Abstract
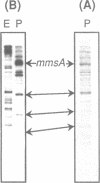
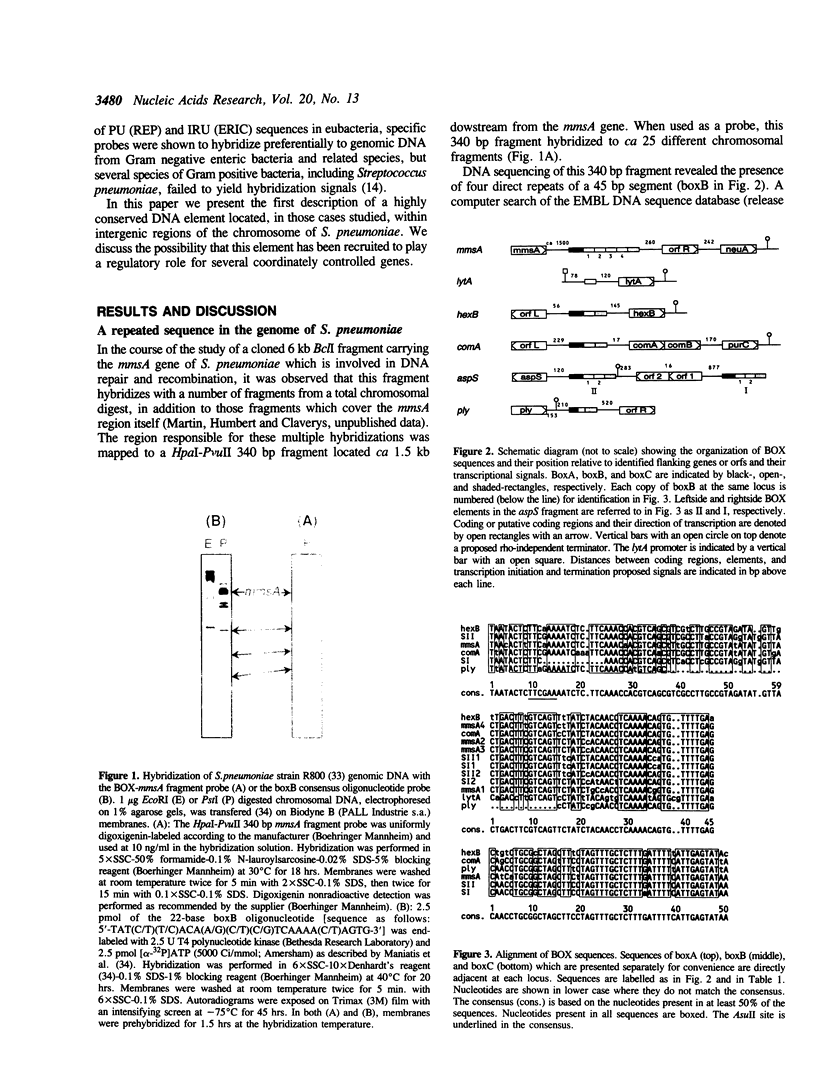
We report the discovery of a group of highly conserved DNA sequences located, in those cases studied, within intergenic regions of the chromosome of the Gram positive Streptococcus pneumoniae. The S. pneumoniae genome contains about 25 of these elements called BOX. From 5' to 3', BOX elements are composed of three subunits (boxA, boxB, and boxC) which are 59, 45 and 50 nucleotides long, respectively. BOX elements containing one, two and four copies of boxB have been observed; boxB alone was also detected in one instance. These elements are unrelated to the two most thoroughly documented families of repetitive DNA sequences present in the genomes of enterobacteria. BOX sequences have the potential to form stable stem-loop structures and one of these, at least, is transcribed. Most of these elements are located in the immediate vicinity of genes whose product has been implicated at some stage in the process of genetic transformation or in virulence of S. pneumoniae. This location raises the intriguing possibility that BOX sequences are regulatory elements shared by several coordinately controlled genes, including competence-specific and virulence-related genes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berry A. M., Lock R. A., Hansman D., Paton J. C. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect Immun. 1989 Aug;57(8):2324–2330. doi: 10.1128/iai.57.8.2324-2330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara M., Mitchell T. J., Andrew P. W., Boulnois G. J. Streptococcus pneumoniae produces at least two distinct enzymes with neuraminidase activity: cloning and expression of a second neuraminidase gene in Escherichia coli. Infect Immun. 1991 Aug;59(8):2856–2858. doi: 10.1128/iai.59.8.2856-2858.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M. S., Morrison D. A. Identification of two proteins encoded by com, a competence control locus of Streptococcus pneumoniae. J Bacteriol. 1988 Jul;170(7):3136–3141. doi: 10.1128/jb.170.7.3136-3141.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. D., Morrison D. A. Modulation of competence for genetic transformation in Streptococcus pneumoniae. J Gen Microbiol. 1987 Jul;133(7):1959–1967. doi: 10.1099/00221287-133-7-1959. [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coykendall A. L. Classification and identification of the viridans streptococci. Clin Microbiol Rev. 1989 Jul;2(3):315–328. doi: 10.1128/cmr.2.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz E., García J. L. Characterization of the transcription unit encoding the major pneumococcal autolysin. Gene. 1990 May 31;90(1):157–162. doi: 10.1016/0378-1119(90)90454-y. [DOI] [PubMed] [Google Scholar]
- García P., García J. L., García E., López R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene. 1986;43(3):265–272. doi: 10.1016/0378-1119(86)90215-5. [DOI] [PubMed] [Google Scholar]
- Gasc A. M., Kauc L., Barraillé P., Sicard M., Goodgal S. Gene localization, size, and physical map of the chromosome of Streptococcus pneumoniae. J Bacteriol. 1991 Nov;173(22):7361–7367. doi: 10.1128/jb.173.22.7361-7367.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Clément J. M., Brutlag D., Hofnung M. A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J. 1984 Jun;3(6):1417–1421. doi: 10.1002/j.1460-2075.1984.tb01986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Perrin D., Hofnung M. DNA polymerase I and a protein complex bind specifically to E. coli palindromic unit highly repetitive DNA: implications for bacterial chromosome organization. Nucleic Acids Res. 1990 Jul 11;18(13):3941–3952. doi: 10.1093/nar/18.13.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Rousset J. P., Clément J. M., Hofnung M. A subfamily of E. coli palindromic units implicated in transcription termination? Ann Inst Pasteur Microbiol. 1986 Nov-Dec;137B(3):259–270. doi: 10.1016/s0769-2609(86)80116-8. [DOI] [PubMed] [Google Scholar]
- Gilson E., Saurin W., Perrin D., Bachellier S., Hofnung M. Palindromic units are part of a new bacterial interspersed mosaic element (BIME). Nucleic Acids Res. 1991 Apr 11;19(7):1375–1383. doi: 10.1093/nar/19.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., McLaren R. S., Newbury S. F. Repetitive extragenic palindromic sequences, mRNA stability and gene expression: evolution by gene conversion? A review. Gene. 1988 Dec 10;72(1-2):3–14. doi: 10.1016/0378-1119(88)90122-9. [DOI] [PubMed] [Google Scholar]
- Hui F. M., Morrison D. A. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J Bacteriol. 1991 Jan;173(1):372–381. doi: 10.1128/jb.173.1.372-381.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulton C. S., Higgins C. F., Sharp P. M. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991 Apr;5(4):825–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Competence for deoxyribonucleic acid uptake and deoxyribonuclease action external to cells in the genetic transformation of Diplococcus pneumoniae. J Bacteriol. 1973 Apr;114(1):152–163. doi: 10.1128/jb.114.1.152-163.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre J. C., Claverys J. P., Sicard A. M. Donor deoxyribonucleic acid length and marker effect in pneumococcal transformation. J Bacteriol. 1979 Apr;138(1):80–86. doi: 10.1128/jb.138.1.80-86.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekalanos J. J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992 Jan;174(1):1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. A., Baker M. F. Competence for genetic transformation in pneumococcus depends on synthesis of a small set of proteins. Nature. 1979 Nov 8;282(5735):215–217. doi: 10.1038/282215a0. [DOI] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Higgins C. F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987 Dec 24;51(6):1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M., Martin B., Mejean V., Claverys J. P. Nucleotide sequence of the Streptococcus pneumoniae hexB mismatch repair gene: homology of HexB to MutL of Salmonella typhimurium and to PMS1 of Saccharomyces cerevisiae. J Bacteriol. 1989 Oct;171(10):5332–5338. doi: 10.1128/jb.171.10.5332-5338.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto H., Tomasz A. Protoplast formation and leakage of intramembrane cell components: induction by the competence activator substance of pneumococci. J Bacteriol. 1975 Jan;121(1):344–353. doi: 10.1128/jb.121.1.344-353.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples G. J., Lloyd R. G. A novel repeated DNA sequence located in the intergenic regions of bacterial chromosomes. Nucleic Acids Res. 1990 Nov 25;18(22):6503–6508. doi: 10.1093/nar/18.22.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M. J., Ames G. F., Smith N. H., Robinson E. C., Higgins C. F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984 Jul;37(3):1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- Stern M. J., Prossnitz E., Ames G. F. Role of the intercistronic region in post-transcriptional control of gene expression in the histidine transport operon of Salmonella typhimurium: involvement of REP sequences. Mol Microbiol. 1988 Jan;2(1):141–152. doi: 10.1111/j.1365-2958.1988.tb00015.x. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Jaeger J. A., Longfellow C. E., Freier S. M., Kierzek R. Improved parameters for prediction of RNA structure. Cold Spring Harb Symp Quant Biol. 1987;52:123–133. doi: 10.1101/sqb.1987.052.01.017. [DOI] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J. R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991 Dec 25;19(24):6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. A., Allen R. L., Falmagne P., Johnson M. K., Boulnois G. J. Molecular cloning, characterization, and complete nucleotide sequence of the gene for pneumolysin, the sulfhydryl-activated toxin of Streptococcus pneumoniae. Infect Immun. 1987 May;55(5):1184–1189. doi: 10.1128/iai.55.5.1184-1189.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ames G. F. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8850–8854. doi: 10.1073/pnas.85.23.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]