Abstract
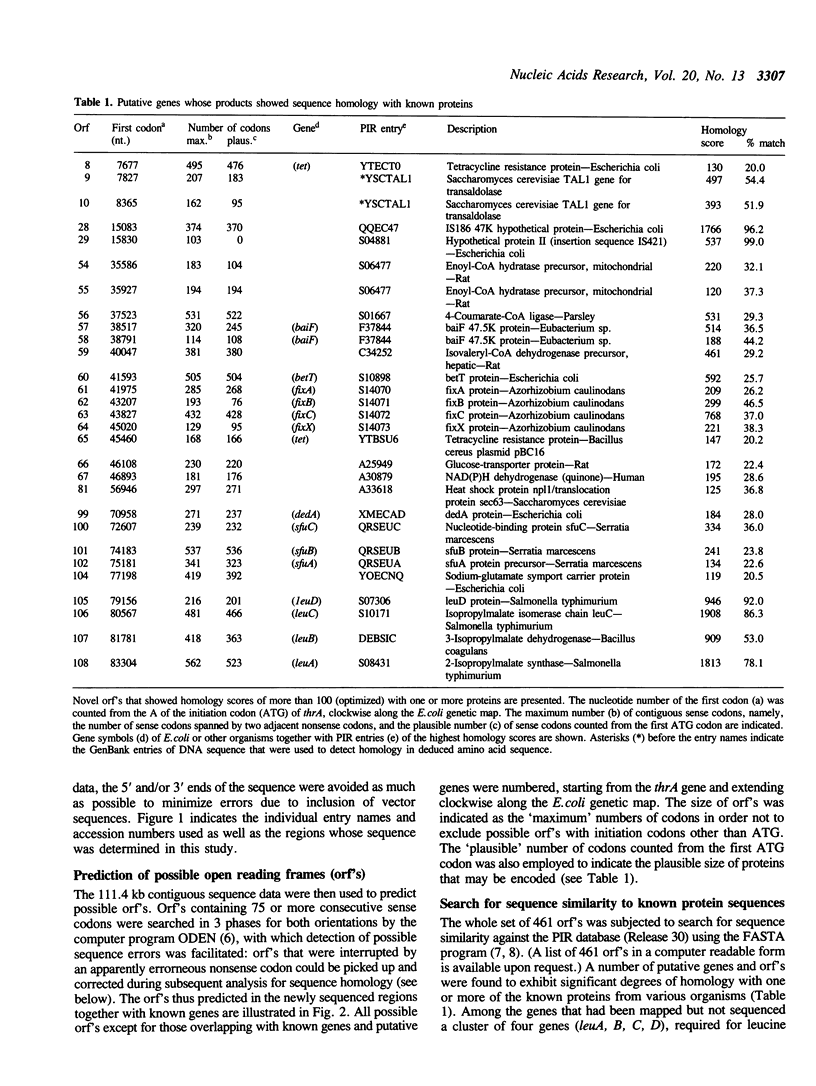
A contiguous 111,402-nucleotide sequence corresponding to the 0 to 2.4 min region of the E. coli chromosome was determined as a first step to complete structural analysis of the genome. The resulting sequence was used to predict open reading frames and to search for sequence similarity against the PIR protein database. A number of novel genes were found whose predicted protein sequences showed significant homology with known proteins from various organisms, including several clusters of genes similar to those involved in fatty acid metabolism in bacteria (e.g., betT, baiF) and higher organisms, iron transport (sfuA, B, C) in Serratia marcescens, and symbiotic nitrogen fixation or electron transport (fixA, B, C, X) in Azorhizobium caulinodans. In addition, several genes and IS elements that had been mapped but not sequenced (e.g., leuA, B, C, D) were identified. We estimate that about 90 genes are represented in this region of the chromosome with little spacer.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angerer A., Gaisser S., Braun V. Nucleotide sequences of the sfuA, sfuB, and sfuC genes of Serratia marcescens suggest a periplasmic-binding-protein-dependent iron transport mechanism. J Bacteriol. 1990 Feb;172(2):572–578. doi: 10.1128/jb.172.2.572-578.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigoni F., Kaminski P. A., Hennecke H., Elmerich C. Nucleotide sequence of the fixABC region of Azorhizobium caulinodans ORS571: similarity of the fixB product with eukaryotic flavoproteins, characterization of fixX, and identification of nifW. Mol Gen Genet. 1991 Mar;225(3):514–520. doi: 10.1007/BF00261695. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong P., Hui I., Loo T., Gillam S. Structural analysis of a new GC-specific insertion element IS186. FEBS Lett. 1985 Nov 11;192(1):47–52. doi: 10.1016/0014-5793(85)80040-5. [DOI] [PubMed] [Google Scholar]
- Friedberg D., Rosenthal E. R., Jones J. W., Calvo J. M. Characterization of the 3' end of the leucine operon of Salmonella typhimurium. Mol Gen Genet. 1985;199(3):486–494. doi: 10.1007/BF00330763. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kröger M., Wahl R., Rice P. Compilation of DNA sequences of Escherichia coli (update 1991). Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2023–2043. doi: 10.1093/nar/19.suppl.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mallonee D. H., White W. B., Hylemon P. B. Cloning and sequencing of a bile acid-inducible operon from Eubacterium sp. strain VPI 12708. J Bacteriol. 1990 Dec;172(12):7011–7019. doi: 10.1128/jb.172.12.7011-7019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami-Ishii N., Taketani S., Osumi T., Hashimoto T. Molecular cloning and sequence analysis of the cDNA for rat mitochondrial enoyl-CoA hydratase. Structural and evolutionary relationships linked to the bifunctional enzyme of the peroxisomal beta-oxidation system. Eur J Biochem. 1989 Oct 20;185(1):73–78. doi: 10.1111/j.1432-1033.1989.tb15083.x. [DOI] [PubMed] [Google Scholar]
- Médigue C., Viari A., Hénaut A., Danchin A. Escherichia coli molecular genetic map (1500 kbp): update II. Mol Microbiol. 1991 Nov;5(11):2629–2640. doi: 10.1111/j.1365-2958.1991.tb01972.x. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis S. H., Kulakauskas S., Krishnan B. R., Hiemstra J., Berg D. E. Transposon Tn5supF-based reverse genetic method for mutational analysis of Escherichia coli with DNAs cloned in lambda phage. J Bacteriol. 1991 Jan;173(2):896–899. doi: 10.1128/jb.173.2.896-899.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricca E., Calvo J. M. The nucleotide sequence of leuA from Salmonella typhimurium. Nucleic Acids Res. 1990 Mar 11;18(5):1290–1290. doi: 10.1093/nar/18.5.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal E. R., Calvo J. M. The nucleotide sequence of leuC from Salmonella typhimurium. Nucleic Acids Res. 1990 May 25;18(10):3072–3072. doi: 10.1093/nar/18.10.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaff I., Hohmann S., Zimmermann F. K. Molecular analysis of the structural gene for yeast transaldolase. Eur J Biochem. 1990 Mar 30;188(3):597–603. doi: 10.1111/j.1432-1033.1990.tb15440.x. [DOI] [PubMed] [Google Scholar]
- Umeda M., Ohtsubo E. Four types of IS1 with differences in nucleotide sequence reside in the Escherichia coli K-12 chromosome. Gene. 1991 Feb 1;98(1):1–5. doi: 10.1016/0378-1119(91)90096-t. [DOI] [PubMed] [Google Scholar]




