Abstract
Chronic alcohol-induced liver disease results in inflammation, steatosis and increased oxidative and nitrosative damage to the mitochondrion. We hypothesized that targeting an antioxidant to the mitochondria would prevent oxidative damage and attenuate the steatosis associated with alcoholic liver disease. To test this we investigated the effects of mitochondria-targeted ubiquinone, MitoQ, (5 & 25 mg/kg/d for 4 weeks) in male Sprague-Dawley rats consuming ethanol using the Lieber-DeCarli diet with pair-fed controls. Hepatic steatosis, 3-nitrotyosine (3-NT), 4-hydroxynonenal (4-HNE), hypoxia inducible factor α (HIF1α) and the activity of the mitochondrial respiratory chain complexes were assessed. As reported previously, ethanol consumption resulted in hepatocyte ballooning, increased lipid accumulation in the form of micro and macrovesicular steatosis and induction of CYP2E1. MitoQ had a minor on the ethanol-dependent decrease in mitochondrial respiratory chain proteins and their activities, it did however decrease hepatic steatosis in ethanol consuming animals and prevented the ethanol-induced formation of 3-NT and 4-HNE. Interestingly, MitoQ completely blocks the increase in HIF1α in all ethanol-fed groups which has previously been demonstrated in cell culture models and shown to be essential in ethanol-dependent hepatosteatosis. These results demonstrate the antioxidant capacity of MitoQ in alleviating alcohol associated mitochondrial ROS and several downstream effects of ROS/RNS production such as inhibiting protein nitration and protein aldehyde formation and specifically ROS-dependant HIF1α stabilization.
Keywords: MitoQ, Oxidative stress, mitochondria, alcohol
Introduction
Chronic alcohol consumption causes a spectrum of liver pathologies ranging from steatosis to steatohepatitis, fibrosis, cirrhosis, and can ultimately progress to hepatocellular carcinoma (1-4). Early stages of the disease are associated with macrovesicular or microvesicular steatosis predominantly in the central and mid-zonal areas of the liver (zones 3 and 2). Prolonged exposure to ethanol elicits secondary pathologies such as inflammation from gut-derived endotoxins and progresses to steatohepatitis which is characterized by hepatocellular ballooning, degeneration and necrosis, Mallory's hyaline body formation and tissue neutrophil infiltration(2, 5). Cirrhosis, the late stage and most severe form of alcoholic liver disease (ALD) is marked by fibrosis, altered liver architecture and decreased function and is often progressive and may eventually lead to organ failure(5, 6). Therefore, it is important to understand the molecular mechanisms that underlie the development of ALD to develop therapies that prevent further disease progression.
Augmented generation of reactive oxygen and nitrogen species (ROS/RNS) through induction of CYP2E1, NADPH oxidase and inducible nitric oxide synthase (iNOS) have been shown to contribute to liver pathology associated with ethanol toxicity in animal models of ALD(7-13). In addition, alcohol metabolism suppresses mitochondrial protein synthesis through its effects on mitochondrial ribosomes and possibily mitochondrial DNA(14, 15). Indeed, the mitochondria has long been recognized as an important target for alcohol mediated toxicity(3, 14, 16).
Chronic alcohol consumption causes marked decreases in respiratory chain enzymes resulting from decreased hepatic mitochondrial DNA (mtDNA) and proteomics studies have demonstrated changes in as many as 40 proteins in response to alcohol(15, 17). In addition to the direct impact of alcohol consumption on mtDNA, and mitochondrial protein synthesis machinery, intra-mitochondrial proteins are irreversibly oxidized by ROS/RNS and reactive lipid species such as 4-HNE (7, 9, 18-21). Functionally, this increases dysregulation of fatty acid metabolism and increases activation of the mitochondrial permeability transition pore (MPTP)(22, 23). Furthermore, endotoxin mediated activation of Kupffer cells also results in nitrosative stress through induction of iNOS(7, 9). Increased generation of nitric oxide then inhibits respiration in mitochondria sensitized by ethanol toxicity and also diet- induced fatty liver indicating commonality in the mechanisms leading to hepatosteatosis in response to metabolic stress(24, 25).
Taken together, these findings have naturally led to the testing of several antioxidant strategies for the prevention of steatohepatitis. Studies using compounds such as vitamin E, ebselen, superoxide dismutase and GSH precursors have demonstrated variable protective effects in models of chronic alcohol toxicity(26-29). Recently, it has been shown that the response to hypoxia plays an essential role in the induction of steatohepatitis(30) and this in turn has been linked to mitochondrial ROS generation(31-34). However none of these therapeutic strategies focus on the mitochondrion. Recently, the recognition that mitochondrial dysfunction is central to a broad range of pathologies has resulted in a number of mitochondria targeted therapies(35). Among the most prominent is the mitochondria targeted derivative of the antioxidant ubiquinone, (MitoQ). Ubiquinone is a naturally occurring electron carrier of mitochondrial respiratory chain. Covalent conjugation of ubiquinone to the lipophilic triphenylphosphonium (TPP) cation, to form MitoQ, results in a several fold accumulation in the cytoplasmic compartment and several-hundred fold accumulation within mitochondria(36) where it is reduced to the active antioxidant ubiquinol by the respiratory chain at complex II(37). It has been demonstrated that MitoQ improves mitochondria-related pathologies such as diabetic nephropathy, organ damage in LPS-induced sepsis, and more recently hepatitis C virus-induced hepatic damage(38-40). From in vivo and cell culture studies the mechanisms through which MitoQ mediates effects largely involve the modulation of mitochondrial ROS formation from the mitochondrion and the consequent effects on cell signaling. For example, it has been that mitochondrial ROS plays a key role in the activation of HIF1α and MitoQ inhibits both mitochondrial ROS formation and the regulatory pathways associated with hypoxia(31, 34, 41). Interestingly, hypoxia has long been recognized as a key feature of ethanol-dependent hepatotoxicity and a key regulator of steatosis leading to the hypothesis that MitoQ could suppress this pathway in vivo (30, 42). Importantly, MitoQ is orally bio-available, with low toxicity and is well tolerated in humans making it a suitable candidate for testing in animal models of alcohol toxicity (40, 43).
Materials and Methods
Chronic ethanol exposure model
Adult male (260-300g) Sprague-Dawley rats (Charles River Laboratories, Hartford, CT) were pair-fed isocaloric Lieber-DeCarli liquid diets (BioServ, NJ) containing 0% or 36% ethanol by caloric content for 4 weeks (44). The feeding regimen was based on a 2 × 3 factorial design with three independent variables: dietary ethanol and two doses of mitochondria-targeted ubiquinone (MitoQ) (5 and 25 mg/kg/d). Based on animal weights throughout the study and the diet consumed measured over 24 hrs, the MitoQ dosage was calculated to give the desired dose of 5 and 25 mg/kg/day. The stability of MitoQ in the liquid diet ± ethanol was established by HPLC and showed no degradation over the course of the experiment (data not shown) and had no effect on the amount of diet consumed in each group (Table 1). Control diets were supplemented with the indicated doses were fed for seven days prior to ethanol exposure. Liver tissues were harvested at the time of sacrifice. All experiments were conducted in accordance with the NIAAA guidelines and approved by the institutional Animal Care and Use Committee at the University of Alabama at Birmingham.
Table 1. Study parameters in control and ethanol-fed rats with and without MitoQ.
| Control | Ethanol | MitoQ 5 mg/kg/d | MitoQ 5 mg/kg/d + Ethanol | MitoQ 25 mg/kg/d | MitoQ 25 mg/kg/d + Ethanol | |
|---|---|---|---|---|---|---|
| Food intake (ml/day) {days 30–35} | 87.4 ± 0.9 | 87.4 ± 0.9 | 87.2 ± 0.6 | 87.2 ± 0.6 | 85.0 ± 0.4 | 85.0 ± 0.4 |
| Liver Weight (g) | 9.6 ± 0.7 | 12.9* ± 1.1 | 9.9 ± 0.7 | 12.7 ± 1.0 | 10.0 ± 1.0 | 11.3 ± .6 |
| Liver/body weight ratio (%) | 2.9 ± 0.2 | 3.7* ± 0.2 | 2.9 ± 0.2 | 3.6* ± 0.1 | 2.6 ± 0.2 | 3.3* ± 0.1 |
| Body weight gain (g/d) | 2.5 ± 0.03 | 2.5 ± 0.3 | 2.5 ± 0.2 | 2.5 ± 0.3 | 2.6 ± 0.2 | 2.0 ± 0.2 |
| Serum ALT, U/l | 60.6 ± 9.0 | 64.7 ± 5.4 | 51.5 ± 4.8 | 60.2 ± 11.8 | 52.1 ± 8.8 | 65.9 ± 6.1 |
Note. Data represent the mean ± SEM for 6 pairs of rats on an ethanol-containing or control diet and 5 pairs of rats fed ethanol and control diets supplemented with MitoQ at 5 and 25 mg/kg/d.
Using the pair-fed feeding model, control animals received the exact of amount of diet as it's ethanol-fed counterpart.
p < 0.05 for pair-fed controls. Statistical analysis was performed using the paired Student's T-Test.
Isolation of rat liver mitochondria
Coupled liver mitochondria were prepared by differential centrifugation of liver homogenates as previously reported (15). Total mitochondria yield from pair-fed controls and animals consuming ethanol was 131 ± 10 and 159 ± 21 mg of protein respectively (n = 6, p = 0.278). Control animals treated with MitoQ (5 and 25 mg/kg/d) had mitochondrial yields of 148 ± 17 and 142 ± 11 mg of protein and animals consuming ethanol treated with MitoQ had 169 ± 13 and 166 ± 9 mg of protein respectively (n = 4-5, p = 0.185 and 0.125).
Histology and immunohistochemistry
Paraformaldehyde fixed liver sections (5μm) were stained with hematoxylin-eosin and quantified. The extent of steatosis was determined by measuring the area of macro- and microsteatotic vesicles separately (six fields per slide, n= 5-6 animals per group) and was quantified using Simple PCI software using the HLS algorithm with specific size exclusion parameters for macro and microsteatosis. Steatotic vesicles larger than the hepatocyte nucleus (7-8μm) and displacing the nucleus from center of the cell were considered macrosteatotic and those which are smaller than the hepatocyte nucleus were characterized as microsteatotic. Immunohistochemistry for 3-NT, HIF1α, iNOS and 4-HNE were performed using antibodies raised against 3-NT (Millipore, Billerica, MA), HIF1α (Epitomics, Burlingame, CA), iNOS (Santa Cruz, Santa Cruz, CA), or 4-HNE (Alpha Diagnostics, San Antonio, Texas) and developed using DAB as substrate. Frozen liver sections (5 μm) were fixed frozen using paraformaldehyde (4%) and stained with osmium tetroxide (0.1%).
Enzyme assays
Mitochondrial function was assessed by measuring the activity levels of NADH-ubiquinone oxidoreductase (complex I), Succinate-ubiquinone oxidoreductase, (complex II), Cytochrome C oxidase (complex IV), ATPase (complex V), citrate synthase and a combined succinate-ubiquinone oxidoreductase/ ubiquinol ferricytochrome c reductase (complex II-III) assay. All assays were measured spectrophotometrically as previously described(7).
Western blotting
Proteins were immunoblotted using antibodies against CYP2E1 (Chemicon, MA), ALDH2 (a kind gift from Dr. Matthew Picklo, University of North Dakota School of Medicine) complexes I (30kD subunit), II (30kD subunit), III (core protein 1, Rieske iron sulphur), and IV (subunits I and IV) (Invitrogen, CA.), phospho AMPK and phospho ACC (Cell Signaling, MA.) and detected using enhanced chemiluminescence (SuperSignal, West-Dura Pierce, IL).
Statistical analysis
All data represents the means ± SEM, n=6 for control and ethanol groups and n=5 for MitoQ treated groups. Statistical significance was determined using Student's TTEST, ANOVA and Newman-Keuls test as post-hoc test. p < 0.05 was taken as significantly different.
Results
MitoQ treatment with and without chronic ethanol consumption
Animals were pair-fed with either ethanol or control liquid diets containing MitoQ (0,5, or 25 mg/kg/day) for 28-35 days. There was no significant difference in body weight gain, however the ethanol group had increased liver weight as compared to pair-fed controls which was prevented by MitoQ although no effect was seen on liver to body weight ratios (Table 1). Consumption of ethanol did not significantly increase serum alanine aminotransferase levels compared with controls. Ethanol consumption resulted in increased serum HDL levels as expected and but was not changed by MitoQ(45). The serum LDL/VLDL also showed no significant changes with alcohol exposure or any treatment (supplementary Figure 1).
MitoQ treatment decreases ethanol-dependent protein nitration and lipid peroxidation
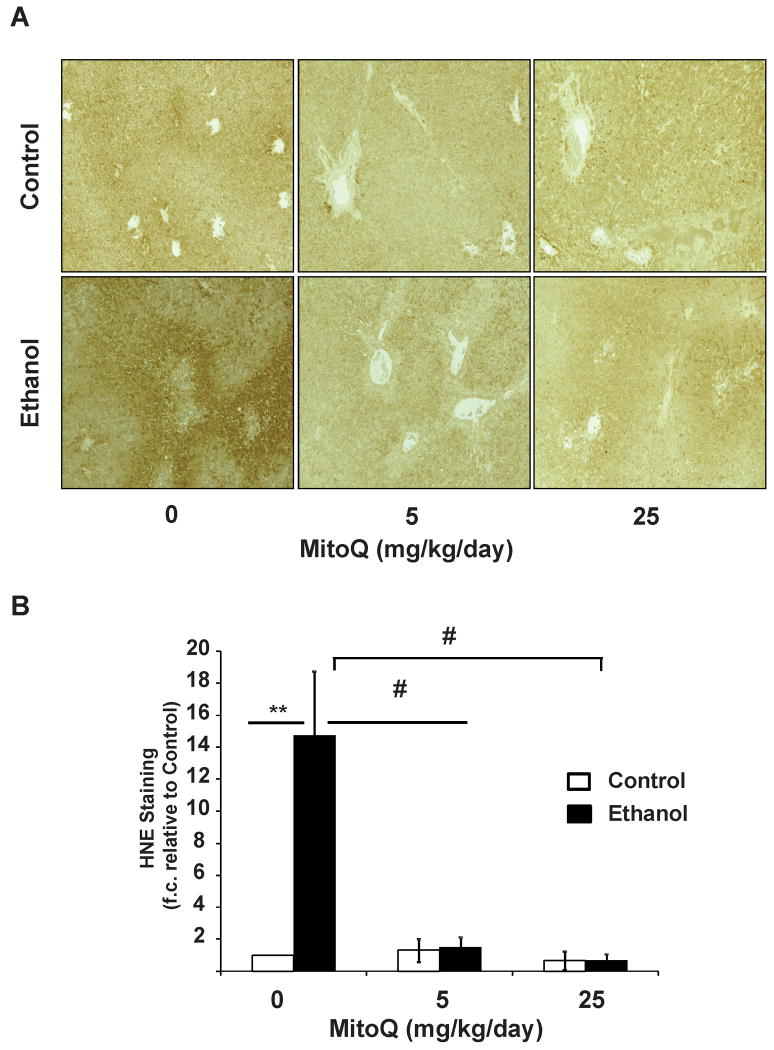
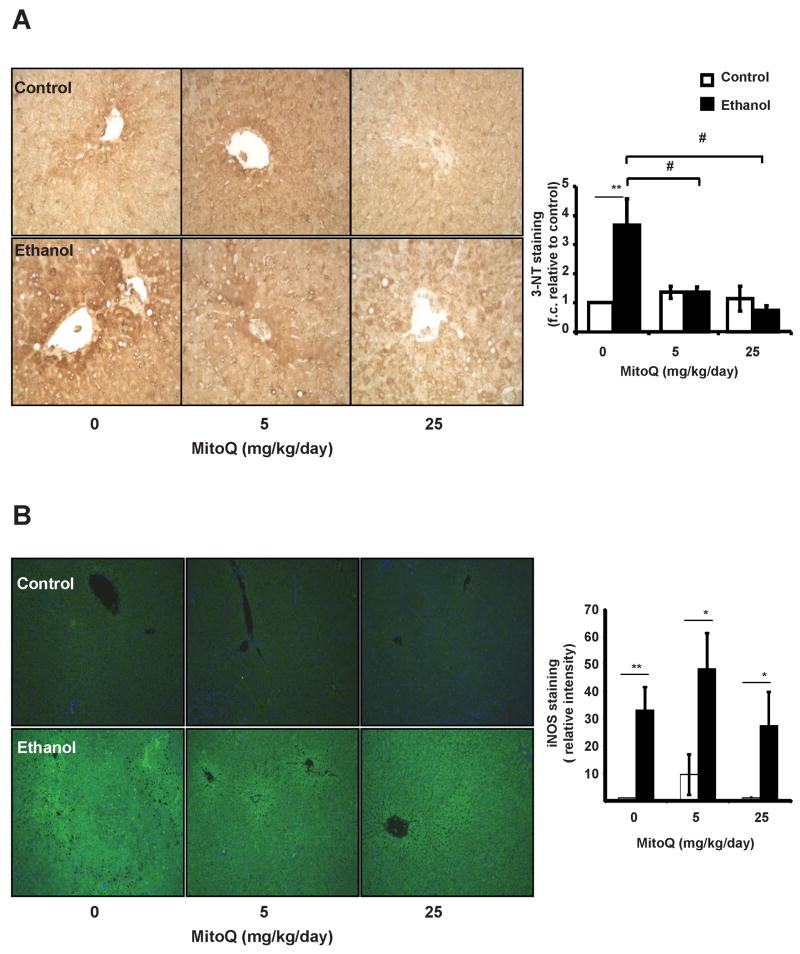
Chronic ethanol consumption is known to increase 4-HNE-protein adducts and iNOS-dependent protein nitration (7, 9, 20, 21). In agreement with these findings liver tissues show intense staining for 4-HNE-protein adducts in chronic ethanol fed animals compared to pair-fed controls (Figure 1A & 1B). HNE immunoreactivity was not uniform with hepatocytes around the central veins showing the most intense staining (zones 2 & 3) and a gradual decline towards the periportal region (zone 1). MitoQ treatment completely abolished ethanol-induced 4-HNE staining in all regions of the liver sections examined. Control experiments show no effect of MitoQ at either dose (Figure 1 A,B) and omission of the primary antibody for HNE resulted in no detectable signal (result not shown). Consistent with previous studies, chronic ethanol feeding increased 3-NT and iNOS staining with highest in zone 3, with intermediate staining in zone 2 (Figure 2A & 2B) (9, 21, 46). MitoQ treatment significantly decreased 3-NT staining in the liver of ethanol fed rats, however it did not have any effect on the induction of iNOS protein. Controls with excess free 3-nitrotyrosine or omission of the primary antibody for 3-NT resulted in no signal (result not shown). It has been shown that MitoQ inhibits mitochondrial ROS and the consequent activation of HIF1α (31, 32, 47). We first confirmed the activation of HIF1α in response to EtOH (Figure 3A)(48, 49). Next we assessed the impact of Mito Q and demonstrated that drug treatment completely suppressed immunoreactivity of HIF1α at both concentrations (Figure 3A,B).
Figure 1. MitoQ prevents ethanol-induced 4-HNE adduct formation in rat liver.
Immunohistochemical detection of HNE adducts was performed on liver sections using anti-HNE antibodies. (A) Representative images from each group (10×). (B) Quantification of each image. **, represents p≤0.01 between ethanol treated group relative to the corresponding non-ethanol group. # represents p≤0.05 among ethanol treated groups.
Figure 2. MitoQ decreases hepatic 3-nitrotyrosine levels, but not iNOS.
Paraffin embedded liver sections were immunostained using anti 3-NT (A) or iNOS (B) antibodies and developed using DAB. Representative sections from each group, 40×; and quantification of each image are depicted. **, represents p≤0.01 between ethanol treated group relative to the corresponding non-ethanol group. # represents p≤0.05 among ethanol treated groups.
Figure 3. MitoQ inhibits ethanol-induced HIF1α accumulation in the liver.
Immunoreactivity of anti-HIF1α antibody was detected in paraffin embedded liver sections (A) Representative images of HIF1α fluorescent staining from each treatment group (10×). (B) Fluorescent intensity of identically stained sections were quantified and expressed as mean±sem. **, represents p≤0.01 between ethanol treated group relative to the corresponding non-ethanol group. # represents p≤0.05 among ethanol treated groups.
MitoQ does not alter AMPK signaling, phosphorylation of ACC nor expression of Cyp2E1 or ALDH2 proteins by chronic ethanol consumption
In order to determine whether MitoQ had an effect on the ethanol metabolizing enzymes, we measured protein expression levels of CYP2E1 and ALDH2 in liver homogenates (Supplementary Figure 2A,B). Consistent with previous reports, CYP2E1 protein expression was increased in all ethanol-fed animals(12). Importantly, CYP2E1 protein expression was similar in ethanol fed control animals indicating similar exposure to ethanol in all treatment groups when compared to treatment with MitoQ. Similarly, ALDH2 protein levels were not affected by ethanol consumption or modified by MitoQ. These data suggest that MitoQ treatment does not affect the key enzymes that are responsible for ethanol metabolism. We next investigated whether MitoQ would alter levels of protein kinase (AMPK) and the level of phosphorylation of its downstream target acetyl CoA carboxylase (ACC) since it has been reported that total AMPK levels are decreased upon chronic ethanol consumption when compared to controls (50). In the current study we observed only modest effects on the AMPK system which only showed significance by MitoQ at 25 mg/kg/d (Supplementary Figure 2C). Furthermore, levels of p-ACC were not different between ethanol-fed animals and their pair-matched controls. MitoQ at 5 mg/kg/d had no affect; however MitoQ at 25 mg/kg/d had a modest however significant effect on the p-ACC/total protein ratio in both the control and ethanol-fed animals (Supplementary Figure 2D).
MitoQ did not attenuate ethanol-dependent damage to the mitochondrial respiratory chain
Chronic ethanol consumption results in decreased activity of mitochondrial respiratory chain proteins coded for by mitochondrial DNA(15, 17). Consistent with previous studies, chronic ethanol consumption resulted in decreases in the activities of complex I, III, IV and V and a small increase in citrate synthase activity in isolated mitochondria but was not changed by MitoQ(Table 2). As previously shown, chronic ethanol consumption decreased complex I (30kDa subunit), Complex IV (Subunits I and IV) and Complex III (Rieske FeS) proteins levels although no effects on complex II or complex III core protein 2 were observed Supplementary Figure 3 (15). Overall, treatment with MitoQ had only a modest effect on Complex I, 30kD subunit and complex IV subunit IV and was only evident at the dose of 5 mg/kg/day MitoQ.
Table 2. MitoQ does not restore decreases in mitochondrial respiratory chain activity.
| Control | Ethanol | MitoQ 5 mg/kg/d | MitoQ 5 mg/kg/d + Ethanol | MitoQ 25 mg/kg/d | MitoQ 25 mg/kg/d + Ethanol | |
|---|---|---|---|---|---|---|
| Complex Ia | 118.14 ± 5.68 | 65.79** ± 5.59 | 111.36 ± 3.87 | 78.56**± 4.91 | 105.83 ± 8.38 | 81.56*± 6.54 |
| Complex IIb | 1.09 ± 0.04 | 1.22 ± 0.04 | 1.00 ± 0.07 | 1.37*± 0.13 | 1.06 ± 0.02 | 1.31± 0.05 |
| Complex II-IIIc | 60.24 ± 3.41 | 22.94**± 1.88 | 57.94 ± 5.09 | 26.24**± 2.24 | 61.10 ± 7.29 | 26.00**± 3.73 |
| Complex IVd | 53.72 ± 1.79 | 30.35**± 3.79 | 56.20 ± 3.06 | 33.10**± 2.81 | 56.99 ± 1.40 | 32.02**± 3.15 |
| Complex Ve (ATPase) | 175.26±17.89 | 149.12±13.78 | 147.77±11.47 | 144.44±8.65 | 181.52±13.52 | 184.58±11.72 |
| Citrate Synthasef | 0.115 ± 0.004 | 0.138*± 0.005 | 0.114 ± 0.005 | 0.138*± 0.006 | 0.110 ± 0.005 | 0.134*± 0.004 |
(nmol NADH/min/mg protein);
nmol DCPIP/min/mg protein;
nmol Cyt.c (reduced)/min/mg protein;
(k/sec)/mg protein;
nmol NADH/min/mg protein;
μmol/min/mg protein
Activities were measured as described in materials and methods. Data represent the mean ± SEM for 6 pairs of rats on an ethanol-containing or control diet and 5 pairs of rats fed ethanol and control diets supplemented with MitoQ at 5 and 25 mg/kg/d. Statistical comparison were performed using one-way ANOVA whereas * and ** represents p≤0.05 and p≤0.01 respectively between ethanol-fed animals and corresponding pair-fed controls.
Hepatic Steatosis in response to chronic ethanol consumption and the impact of MitoQ
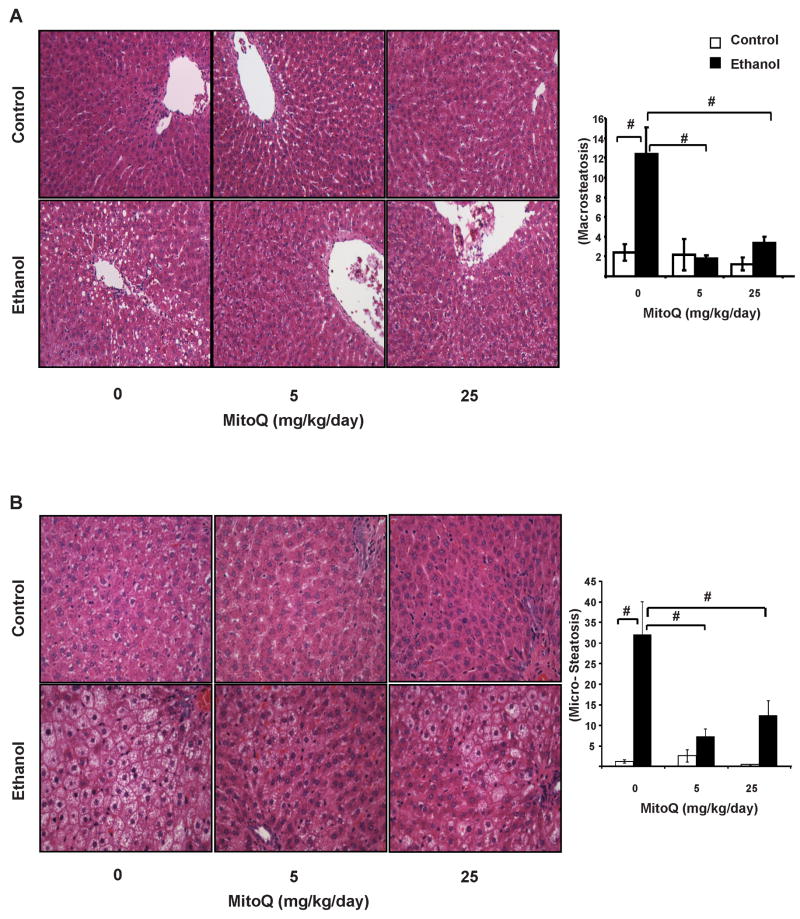
Chronic ethanol consumption increased hepatic macro- and microvesicular steatosis compared to the pair-fed controls (Figure 4). Macrosteatototic vesicles distributed around the pericentral region, in contrast, microvesicular steatosis is predominantly present around the portal tract (zone 1) and to a lesser extent in the pericentral region(Figure 4A). MitoQ (5 and 25 mg/kg/day) significantly decreased macro- and microsteatosis in ethanol -fed rats. In contrast to macrosteatosis, MitoQ did not demonstrate complete protection of microsteatosis at 25 mg/kg/day (Figure 4B). MitoQ alone at either dose had no effect on steatosis in the control animals. In order to confirm the presence of lipids in the liver and to identify the type of lipid accumulated, frozen tissue sections were stained with osmium tetroxide which selectively reacts with unsaturated lipids. Consistent with the data shown in Figure 4 chronic alcohol consumption resulted in extensive accumulation of osmium tetroxide stained lipids in macrovesicular steatotic vesicles (Figure 5A) around the central vein and as microvesicular steatosis in the periportal region. MitoQ treatment decreased the number and size of steatotic vesicles containing unsaturated lipids in ethanol-fed animals as reflected in the quantification of area of osmium tetroxide staining (Figure 5B).
Figure 4. MitoQ prevents hepatic macrosteatosis and microsteastosis in ethanol-fed animals.
Hepatic steatosis was assessed using H&E stained liver sections (A) Representative images of macrosteatotic regions from each group, 40×, and steatosis quantification (B) Representative images of microsteatosis in the periportal region from each group (40×) and quantification. # represents p≤0.05 between ethanol treated group relative to the corresponding non-ethanol group. # represents p≤0.05 among ethanol treated groups.
Figure 5. MitoQ prevents the accumulation of unsaturated lipids in rat liver.
Hepatic steatosis was assessed using osmium tetroxide stained frozen liver sections. (Panel A) Representative images and (B) Quantitation of each set. ** represents p≤0.05 between ethanol treated group relative to the corresponding non-ethanol group. # represents p≤0.05 among ethanol treated groups.
Discussion
Alcohol-induced fatty liver enhances the susceptibility of the liver to develop steatohepatitis, fibrosis, cirrhosis, and hepatocellular carcinoma(1, 5, 6). It has recently been shown that steatosis is in response to ethanol consumption is modulated by the regulation of hypoxia in the liver and this pathway is known to be responsive to mitochondrial function (30, 34, 41). Since mitochondria are both a source and target for reactive oxygen and nitrogen species (ROS/RNS) it is not surprising that they are thought to play a central role in the pathophysiology of ethanol-dependent hepatotoxicity(3, 4, 13, 14, 51, 52) but the link to hypoxia in this pathology is not clear. Taken together these data and other studies have closely linked the production of ROS/RNS to mitochondrial DNA and protein damage and alcohol-dependent metabolic derangements in the liver. On the basis of these findings we hypothesized that a mitochondrial targeted antioxidant could potentially alleviate pathological changes which occur in response to chronic alcohol consumption.
To test this we used oral treatment of MitoQ, an amphipathic conjugate of ubiquinone with the triphenylphosphonium cation (TPP+) which has been shown to be non-toxic and orally bioavailable in animal models and humans (32, 43, 53). Recent reports demonstrate MitoQ mediated protection against cardiac ischemia-reperfusion injury, diabetic nephropathy, adriamycin induced cardiotoxicity and hepatitis C induced liver injury (38, 40, 54, 55). The TPP+ moiety targets the quinone functional group to the mitochondrion where it is reduced to the quinol form by complex II, unlike the endogenous co-enzyme Q, it interacts poorly with mitochondrial respiratory chain complexes I and III(56). Overall this allows MitoQ to act as a source of reducing equivalents in this portion of the respiratory chain without greatly impacting the normal electron transfer process. This increased concentration of MitoQ in the mitochondrial inner membrane can act as an inhibitor of lipid peroxidation that generates 4-HNE and can also prevent peroxynitrite mediated protein modification or potentially scavenge peroxynitrite directly (36, 37). Importantly, alcohol-induced 4-HNE modification of key proteins has been reported in the mitochondria including cytochrome c oxidase and aconitase which are associated with the severity of steatosis in human subjects (1, 19, 57). The mechanism by which MitoQ-mediated inhibition of lipid peroxidation and protein nitration acts most likely through its direct antioxidant action concomitant with recent reports which propose antioxidant, anti-inflammatory properties and anti-hypoxic properties for this compound (31, 32, 34, 56, 58).
In support of our hypothesis, we found MitoQ inhibited the formation 4-HNE protein adduct formation and 3-NT levels, indicators of the antioxidant action of MitoQ (Figure 1,2). The pattern of 4-HNE and 3-NT staining demonstrate a strong gradient extending from the pericentral region deep into the periportal region of the liver and is consistent with similar studies of ethanol-induced liver injury(59). The enhancement of the oxidative/nitrosative stress gradient is linked to several factors including exacerbation of the hypoxic gradient developed in the liver acini and LPS-induced cytokine production in chronic ethanol consumption. MitoQ treatment in LPS-induced inflammation has been shown to involve decreases in proinflammatory cytokines such as IL-1β, IL-6, and IL-8 and increase in the anti-inflammatory cytokine IL-10 levels (60). However, in the present study we found that MitoQ did not significantly change the expression of iNOS suggesting that its mode of action is downstream of cytokine signaling (Figure 2B).
Since it has been shown that mitochondria and specifically MitoQ can modify the cellular response to hypoxia we next examined this pathway (31, 34, 41). Induction of tissue hypoxia and HIF1α in the liver is a hallmark of alcohol-induced liver disease(30, 61). Furthermore iNOS derived NO has been shown to inhibit prolyl hydroxylase enzyme activity by competing for the iron(II) in the catalytic site of the enzymes during normoxia and changes mitochondrial function with increased ROS formation (7, 51, 62). Our data are consistent with this literature since we found chronic ethanol induced HIF1α expression/stabilization (Figure 3A), increased ROS and increased iNOS (Figure 2B). MitoQ treatment inhibited ethanol-induced HIF1α expression in the liver (Figure 3A) whereas iNOS expression remained unaltered (Figure 2B). Recently it has been shown that HIF1α in heptocytes is a major determinant in the pathogenesis of alcoholic steatosis(30). Taken together we propose that MitoQ inhibits the mitochondrial-dependent induction of HIF1α through suppression of increased mitochondrial ROS in response to NO exposure or damage to the mitochondrion by peroxynitrite. To form peroxynitrite superoxide must also be formed in response to ethanol consumption and could come from a number of sources including the mitochondrion or NADPH oxidase(10, 14). Since it has been shown that MitoQ can directly scavenge peroxynitrite this is a likely mechanism through which activation of HIF1α is prevented(37).
Ethanol feeding leads to inhibition of mitochondrial protein synthesis, which is largely responsible for the changes in the activities of the mitochondrial respiratory chain complexes(4, 15, 52, 63). Moreover, MitoQ does not change ALDH2 levels and has no known interaction with this enzyme therefore it is unlikely that it would have a major impact on this aspect of ethanol-dependent hepatotoxicity. Since the effects of MitoQ on respiratory chain proteins or activities are minor this is unlikely to be a major contributor to the mechanism of MitoQ-mediated prevention of steatosis.
Accumulation of lipids as micro and macrovesicles and the distinctive localization of lipid vesicles demonstrate the characteristic tissue pathology induced by ethanol(64). MitoQ mediated inhibition of both micro and macrosteatosis at a dose of 5 mg/kg/day suggest direct or indirect interference with the pathways leading to lipid accumulation in the liver most likely through the prevention of oxidative and nitrative stress as discussed above (Figure 4). Interestingly, MitoQ treatment at 25 mg/kg/d increased the level of microsteatosis in the periportal areas of the liver similar to that observed in drug-induced liver injury through mechanisms that are unclear at the present time.
In summary, we have shown that although MitoQ did not improve ethanol induced inhibition of mitochondrial respiration or enzyme activities, but clearly protected the liver against ethanol-induced oxidant damage and steatosis through a mechanism which involves scavenging of ROS/RNS and the suppression of HIF1α activation. As MitoQ can be safely administered long-term to humans [35, 36] and has been shown to decrease liver damage in Hepatitis C patients [35] it may have potential to ameliorate the initial stages of fatty liver disease in patients with alcoholic and non-alcoholic liver disease.
Supplementary Material
Acknowledgments
The authors thank Jeff Dubuisson for excellent technical assistance.
Abbreviations
- MitoQ
mitochondria-targeted ubiquinone
- LPS
lipopolysaccharide
- ROS/RNS
reactive oxygen and reactive nitrogen species
- iNOS
inducible nitric oxide synthase
- 4-HNE
4-hydroxynonenal
- 3-NT
3-nitrotyrosine
- CYP2E1
cytochrome P450 2E1
- mtDNA
mitochondrial DNA
- ALT
alanine aminotransferase
- HIF1α
hypoxia inducible factor α
Footnotes
Disclosures: Dr. Michael P. Murphy holds patents related to the area of mitochondria-targeted antioxidants as therapies and serves on the scientific advisory board of Antipodean Pharmaceuticals Inc. that develops and commercializes MitoQ.
References
- 1.Lieber CS. Alcoholic fatty liver: its pathogenesis and mechanism of progression to inflammation and fibrosis. Alcohol. 2004;34:9–19. doi: 10.1016/j.alcohol.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Hoek JB, Pastorino JG. Ethanol, oxidative stress, and cytokine-induced liver cell injury. Alcohol. 2002;27:63–68. doi: 10.1016/s0741-8329(02)00215-x. [DOI] [PubMed] [Google Scholar]
- 3.Hoek JB, Cahill A, Pastorino JG. Alcohol and mitochondria: a dysfunctional relationship. Gastroenterology. 2002;122:2049–2063. doi: 10.1053/gast.2002.33613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahill A, Cunningham CC, Adachi M, Ishii H, Bailey SM, Fromenty B, Davies A. Effects of alcohol and oxidative stress on liver pathology: the role of the mitochondrion. Alcohol Clin Exp Res. 2002;26:907–915. [PMC free article] [PubMed] [Google Scholar]
- 5.Breitkopf K, Nagy LE, Beier JI, Mueller S, Weng H, Dooley S. Current experimental perspectives on the clinical progression of alcoholic liver disease. Alcohol Clin Exp Res. 2009;33:1647–1655. doi: 10.1111/j.1530-0277.2009.01015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arteel G, Marsano L, Mendez C, Bentley F, McClain CJ. Advances in alcoholic liver disease. Best Pract Res Clin Gastroenterol. 2003;17:625–647. doi: 10.1016/s1521-6918(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 7.Venkatraman A, Shiva S, Wigley A, Ulasova E, Chhieng D, Bailey SM, Darley-Usmar VM. The role of iNOS in alcohol-dependent hepatotoxicity and mitochondrial dysfunction in mice. Hepatology. 2004;40:565–573. doi: 10.1002/hep.20326. [DOI] [PubMed] [Google Scholar]
- 8.Venkatraman A, Landar A, Davis AJ, Ulasova E, Page G, Murphy MP, Darley-Usmar V, et al. Oxidative modification of hepatic mitochondria protein thiols: effect of chronic alcohol consumption. Am J Physiol Gastrointest Liver Physiol. 2004;286:G521–527. doi: 10.1152/ajpgi.00399.2003. [DOI] [PubMed] [Google Scholar]
- 9.McKim SE, Gabele E, Isayama F, Lambert JC, Tucker LM, Wheeler MD, Connor HD, et al. Inducible nitric oxide synthase is required in alcohol-induced liver injury: studies with knockout mice. Gastroenterology. 2003;125:1834–1844. doi: 10.1053/j.gastro.2003.08.030. [DOI] [PubMed] [Google Scholar]
- 10.Wheeler MD, Kono H, Yin M, Nakagami M, Uesugi T, Arteel GE, Gabele E, et al. The role of Kupffer cell oxidant production in early ethanol-induced liver disease. Free Radic Biol Med. 2001;31:1544–1549. doi: 10.1016/s0891-5849(01)00748-1. [DOI] [PubMed] [Google Scholar]
- 11.Wu D, Cederbaum AI. Oxidative stress and alcoholic liver disease. Semin Liver Dis. 2009;29:141–154. doi: 10.1055/s-0029-1214370. [DOI] [PubMed] [Google Scholar]
- 12.Lu Y, Cederbaum AI. CYP2E1 and oxidative liver injury by alcohol. Free Radic Biol Med. 2008;44:723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey SM. A review of the role of reactive oxygen and nitrogen species in alcohol-induced mitochondrial dysfunction. Free Radic Res. 2003;37:585–596. doi: 10.1080/1071576031000091711. [DOI] [PubMed] [Google Scholar]
- 14.Bailey SM, Cunningham CC. Contribution of mitochondria to oxidative stress associated with alcoholic liver disease. Free Radic Biol Med. 2002;32:11–16. doi: 10.1016/s0891-5849(01)00769-9. [DOI] [PubMed] [Google Scholar]
- 15.Venkatraman A, Landar A, Davis AJ, Chamlee L, Sanderson T, Kim H, Page G, et al. Modification of the mitochondrial proteome in response to the stress of ethanol-dependent hepatotoxicity. J Biol Chem. 2004;279:22092–22101. doi: 10.1074/jbc.M402245200. [DOI] [PubMed] [Google Scholar]
- 16.Mantena SK, King AL, Andringa KK, Landar A, Darley-Usmar V, Bailey SM. Novel interactions of mitochondria and reactive oxygen/nitrogen species in alcohol mediated liver disease. World J Gastroenterol. 2007;13:4967–4973. doi: 10.3748/wjg.v13.i37.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sykora P, Kharbanda KK, Crumm SE, Cahill A. S-adenosyl-L-methionine co-administration prevents the ethanol-elicited dissociation of hepatic mitochondrial ribosomes in male rats. Alcohol Clin Exp Res. 2009;33:1–9. doi: 10.1111/j.1530-0277.2008.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem Res Toxicol. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Robinson NC, Schenker S, Frosto TA, Henderson GI. Formation of 4-hydroxynonenal adducts with cytochrome c oxidase in rats following short-term ethanol intake. Hepatology. 1999;29:1792–1798. doi: 10.1002/hep.510290611. [DOI] [PubMed] [Google Scholar]
- 20.Lee JH, Yang ES, Park JW. Inactivation of NADP+-dependent isocitrate dehydrogenase by peroxynitrite. Implications for cytotoxicity and alcohol-induced liver injury. J Biol Chem. 2003;278:51360–51371. doi: 10.1074/jbc.M302332200. [DOI] [PubMed] [Google Scholar]
- 21.Baraona E, Zeballos GA, Shoichet L, Mak KM, Lieber CS. Ethanol consumption increases nitric oxide production in rats, and its peroxynitrite-mediated toxicity is attenuated by polyenylphosphatidylcholine. Alcohol Clin Exp Res. 2002;26:883–889. [PubMed] [Google Scholar]
- 22.King AL, Swain TM, Dickinson DA, Lesort MJ, Bailey SM. Chronic ethanol consumption enhances sensitivity to Ca(2+)-mediated opening of the mitochondrial permeability transition pore and increases cyclophilin D in liver. Am J Physiol Gastrointest Liver Physiol. 299:G954–966. doi: 10.1152/ajpgi.00246.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pastorino JG, Marcineviciute A, Cahill A, Hoek JB. Potentiation by chronic ethanol treatment of the mitochondrial permeability transition. Biochem Biophys Res Commun. 1999;265:405–409. doi: 10.1006/bbrc.1999.1696. [DOI] [PubMed] [Google Scholar]
- 24.Mantena SK, Vaughn DP, Andringa KK, Eccleston HB, King AL, Abrams GA, Doeller JE, et al. High fat diet induces dysregulation of hepatic oxygen gradients and mitochondrial function in vivo. Biochem J. 2009;417:183–193. doi: 10.1042/BJ20080868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkatraman A, Shiva S, Davis AJ, Bailey SM, Brookes PS, Darley-Usmar VM. Chronic alcohol consumption increases the sensitivity of rat liver mitochondrial respiration to inhibition by nitric oxide. Hepatology. 2003;38:141–147. doi: 10.1053/jhep.2003.50293. [DOI] [PubMed] [Google Scholar]
- 26.Lieber CS. S-Adenosyl-L-methionine and alcoholic liver disease in animal models: implications for early intervention in human beings. Alcohol. 2002;27:173–177. doi: 10.1016/s0741-8329(02)00230-6. [DOI] [PubMed] [Google Scholar]
- 27.Bailey SM, Robinson G, Pinner A, Chamlee L, Ulasova E, Pompilius M, Page GP, et al. S-adenosylmethionine prevents chronic alcohol-induced mitochondrial dysfunction in the rat liver. Am J Physiol Gastrointest Liver Physiol. 2006;291:G857–867. doi: 10.1152/ajpgi.00044.2006. [DOI] [PubMed] [Google Scholar]
- 28.Kono H, Arteel GE, Rusyn I, Sies H, Thurman RG. Ebselen prevents early alcohol-induced liver injury in rats. Free Radic Biol Med. 2001;30:403–411. doi: 10.1016/s0891-5849(00)00490-1. [DOI] [PubMed] [Google Scholar]
- 29.Loguercio C, Federico A. Oxidative stress in viral and alcoholic hepatitis. Free Radic Biol Med. 2003;34:1–10. doi: 10.1016/s0891-5849(02)01167-x. [DOI] [PubMed] [Google Scholar]
- 30.Nath B, Levin I, Csak T, Petrasek J, Mueller C, Kodys K, Catalano D, et al. Hepatocyte-specific hypoxia inducible factor-1α is a determinant of lipid accumulation and liver injury in alcohol-induced steatosis in mice. Hepatology:n/a-n/a. doi: 10.1002/hep.24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bell EL, Klimova TA, Eisenbart J, Moraes CT, Murphy MP, Budinger GR, Chandel NS. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J Cell Biol. 2007;177:1029–1036. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanjuan-Pla A, Cervera AM, Apostolova N, Garcia-Bou R, Victor VM, Murphy MP, McCreath KJ. A targeted antioxidant reveals the importance of mitochondrial reactive oxygen species in the hypoxic signaling of HIF-1alpha. FEBS Lett. 2005;579:2669–2674. doi: 10.1016/j.febslet.2005.03.088. [DOI] [PubMed] [Google Scholar]
- 33.Pan Y, Mansfield KD, Bertozzi CC, Rudenko V, Chan DA, Giaccia AJ, Simon MC. Multiple factors affecting cellular redox status and energy metabolism modulate hypoxia-inducible factor prolyl hydroxylase activity in vivo and in vitro. Mol Cell Biol. 2007;27:912–925. doi: 10.1128/MCB.01223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon MC. Mitochondrial reactive oxygen species are required for hypoxic HIF alpha stabilization. Adv Exp Med Biol. 2006;588:165–170. doi: 10.1007/978-0-387-34817-9_15. [DOI] [PubMed] [Google Scholar]
- 35.Murphy MP. Targeting lipophilic cations to mitochondria. Biochim Biophys Acta. 2008;1777:1028–1031. doi: 10.1016/j.bbabio.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 36.Kelso GF, Porteous CM, Coulter CV, Hughes G, Porteous WK, Ledgerwood EC, Smith RA, et al. Selective targeting of a redox-active ubiquinone to mitochondria within cells: antioxidant and antiapoptotic properties. J Biol Chem. 2001;276:4588–4596. doi: 10.1074/jbc.M009093200. [DOI] [PubMed] [Google Scholar]
- 37.James AM, Cocheme HM, Smith RA, Murphy MP. Interactions of mitochondria-targeted and untargeted ubiquinones with the mitochondrial respiratory chain and reactive oxygen species. Implications for the use of exogenous ubiquinones as therapies and experimental tools. J Biol Chem. 2005;280:21295–21312. doi: 10.1074/jbc.M501527200. [DOI] [PubMed] [Google Scholar]
- 38.Chacko B, Reily C, Srivastava A, Johnson MS, Ulasova E, Agarwal A, Zinn K, et al. Prevention of diabetic nephropathy in Ins2+/-AkitaJ mice by the mitochondria-targeted therapy Mito Q. Biochem J. doi: 10.1042/BJ20100308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Supinski GS, Murphy MP, Callahan LA. MitoQ administration prevents endotoxin-induced cardiac dysfunction. Am J Physiol Regul Integr Comp Physiol. 2009;297:R1095–1102. doi: 10.1152/ajpregu.90902.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gane EJ, Weilert F, Orr DW, Keogh GF, Gibson M, Lockhart MM, Frampton CM, et al. The mitochondria-targeted anti-oxidant mitoquinone decreases liver damage in a phase II study of hepatitis C patients. Liver Int. 30:1019–1026. doi: 10.1111/j.1478-3231.2010.02250.x. [DOI] [PubMed] [Google Scholar]
- 41.Simon MC. Coming up for air: HIF-1 and mitochondrial oxygen consumption. Cell Metab. 2006;3:150–151. doi: 10.1016/j.cmet.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Arteel GE, Thurman RG, Raleigh JA. Reductive metabolism of the hypoxia marker pimonidazole is regulated by oxygen tension independent of the pyridine nucleotide redox state. Eur J Biochem. 1998;253:743–750. doi: 10.1046/j.1432-1327.1998.2530743.x. [DOI] [PubMed] [Google Scholar]
- 43.Snow BJ, Rolfe FL, Lockhart MM, Frampton CM, O'Sullivan JD, Fung V, Smith RA, et al. A double-blind, placebo-controlled study to assess the mitochondria-targeted antioxidant MitoQ as a disease-modifying therapy in Parkinson's disease. Mov Disord. 25:1670–1674. doi: 10.1002/mds.23148. [DOI] [PubMed] [Google Scholar]
- 44.Lieber CS, DeCarli LM. The feeding of alcohol in liquid diets: two decades of applications and 1982 update. Alcohol Clin Exp Res. 1982;6:523–531. doi: 10.1111/j.1530-0277.1982.tb05017.x. [DOI] [PubMed] [Google Scholar]
- 45.Park SH, Choi MS, Park T. Changes in the hepatic gene expression profile in a rat model of chronic ethanol treatment. Food Chem Toxicol. 2008;46:1378–1388. doi: 10.1016/j.fct.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 46.Arteel GE, Kadiiska MB, Rusyn I, Bradford BU, Mason RP, Raleigh JA, Thurman RG. Oxidative stress occurs in perfused rat liver at low oxygen tension by mechanisms involving peroxynitrite. Mol Pharmacol. 1999;55:708–715. [PubMed] [Google Scholar]
- 47.Zhou J, Damdimopoulos AE, Spyrou G, Brune B. Thioredoxin 1 and thioredoxin 2 have opposed regulatory functions on hypoxia-inducible factor-1alpha. J Biol Chem. 2007;282:7482–7490. doi: 10.1074/jbc.M608289200. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Chen SH, Zhang Y, Yu CH, Li SD, Li YM. Is the hypoxia-inducible factor-1 alpha mRNA expression activated by ethanol-induced injury, the mechanism underlying alcoholic liver disease? Hepatobiliary Pancreat Dis Int. 2006;5:560–563. [PubMed] [Google Scholar]
- 49.French SW. The role of hypoxia in the pathogenesis of alcoholic liver disease. Hepatol Res. 2004;29:69–74. doi: 10.1016/j.hepres.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 50.Tomita K, Tamiya G, Ando S, Kitamura N, Koizumi H, Kato S, Horie Y, et al. AICAR, an AMPK activator, has protective effects on alcohol-induced fatty liver in rats. Alcohol Clin Exp Res. 2005;29:240S–245S. doi: 10.1097/01.alc.0000191126.11479.69. [DOI] [PubMed] [Google Scholar]
- 51.Shiva S, Oh JY, Landar AL, Ulasova E, Venkatraman A, Bailey SM, Darley-Usmar VM. Nitroxia: the pathological consequence of dysfunction in the nitric oxide-cytochrome c oxidase signaling pathway. Free Radic Biol Med. 2005;38:297–306. doi: 10.1016/j.freeradbiomed.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 52.Cunningham CC, Bailey SM. Ethanol consumption and liver mitochondria function. Biol Signals Recept. 2001;10:271–282. doi: 10.1159/000046892. [DOI] [PubMed] [Google Scholar]
- 53.Smith RA, Murphy MP. Animal and human studies with the mitochondria-targeted antioxidant. MitoQ Ann N Y Acad Sci. 1201:96–103. doi: 10.1111/j.1749-6632.2010.05627.x. [DOI] [PubMed] [Google Scholar]
- 54.Adlam VJ, Harrison JC, Porteous CM, James AM, Smith RA, Murphy MP, Sammut IA. Targeting an antioxidant to mitochondria decreases cardiac ischemia-reperfusion injury. Faseb J. 2005;19:1088–1095. doi: 10.1096/fj.05-3718com. [DOI] [PubMed] [Google Scholar]
- 55.Chandran K, Aggarwal D, Migrino RQ, Joseph J, McAllister D, Konorev EA, Antholine WE, et al. Doxorubicin inactivates myocardial cytochrome c oxidase in rats: cardioprotection by Mito-Q. Biophys J. 2009;96:1388–1398. doi: 10.1016/j.bpj.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.James AM, Sharpley MS, Manas AR, Frerman FE, Hirst J, Smith RA, Murphy MP. Interaction of the mitochondria-targeted antioxidant MitoQ with phospholipid bilayers and ubiquinone oxidoreductases. J Biol Chem. 2007;282:14708–14718. doi: 10.1074/jbc.M611463200. [DOI] [PubMed] [Google Scholar]
- 57.Seki S, Kitada T, Sakaguchi H, Nakatani K, Wakasa K. Pathological significance of oxidative cellular damage in human alcoholic liver disease. Histopathology. 2003;42:365–371. doi: 10.1046/j.1365-2559.2003.01599.x. [DOI] [PubMed] [Google Scholar]
- 58.Cocheme HM, Kelso GF, James AM, Ross MF, Trnka J, Mahendiran T, Asin-Cayuela J, et al. Mitochondrial targeting of quinones: therapeutic implications. Mitochondrion. 2007;7(Suppl):S94–102. doi: 10.1016/j.mito.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 59.Kurose I, Higuchi H, Kato S, Miura S, Watanabe N, Kamegaya Y, Tomita K, et al. Oxidative stress on mitochondria and cell membrane of cultured rat hepatocytes and perfused liver exposed to ethanol. Gastroenterology. 1997;112:1331–1343. doi: 10.1016/s0016-5085(97)70147-1. [DOI] [PubMed] [Google Scholar]
- 60.Lowes DA, Thottakam BM, Webster NR, Murphy MP, Galley HF. The mitochondria-targeted antioxidant MitoQ protects against organ damage in a lipopolysaccharide-peptidoglycan model of sepsis. Free Radic Biol Med. 2008;45:1559–1565. doi: 10.1016/j.freeradbiomed.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 61.Arteel GE, Raleigh JA, Bradford BU, Thurman RG. Acute alcohol produces hypoxia directly in rat liver tissue in vivo: role of Kupffer cells. Am J Physiol. 1996;271:G494–500. doi: 10.1152/ajpgi.1996.271.3.G494. [DOI] [PubMed] [Google Scholar]
- 62.Fong GH, Takeda K. Role and regulation of prolyl hydroxylase domain proteins. Cell Death Differ. 2008;15:635–641. doi: 10.1038/cdd.2008.10. [DOI] [PubMed] [Google Scholar]
- 63.Young TA, Cunningham CC, Bailey SM. Reactive oxygen species production by the mitochondrial respiratory chain in isolated rat hepatocytes and liver mitochondria: studies using myxothiazol. Arch Biochem Biophys. 2002;405:65–72. doi: 10.1016/s0003-9861(02)00338-7. [DOI] [PubMed] [Google Scholar]
- 64.Zeng T, Xie KQ. Ethanol and liver: recent advances in the mechanisms of ethanol-induced hepatosteatosis. Arch Toxicol. 2009 doi: 10.1007/s00204-009-0457-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.