Abstract
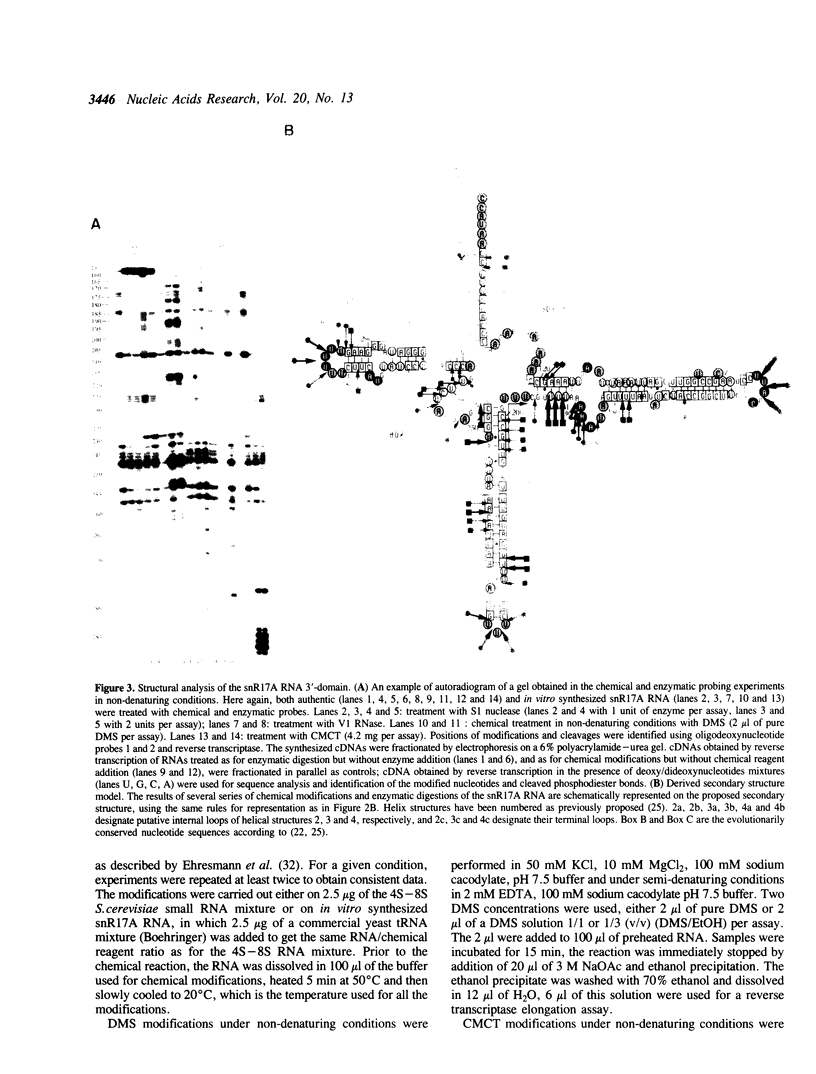
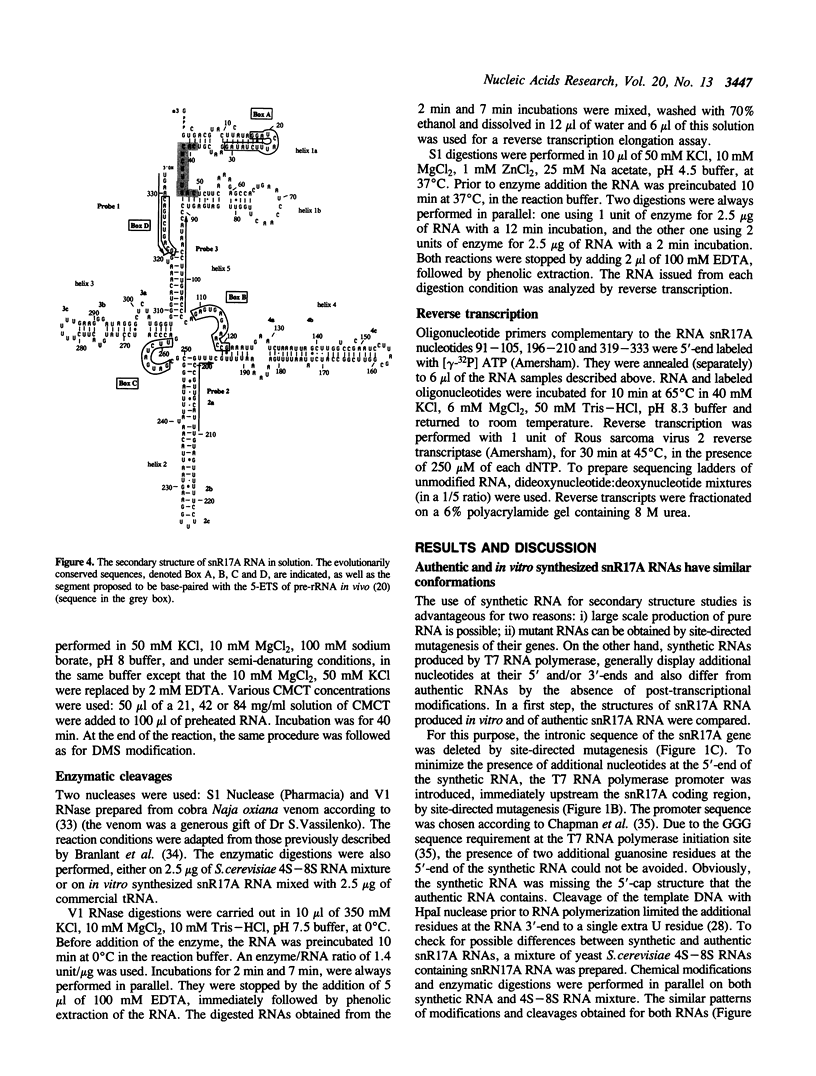
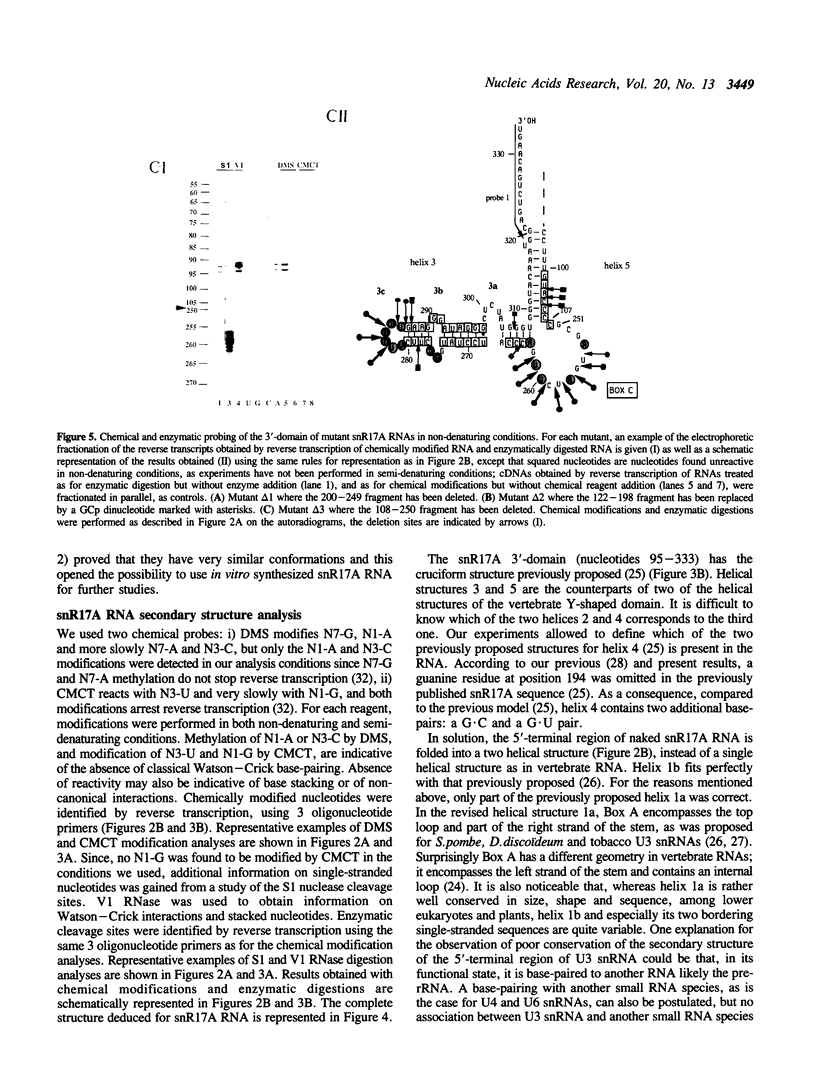
The conformation of Saccharomyces cerevisiae U3 snRNA (snR17A RNA) in solution was studied using enzymatic and chemical probes. In vitro synthesized and authentic snR17A RNAs have a similar conformation in solution. The S. cerevisiae U3 snRNA is folded in two distinct domains. The 5'-domain has a low degree of compactness; it is constituted of two stem-loop structures separated by a single-stranded segment, which has recently been proposed to basepair with the 5'-ETS of pre-ribosomal RNA. We demonstrate that, as previously proposed, the 5'-terminal region of U3 snRNA has a different structure in higher and lower eukaryotes and that this may be related to pre-rRNA 5'-ETS evolution. The S. cerevisiae U3 snRNA 3'-domain has a cruciform secondary structure and a compact conformation resulting from an higher order structure involving the single-stranded segments at the center of the cross and the bottom parts of helices. Compared to tRNA, where long range interactions take place between terminal loops, this represents another kind of tertiary folding of RNA molecules that will deserve further investigation, especially since the implicated single-strands have highly evolutionarily conserved primary structures that are involved in snRNP protein binding.
Full text
PDF
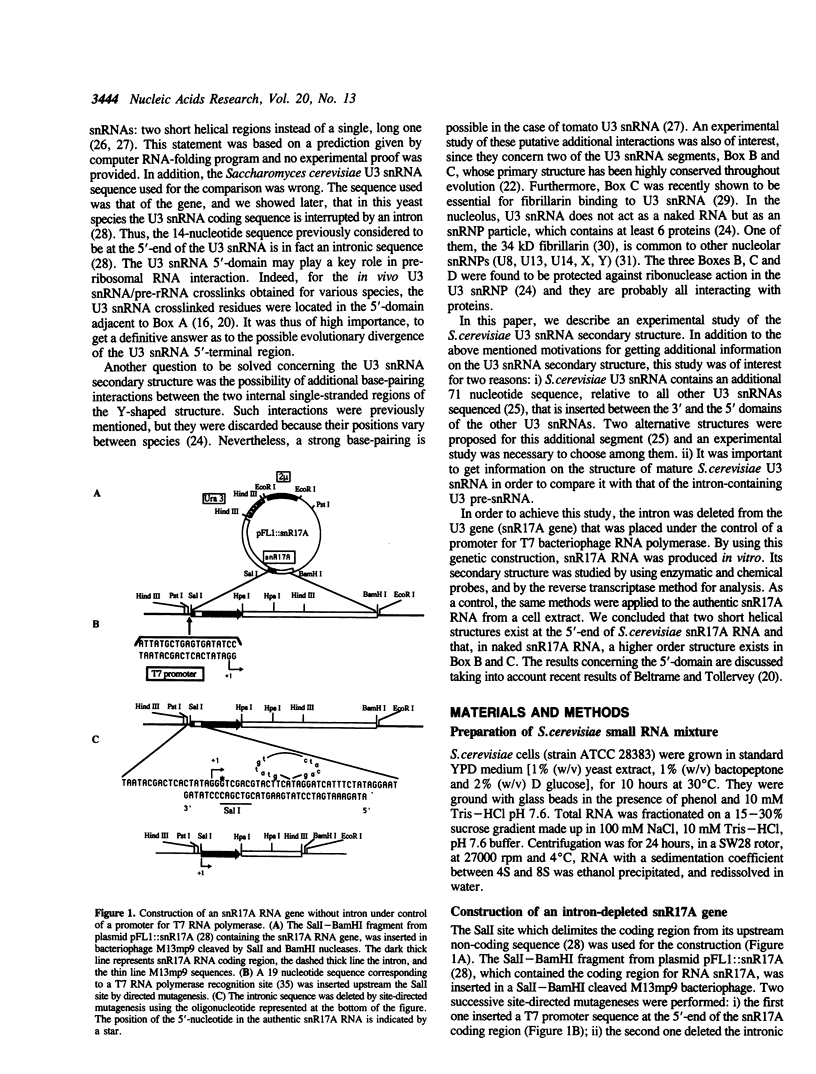
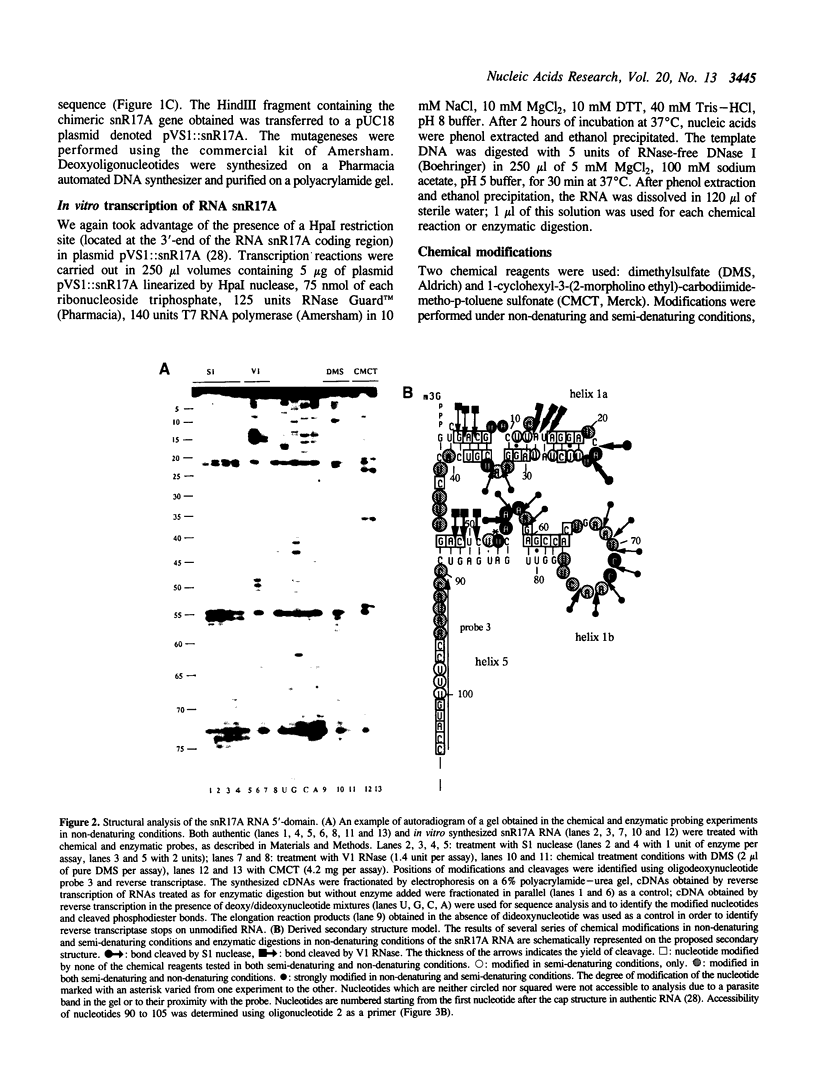



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachellerie J. P., Michot B., Raynal F. Recognition signals for mouse pre-rRNA processing. A potential role for U3 nucleolar RNA. Mol Biol Rep. 1983 May;9(1-2):79–86. doi: 10.1007/BF00777477. [DOI] [PubMed] [Google Scholar]
- Baudin F., Romaniuk P. J., Romby P., Brunel C., Westhof E., Ehresmann B., Ehresmann C. Involvement of "hinge" nucleotides of Xenopus laevis 5 S rRNA in the RNA structural organization and in the binding of transcription factor TFIIIA. J Mol Biol. 1991 Mar 5;218(1):69–81. doi: 10.1016/0022-2836(91)90874-6. [DOI] [PubMed] [Google Scholar]
- Beltrame M., Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992 Apr;11(4):1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein L. B., Mount S. M., Weiner A. M. Pseudogenes for human small nuclear RNA U3 appear to arise by integration of self-primed reverse transcripts of the RNA into new chromosomal sites. Cell. 1983 Feb;32(2):461–472. doi: 10.1016/0092-8674(83)90466-x. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol A., Ebel J. P., Gallinaro H., Lazar E., Jacob M. The conformation of chicken, rat and human U1A RNAs in solution. Nucleic Acids Res. 1981 Feb 25;9(4):841–858. doi: 10.1093/nar/9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K. A., Burgess R. R. Construction of bacteriophage T7 late promoters with point mutations and characterization by in vitro transcription properties. Nucleic Acids Res. 1987 Jul 10;15(13):5413–5432. doi: 10.1093/nar/15.13.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch R. J., Kanaya S., Earl P. L. A model for the involvement of the small nucleolar RNA (U3) in processing eukaryotic ribosomal RNA. Mol Biol Rep. 1983 May;9(1-2):75–78. doi: 10.1007/BF00777476. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein P., Reddy R., Busch H. Multiple states of U3 RNA in Novikoff hepatoma nucleoli. Biochemistry. 1984 Nov 6;23(23):5421–5425. doi: 10.1021/bi00318a007. [DOI] [PubMed] [Google Scholar]
- Gurney T., Jr Characterization of mouse 45S ribosomal RNA subspecies suggests that the first processing cleavage occurs 600 +/- 100 nucleotides from the 5' end and the second 500 +/- 100 nucleotides from the 3' end of a 13.9 kb precursor. Nucleic Acids Res. 1985 Jul 11;13(13):4905–4919. doi: 10.1093/nar/13.13.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. M., Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5' external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991 Dec;10(13):4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. M., Konings D. A., Cesareni G. The yeast homologue of U3 snRNA. EMBO J. 1987 Jul;6(7):2145–2155. doi: 10.1002/j.1460-2075.1987.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Craig N., Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol Cell Biol. 1987 Aug;7(8):2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Tyc K., Steitz J. A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990 Mar 23;60(6):897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Kiss T., Solymosy F. Molecular analysis of a U3 RNA gene locus in tomato: transcription signals, the coding region, expression in transgenic tobacco plants and tandemly repeated pseudogenes. Nucleic Acids Res. 1990 Apr 25;18(8):1941–1949. doi: 10.1093/nar/18.8.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A., Westhof E., Bach M., Lührmann R., Ebel J. P., Carbon P. Solution structure of human U1 snRNA. Derivation of a possible three-dimensional model. Nucleic Acids Res. 1990 Jul 11;18(13):3803–3811. doi: 10.1093/nar/18.13.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lührmann R., Kastner B., Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim Biophys Acta. 1990 Nov 30;1087(3):265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- Maser R. L., Calvet J. P. U3 small nuclear RNA can be psoralen-cross-linked in vivo to the 5' external transcribed spacer of pre-ribosomal-RNA. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6523–6527. doi: 10.1073/pnas.86.17.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P. Secondary structure of the 5' external transcribed spacer of vertebrate pre-rRNA. Presence of phylogenetically conserved features. Eur J Biochem. 1991 Feb 14;195(3):601–609. doi: 10.1111/j.1432-1033.1991.tb15743.x. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- Myslinski E., Ségault V., Branlant C. An intron in the genes for U3 small nucleolar RNAs of the yeast Saccharomyces cerevisiae. Science. 1990 Mar 9;247(4947):1213–1216. doi: 10.1126/science.1690452. [DOI] [PubMed] [Google Scholar]
- Ochs R. L., Lischwe M. A., Spohn W. H., Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell. 1985;54(2):123–133. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Parker K. A., Bruzik J. P., Steitz J. A. An in vitro interaction between the human U3 snRNP and 28S rRNA sequences near the alpha-sarcin site. Nucleic Acids Res. 1988 Nov 25;16(22):10493–10509. doi: 10.1093/nar/16.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker K. A., Steitz J. A. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol Cell Biol. 1987 Aug;7(8):2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter G. L., Brennwald P. J., Holm K. A., Wise J. A. The sequence of U3 from Schizosaccharomyces pombe suggests structural divergence of this snRNA between metazoans and unicellular eukaryotes. Nucleic Acids Res. 1988 Nov 11;16(21):10131–10152. doi: 10.1093/nar/16.21.10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestayko A. W., Tonato M., Busch H. Low molecular weight RNA associated with 28 s nucleolar RNA. J Mol Biol. 1970 Feb 14;47(3):505–515. doi: 10.1016/0022-2836(70)90318-9. [DOI] [PubMed] [Google Scholar]
- Romby P., Moras D., Bergdoll M., Dumas P., Vlassov V. V., Westhof E., Ebel J. P., Giegé R. Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J Mol Biol. 1985 Aug 5;184(3):455–471. doi: 10.1016/0022-2836(85)90294-3. [DOI] [PubMed] [Google Scholar]
- Savino R., Gerbi S. A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990 Jul;9(7):2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroke I. L., Weiner A. M. The 5' end of U3 snRNA can be crosslinked in vivo to the external transcribed spacer of rat ribosomal RNA precursors. J Mol Biol. 1989 Dec 5;210(3):497–512. doi: 10.1016/0022-2836(89)90126-5. [DOI] [PubMed] [Google Scholar]
- Tague B. W., Gerbi S. A. Processing of the large rRNA precursor: two proposed categories of RNA-RNA interactions in eukaryotes. J Mol Evol. 1984;20(3-4):362–367. doi: 10.1007/BF02104742. [DOI] [PubMed] [Google Scholar]
- Tiollais P., Galibert F., Boiron M. Evidence for the existence of several molecular species in the "45S fraction" of mammalian ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1971 Jun;68(6):1117–1120. doi: 10.1073/pnas.68.6.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987 Dec 20;6(13):4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K., Steitz J. A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989 Oct;8(10):3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilenko S. K., Babkina G. T. Vydelenie i svoistva ribonukleazy iz iada kobry. Biokhimiia. 1965 Jul-Aug;30(4):705–712. [PubMed] [Google Scholar]
- Wise J. A., Weiner A. M. Dictyostelium small nuclear RNA D2 is homologous to rat nucleolar RNA U3 and is encoded by a dispersed multigene family. Cell. 1980 Nov;22(1 Pt 1):109–118. doi: 10.1016/0092-8674(80)90159-2. [DOI] [PubMed] [Google Scholar]
- Zieve G., Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976 May;8(1):19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]