 1994;242:369–373. doi: 10.1007/BF00281785. [DOI] [PubMed] [Google Scholar]
- Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- Blank CE, Kessler PS, Leigh JA. Genetics in methanogens: transposon insertion mutagenesis of a Methanococcus maripaludis nifH gene. J Bacteriol. 1995;177:5773–5777. doi: 10.1128/jb.177.20.5773-5777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck A, Thanbichler M, Rother M, Resch A. Selenocysteine. In: Ibba M, Francklyn CS, Cusack S, editors. Aminoacyl-tRNA Synthetases. Georgetown, Washington D.C: Landes Bioscience; 2005. pp. 320–327. [Google Scholar]
- Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc Natl Acad Sci USA. 2004;101:12848–12853. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban B, Ng SY, Kanbe M, Saltzman I, Nimmo G, Aizawa S, Jarrell KF. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol Microbiol. 2007;66:596–609. doi: 10.1111/j.1365-2958.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Ching WM. Occurrence of selenium-containing tRNAs in mouse leukemia cells. Proc Natl Acad Sci USA. 1984;81:3010–3013. doi: 10.1073/pnas.81.10.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching WM, Wittwer AJ, Tsai L, Stadtman TC. Distribution of two selenonucleosides among the selenium-containing tRNAs from Methanococcus vannielii. Proc Natl Acad Sci USA. 1984;81:57–60. doi: 10.1073/pnas.81.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching WM, Alzner-DeWeerd B, Stadtman TC. A selenium-containing nucleoside at the first position of the anticodon in seleno-tRNAGlu from Clostridium sticklandii. Proc Natl Acad Sci USA. 1985;82:347–350. doi: 10.1073/pnas.82.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki SM. A modular set of Flp, FRT and lacZ fusion vectors for manipulating genes by site-specific recombination. Gene. 1996;171:197–201. doi: 10.1016/0378-1119(96)00035-2. [DOI] [PubMed] [Google Scholar]
- Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flohe L, Gunzler WA, Schock HH. Glutathione peroxidase: a selenoenzyme. FEBS Lett. 1973;32:132–134. doi: 10.1016/0014-5793(73)80755-0. [DOI] [PubMed] [Google Scholar]
- Forchhammer K, Leinfelder W, Böck A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989;342:453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- Forchhammer K, Böck A. Selenocysteine synthase from Escherichia coli. Analysis of the reaction sequence. J Biol Chem. 1991;266:6324–6328. [PubMed] [Google Scholar]
- Hendrickson EL, Kaul R, Zhou Y, Bovee D, Chapman P, Chung J, et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J Bacteriol. 2004;186:6956–6969. doi: 10.1128/JB.186.20.6956-6969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildenbrand C, Stock T, Lange C, Rother M, Soppa J. Genome copy numbers and gene conversion in methanogenic archaea. J Bacteriol. 2011;193:734–743. doi: 10.1128/JB.01016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser JT, Gromadski K, Rother M, Engelhardt H, Rodnina MV, Wahl MC. Structural and functional investigation of a putative archaeal selenocysteine synthase. Biochemistry. 2005;44:13315–13327. doi: 10.1021/bi051110r. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Gladyshev VN. The prokaryotic selenoproteome. EMBO Rep. 2004;5:538–543. doi: 10.1038/sj.embor.7400126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinfelder W, Forchhammer K, Veprek B, Zehelein E, Böck A. In vitro synthesis of selenocysteinyl-tRNA(UCA) from seryl-tRNA(UCA): involvement and characterization of the selD gene product. Proc Natl Acad Sci USA. 1990;87:543–547. doi: 10.1073/pnas.87.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyznik LA, Rao KV, Hodges TK. FLP-mediated recombination of FRT sites in the maize genome. Nucleic Acids Res. 1996;24:3784–3789. doi: 10.1093/nar/24.19.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BC, Leigh JA. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J Bacteriol. 2005;187:972–979. doi: 10.1128/JB.187.3.972-979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother M, Resch A, Wilting R, Böck A. Selenoprotein synthesis in archaea. Biofactors. 2001;14:75–83. doi: 10.1002/biof.5520140111. [DOI] [PubMed] [Google Scholar]
- Rother M, Mathes I, Lottspeich F, Böck A. Inactivation of the selB gene in Methanococcus maripaludis: effect on synthesis of selenoproteins and their sulfur-containing homologs. J Bacteriol. 2003;185:107–114. doi: 10.1128/JB.185.1.107-114.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother M, Metcalf WW. Genetic technologies for Archaea. Curr Opin Microbiol. 2005;8:745–751. doi: 10.1016/j.mib.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Rotruck JT, Pope AL, Ganther HE, Swanson AB, Hafeman DG, Hoekstra WG. Selenium: biochemical role as a component of glutathione peroxidase. Science. 1973;179:588–590. doi: 10.1126/science.179.4073.588. [DOI] [PubMed] [Google Scholar]
- Sadowski PD. The Flp recombinase of the 2-microns plasmid of Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1995;51:53–91. [PubMed] [Google Scholar]
- Sandbeck KA, Leigh JA. Recovery of an integration shuttle vector from tandem repeats in Methanococcus maripaludis. Appl Env Microbiol. 1991;57:2762–2763. doi: 10.1128/aem.57.9.2762-2763.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, et al. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- Schweizer HP. Applications of the Saccharomyces cerevisiae Flp-FRT system in bacterial genetics. J Mol Microbiol Biotechnol. 2003;5:67–77. doi: 10.1159/000069976. [DOI] [PubMed] [Google Scholar]
- Sherrer RL, Ho JM, Soll D. Divergence of selenocysteine tRNA recognition by archaeal and eukaryotic O-phosphoseryl-tRNASec kinase. Nucleic Acids Res. 2008a;36:1871–1880. doi: 10.1093/nar/gkn036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrer RL, O’Donoghue P, Söll D. Characterization and evolutionary history of an archaeal kinase involved in selenocysteinyl-tRNA formation. Nucleic Acids Res. 2008b;36:1247–1259. doi: 10.1093/nar/gkm1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos C, Kim W, Li T, Anderson I, Deutsch B, Palioura S, et al. Cysteinyl-tRNA synthetase is not essential for viability of the archaeon Methanococcus maripaludis. Proc Natl Acad Sci USA. 2001;98:14292–14297. doi: 10.1073/pnas.201540498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock T, Selzer M, Rother M. In vivo requirement of selenophosphate for selenoprotein synthesis in archaea. Mol Microbiol. 2010;75:149–160. doi: 10.1111/j.1365-2958.2009.06970.x. [DOI] [PubMed] [Google Scholar]
- Sun J, Klein A. A lysR-type regulator is involved in the negative regulation of genes encoding selenium-free hydrogenases in the archaeon Methanococcus voltae. Mol Microbiol. 2004;52:563–571. doi: 10.1111/j.1365-2958.2004.03998.x. [DOI] [PubMed] [Google Scholar]
- Tumbula DL, Makula RA, Whitman WB. Transformation of Methanococcus maripaludis and identification of a Pst I-like restriction system. FEMS Microbiol Lett. 1994;121:309–314. [Google Scholar]
- VanDyke DJ, Wu J, Logan SM, Kelly JF, Mizuno S, Aizawa S, Jarrell KF. Identification of genes involved in the assembly and attachment of a novel flagellin N-linked tetrasaccharide important for motility in the archaeon Methanococcus maripaludis. Mol Microbiol. 2009;72:633–644. doi: 10.1111/j.1365-2958.2009.06671.x. [DOI] [PubMed] [Google Scholar]
- Welander PV, Metcalf WW. Mutagenesis of the C1 oxidation pathway in Methanosarcina barkeri: new insights into the Mtr/Mer bypass pathway. J Bacteriol. 2008;190:1928–1936. doi: 10.1128/JB.01424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Tumbula DL, Yu JP, Kim W. Development of genetic approaches for the methane-producing archaebacterium Methanococcus maripaludis. Biofactors. 1997;6:37–46. doi: 10.1002/biof.5520060105. [DOI] [PubMed] [Google Scholar]
- Wittwer AJ. Specific incorporation of selenium into lysine- and glutamate- accepting tRNAs from Escherichia coli. J Biol Chem. 1983;258:8637–8641. [PubMed] [Google Scholar]
- Wood GE, Haydock AK, Leigh JA. Function and regulation of the formate dehydrogenase genes of the methanogenic archaeon Methanococcus maripaludis. J Bacteriol. 2003;185:2548–2554. doi: 10.1128/JB.185.8.2548-2554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, et al. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Palioura S, Salazar JC, Su D, O’Donoghue P, Hohn MJ, et al. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc Natl Acad Sci USA. 2006;103:18923–18927. doi: 10.1073/pnas.0609703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Hohn MJ, Sherrer RL, Palioura S, Su D, Söll D. A tRNA-dependent cysteine biosynthesis enzyme recognizes the selenocysteine-specific tRNA in Escherichia coli. FEBS Lett. 2010;584:2857–2861. doi: 10.1016/j.febslet.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.