Abstract
Present-day members of the Chlamydiaceae contain parasitic bacteria that have been co-evolving with their eukaryotic hosts over hundreds of millions of years. Likewise, a type III secretion system encoded within all genomes has been refined to complement the unique obligate intracellular niche colonized so successfully by Chlamydia spp. All this adaptation has occurred in the apparent absence of the horizontal gene transfer responsible for creating the wide range of diversity in other Gram-negative, type III-expressing bacteria. The result is a system that is, in many ways, uniquely chlamydial. A critical mass of information has been amassed that sheds significant light on how the chlamydial secretion system functions and contributes to an obligate intracellular lifestyle. Although the overall mechanism is certainly similar to homologous systems, an image has emerged where the chlamydial secretion system is essential for both survival and virulence. Numerous apparent differences, some subtle and some profound, differentiate chlamydial type III secretion from others. Herein, we provide a comprehensive review of the current state of knowledge regarding the Chlamydia type III secretion mechanism. We focus on the aspects that are distinctly chlamydial and comment on how this important system influences chlamydial pathogenesis. Gaining a grasp on this fascinating system has been challenging in the absence of a tractable genetic system. However, the surface of this tough nut has been scored and the future promises to be fruitful and revealing.
Keywords: secretion, virulence, intracellular
Introduction
The Family Chlamydiaceae contains obligate intracellular, Gram-negative bacteria capable of infecting an impressive array of hosts including reptiles, amphibians, and animals. Three species, C. trachomatis, C. pneumoniae, and the zoonotically acquired C. psittaci, are significantly associated with human disease. Ocular (serovars A–C) or genital (serovars D–K, L1–L3) infections with C. trachomatis represent leading causes of infectious blindness and sexually transmitted disease, respectively. C. pneumoniae is a common respiratory pathogen associated with relatively mild, community-acquired pneumonias (Kuo et al., 1995), whereas C. psittaci respiratory infections, acquired rarely from an avian reservoir, cause an atypical pneumonia which can be fatal (Sessa et al., 2009). The prevalence of Chlamydia-mediated human disease, the overall broad host range, and the identification of environmental Chlamydia-like organisms has led to the assertion that the Chlamydiales represent a group of successful parasites that are virtually ubiquitous in nature.
Indeed, considerable effort has been invested to elucidate molecular mechanisms that contribute to this overall success. As obligate intracellular pathogens expressing a minimal genome, many distinct factors likely orchestrate an intricate and highly interdependent manipulation of host cell biology to culminate in successful pathogenesis. The type III secretion system (T3SS) has emerged as one mechanism capable of promoting chlamydial virulence. Genes encoding the virulence-associated T3SS were first identified in Yersinia spp. (Hueck, 1998) and subsequent characterization of the archetype Ysc-Yop system revealed an intricate bacterial mechanism to subvert eukaryotic hosts. T3SSs have since been identified in a wide variety of medically significant Gram-negative pathogens where they are essential for virulence (He et al., 2004). Intriguingly, the T3SS has also been shown to mediate symbiotic relationships and more recently, several non-pathogenic organisms have been found to contain T3SS genes (Krishnan et al., 2003; Tampakaki, 2004; Pallen et al., 2005). We will employ the term “non-flagellar T3SS (NF-T3SS)” throughout this review to differentiate from the process of flagellar biogenesis and in reference to the broader capacity of T3S to mediate interactions between bacteria and a respective eukaryotic host cell (Pallen et al., 2005).
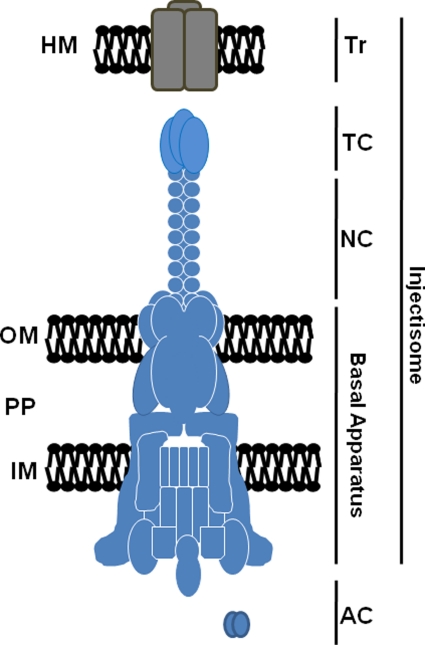
The NF-T3SS is often referred to as acting like a “molecular syringe” to deliver anti-host bacterial “effector” proteins directly into a host cell in a contiguous process (Hueck, 1998). Essential components of the NF-T3SS (Figure 1) include (i) ancillary components (AC) functioning within the bacterial cytoplasm and represented by a set of chaperones and regulatory factors, (ii) membrane-associated gene products that form a multipartite core secretory apparatus that is highly conserved among systems, (iii) proteins comprising an extended needle and tip complex (NC and TC, respectively) which bridges the space between bacterium and host membranes, and (iv) secreted translocon (Tr) proteins required to form pores in eukaryotic membranes through which secretion substrates gain access to the host cytosol (Ghosh, 2004; Tampakaki, 2004). Collectively referred to as the “injectisome”, secretion activity of these machines is tightly regulated and stimulated only upon contact with target host molecules. The reader is referred to several recent reviews for a detailed overview of the T3S mechanism in other bacteria (Moraes et al., 2008; Mueller et al., 2008; Marlovits and Stebbins, 2009; Beeckman and Vanrompay, 2010).
Figure 1.
Schematic representation of a prototypical T3S injectisome. Components including the translocator (Tr), tip (TC), and needle (NC) complexes, basal apparatus, and cytoplasmic ancillary proteins (AC) are shown. The basal apparatus spans the bacterial inner membrane (IM), periplasm (PP), and outer membrane (OM). Secreted translocon components are shown localized to a host membrane (HM). Correct stoichiometry of multimeric proteins is not indicated.
In 1997, Hsia et al. identified a locus of four open reading frames (ORFs) in C. psittaci with homology to Yersinia T3S proteins YscU, LcrD/YscV, YopN, and SycE respectively (Hsia et al., 1997). Initial genome sequencing projects then confirmed an entire complement of conserved, putative T3SS genes in C. trachomatis serovar D and C. pneumoniae (Stephens et al., 1998; Kalman et al., 1999). Genome sequencing projects have subsequently identified ORFs capable of encoding a complete T3SS in all sequenced members of the Chlamydiaceae family (Read et al., 2000, 2003; Thomson et al., 2005; Azuma et al., 2006) and more recently Chlamydia-related bacteria infecting amoebae (Bertelli et al., 2010). The T3S mechanism is therefore clearly entrenched deeply in the biology of these fascinating bacteria. As expected, questions regarding how the T3SS functions and contributes to chlamydial survival have been slow to find answers. This is largely due to the genetic intractability of Chlamydia spp. and a complex parasitic developmental cycle. In recent years, however, there has been a significant increase in our understanding of the role of the NF-T3SS in chlamydial pathogenesis and development. Like other NF-T3SSs, the basic secretion mechanism is likely conserved. Yet variations, consistent with Chlamydia's unique biology, have been noted. Certainly, the emerging complement of niche-specific, secreted effector proteins provides a significant basis for distinctive biology. In fact, several recent reviews provide excellent overviews of chlamydial effector proteins deployed by the T3S mechanism (Valdivia, 2008; Betts et al., 2009; Cocchiaro and Valdivia, 2009). In this review, we will focus on the chlamydial T3S apparatus and comprehensively examine current knowledge regarding the mechanics of this remarkable nanomachine in Chlamydia spp. Rather than focus solely on how the chlamydial T3SS is the same as prototypical systems, we will emphasize variations found in Chlamydia (summarized in Table 1). We endeavor herein to consider how these variable attributes respond to, govern, or otherwise contribute to the successful obligate intracellular lifestyle exemplified by Chlamydia spp.
Table 1.
Distinguishing features of Chlamydia T3SS componentsa.
| Chlamydial T3SS componentb | Yersinia counterpart | Putative function | Chlamydia-specific properties | |||
|---|---|---|---|---|---|---|
| CHAPERONES AND REGULATORY | ||||||
| Scc2 (CT576) | SycD | Class II chaperone for CopB | Basic PI, unique N-terminal consisting of 60 residues which do not share homology with other proteins in the data base or Scc3 | |||
| Scc3 (CT862) | SycD | Class II chaperone CopB2 | Neutral PI, unique 60 residue N-terminal region containing a canonical amphipathic helix absent in Scc2. Interacts with C-terminal domain of CopN in addition to CopB2 | |||
| CT274 | SycD? | Putative Class II chaperone by secondary structure analysis, interacts with CT161 and CT668 | “Stand alone” ORFc. Putative Class II TPR containing chaperone interacts with putative T3S effectors. | |||
| CdsO (CT670) | YscO | Chaperone for putative molecular ruler, CdsP | May interact with CdsN in C. pneumoniae. | |||
| Mscs (CT260) | Multi-cargo effector chaperone, docks with inner membrane component CdsQ to presumably facilitate delivery of effectors to CdsN for secretion. | “Stand alone” ORF. Able to bind CdsQ directly leading to speculation that Mscs dimers may be pre-docked at the cytoplasmic portion of the inner membrane C-ring. | ||||
| CT663 | SycH? | Putative Class I chaperone, may be involved in negative regulation | Class I chaperone able to exert negative regulatory effect on σ66-dependent transcription. | |||
| CT043 | Putative Class I chaperone | “Stand alone” ORF | ||||
| CopN (CT089) | YopN | T3SS “Plug”. Involved in regulation of secretion by maintaining the secretion system in an inactive state prior to the presence of an inductive signal. | Required to support intracellular growth of C. pneumoniae in vitro. Interacts CdsN. | |||
| SECRETION APPARATUS | ||||||
| CdsL (CT561) | YscL | Basal apparatus component interacts with CdsQ. Based on homology likely to be involved in regulation of CdsN by inhibiting ATPase activity. May also be involved in assembly of the apparatus | Capable of interacting with the C-terminus of CdsD. Also interacts with FliI and FlhA homologues in the Bacterial 2-hybrid system and pull-down assays | |||
| CdsN (CT669) | YscN | ATPase involved in release of effectors from cognate chaperones and unfolding effectors to render them secretion competent | Capable of binding with CdsD | |||
| CdsQ (CT672) | YscQ | Cytoplasmic C-ring component of apparatus, docks chaperone-effector complexes and presumably facilitates effector delivery to CdsN. | Interacts with multiple apparatus components including “flagellar homologs” | |||
| CdsD (CT664) | YscD | Likely forms the inner membrane ring with CdsJ | Has a unique N-terminus containing an additional FHA domain which along with its FHA-2 domain undergoes phosphorylation by cytosolic PknD in vitro. Capable of interacting with a novel subset of inner membrane proteins. | |||
| CdsC (CT674) | YscC | Outer membrane secretin | Large unique N-terminal region comprising a distinctive hydrophilic domain that is variable between Chlamydia spp. Lacks an obvious lipoprotein pilot required in other NF-T3SSs for secretin oligomerization and membrane association. | |||
| TRANSLOCATION MACHINERY | ||||||
| CdsF (CT666) | YscF | Needle subunit protein | Contains two cysteine residues at the N-terminus | |||
| CT584 | LcrV | Putative needle tip protein, may be involved in host-cell sensing and contact dependent activation of the secretion system | “Stand alone” ORF. Not recognized by human serum from Chlamydia infected patients | |||
| CopB2 (CT861) | YopB | Possible additional translocator | Present in the host-cell cytosol | |||
aOnly components that have features that clearly distinguish them from homologs in other T3SSs are listed.
bComponent genomic designations are indicated based on the annotated genome of C. trachomatis serovar D (Stephens, 1998).
c“Stand alone” ORF designation indicates the respective gene is encoded outside of T3SS-associated loci.
A thorough consideration of the chlamydial T3SS cannot proceed without comment on key aspects of chlamydial physiology and development. For example, Chlamydia spp. possess an atypical peptidoglycan layer (McCoy et al., 2006). Osmotic stability of infectious particles is instead likely conferred predominantly by disulfide bonding of cysteine-rich outer membrane (OM) and periplasmic proteins (Hatch, 1996). Therefore, the chlamydial T3SS need not contend with remodeling a dense peptidoglycan layer but must accommodate a disulfide linked envelope that is reduced during vegetative growth and oxidized in infectious particles. All Chlamydia spp. exhibit a unique bi-phasic developmental cycle which occurs within parasitophorous membrane bound vesicles, termed inclusions. Development is manifested when infectious, metabolically inert elementary bodies (EBs) invade epithelial cells and begin to differentiate into vegetative, non-infectious, metabolically active reticulate bodies (RBs). RBs replicate within the expanding inclusion before they differentiate back into EBs that are subsequently released, completing the developmental cycle (Abdelrahman and Belland, 2005). As emphasized below, the chlamydial T3SS is present and functional at each stage of this complex cycle.
The unique physiology noted above results in salient and subtle differences between the chlamydial T3S machinery and other systems. In the following section we will discuss characterized components of the chlamydial T3SS and, where appropriate, speculate on latent candidates. A T3SS-specific nomenclature based on the characterized Yersinia system was suggested by Hsia et al. (1997) where components of the basal apparatus are denoted with contact-dependent secretion (Cds), secreted or mobile components as Chlamydia outer protein (Cop), and cytoplasmic chaperones as specific Chlamydia chaperone (Scc). We will continue to use this nomenclature herein. To maintain conciseness, only designations found in the C. trachomatis serovar D genome database1 will be employed when function is ambiguous or names have not been formally assigned.
Origin and Function of the Chlamydial T3SS
Generally, genes encoding components of the T3SS apparatus are clustered together in pathogenicity islands (PAIs) on a plasmid or the bacterial chromosome. Nucleotide content of T3SS coding sequences typically differ from that of the overall genome and are often denoted by a low G + C ratio (Tampakaki, 2004). In contrast, chlamydial genome analyses indicate a homogenous G+C content for T3SS-encoding genes that is similar to the roughly 40% level found in the remainder of respective genomes (Stephens et al., 1998). Moreover, T3SS genes are disseminated similarly throughout chlamydial genomes in four main clusters (Fields and Hackstadt, 2006) arranged into multiple operons (Hefty and Stephens, 2007). Although some of the clusters are flanked by tRNAs, insertion elements, and gene arrangements consistent with horizontal gene transfers or PAIs are lacking. These observations have provided the basis for hypotheses proposing that the NF-T3SS found in Chlamydia spp. represents an ancestral or primordial system (Stephens et al., 1998; Kim, 2001). This notion is supported by the identification of T3S genes in the amoeba symbiont Parachlamydia sp. UWE25, indicating that T3SS genes were present when symbiotic and pathogenic Chlamydia diverged some 700 million years ago (Horn et al., 2004; Pallen et al., 2005). Chlamydial genomes also contain several T3S components having more robust similarity to the flagellar T3SS, raising the possibility that chlamydiae represent a missing link in the transformation of flagellar to non-flagellar T3SSs (Kim, 2001). However, multiple groups have since made compelling arguments that flagellar and non-flagellar systems evolved divergently from a common ancestor (Gophna et al., 2003; Pallen et al., 2005). In addition, it was noted that similar to Chlamydia spp., genes encoding one of the NF-T3SSs found in the proteobacterium Chromobacterium violaceum also have G + C content similar to the remaining genome and some apparatus components are encoded in a separated loci (Betts et al., 2004). Therefore, the evolutionary history of T3SSs remains clouded. Regardless of precise lineage, it is clear that the chlamydial T3SS has evolved in isolation for an evolutionarily significant span of time. This is in agreement with phylogenetic analyses that typically sort the chlamydial T3SS to a distinct family (Troisfontaines and Cornelis, 2005). One can therefore anticipate a great deal of T3S biology that is distinctly chlamydial.
Functionality of the chlamydial T3SS was first demonstrated indirectly. The chlamydial homolog of Yersinia YopN termed CopN was secreted by a heterologous T3SS when ectopically expressed in Yersinia, and immunolocalization data indicated secretion of the endogenous protein during chlamydial infection (Fields and Hackstadt, 2000). It is now doubtless that the primary function of the chlamydial T3SS is to translocate effector proteins into host cells for manipulation of eukaryotic cellular processes. Subsequent studies have further exploited the use of heterologous secretion systems to demonstrate an ability to secrete effector proteins (Subtil et al., 2001; Fields et al., 2003; Lugert et al., 2004; Ho and Starnbach, 2005). Furthermore, the development of small molecule inhibitors of T3S has now enabled more direct demonstration of T3SS-mediated secretion of endogenous chlamydial proteins (Nordfelth et al., 2005; Muschiol et al., 2006; Wolf et al., 2006; Bailey et al., 2007). We have speculated that C. trachomatis may secrete as many as 80 anti-host proteins (Betts et al., 2009). This number would be in line with an absolute dependence on host cell biology and underscores the importance of T3S in chlamydial survival. Combined with 20–30 apparent and predicted apparatus components, this T3SS gene content would represent roughly 10% of the 894 ORFs predicted within the C. trachomatis serovar D genome. This level of genomic commitment raises the possibility that functions beyond secretion of anti-host effector proteins likely exist for the chlamydial T3S apparatus. Several possibilities where the apparatus itself could contribute directly to chlamydial pathogenesis will be elaborated on below.
Assembling the T3SS
The apparatus scaffold
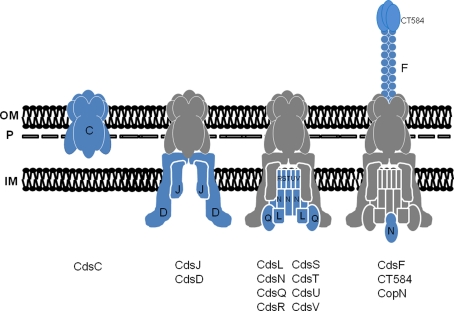
Current evidence from genetically tractable systems indicate that apparatus assembly is initiated by three proteins: an OM ring protein, a lipoprotein forming the inner membrane (IM) ring, and an IM-associated protein connecting the two rings (Yip et al., 2005; Spreter et al., 2009; Diepold et al., 2010). Based on sequence similarity, these proteins would correspond to chlamydial CdsC, CdsJ, and CdsD, respectively (Figure 2). It is likely that a similar outside–in assembly is conserved in Chlamydia spp. Similar to their homologs, CdsC and CdsJ possess predicted Sec-dependent secretion signals that would be necessary for membrane access prior to injectisome assembly. Moreover, both CdsC and CdsJ partition as integral membrane proteins after Triton X-114 extraction of EBs (Fields et al., 2003). As predicted, CdsC is detectible in chlamydial OM complexes (Betts et al., 2008; Birkelund et al., 2009; Liu et al., 2010), whereas most of CdsJ is absent from this fraction (Betts et al., 2008). Aside from modeling predictions that indicate CdsJ is a lipoprotein, direct characterization of this protein is yet forthcoming. Consideration of CdsC and CdsD, however, reveals potentially interesting insights into the chlamydial system.
Figure 2.
A working model for the temporal assembly and composition of the chlamydial T3S apparatus. Stepwise addition of proteins is indicated with newly added components shown in blue and previously assembled components in gray (shown with Cds letter designation only). The basal apparatus spans the chlamydial inner membrane (IM), OmcB-containing P-layer (P), and outer membrane (OM). Flagellar homologs are omitted for clarity.
Based on sequence similarity, CdsC is a member of the PulD family of OM secretins (Hueck, 1998). Interestingly, primary sequence analysis reveals that the N-terminal 250 residues of CdsC comprise a distinctive hydrophilic domain that is unique to chlamydial CdsC yet divergent in primary sequence among chlamydial species. What additional function could be conveyed by this domain remains unclear as no significant homologies are detected via PSI-BLAST. Direct evidence for the predicted multimerization is lacking, and a lipoprotein pilot commonly required for other secretins to oligomerize and become membrane associated (Hueck, 1998) has not been identified for Chlamydia spp. This is perhaps not surprising given that pilot proteins typically display <15% sequence similarity among NF-T3SSs (Okon et al., 2008). However, structures formed by homologs of CdsC are similar in morphology to the multimeric, “rosette-like” structures first observed on C. psittaci EBs (Matsumoto, 1982b). Other candidates such as C. trachomatis PmpD have been proposed for these structures (Crane et al., 2006), yet it seems equally reasonable that they will correspond to the T3SS apparatus. Biochemical extraction data indicate that portions of CdsC are almost certainly surface-exposed (Betts et al., 2008) and Peters et al., (2007) pointed out that, in addition to looking very much like other T3SS rings in electron micrographs, the apparent inner diameter of the rosettes is similar to those of OM rings in Yersinia and Salmonella.
Chlamydial CdsD represents a novel member of the YscD family of proteins, which in Yersinia forms the IM ring in conjunction with YscJ (Silvia-Herzog et al., 2008; Diepold et al., 2010). Interestingly, only the ca. 400 C-terminal residues of CdsD contain regions of similarity to members of the YscD family including putative transmembrane and phospholipid binding (BON) domains (Johnson et al., 2008). YscD family proteins also contain an forkhead associated (FHA)-like domain at the N-terminus (Pallen et al., 2005). FHA domains are often involved in mediating phosphorylation dependent protein-protein interactions via phospho-serine or phospho-threonine peptides (Durocher et al., 2000). CdsD contains two predicted FHA domains, FHA-1 within the first 100 residues and FHA-2 within the C-terminal region that contains YscD similarity. It should be pointed out that early studies suggested that CdsD may be surface exposed on EBs or even secreted during infection (Tanzer and Hatch, 2001; Herrmann et al., 2006). However, the weight of evidence now casts doubt on those interpretations. For example, protein interaction studies (Figure 3) have provided evidence for the C-terminus of CdsD interacting with IM-associated T3S proteins CdsL, CdsQ (Johnson et al., 2008), and CdsN (Stone et al., 2008). These data would be consistent with at least some portion of CdsD being exposed to the chlamydial cytoplasm. More direct evidence for this exposure is apparent since both FHA domains have recently been shown (Johnson and Mahony, 2007) in vitro to be substrates for protein kinasae D (PknD). PknD is one of two currently identified chlamydial eukaryotic-like, IM-associated serine/threonine protein kinases (STPK) having confirmed activity (Verma and Maurelli, 2003). Moreover, PknD may play a role in chlamydial replication (Johnson et al., 2009). Unfortunately technical obstacles have so far precluded corroboration of CdsD phosphorylation in vivo so it is unclear how this may correspond to T3SS activity. Regardless, CdsD represents the first example of a T3SS apparatus protein that has been shown to undergo phosphorylation (Virok et al., 2005; Johnson and Mahony, 2007; Johnson et al., 2009). Considerable questions remain regarding CdsD contribution to chlamydial T3S. Although a single transmembrane domain is predicted, Triton X-114 solubility is inconsistent with CdsD being an integral membrane protein (Johnson et al., 2008). The interaction with cytoplasmic proteins renders it difficult to then understand how CdsD could interact with CdsC to serve its presumed scaffolding function in the IM ring. Clearly, much more information is required.
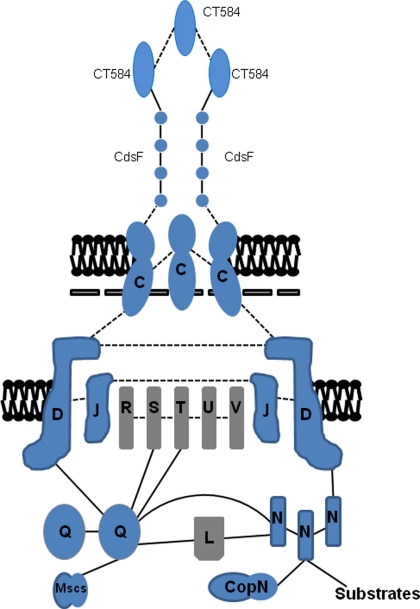
Figure 3.
Summary model of protein–protein interactions among chlamydial T3SS components. Respective proteins are placed based on homology to components in other T3SSs (gray) or based on published in vitro or in vivo data in Chlamydia (blue). Interactions are depicted as dashed lines for predicted interactions or solid lines for those that have been established experimentally. Flagellar homologs are omitted for clarity.
Inner membrane components
Presently, there are only a handful of studies regarding additional components that form the multipartite IM core of the chlamydial NF-T3SS. Homology searches indicate that the composition is similar to the Yersinia apparatus with integral IM components consisting of Cds R/S/T/U/V and peripherally IM-associated components Cds L/N/Q (Fields and Hackstadt, 2006). Recent studies relating directly to protein–protein interactions of the chlamydial apparatus components have been particularly illuminating (Figure 3). Consistent with other NF-T3SSs, these studies indicated that CdsQ likely forms an important platform for structural integrity and secretion activity of the chlamydial T3SS. CdsQ homologs interact with other IM components of the apparatus to form the inner C-ring complex (Jackson and Plano, 2000; Fadouloglou et al., 2004; Gonzalez-Pedrajo et al., 2006; Morita-Ishihara et al., 2006; Riordan and Schneewind, 2008; Diepold et al., 2010). In Chlamydia, association with the IM is apparently mediated by multiple proteins. A combination of GST-fusion co-precipitations and two-hybrid studies using C. trachomatis and C. pneumoniae proteins were used to identify direct CdsQ interactions with IM components CdsD (Johnson et al., 2008), CdsS, and CdsT (Spaeth et al., 2009). An interaction between CdsQ and CdsV was not detected but instead CdsQ interacted with CT060, the flagella homolog of CdsV (Spaeth et al., 2009). In accordance with its counterparts in Yersinia and Shigella, Chlamyida CdsQ was found to interact with itself (Johnson et al., 2008) and the peripheral IM component CdsL (Johnson et al., 2008; Spaeth et al., 2009). Interestingly, a direct interaction of CdsQ was also detected with CdsN (Stone et al., 2008), a secretion chaperone (discussed below), and currently undefined proteins (Spaeth et al., 2009), further supporting the importance of this protein as an integral linchpin in the chlamydial NF-T3S process.
The close association of CdsL and CdsN warrants further exploration. Chlamydia CdsN is homologous to the T3S family of ATPases having similarity to the β subunit of the F0F1 ATPases (Hueck, 1998). This class of ATPases plays an important role in release of effectors from their chaperones and unfolding effectors at the IM to render them secretion competent (Akeda and Galan, 2004). C. pneumoniae CdsN oligomerizes in solution and hydrolyzes ATP in a time- and dose-dependent manner (Stone et al., 2008). CdsN interaction with CdsL likely facilitates CdsL-mediated regulation of CdsN by inhibiting its ATPase activity (Blaylock et al., 2006; Stone et al., 2008). In addition to interactions with CdsL and CdsQ, potentially novel interactions (Stone et al., 2008) have been detected with apparatus components CdsD, CdsO, and CopN (discussed below). In accordance with putative function in modulating effector folding, a weak interaction between C. trachomatis CdsN and the apparent effector protein CT621 (Hobolt-Pederson et al., 2009) was detected in yeast two-hybrid studies (Spaeth et al., 2009). These interactions are likely dynamic in vivo and would orchestrate regulated donation of secretion substrates to the T3S apparatus.
Many of the IM components of the virulence-associated T3SS are homologous with IM components of the flagellar T3SS. Intriguingly a recent study has demonstrated interactions between putative flagella homologs and several of the IM components of the C. pneumoniae T3SS (Stone et al., 2010). The presence of a handful of flagella genes in chlamydial genomes represents an enigma (Fields and Hackstadt, 2006; Peters et al., 2007). In C. trachomatis, CT060 (CdsV paralog) encodes an ortholog of FlhA, an essential integral membrane component of the flagella basal body ring. CT061 is orthologous to FliA, the flagella sigma factor (Macnab, 2004). A second small cluster comprises CT717–CT719 encoding orthologs of FliI (CdsN paralog), the flagella ATPase, a homolog of FliH (CdsL paralog) which is a negative regulator of FliI, and FliF (CdsJ paralog) the MS ring protein, respectively (Macnab, 2004; Peters et al., 2007; Stone et al., 2010). In C. pneumoniae several interactions between putative flagella components were observed (Stone et al., 2010). FliI was found to have ATPase activity similar to CdsN, and interacted with the cytoplasmic domain of FlhA. FlhA in turn interacted with the C-terminal region of FliF. Cpn0859 (a FliH homolog by PSI-BLAST analysis) was found to homodimerize and interact with FliI and FlhA in pull-down assays. These findings are all consistent with counterparts from other flagella systems. Although an interaction between the putative FliH homolog Cpn0859 and the ATPase FliI was established, the authors concluded that Cpn0859 is not a FliH ortholog since it was not found to have regulatory function. However, rigorous PSI-BLAST scores argue for the FliH designation and lack of activity could simply be due to technical failure of an in vitro system. Certainly the most interesting aspect of this study is derived from interactions identified between the flagella orthologs and IM components of the NF-T3SS. Using the Bacterial-2-hybrid system and co-purification assays, identified interaction partners included FliI-CdsL, FlhA-CdsL, FliI-CopN, FlhA-CdsU (Stone et al., 2010), and FlhA-CdsQ (Spaeth et al., 2009). The fact that Chlamydia are non-motile and do not possess a full repertoire of flagella genes is an exciting conundrum (Abdelrahman and Belland, 2005; Peters et al., 2007; Stone et al., 2010), and a minimalist genome demands function for retained genes. We favor a model where a hybrid apparatus is formed incorporating these flagella components into the basal apparatus. Thus, interchanging key apparatus components could result in functionally distinct machinery that correlates with or is dependent on the chlamydial developmental cycle (Stone et al., 2010). This hypothesis is consistent with the observations that “flagellar” genes are consistently transcribed earlier in C. trachomatis (Belland et al., 2003) and C. pneumoniae (Maurer et al., 2007) development than corresponding paralogs. Clearly, this area represents another exciting facet of the chlamydial T3SS that requires and warrants further study.
The extra-cellular injectisome
Once a secretion-competent basal apparatus is completed, components can be exported that form exterior structures. Extra-cellular components of a prototypical T3SS contain a needle, tip, and a pair of translocator proteins (Mueller et al., 2008). Under non-secreting conditions the needle is “capped” by a tip protein that is likely involved in sensing contact with the host cell and subsequently triggers active secretion. Upon activation, translocator proteins are the first to be exported and are required to form a pore in the cytoplasmic membrane consequently allowing direct translocation of the effector substrates (Mota, 2006; Broz et al., 2007). Bioinformatic and biochemical analyses were used to demonstrate that surface-localized CdsF polymerizes to form the Chlamydia needle (Betts et al., 2008). CdsF is highly conserved among chlamydial species and, similar to other NF-T3S needle proteins, is comprised of two predicted helices connected by a loop region. In addition, CdsF interacts with a heterodimeric chaperone (described below) and C. trachomatis CT584 (Spaeth et al., 2009). Although the primary sequence of CT584 lacks similarity to other tip proteins, biophysical characterization and structural modeling indicate that CT584 likely encodes the chlamydial tip protein (Markham et al., 2009). This would be consistent with the interaction with CdsF. Importantly, needle and tip proteins from other systems are antigenic and specific antibodies often confer protection from lethal challenge in animal models (Leary et al., 1995; Matson et al., 2005). However, current studies indicate that neither CdsF nor CT584 induce significant antibody responses in C. trachomatis infected humans (Wang et al., 2010). While it remains to be tested whether antibodies that do arise have neutralizing potential, it is entirely possible that, given an absolute dependence on T3SS for growth, chlamydial components have evolved to be less antigenic than counterparts in facultative and extra-cellular organisms.
The T3SS apparatus is completed by translocator proteins which presumably oligomerize to form a hetero-oligomeric pore in the host cell cytoplasmic membrane. In the case of Chlamydia this would also include the inclusion membrane. Chlamydia spp. potentially encode two sets of translocators and this may make sense if functionally distinct T3SSs occur in chlamydiae as discussed above. Gene products CopB and CopB2 are both secreted via a T3S mechanism (Fields et al., 2005; Subtil et al., 2005). In addition, they resemble translocator proteins in predicted structure, and are encoded downstream of genes encoding homologous chaperones (Scc2 and Scc3) and proteins (CopD and CopD2) having characteristics common to additional translocator proteins (Pallen et al., 1997, 2005). CopB would be predicted to act as a translocator with secreted CopD (Ho and Starnbach, 2005) whereas CopB2 would presumably pair with CopD2. Intuitively a second translocator set would be required since CopB is not transcribed until late-cycle (discussed below). Although CopB could likely enable translocation during invasion, it is unclear how it could function as RBs begin to divide. Unfortunately, endogenous CopB2 (unlike CopB) is not detectable in either C. trachomatis (Fields et al., 2005) or C. pneumoniae (Lugert et al., 2004) inclusion membranes and does not fractionate as an integral membrane protein (Fields, unpublished). If CopB2 has translocator activity, it is certainly atypical. Some translocators in other systems have been shown to have additional effector functions (Hamaguchi et al., 2008; Hamada et al., 2010) and it is also possible that CopB2 has diverged to accomplish this role. This hypothesis would be supported by apparent co-localization of ectopically expressed C. pneumoniae CopB2 with host endoplasmic reticulum (Muller et al., 2008).
Chaperones
Molecular chaperones play a pivotal role in T3S via their interaction with translocator and effector molecules in the bacterial cytosol. They are involved in prevention of substrate degradation prior to secretion, delivery of the substrate to the apparatus, prevention of substrate aggregation and premature interaction, and transcriptional regulation of T3S genes (Feldman and Cornelis, 2003). In addition it has been postulated that they are required to maintain substrates in a secretion competent state and may also be involved in coordinating secretion to ensure secreted components are released in the correct order (Akeda and Galan, 2005). T3SS chaperones can be separated into several general classes based on the nature of their cognate substrates and structural conservation. For example Class IA chaperone one effector, Class IB chaperone multiple effectors and both adopt similar homo-dimeric structural folds (Parsot et al., 2003). Class II chaperone translocator proteins are structurally distinct, typically containing three tetratricopeptide repeat (TPR) motifs (Pallen et al., 2003). Recently, a third class (Class III) has been recognized that chaperone extra-cellular filament proteins and assume an extended alpha helical structure (Quinaud et al., 2005; Yip et al., 2005; Quinaud et al., 2007; Betts et al., 2008). Studies identifying a number of T3S chaperones have begun to reveal a uniquely complex picture regarding chlamydial T3SS chaperones. Demonstrated or potential C. trachomatis T3SS chaperones include: Scc1, Scc2, Scc3, CdsE, CdsG, CT043, CT260, CT274, and CT663 (Fields and Hackstadt, 2006; Spaeth et al., 2009). This is a suspiciously small number of potential chaperones given the number of predicted secretion substrates. However, it is clear from other systems that not all effectors require chaperones (Page and Parsot, 2002; Feldman and Cornelis, 2003) and a picture is emerging in Chlamydia that a single chaperone can indeed associate with multiple T3S substrates.
According to similarities in primary sequence, predicted secondary structure, and deduced biochemical parameters with prototypical chaperones, CT043, Scc1, and CT663 would represent chlamydial Class IA chaperones (Fields and Hackstadt, 2006). Currently, no interaction partners for CT043 have been identified. Generally effectors are encoded close to their chaperones, yet CT043 is flanked by genes involved in glycogen metabolism and DNA replication. However, since several of the chlamydial effectors as well as other chaperones (CT274 and CT260) apparently “stand alone” in the genome, this cannot be considered anomalous. On the contrary, the scattering of type-III related ORFs is certainly one of the distinguishing features of the chlamydial T3SS. It is possible that CT043 represents a multi-cargo effector chaperone, and like CesT's involvement with at least seven E. coli effectors, may be capable of interacting with a significant number of T3S substrates (Thomas et al., 2005). Analyses of Scc1 and CT663 confound typical classification. Instead of interacting with secretion substrates, yeast two-hybrid data indicate they interact with each other (Spaeth et al., 2009). Hence Scc1 and CT663 likely constitute a heterodimeric chaperone. Although a corresponding secretion substrate awaits identification, CT663 may play an additional role in regulation (discussed below) through an interaction with RNA polymerase (Rao et al., 2009).
An extensive yeast two-hybrid screen identified a unique chlamydial protein that represents a new Class IB T3S chaperone capable of binding several effectors (Spaeth et al., 2009). CT260/multiple cargo secretion chaperone (Mcsc) is at the center of a protein–protein interaction hub involving three inclusion membrane proteins, Cap1, CT618, and CT225, respectively and the cytoplasmic C-ring of the apparatus, CdsQ. The data suggest that Mcsc is required to stabilize Cap1 and CT618 in the bacterial cytosol and may facilitate their delivery to the ATPase of the secretion apparatus via a docking interaction with CdsQ (Spaeth et al., 2009). The Mcsc-effector-CdsQ interaction is not unique to Chlamydia since other CdsQ homologs have been shown to be capable of recognizing chaperone-effector complexes (Gonzalez-Pedrajo et al., 2006; Morita-Ishihara et al., 2006; Spaeth et al., 2009). What is unique is that Mcsc alone was able to bind to CdsQ, raising the interesting possibility that Mcsc may be pre-docked at the T3S apparatus. Indeed, Mcsc partitioned within the EB IM in the absence of any effector cargo, and Cap1 and CT618 are synthesized at different time points post-invasion (Spaeth et al., 2009). Speath et al. speculate that either Mcsc dimers detach from CdsQ to stabilize newly synthesized Cap1 and CT618, or more intriguingly, that the transcribing ribosome itself is recruited to the C-ring. If the latter proved to be the case it would be the first time that translational machinery has been shown to directly associate with the T3S apparatus.
Potential chlamydial Class II chaperones include CT274, Scc2, and Scc3 (Fields and Hackstadt, 2006). Despite conclusive sequence similarity with Yersinia SycD and the presence of TPR domains, in silico analyses reveal some distinguishing features for these proteins. It has previously been noted that T3SS chaperones are often small with an acidic pI (Feldman and Cornelis, 2003). Like homologs in other systems, CT274 is small (15.5 kDa) and has an acidic pI of 4.45, yet Scc2 and Scc3 do not fit either of these criteria at 26 and 23 kDa with pIs of 9.58 and 6.71, respectively. The larger mass corresponds to unique N-terminal regions of approximately 60 residues that share little sequence similarity to each other or other chaperones. Despite these differences, in vitro studies demonstrated that both Scc2 and Scc3 were capable of interacting with the Yersinia SycD substrate YopD, supportive of a conserved function. Work indicates an interaction of Scc2 with putative translocator proteins CopB (Fields et al., 2005; Spaeth et al., 2009) and CopD (Spaeth et al., 2009). Scc3 has been found to interact with CopB2 (Fields et al., 2005) and with the C-terminal domain of CopN from C. trachomatis, C. pneumoniae and C. pscittaci, respectively (Slepenkin et al., 2005). Similar to Scc2 and Scc3, PSI-BLAST analysis indicates a characteristic TPR domain within CT274. In contrast, specific homology with known T3SS chaperones is not apparent. Work by Spaeth et al. (2009) has identified an interaction of CT274 with CT668 and CT161. CT161 represents a Chlamydia-specific hypothetical ORF, whereas the genomic positioning of CT688 would suggest a role in the T3S apparatus, perhaps corresponding to Yersinia YscH/YopR (Blaylock et al., 2010). In support of this prediction and the need for a chaperone, the gene product corresponding to CT668 in C. pneumoniae is secreted (Subtil et al., 2005; Muller et al., 2008).
Finally, chlamydial T3SS chaperones also include the recently identified Class III needle subunit co-chaperones CdsE and CdsG (Betts et al., 2008). CdsE and CdsG form a heteromeric complex (Betts et al., 2008; Spaeth et al., 2009) required for CdsF stability. Secondary structure analysis of CdsE and CdsG demonstrated conserved features with other needle chaperones and gene order was conserved with Yersinia, Pseudomonas, and Bordetella (Betts et al., 2008). This is thus another example of chlamydial sequence divergence with functional and “genetic neighborhood” conservation (Betts et al., 2008; Lorenzini et al., 2010). This theme is further exemplified by the recently crystallized CT670, a Chlamydia-specific protein with no identifiable homologs outside of Chlamydia spp. (Gupta and Griffiths, 2006; Lorenzini et al., 2010). Located in the same T3SS gene cluster as CdsD/E/F/G, gene order conservation suggests CT670 may be analogous to YscO, an essential multi-functional component of the Yersinia T3SS that is required for effector export and secretion of the “molecular ruler” protein YscP (Journet et al., 2003; Riordan et al., 2008). Lorenzini et al. provided compelling in silico and in vitro evidence, including protein crystallization, that CT670 is indeed a YscO homolog and should now be considered CdsO. In addition, a chaperone function for CdsO is supported by the demonstration of CdsO interaction with the secreted protein CT671 (Subtil et al., 2005) which likely represents the homolog of Yersinia YscP. CdsO was also found to interact with itself in C. pneumoniae (Stone et al., 2008); thus being consistent with other T3S chaperones which typically dimerize (Page and Parsot, 2002). Interestingly, an interaction between CdsO and the ATPase CdsN was demonstrated in C. pneumoniae. This lead to speculation that C. pneumoniae CdsO may also act as a chaperone for CdsN (Stone et al., 2008). However, since there is considerable evidence that Yersinia YscO is also capable of interactions with IM T3S components (Riordan and Schneewind, 2008) and co-purified with a complex containing the Yersinia ATPase YscN (Riordan et al., 2008), it is possible that this interaction represents additional functional roles for CdsO distinct from chaperone activity.
Regulation of T3SS
Regulation of gene expression
Chlamydial gene transcription is temporally regulated by incompletely defined mechanisms and is generally classified as early-, mid-, and late-cycle. These times correspond roughly to key developmental events such as EB to RB conversion, active RB replication, and differentiation of RBs back to EBs, respectively (Abdelrahman and Belland, 2005). An apparent paradox in the early days of chlamydial T3S was the fact that de novo synthesis of apparatus components generally does not begin until mid-cycle. Moreover, genes encoding Scc2, CopB, and CopD are not transcribed until late-cycle (Fields and Hackstadt, 2006). Considerable work, including identification of EB-localized T3S proteins (Vandahl et al., 2001; Fields et al., 2003) and secretion of T3S effectors such as TARP (Clifton et al., 2004) and CT694 (Hower et al., 2009) during invasion, indicate that a T3SS is present and functional throughout chlamydial development. A reasonable model predicts that pre-formed T3SSs in EBs mediate effector secretion during invasion and early development until replaced by de novo synthesized systems as RBs begin to divide.
Clues are beginning to emerge regarding how modulation of T3S gene expression is accomplished. RT-PCR transcriptional linkage analysis profiling of six disseminated T3SS-encoding loci revealed seven polycistronic operons preceded by predicted E. coli σ70-like promoter elements (Hefty and Stephens, 2007). Interestingly, quantitative gene expression provided evidence for three internal operons also possessing putative σ70-like promoters. These internal operons would result in expression of (i) cdsEFGHNOPQ, (ii) cdsJLRST, and (iii) cdsC independent from CT663-cdsD, lpdA-lipA, and pkn5, respectively. It is currently unclear what would necessitate this separation. Transcriptional regulation of T3S genes in other organisms is primarily controlled by AraC-like transcriptional activators which act in response to environmental cues encountered during infection; orthologs of which are conspicuously absent from the chlamydial genome (Darwin and Miller, 1999; Rakeman et al., 1999; Mavris et al., 2002; Akbar et al., 2003; Case et al., 2010). Recent data suggests that there are no explicit transcriptional activators of chlamydial T3S operons and that their temporal expression is under the control of general mechanisms. Specifically it was found that, as predicted by previous studies (Hefty and Stephens, 2007), promoters for all ten of the T3SS operons were transcribed in vitro by σ66, the major form of chlamydial RNA polymerase. Furthermore, these promoters were differentially sensitive to alterations in DNA supercoiling that corresponded to in vivo temporal expression patterns (Di Russo Case et al., 2010). Transcription of T3S mid-cycle promoters was found to be activated in response to an increase in DNA supercoiling in a similar manner to other chlamydial mid-cycle promoters. Conversely, late cycle T3S promoters were insensitive to DNA supercoiling in accordance with several other supercoiling-independent, σ66-regulated chlamydial late genes (Case et al., 2010). This apparent lack of T3SS-specific transcriptional activators certainly makes intuitive sense for the coordination of T3SS expression with the overall Chlamydia developmental cycle.
Regulation of secretion activity
The chlamydial T3SS is activated within minutes of contact with a eukaryotic cell (Clifton et al., 2004) and likely remains active so long as chlamydiae remain attached to the inclusion membrane (Fields and Hackstadt, 2006). Similar to other T3SSs (Hueck, 1998), the chlamydial system can therefore correctly be referred to as contact-dependent. But what factors directly govern secretion activity? Several clues now exist, and initial attachment to the host plasma membrane likely represents the activation signal. In direct support of this notion, an in vitro release assay was adapted that was capable of stimulating release, albeit inefficiently, of invasion-related Tarp from cell-free EBs (Jamison and Hackstadt, 2008; Spaeth et al., 2009). Importantly, the stimuli, including the Ca2+ chelator EGTA, are also functional for other contact-activated T3SSs (Kim et al., 2005). It should be noted that activation of the chlamydial T3SS is likely more complicated given the metabolic inactivity and disulfide-bonded envelope of EBs. For example, we have emphasized that the presence of cysteine residues in CdsF is a feature unique to the chlamydial T3SS (Betts et al., 2008). Indeed, we find that EB, but not RB, CdsF cysteines are disulfide bonded (Betts-Hampikian, unpublished). If these bonds interfere with secretion, reduction of EB-localized bonds would likely be required before full activity could be achieved.
Activation of secretion would also likely require concomitant secretion of CopN. Although secretion to the inclusion membrane is evident later in development (Fields and Hackstadt, 2000), complications in immunolocalization sensitivity currently confound efforts to demonstrate early export. CopN contains domains similar to Yersinia YopN and TyeA, two proteins involved in negative regulation of T3S. According to one plausible Yersinia model (Joseph and Plano, 2007), YopN is directed to the injectisome by the heterodimeric chaperone SycN/YscB but its secretion is prevented by interaction with the apparatus through TyeA. Once the appropriate stimuli are detected by the needle/tip complex, the interaction with TyeA is dissolved, YopN is secreted, and the apparatus is competent for secretion. The interaction of Scc3 with CopN noted above occurs through the domain having similarity to TyeA. Therefore, this could mediate CopN mobilization to the T3S apparatus or other, yet to be identified chaperones similar to SycN/YscB, could be involved. It is formally possible that the interaction of CopN with CdsN could also play a role (Stone et al., 2008), but we favor an interpretation that this interaction merely reflects CdsN association with secretion substrates. Once CopN is secreted, it is unclear if substrate switching would then occur in Chlamydia spp. Although chlamydial genomes contain CdsU, it has not been determined whether this protein is cleaved similarly to Yersinia YscU to allow secretion of effector proteins (Riordan and Schneewind, 2008).
Secretion of mid- and late-cycle effectors (Valdivia, 2008; Betts et al., 2009) indicate secretion activity that is sustained presumably until an RB dissociates from the inclusion membrane during differentiation to an EB. The environmental cues that result in this event remain undefined, but Wilson et al. (2009) have suggested that something as simple as space constraints within the inclusion dictate dissociation. Electron micrographs from multiple groups (Matsumoto, 1982a; Nichols et al., 1985) contain images of cylindrical surface projections emanating from RBs and forming direct connection with the inclusion membrane. Presuming that these structures correspond to CdsF-containing T3S filaments, enumeration studies suggest that intra-inclusion space constraints result in fewer T3SSs making contact with a eukaryotic membrane (Wilson et al., 2006). This possibility raises a multitude of questions. For example, does CT671 correspond to a molecular ruler similar to Yersinia YscP (Journet et al., 2003; Riordan et al., 2008) and could it be involved in retracting CdsF-containing needles? Alternatively, is a more direct physical dissociation responsible for loss of contact? Since CT584 is a late-cycle gene product, does its de novo production result in capping of needle filaments to cease secretion activity? Is secretion activity responding to dissociation or causing it? Are the uniquely late-cycle Scc2, CopB, and CopD involved in the process? Regardless of the precise mechanism, T3SS activity clearly corresponds to chlamydial development and represents an active area of current investigation.
Additional roles in pathogenesis
As already noted, one way the T3S apparatus contributes directly to chlamydial pathogenesis is through secretion of effector proteins affecting host cellular processes (Valdivia, 2008; Betts et al., 2009) and immunostimulatory capability (Prantner and Nagarajan, 2009). The secretion system may contribute indirectly to the host immune response, and therefore to immunopathology, since injectisome components such as CopN elicit significant antibody responses during natural infection (Wang et al., 2010). It is additionally postulated, and highly likely, that the chlamydial T3SS is intimately involved in mediation of intracellular development and absolutely essential to the developmental cycle; a concept that is in contrast to other T3SS containing organisms whereby the secretion system is essential for virulence but dispensable for survival (Wilson et al., 2006; Peters et al., 2007). We acknowledge that separation of direct apparatus-mediated affects from developmental affects resulting from downstream responses to activities of secreted effectors will be complex. The impact(s) of secretion and substrate activities are likely intimately entwined in the chlamydial system. Possibilities for direct roles of the apparatus do, however, exist. First, it is possible that the T3S apparatus contributes to maintaining attachment of chlamydiae to eukaryotic membranes. We feel that it is unlikely that the T3SS is the sole mediator of initial attachment to plasma membranes. This concept is rare for T3SSs (Lara-Tejero and Galan, 2009) and evidence for a plethora of chlamydial attachment factors is abundant in the literature (Campbell and Kuo, 2006). Moreover, distinct variations exist among attachment requirements of different Chlamydia serovars and species (Dautry-Varsat et al., 2005) while surface-exposed T3S components are highly conserved. However, it is much more likely that the T3S apparatus is directly involved in stable association with the inclusion membrane. Structures likely corresponding to T3S needles (Matsumoto, 1982a; Nichols et al., 1985) conspicuously protrude through the inclusion membrane to potentially tether RBs. As noted above, these observations led to the hypothesis that T3S activity directly controls differentiation of RBs back into infectious EBs (Peters et al., 2007). In addition to the discussed mathematical models (Wilson et al., 2006), there is direct evidence to suggest that this may be the case. The recent discovery that certain acylated hydrazones of salicylaldehydes represent specific, small molecule inhibitors of T3SSs (Keyser et al., 2008) has provided a potentially valuable tool to investigate contributions of T3S to chlamydial development. We (Wolf et al., 2006) and others (Muschiol et al., 2006; Bailey et al., 2007; Slepenkin et al., 2007) have shown that treatment of Chlamydia infected cultures with these inhibitors interferes with T3S and inhibits productive development of chlamydiae. It is not currently possible to rule out potential T3SS-independent effects on Chlamydia spp., and indirect effects such as chelation of iron likely contribute to the ability of inhibitors to interfere with chlamydial growth (Slepenkin et al. 2007). However, these studies are consistent in that drug treatment prevents conversion of RBs to EBs, reinforcing the possibility for a direct link between the T3SS and development. In addition, independent observations that (i) ectopic expression of the T3S effector IncA blocks development of C. caviae (Alzhanov et al., 2004), (ii) microinjection of neutralizing antibodies specific for the T3S effector CT229 arrest C. trachomatis development (Hackstadt et al., 1999; Rzomp et al., 2006), and (iii) inhibition of CopN interferes with C. pneumoniae growth (Huang et al., 2008) are all consistent with a predominant and direct role of T3S in developmental progression. Finally, findings that the putative Class I chaperone CT663 can exert a negative regulatory effect on σ66-dependent transcription (Rao et al., 2009) provide potential mechanisms by which secretion activity could feed-back to govern chlamydial gene expression on a global scale.
Concluding Remarks
One may reasonably question the logic and sanity of pursuing characterization of a complex secretion system in a genetically intractable organism. We have endeavored herein to provide an overview of current data describing the chlamydial T3SS and make forward-looking interpretations regarding unique biology. We certainly acknowledge that some of the discussed work represents preliminary characterization and the overall picture is far from complete. Indeed some studies, particularly those involving interaction studies of ectopically expressed proteins, should be viewed with a level of caution. Given the unique physiology involved it is probably not possible to reconstitute the chlamydial T3SS in an amenable host as was done for the Yersinia system (Bartra et al., 2006). As the field continues to mature however, the weight of accumulated evidence will doubtless validate findings that are physiologically relevant. Clearly, qualities regarding the chlamydial T3S apparatus remain to be elucidated. For example, several components of the basal apparatus are conspicuously absent from the chlamydial T3SS. A homolog of YscK and the inner rod protein YscI are not readily apparent. Whether these absences represent proteins that are present but remain to be discovered or are further examples of divergence of the chlamydial system remains to be seen. However, the most interesting studies will likely reveal how this intricate system directly responds to and/or controls chlamydial development and therefore overall virulence.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. G. Plano for helpful discussions and comments. We also thank H. Maslowski and M. McKuen for critical reading of the manuscript. K.A. Fields and H.J. Betts-Hampikian are supported by a Public Health Service grant from the National Institutes of Health, NIAID AI065530 to K. A. Fields H.J. Betts-Hampikian is partially supported by an award from the Stanley J. Glaser Foundation.
Footnotes
References
- Abdelrahman Y. M., Belland R. J. (2005). The chlamydial developmental cycle. FEMS Microbiol. Rev. 29, 949–959 10.1016/j.femsre.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Akbar S., Schechter L. M., Lostroh C. P., Lee C. A. (2003). AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 47, 715–728 10.1046/j.1365-2958.2003.03322.x [DOI] [PubMed] [Google Scholar]
- Akeda Y., Galan J. E. (2004). Genetic analysis of the Salmonella enterica type III secretion-associated ATPase InvC defines discrete functional domains. J. Bacteriol. 186, 2402–2412 10.1128/JB.186.8.2402-2412.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akeda Y., Galan J. E. (2005). Chaperone release and unfolding of substrates in type III secretion. Nature 437, 911–915 10.1038/nature03992 [DOI] [PubMed] [Google Scholar]
- Alzhanov D., Barnes J., Hruby D., Rockey D. D. (2004). Chlamydial development is blocked in host cells transfected with Chlamydophila caviae incA. BMC Microbiol. 4, 1–10 10.1186/1471-2180-4-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y., Hirakawa H., Yamashita A., Cai Y., Rahman M. A., Suzuki H., Mitaku S., Toh H., Goto S., Murakami T., Sugi K., Hayashi H., Fukushi H., Hattori H., Kuhara S., Shirai M. (2006). Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res. 13, 15–23 10.1093/dnares/dsi027 [DOI] [PubMed] [Google Scholar]
- Bailey L., Gylfe A., Sundin C., Muschiol S., Elofsson M., Nordstrom P., Henriques-Normark B., Lugert R., Waldenstrom A., Wolf-Watz H., Bergstrom S. (2007). Small molecule inhibitors of type III secretion in Yersinia block the Chlamydia pneumoniae infection cycle. FEBS Lett. 58, 587–595 10.1016/j.febslet.2007.01.013 [DOI] [PubMed] [Google Scholar]
- Bartra S. S., Jackson M. W., Ross J. A., Plano G. V. (2006). Calcium-regulated type III secretion of Yop proteins by an Escherichia coli hha mutant carrying a Yersinia pestis pCD1 virulence plasmid. Infect. Immun. 74, 1381–1386 10.1128/IAI.74.2.1381-1386.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman D. S., Vanrompay D. C. (2010). Bacterial secretion systems with an emphasis on the chlamydial Type III secretion system. Curr. Issues Mol. Biol. 12, 17–41 [PubMed] [Google Scholar]
- Belland R. J., Zhong G., Crane D. D., Hogan D., Sturdevant D., Sharma J., Beatty W. L., Caldwell H. D. (2003). Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U.S.A. 100, 8478–8483 10.1073/pnas.1331135100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C., Collyn F., Croxatto A., Ruckert C., Polkinghorne A., Kebbi-Beghdadi C., Goesmann A., Vaughan L., Greub G. (2010). The Waddlia genome: a window into chlamydial biology. PLoS One 5, 1–10 10.1371/journal.pone.0010890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts H. J., Chaudhuri R. R., Pallen M. J. (2004). Analysis of type III secretion gene clusters in Chromobacterium violaceum. Trends Microbiol. 12, 476–482 10.1016/j.tim.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Betts H. J., Twiggs L. E., Sal M. S., Wyrick P. B., Fields K. A. (2008). Bioinformatic and biochemical evidence for the identification of the type III secretion system needle protein of Chlamydia trachomatis. J. Bacteriol. 190, 1680–1690 10.1128/JB.01671-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts H. J., Wolf K., Fields K. A. (2009). Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal. Curr. Opin. Microbiol. 12, 81–87 10.1016/j.mib.2008.11.009 [DOI] [PubMed] [Google Scholar]
- Birkelund S., Morgan-Fisher M., Timmerman E., Gevaert K., Shaw A. C., Christiansen G. (2009). Analysis of proteins in Chlamydia trachomatis L2 outer membrane complex, COMC. FEMS Immunol. Med. Microbiol. 55, 187–195 10.1111/j.1574-695X.2009.00522.x [DOI] [PubMed] [Google Scholar]
- Blaylock B., Berube B. J., Schneewind O. (2010). YopR impacts type III needle polymerization in Yersinia species. Mol. Microbiol. 221–229 10.1111/j.1365-2958.2009.06988.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaylock B., Riordan K. E., Missiakas D. M., Schneewind O. (2006). Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J. Bacteriol. 188, 3525–3534 10.1128/JB.188.10.3525-3534.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P., Mueller C. A., Muller S., Philippsen A., Sorg I., Engel A., Cornelis G. (2007). Function and molecular architecture of the Yersinia injectisome tip complex. Mol. Microbiol. 65, 1311–1320 10.1111/j.1365-2958.2007.05871.x [DOI] [PubMed] [Google Scholar]
- Campbell L. A., Kuo C. C. (2006). “Interactions of Chlamydia with host cells that mediate attachment and uptake,” in: Chlamydia: Genomics and Pathogenesis eds Bavoil P., Wyrick P. B. (Norfolk, UK: Horizon Press; ), 505–522 [Google Scholar]
- Case E. D., Peterson E. M., Tan M. (2010). Promoters for Chlamydia type III secretion genes show a differential response to DNA supercoiling that correlates with temporal expression pattern. J. Bacteriol. 192, 2569–2574 10.1128/JB.00068-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton D. R., Fields K. A., Grieshaber S. S., Dooley C. A., Fischer E. R., Mead D. J., Carabeo A., Hackstadt T. (2004). A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U.S.A. 101, 10166–10171 10.1073/pnas.0402829101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchiaro J. L., Valdivia R. H. (2009). New insights into Chlamydia intracellular survival mechanisms. Cell. Microbiol. 11, 1571–1578 10.1111/j.1462-5822.2009.01364.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane D. D., Carlson J. H., Fischer E. R., Bavoil P., Hsia R., Tan C., Kuo C., Caldwell H. D. (2006). Chlamydia trachomatis polymorphic membrane protein D is a species-common pan-neutralizing antigen. PNAS 103, 1894–1899 10.1073/pnas.0508983103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin K. H., Miller V. L. (1999). InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautry-Varsat A., Subtil A., Hackstadt T. (2005). Recent insights into the mechanisms of Chlamydia entry. Cell. Microbiol. 7, 1714–1722 [DOI] [PubMed] [Google Scholar]
- Di Russo Case E., Peterson E. M., Tan M. (2010). Promoters for Chlamydia type III secretion genes show a differential response to DNA supercoiling that correlates with temporal expression pattern. J. Bacteriol. 192, 2569–2574 10.1128/JB.00068-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A., Amstutz M., Abel S., Sorg I., Jenal U., Cornelis G. (2010). Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 29, 1928–1940 10.1038/emboj.2010.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D., Taylor I. A., Sarbassova D., Haire L. F., Westcott S. L., Jackson S. P., Smerdon S. J., Yaffe M. B. (2000). The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6, 1169–1182 10.1016/S1097-2765(00)00114-3 [DOI] [PubMed] [Google Scholar]
- Fadouloglou V. E., Tampakaki A. P., Glykos M. N., Bastaki J. M., Hadden S. E., Phillips N., Panopoulos N. J., Kokkinidis M. (2004). Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc. Natl. Acad. Sci. U.S.A. 101, 70–75 10.1073/pnas.0304579101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M. F., Cornelis G. R. (2003). The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol. Lett. 219, 151–158 10.1016/S0378-1097(03)00042-9 [DOI] [PubMed] [Google Scholar]
- Fields K. A., Fischer E. R., Mead D. J., Hackstadt T. (2005). Analysis of putative Chlamydia trachomatis chaperones Scc2 and Scc3 and their use in the identification of type III secretion substrates. J. Bacteriol. 187, 6466–6478 10.1128/JB.187.18.6466-6478.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields K. A., Hackstadt T. (2000). Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 38, 1048–1060 10.1046/j.1365-2958.2000.02212.x [DOI] [PubMed] [Google Scholar]
- Fields K. A., Hackstadt T. (2006). “The chlamydia type III secretion system: structure and implications for pathogenesis,” in: Chlamydia: Genomics and Pathogenesis, eds Bavoil P., Wyrick P. B. (Norfolk, UK: Horizon Press; ), 220–233 [Google Scholar]
- Fields K. A., Mead D. J., Dooley C. A., Hackstadt T. (2003). Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48, 671–683 10.1046/j.1365-2958.2003.03462.x [DOI] [PubMed] [Google Scholar]
- Ghosh P. (2004). Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68, 771–795 10.1128/MMBR.68.4.771-795.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pedrajo B., Minamino T., Kihara M., Namba K. (2006). Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol. Microbiol. 60, 984–998 10.1111/j.1365-2958.2006.05149.x [DOI] [PubMed] [Google Scholar]
- Gophna U., Ron E., Graur D. (2003). Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312, 151–163 [DOI] [PubMed] [Google Scholar]
- Gupta R. S., Griffiths E. (2006). Chlamydiae-specific proteins and indels: novel tools for studies. Trends Microbiol. 14, 527–535 10.1016/j.tim.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Hackstadt T., Scidmore-Carlson M. A., Dooley C. A. (1999). Chlamydia trachomatis inclusion membrane protein required for intracellular development. Mol. Biol. Cell 10(Suppl. S), 182A.17 [Google Scholar]
- Hamada D., Hamaguchi M., Suzuki K. N., Sakata I., Yanagihara I. (2010). Cytoskeleton-modulating effectors of enteropathogenic and enterohemorrhagic Escherichia coli: a case for EspB as an intrinsically less-ordered effector. FEBS J. 277, 2409–2415 10.1111/j.1742-4658.2010.07655.x [DOI] [PubMed] [Google Scholar]
- Hamaguchi M., Hamada D., Suzuki K. N., Sakata I., Yanagihara I. (2008). Molecular basis of actin reorganization promoted by binding of enterohaemorrhagic Escherichia coli EspB to alpha-catenin. FEBS J. 275, 6260–6260 10.1111/j.1742-4658.2008.06750.x [DOI] [PubMed] [Google Scholar]
- Hatch T. P. (1996). Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J. Bacteriol. 178, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S. Y., Nomura K., Whittam T. S. (2004). Type III protein secretion mechanism in mammalian and plant pathogens. Biochem. Biophys. Acta 1694, 181–206 10.1016/j.bbamcr.2004.03.011 [DOI] [PubMed] [Google Scholar]
- Hefty P. S., Stephens R. S. (2007). Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J. Bacteriol. 189, 198–206 10.1128/JB.01034-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann M., Schuhmacher A., Muhldorfer I., Melchers K., Prothmann C., Dammeier S. (2006). Identification and characterization of secreted effector proteins of Chlamydophila pneumoniae TW183. Res. Microbiol. 157, 513–524 10.1016/j.resmic.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Ho T. D., Starnbach M. N. (2005). The Salmonella enterica serovar typhimurium-encoded type III secretion systems can translocate Chlamydia trachomatis proteins into the cytosol of host cells. Infect. Immun. 73, 905–911 10.1128/IAI.73.2.905-911.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobolt-Pederson A. S., Christiansen G., Timmerman E., Gevaert K., Birkelund S. (2009). Identification of Chlamydia trachomatis CT621, a protein delivered through the type III secretion system to the host cell cytoplasm and nucleus. FEMS Immunol. Med. Microbiol. 57, 46–58 10.1111/j.1574-695X.2009.00581.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M., Collingro A., Schmitz-Esser S., Beier C. L., Purkhold U., Fartmann B., Brandt P., Nyakatura G. J., Droege M., Frishman D., Rattei T., Mewes H., Wagner M. (2004). Illuminating the evolutionary history of Chlamydiae. Science 304, 728–730 10.1126/science.1096330 [DOI] [PubMed] [Google Scholar]
- Hower S., Wolf K., Fields K. A. (2009). Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol. Microbiol. 72, 1423–1437 10.1111/j.1365-2958.2009.06732.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia R. C., Pannekoek Y., Ingerowski E., Bavoil P. M. (1997). Type III secretion genes identify a putative virulence locus of Chlamydia. Mol. Microbiol. 25, 351–359 10.1046/j.1365-2958.1997.4701834.x [DOI] [PubMed] [Google Scholar]
- Huang J., Lesser C. F., Lory S. (2008). The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature 456, 112–115 10.1038/nature07355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck C. J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. W., Plano G. V. (2000). Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol. Lett. 186, 85–90 10.1111/j.1574-6968.2000.tb09086.x [DOI] [PubMed] [Google Scholar]
- Jamison W. P., Hackstadt T. (2008). Induction of type III secretion by cell-free Chlamydia trachomatis elementary bodies. Microb. Pathog. 45, 435–440 10.1016/j.micpath.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. L., Mahony J. B. (2007). Chlamydophila pneumoniae PknD exhibits dual amino acid specificity and phosphorylates Cpn0712, a putative type III secretion YscD homolog. J. Bacteriol. 189, 7549–7555 10.1128/JB.00893-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. L., Stone C. B., Bulir D. C., Coombes B. K., Mahony J. B. (2009). A novel inhibitor of Chlamydophila pneumoniae protein kinase D (PknD) inhibits phosphorylation of CdsD and suppresses bacterial replication. BMC Microbiol. 9, 218. 10.1186/1471-2180-9-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. L., Stone C. B., Mahony J. B. (2008). Interactions between CdsD, CdsQ, and CdsL, three putative Chlamydophila pneumoniae type III secretion proteins. J. Bacteriol. 190, 2972–2980 10.1128/JB.01997-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. S., Plano G. V. (2007). Identification of TyeA residues required to interact with YopN and to regulate Yop secretion. Adv. Exp. Med. Biol. 603, 235–245 10.1007/978-0-387-72124-8_21 [DOI] [PubMed] [Google Scholar]
- Journet L., Agrain C., Broz P., Cornelis G. R. (2003). The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302, 1757–1760 10.1126/science.1091422 [DOI] [PubMed] [Google Scholar]
- Kalman S., Mitchell W., Marathe R., Lammel C., Fan J., Hyman R. W., Olinger L. J., Grimwood R., Davis W., Stephens R. S. (1999). Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21, 385–389 10.1038/7716 [DOI] [PubMed] [Google Scholar]
- Keyser P., Elofsson M., Rosell S., Wolf-Watz H. (2008). Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 264, 17–29 10.1111/j.1365-2796.2008.01941.x [DOI] [PubMed] [Google Scholar]
- Kim J., Ahn K., Min S., Jia J., Ha U., Wu D., Jin S. (2005). Factors triggering type III secretion in Pseudomonas aeruginosa. Microbiology 151, 3575–3587 10.1099/mic.0.28277-0 [DOI] [PubMed] [Google Scholar]
- Kim J. F. (2001). Revisiting the chlamydial type III protein secretion system: clues to the origin of type III protein secretion. Trends Genet. 17, 65–69 10.1016/S0168-9525(00)02175-2 [DOI] [PubMed] [Google Scholar]
- Krishnan H. B., Lorio J., Kim W. S., Jiang G., Kim K. Y., DeBoer M., Pueppke S. G. (2003). Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant Microbe Interact. 16, 617–625 10.1094/MPMI.2003.16.7.617 [DOI] [PubMed] [Google Scholar]
- Kuo C. C., Jackson L. A., Campbell L. A., Grayston J. T. (1995). Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8, 451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Tejero M., Galan J. E. (2009). Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect. Immun. 77, 2635–2642 10.1128/IAI.00077-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary S. E., Williamson E. D., Griffin K. F., Russell P., Eley S. M., Titball R. W. (1995). Active immunization with recombinant V antigen from Yersinia pestis protects mice against plague. Infect. Immun. 63, 2854–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Afrane M., Clemmer D. E., Zhong G., Nelson D. E. (2010). Identification of Chlamydia trachomatis outer membrane complex proteins by differential proteomics. J. Bacteriol. 192, 2852–2860 10.1128/JB.01628-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzini E., Singer A., Singh B., Lam R., Skarina T., Chirgadze N. Y., Savchenko A., Gupta R. S. (2010). Structure and protein-protein interaction studies on Chlamydia trachomatis protein CT670 (YscO Homolog). J. Bacteriol. 192, 2746–2756 10.1128/JB.01479-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugert R., Kuhns M., Polch T., Gross U. (2004). Expression and localization of type III secretion-related proteins of Chlamydia pneumoniae. Med. Microbiol. Immunol. 193, 163–171 10.1007/s00430-003-0206-x [DOI] [PubMed] [Google Scholar]
- Macnab R. M. (2004). Type III flagellar protein export and flagellar assembly. Biochem. Biophys. Acta 1694, 207–217 10.1016/j.bbamcr.2004.04.005 [DOI] [PubMed] [Google Scholar]
- Markham A. P., Jaafar Z. A., Kemege K. E., Middaugh C. R., Hefty P. S. (2009). Biophysical characterization of Chlamydia trachomatis CT584 supports its potential role as a type III secretion needle tip protein. Biochemistry 48, 10353–10361 10.1021/bi901200y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits T. C., Stebbins C. E. (2009). Type III secretion systems shape up as they ship out. Curr. Opin. Microbiol. 13, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson J. S., Durick K. A., Bradley D. S., Nilles M. L. (2005). Immunization of mice with YscF provides protection from Yersinia pestis infections. BMC Microbiol. 24, 38–48 10.1186/1471-2180-5-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A. (1982a). Electron microscopic observations of surface projections on Chlamydia psittaci reticulate bodies. J. Bacteriol. 150, 358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A. (1982b). Surface projections of Chlamydia psittaci elementary bodies as revealed by freeze-deep-etching. J. Bacteriol. 151, 1040–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer A. P., Mehlitz A., Mollenkopf H. J., Meyer T. F. (2007). Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathog. 3, 752–769 10.1371/journal.ppat.0030083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavris M., Page A. L., Tournebize R., Demers B., Sansonetti P., Parsot C. (2002). Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 43, 1543–1553 10.1046/j.1365-2958.2002.02836.x [DOI] [PubMed] [Google Scholar]
- McCoy A. J., Adams N. E., Hudson A. O., Gilvarg C., Leustek T., Maurelli A. T. (2006). L,L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. PNAS 103, 17909–17914 10.1073/pnas.0608643103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes T. F., Spreter T., Strynadka N. C. (2008). Piecing together the type III injectisome of bacterial pathogens. Curr. Opin. Struct. Biol. 18, 258–256 [DOI] [PubMed] [Google Scholar]
- Morita-Ishihara T., Ogawa M., Sagara H., Yoshida M., Katayama E., Sasakawa C. (2006). Shigella Spa33 is an essential C-ring component of type III secretion machinery. J. Biol. Chem. 281, 599–607 10.1074/jbc.M509644200 [DOI] [PubMed] [Google Scholar]
- Mota L. J. (2006). Type III secretion gets an LcrV tip. Trends Microbiol. 14, 197–200 10.1016/j.tim.2006.02.010 [DOI] [PubMed] [Google Scholar]
- Mueller C. A., Broz P., Cornelis G. (2008). The type III secretion system tip complex and translocon. Mol. Microbiol. 68, 1085–1095 10.1111/j.1365-2958.2008.06237.x [DOI] [PubMed] [Google Scholar]
- Muller N., Sattelmacher F., Lugert R., Gross U. (2008). Characterization and intracellular localization of putative Chlamydia pneumoniae effector proteins. Med. Microbiol. Immunol. 197, 387–396 10.1007/s00430-008-0097-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschiol S., Bailey L., Gylfe A., Sundin C., Hultenby K., Bergstrom S., Elofsson M., Wolf-Watz H., Normark S., Henriques-Normark B. (2006). A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U.S.A. 103, 14566–14571 10.1073/pnas.0606412103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols B. A., Setzer P. Y., Pang F., Dawson C. R. (1985). New view of the surface projections of Chlamydia trachomatis. J. Bacteriol. 164, 344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordfelth R., Kauppi A. M., Norberg H. A., Wolf-Watz H., Elofsson M. (2005). Small molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73, 3104–3114 10.1128/IAI.73.5.3104-3114.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okon M., Moraes T. F., Lario P. I., Creagh A. L., Haynes C. A., Strynadka N. C., McIntosh L. P. (2008). Structural characterization of the type-III pilot-secretin complex from Shigella flexneri. Structure 16, 1544–1554 10.1016/j.str.2008.08.006 [DOI] [PubMed] [Google Scholar]
- Page A. L., Parsot C. (2002). Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 46, 1–11 10.1046/j.1365-2958.2002.03138.x [DOI] [PubMed] [Google Scholar]
- Pallen M. J., Beatson S. A., Bailey C. M. (2005). Bioinformatics, genomics and evolution of non-flagellar type III secretion systems: a Darwinian perspctive. FEMS Microbiol. Rev. 29, 201–229 10.1016/j.femsre.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Pallen M. J., Dougan G., Frankel G. (1997). Coiled-coil domains in proteins secreted by type III secretion systems. Mol. Microbiol. 25, 423–425 10.1046/j.1365-2958.1997.4901850.x [DOI] [PubMed] [Google Scholar]
- Pallen M. J., Francis M. S., Futterer K. (2003). Tetratricopeptide-like repeats in type III-secretion chaperones and regulators. FEMS Microbiol. Lett. 223, 53–60 10.1016/S0378-1097(03)00344-6 [DOI] [PubMed] [Google Scholar]
- Parsot C., Hamiaux C., Page A. (2003). The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6, 7–14 10.1016/S1369-5274(02)00002-4 [DOI] [PubMed] [Google Scholar]
- Peters J., Wilson D. P., Myers G., Timms P., Bavoil P. (2007). Type III secretion a la Chlamydia. Trends Microbiol. 15, 241–251 10.1016/j.tim.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Prantner D., Nagarajan U. M. (2009). Role for the chlamydial type III secretion apparatus in host cytokine expression. Infect. Immun. 77, 76–84 10.1128/IAI.00963-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinaud M., Chabert J., Faudry E., Neumann E., Lemaire D., Pastor A., Elsen S., Dessen A., Attree I. (2005). The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J. Biol. Chem. 280, 36293–36300 10.1074/jbc.M508089200 [DOI] [PubMed] [Google Scholar]
- Quinaud M., Ple S., Job V., Contreras-Martel C., Simorre J. P., Attree I., Dessen A. (2007). Structure of the heterotrimeric complex that regulates type III secretion needle formation. PNAS 104, 7803–7808 10.1073/pnas.0610098104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakeman J. L., Bonifield H. R., Miller S. I. (1999). A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181, 3096–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao X., Deighan P., Hua Z., Wang J., Luo M., Wang J., Liang Y., Zhong G., Hochschild A., Shen L. (2009). A regulator from Chlamydia trachomatis modulates the activity of RNA polymerase through direct interaction with the beta subunit and the primary sigma subunit. Genes Dev. 23, 1818–1829 10.1101/gad.1784009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read T., Myers G., Brunham R., Nelson W., Paulsen I., Heidelberg J., Khouri H. E., Federova N., Carty H., Umayam L., Haft D., Peterson J., Beanan M., White O., Salzberg S., Hsia R., McClarty G., Rank R., Bavoil P., Fraser C. (2003). Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nuc. Acid Res. 31, 2134–2147 10.1093/nar/gkg321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read T. D., Brunham R. C., Shen C., Gill S. R., Heidelberg J. F., White O., Hickey E. K., Peterson J., Utterback T., Berry S., Bass K., Linher J., Weidman H., Khouri B., Craven C., Bowman R., Dodson M., Gwinn W., Nelson R., DeBoy J., Kolonay G., McClarty, Salzberg S. L., Eisen J., Fraser C. M. (2000). Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28, 1397–1406 10.1093/nar/28.6.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan K. E., Schneewind O. (2008). YscU cleavage and the assembly of Yersinia type III secretion machine complexes. Mol. Microbiol. 68, 1485–1501 10.1111/j.1365-2958.2008.06247.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan K. E., Sorg I., Berube B. J., Schneewind O. (2008). Impassable YscP substrates and their impact on the Yersinia enterocolitica type III secretion pathway. J. Bacteriol. 190, 6204–6216 10.1128/JB.00467-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzomp K. A., Moorhead A. R., Scidmore M. A. (2006). The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect. Immun. 74, 5362–5373 10.1128/IAI.00539-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa R., Nicoletti M., Di Pierto M., Schiavoni G., Santino I., Zagaglia C., Del Piano M., Cipriani P. (2009). Chlamydia pneumoniae and atherosclerosis: current state and future prospectives. Int. J. Immunopathol. Pharmacol. 22, 9–14 [DOI] [PubMed] [Google Scholar]
- Silvia-Herzog E., Ferracci F., Jackson M. W., Joseph S. S., Plano G. V. (2008). Membrane localization and topology of the Yersinia pestis YscJ lipoprotein. Microbiology 154, 593–607 10.1099/mic.0.2007/013045-0 [DOI] [PubMed] [Google Scholar]
- Spaeth K. E., Chen Y. S., Valdivia R. H. (2009). The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 5: e1000579. 10.1371/journal.ppat.1000579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreter T., Uyip C. K., Sanowar S., Andre I., Kimbrough T. G., Vuckovic M., Pfuetzner R. A., Deng W., Yu A. C., Finlay B. B., Baker D., Miller S. I., Strynadka N. C. (2009). A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 16, 468–476 10.1038/nsmb.1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R. S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Tatusov R. L., Zhao Q., Koonin E. V., Davis R. W. (1998). Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759 10.1126/science.282.5389.754 [DOI] [PubMed] [Google Scholar]
- Stone C. B., Bulir D. C., Gilchrist J. D., Toor R. K., Mahony J. B. (2010). Interactions between flagellar and type III secretion proteins in Chlamydia pneumoniae. BMC Microbiol. 10, 1471–2180 10.1186/1471-2180-10-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone C. B., Johnson D. L., Bulir D. C., Gilchrist J. D., Mahony J. B. (2008). Characterization of the putative type III secretion ATPase CdsN (Cpn0707) of Chlamydophila pneumoniae. J. Bacteriol. 190, 6580–6588 10.1128/JB.00761-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil A., Delevoye C., Balana M. E., Tastevin L., Perrinet S., Dautry-Varsat A. (2005). A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol. Microbiol. 56, 1636–1647 10.1111/j.1365-2958.2005.04647.x [DOI] [PubMed] [Google Scholar]
- Subtil A., Parsot C., Dautry-Varsat A. (2001). Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol. Microbiol. 39, 792–800 10.1046/j.1365-2958.2001.02272.x [DOI] [PubMed] [Google Scholar]
- Tampakaki A. P. (2004). Conserved features of type III secretion. Cell. Microbiol. 6, 805–816 10.1111/j.1462-5822.2004.00432.x [DOI] [PubMed] [Google Scholar]
- Tanzer R. J., Hatch T. P. (2001). Characterization of outer membrane proteins in Chlamydia trachomatis LGV serovar L2. J. Bacteriol. 183, 2686–2690 10.1128/JB.183.8.2686-2690.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N. A., Deng W., Puente J. L., Frey E. A., Yip C. K., Strynadka N. C., Finlay B. B. (2005). CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol. Microbiol. 57, 1762–1779 10.1111/j.1365-2958.2005.04802.x [DOI] [PubMed] [Google Scholar]
- Thomson N. R., Yeats C., Bell K., Holden M. T., Bentley S. D., Linvingstone M., Cerdeno-Tarraga A. M., Harris B., Doggett J., Ormond D., Mungall K., Clarke K., Feltwell T., Hance Z., Sanders M., Quail M. A., Price C., Barrell B. G., Parkhill J., Longbottom D. (2005). The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15, 629–640 10.1101/gr.3684805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troisfontaines P., Cornelis G. (2005). Type III secretion: more systems than you think. Physiology 20, 326–339 10.1152/physiol.00011.2005 [DOI] [PubMed] [Google Scholar]
- Valdivia R. H. (2008). Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 11, 53–59 10.1016/j.mib.2008.01.003 [DOI] [PubMed] [Google Scholar]
- Vandahl B. B., Birkelund S., Demol H., Hoorelbeke B., Christiansen G., Vandekerckhove J., Gevaert K. (2001). Proteome analysis of the Chlamydia pneumoniae elementary body. Electrophoresis 22, 1204–1223 [DOI] [PubMed] [Google Scholar]
- Verma A., Maurelli A. T. (2003). Identification of two eukaryote-like serine/threonine kinases encoded by Chlamydia trachomatis serovar L2 and characterization of interacting partners of Pkn1. Infect. Immun. 71, 5772–5784 10.1128/IAI.71.10.5772-5784.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virok D. P., Nelson D. E., Whitmire W. M., Crane D. D., Goheen M. M., Caldwell H. D. (2005). Chlamydial infection induces pathobiotype-specific protein tyrosine phosphorylation in epithelial cells. Infect. Immun. 73, 1939–1946 10.1128/IAI.73.4.1939-1946.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang Y. X., Lu C., Lei L., Yu P., Zhong G. (2010). A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J. Immunol. 185, 1670–1680 10.4049/jimmunol.1001240 [DOI] [PubMed] [Google Scholar]
- Wilson D. P., Timms P., McElwain D. L., Bavoil P. (2006). Type III secretion, contact-dependent model for the intracellular development of Chlamydia. Bull. Math. Biol. 68, 161–178 10.1007/s11538-005-9024-1 [DOI] [PubMed] [Google Scholar]
- Wilson D. P., Whittum-Hudson J. A., Timms P., Bavoil P. (2009). Kinematics of intracellular Chlamydiae provide evidence for contact-dependent development. J. Bacteriol. 191, 57834–55742 10.1128/JB.00293-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Betts H. J., Chellas-Gery B., Hower S., Linton C. L., Fields K. A. (2006). Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61, 1543–1555 10.1111/j.1365-2958.2006.05347.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip C. K., Kimbrough T. G., Felise H. B., Vuckovic M., Thomas N. A., Pfuetzner R. A., Frey E. A., Finlay B. B., Miller S. I., Strynadka N. C. (2005). Structural characterization of the molecular platform for type III secretion system assembly. Nature 435, 702–707 10.1038/nature03554 [DOI] [PubMed] [Google Scholar]