Abstract
The alterations occurring in yeast genomic expression during early response to acetic acid and the involvement of the transcription factor Haa1p in this transcriptional reprogramming are described in this study. Haa1p was found to regulate, directly or indirectly, the transcription of approximately 80% of the acetic acid-activated genes, suggesting that Haa1p is the main player in the control of yeast response to this weak acid. The genes identified in this work as being activated in response to acetic acid in a Haa1p-dependent manner include protein kinases, multidrug resistance transporters, proteins involved in lipid metabolism, in nucleic acid processing, and proteins of unknown function. Among these genes, the expression of SAP30 and HRK1 provided the strongest protective effect toward acetic acid. SAP30 encode a subunit of a histone deacetylase complex and HRK1 encode a protein kinase belonging to a family of protein kinases dedicated to the regulation of plasma membrane transporters activity. The deletion of the HRK1 gene was found to lead to the increase of the accumulation of labeled acetic acid into acid-stressed yeast cells, suggesting that the role of both HAA1 and HRK1 in providing protection against acetic acid is, at least partially, related with their involvement in the reduction of intracellular acetate concentration.
Introduction
The Saccharomyces cerevisiae transcriptional activator Haa1p was first included into a family of copper-regulated transcription factors, based on the identification of a putative copper-regulatory domain within its DNA binding domain (Keller et al., 2001). However, unlike its homologous proteins, the function of Haa1p is independent of the copper-status of the cell (Keller et al., 2001) and a biological role for Haa1p was related with yeast resistance to acetic acid and propionic acid (Fernandes et al., 2005). The expression of the HAA1 gene was shown to lead to the reduction of the duration of the adaptation period of a yeast cell population suddenly exposed to toxic concentrations of these low chain carboxylic acids, by decreasing the loss of cell viability occurring during the phase of latency (Fernandes et al., 2005). More recently, Haa1p was also described as a determinant of yeast resistance to lactic acid (Abbott et al., 2008). Acetic and propionic acids are widely used by food and beverage industries to prevent microbial spoilage often caused by yeasts and other fungi at low pH. However, some strains can adapt and proliferate even in the presence of concentrations of these chemicals, which are close to the legally permitted values, causing serious economic losses (Fleet, 2007). Acetic acid and propionic acid are also bypass products of Saccharomyces cerevisiae alcoholic fermentation, and their accumulation in the growth medium to toxic levels contributes to decreased ethanol yields and may lead to stuck and sluggish fermentations (Gibson et al., 2007). Acetic acid is also one of the inhibitory compounds resulting from the pretreatment of lignocellulosic substrates used for bioethanol production (Almeida et al., 2007). It is thus expected that the understanding of the molecular mechanisms underlying yeast resistance to acetic acid and propionic acid may be successfully used to guide the design of more efficient weak acid-based food preservation strategies and of more robust industrial yeast strains.
During the early response of unadapted yeast cell populations to acetic acid, propionic acid, or lactic acid, the rapid transcriptional activation of a small set of genes, previously shown to be transcriptionally regulated in the dependence of HAA1 expression (Keller et al., 2001), was found to occur (Abbott et al., 2008; Fernandes et al., 2005). These genes include: (1) TPO2 and TPO3, encoding two plasma membrane multidrug resistance (MDR) transporters of the major facilitator superfamily involved in the efflux of polyamines (Albertsen et al., 2003); (2) YGP1, encoding a cell wall glycoprotein expressed under nutrient limitation (Destruelle et al., 1994); (3) other genes encoding proteins of unknown or poorly characterized function (Abbott et al., 2008; Fernandes et al., 2005). Among these genes, only the expression of TPO2, TPO3, and YGP1 exerts a moderate protection against acetic acid and propionic acid, but the level of protection is much below the one exerted by Haa1p itself (Fernandes et al., 2005). The protective effect of both TPO3 and HAA1 genes was related with their involvement in the reduction of the intracellular concentration of acetate during acetic acid challenge, being the effect exerted by Haa1p much more evident than the one associated to the expression of its target-gene TPO3 (Fernandes et al., 2005). The much higher susceptibility of Δhaa1 cells to acetic acid and lactic acid, compared to the susceptibility exhibited by cells devoid of any of its initially described targets (Abbott et al., 2008; Fernandes et al., 2005; Keller et al., 2001), suggests that the Haa1p-regulon active under stress imposed by those weak acids includes other unidentified genes (Keller et al., 2001). Consistent with this idea, the SPI1 gene was recently identified as a novel weak acid-responsive gene dependent on HAA1 expression (Simões et al., 2006). SPI1 encodes a GPI-anchored cell wall protein required for the weak acid-induced remodeling of cell wall structure (Simões et al., 2006) but, contrasting with Haa1p, the protection exerted by Spi1p against acetic acid is negligible, its protective action being more evident toward the more lipophilic weak acids (Simões et al., 2006). In the present study we explored a transcriptomic approach to search for the complete Haa1p-regulon acting in yeast cells following sudden exposure to acetic acid stress. A number of novel genes regulated directly or indirectly by Haa1p, are here described for the first time, some of which play a role in yeast resistance to acetic acid.
Materials and Methods
Strains and growth media
The parental strain Saccharomyces cerevisiae BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and the derived mutant strains used in this work were obtained from the Euroscarf collection. Cells were batch-cultured at 30°C, with orbital agitation (250 rpm), in MM4 liquid medium that contains, per liter: 1.7 g yeast nitrogen base without amino acids or NH4+ (Difco Laboratories, Detroit, MI, USA), 20 g glucose (Merck, West Point, PA, USA), 2.65 g (NH4)2SO4 (Merck), 20 mg methionine, 20 mg histidine, 60 mg leucine, 20 mg uracil, all from Sigma (Spain). Yeast Peptone Dextrose (YPD) solid medium, containing, per liter, 20 g glucose, 20 g bactopeptone (Difco), 10 g yeast extract (Difco), and 20 g/L of agar (Iberagar, Barreiro, Portugal), was used to assess viable cell concentration.
Acetic acid susceptibility assays
The susceptibility to acetic acid of the parental strain BY4741 and selected deletion mutants was assessed based on the comparison of the growth curves in liquid MM4 medium supplemented with 60 mM acetic acid (acidified to pH 4.0 with HCl). Cells were cultivated in unsupplemented MM4 (pH 4.0) growth medium until mid-exponential phase (standard OD600 nm 0.4) and used to reinoculate (at an initial OD600 nm of 0.05 ± 0.005) the fresh growth medium supplemented with the acid. Growth was followed by measuring culture OD600 nm.
Effect of Haa1p in yeast transcriptional response to acetic acid
The effect of Haa1p in yeast response to acetic acid stress was carried out by comparing the transcriptomes of cells of S. cerevisiae BY4741 and of the mutant BY4741_Δhaa1 in the presence or absence of acetic acid. Cells were cultivated in MM4 growth medium (at pH 4.0) until mid-exponential phase (standard OD600 nm 0.8) and then reinoculated into fresh medium either or not supplemented with 50 mM of acetic acid (at pH 4.0). After 30 min of incubation in this medium in the absence (sample A) or presence (sample B) of acetic acid, cells were harvested, immediately frozen in liquid nitrogen, and kept at −80°C until total RNA extraction. Total RNA extraction was performed according to the hot phenol method. Concentration and purity was determined by spectrophotometry and integrity was confirmed using an Agilent 2100 Bioanalyzer with a RNA 6000 Nano Assay (Agilent Technologies, Palo Alto, CA, USA). RNA was processed for use on Affymetrix (Santa Clara, CA, USA) GeneChip Yeast Genome S98 Arrays, according to the manufacturer's One-Cycle Target Labeling Assay. Briefly, 5 μg of total RNA containing spiked in Poly-A RNA controls (GeneChip Expression GeneChip Eukaryotic Poly-A RNA Control Kit; Affymetrix) was used in a reverse transcription reaction (One-Cycle DNA synthesis kit; Affymetrix) to generate first-strand cDNA. After second-strand synthesis, double-stranded cDNA was used in an in vitro transcription (IVT) reaction to generate biotinylated cRNA (GeneChip Expression 3′-Amplification Reagents for IVT-Labeling; Affymetrix). Size distribution of the cRNA and fragmented cRNA, respectively, was assessed using an Agilent 2100 Bioanalyzer with a RNA 6000 Nano Assay. A total of 15 μg of fragmented cRNA was used in a 300-μL hybridization containing added hybridization controls. Mixture (200 μL) was hybridized on arrays for 16 h at 45°C. Standard posthybridization wash and double-stain protocols (EukGE-WS2v5) were used on an Affymetrix GeneChip Fluidics Station 400. Arrays were scanned on an Affymetrix GeneChip scanner 3000. To ensure the reliability of the analysis, two biological replicates were used and the replicate data for the same sample was weighted gene-wise using inverse square SE as weights. The scanned arrays were analyzed first with Affymetrix MAS 5.0 software to obtain Absent/Present calls and subsequently with DNA-Chip Analyzer (dChip) 2006 (http://www.dchip.org, Wong Lab, Harvard). The arrays were normalized to a baseline array with median CEL intensity by applying an Invariant Set Normalization Method (Li and Hung Wong, 2001). Normalized CEL intensities of the eight arrays were used to obtain model-based gene expression indices based on a PM (Perfect Match)-only model (Li and Hung Wong, 2001). Genes compared were considered to be differentially expressed if they were called Present in at least one of the arrays and if the 90% lower confidence bound of the fold change between experiment and baseline was above 1.2. For downstream analysis of acetic acid-responsive genes only the genes whose transcriptional change was above or below 1.5-fold the values registered in control cells were considered to be altered. Clustering of the up- or downregulated genes, based on biological function, was performed using the MIPS Functional Catalogue (http://mips.gsf.de/projects/funct) and the description of gene function is based on the information available in SGD (www.yeastgenome.org).
To confirm some of the results obtained in the microarray analysis carried out, the transcript levels of selected acetic acid-responsive genes were compared by real-time RT-PCR in cells of the parental strain BY4741 and in the Δhaa1 mutant. For this, cells were cultivated in conditions similar to those used for the microarray analysis and total RNA extraction was performed using the same protocol. Synthesis of cDNA from total RNA samples was performed using the MultiscribeTM reverse transcriptase kit and the subsequent real time RT-PCR step was carried out using SYBR® Green reagents 7500 RT-PCR in a Thermal Cycler Block (Applied Biosystems, Bedford, MA, USA). Specific primers for the amplification of the selected genes and of the ACT1 gene, used as an internal control, were designed using Primer Express Software (Applied Biosystems).
Extracellular acidification curves promoted by yeast cells in the presence or absence of acetic acid
To compare the in vivo active export of protons from cells of the parental strain BY4741 or the Δhrk1 mutant in the presence of increasing concentrations of acetic acid, the values of pH of the aqueous cell suspension medium was followed for 10 min. Cells of both strains were grown in MM4 medium (at pH 4.0) and harvested by centrifugation (8,000 rpm, 5 min, 4°C) when the culture reached a standardized OD600 nm of 0.5–0.6, in mid-exponential phase. Cells were washed twice with ice-cold water and then incubated in sorbitol (20 g/liter) for 30 min to deenergize the cells and deactivate plasma membrane H+-ATPase (Portillo and Serrano, 1989). After this, cells were washed and resuspended in distilled water to obtain a dense cell suspension (OD600 nm of 20.0 ± 2.0). Acidification experiments were carried out in a water-jacketed cell of 10-mL capacity, at 30°C, containing 3.0 mL of water with acetic acid (0, 0.4, 0.8, and 1.2 mM) and 1 mL of the above-described cellular suspension. The pH of the resulting suspension was adjusted to 4.0 ± 0.05 and 1 mL of 100 g/L glucose (at pH 4.0) was added (to obtain a final concentration of 20 g/L). The variation of extracellular medium pH was followed by potentiometry using a pH microelectrode attached to a pH meter (Metrohm 605).
[1-14C]-acetic acid accumulation assays
The accumulation ratio of [1-14C]-acetic acid (defined as the ratio between intracellular and extracellular concentrations of the radiolabeled chemical) in cells of the parental strain BY4741 or the mutant Δhrk1 was followed for 30 min during cultivation in MM4 growth medium (at pH 4.0) supplemented with 60 mM of cold acetic acid. Cells used to perform this assay were grown until mid-exponential phase (at the standardized OD600 nm = 0.5 ± 0.05) in MM4 growth medium (pH 4.0) and harvested by filtration (Supor®200 membranes, 0.2 μm). Following harvesting, cells were washed with fresh MM4 growth medium (pH 4.0) and resuspended in 5 mL of the same growth medium to obtain a cell suspension with OD600 nm = 0.7 ± 0.05. After 5 min of incubation at 30°C with agitation (150 rev/min) for temperature equilibration, 60 mM of cold acetic acid and 10 μM of [1-14C]-acetic acid (sodium salt from GE Healthcare, Piscataway, NJ; 9.25 MBq) were added to the culture medium and incubation proceeded for 30 min. During this period of incubation, the intracellular accumulation of labeled acetic acid was followed by filtering 200 μL of cell suspensions, at adequate intervals, through prewetted glass microfiber filters (Whatman GF/C). The filters were washed with ice-cold water and the radioactivity was measured in a Beckman LS 5000TD scintillation counter. Extracellular [1-14C]-acetic acid was estimated by measuring the radioactivity of 50 μL of the supernatant. The nonspecific [1-14C]-acetic acid adsorption to the filters and to the cells (<5% of the total bound-radioactivity) was assessed and taken into consideration. To calculate the intracellular concentration of [1-14C]-acetic acid, the internal cell volume (Vi) of the different strains was considered constant and equal to 2.5 μL (mg dry weight)−1 (Carmelo et al., 1997).
Results
Genome-wide transcriptional response to acetic acid stress controlled by Haa1p
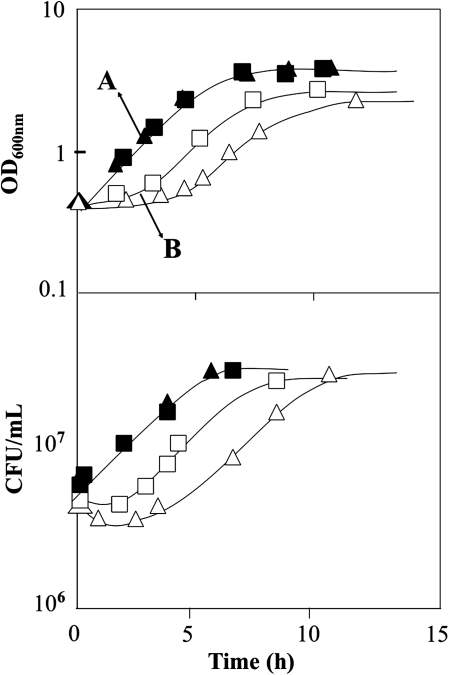
The genome-wide transcription alterations occurring in yeast cells during early response to acetic acid stress and the effect of HAA1 expression in this genomic expression program were examined in this study, based on the DNA microarray profiling of the transcriptomes of unadapted cells of S. cerevisiae BY4741 and of the Δhaa1 mutant cultivated for 30 min in the absence or presence of acetic acid (50 mM at pH 4.0). The growth curves of the yeast populations examined are shown in Figure 1. During cultivation in acetic acid-supplemented growth medium, a latency period of 2 or 5 h was observed for the parental strain and the Δhaa1 mutant, respectively, although the reinoculation of the unsupplemented growth medium with the same inoculum caused no detectable growth delay (Fig.1). During the adaptation period, the concentration of the viable cell population was maintained for the parental strain and slightly reduced for the Δhaa1 population (Fig. 1). Following adaptation to the acetic acid concentration tested, both populations resumed exponential growth, although with a different delay (Fig.1). These observations suggest that the role of Haa1p in reducing the acetic acid-induced loss of cell viability is essentially exerted during the early adaptation period, as proposed before (Fernandes et al., 2005), therefore justifying the short exposure of 30 min to the acid, during the acute phase of stress, that was selected to perform the microarray experiments. Moreover, an acetic acid concentration that had a slight or moderate effect in the growth of the parental strain and of the Δhaa1 mutant growth was chosen to avoid a marked deleterious effect of the acid toward the very susceptible mutant population, which could interfere with the identification of the genes, directly or indirectly, regulated by Haa1p.
FIG. 1.
Growth curves of S. cerevisiae BY4741 (▪, □) and of the deletion mutant Δhaa1 (▴, ▵) in MM4 medium (pH 4.0) (▪, ▴) or in this same basal medium supplemented with 50 mM acetic acid (pH 4.0) (□,▵). The arrows indicate the time of cultivation at which cell samples were harvested to compare their transcriptomes. Growth was followed by measuring culture OD600 nm and the concentration of viable cells was assessed as the number of colony-forming units/mL of cell culture (CFU mL−1). The growth curves shown are means of, at least, three independent growth curves that gave rise to the same growth pattern.
The concentration of acetic acid tested led to the upregulation (above 1.5-fold) of 112 genes in the parental strain S. cerevisiae BY4741 and to the decreased expression (below 1.5-fold) of 83 genes (Supplementaryl Tables S1 and S2 of the supplementary material). The acetic acid-responsive genes were clustered based on the biological process in which they are involved and the most significant functional classes are shown in Supplementary Figure S1 of supplementary material. In our dataset of acetic acid transcriptionally activated genes, the functional classes enriched in our dataset are: “Stress response,” “Signal transduction,” “Regulation of carbohydrate metabolism,” and “Drug transport,” whereas the “Amino acid catabolism” and “Ion transport” functional classes are the enriched functional classes among the downregulated genes (Fig. S1 of the supplementary material).
In the absence of acetic acid stress, the elimination of HAA1 gene only had a slight effect on the yeast transcriptome: 11 genes exhibited an increased expression level in Δhaa1 cells and the level of 15 transcripts was decreased in this mutant strain (using 1.5-fold as the threshold cut level) (Supplementary Table S3 of the supplementary material).
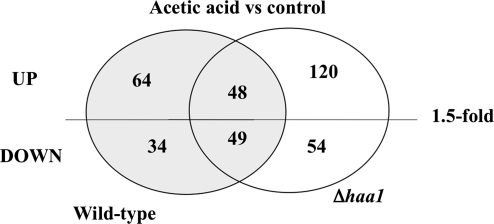
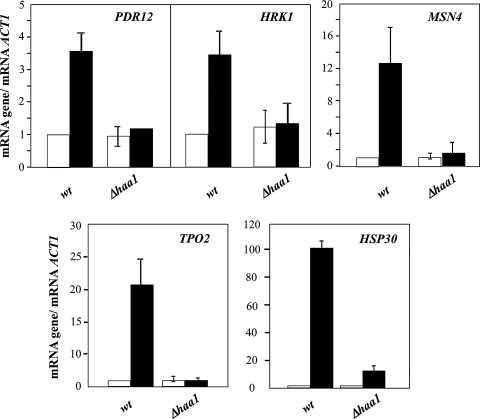
Contrasting with the subtle effect registered in the transcriptome of cells cultivated in the absence of acetic acid, the expression of HAA1 had a major impact in the alterations occurring in yeast genomic expression during early adaptive response to the acid (Fig. 2) The elimination of HAA1 gene reduced the transcription of 85 out of the 112 acetic acid-activated genes in the parental strain, corresponding to approximately 80% of the upregulated genes that are dependent of HAA1 expression. Remarkably, 120 genes were only found to be upregulated (above 1.5-fold) in acetic acid-challenged Δhaa1 cells but not in the parental strain (Fig. 2), presumably as a result of the higher concentration of acetic acid accumulated inside Δhaa1 cells, compared to the parental strain (Fernandes et al., 2005). The Haa1p-regulated genes could be separated in two classes. The first class includes the 64 genes whose acetic acid-induced transcriptional activation was fully dependent on Haa1p (listed in Table 1 and clustered in Fig. 2 as “wild-type only” activated genes), these genes not being considered upregulated in Δhaa1 cells, at least above the threshold level of 1.5-fold. The other class includes the 21 genes whose acetic acid-induced transcriptional activation was only partially dependent on Haa1p (listed in Table 2 and clustered in Fig. 2 as genes activated both in “wild-type and Δhaa1”), the transcription of these genes being activated in Δhaa1 acetic acid-stressed cells (above 1.5-fold) but their transcript levels in the mutant strain is below the one registered in the parental strain. However, only the genes whose transcription was reduced in acetic acid-challenged Δhaa1 cells by more than 25%, compared to wild-type cells, are shown in Table 2. The full list of genes is available in Supplementary Table S4. The effect of HAA1 expression over the transcription of a few selected genes (TPO2, MSN4, HRK1, HSP30, and PDR12) was confirmed by real-time RT-PCR, thus validating the results of the microarray analysis. The more evident effect of HAA1 elimination in the transcription level of TPO2 (16-fold), MSN4 (7-fold), and HSP30 (6.8-fold) genes (Fig. 3) is also in agreement with the microarray data.
FIG. 2.
Venn diagram showing the number of genes whose transcription is altered in response to acetic acid in cells of the parental strain S. cerevisiae BY4741 and/or in the Δhaa1 mutant. Genes whose transcript level was increased (or decreased) above the threshold level of 1.5-fold in both strains cultivated in the presence of the acid, compared to the values registered in unstressed cells (control), were selected for this analysis.
Table 1.
List of Genes Whose Acetic Acid-Induced Transcriptional Activation Registered in Cells of S. cerevisiae BY4741 Is Abolished in the Absence of HAA1 Expression
| |
|
(mRNA wt )ac |
(mRNA Δhaa1 )ac |
(mRNA wt )ac |
|
Induced by |
|||
|---|---|---|---|---|---|---|---|---|---|
| Gene or ORF | Biological function | (mRNA wt)c | (mRNA Δhaa1)c | (mRNA Δhaa1)ac | Acetic acid susceptibility | Actld. | Prop. | Sorb. | Lact. |
| TPO2 | Plasma membrane polyamine transporter of the major facilitator superfamily | 14.9 | 1.3 | 18.7 | + | ||||
| YLR297w | Putative protein of unknown function | 11.3 | 1.2 | 10.5 | − | ||||
| STP3 | Zinc-finger protein of unknown function | 2.6 | −1.4 | 8.1 | + | ||||
| YRO2 | Putative protein of unknown function | 11.1 | 1.4 | 7.8 | − | ||||
| YAR029w | Unknown function | 3.7 | −1.1 | 7.6 | − | ||||
| TOS3 | Protein kinase required for phosphorylation and activation of Snf1p; glucose metabolism | 5.7 | 1.1 | 6.2 | + | ||||
| YIR035c | Unknown function | 5.5 | 1.2 | 6.2 | − | ||||
| YGP1 | Cell wall-related secretory glycoprotein | 3.0 | 1.1 | 4.8 | + | ||||
| PCL10 | Cyclin involved in regulation of activity of protein kinase Pho85p | 5.0 | 1.1 | 4.3 | + | ||||
| YPR127w | Unknown function | 3.3 | 1.0 | 4.1 | − | ||||
| DSD1 | D-serine dehydratase | 3.2 | −1.1 | 4.0 | − | ||||
| MSN4 | Transcription factor activated in stress conditions | 6.1 | 1.4 | 3.8 | − | ||||
| YJR096w | Putative xylose and arabinose reductase | 2.7 | −1.1 | 3.7 | − | ||||
| SPI1 | GPI-anchored cell wall protein; involved in weak acid resistance | 5.1 | 1.4 | 3.5 | − | ||||
| HOR2 | Glycerol phosphatase involved in glycerol biosynthesis; induced in response to stress | 4.8 | −1.4 | 3.4 | + | ||||
| YKR075c | Unknown function | 2.9 | 1.1 | 3.3 | − | ||||
| SUR2 | Sphinganine C4-hydroxylase involved in sphingolipid metabolism | 3.5 | 1.2 | 3.0 | ++ | ||||
| ICY1 | Unknown function | 2.7 | 1.0 | 3.0 | + | ||||
| INM1 | Inositol monophosphatase involved in biosynthesis of inositol | 3.4 | 1.3 | 2.6 | + | ||||
| SAP30 | Subunit of a histone deacetylase complex | 3.2 | 1.3 | 2.6 | +++ | ||||
| YNL200c | Unknown function | 2 | −1.2 | 2.4 | ++ | ||||
| STF2 | Protein involved in the regulation of F1F0ATPase | 2.0 | −1.3 | 2.2 | − | ||||
| SYC1 | Subunit of the APT subcomplex of cleavage and polyadenylation factor | 2.9 | 1.1 | 2.2 | + | ||||
| YLR326w | Unknown function | 2.7 | 1.3 | 2.2 | − | ||||
| YAR028w | Unknown function | 4.0 | 1.1 | 2.1 | − | ||||
| YNL024c | Unknown function | 2 | 1.0 | 2.1 | − | ||||
| YNR034w-a | Unknown function | 2.2 | 1.3 | 2 | − | ||||
| GPG1 | Subunit of the heterotrimeric G protein that interacts with the receptor Grp1p; presumably involved in pseudohyphal growth | 2.2 | 1.1 | 2 | − | ||||
| PDE1 | Low afinitity cyclic AMP phosphodiesterase | 1.7 | 1.1 | 2.0 | − | ||||
| ADI1 | Acireductone dioxigenase involved in methionine salvage pathway | 1.9 | 1.0 | 2.0 | − | ||||
| YNL217w | Unknown function | 2.3 | 1.4 | 1.9 | − | ||||
| NRG1 | Transcriptional repressor involved in response to alkaline pH and glucose catabolic repression | 2.2 | 1.4 | 1.9 | ++ | ||||
| YPL071c | Unknown function | 2.3 | 1.3 | 1.8 | − | ||||
| MUK1 | Unknown function | 2.1 | 1.2 | 1.8 | − | ||||
| PDR12 | Plasma membrane ABC transporter involved in efflux of benzoate and sorbate anions | 4.2 | 1.3 | 1.8 | − | ||||
| TMA10 | Unknown function | 2.8 | 1.4 | 1.7 | − | ||||
| GRX8 | Glutaredoxin enzyme | 2.0 | 1.2 | 1.7 | − | ||||
| PFK27 | 6-phosphofructo-2-kinase | 2.1 | −1.4 | 1.7 | − | ||||
| FKH2 | Transcription factor involved in the expression of G2/M phase genes | 2 | 1.0 | 1.7 | − | ||||
| EEB1 | Acyl-coenzymeA:ethanol O-acyltransferase; presumably involved in lipid metabolism | 2.2 | 1.0 | 1.6 | − | ||||
| YLR346c | Putative protein of unknown function | 1.9 | 1.4 | 1.6 | − | ||||
| QCR10 | Subunit of the ubiqunol-cytochrome c oxidoreductase complex which comprises part of the mitochondrial respiratory chain | 1.6 | −1.1 | 1.5 | − | ||||
| ATG8 | Component of autophagosomes and Cvt vesicles; involved in intracellular trafficking | 1.6 | −1.1 | 1.5 | − | ||||
| YER188w | Unknown function | 1.7 | 1.4 | 1.5 | − | ||||
This table was prepared based on the comparative analysis of the transcriptome of cells of the parental strain BY4741 (wt) and of the mutant BY4741_Δhaa1 (Δhaa1) cultivated in MM4 medium (pH 4.0) supplemented with 50 mM acetic acid. The magnitude of the transcriptional activation registered in response to acetic acid stress in the parental strain (wtac/wtc) is shown as well as the susceptibility toward acetic acid of mutants devoid of Haa1p-target genes (−susceptibility identical to the parental strain;+,++,+++−increasing levels of susceptibility to the acid). The activation of Haa1p-target genes transcription in response to acetaldehyde (Actld), propionic acid (Prop), sorbic acid (Sorb), and lactic acid (Lact) is also indicated (shaded in gray) based on results reported from Aranda and del Olmo (2004), Mira et al. (2009), Schuller et al. (2004), and Abbot et al. (2008), respectively. Genes are sorted based on the ratio of mRNA gene level between the wild-type and the Δhaa1 mutant.
Table 2.
List of Genes Whose Acetic Acid-Induced Transcriptional Activation Registered in S. cerevisiae BY4741 (Above 1.5-Fold) Is Reduced, But Not Eliminated, in the ΔHAA1 Background
| |
|
(mRNA wt )ac |
(mRNA Δhaa1 )ac |
(mRNA wt )ac |
|
Induced by |
|||
|---|---|---|---|---|---|---|---|---|---|
| Gene or ORF | Biological function | (mRNA wt)c | (mRNA Δhaa1)c | (mRNA Δhaa1)ac | Acetic acid susceptibility | Actld. | Prop. | Sorb. | Lact. |
| YPR157w | Unknown function | 32.6 | 2.2 | 19.2 | − | ||||
| STP4 | Putative transcription factor | 12.6 | 2.2 | 12.7 | − | ||||
| YDR222w | Unknown function | 10.9 | 1.6 | 9.5 | − | ||||
| YPL014w | Unknown function | 10.3 | 1.8 | 7.3 | − | ||||
| PHM8 | Unknown function; expression regulated by phosphate availability | 15.6 | 2.8 | 6.3 | − | ||||
| YER130c | Unknown function | 10.9 | 2.4 | 4.7 | − | ||||
| TPO3 | Plasma membrane polyamine transporter of the major facilitator superfamily | 4.8 | 1.8 | 3.7 | ++ | ||||
| HSP30 | Putative negative regulator of plasma membrane H+-ATPase activity; expression induced under stress | 86.9 | 54.1 | 3.5 | − | ||||
| YPC1 | Alkaline ceramidase involved in sphingolipid and ceramide metabolism | 9.3 | 2.6 | 3.4 | + | ||||
| SSE2 | Heat-shock protein involved in protein folding; expression induced under stress | 6.8 | 3.1 | 2.9 | + | ||||
| SCM4 | Putative regulator of Cdc4p activity required for G1/S phase transition | 3.8 | 2.1 | 2.7 | − | ||||
| HSP26 | Heat-shock protein involved in protein folding; expression induced under stress | 2.7 | 1.9 | 2.5 | − | ||||
| YGR250c | Unknown function | 6.5 | 3.5 | 2.3 | − | ||||
| SKS1 | Putative serine/threonine protein kinase involved in adaptation to glucose starvation | 4.4 | 2.3 | 2.3 | − | ||||
| MTH1 | Negative regulator of the glucose-sensing signal transduction pathway | 3.1 | 1.7 | 2.1 | − | ||||
| FMP48 | Putative mithocondrial protein; unknown function | 28.2 | 16.1 | 2 | ++ | ||||
| RGS2 | Negative regulator of glucose-induced cAMP signaling pathway | 5.2 | 2.9 | 2 | − | ||||
| ECM13 | Unknown function | 6.8 | 3.0 | 2 | − | ||||
| HRK1 | Protein kinase implicated in activation of the plasma membrane H+-ATPase in response to glucose metabolism | 2.1 | 1.8 | 1.9 | +++ | ||||
| CUR1 | Unknown function | 2.3 | 1.5 | 1.7 | − | ||||
| PDR16 | Phosphatidylinositol transfer protein | 2.3 | 1.6 | 1.6 | − | ||||
The genes indicated in this table are only those whose transcript level in Δhaa1 acetic acid-stressed cells is 25% below the values attained in the parental strain, as for Table 1 legend.
FIG. 3.
Comparison of the transcript levels, estimated by real-time RT-PCR, of PDR12, HSP30, HRK1, MSN4, and TPO2 genes produced in cells of the parental strain BY4741 (wt) or the Δhaa1 mutant during cultivation for 30 min in MM4 growth medium (at pH 4.0) (white bars) or in this same growth medium supplemented with 50 mM acetic acid (dark bars). ACT1 mRNA was used as internal control. For each gene the value registered in parental strain control cells (corresponding to cells cultivated in unsupplemented growth medium) were set as 1 and the induction ratios are relative to this control. mRNA values shown are representative of results obtained, at least, in two independent incubation experiments.
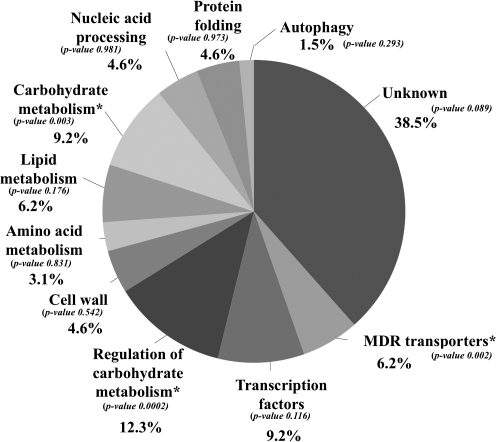
Eleven genes whose transcription was previously reported in the literature to be dependent on the presence of Haa1p—PHM8, TPO2, YRO2, TPO3, YGP1, YER130c, YIR035c, YLR297w, YPR157w, YPR127w, and SPI1 (Abbott et al., 2008; Fernandes et al., 2005; Keller et al., 2001; Simões et al., 2006)—were confirmed by our microarray analysis to have a reduced expression in Δhaa1 acetic acid-challenged cells (Table 1). The remaining 73 genes identified in our work as being activated by Haa1p in response to acetic acid are novel, direct or indirect, targets of this transcription factor. The Haa1p-dependent genes encode proteins belonging to the following functional classes: “Transcription factors,” “Multidrug resistance transporters,” “Cell wall,” “Lipid metabolism,” “Regulation of carbohydrate metabolism,” “Protein folding,” “Carbohydrate metabolism,” “Amino acid metabolism,” “Nucleic acid processing,” and “Uncharacterized biological function” (Fig. 4 and Tables 1 and 2). This last class comprises approximately 40% of the dataset. The functional classes considered enriched in the dataset (p-values below 0.01) are “Multidrug resistance transporters,” “Carbohydrate metabolism,” and “Regulation of carbohydrate metabolism” (Fig. 4).
FIG. 4.
Clustering, based on biological function, of genes activated in response to acetic acid stress in a Haa1p-dependent way. Genes whose transcript level in yeast cells challenged with acetic acid was reduced by more than 25% in the Δhaa1 mutant, compared to the levels attained in the parental strain, were grouped according to their biological functions described on MIPS functional catalog. The relative percentage of each functional class was calculated taking into consideration the number of genes in each class compared to the total of Haa1p-regulated genes (85) considered and the p-values of each functional class. Enriched classes (p-value below 0.01) are marked with*.
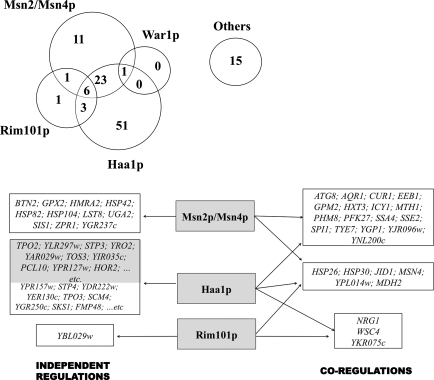
Using the YEASTRACT database (Teixeira et al., 2006b), the genes transcriptionally activated in response to acetic acid were clustered based on documented regulatory associations with the transcription factors Rim101p, War1p, Msn2p/Msn4p, and Haa1p that control the so far described regulatory pathways involved in yeast response to weak acid stress (Fernandes et al., 2005; Mira et al., 2009; Schüller et al., 2004). This analysis indicates that Haa1p specifically regulates almost one-half of the acetic acid-induced genes (51 out of 112 genes), whereas the number of genes specifically dependent on Msn2p/Msn4p, Rim101p, or War1p is much lower (Fig. 5). Remarkably, the sole War1p-target gene, PDR12, was also identified among the genes activated by Haa1p in response to acetic acid stress. An overlap between the Haa1p and the Msn2p/Msn4p regulons was also registered 23 acetic acid-activated genes being coregulated by these two regulatory pathways (Fig. 5). Three genes (NRG1, YKR075c, WSC4) are common to both the Haa1p- and Rim101p-dependent regulons (Fig. 5) and six genes (MSN4, YPL014w, JID1, HSP26, MDH2, and HSP30) are shared by the Rim101p, Msn2p/Msn4p, and Haa1p-regulatory pathways (Fig. 5). The 13 remaining acetic acid-responsive genes have no documented regulatory associations with any of the above referred transcription factors and were clustered as “Others” (Fig. 5). Most of them are documented targets of Yap1p and Hsf1p transcription factors, also involved in yeast response to stress (Hahn et al., 2004; Rodrigues-Pousada et al., 2004).
FIG. 5.
Venn diagram illustrating the crosstalk between the regulatory systems controlled by the weak acid-responsive transcription factors Haa1p, Rim101p, Msn2/Msn4p, and War1p in yeast response to acetic acid stress. The 112 genes found to be upregulated in response to acetic acid (50 mM) in the parental strain were clustered among the referred regulons, based on the existence of documented regulatory associations with the indicated transcription factors, according with the information available in the YEASTRACT database (www.yeastract.com) (Teixeira et al., 2006b). The number of genes in each cluster is indicated and the full or the partial list of genes belonging to each regulon is shown in the schematic representation below the diagram, emphasizing the genes specific of each regulon and the coregulated genes. In the case of the genes specifically dependent on Haa1p, 10 genes were selected from those listed in Tables 1 (indicated in gray background) and 2 (indicated in a white background). These genes exhibit the highest reduction in transcript level in Δhaa1 cells, compared to the parental strain.
Involvement of genes of the Haa1p regulon in yeast tolerance to acetic acid
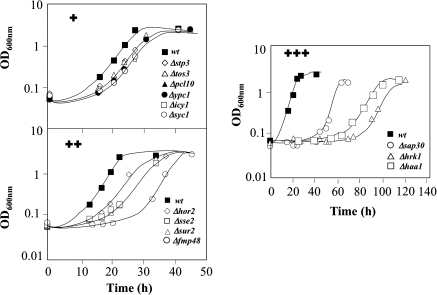
The susceptibility phenotype of Δhaa1 cells toward acetic acid is, presumably, due to the lack of activation of Haa1p target genes required for yeast protection against the acid. Among the group of genes whose transcription was initially described by Keller et al. (2001) to be dependent on a functional Haa1p only TPO2, TPO3, and YGP1 genes provided protection against acetic acid. However, their protective effect was much less evident than the one exerted by Haa1p (Fernandes et al., 2005), which prompted us to examine the role in yeast tolerance to acetic acid of the novel Haa1p-targets uncovered in the present work. The expression of 20% of these genes, although at different levels, contributed to a reduced duration of latency period observed under acetic acid stress (Fig. 6 and Tables 1 and 2). In the absence of acetic acid, the elimination of these genes had no detectable in the growth curve (results not shown). The genes of the Haa1p-regulon conferring protection toward acetic acid include: HOR2 and SSE2, two genes of the environmental stress response (Gasch et al., 2000) that encode a glycerol-3-phosphatase involved in glycerol biosynthesis and a heat shock protein, respectively; NRG1, HRK1, TOS3, and PCL10, whose function is related with the regulation of carbohydrate metabolism; and SYC1 and SAP30, which encode two proteins involved in nucleic acid processing. The other genes that are determinants of resistance to acetic acid and that are activated in response to acetic acid stress in the dependence of HAA1 expression encode proteins involved in lipid metabolism. This is the case of YPC1 and SUR2, encoding two enzymes that catabolyze ceramide to the C26-sphingolipid phytosphingosine and INM1 whose product is a monophosphatase involved in inositol biosynthesis.
FIG. 6.
Comparison of the susceptibility toward acetic acid of mutants devoid of the expression of genes whose transcription is activated in response to the acid in a Haa1p-dependent manner. Cells of the parental strain (wt) and of the indicated deletion mutants were grown until mid-exponential phase in liquid MM4 medium (at pH 4.0) and then used to inoculate the same basal medium either supplemented or not supplemented with 60 mM acetic acid (at pH 4.0; initial OD = 0.05 ± 0.005). Cells were batch cultured at 30°C and growth was monitored based on OD600 nm. Three levels of susceptibility phenotypes were considered based on the level of inhibition of the growth curves of the parental strain and of the different deletion mutants. The growth curves shown are representative of at least four independent experiments that gave essentially the same results.
The elimination of HRK1 and, to a lower extent, of SAP30 gene, led to the strongest susceptibility phenotypes to acetic acid (Fig. 6). HRK1 encodes a protein kinase involved in the activation of plasma membrane H+-ATPase activity in response to glucose metabolism (de la Fuente et al., 1997; Goossens et al., 2000) and SAP30 encodes a component of the Rpd3L histone deacetylase complex (Carrozza et al., 2005). SAP30 was identified before as a determinant of resistance to propionic acid (Mira et al., 2009) and to a variety of other chemicals (Hillenmeyer et al., 2008). Interestingly, the elimination of SDS3, SIN3, and DEP1, encoding, respectively, two subunits of the Rpd3L complex and a protein associated to this complex (Carrozza et al., 2005), also led to an increase in yeast susceptibility to acetic acid (Supplementary Fig. S2).
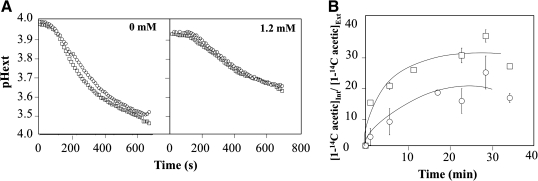
The role of Hrk1p in providing protection against acetic acid involves the reduction of intracellular acetate concentration
Hrk1p was proposed to be a positive regulator of plasma membrane (PM) H+-ATPase (Pma1p) activity in response to glucose metabolism, although less important than its functional homologue, the Ptk2p kinase (de la Fuente et al., 1997; Goossens et al., 2000). Taking into account the essential role played by PM-H+-ATPase activity in yeast response and resistance to weak acid stress (Holyoak et al., 1997; Viegas et al., 1998), the putative effect of Hrk1p in acetic acid-induced activation of Pma1p was hypothesized. Considering that the result of both the in vivo activity of this H+-pump and the passive H+ influx through the plasma membrane can be roughly assessed based on the acidification rate of the external environment induced by glucose (Portillo and Serrano, 1989), the acidification curves promoted by cells of the parental strain and of the Δhrk1 mutant following addition of glucose, either in the presence or absence of acetic acid, were compared (Fig. 7A). In the absence of acetic acid, glucose supplementation triggered the activation of the PM-H+-ATPase and the rapid extrusion of H+ to the external environment was registered (Fig. 7A). Compared with the parental strain, the kinetics of growth medium acidification was inhibited in the Δhrk1 mutant, although the effect is mild, probably as the result of Ptk2p activity (Goossens et al., 2000) (Fig. 7A). The presence of increasing concentrations of acetic acid led to the reduction of the rate of extracellular acidification promoted by yeast cells, consistent with the nonspecific permeabilization of plasma membrane induced by the acid and the consequent increase of the H+-passive influx (Fig. 7A). Under acetic acid stress, the expression of HRK1 had little or no detectable effect in the active efflux of protons (Fig. 7A, and other results not shown) indicating that it is unlikely that the role of this kinase in yeast tolerance to acetic acid is exerted through Pma1p activation. Because Hrk1p belongs to a family of protein kinases dedicated to the regulation of plasma membrane transporters (Goossens et al., 2000), it was hypothesized that Hrk1p phosphorylation targets might include putative transporters involved in the reduction of acetate concentration into acetic acid-challenged cells. This mechanism was considered because the concentration of acetate is higher in Δhaa1 cells (Fernandes et al., 2005), and this effect could not be fully attributed to other candidate targets of Haa1p, specifically to the plasma membrane drug-H+ antiporters Tpo2p and Tpo3p (Fernandes et al., 2005). Consistent with the hypothesized mechanism, the accumulation of labeled acetic acid in Δhrk1 cells attained higher levels than those registered in the parental strain (Fig. 7B).
FIG. 7.
(A) Comparison of the acetic acid-induced inhibition of external medium acidification rate promoted by energized cells of the parental strain BY4741 (□) and of the deletion mutant Δhrk1 (○). Deenergized cell suspensions of mid-exponential phase cells of both strains were either exposed or not to acetic acid (0 or 1.2 mM at pH 4.0) and then energized with a pulse of glucose (2%) to assess plasma membrane H+-ATPase activity, associated to the external acidification rate of the growth medium. The values shown are means of, at least, three independent experiments that gave rise to similar results. (B) Time course of the accumulation ratio of [1-14C]-acetic acid (corresponding to the ratio between intracellular (Int) and extracellular (Ext) [1-14C]-acetic acid) during cultivation of cells of the parental strain BY4741 (□) or the Δhrk1 mutant (○) in MM4 growth medium supplemented with 60 mM acetic acid (at pH 4.0). Cells used to perform the assay were grown in unsupplemented MM4 medium (at pH 4.0), harvested by centrifugation and reinoculated in fresh growth medium supplemented with radiolabeled and cold acetic acid as described in Materials and Methods.
Discussion
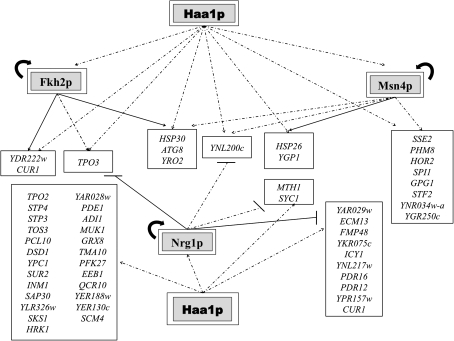
The alterations occurring in yeast genomic expression during early response to acetic acid and the involvement of the transcription factor Haa1p in this transcriptional reprogramming is described here for the first time. Results indicate that Haa1p regulates, directly or indirectly, the transcription of approximately 80% of the acetic acid-responsive genes (85 out of 112 genes). This percentage is much higher than the percentage of acetic acid-responsive genes that is regulated by the other weak acid-responsive regulatory systems described so far, controlled by Rim101p (9%), Msn2p/Msn4p (29%), and War1p (the PDR12 gene only), according to the information available in the YEASTRACT database (Teixeira et al., 2006b). These observations sustain the idea that Haa1p is the main player in the direct or indirect control of yeast transcriptional response to acetic acid stress. Approximately 52% of the Haa1p upregulated genes are presumably direct targets of this transcription factor as they exhibit in their promoter region a Haa1p DNA binding motif (our unpublished results). Among them are TPO3, YPR157w, and YGP1 genes, previously proposed as direct Haa1p targets (Keller et al., 2001). The effect of HAA1 deletion over the transcriptome of acetic acid-stressed cells may also result from the indirect effect of Haa1p over the transcription of genes encoding other transcriptional regulators, in particular, MSN4, NRG1, FKH2, STP3, and STP4. The putative Haa1p-dependent transcriptional regulatory network presumably involved in the control of the early transcriptional response to acetic acid is proposed in Figure 8, based on the information gathered from the present study and in the literature (Teixeira et al., 2006b). This putative regulatory network reveals that a large number of genes whose transcription is reduced in Δhaa1 cells are documented targets of Nrg1p and Msn4p transcription factors (Fig. 8), suggesting that some crosstalk between these regulons may exist. Msn4p and its close homolog Msn2p are known to mediate yeast transcriptional response to environmental stress and to cooperate with other transcription factors under stress (Bar-Joseph et al., 2003; Gasch et al., 2000; Gorner et al., 1998; Rep et al., 1999; Schüller et al., 2004; Vyas et al., 2001). Nrg1p is a transcriptional repressor involved in glucose-dependent regulation of gene expression, in the negative regulation of filamentous growth, in alkaline pH response, and in the repression of stress-responsive genes (Kuchin et al., 2002; Vyas et al., 2005; Zhou and Winston, 2001). It is likely that under acetic acid stress Haa1p may act over the transcription of Nrg1p-target genes to alleviate the transcriptional repression exerted by this transcription factor, as proposed before to occur with Msn2p/Msn4p (Vyas et al., 2005) and with the Candida albicans transcription factor Mnl1p (Ramsdale et al., 2008). Such mechanism is thought to fine-tune the regulation of Msn2p/Msn4p- and Mnl1p-dependent genes to prevent unwanted energy expenses (Ramsdale et al., 2008; Vyas et al., 2005). Remarkably, Nrg1p was identified in this study as a determinant of resistance to acetic acid.
FIG. 8.
Transcriptional regulatory associations that may be governing the Haa1p-dependent response in yeast cells under acetic acid stress. The model is based on the microarray data obtained in this study and on the information available in the YEASTRACT database (December 2009). Genes whose transcription is activated in response to acetic acid in a Haa1p-dependent manner were selected for this analysis. The regulatory network is based on the documented regulatory associations between transcription factors (TF) whose expression is activated by Haa1p (the encoding genes are shown inside double boxes) and the other genes regulated by Haa1p, according with the microarray analysis carried out. Positive regulations between activators and target genes are represented by an arrow (→) and negative regulations between repressors and target genes are represented by lines (—). Documented associations are based on immunoprecipitation experiments (closed arrows or lines; → or —) or on transcription analysis approaches with no demonstration of the binding of the TF to the promoter region of target genes (dashed arrows or lines, -- -▸ or - · - · -).
Several genes transcriptionally controlled by HAA1 expression in response to acetic acid stress were implicated in this study, for the first time, in yeast tolerance to this weak acid but the level of protection exerted could not be directly correlated with the level of transcriptional activation induced by the acid, as reported before in studies dedicated to sorbic acid, citric acid, and to the lipophilic acid herbicide 2,4-dichlorophenoxyacetic acid (Lawrence et al., 2004; Schüller et al., 2004; Teixeira et al., 2006a). For example, the more evident susceptibility phenotypes were observed with mutants deleted for genes whose activation was around twofold only (Δhrk1, Δsap30, and Δfmp48).
With the exception of the drug:H+ antiporters Tpo2p, Tpo3p, and of the cell-wall related proteins Ygp1p and Spi1p, previously implicated in yeast tolerance to acetic acid (Fernandes et al., 2005; Simões et al., 2006), all the other genes of the Haa1p regulon that are responsive to acetic acid and determinants of resistance to acetic acid are described here for the first time. Three of these genes encode proteins involved in lipid metabolism: YPC1 and SUR2 involved in sphingolipid metabolism, and INM1 involved in inositol metabolism. Assembly of the vacuolar membrane H+-ATPase (V-ATPase) is known to depend on sphingolipid metabolism (Chung et al., 2003), and C26-based sphingolipids also have a role in intracellular trafficking to the plasma membrane of nutrient transporters and of the H+-ATPase Pma1p (Chung et al., 2001; Gaigg et al., 2006). Proper vacuolar acidification under acetic acid stress was demonstrated to depend on the signaling molecule phosphatidylinositoide PtdIns(3,5)P2 (Mollapour et al., 2006), produced from inositol. The activities of plasma membrane H+-ATPase (PM-ATPase) and vacuolar membrane H+-ATPase (V-ATPase) are essential for yeast adaptation and resistance to weak acid-induced stress, being required for maintenance of intracellular pH homeostasis, nutrient compartmentalization, and protein sorting (Carmelo et al., 1997; Fernandes et al., 2003; Makrantoni et al., 2007; Martinez-Munoz and Kane, 2008; Mira et al., 2009; Viegas et al., 1998). Given this, it is likely that the Haa1p-dependent increased transcription of SUR2, YPC1, and INM1 genes in acetic acid-challenged cells may contribute to optimal activity of these two proton pumps, and thus to increased tolerance towards acetic acid.
Haa1p-mediated resistance to acetic acid was also found to involve the expression of SAP30 gene, which encodes a component of the histone deacetylase Rpd3L complex. Indeed, SAP30 expression provided one of the highest levels of protection against acetic acid identified in our study. The deletion of SIN3 and SDS3 genes, which encode two proteins required for the structural integrity and catalytic activity of the Rpd3L complex and whose expression is not Haa1p-dependent, also leads to a higher susceptibility to acetic acid. Surprisingly, the elimination of RPD3 gene, encoding a histone deacetylase of the complex, does not significantly increase yeast susceptibility to acetic acid. It is possible that the reduced susceptibility phenotype of Δrpd3 cells to acetic acid results from a functional redundancy between Rpd3p and Hos2p, another histone deacetylase that was recently considered part of the Rpd3L complex (Shevchenko et al., 2008). Acetylation and deacetylation of histones are among the complex mechanisms involved in the regulation of gene expression in eukaryotes by modulating the accessibility of transcription factors to their target promoter regions (Kurdistani and Grunstein, 2003). It is therefore conceivable that, through the Rpd3L complex, Sap30p modulates the yeast transcriptional response to acetic acid by affecting the activity of transcription factors involved in such genomic expression program. Consistent with this hypothesis, it was recently demonstrated that the Rpd3L complex affects the activity of the general stress responsive transcription factors Msn2p and Msn4p (Alejandro-Osorio et al., 2009), which govern the transcriptional response to acetic acid-induced stress. Interestingly, the activity of Haa1p was also suggested to depend on histone acetylation (Guo et al., 2006) and a protein–protein interaction of Haa1p with the histone deacetylase Hos2p was demonstrated (Beltrao et al., 2009).
The other major determinant of resistance to acetic acid stress that is transcriptionally controlled by Haa1p is the HRK1 gene. Notably, the elimination of HRK1 leads to an increased accumulation of acetate into yeast cells challenged with acetic acid, as reported to occur in Δhaa1 cells (Fernandes et al., 2005). Because Hrk1p belongs to a group of protein kinases thought to be involved in the regulation of the activity of plasma membrane transporters (Goossens et al., 2000), it is possible to speculate that this protein kinase may be involved in the reduction of the intracellular acetate concentration through the activation, by phosphorylation, of putative(s) plasma membrane acetate exporter(s). Due to its negative charge, acetate tends to accumulate in the cytosol following dissociation of the lipophilic acid form, and the active expulsion of the counterion from the cell interior contributes to the cell protection against acetic acid. So far, only the plasma membrane drug:H+ antiporters Tpo2p, Tpo3p, Aqr1p, and Azr1p were suggested to play a role in acetic acid resistance (Fernandes et al., 2005; Tenreiro et al., 2000, 2002). According to our microarray analysis, the transcription of TPO3, TPO2, and AQR1 genes is activated by acetic acid under the dependence of Haa1p. The AQR1 gene is not listed in Table 2 because the difference in the mRNA levels of this gene produced in the wild-type strain and in Δhaa1 cells was below 25%, but it is included in Supplementary Table S4. A large scale screening of the yeast kinome, performed using cells cultivated in the absence of stress, in rich growth medium, led to the identification of a few Hrk1p targets although none of them is a plasma membrane transporter (Ptacek et al., 2005). One of the identified Hrk1p targets is a cytosolic enzyme, Gph1p, involved in the catabolism of glycogen, the expression of GPH1 providing protection against acetic acid (our unpublished results). This suggests that other Hrk1p targets relevant for acetic acid tolerance may exist besides plasma membrane transporters. The ongoing analysis of the phosphoproteome of yeast cells under acetic acid stress, either expressing or not HRK1, is expected to bring light into the biological role of Hrk1p as a major determinant of yeast resistance to acetic acid. In previous studies, Hrk1p was proposed to stimulate the activity of the plasma membrane proton pump Pma1p (de la Fuente et al., 1997), but according to our results, there is no positive effect of HRK1 expression in the in vivo activity of PM-H+-ATPase under acetic acid stress. However, a possible role for Hrk1p in counteracting the weak acid-induced decrease of plasma membrane electrochemical potential, essential to secondary nutrient uptake and acetate export, is supported by the increased susceptibility of Δhrk1 cells to hygromycine B (Goossens et al., 2000), a compound known to disrupt plasma membrane electrochemical potential. Remarkably, Hal4p and Hal5p protein kinases, which modulate the activity of the potassium uptake transporters Trk1p and Trk2p, thereby playing a key role in the maintenance of plasma membrane electrochemical potential, belong to the same family of protein kinases as Hrk1p (Goossens et al., 2000).
Haa1p was also implicated in yeast transcriptional response to lactic acid, propionic acid, and acetaldehyde (Abbott et al., 2008; Fernandes et al., 2005; Mira et al., 2009). The analysis of the published data shows that most of the genes identified in our study as being transcriptionally activated in response to acetic acid, under the direct or indirect dependence of Haa1p, are also upregulated in response to those compounds (Tables 2 and 3), whereas only a small group of these genes is activated in response to the more lipophilic sorbic acid (Tables 2 and 3). These results are consistent with the concept that the Haa1p-dependent regulatory pathway is particularly responsive to less lipophilic chemicals.
In summary, the present study has characterized the key role played by Haa1p and by genes of the full Haa1p-regulon in yeast response to acetic acid. Only 20% of the genes whose transcript level is increased in response to acetic acid under the direct or indirect dependence of Haa1p have a significant role in yeast protection against the acid. These genes are involved in lipid metabolism, in the regulation of gene expression by modulation of chromatin accessibility, and in the reduction of the intracellular acetate concentration. Microbial production of acetic acid or lactic acid using yeasts as cells factories is a very active area of research in the present days (Sauer et al., 2008). Acetic acid is also one of the inhibitory compounds resulting from the pretreatment of lignocellulosics (Almeida et al., 2007) and from yeast alcoholic fermentation, its accumulation contributing to a decreased performance of wine and bioethanol production processes. The present study therefore provides useful insights to guide the engineering of more robust yeast strains suited to overproduce acetic acid or lactic acid and to conduct alcoholic fermentation envisaging bioethanol and wine production. Because the fungicidal action of fluconazole is elicited by short-chain weak acids, in particular by acetic acid (Moosa et al., 2004), and considering that Haa1p and the proteins encoded by its target genes have robust homologs in Candida glabrata, the indications emerging from this study might also be useful to develop suitable strategies to control this pathogenic yeast.
Supplementary Material
Acknowledgements
This research was supported by Fundação para a Ciência e a Tecnologia (FCT), FEDER and PTDC Programmes (Contracts: PTDC/EIA/67722/2006, PTDC/BIO/66151/2006 and PTDC/AGR-ALI/102608/2008) and a postdoctoral grant to NPM (SFRH/BPD/46982/2008).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Abbott D.A. Suir E. Van Maris A.J.A. Pronk J.T. Physiological and transcriptional responses to high concentrations of lactic acid in anaerobic chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol. 2008;74:5769–5768. doi: 10.1128/AEM.01030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsen M. Bellahn I. Kramer R. Waffenschmidt S. Localization and function of the yeast multidrug transporter Tpo1p. J Biol Chem. 2003;278:12820–12825. doi: 10.1074/jbc.M210715200. [DOI] [PubMed] [Google Scholar]
- Alejandro-Osorio A.L. Huebert D.J. Porcaro D.T. Sonntag M.E. Nillasithanukroh S. Will J.L., et al. The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol. 2009;10:R57. doi: 10.1186/gb-2009-10-5-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J.R.M. Tobias M. Anneli P. Bärbel H.-H. Gunnar L. Marie F.G.-G. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol. 2007;82:340–349. [Google Scholar]
- Bar-Joseph Z. Gerber G.K. Lee T.I. Rinaldi N.J. Yoo J.Y. Robert F., et al. Computational discovery of gene modules and regulatory networks. Nat Biotechnol. 2003;21:1337–1342. doi: 10.1038/nbt890. [DOI] [PubMed] [Google Scholar]
- Beltrao P. Trinidad J.C. Fiedler D. Roguev A. Lim W.A. Shokat K.M., et al. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmelo V. Santos H. Sa-Correia I. Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim Biophys Acta. 1997;1325:63–70. doi: 10.1016/s0005-2736(96)00245-3. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J. Florens L. Swanson S.K. Shia W.J. Anderson S. Yates J., et al. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim Biophys Acta. 2005;1731:77–87. doi: 10.1016/j.bbaexp.2005.09.005. discussion 75–76. [DOI] [PubMed] [Google Scholar]
- Chung J.-H. Lester R.L. Dickson R.C. Sphingolipid requirement for generation of a functional V1 component of the vacuolar ATPase. J Biol Chem. 2003;278:28872–28881. doi: 10.1074/jbc.M300943200. [DOI] [PubMed] [Google Scholar]
- Chung N. Mao C. Heitman J. Hannun Y.A. Obeid L.M. Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J Biol Chem. 2001;276:35614–35621. doi: 10.1074/jbc.M105653200. [DOI] [PubMed] [Google Scholar]
- De la Fuente N. Maldonado A.M. Portillo F. Yeast gene YOR137c is involved in the activation of the yeast plasma membrane H+-ATPase by glucose. FEBS Lett. 1997;420:17–19. doi: 10.1016/s0014-5793(97)01478-6. [DOI] [PubMed] [Google Scholar]
- Destruelle M. Holzer H. Klionsky D.J. Identification and characterization of a novel yeast gene: the YGP1 gene product is a highly glycosylated secreted protein that is synthesized in response to nutrient limitation. Mol Cell Biol. 1994;14:2740–2754. doi: 10.1128/mcb.14.4.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A.R. Durão P.J. Santos P.M. Sá-Correia I. Activation and significance of vacuolar H+-ATPase in Saccharomyces cerevisiae adaptation and resistance to the herbicide 2,4-dichlorophenoxyacetic acid. Biochem Biophys Res Comm. 2003;312:1317–1324. doi: 10.1016/j.bbrc.2003.11.072. [DOI] [PubMed] [Google Scholar]
- Fernandes A.R. Mira N.P. Vargas R.C. Canelhas I. Sá-Correia I. Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem Biophys Res Comm. 2005;337:95–103. doi: 10.1016/j.bbrc.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Fleet G. Yeasts in foods and beverages: impact on product quality and safety. Curr Opin Biotechnol. 2007;18:170–175. doi: 10.1016/j.copbio.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Gaigg B. Toulmay A. Schneiter R. Very long-chain fatty acid-containing lipids rather than sphingolipids per se are required for raft association and stable surface transport of newly synthesized plasma membrane ATPase in yeast. J Biol Chem. 2006;281:34135–34145. doi: 10.1074/jbc.M603791200. [DOI] [PubMed] [Google Scholar]
- Gasch A.P. Spellman P.T. Kao C.M. Carmel-Harel O. Eisen M.B. Storz G., et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B.R. Lawrence F.M. Leclaire J.P.R. Powell C.D. Smart K.A. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol Rev. 2007;31:535–569. doi: 10.1111/j.1574-6976.2007.00076.x. [DOI] [PubMed] [Google Scholar]
- Goossensm A. De la Fuente N. Forment J. Serrano R. Portillo F. Regulation of yeast H+-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters. Mol Cell Biol. 2000;20:7654–7661. doi: 10.1128/mcb.20.20.7654-7661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W. Durchschlag E. Martinez-Pastor M.T. Estruch F. Ammerer G. Hamilton B., et al. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Gene Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. Tatsuoka K. Liu R. Histone acetylation and transcriptional regulation in the genome of Saccharomyces cerevisiae. Bioinformatics. 2006;22:392–399. doi: 10.1093/bioinformatics/bti823. [DOI] [PubMed] [Google Scholar]
- Hahn J.-.S. Hu Z. Thiele D.J. Iyer V.R. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol. 2004;24:5249–5256. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M.E. Fung E. Wildenhain J. Pierce S.E. Hoon S. Lee W., et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holyoak C.D. Stratford M. McMullin Z. Cole M.B. Crimmins K. Brown A.J., et al. Activity of the plasma membrane H+-ATPase and optimal glycolytic flux are required for rapid adaptation and growth of Saccharomyces cerevisiae in the presence of the weak-acid preservative sorbic acid. Appl Environ Microb. 1997;62:3158–3164. doi: 10.1128/aem.62.9.3158-3164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. Ray E. Brown P.O. Winge D.R. Haa1, a protein homologous to the copper-regulated transcription factor Ace1, is a novel transcriptional activator. J Biol Chem. 2001;276:38697–38702. doi: 10.1074/jbc.M107131200. [DOI] [PubMed] [Google Scholar]
- Kuchin S. Vyas V.K. Carlson M. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol Cell Biol. 2002;22:3994–4000. doi: 10.1128/MCB.22.12.3994-4000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani S.K. Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- Lawrence C.L. Botting C.H. Antrobus R. Coote P.J. Evidence of a new role for the high-osmolarity glycerol mitogen-activated protein kinase pathway in yeast: regulating adaptation to citric acid stress. Mol Cell Biol. 2004;24:3307–3323. doi: 10.1128/MCB.24.8.3307-3323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. Hung Wong W.H. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrantoni V. Dennison P. Stark M.J. Coote P.J. A novel role for the yeast protein kinase Dbf2p in vacuolar H+-ATPase function and sorbic acid stress tolerance. Microbiology. 2007;153:4016–4026. doi: 10.1099/mic.0.2007/010298-0. [DOI] [PubMed] [Google Scholar]
- Martinez-Munoz G.A. Kane P. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J Biol Chem. 2008;283:20309–20319. doi: 10.1074/jbc.M710470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira N.P. Lourenço A.B. Fernandes A.R. Sá-Correia I. The RIM101 pathway has a role in yeast adaptation and resistance to propionic acid and other weak acids. FEMS Yeast Res. 2009;9:202–216. doi: 10.1111/j.1567-1364.2008.00473.x. [DOI] [PubMed] [Google Scholar]
- Mollapour M. Phelan J.P. Millson S.H. Piper P.W. Cooke F.T. Weak acid and alkali stress regulate phosphatidylinositol bisphosphate synthesis in Saccharomyces cerevisiae. Biochem J. 2006;395:73–80. doi: 10.1042/BJ20051765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosa M.-Y.S. Sobel J.D. Elhalis H. Du W. Akins R.A. Fungicidal activity of fluconazole against Candida albicans in a synthetic vagina-simulative medium. Antimicrob Agents Chemother. 2004;48:161–167. doi: 10.1128/AAC.48.1.161-167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portillo F. Serrano R. Growth control strength and active site of yeast plasma membrane ATPase studied by site-directed mutagenesis. Eur J Biochem. 1989;186:501–507. doi: 10.1111/j.1432-1033.1989.tb15235.x. [DOI] [PubMed] [Google Scholar]
- Ptacek J. Devgan G. Michaud G. Zhu H. Zhu X. Fasolo J., et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Ramsdale M. Selway L. Stead D. Walker J. Yin Z. Nicholls S.M., et al. MNL1 Regulates weak acid-induced stress responses of the fungal pathogen Candida albicans. Mol Biol Cell. 2008;19:4393–4403. doi: 10.1091/mbc.E07-09-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M. Reiser V. Gartner U. Thevelein J.M. Hohmann S. Ammerer G., et al. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol. 1999;19:5474–5485. doi: 10.1128/mcb.19.8.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Pousada C.A. Tracy N. Regina M. Dulce A. Jorge P. Catarina A. Yeast activator proteins and stress response: an overview. FEBS Lett. 2004;567:80–85. doi: 10.1016/j.febslet.2004.03.119. [DOI] [PubMed] [Google Scholar]
- Sauer M. Porro D. Mattanovich D. Branduardi P. Microbial production of organic acids: expanding the markets. Trends Biotechnol. 2008;26:100–108. doi: 10.1016/j.tibtech.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Schüller C. Mamnun Y.M. Mollapour M. Krapf G. Schuster M. Bauer B., et al. Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:706–720. doi: 10.1091/mbc.E03-05-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A. Roguev A. Schaft D. Buchanan L. Habermann B. Sakalar C., et al. Chromatin central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 2008;9:R167. doi: 10.1186/gb-2008-9-11-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões T. Mira N.P. Fernandes A.R. Sá-Correia I. The SPI1 gene, encoding a glycosylphosphatidylinositol (GPI)-anchored cell wall protein, plays a prominent role in the development of yeast resistance to lipophilic weak acids food preservatives. Appl Environ Microbiol. 2006;72:7168–175. doi: 10.1128/AEM.01476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M.C. Fernandes A.R. Mira N.P. Becker J. Sá-Correia I. Early transcriptional response of Saccharomyces cerevisiae to stress imposed by the herbicide 2,4-dichlorophenoxyacetic acid. FEMS Yeast Res. 2006a;6:230–248. doi: 10.1111/j.1567-1364.2006.00041.x. [DOI] [PubMed] [Google Scholar]
- Teixeira M.C. Monteiro P. Jain P. Tenreiro S. Fernandes A.R. Mira N.P., et al. The YEASTRACT database: a tool for the analysis of transcriptional regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006b;34:D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenreiro S. Nunes P.A. Viegas C.A. Neves M.S. Teixeira M.C. Cabral M.G., et al. AQR1 gene (ORF YNL065w) encodes a plasma membrane transporter of the major facilitator superfamily that confers resistance to short-chain monocarboxylic acids and quinidine in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2002;292:741–748. doi: 10.1006/bbrc.2002.6703. [DOI] [PubMed] [Google Scholar]
- Tenreiro S. Rosa P.C. Viegas A.C. Sá-Correia I. Expression of the AZR1 gene (ORF YGR224w), encoding a plasma membrane transporter of the major facilitator superfamily, is required for adaptation to acetic acid and resistance to azoles in Saccharomyces cerevisiae. Yeast. 2000;16:1469–1481. doi: 10.1002/1097-0061(200012)16:16<1469::AID-YEA640>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Viegas A.C. Almeida P.F. Cavaco M. Sá-Correia I. The H+-ATPase in the plasma membrane of Saccharomyces cerevisiae is activated during growth latency in octanoic acid-supplemented medium accompanying the decrease in intracellular pH and cell viability. Appl Environ Microbiol. 1998;64:779–783. doi: 10.1128/aem.64.2.779-783.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas V.K. Kuchin S. Carlson M. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics. 2001;158:563–572. doi: 10.1093/genetics/158.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas V.K. Berkey C.D. Miyao T. Carlson M. Repressors Nrg1 and Nrg2 regulate a set of stress-responsive genes in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1882–1891. doi: 10.1128/EC.4.11.1882-1891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. Winston F. NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. BMC Genetics. 2001;2:5. doi: 10.1186/1471-2156-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.