Abstract
Entamoeba histolytica, a protozoan parasite, is an important cause of diarrhea and colitis in the developing world. Amebic colitis is characterized by ulceration of the intestinal mucosa. We performed microarray analysis of intestinal biopsies during acute and convalescent amebiasis in order to identify genes potentially involved in tissue injury or repair. Colonic biopsy samples were obtained from 8 patients during acute E. histolytica colitis and again 60 days after recovery. Gene expression in the biopsies was evaluated using microarray, and confirmed by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR). REG 1A and REG 1B were the most up-regulated of all genes in the human intestine in acute versus convalescent E. histolytica disease: as determined by microarray, the levels of induction were 7.4-fold and 10.7 fold for REG1A and B; p=0.003 and p=0.006 respectively. Increased expression of REG 1A and REG 1B protein in the colonic crypt epithelial cells during acute amebiasis was similarly observed by immunohistochemistry. Because REG 1 protein is anti-apoptotic and pro-proliferative, and since E. histolytica induces apoptosis of the intestinal epithelium as part of its disease process, we next tested if REG 1 might be protective during amebiasis by preventing parasite-induced apoptosis. Intestinal epithelial cells from REG 1 -/- mice were found to be more susceptible to spontaneous, and parasite-induced, apoptosis in vitro (p=0.03). We concluded that REG 1A and REG 1B were upregulated during amebiasis and may function to protect the intestinal epithelium from parasite-induced apoptosis.
Keywords: Entamoeba histolytica, Regenerating gene (REG) 1, apoptosis
1. Introduction
Entamoeba histolytica, the etiologic pathogen of amebiasis, is an important cause of diarrhea and dysentery worldwide. In children, E. histolytica diarrhea is associated with malnourishment and stunting [1]. It is estimated that 50,000 cases of invasive E. histolytica disease occur each year [2], however the number of intestinal infections is even greater since most are asymptomatic. The host and pathogen factors which determine whether mucosal invasion and symptomatic disease occur are not completely understood.
The prevalence of E. histolytica is greatest in areas with inadequate sanitation [3]. Intestinal infection with E. histolytica typically occurs after ingestion of amebic cysts in fecally contaminated food or water. When the cysts reach the terminal ileum or colon, excystation takes place. The resulting trophozoites can re-encyt in the colon and be excreted as infectious cysts in the stool, or may invade through the mucosa and into the submucosal tissues, forming discrete ulcers. It is known that E. histolytica trophozoites can cause host cell death as part of the disease process through both cytolysis and induction of intestinal epithelial cell apoptosis [3,4].
REG1A and REG1B share 89% amino acid homology [5]. REG 1A (islet of langerhans regenerating protein; lithostathine-1-alpha; pancreatic stone protein; pancreatic thread protein; protein-X; regenerating islet-derived protein 1-alpha) is the best studied of the two human REG 1 genes and is known to be expressed in a number of tissues within the gastrointestinal tract [6,7]. In addition to proliferative effects, REG1A is anti-apoptotic, which may be important for gastrointestinal mucosal regeneration [8-10]. To investigate a potential protective role for REG 1 in E. histolytica colitis, here we tested its expression in humans with amebic colitis, and assessed its role in protection of intestinal epithelium from E. histolytica-induced apoptosis.
2. Materials and methods
2.1 Colonic biopsy samples
Colon biopsy samples for microarray, RT-qPCR, and immunohistochemistry (IHC) were obtained from eight patients, ranging in age from 17 to 40, who came to the International Centre for Diarrhoeal Diseases, Bangladesh, with acute E. histolytica diarrhea. Patients with acute disease were stool antigen positive for E. histolytica, had blood and mucous in their stools, and were culture negative for other common diarrheal agents. Colonic biopsies were obtained from an ulcerative region of the sigmoid colon in subjects with an acute episode of E. histolytica diarrhea, and patients were treated with a 10 day course of metronidazole. A follow-up biopsy was obtained from an area of normal sigmoid colon 60 days later during convalescence. Biopsy specimens were placed in RNAlater (Qiagen, CA) and stored at -80°C until isolation of RNA for microarray and RT-qPCR, or collected in Histocon (Histolab, Gutenberg, Sweden) and snap frozen in liquid nitrogen for IHC.
The studies were reviewed and approved by the Institutional Review Board at the University of Virginia and the Ethical Review Committee of the International Centre for Diarrhoeal Diseases Research, Bangladesh.
2.2 Microarray
Samples were prepared according to Affymetrix protocols (Affymetrix, Inc., Santa Clara, CA). RNA quality and quantity was ensured using the Bioanalyzer (Agilent, Inc) and NanoDrop (Thermo Scientific, Inc) respectively. Per RNA labeling, 300 nanograms of total RNA was used in conjunction with the Affymetrix recommended protocol for the GeneChip 1.0 ST chips. The hybridization cocktail containing the fragmented and labeled cDNAs were hybridized to The Affymetrix Human GeneChip® 1.0 ST chips. The chips were washed and stained by the Affymetrix Fluidics Station using the standard format and protocols as described by Affymetrix. The probe arrays were stained with streptavidin phycoerythrin solution (Molecular Probes, Carlsbad, CA) and enhanced by using an antibody solution containing 0.5 mg/mL of biotinylated anti-streptavidin (Vector Laboratories, Burlingame, CA). An Affymetrix Gene Chip Scanner 3000 was used to scan the probe arrays. Gene expression intensities were calculated using GeneChip® Command Console® Software (AGCC) <http://www.affymetrix.com/products_services/software/specific/command_console_software.affx> and Expression Console(tm) Software <http://www.affymetrix.com/products_services/software/specific/expression_console_software.affx>. Cel files generated by the Affymetrix AGCC program were imported in the Affymetrix Expression Consol software and RMA (Robust Multichip Analysis) normalization was performed to generate the .Chp files. The .Chp files are normalized, log2 transformed and summarized. Pairwise comparisons within GeneSifter software (VizX Labs, Seattle, WA) were performed on the .Chp files and t-test and Benjamin and Hochberg correction were applied.
All expression microarray data have been deposited in the Gene Expression Omibus (GEO) database at NCBI at http://www.ncbi.nlm.nih.gov/geo/.
2.3 Real-time polymerase chain reaction
Total RNA from colonic tissue was isolated using TRIzol reagent (Invitrogen, Carlsabad, CA) and reverse transcribed into cDNAs using Superscript II reverse transcriptase. Real-time PCR was performed in duplicate using a standard Quantifast SYBR Green protocol per manufacturer's instructions, Quantifast PCR master mix, and Quantitect PCR primers for REG 1A and REG 1B and the housekeeping genes Peptidylprolyl isomerase A (PPIA) and hypoxanthine phosphoribosyltransferase (HPRT1) (Qiagen, Valencia, CA).
A mixed model repeated measure (MMRM) with a compound symmetry covariance structure was performed using all available C(t) values. The model includes a variable of gene (4 levels for the 4 genes), a variable of day (2 levels for day 1 and day 60) and the interaction between the two variables. This regression model accounts for the correlation between C(t) measures from the same biopsy samples for the 4 genes and between the duplicates. Contrasts were constructed to estimate the means and standard errors of the means for the normalized C(t) for the two study genes, REG1A and REG1B, at day 1 and day 60 and to examine the expression difference between the two study genes.
2.4 Immunohistochemical staining
Immunohistochemistry was performed for the detection of REG 1A and REG 1B in the acute and convalescent colonic biopsy samples. Tissue sections were deparaffined and rehydrated prior to antigen retrieval using the Pascal Pressure Chamber with TRS buffer for 30 seconds at 125°C and 22 psi. Slides were stained using the Dako Autostainer Universal Staining system (DAKO, Denmark). Sections were incubated for 10 minutes with DAKO Dual Endogenous Enzyme Block prior to a 30-minute incubation with the primary antibody, either Rat anti-human REG 1A monoclonal antibody at a 1:150 dilution, or goat anti-human REG 1B polyclonal antibody at a 1:200 dilution (R&D systems, Minneapolis, MN). Sections were subsequently incubated for 30 minutes with rabbit anti-rat biotinylated antibody at 1:200 dilution (DakoCytomation) or anti-goat biotinylated antibody (Vector laboratories, Burlingame, CA) respectively. The sections were then incubated with a 1:200 dilution of horseradish peroxidase streptavidin (Vector laboratories) prior to incubation with chromagen substrate diaminobenzidine tetrahydrochloride (DAB) for 10 minutes. The slides were counterstained with hematoxylin and mounted. Negative controls were performed by omitting the primary antibody. Colon tissue from subjects with ulcerative colitis was used as a positive control. IgG isotype controls were additionally performed.
2.5 Animals
Regenerating gene 1 (REG 1) knock-out mice in the ICR (CD1) background were previously generated by Unno et al [11] by disruption of the exon 2 of REG 1 gene with a targeting vector and subsequent germ-line transmission in chimeric mice. Brother-sister mating of REG 1 +/- mice was carried out in our colony to generate REG 1 -/- and wild type mice.
2.6 Intestinal cell isolation, staining, and sorting
The ceca were removed from 4 wild type littermates and 4 REG 1 -/- mice, placed in 5% Fetal Bovine Serum(FBS)-Hanks media, and separated into pieces prior to incubation in 5% FBS-Hanks/2mM ethylenediaminetetraacetic acid (EDTA) at 37°C for 45 minutes at 250 rpm. The solution was then passed through a column and pelleted by centrifugation. Cells were resuspended in 5% FBS-Hanks and washed. Then cells were incubated with mouse BD Fc block (BD pharminogen, San Jose, CA) prior to incubation with APC anti-mouse CD45 antibody and PE anti-mouse CD49f antibody (Ebiosciences, San Diego, CA). The cells were washed and resuspended in sorting media. DAPI viability dye was added prior to sorting on a Fluorescence-activated cell sorter (FACS) Vantage to obtain CD49f+/CD45- live cells.
2.7 Measurement of apoptosis
Cells obtained at sorting from REG 1 -/- or wild type mice were resuspended at 1×106 cells/mL and aliquotted at 100 l of cells per well of a microtiter plate. E. histolytica were washed 3 times in phosphate buffered saline (PBS), resuspended in homogenization buffer (10mM Tris pH 7.4 and 0.25M sucrose) at 1×106 cells/mL, and dounce homogenized until no intact trophozoites remained. Then 50 l of this E. histolytica lysate, or homogenization buffer without lysate as a control, was added per well of the microtiter plate and incubated for 1 hour. Caspase reagent (Apo-ONE Homogeneous Caspase-3/7 Assay, Promega, Madison, WI) was aliquotted at 50 l per well to a new microtiter plate and 50 l of the cell and lysate, or cell and buffer, mixture was transferred to each well containing caspase reagent. The plate was incubated overnight at room temperature in the dark prior to reading fluorescence (485excitation/527emission). Lysate with cell media and homogenization buffer with cell media were also measured to obtain background fluorescence. All samples were performed in triplicate.
Unpaired t-tests were used to compare differences in apoptosis among intestinal epithelial cells from REG 1-/- mice with or without E. histolytica lysate. One-way analysis of variance (ANOVA) was used to compare differences between REG 1 -/- and wild type intestinal epithelial cells with or without E. histolytica lysate.
3. Results
3.1 Microarray
Colon biopsies from patients with acute E. histolytica colitis were obtained from the sigmoid colon in an area of ulceration. Convalescent biopsies for comparison were obtained from the same patient, from an area of normal sigmoid colon, 60 days later. We analyzed gene transcripts using the Affymetrix Human GeneChip® 1.0 ST chips. REG 1A and REG 1B were the most upregulated genes in the human intestine during acute E. histolytica disease compared to convalescence (p=<0.003 and p=0.006 respectively) (Table 1). The fold change for day 1 over day 60 samples was 7.4 for REG1A and 10.65 for REG1B. The REG 1 receptor (Exostoses-like 3) was not significantly changed. The ten genes with the greatest change in expression in the human colon during acute E. histolytica colitis compared to convalescence can be found in table 2. All microarray expression data has been deposited in Gene Expression Omnibus (GEO), accession number GSE23750.
Table 1.
Colon biopsies from patients with acute E. histolytica colitis were obtained from the sigmoid colon in an area of ulceration. Convalescent biopsies for comparison were obtained from the same patient 60 days later. RNA from colonic biopsy samples was isolated simultaneously from the same sample for microarray and RT-qPCR. (A) Comparison of microarray and (B) RT-qPCR REG 1A and REG 1B mRNA expression between colon biopsy samples from individuals with acute E. histolytica diarrhea (day 1) versus convalescence (day 60)
| Day 1 mean expression (Log2) (±SEM) | Day 60 mean expression (Log2) (±SEM) | Fold change | p-value | |
|---|---|---|---|---|
| REG 1A | 10.69 (0.76) | 7.8 (0.22) | 7.4 | 0.003 |
| REG 1B | 9 (1.02) | 5.59 (0.23) | 10.65 | 0.006 |
| (A) |
| Day 1 normalized CT mean (±SEM) | Day 60 normalized CT mean (±SEM) | CT change mean (SEM) | Expression Ratio (Day 1 /Day 60) | p-value | |
|---|---|---|---|---|---|
| REG 1A | 3.93 (0.86) | 9.98 (0.96) | -6.05(1.29) | 66.3 | <0.001 |
| REG 1B | 4.07 (0.86) | 9.04 (0.96) | -4.97 (1.29) | 31.3 | 0.001 |
| (B) |
Table 2.
Listing of the 10 genes with the greatest change in expression in the human colon during acute E. histolytica colitis compared to convalescence. RNA was isolated from colonic biopsy samples, and gene transcripts were analyzed for microarray using the Affymetrix Human GeneChip® 1.0 ST chips.
| Day 1 mean expression (Log2) (±SEM) | Day 60 mean expression (Log2) (±SEM) | Fold change | p-value | |
|---|---|---|---|---|
| REG1B | 9 (1.02) | 5.59 (0.23) | 10.65 | 0.006 |
| REG1A | 10.69 (0.76) | 7.8 (0.22) | 7.4 | 0.003 |
| MMP3 | 8.04 (0.73) | 5.49 (0.17) | 5.86 | 0.004 |
| S100A8 | 8.28 (0.87) | 6.08 (0.17) | 4.59 | 0.026 |
| MMP1 | 8.85 (0.62) | 6.8 (0.26) | 4.14 | 0.008 |
| DMBT1 | 11.43 (0.22) | 9.51 (0.46) | 3.8 | 0.002 |
| CHI3L1 | 8.3 (0.50) | 6.48 (0.19) | 3.54 | 0.004 |
| AQP8 | 8.89 (0.67) | 10.6 (0.33) | 3.28 | 0.039 |
| SLC26A2 | 9.59 (0.56) | 11.11 (0.17) | 2.88 | 0.022 |
| OLFM4 | 12.16 (0.25) | 10.76 (0.38) | 2.64 | 0.009 |
3.2 PCR
RNA from colonic biopsy samples was isolated simultaneously from the same sample for microarray and PCR. Quantitative PCR confirmed the microarray results that REG1A and REG1B expression are significantly increased in colonic biopsy samples during acute E. histolytica disease compared to convalescence (p=<0.001 and p=0.001 respectively) (Table 1). The expression ratio for day 1 over day 60 samples was 66.3 for REG1A and 31.3 for REG1B.
3.3 Immunohistochemistry
REG 1A and REG1B staining was observed in the epithelial cells of the intestinal crypts during acute E. histolytica disease. The staining was nearly absent during convalescence (Figure 1). The staining was most intense in the crypts with a gradient decreasing as the cells migrated to the apical surface. Predominant staining of the intestinal crypt epithelial cells during inflammatory colitis has also been noted by others [10,12]. REG 1A and REG 1B staining were not detected in the negative controls when primary antibody was excluded, or with the IgG isotype controls (data not shown).
Figure 1.
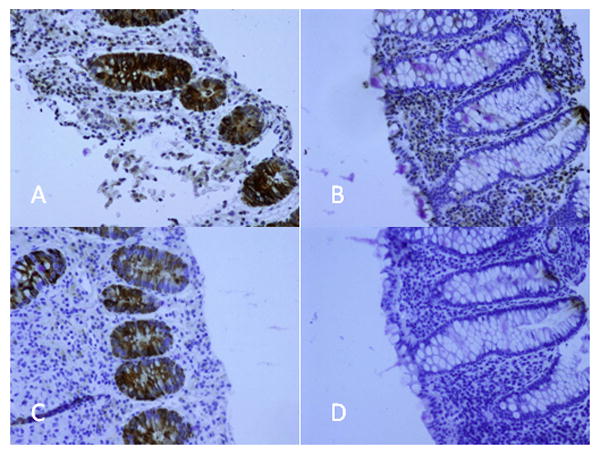
REG 1 immunohistochemical staining of human biopsy samples taken from the sigmoid colon in an area of active colitis during acute E. histolytica diarrhea (day 1) and normal sigmoid colon during convalescence (day 60). A) REG 1A staining of an ulcer region during acute E. histolytica diarrhea. B) REG 1A staining during convalescence. C) REG 1B staining of an ulcer region during acute E. histolytica diarrhea. D) REG 1B staining during convalescence.
3.4 Apoptosis of intestinal epithelial cells
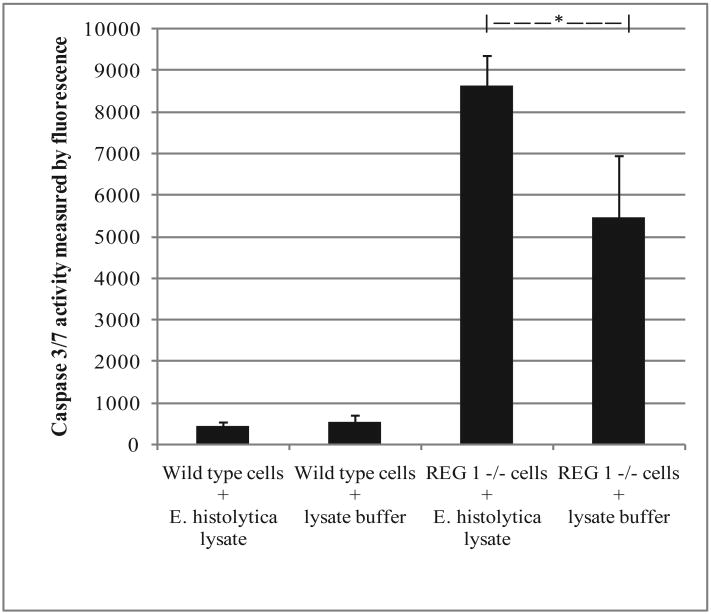
To determine if REG 1 might be protective during amebiasis by preventing parasite-induced apoptosis, we evaluated E. histolytica-induced apoptosis of intestinal epithelial cells in REG 1 -/- or wild type mice. E. histolytica lysate was used to induce apoptosis instead of E. histolytica trophozoites, as the latter are able to engulf apoptotic cells, limiting the ability to measure apoptotic cells. Intestinal cells were isolated from REG 1 -/- or wild type mice and sorted by FACS for CD49f+/CD45- live cells to obtain epithelial cells. The intestinal epithelial cells were then incubated with E. histolytica lysate or buffer without lysate prior to adding caspase reagent for determination of Caspase 3/7 activity by fluorescence. Intestinal epithelial cells from REG 1 -/- mice had increased apoptosis compared to cells from wild type mice (p=<0.001). E. histolytica lysate enhanced apoptosis of intestinal epithelial cells from REG 1 -/-mice (p=0.03) (Figure 2).
Figure 2.
Comparison of in vitro apoptosis between intestinal epithelial cells from REG 1 +/+ (wild type) or REG 1 -/- mice, with E. histolytica lysate or with lysate buffer only (without E. histolytica). Intestinal cells were isolated from REG 1 -/- or wild type mice and sorted by FACS for CD49f+/CD45- live cells to obtain epithelial cells. The intestinal epithelial cells were then incubated with E. histolytica lysate or lysate buffer only prior to adding caspase reagent for determination of Caspase 3/7 activity by fluorescence. Intestinal epithelial cells from REG 1 -/-mice had increased apoptosis compared to cells from wild type mice (p=<0.001). E. histolytica lysate enhanced apoptosis of intestinal epithelial cells from REG 1 -/- mice (*p=0.03).
4. Discussion
We identified genes with differential expression within the human colon during acute and convalescent amebic colitis. REG 1A and REG1B were the most upregulated genes during acute amebiasis, and may provide protection for the intestinal epithelium. Evidence suggests REG 1 is involved in tissue repair [13,14], cell growth and differentiation [9,15 -16]. In addition, REG 1 expression occurs in areas of cell renewal, including intestinal crypt epithelial cells [7,10].
We confirmed the significant differences between acute and convalescent REG 1A and REG 1B expression in colonic biopsy samples by real-time PCR. These findings are in keeping with other studies which have found REG 1 expression is increased in the colon in the setting of inflammation. One study found upregulation of REG1A mRNA in epithelium from inflamed colonic tissue compared to inactive inflammatory bowel disease (IBD) or normal colonic tissue [17]. Other studies have also found increased REG1A mRNA expression in colonic tissue affected by ulcerative colitis [12] or Crohn's disease [10] compared to normal colonic tissue, with the level of expression positively correlated with the severity of inflammation [10].
Studies evaluating REG1A expression by immunohistochemistry have found that REG1A protein expression in normal colonic tissue is detectable only among epithelial cells in the basal portion of crypts. The intensity and number of positively stained crypt cells increased within ulcerative colitis tissue [10,12]. A study using in situ hybridization showed REG1A to be localized to epithelial cells of the intestinal crypts in the setting of active colitis, with minimal expression in mesenchymal cells, and no expression in normal colonic tissue [17].
In the current study, REG 1A and REG 1B staining was intense during the acute stage of E. histolytica colitis and present throughout the intestinal cells of the crypt epithelium. In contrast, both REG 1A and REG 1B staining were limited to the basal portion of the colonic crypts after recovery. These findings are similar to other studies assessing REG 1A expression in IBD or normal colon [10,12], and also correspond with our microarray and PCR data showing markedly increased REG 1A and REG 1B expression during acute E. histolytica colitis. REG 1A, but not REG 1B, staining was also detectable in the lamina propria of both acute and convalescent samples. REG 1A and REG 1B staining were not detected in the negative controls when primary antibody was excluded, or with the IgG isotype controls.
REG1A has been shown to have anti-apoptotic effects in gastric and colon cancer lines [8-10] through activation of the Akt/Bad/Bcl-xL pathway [18]. In contrast, E. histolytica is able to induce apoptosis of the intestinal epithelium [4]. Host intestinal cell apoptosis by E. histolytica occurs by activation of host caspase-3 in a caspase-8 and -9 independent manner [19]. To test the hypothesis that REG 1 may be protective in E. histolytica disease due to its anti-apoptotic effects, we evaluated E. histolytica-induced apoptosis of intestinal epithelial cells from REG 1 -/- or REG 1 +/+ mice. We observed increased apoptosis of intestinal epithelial cells from REG 1 -/- mice compared to intestinal epithelial cells from REG1 +/+ mice. E. histolytica lysate further enhanced apoptosis in the REG 1-/- epithelium.
This study is the first to assess differential gene expression during acute and convalescent amebic colitis outside of an animal model. Of the ten genes most significantly changed in the human colon between acute and convalescent E. histolytica disease, including REG 1A and REG 1B, eight were upregulated during acute disease. Matrix metalloproteinases-1 and -3 (MMP-1, MMP-3) are involved in tissue injury and repair, S100 calcium binding protein A8 (S100A8) is expressed in neutrophil and monocyte cytosol and increased with inflammation [20], Deleted in malignant brain tumors 1 (DMBT1) is involved in epithelial cell differentiation [21] and innate immunity [22], and Olfactomedin 4 (OLFM4) has a role in cell adhesion [23] and anti-apoptosis [24]. The apical water channel, Aquaporin 8 (AQP8), and Solute carrier family 26, member 2 (SLC26A2), were decreased during acute disease.
Many of the genes which were differentially expressed during amebiasis are similarly involved in other inflammatory intestinal diseases. The top 5 most upregulated genes during acute amebiasis in the current study (REG1A, REG1B, MMP-1, S100A8, and MMP-3) were found, in a study by Dieckgraefe et al which used genome wide expression analysis, to be more abundant in colonic tissue during active ulcerative colitis as compared to non-inflamed control tissue [25]. In another study, DMBT1 expression was found to be increased in inflammatory bowel disease tissue compared to controls, correlated with disease activity, and predominately occurred within intestinal epithelial and Paneth cells [26]. Aquaporin 8 was also shown to be downregulated in three experimental models of colitis in mice, and in affected colonic tissue of patients with inflammatory bowel disease compared to nonaffected colon [27].
A mouse model has been used to compare transcriptional responses within human colonic xenografts at four and twenty-four hours post-infection with either Shigella flexneri or E. histolytica [28]. Several of the genes elevated post-infection with either organism, similar to the current study, encoded proteins involved in tissue injury and repair including matrix metalloproteinases, tissue inhibitor of metalloproteinase 1, and cystine-rich angiogenic inducer. Unlike the current study, increased expression of several leukotrienes (1β, -6, -8, -11) and monocyte chemotactic proteins (-1, -2, -3) were also found. Notably, a mouse model of Citrobacter rodentium, a murine attaching and effacing pathogen similar to enteropathogenic and enterohaemorrhagic Escherichia coli, found that genes differentially expressed between susceptible and resistant strains of mice were greatly enriched for transport activity [29]. More transport genes than immune-related genes were demonstrated to determine susceptibility to infection. We also found differential expression of several transport molecules during acute and convalescent amebic colitis, including seven solute family members and aquaporin 8.
There were limitations in this study. The increased spontaneous apoptosis in REG 1 -/-epithelial cells found in vitro requires confirmation in vivo in uninfected and E. histolytica-infected mice. In addition, the incremental increase in apoptosis seen with amebic lysate should be tested to see if it is specific to E. histolytica, since lysate from other parasites or pathogens was not tested. Other potential mechanisms of REG 1 protection during amebiasis, such as its pro-proliferative effects, should also be evaluated. Further studies in vivo are planned to assess the susceptibility and severity of amebic disease in REG 1 -/- and wild type mice.
In summary, we found that REG 1A and REG 1B mRNA and protein expression were significantly increased in the human intestine during acute E. histolytica colitis. We also found that apoptosis was increased in REG 1 -/- intestinal epithelial cells compared to REG 1 +/+ cells, and E. histolytica lysate further enhanced apoptosis. Therefore, we hypothesize that increased REG 1 expression may be beneficial during E. histolytica colitis through a protective role against E. histolytica-induced apoptosis.
Supplementary Material
Acknowledgments
We thank Patcharin Pramoonjago for her assistance with immunohistochemistry. This work was supported by grant AI-43596 (to WP) and K08A1072470 (to KP) from the National Institutes of Health, and also funded in part by the intramural division of the National Human Genome Research Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mondal D, Petri WAJ, Sack RB, Haque R. Entamoeba histolytica-associated diarrheal illness is negatively associated with the growth of preschool children: Evidence from a prospective study. Trans R Soc Trop Med Hyg. 2006;100:1032–8. doi: 10.1016/j.trstmh.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin Microbiol Rev. 2007;20(3):511–32. doi: 10.1128/CMR.00004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanley SL., Jr Amoebiasis. Lancet. 2003;361:1025–34. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- 4.Blazquez S, Rigothier M, Huerre M, Guillén N. Initiation of inflammation and cell death during liver abscess formation by Entamoeba histolytica depends on activity of galactose/N-acetyl-d-galactosamine lectin. Int J Parasitol. 2007;37(3-4):425–33. doi: 10.1016/j.ijpara.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez D, Figarella C, Marchand-Pinatel S, Bruneau N, Guy-Crotte O. Preferential expression of reg I beta gene in human adult pancreas. Biochem Biophys Res Commun. 2001;284(3):729–37. doi: 10.1006/bbrc.2001.5033. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe T, Yonekura H, Terazono K, Yamamoto H, Okamoto H. Complete nucleotide sequence of human reg gene and its expression in normal and tumoral tissues. The reg protein, pancreatic stone protein, and pancreatic thread protein are one and the same product of the gene. J Biol Chem. 1990;265(13):7432–9. [PubMed] [Google Scholar]
- 7.Senegas-Balas FO, Figarella CG, Amouric MA, Guy-Crotte OM, Bertrand CA, Balas DC. Immunocytochemical demonstration of a pancreatic secretory protein of unknown function in human duodenum. J Histochem Cytochem. 1991;39(7):915–9. doi: 10.1177/39.7.1865108. [DOI] [PubMed] [Google Scholar]
- 8.Dieckgraefe BK, Crimmins DL, Landt V, Houchen C, Anant S, Porche-Sorbet R, et al. Expression of the regenerating gene family in inflammatory bowel disease mucosa: Reg 1alpha upregulation, processing, and antiapoptotic activity. J Investig Med. 2002;50(6):421–34. doi: 10.1136/jim-50-06-02. [DOI] [PubMed] [Google Scholar]
- 9.Sekikawa A, Fukui H, Fujii S, Takeda J, Nanakin A, Hisatsune H, et al. REG 1 alpha protein may function as a trophic and/or anti-apoptotic factor in the development of gastric cancer. Gastroenterology. 2005;128(3):642–53. doi: 10.1053/j.gastro.2004.12.045. [DOI] [PubMed] [Google Scholar]
- 10.Sekikawa A, Fukui H, Fujii S, Nanakin A, Kanda N, Uenoyama Y, et al. Possible role of REG 1alpha protein in ulcerative colitis and colitic cancer. 2005;54(10):1437–44. doi: 10.1136/gut.2004.053587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unno M, Nata K, Noguchi N, Narushima Y, Akiyama T, Ikeda T, et al. Production and characterization of REG knockout mice: Reduced proliferation of pancreatic beta-cells in REG knockout mice. Diabetes. 2002;51(suppl 3):S478–83. doi: 10.2337/diabetes.51.2007.s478. [DOI] [PubMed] [Google Scholar]
- 12.Sekikawa A, Fukui H, Suzuki K, Karibe T, Fujii S, Ichikawa K, et al. Involvement of the IL-22/REG I alpha axis in ulcerative colitis. Lab Invest. 2010;90(3):496–505. doi: 10.1038/labinvest.2009.147. [DOI] [PubMed] [Google Scholar]
- 13.Kawanami C, Fukui H, Kinoshita Y, Nakata H, Asahara M, Matsushima Y, et al. Regenerating gene expression in normal gastric mucosa and indomethacin-induced mucosal lesions of the rat. J Gastroenterol. 1997;32(1):12–8. doi: 10.1007/BF01213290. [DOI] [PubMed] [Google Scholar]
- 14.Fukuhara H, Kadowaki Y, Ose T, Monowar A, Imaoka H, Ishihara S, et al. In vivo evidence for the role of RegI in gastric regeneration: Transgenic overexpression of RegI accelerates the healing of experimental gastric ulcers. Lab Invest. 2010;90(4):556–65. doi: 10.1038/labinvest.2010.42. [DOI] [PubMed] [Google Scholar]
- 15.Gross DJ, Weiss L, Reibstein I, van den Brand J, Okamoto H, Clark A, et al. Amelioration of diabetes in nonobese diabetic mice with advanced disease by linomide-induced immunoregulation combined with reg protein treatment. Endocrinology. 1998;139(5):2369–74. doi: 10.1210/endo.139.5.5997. [DOI] [PubMed] [Google Scholar]
- 16.Fukui H, Kinoshita Y, Maekawa T, Okada A, Waki S, Hassan S, et al. Regenerating gene protein may mediate gastric mucosal proliferation induced by hypergastrinemia in rats. Gastroenterology. 1998;115:1483–93. doi: 10.1016/s0016-5085(98)70027-7. [DOI] [PubMed] [Google Scholar]
- 17.Shinozaki S, Nakamura T, Limura M, Kato Y, Lizuka B, Kobayashi M, et al. Upregulation of reg 1alpha and GW112 in the epithelium of inflamed colonic mucosa. Gut. 2001;48:623–9. doi: 10.1136/gut.48.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekikawa A, Fukui H, Fujii S, Ichikawa K, Tomita S, Imura J, et al. REG 1 alpha protein mediates an anti-apoptotic effect of STAT3 signaling in gastric cancer cells. Carcinogenesis. 2008;29(1):76–83. doi: 10.1093/carcin/bgm250. [DOI] [PubMed] [Google Scholar]
- 19.Huston CD, Houpt ER, Mann BJ, Hahn CS, Petri WA., Jr Caspase-3 dependent killing of host cells by the parasite Entamoeba histolytica. Cell Microbiol. 2000;2:617. doi: 10.1046/j.1462-5822.2000.00085.x. [DOI] [PubMed] [Google Scholar]
- 20.Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier P. Proinflammatory activities of S100: Proteins S100A8, S100A9, and S100 A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. 2003;170:3233–42. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- 21.Takito J, Al-Awqati Q. Conversion of ES cells to columnar epithelia by hensin and to squamous epithelia by laminin. J Cell Biol. 2004;166(7):1093–102. doi: 10.1083/jcb.200405159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madsen J, Tornøe I, Nielsen O, Lausen M, Krebs I, Mollenhauer J, et al. CRP-ductin, the mouse homologue of gp-340/deleted in malignant brain tumors 1 (DMBT 1), binds gram-positive and gram-negative bacteria and interacts with lung surfactant protein D. Eur J Immunol. 2003;33:2327–36. doi: 10.1002/eji.200323972. [DOI] [PubMed] [Google Scholar]
- 23.Grover PK, Hardingham JE, Cummins AG. Stem cell marker olfactomedin 4: Critical appraisal of its characteristics and role in tumorigenesis. Cancer Metastasis Rev. 2010;29(4):761–75. doi: 10.1007/s10555-010-9262-z. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Huang Q, Yang Z, Li Y, Li CY. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer Res. 2004;64:2474–81. doi: 10.1158/0008-5472.can-03-3443. [DOI] [PubMed] [Google Scholar]
- 25.Dieckgraefe BK, Stenson WF, Korzenik JR, Swanson PE, Harrington CA. Analysis of mucosal gene expression in inflammatory bowel disease by parallel oligonucleotide arrays. Physiol Genomics. 2000;4:1–11. doi: 10.1152/physiolgenomics.2000.4.1.1. [DOI] [PubMed] [Google Scholar]
- 26.Renner M, Bergmann G, Krebs I, End C, Lyer S, Hilberg F, et al. DMBT1 confers mucosal protection in vivo and detection variant is associated with crohn's disease. Gastroenterology. 2007;133:1499–509. doi: 10.1053/j.gastro.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Te Velde AA, Pronk I, de Kort F, Stokkers PC. Glutathione peroxidase 2 and aquaporin 8 as new markers for colonic inflammation in experimental colitis and inflammatory bowel diseases: An important role for H2O2? Eur J Gastroenterol Hepatol. 2008;20(6):555–60. doi: 10.1097/MEG.0b013e3282f45751. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Stanley SL., Jr Stereotypic and specific elements of the human colonic response to Entamoeba histoltyica and Shigella flexneri. Cell Microbiol. 2004;6(6):535–54. doi: 10.1111/j.1462-5822.2004.00381.x. [DOI] [PubMed] [Google Scholar]
- 29.Borenshtein D, Fry RC, Groff EB, Nambiar PR, Carey VJ, Fox JG, et al. Diarrhea as a cause of mortality in a mouse model of infectious colitis. Genome Biol. 2008;9(8):R122. doi: 10.1186/gb-2008-9-8-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.