Abstract
One approach to the growing health problem of antibiotic resistant bacteria is the development of antimicrobial peptides (AMPs) as alternative treatments. The mechanism by which these AMPs selectively attack the bacterial membrane is not well understood, but is believed to depend on differences in membrane lipid composition. N-acylation of the small amidated hexapeptide, RRWQWR-NH2 (LfB6) derived from the 25 amino acid bovine lactoferricin (LfB25) can be an effective means to improve its antimicrobial properties. Here, we investigate the interactions of C6-LfB6, N-acylated with a 6 carbon fatty acid, with model lipid bilayers with two distinct compositions: 3:1 POPE:POPG (negatively charged) and POPC (zwitterionic). Results from solid-state 2H and 31P NMR experiments are compared with those from an ensemble of all-atom molecular dynamics simulations running in aggregate more than 8.6 microseconds. 2H NMR spectra reveal no change in the lipid acyl chain order when C6-LfB6 is bound to the negatively charged membrane and only a slight decrease in order when it is bound to the zwitterionic membrane. 31P NMR spectra show no significant perturbation of the phosphate headgroups of either lipid system in the presence of C6-LfB6. Molecular dynamics simulations show that for the negatively charged membrane, the peptide’s arginines drive the initial association with the membrane, followed by attachment of the tryptophans at the membrane-water interface, and finally by the insertion of the C6 tails deep into the bilayer. In contrast, the C6 tail leads the association with the zwitterionic membrane, with the tryptophans and arginines associating with the membrane-water interface in roughly the same amount of time. We find similar patterns in the order parameters from our simulations. Moreover, we find in the simulations that the C6 tail can insert 1–2 Å more deeply into the zwitterionic membrane and can exist in a wider range of angles than in the negatively charged membrane. We propose this is due to the larger area per lipid in the zwitterionic membrane, which provides more space for the C6 to insert and assume different orientations.
1. Introduction
An increase in bacterial resistance to conventional antibiotics has led to an intense search for alternative treatments. The hexapeptide lactoferricin B6 (LfB6; RRWQWR-NH2) is the minimal active sequence of bovine lactoferrin[1] that retains broad spectrum antimicrobial activity, and is therefore an attractive candidate for drug development. Antimicrobial peptides (AMPs), produced in all animal kingdoms, are vital components of the innate immune response, acting as a first line of defense against diverse pathogens[2]. They are relatively short peptides, consisting of fewer than 50 amino acids, generally with amphipathic α-helical, β-sheet, or loop conformations. Most have a net positive charge due to an excess of the cationic amino acids lysine (K; Lys), arginine (R; Arg), and/or histidine (H; His), and more than 50% hydrophobic amino acids[3]. Unlike conventional antibiotics that target specific proteins, these peptides exert their effects directly on cellular membrane lipids, and therefore are less susceptible to inducing bacterial resistance[4]. In the case of LfB6, the presence of three arginines is thought to promote selective interaction with bacterial cell membranes while the two tryptophans (W; Trp) prefer to reside in the membrane-water interface.
Although many questions regarding the exact mechanism by which AMPs inhibit the growth (bacteriostatic) or kill (bactericidal) bacterial pathogens remain unanswered, it is nevertheless believed that it involves an initial electrostatic interaction between the cationic peptide and negative charges on the bacterial membrane. The interfacial activity of an AMP, which determines how well it binds to and partitions into the membrane-water interface, frequently altering the packing and organization of the lipids, depends on an intricate balance of hydrophobic and electrostatic interactions between the peptides, water and the membrane lipids [5]. The combination, in addition to the relative position, of the cationic and hydrophobic amino acids present in AMPs promote interaction with the anionic enriched surfaces of bacterial membranes [6]. In contrast to mammalian cell membranes that are comprised primarily of zwitterionic lipids, bacterial membranes generally contain a significant fraction (≈20–25%) of negatively charged lipids, such as phosphatidylglycerol (PG). The distinct lipid compositions of bacterial and eukaryotic cell membranes therefore play key roles in modulating interactions between AMPs and membranes[7, 8, 9]. For instance, the natural antimicrobial pepsin cleavage product of lactoferrin, LfB25 (FKCRRWQWRMKKLGAPSITCVRRAF), that contains the sequence of LfB6 (underlined), was shown to partition with higher affinity to acidic phospholipid bilayers, causing the formation of membrane pore defects[10]. Although most AMPs interact initially with microbial membranes, the membrane may not always be the site of lethal action. Some AMPs are also thought to exert their effects upon binding to intracellular targets after traversing the microbial membrane(s)[11].
Modification of AMPs by N-acylation can be an effective method to increase their interaction with microbial membranes and thereby improve their biological activity[12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22]. We have recently reported that the activity of LfB6 peptides can be enhanced by a combination of N-acylation and Trp-methylation[12]. Here we report on the differential effects of the LfB6 peptide aminoacylated with a 6-carbon chain (C6-LfB6; CH3(CH2)4CO-RRWQWR-NH2) on model bacterial-like and mammalian-like membranes using all-atom molecular dynamics simulations and solid-state 2H and 31P NMR spectroscopy. To mimic bacterial membranes, a 3:1 mixture of zwitterionic POPE (1-palmitoyl-2-oleoyl phosphatidylethanolamine) and anionic POPG (1-palmitoyl-2-oleoyl phosphatidylglycerol) lipids was used. We used POPC (1-palmitoyl-2-oleoyl phosphatidylcholine) as a model for a mammalian-like membrane.
We examined lipid order parameters in both types of membranes using molecular dynamics simulations and solid-state 2H NMR methods. Additionally, the 31P chemical shift anisotropy (CSA) was monitored as a possible indicator of peptide interactions with lipid head groups. The MD simulations are consistent with the experimental data, and in addition reveal details of the peptide’s binding mechanism. Our findings provide insights into the subtle but fundamental differences by which short acylated cationic peptides interact with anionic versus zwitterionic membranes.
2. Methods
2.1. Peptide Synthesis
The C6-LfB6 peptide was synthesized using solid-phase Fmoc methods on an Applied Biosystems (Foster City, CA) 433A peptide synthesizer using modified FastMoc® chemistry according to methods described in Greathouse et al.[12] Peptide purity was assessed by reversed phase HPLC using a 4.6 × 50 mm Zorbax SB-C8 column packed with 3.5 μl octylsilica (Agilent Technologies, Santa Clara CA), at 1 mL/min using a methanol/water gradient (including 0.1% TFA) from 10% to 60% over 8 min followed by a hold at 60% for 5 min. Chromatograms revealed a single major peak and mass spectrometry confirmed the correct mass (1084); therefore, the peptide was used without further purification. Peptide concentrations were quantified based upon UV absorbance at 280 nm, using an extinction coefficient of 5,600 M−1cm−1 per Trp.
2.2. Antimicrobial Assay
The antimicrobial activity of LfB6 and C6-LfB6 was assayed against E. coli (ATCC 25922) and S. aureus (ATCC 29213). The MIC of each peptide was determined using standard microdilution broth assay methods[23], adapted for cationic peptides by Hancock[24]. Briefly, 500 μl of overnight cultures in Mueller Hinton broth (MHB) was diluted in 25 ml fresh MHB, incubated at 37° C to exponential phase (optical density at 600 nm of 0.6), and then diluted 1 × 10−5 in fresh MHB. Serial dilutions (200, 100, 50, 25, 12.5, 6.25, 3.125, 1.6 μg/ml) of each peptide were made in 0.2% bovine serum albumin-0.01% acetic acid solution. Each well of a 96-well polypropylene microtiter plate was inoculated with a total volume of 100 μl (approximately 1 × 108 colony forming units/ml). The MIC was taken as the lowest peptide concentration at which growth was inhibited after 24 h of incubation at 37° C as determined by measuring optical density at 600 nm. The MBC was confirmed when no bacterial colonies were observed after 100 μl from the 96-well plate was spread onto an MHB agar plate and incubated overnight at 37° C. Each peptide was assayed at least twice.
2.3. Sample Preparation for Solid-State NMR Spectroscopy
Mechanically aligned samples for solid-state NMR spectroscopy were prepared by combining C6-LfB6 (peptide/lipid, 1/100) with mixtures of non-deuterated and sn-1 chain perdeuterated (Avanti Polar Lipids, Alabaster, AL) POPE-POPE-d31-POPG (2:1:1), POPE-POPG-d31 (3:1) and POPC:POPC-d31(85:15) based on the stacked glass plate procedure outlined by van der Wel et al[25]. Briefly, from a stock solution (acetonitrile/water, 50/50), 0.25 μmol peptide was added to a 25 ml glass vial. After removal of the solvent under a stream of N2 gas, 25 μmol total lipid (POPC or POPE:POPG) was added from chloroform, and the solvent was again removed, first with N2 gas and then under high vacuum (10 mtorr) over night. The peptide-lipid films were resuspended in methanol (400 μl), chloroform (400 μl), and water (50 μl). Using a glass syringe, the solution was applied in increments of 10 μl to 40 glass slides (4.8 × 23 × 0.07 mm; Marienfeld, Lauda-Konigshofen, Germany) in two glass Petri dishes. The slides were dried in a dessicator under high vacuum for 48 hours to remove all traces of solvent, and the peptide-lipid films were hydrated with excess water (50%, w/w; ≈ 50 molar equivalents of water per lipid molecule), by applying deuterium-depleted water (Cambridge Isotope Labs, Andover, MA) to each of the 40 slides in 3 to 4 small drops. The slides were stacked in increments of 5–6, and slight pressure was applied before each stack was inserted into a glass cuvette. After filling any remaining space with blank glass slides, the cuvette was capped with glass, sealed with epoxy, and equilibrated in a 40° C heating block for at least 48 hours to allow bilayers to form. To avoid artifacts associated with lipid hydrolysis, which was observed in some samples after 4–6 weeks, NMR spectra were acquired within 2 weeks of sample preparation. Lipid-only samples were prepared in the same manner without the addition of peptide. The experimental temperature and hydration levels were maintained to ensure that POPC, POPG, and POPE were above their respective gel-to-liquid crystalline transition temperatures of −2, −2, and 25° C, respectively[26].
Large multilamellar vesicles (MLVs) for 31P NMR were prepared by codis-solving lipids (40 μmol POPE:POPG (3:1) or POPC) from chloroform stock solution with C6-LfB6 (0.4 μmol), from water/acetonitrile stock solution (50/50). Solvents were removed under a stream of N2 gas and then under high vacuum (10 mtorr) over night. After resuspension in 0.4 ml 25 mM Hepes pH 7.4, the dried peptide/lipid film was subjected to several heating cycles (5, 10, 20, 30, and 60 min) at 50° C with 1 min vortexing in between. The milky white peptide/lipid suspension was transferred to 5 mm glass tubes and centrifuged at 14,000 rpm for 2 hours at 4° C. The supernatant was removed, the fully hydrated pellet (excess water) was flushed with N2 gas, and the tube was sealed with a rubber stopper and epoxy. Lipid only MLVs were prepared in the same manner except without the addition of peptide. Phosphorus NMR was run within 18 hours of preparing the MLVs.
2.4. Solid-State NMR Spectroscopy
Deuterium NMR spectra were recorded on a Bruker Avance 300 spectrometer using a quadrupolar echo pulse sequence with full-phase cycling[27]. An echo delay of 105 μs, a pulse length of 3 μs, and 300,000 scans were acquired for each sample with a 120 ms recycle delay time. Spectra were recorded with the lipid bilayer normal aligned either parallel to the magnetic field (H0), β = 0°; or perpendicular, β = 90° The samples were run at 303 K and 323 K. An exponential weighting function with 100 Hz line broadening was applied prior to Fourier transformation. Quadrupolar splittings (Δνq) were measured as the distances between corresponding peak maxima.
Phosphorus NMR spectra were acquired using the Bruker zgpg pulse program with 256 scans, a 6 μs 90° pulse, and a recycle delay time of 5 s. Measurements were performed in a Doty 8 mm wideline probe (Doty Scientific Inc., Columbia, SC) with broadband 1H decoupling at 50° C. Before Fourier transformation, an exponential weighting function with 100 Hz line broadening was applied. The chemical shift was referenced externally to 85% phosphoric acid at 0 ppm.
2.5. Simulations
Two different membrane systems were used. The first represents a putative bacterial (anionic) membrane and consists of a bilayer with 100 lipids per leaflet with a ratio of 3:1 POPE to POPG. The second represents a putative mammalian membrane and consists of a bilayer with 90 lipids per leaflet of POPC. The construction of the neat membrane has been described previously[28]. In brief, eight POPE:POPG neat membrane systems were constructed, solvated; neutralizing sodium ions were added using a previously described procedure[29].
The C6-LfB6 peptides were constructed in an extended conformation using PyMOL[30]. The topology for the C6 (hexanoic acid) was made by truncating the topology for palmitoyl to 6 carbons, and standard peptide parameters were used for the linkage. The resulting peptide was then solvated and neutralizing ions were added using VMD[31]. This system was then heated to 500K to produce a set of extended chain conformations. Two C6-LfB6 structures were randomly selected from this set and placed several angstroms above a neat membrane. In the POPE:POPG systems, a total of 7,900 waters were used to solvate the system to approximately 50% water by weight. To neutralize the peptide, 6 Cl− ions were added to each system, plus enough additional salt resulting in a free salt concentration of 50 mM (6 Cl− and 50 Na+ in the POPE:POPG simulations). The total number of atoms simulated was 49,172. A total of 8 independent systems were constructed. The POPC systems used 7,850 explicit water molecules with 21 Cl− and 15 Na+ for a total of 48,022 atoms. Four independent POPC systems were simulated.
All MD simulations used the CHARMM27[32, 33] lipid and cholesterol parameters, and CHARMM22[34, 35, 36] with CMAP for the peptide. In addition, SPME[37] was as used for the electrostatics along with a 10 Å vdW cutoff. The temperature was held at 323K using Langevin dynamics, and a 2 fs step-size was used with bonds constrained to their equilibrium lengths using RATTLE[38]. The system was simulated in the NPγT (constant number of molecules, normal pressure, constant surface tension and temperature) ensemble using NAMD[39] on the BlueGene/P supercomputer located at the University of Rochester. The POPE:POPG systems were simulated for an average length of 426 ns and the POPC systems were simulated for an average length of 643 ns (summarized in Table 2).
Table 2.
List of all simulations. The first 100 ns of each simulation is considered as equilibration and excluded from all calculations. POPE:POPG is in a 3:1 ratio.
| Membrane | Type | Tension (dyn/cm) | Length (ns) | Average Length | Avg Area/Lipid (Å 2) | Average Area |
|---|---|---|---|---|---|---|
| POPE:POPG | Neat | 32.5 | 241.8 | 239.15 | 65.4 | 65.2 ± 0.4 |
| POPE:POPG | Neat | 32.5 | 236.5 | 64.9 | ||
|
| ||||||
| POPE:POPG | Neat | 35 | 238.0 | 242.2 | 66.8 | 66.8 ± 0.5 |
| POPE:POPG | Neat | 35 | 244.8 | 66.1 | ||
| POPE:POPG | Neat | 35 | 243.0 | 67.4 | ||
| POPE:POPG | Neat | 35 | 243.0 | 66.8 | ||
|
| ||||||
| POPE:POPG | Neat | 37.5 | 241.8 | 242.8 | 68.2 | 68.3 ± 0.1 |
| POPE:POPG | Neat | 37.5 | 243.8 | 68.4 | ||
|
| ||||||
| POPE:POPG | C6-LfB6 | 32.5 | 535.7 | 429.6 | 65.5 | 65.5 ± 0.4 |
| POPE:POPG | C6-LfB6 | 32.5 | 531.7 | 66.3 | ||
| POPE:POPG | C6-LfB6 | 32.5 | 530.2 | 65.6 | ||
| POPE:POPG | C6-LfB6 | 32.5 | 529.8 | 65.4 | ||
| POPE:POPG | C6-LfB6 | 32.5 | 350.2 | 65.5 | ||
| POPE:POPG | C6-LfB6 | 32.5 | 344.7 | 65.1 | ||
| POPE:POPG | C6-LfB6 | 32.5 | 333.4 | 65.4 | ||
| POPE:POPG | C6-LfB6 | 32.5 | 280.8 | 65.3 | ||
|
| ||||||
| POPC | Neat | 32.5 | 347.6 | 346.2 | 70.4 | 69.4 ± 1.5 |
| POPC | Neat | 32.5 | 344.7 | 68.3 | ||
|
| ||||||
| POPC | C6-LfB6 | 32.5 | 584.6 | 634.0 | 71.1 | 71.1 |
| POPC | C6-LfB6 | 32.5 | 672.1 | 71.1 | ||
| POPC | C6-LfB6 | 32.5 | 663.6 | 71.1 | ||
| POPC | C6-LfB6 | 32.5 | 651.8 | 71.1 | ||
Because the CHARMM27 lipid potential has a tendency to form gel phase under ambient conditions, it is typical to run the simulations in the NPAγT ensemble, with a tension applied in the x-y plane; the best choice of the tensions appears to depend on a number of factors, including system size, composition, and temperature. Accordingly, a range of tensions from 32.5 dyn/cm to 37.5 dyn/cm were used to determine a value that best matched the experimentally observed order parameters. Each system was simulated for approximately 250 ns (see Table 2). A tension of 32.5 dyn/cm was found to be optimal, and a set of structures from the combined 497 ns simulation were selected to form the membranes used for inserting the acyl-peptide. The neat POPC membrane systems were constructed similarly, with 4 independent systems using with a tension of 32.5 dyn/cm, which was judged to be sufficiently close to the experimental results. The recently published C36 parameter[40] set appears to fix the need to apply a tension to the membrane, but the present simulations were performed prior to its release.
2.6. Simulation Analysis
All analyses of the molecular dynamics simulations were performed using the LOOS library and tools[41] and custom software built using the LOOS library. LOOS is an object-oriented library implemented in C++ and Boost and provides a lightweight but powerful library for rapidly creating new analytical tools for molecular dynamics. Included in the library is a “selection expression” parser that makes it trivial for tools to allow the user to select which atoms from a model to use. LOOS supports the native file formats for most major MD packages, such as CHARMM[34]/NAMD[39], Amber[42], Gromacs[43], and Tinker[44]. LOOS also comes bundled with over 50 different tools.
2.6.1. Fractional Contacts Analysis
Analysis of inter-molecular contacts was performed using LOOS. For each atom in a given “query” molecule—in this case, the C6-LfB6 or some component of it—all atoms from a set of “targets” are searched to find the number from each target that are within a 5 Å distance of the query atom. The number of contacts for each target is then normalized by the total number contacts across all targets giving the fraction of contacts made between the query molecule and each target. Hydrogen atoms were excluded from the contacts calculation. Intermolecular C6-LfB6 contacts were also excluded since contacts between the two C6-LfB6 molecules occurred only briefly in one trajectory.
2.6.2. Density Distribution Analysis
The density distribution for different components of the system was determined using the electron density as computed by LOOS. The density histogram was built using 1 Å bins and symmetrized about the membrane center. Each density histogram was normalized to 1 for each component, otherwise the peaks for the peptide components would be difficult to resolve compared to the scale of the lipid head groups. Since there are no restraints to prevent the peptides from migrating through the periodic boundaries to either leaflet, the system is treated as symmetric about the center of the membrane. The first 100 ns of each simulation was excluded from the calculation so that the distributions only reflect the bound peptide, as determined by the contact plots.
2.6.3. Hydrogen Bond Analysis
Hydrogen bonding patterns were analyzed using LOOS[41]. The criteria for determining a possible hydrogen bond was a donor-proton to acceptor atom distance of less than 3 Å with a donor–proton–acceptor angle greater than 140°. Correlation functions for hydrogen bonding were also computed using LOOS. Briefly, a contact matrix between the acyl-peptide and the membrane lipids was constructed. Whenever a putative hydrogen bond was found, a 1 was placed in the matrix with 0 otherwise. A correlation function was computed for any column that has a hydrogen bond. These correlation functions were then averaged together.
3. Results and Discussion
3.1. Antimicrobial Activity
The antimicrobial activities of LfB6 and C6-LfB6 were tested against a Gram negative (E. coli ATCC 25922) and a Gram positive (S. aureus ATCC 29213) bacterial strain using a standard microdilution Mueller Hinton broth assay adapted for cationic AMPs by Hancock, et al.[24] This assay provides a stringent test, and therefore more clinically relevant results, for the activity of cationic antimicrobial peptides due to the high ionic strength of Mueller Hinton medium that can inhibit the activity of such peptides.[45] The results, shown in Table 1, reveal that addition of the C6 acyl chain to the N-terminal of LfB6 did not increase the activity against Gram negative E. coli; whereas the activity against the Gram positive strain S. aureus was increased 3-fold compared to LfB6. Additionally, although both LfB6 and C6-LfB6 inhibited the growth of E. coli (MIC 150 μg/ml), neither peptide was bactericidal. By contrast, both LfB and C6-LfB were bactericidal against S. aureus at their respective MIC values of 150 and 50 μg/ml. Acylation of LF12, a 12 amino acid fragment from human lactoferricin, was found to increase the activity against both Gram positive and negative bacteria, however the improvement was higher for S. aureus than for E. coli[14]. The same study also found that the antimicrobial activity of the acylated peptides depended on the composition of the bacterial culture medium, with higher MIC values (lower activity) observed for assays performed with complex LB medium compared to those with low ionic strength buffer. Direct comparisons of the antimicrobial activities reported from various studies, therefore, are difficult since different bacterial strains and experimental protocols are frequently used.[46] The lack of an outer membrane in Gram positive bacteria might explain the enhanced activity of the acylated peptide, C6-LfB6, against S. aureus. The outer cell membrane of Gram negative bacteria poses an additional barrier for AMPs to gain access to the periplasmic space and inner cytoplasmic membrane. A systematic investigation into the effects of acylation on dermaseptin, a 13 amino acid AMP isolated from the South American tree frog of the Phyllomedusa genus, revealed complex antimicrobial behavior.[47] Enhanced activity against Gram positive S. aureus was observed for intermediate chain length acyl (C6-C12) derivatives, whereas all acyl derivatives were detrimental to activity against E. coli. It has been proposed that some cationic AMPs are retained on the outer membrane of Gram negative bacteria upon binding to lipopolysaccharide (LPS), thus impeding or precluding their access to the inner membrane.[48, 49] LPS-binding motifs consisting of two positively charged amino acid residues separated from the third by a short hydrophobic, aromatic stretch[50] have been identified in human and bovine lactoferricin peptides.[51, 14] Limited access to the cytoplasmic membrane due to the absence of an outer membrane might also explain why both LfB6 and C6-LfB6 are bactericidal at their MIC values against S. aureus, but not against E. coli.
Table 1.
Minimal inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of LfB6 and C6-LfB6 (μg/ml)
| Peptide | E. coli ATCC 25922 | S. aureus ATCC 29213 | ||
|---|---|---|---|---|
| MIC | MBC | MIC | MBC | |
| LFB6 | 150 | >200 | 150 | 150 |
| C6-LfB6 | 150 | >200 | 50 | 50 |
3.2. Solid-State 2H NMR Spectroscropy
Solid-state 2H-NMR is a well-established method for characterizing the liquid crystalline (Lα) phase of lipid bilayers containing perdeuterated lipids because the segmental order parameters (SCD) along the acyl chain C-D bonds can be estimated from the experimentally determined quadrupolar splittings (Δνq). For samples aligned at the β = 0 orientation the quadrupolar splitting and segmental order parameter are related by[52, 53]:
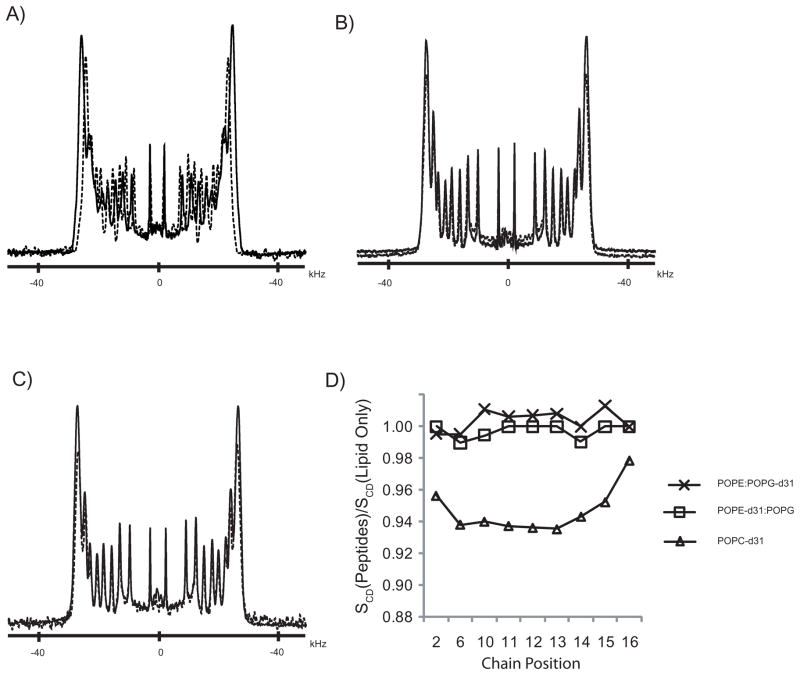
where e2qQ/h is the quadrupolar coupling constant (QCC), which is ≈168 kHz for aliphatic C–D bond[54, 55]. The lipid acyl chain order of mechanically oriented lipid bilayers composed of sn-1 chain perdeuterated zwitterionic (POPC) or mixed anionic:neutral (POPE:POPG) lipids was examined by solid-state 2H NMR in the presence or absence of C6-LfB6. The 2H spectra, shown in Figure 1, are characteristic of phospholipids in liquid crystalline bilayers, consisting of a series of doublet resonances resulting from the different CD2 segments along the lipid acyl chain[52]. Rapid rotation of the terminal methyl group, in combination with its tetrahedral geometry results in low order parameters. The smallest quadru-polar splittings were therefore assigned to the terminal methyl group.[56]. The remaining 2H resonances were assigned to carbons in decreasing order along the phospholipid chain. The quadrupolar splittings vary from a minimum of about 5 kHz for the terminal methyl groups to a maximum of about 50 and 54 kHz for deuterons near the head group carboxyls of POPC and POPE:POPG, respectively. In the presence of 1 mol % C6-LfB6, the 2H spectral width observed from POPC-d31 bilayers was slightly reduced compared to POPC-d31 bilayers without peptide (Figure 1A). Indeed the 2H quadrupolar splitting was reduced at each position along the acyl chain, except for the terminal methyl group which was essentially unchanged; namely, the methyl group Δνq was 4.7 kHz in the absence of peptide compared to 4.6 kHz in the presence of C6-LfB6. The maximum quadrupolar splitting in the plateau region resulting from the C2 position closest to the carbonyl group was reduced by approximately 2 kHz in the presence of C6-LfB6. Surprisingly, no changes were observed in the quadrupolar splittings for either POPE-d31 or POPG-d31 in the presence of C6-LfB6, as shown in Figures 1B and 1C, respectively. For the mixed POPE:POPG bilayers, the spectra are virtually superimposable in the presence and absence of peptide, regardless of which lipid is deuterated.
Figure 1.
2H spectra of mechanically aligned bilayers composed of a) POPC-d31, b) POPE:POPG-d31 (3:1), and c) POPE-d31:POPG (3:1) at 50° C and β = 0. Solid lines are of pure lipid, dashed lines are in the presence of 1 mol % C6-LfB6. The 2H order parameter profiles calculated relative to pure lipid samples are shown in (d).
The segmental order parameters along the lipid acyl chain, estimated from the quadrupolar splittings and plotted relative to the peptide-free control samples, are shown in Figure 1D. As noted above, there is little change in the order parameter profile for either the zwitterionic POPE or the anionic POPG when C6-LfB6 (1 mol %) is added to the mixed POPE:POPG (3:1) membranes The reduction in order induced in the zwitterionic POPC acyl chains by C6-LfB6 is largest near the lipid head group, extends into the plateau region toward the center of the bilayer, and tapers off near the terminal methyl group.
3.3. Solid-State 31P NMR Spectroscopy
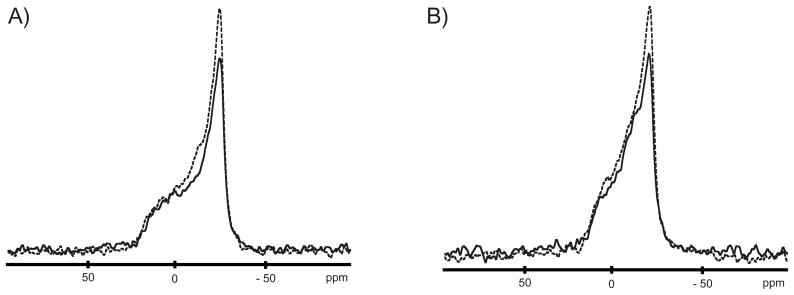
To test whether C6-LfB6 influences the lipid phosphate head groups, the 31P NMR chemical shift anisotropy (CSA) of multilamellar vesicles (MLVs) composed of POPC and POPE:POPG (3:1) was monitored in the presence and absence of peptide. The static 31P spectra of POPC and POPE:POPG (3:1) bilayers, shown in Figures 2A and 2B, respectively, are characteristic of MLVs in the liquid crystalline phase (Lα)[57, 58]. The CSAs of POPC (≈ 47 ppm ±1 ppm) and POPE:POPG (3:1) (≈ 37 ppm ±1 ppm) are furthermore unchanged in the presence of 1 mol % C6-LfB, indicating little observable perturbation of either the neutral or anionic lipid head groups. These findings support our previous results that acylated and Trp-methylated LfB peptides have little effect on the phosphate head groups of DMPC and DMPC:DMPG (3:1) bilayers even at 4 mol %[12]. Moreover, the MD simulations also show no change.
Figure 2.
Static 31P spectra of MLVs composed of a) POPC, b) POPE:POPG (3:1) at 50° C. Solid lines are of pure lipid, dashed lines are in the presence of 1 mol % C6-LfB6.
3.4. Membrane Order
The acyl C–H bond orientation relative to the membrane normal is expressed by the order parameter:
These order parameters can be measured experimentally by deuterium quadru-polar splitting in solid state NMR. The NMR experiment, however, cannot specify which order parameter is associated with which carbon, so the order parameters are typically sorted in decreasing order and it is assumed that this corresponds to increasing carbon number. When calculated from an MD simulation, it is known exactly which carbon is associated with which order parameter. In order to compare with the experimental results however, it is necessary to sort the simulation order parameters by decreasing magnitude.
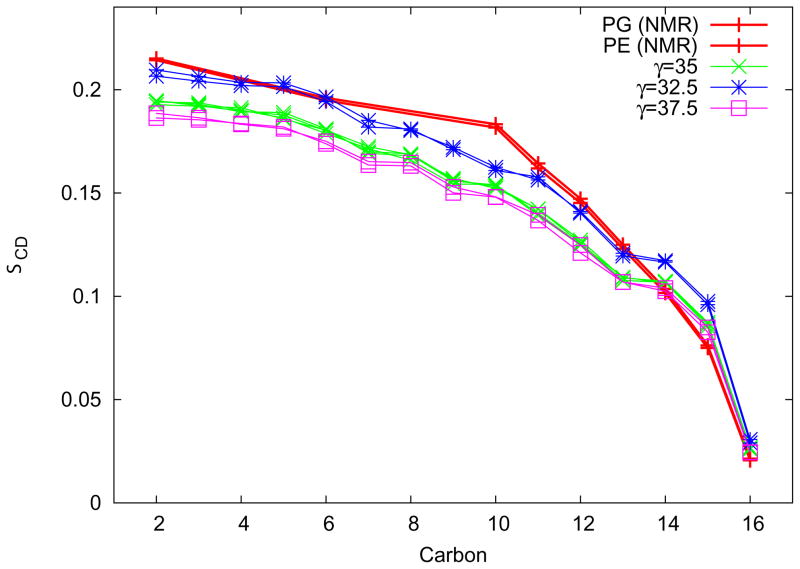
The results of the tension titration on order parameters for the POPE:POPG neat system are shown in Figure 3. Although the difference between 35.0 dyn/cm and 37.5 dyn/cm is minor, there is a greater difference with the 32.5 dyn/cm simulations. The 32.5 dyn/cm tension order parameters match the experimentally determined order parameters reasonably well. This tension was then used in all subsequent simulations. It is important to note that convergence of the area per lipid under a new tension (used as a criterion for determining the end of tension equilibration) took on average 100 ns; the standard deviation of the average areas for 200 ns trajectories was as large as 0.5 Å 2.
Figure 3.
Order parameters for the simulation of neat POPE:POPG membranes at different tensions are shown here compared with the experimentally determined order parameters. The simulation order parameters are sorted to correspond to the experimental data. The first 100 ns for each simulation is excluded from the calculation.
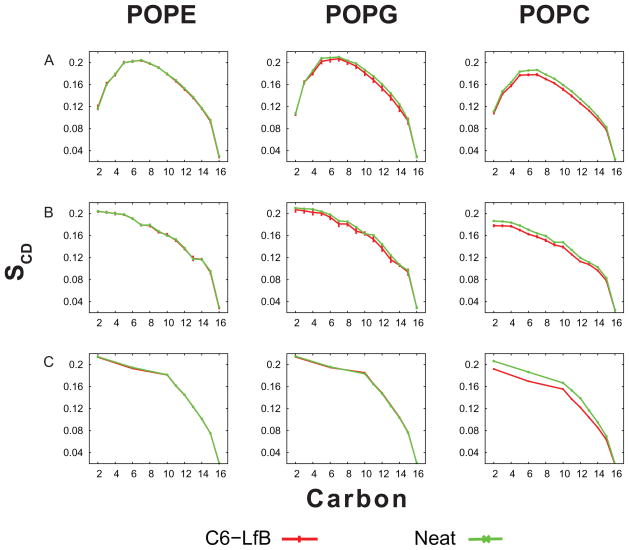
The order parameters for both neat and C6-LfB6 containing simulations are shown in Figure 4. Row A shows the order parameters calculated from the simulation in their natural order while Row B shows them sorted for comparison against the experimental data, shown in Row C. In the POPE:POPG experiments, the POPE and POPG were deuterated separately and the individual order parameters are shown in the respective columns although the two lipids produce nearly identical values.
Figure 4.
Order parameters for the different systems. Row A shows the order parameters in their “natural” order from the simulation. In Row B, they are sorted in decreasing order to match the NMR results. Row C shows the experimentally determined order parameters.
Comparing the order parameters from the simulation of POPE:POPG and the experimental data show that the profiles are very similar. Moreover, the experimental data shows virtually no change in order parameters upon C6-LfB6 binding—a result that is replicated in the simulations. This is perhaps a result of the low concentrations of acyl-peptides used.
Interestingly, the order parameters for the putative mammalian membrane (POPC) decrease noticeably upon association of C6-LfB6 in both the experiment and the simulation. This result seems counter-intuitive since AMPs typically affect mammalian membranes less than bacterial. This is surprising because one would expect the anionic “bacterial” POPE:POPG membrane to be more sensitive to peptide effects than the “mammalian” POPC membrane. Comparing the area per lipid between the POPE:POPG and POPC simulation (Table 2), the POPC membranes have a nearly 5 Å 2 greater area per lipid than the POPE:POPG membranes, perhaps as a result of the larger lipid head groups or the absence of inter-headgroup hydrogen bonds. The larger spacing between lipids could provide more opportunities for the C6-LfB6 to insert deep enough into the membrane to alter chain structure. The concentration of peptide used here (1:100) is far lower than what is needed to lyse membranes.
3.5. Acyl-Peptide–Membrane Association
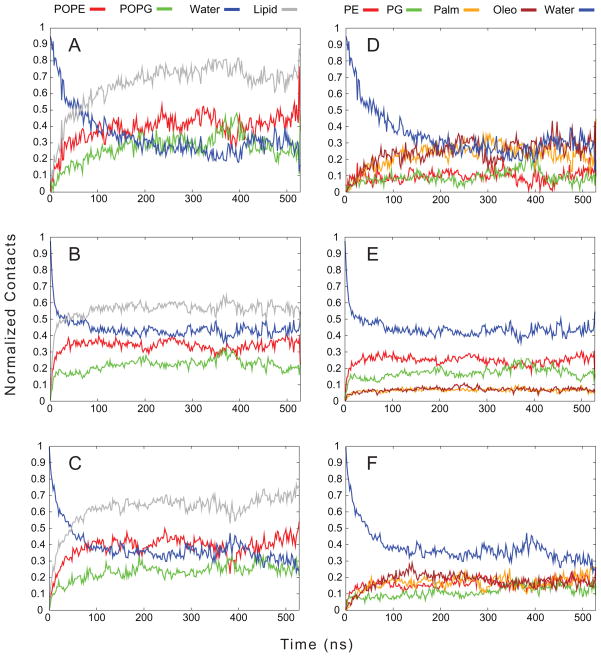
Examining the contacts made between the different components of the acyl-peptide, such as the C6 tail, the arginines, and the tryptophans, and the system provides a quantitative measure of how the acyl-peptide associates with the membrane and where it is positioned once it is associated. Figure 5 shows the contacts made between C6-LfB6 and the POPE:POPG system. In panels A–C, the fractional contacts between each C6-LfB6 component and all atoms of POPE, of POPG, and all waters are shown, averaged over all C6-LfB6 molecules, i.e. 2 peptides × 4 repeats for POPC and 2×8 for POPE:POPG. The replicates for each simulation are needed to allow us to confidently discuss the mechanisms of binding, as any single trajectory may not have all of these features. In addition, the total lipid contact (sum of POPE and POPG contacts) is also shown. We started from an out-of-equilibrium state–no peptide bound–and noted the approach mechanisms. We define membrane association as the point where the lipid and water contacts are equal. Panel A shows the association of the C6 tails with the membrane occurring after approximately 75 ns. Panel B shows that the arginines associated very rapidly, within the first 25–50 ns. This is followed by the tryptophans, at approximately 50 ns. What is striking in these panels is that while the system consisted of a 3:1 ratio of POPE:POPG, the contacts made between C6-LfB6 and POPE are closer to twice that of POPG and are often almost equal, suggesting a preferential interaction between the peptide and PG. There is also only one instance of contact between two C6-LfB6 molecules, and it lasts for less than half of one trajectory, indicating that dimer formation is unlikely, at least at these concentrations. Overall, the C6 tails show the highest degree of contact with lipids (≈ 0.7), followed by the tryptophans (≈ 0.65) and the arginines (≈ 0.55).
Figure 5.
Normalized contacts made between different parts of the C6-LfB and either water or lipid. The C6 tail is shown in Panels A and D, the Arginines in Panels B and E, and the Tryptophans in Panels C and F. The total lipid in Panels A, B, and C is the sum of the contacts with both POPE and POPG. In Panels D, E, and F, the membrane is broken down into the lipid head group (PE and PG) and fatty acid tails (Palm and Oleo).
Panels D–F increase the detail of the contacts by breaking the lipids into their head groups (PE and PG) and the tails as separate entities, along with solvent. Panel D shows that the C6 tail inserts into the membrane and makes considerably more contacts with the acyl chains than the lipid head groups, indicating it is buried in the membrane. In contrast, the arginines, shown in Panel E, prefer polar contacts, remaining well solvated, and also making contacts with the PE and PG head groups as opposed to the acyl chains. This indicates that the arginines stay near the membrane interface. The tryptophans have nearly equal contacts with all components of the membrane and a lower solvent contact than the arginines. This suggests that the tryptophans can bury themselves slightly into the membrane, though not to the extent that the C6 tail inserts, and stay more near the membrane–water interface. This is consistent with much previous work suggesting tryptophans prefer to reside in the interface[59, 60, 61, 62].
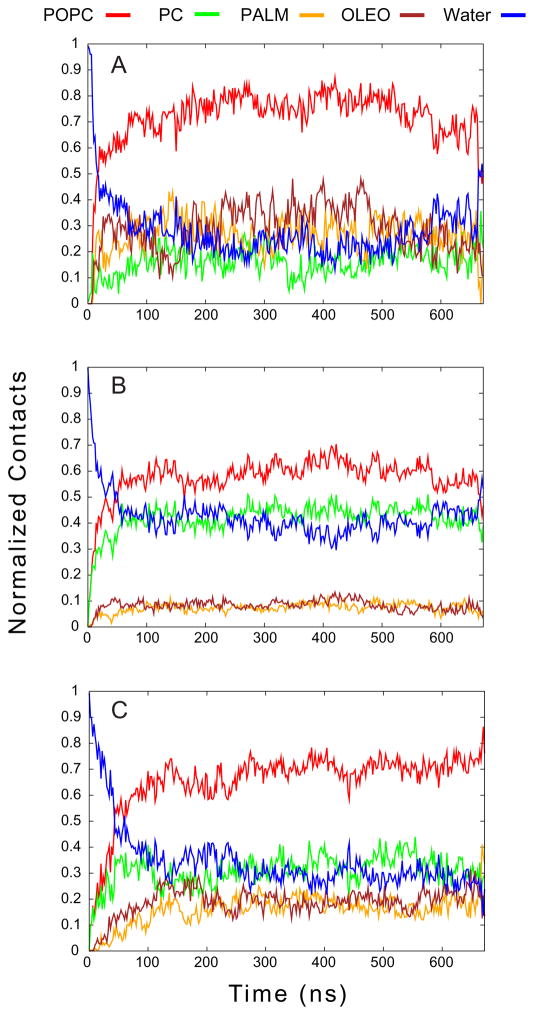
The contacts for C6-LfB6 in POPC are shown in Figure 6. Here, it is the C6 tail (Panel A) that rapidly associates with the membrane after approximately 25 ns, followed by the arginines (Panel B) and tryptophans (Panel C) at 50 ns. The overall pattern of contacts between parts of the C6-LfB6 and different components of the membrane are similar between the POPC and POPE:POPG systems. In both cases, the C6 inserts deeply into the membrane while the arginines remain near the membrane-solvent interface. The tryptophans also insert into the membrane, but not as deeply as the C6 tails. While it appears in Figure 6C that the tryptophans make more contacts with the PC head groups than the PE and PG headgroups in Figure 5F, the contacts in the latter are divided between two different headgroup species; comparing total head-group contacts, irrespective of whether it is with PE or PG, the pattern is similar between the POPC and POPE:POPG systems.
Figure 6.
Contacts between different parts of the C6-LfB and either water or lipid. The C6 tail is shown in Panel A, the Arginines in Panel B, and the Tryptophans in Panel C.
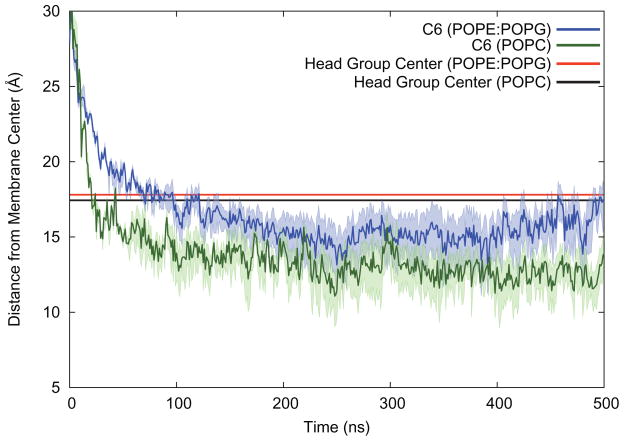
Althought the contact plots suggest that the C6-LfB6 resides near the membrane-solvent interface and that the C6 tail reaches down into the membrane, it is not clear how far down the tail resides. A plot of the average centroid for the PE, PG, and PC head groups and the centroid of the C6 tails is shown in Figure 7. The C6 tail inserts into the POPC membrane far faster than in the POPE:POPG mixture. Moreover, while the average location of the center of the lipid head groups is unchanged between POPC and POPE:POPG, the C6 tail is 1–2 Å deeper in the membrane in the POPC simulations. The greater depth of the C6 in POPC is consistent with there being more space between the lipids for the C6, and indeed the whole C6-LfB6.
Figure 7.
Average distance from the membrane center (Z-coordinate) for the lipid head groups and the C6 “tail”. The tail is buried inside the membrane once C6-LfB binds. The wide bands show the average distance from the membrane center for C1 and C6 of the tail.
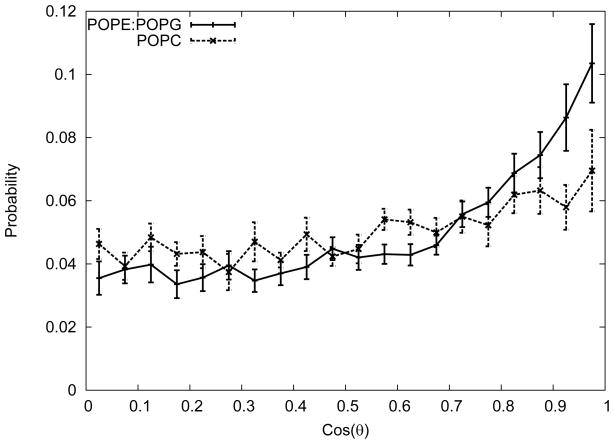
There is a significant difference in the orientation of the C6 tails in the membrane for POPC compared with POPE:POPG. The orientation can be quantified by computing the principal axes for the heavy atoms of the C6. The first principal axis points along the direction of the tail, so we computed the cosine of the angle between it and the membrane normal. The probability distribution for the tail orientations in both POPC and POPE:POPG are shown in Figure 8. In POPE:POPG, the C6 tail distribution is fairly flat until at about 0.6 (corresponding to 53°) leading to a peak at nearly 1 (or parallel to the membrane normal). In contrast, the POPC distribution is largely flat with a peak in the vertical orientation some 30% smaller than POPE:POPG. This is again consistent with there being greater area per lipid in POPC, providing more space for the C6 to adopt a larger range of orientations.
Figure 8.
Probability distribution of the cosine of the angle between the C6 tail and the membrane normal. The error bars shown are the standard errors of the angles of individual peptides.
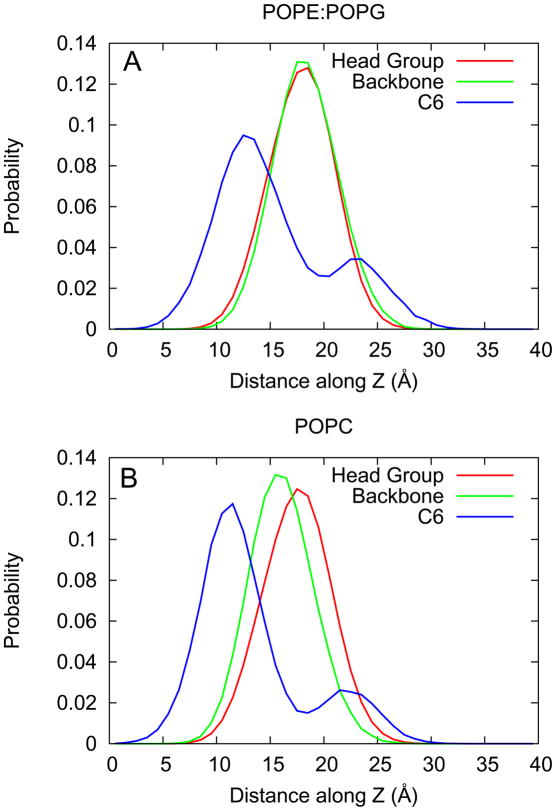
The distribution of the electron density for both the peptide backbone and the C6 tails is shown in Figure 9. The density for the lipid head groups are shown as well: PE and PG in Figure 9A and PC in Figure 9B. In the POPC system, the peptide backbone resides approximately 2 Å deeper than in the POPE:POPG system. The C6 tail also resides approximately 2 Å deeper in the POPC system. In both systems, the tail can also reside at the membrane-water interface. Comparing the area under the peaks, the probability of finding the tail buried is three times as likely as at the interface in both POPC and POPE:POPG.
Figure 9.
Probability distribution for the location of the lipid head groups, C6-LfB6 backbone, and C6 tail. Panel A shows the results for the POPE:POPG simulations while panel B shows the results for the POPC simulations.
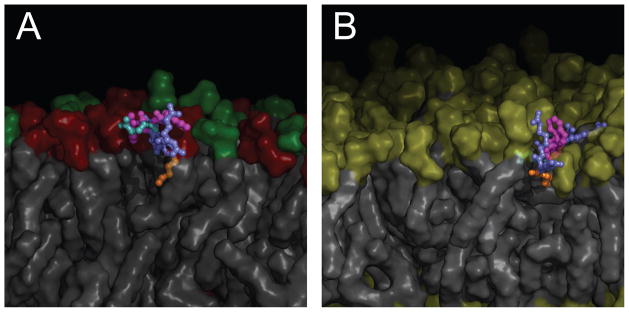
Representative conformations of one of the bound peptides are shown in Figure 10 for the POPE:POPG system (Panel A) and POPC (Panel B), taken from approximately 300 ns into the respective simulations. In both cases, the C6 tail is inserted into the membrane with the arginines and tryptophans remaining near the head groups/membrane-water interface.
Figure 10.
Representative conformations of bound C6-LfB6 in POPE:POPG (Panel A) and in POPC (Panel B). Membrane lipids are shown as gray surfaces with the head groups colored red (PE), green (PG), and yellow (PC). The peptide residues are colored slate (Arg), magenta (Trp), light blue (Gln), and orange (Hexanoic). In both panels, the membrane has been cut away to reveal the bound peptide.
3.6. Hydrogen Bonding
We looked at the fraction of time spent with a hydrogen bond between various possible donor nitrogens moeities in the acyl-peptide with the lipid carbonyls, lipid phosphates, solvent, and with the acyl-peptide itself. We found no significant change in hydrogen bonding patterns between the POPE:POPG system and the POPC system. On average, there is a hydrogen bond between the lipid phosphates and acyl-peptides one third of the time and with solvent another third of the time. The greatest contact between the acyl-peptides and the lipid phosphates occurred between the arginines (30%–40% of the time), followed by the tryptophans (18%–20%), and the glutamine (≈ 18%).
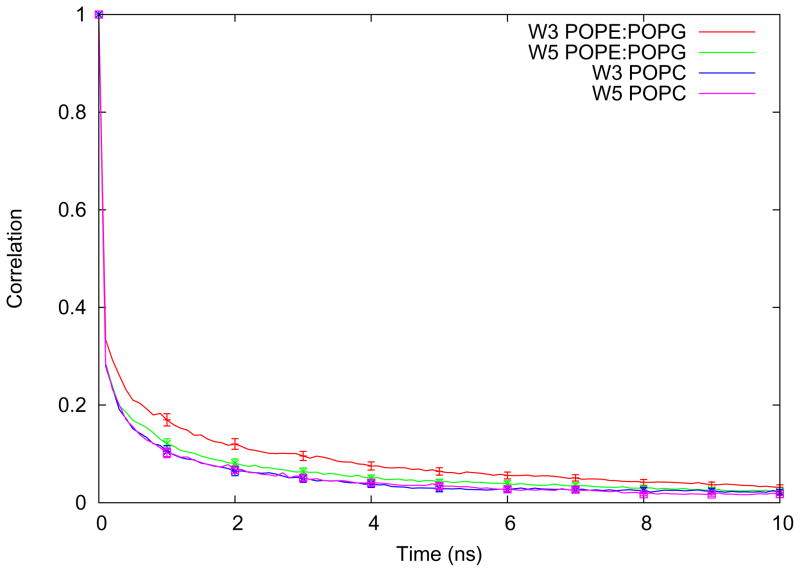
There are no significant changes in lifetimes found with the exception of the hydrogen bond between the indole-nitrogen of tryptophan 3 (closest to the C6) and the lipid carbonyls, shown in Figure 11. In this case, there is a slightly longer lifetime for W3 in POPE:POPG than W5. The lifetime for the latter is also comparable to the lifetimes of both W3 and W5 in POPC.
Figure 11.
Time correlation function for hydrogen bonding between the two different indole-nitrogens of the acyl-peptide and the lipid carbonyl oxygens is shown for both the POPE:POPG and POPC systems. The error bars represent the standard errors of the averages from the individual peptides.
Conclusions
We compared the activities of LfB6 and C6-LfB6 against Gram positive and Gram negative bacterial strains commonly used for antimicrobial assays. The assays demonstrated that the activity of C6-LfB6 was enhanced 3-fold against Gram positive S. aureus compared to LfB6 and that both peptides were bactericidal at their respective MIC values. In contrast, no improvement in activity was observed upon acylation of LfB6 against Gram negative E. coli, and neither LfB6 nor C6-LfB6 was bactericidal against E. coli at their MICs. The interaction of cationic AMPs with bacterial cell membranes depends on a complex balance of electrostatic and hydrophobic interactions. The highly anionic lipopolysaccharide-rich external membrane of Gram negative bacteria can present a barrier that obstructs the translocation of some cationic AMPS. Our results suggest that modification of AMPs by N-acylation can provide a means to enhance their antimicrobial selectivity, as well as their activity.
We have used a total of 8.6 μs of all-atom molecular dynamics simulations in concert with solid state NMR experiments to characterize the associations of the small acylated hexapeptide C6-LfB6 with both a model bacterial membrane (POPE:POPG in 3:1 ratio) and a model mammalian membrane (POPC). Carefully adjusting the applied surface tension in the membrane simulation has enabled us to closely match the conditions present in the NMR experiment. In the bacterial membrane, we have shown that the positively charged arginines associate first, followed by the tryptophans, and finally the C6 tail. Our simulation order parameters agree closely with the NMR experiment and show negligible change upon the introduction of C6-LfB6 to the POPE:POPG membrane. Interestingly the intermolecular contacts between the C6-LfB6 and the membrane suggest a preference for POPG contact with a probability of 3/2, implying a difference of ≈ 0.26 kcal/mol.
In contrast, the association between the model mammalian membrane (POPC) and C6-LfB6 is initiated with the C6 tail and there is no preference between the arginines and tryptophans. The NMR data indicates a decrease in lipid order parameters upon association with C6-LfB6 that is replicated in the simulation. The simulation results also show that the C6 buries more deeply into the membrane and adopts a wider range of angles with respect to the membrane normal in POPC than in the POPE:POPG membrane. It is likely that the larger area per lipid of the POPC membrane allows more opportunities for the C6-LfB6 to insert as well as more space for it to reorient, hence the larger affect on the lipid order parameters than in POPE:POPG.[53, 63] While this larger affect on POPC may seem contradictory, the overall change in order parameters is small in both POPE:POPG and POPC. Moreover, the mechanism of binding does not necessarily correlate to lysing and hence antimicrobial activity.
This work illustrates the benefits of carefully combining experiment with simulation in order to gain a better understanding of the mechanisms involved in biologically relevant systems. This understanding will lead to the design of better antimicrobial peptides.
Acknowledgments
We are grateful to the Center for Research Computing at the University of Rochester for providing the necessary computing systems and personnel to enable the research presented in this manuscript. Portions of the work at the University of Arkansas were supported by NIH grants RR031154 and RR016460, and by the Arkansas Biosciences Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tomita M, Takase M, Bellamy W, Shimamura S. A review: the active peptide of lactoferrin. Acta Paediatr Jpn. 1994;36:585–91. doi: 10.1111/j.1442-200x.1994.tb03250.x. [DOI] [PubMed] [Google Scholar]
- 2.Boman HG. Peptide antibiotics and their role in innate immunity. Annu Rev Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 3.Hancock R. Cationic peptides: effectors in innate immunity and novel antimicrobials. The Lancet infectious diseases. 2001;1:156–64. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- 4.Jenssen H, Hamill P, Hancock REW. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rathinakumar R, Walkenhorst WF, Wimley WC. Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: The importance of interfacial activity. J Am Chem Soc. 2009;131:7609–7617. doi: 10.1021/ja8093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol. 2010;5:905–17. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohner K. Development of Novel Antimicrobial Agents: Emerging Strategies. Horizon Scientific Press; Wymondham, Norfolk, UK: pp. 149–65. [Google Scholar]
- 8.Ishitsuka Y, Pham DS, Waring AJ, Lehrer RI, Lee KYC. Insertion selectivity of antimicrobial peptide protegrin-1 into lipid monolayers: effect of head group electrostatics and tail group packing. Biochim Biophys Acta. 2006;1758:1450–60. doi: 10.1016/j.bbamem.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Blazyk J, Lorigan GA. Exploring membrane selectivity of the antimicrobial peptide KIGAKI using solid-state NMR spectroscopy. Biochim Biophys Acta. 2006;1758:1303–13. doi: 10.1016/j.bbamem.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Umeyama M, Kira A, Nishimura K, Naito A. Interactions of bovine lactoferricin with acidic phospholipid bilayers and its antimicrobial activity as studied by solid-state nmr. BBA - biomembranes. 2006;1758:1523–8. doi: 10.1016/j.bbamem.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Hale JDF, Hancock REW. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev Anti Infect Ther. 2007;5:951–9. doi: 10.1586/14787210.5.6.951. [DOI] [PubMed] [Google Scholar]
- 12.Greathouse D, Vostrikov V, Mcclellan N, Chipollini J, Lay J, Liyanage R, Ladd T. Lipid interactions of acylated tryptophan-methylated lactoferricin peptides by solid-state NMR. J Pept Sci. 2008;14:1103–1110. doi: 10.1002/psc.1047. [DOI] [PubMed] [Google Scholar]
- 13.Japelj B, Zorko M, Majerle A, Pristovšek P. The acyl group as the central element of the structural organization of antimicrobial lipopeptide. J Am Chem Soc. 2007;129:1022–1023. doi: 10.1021/ja067419v. [DOI] [PubMed] [Google Scholar]
- 14.Majerle A, Kidrič J, Jerala R. Enhancement of antibacterial and lipopolysaccharide binding activities of a human lactoferrin peptide fragment by the addition of acyl chain. J Antimicrob Chemother. 2003;51:1159–65. doi: 10.1093/jac/dkg219. [DOI] [PubMed] [Google Scholar]
- 15.Malina A, Shai Y. Conjugation of fatty acids with different lengths modulates the antibacterial and antifungal activity of a cationic biologically inactive peptide. Biochem J. 2005;390:695–702. doi: 10.1042/BJ20050520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porat Y, Marynka K, Tam A, Steinberg D, Mor A. Acyl-substituted dermaseptin S4 derivatives with improved bactericidal properties, including on oral microflora. Antimicrob Agents Chemother. 2006;50:4153–60. doi: 10.1128/AAC.00750-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Straus SK, Hancock REW. Mode of action of the new antibiotic for gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim Biophys Acta. 2006;1758:1215–23. doi: 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Wakabayashi H, Matsumoto H, Hashimoto K, Teraguchi S, Takase M, Hayasawa H. N-Acylated and D enantiomer derivatives of a nonamer core peptide of lactoferricin B showing improved antimicrobial activity. Antimicrob Agents Chemother. 1999;43:1267–9. doi: 10.1128/aac.43.5.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zweytick D, Pabst G, Abuja PM, Jilek A, Blondelle SE, Andrä J, Jerala R, Monreal D, de Tejada GM, Lohner K. Influence of N-acylation of a peptide derived from human lactoferricin on membrane selectivity. Biochim Biophys Acta. 2006;1758:1426–35. doi: 10.1016/j.bbamem.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 20.Andrä J, Lohner K, Blondelle SE, Jerala R, Moriyon I, Koch MHJ, Garidel P, Brandenburg K. Enhancement of endotoxin neutralization by coupling of a C12-alkyl chain to a lactoferricin-derived peptide. Biochem J. 2005;385:135–43. doi: 10.1042/BJ20041270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh HS, Kim S, Cho H, Lee KH. Development of novel lipid-peptide hybrid compounds with antibacterial activity from natural cationic antibacterial peptides. Bioorg Med Chem Lett. 2004;14:1109–13. doi: 10.1016/j.bmcl.2003.12.085. [DOI] [PubMed] [Google Scholar]
- 22.Radzishevsky IS, Rotem S, Zaknoon F, Gaidukov L, Dagan A, Mor A. Effects of acyl versus aminoacyl conjugation on the properties of antimicrobial peptides. Antimicrob Agents Chemother. 2005;49:2412–20. doi: 10.1128/AAC.49.6.2412-2420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amsterdam D. Laboratory Medicine. Lippincott Williams & Wilkins; 1996. [Google Scholar]
- 24.Hancock RE. Hancock Laboratory Methods. 2011 http://cmdr.ubc.ca/bobh/methods/MODIFIEDMIC.html.
- 25.van der Wel PCA, Strandberg E, Killian JA, Koeppe RE. Geometry and intrinsic tilt of a tryptophan-anchored transmembrane alpha-helix determined by (2)H NMR. Biophys J. 2002;83:1479–88. doi: 10.1016/S0006-3495(02)73918-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silvius JR. Lipid-Protein Interactions. John Wiley & Sons, Inc; New York, NY: 1982. Thermotropic phase transitions of pure lipids in model membranes and their modifications by membrane proteins. [Google Scholar]
- 27.Davis J, Jeffrey K, Bloom M, Valic M. Quadrupolar echo deuteron magnetic resonance spectroscopy in ordered hydrocarbon chains. Chem Phys Lett. 1976;42:390–394. [Google Scholar]
- 28.Grossfield A, Pitman MC, Feller SE, Soubias O, Gawrisch K. Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. J Mol Biol. 2008;381:478–86. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grossfield A, Feller SE, Pitman MC. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proc Natl Acad Sci USA. 2006;103:4888–93. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The PyMOL Molecular Graphics System, Version 1.3. Schrödinger, LLC; 2010. [Google Scholar]
- 31.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Molec Graphics. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 32.Feller SE, Gawrisch K, MacKerell AD. Polyunsaturated fatty acids in lipid bilayers: intrinsic and environmental contributions to their unique physical properties. J Am Chem Soc. 2002;124:318–26. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 33.Klauda JB, Brooks BR, MacKerell AD, Venable RM, Pastor RW. An ab initio study on the torsional surface of alkanes and its effect on molecular simulations of alkanes and a dppc bilayer. J Phys Chem B. 2005;109:5300–11. doi: 10.1021/jp0468096. [DOI] [PubMed] [Google Scholar]
- 34.Brooks B, Bruccoleri R, Olafson B, States D, Swaminathan S, Karplus M. Charmm: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 35.MacKerell AD, Jr, Brooks CL, III, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: The Energy Function and Its Parameterization with an Overview of the Program. Vol. 1. John Wiley and Sons; Chichester: pp. 271–277. [Google Scholar]
- 36.MacKerell JAD, Feig M, Brooks ICL. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformaitonal distributions in molecular dynamics simulations. J Comp Chem. 2004;25:1400–15. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 37.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 38.Andersen HC. A ”velocity” version of the shake algorithm for molecular dynamics calculations. J Comput Chem. 1983;52:24–34. [Google Scholar]
- 39.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klauda JB, Venable RM, Freites JA, OConnor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114:7830–43. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romo TD, Grossfield A. Loos: An extensible platform for the structural analysis of simulations. Conf Proc IEEE Eng Med Biol Soc. 2009;1:2332–5. doi: 10.1109/IEMBS.2009.5335065. [DOI] [PubMed] [Google Scholar]
- 42.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spoel DVD, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–18. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 44.Ponder J. Tinker 4.2. 2004 http://dasher.wustl.edu/tinker.
- 45.Hancock R. Synergistic interactions between mammalian antimicrobial defense peptides. Antimicrobial agents and chemotherapy. 2001;45:1558–60. doi: 10.1128/AAC.45.5.1558-1560.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strøm MB, Rekdal O, Svendsen JS. The effects of charge and lipophilicity on the antibacterial activity of undecapeptides derived from bovine lactoferricin. J Pept Sci. 2002;8:36–43. doi: 10.1002/psc.365. [DOI] [PubMed] [Google Scholar]
- 47.Radzishevski I, Rotem S, Zaknoon F, Gaidukov L, Dagan A, Mor A. Effects of acyl versus aminoacyl conjugation on the properties of antimicrobial peptides. Antimicrob Agents Chemother. 2005;49:2412–20. doi: 10.1128/AAC.49.6.2412-2420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papo N, Shai Y. A molecular mechanism for lipopolysaccharide protection of Gram-negative bacteria from antimicrobial peptides. J Biol Chem. 2005;280:10378–87. doi: 10.1074/jbc.M412865200. [DOI] [PubMed] [Google Scholar]
- 49.Epand RM, Epand RF, Arnusch CJ, Papahadjopoulos-Sternberg B, Wang G, Shai Y. Lipid clustering by three homologous arginine-rich antimicrobial peptides is insensitive to amino acid arrangement and induced secondary structure. Biochimica et Biophys Acta. 2010;1798:1272–80. doi: 10.1016/j.bbamem.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 50.Dathe M, Nikolenko H, Klose J, Bienert M. Cyclization increases the antimicrobial activity and selectivity of arginine- and tryptophan-containing hexapeptides. Biochemistry. 2004;43:9140–50. doi: 10.1021/bi035948v. [DOI] [PubMed] [Google Scholar]
- 51.Elass-Rochard E, Roseanu A, Legrand D, Trif M, Salmon V, Motas C, Montreuil J, Spik G. Lactoferrin- lipopolysaccharide interaction: involvement of the 28–34 loop region of human lactoferrin in the high-affinity binding to Escherichia coli 055B5 lipopolysaccharide. Biochem J. 1995;312:839–45. doi: 10.1042/bj3120839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lafleur M, Fine B, Sternin E, Cullis PR, Bloom M. Smoothed orientational order profile of lipid bilayers by 2H-nuclear magnetic resonance. Biophys J. 1989;56:1037–41. doi: 10.1016/S0006-3495(89)82749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seelig J. Deuterium magnetic resonance: theory and application to lipid membranes. Q Rev Biophys. 1977;10:353–418. doi: 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- 54.Gall C, DiVerdi J, Opella S. Phenylalanine ring dynamics by solid-state deuterium NMR. J Am Chem Soc. 1981;103:5039–43. [Google Scholar]
- 55.Burnett LJ, Müller BH. Deuteron quadrupole coupling constants in three solid deuterated paraffin hydrocarbons C2D6, D4D10, C6D14. J Chem Phys. 1971;55:5829–31. [Google Scholar]
- 56.Dave PC, Tiburu EK, Damodaran K, Lorigan GA. Investigating structural changes in the lipid bilayer upon insertion of the transmembrane domain of the membrane-bound protein phospholamban utilizing 31P and 2H solid-state NMR spectroscopy. Biophys J. 2004;86:1564–73. doi: 10.1016/S0006-3495(04)74224-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seelig J. 31P nuclear magnetic resonance and the head group structure of phospholipids in membranes. Biochim Biophys Acta. 1978;515:105–40. doi: 10.1016/0304-4157(78)90001-1. [DOI] [PubMed] [Google Scholar]
- 58.Smith ICP, Ekiel IH. Phosphorus-31 NMR of phosphlipids in membranes. In: Gorenstein DG, editor. Phosphorus-31 NMR: principals and applications. Academic Press; Orlando, FL: 1984. [Google Scholar]
- 59.de Planque MR, Kruijtzer JA, Liskamp RM, Marsh D, Greathouse DV, Koeppe RE, de Kruijff B, Killian JA. Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane alpha-helical peptides. J Biol Chem. 1999;274:20839–46. doi: 10.1074/jbc.274.30.20839. [DOI] [PubMed] [Google Scholar]
- 60.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–8. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 61.Grossfield A, Woolf T. Interaction of tryptophan analogs with POPC lipid bilayers investigated by molecular dynamics calculations. Langmuir. 2002;18:198–210. [Google Scholar]
- 62.Norman KE, Nymeyer H. Indole localization in lipid membranes revealed by molecular simulation. Biophys J. 2006;91:2046–54. doi: 10.1529/biophysj.105.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mely B, Charvolin J, Keller P. Disorder of lipid chains as a function of their lateral packing in lyotropic liquid crystals. Chem and Phys of Lipids. 1975;15:161–73. [Google Scholar]