Abstract
It has been proposed that human cooperation is unique among animals for its scale and complexity, its altruistic nature and its occurrence among large groups of individuals that are not closely related or are even strangers. One potential solution to this puzzle is that the unique aspects of human cooperation evolved as a result of high levels of lethal competition (i.e. warfare) between genetically differentiated groups. Although between-group migration would seem to make this scenario unlikely, the plausibility of the between-group competition model has recently been supported by analyses using estimates of genetic differentiation derived from contemporary human groups hypothesized to be representative of those that existed during the time period when human cooperation evolved. Here, we examine levels of between-group genetic differentiation in a large sample of contemporary human groups selected to overcome some of the problems with earlier estimates, and compare them with those of chimpanzees. We find that our estimates of between-group genetic differentiation in contemporary humans are lower than those used in previous tests, and not higher than those of chimpanzees. Because levels of between-group competition in contemporary humans and chimpanzees are also similar, these findings suggest that the identification of other factors that differ between chimpanzees and humans may be needed to provide a compelling explanation of why humans, but not chimpanzees, display the unique features of human cooperation.
Keywords: chimpanzees, Pan troglodytes, group competition, hunter–gatherer, altruism, warfare
1. Introduction
Human cooperation is apparently unique among vertebrates in its combination of three features: the large number of individuals that can cooperate together, the high frequency of cooperation that involves individuals incurring a cost to their personal reproduction (i.e. ‘altruistic’ cooperation), and its occurrence within such large groups that cooperators are not closely related or are even strangers [1–4]. Thus, the long-term social relationships based on kin selection and reciprocity that underlie cooperation in many other animals, particularly other primates, appear to be insufficient to account for the evolution of cooperation in humans [1–4].
One potential solution to this puzzle is that high levels of lethal competition between groups (i.e. warfare) may have played a key role in the evolution of the unique facets of human cooperation [5–8]. A large body of empirical research suggests that humans pay special attention to in-group membership when cooperating [9–11], while theory suggests that altruistic cooperation can evolve via between-group competition provided groups containing a higher proportion of altruists out-reproduce groups with fewer altruists more quickly than non-altruists out-reproduce altruists within groups [12]. For this process to occur, there should be sufficiently high levels of genetic differentiation between groups so that there are large differences among groups in the fraction of altruists that they contain. While the homogenizing effects of between-group migration would appear to make this scenario unlikely [13,14], it is only recently that attempts have been made to assess the role of between-group competition in the evolution of human cooperation using empirical data. Bowles [15,16] estimated levels of mortality owing to between-group competition as well as levels of between-group genetic differentiation in contemporary and recently living hunter–gatherers, and concluded that altruistic cooperation could evolve in humans if similar conditions applied during the period when this behaviour evolved (presumably the Late Pleistocene).
However, there are several limitations to our understanding of between-group genetic differentiation in humans that limit our ability to evaluate the role of between-group competition in the evolution of human cooperation. First, as his benchmark value of between-group genetic differentiation, Bowles [15] used the median FST values from a number of studies that assayed genetic variation using a variety of different marker systems, including Y-chromosome and mitochondrial DNA. In contrast to biparentally inherited autosomal markers, such uniparentally inherited markers can be very poor indicators of patterns of genome-wide genetic differentiation, and thus of the assortment of altruistic alleles within and between groups, if between-group migration is female- or male-biased, respectively, as typically occurs in human societies [17,18]. Second, most groups in these studies were separated by such large distances that they were very unlikely to have interacted. However, groups tend to be further away from one another and exchange fewer migrants the longer the time since they split from a common ancestral population, and so the amount of between-group genetic differentiation usually increases with geographical distance [19–21]. Thus, it is currently unknown if levels of between-group genetic differentiation measured at the more local scale at which most between-group competition occurs are sufficiently high for the evolution of altruistic cooperation in humans. Third, we have very little understanding of how levels of genetic differentiation between local competing human groups compare to those of other group-living animals. This comparison is important because any compelling explanation of the evolution of human cooperation must also explain why other animals do not display the unique features of human cooperation.
Chimpanzees represent a particularly relevant test for the human between-group competition model, as they are one of humanity's two closest living relatives and represent the base-level of relatively simple, reciprocity- and kinship-based cooperation from which human cooperation evolved [22,23]. Like humans, chimpanzees are one of the few species in which members of one group make lethal coalitionary attacks against members of other groups, a behaviour that has long drawn attention for its similarity to warfare or raiding in traditional human societies [24,25]. Although direct comparisons are difficult, the available evidence suggests that the fitness consequences of between-group competition are as high in chimpanzees as they are in humans; for example, the fraction of adult mortality owing to between-group violence in chimpanzees may match [26] or even exceed [27] that of humans living in traditional societies. However, whether levels of genetic differentiation between competing groups are higher in humans than chimpanzees is unknown, as almost all studies on genetic differentiation in chimpanzees have been conducted at broad geographical scales [28,29] or have used uniparentally inherited markers [30,31], and thus suffer from the same limitations as the data used in Bowles' [15,16] models.
There are several reasons to suspect that levels of genetic differentiation between competing groups may be higher in humans than in other primates. The first stems from the fact that in contrast to most non-human primates, humans have a hierarchical social structure, where multiple local groups are subsumed within a larger ethnolinguistic group that shares a common language, culture and ethnic identity [32]. Ethnographic evidence suggests that most people marry within their ethnolinguistic group [33], and genetic evidence indicates that ethnolinguistic identity predicts genetic differentiation between groups independently of the effects of geographical distance and barriers [19–21]. The second is that while dispersal in non-human primates usually involves a single individual or small number of individuals dispersing from their natal group to join a nearby, established group [34], in humans whole groups can engage in long-distance migrations to settle new lands. This process can lead to competition between neighbouring groups whose genetic differentiation is elevated owing to the previous long-distance geographical separation between them. While a similar phenomenon occurs in chimpanzees when the extinction of geographically intermediate groups brings previously separated groups into competition [35,36], its frequency and scale throughout evolutionary history is probably lower than in humans.
Here, we determine whether levels of autosomal genetic differentiation between local human groups reach the levels previously suggested [15,16] as sufficient to allow the evolution of unique facets of human cooperation via group competition, and further examine whether values in humans exceed those in chimpanzees. In an attempt to compensate for the necessity of using samples of contemporary humans to infer levels of between-group genetic differentiation that existed during the time period when human cooperation evolved, we examined between-group genetic differentiation in a large sample of many different types of human societies across the world. While previous studies [15,16] only considered hunter–gatherers, it has been argued that recent hunter–gatherers live in more marginal habitats than those of Pleistocene hunter–gatherers, whose resource-rich habitats (e.g. oceanic coasts) may have resulted in higher levels of sedentism, population density, polygyny and endogamy that are more similar to those of contemporary food-producing societies [37,38]. Thus, rather than limiting our comparisons to hunter–gatherers, we also examined levels of between-group genetic differentiation in traditional (i.e. non-industrialized) food-producing human societies. As a further step towards ensuring that our sample of contemporary human groups was representative of the full range of between-group genetic differentiation values possibly characteristic of Pleistocene hunter–gatherers, we also performed additional tests where we limited comparisons to pairs of human groups that belonged to different ethnolinguistic groups and spoke languages belonging to different language families.
2. Material and methods
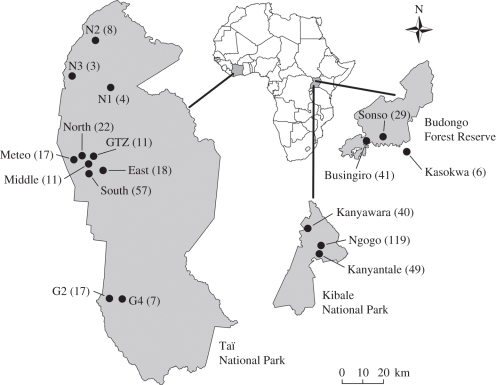
We used autosomal microsatellite genotypes to estimate levels of genetic differentiation between potentially competing groups of chimpanzees and humans. Using DNA extracted from faeces [39], we genotyped 19 autosomal loci in 486 individuals from 18 chimpanzee groups from three locations (figure 1). Genotypes from five chimpanzee groups were previously published [22,40–42], while genotypes for 13 groups were newly generated for this study. We used a two-step amplification method, where all 19 loci were combined with template DNA in an initial multiplex PCR reaction, with dilutions of the resultant PCR products subsequently amplified in singleplex PCR reactions using fluorescently labelled forward primers and unlabelled nested reverse primers [43]. We performed the necessary number of PCR replications to produce error rates of less than 1 per cent, as established in previous work [43]. Eleven of the chimpanzee groups were habituated or semi-habituated to human observation, facilitating the collection of faecal samples from identified adult individuals. In the remaining seven unhabituated chimpanzee groups, genotypes were assigned to individuals and individuals assigned to groups as described elsewhere [42]. We classified as potentially competing 25 pairs of chimpanzee groups that were separated from one another by less than or equal to 20 km, as determined by the centres of their sampling locations (unhabituated groups) or the centres of their territories (habituated groups).
Figure 1.
Locations of chimpanzee groups. The number of individuals genotyped per group is shown in brackets.
We used published autosomal microsatellite genotypes to measure levels of genetic differentiation between potentially competing human groups (Africans [44,45], Aboriginal Australians [46], Pacific islanders [47] and Native Americans [48]). Details of the laboratory procedures followed to produce autosomal genotypes are provided in the individual publications. Because the seafaring technology that would have allowed frequent competition between groups separated by oceans probably did not evolve until late in the Holocene, we only considered human groups that occupied the same land mass. Distances between human groups were determined by their sampling location, or if unavailable, the centre of their traditional territory. As the geographical scale at which most between-group competition occurred during the period when human altruism evolved is unknown, we examined several different cut-off points for potentially competing human groups: those separated by ≤100, ≤200, ≤300, ≤400 and ≤500 km. As expected, we found the highest levels of between-group genetic differentiation among potentially competing groups when we included pairs separated by up to 500 km. As we found that between-group genetic differentiation was not higher in humans than chimpanzees even when including human groups separated by up to 500 km (see §3), we do not present the results for the more geographically restrictive classifications of potentially competing human groups, even if it is more probable that most competition occurs at these more local scales, particularly among direct neighbours.
Genetic differentiation between groups was calculated using D [49] and FST [50]. D measures the actual relative degree of differentiation of allele frequencies among the groups of a population. FST, by contrast, was designed to estimate one of the causes of differences in allele frequencies between groups, the amount of migration (other factors include mutation rate, bottlenecks, founder effects, etc.). Unlike FST, which is mathematically bound by the amount of within-population diversity, D increases monotonically with increasing levels of allelic differentiation. Thus, D is a more appropriate measure to approximate the assortment of altruistic alleles within- and between-groups, and we used D-values for statistical comparisons of levels of between-group genetic differentiation in chimpanzees versus humans.
We used a bootstrapping procedure to assess the statistical significance of differences in the means of pairwise between-group D-values of chimpanzees and humans. Here, we generated 95% confidence intervals (CIs) by calculating means based on 10 000 resamples (with replacement) of the pairwise D-values, and determined the statistical significance of differences by examination of the overlap of the 95% CIs. We also repeated all of our analyses with FST, but as they did not qualitatively change any of our conclusions concerning average levels of between-group genetic differentiation in chimpanzees versus humans, we only report FST when making comparisons with the FST values used by Bowles in his earlier work on this topic. In addition to comparing average pairwise FST, we also examined the percentage of pairwise FST values in chimpanzees and our newly assembled human datasets that are as large or larger than the benchmark value Bowles used in his original work on this topic (0.076 [15]), as well as the minimum value he considered in subsequent work (0.022 [16]). We made these comparisons because it is possible that although chimpanzees and humans do not differ in average pairwise genetic differentiation, values that are sufficiently high for the evolution of altruism may occur more frequently in humans than in chimpanzees.
We compared levels of between-group genetic differentiation in chimpanzees with three sets of human groups: (i) both groups in a dyadic comparison are hunter–gatherers (HG–HG comparisons), (ii) both groups are food producers (FP–FP), and (iii) one group is a hunter–gatherer and the other group is a food producer (HG–FP). We repeated each of these comparisons with restricted human datasets that only included pairs of groups that belonged to different ethnolinguistic groups and spoke languages belonging to different language families.
The ethnolinguistic identities, language families and subsistence systems of human groups were determined from information reported in the original publications from which we obtained the genetic data, and along with human and chimpanzee D and FST values, are reported in the electronic supplementary material. Despite the fact that African Pygmies typically speak languages that combine their native tongues with those of their immediate non-Pygmy neighbours [51], we classified all pairs of African Pygmy groups as having languages of the same language family, and all Pygmy/non-Pygmy pairs as having languages of different language families, as we felt that this classification would more closely reflect the purpose of the language family variable, namely, to assay levels of genetic differentiation between groups where large cultural differences may inhibit between-group migration.
3. Results
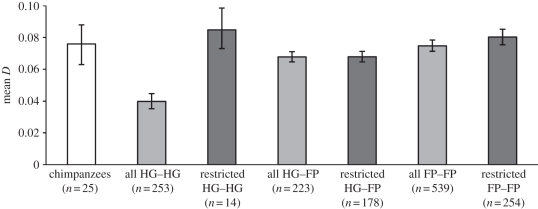
Overall, we found that genetic differentiation was the same or greater between pairs of chimpanzee groups than between human groups (figure 2). Using D, the most appropriate measure of genetic differentiation for assaying the assortment of altruistic alleles within- and between-groups, we found that average genetic differentiation was significantly higher in chimpanzees (D = 0.076, 95% CI = 0.063–0.088, n = 25 pairs of groups) than in hunter–gatherers (D = 0.040, 95% CI = 0.035–0.045, n = 253). The average D of hunter–gatherers doubled to 0.085 when comparisons were made only between groups with different ethnolinguistic affiliations and speaking languages belonging to different families, but did not significantly differ (95% CI = 0.073–0.099, n = 14) from that of chimpanzees. Similarly, although genetic differentiation was higher in HG–FP (D = 0.068, 95% CI = 0.065–0.071, n = 223) and FP–FP (D = 0.075, 95% CI = 0.071–0.078, n = 539) comparisons than in HG–HG comparisons, in neither of these sets of groups were average D-values significantly higher than in chimpanzees. Unlike in the HG–HG comparisons, average D-values among HG–FP (D = 0.068, 95% CI = 0.065–0.071, n = 178) and FP–FP (D = 0.080, 95% CI = 0.076–0.085, n = 254) comparisons did not substantially increase when restricted to comparisons of groups with different ethnolinguistic affiliations and speaking languages from different families.
Figure 2.
Average pairwise D-values (and 95% CIs) of chimpanzee (empty bar) and human groups (grey bars) with different combinations of subsistence systems. Sample sizes (number of pairs of groups) are shown in brackets. For humans, light grey bars represent values for all pairs of groups, and dark grey bars represent values for the restricted sample consisting only of pairs of groups belonging to different ethnolinguistic groups and speaking languages belonging to different language families. HG, hunter–gatherer; FP, food producer.
Very similar results were obtained with the more widely used estimator of genetic differentiation, FST (table 1). The average genetic differentiation of hunter–gatherers was once again low, and values for none of the sets of human groups were higher than among chimpanzees. Notably, human pairwise FST values rarely reached levels used in prior assessment of the models exploring the potential for the evolution of human cooperation via between-group competition [15,16] (table 1). It was actually more common for chimpanzees to reach the minimum pairwise FST value (0.022), recently suggested necessary for the evolution of altruism [16], than it was for HG–HG and HG–FP pairs. Only in FP–FP pairs was the percentage of pairwise comparisons that met the minimum value of 0.022 higher than in chimpanzees.
Table 1.
FST values in chimpanzee and human groups practising different forms of subsistence. (Shown are the sample sizes (number of pairs of groups) and averages of the pairwise FST values, along with the percentage of pairwise FST values that reach those used in Bowles' models. For humans, values are shown for all pairs of groups and for the restricted sample consisting only of pairs of groups belonging to different ethnolinguistic groups and speaking languages belonging to different language families. HG, hunter–gatherer; FP, food producer.)
| n | mean FST | % FST ≥ 0.022 | % FST ≥ 0.076 | |
|---|---|---|---|---|
| chimpanzees | 25 | 0.014 | 8.0 | 0 |
| all HG–HG | 253 | 0.005 | 1.2 | 0.4 |
| restricted HG–HG | 14 | 0.013 | 0 | 0 |
| all HG–FP | 223 | 0.011 | 0.9 | 0.9 |
| restricted HG–FP | 178 | 0.010 | 0 | 0 |
| all FP–FP | 539 | 0.015 | 23.6 | 0.2 |
| restricted FP–FP | 254 | 0.015 | 23.2 | 0 |
4. Discussion
Using the measure of genetic differentiation (D) most appropriate for interpopulation and interspecies comparisons, we showed that average levels of small-scale genetic differentiation between human groups, even when limited to groups exhibiting marked cultural differences, are not higher than levels observed in chimpanzees. In addition, while individual pairwise estimates of FST infrequently reached threshold levels deemed sufficient for the evolution of cooperation via group competition [15,16], this occurred in both humans and chimpanzees with no consistent difference between the two species. The apparent lack of higher local genetic differentiation in humans relative to chimpanzees is surprising given our expectations based on how cultural barriers to between-group migration could lead to higher levels of genetic differentiation between local competing groups of humans than chimpanzees. However, it is also important to consider how other differences between the species could produce the opposite effect. Of particular relevance in this regard is the lower level of autosomal genetic variation in humans than chimpanzees, probably owing to a bottleneck at the recent origin of Homo sapiens some 200 ka, which may limit the extent of differentiation in allele frequencies between groups that have all recently diverged from the same source population [29].
Our results, while suggesting that between-group genetic differentiation in contemporary humans is not greater than in chimpanzees, do not necessarily disprove the hypothesis that high levels of competition between genetically differentiated groups led to the evolution of the unique aspects of human cooperation. Although we found that the frequency of pairwise genetic differentiation values thought to be sufficient for the evolution of altruistic cooperation was not markedly higher in contemporary humans than in chimpanzees, even when comparisons were limited to the most genetically differentiated types of human groups, we cannot definitively rule out the possibility that altruistic cooperation in humans might have evolved owing to the existence of occasional or even single instances of high genetic differentiation of an isolated population. In addition, although we have attempted to infer levels of between-group genetic differentiation present at the critical time of the evolution of human cooperation in the Late Pleistocene by examining genetic differentiation in a large and diverse sample of contemporary human groups, there is currently no way of knowing how successful we have been in this regard. This situation may change in the future as improvements in the ability to extract reliable DNA sequence information from ancient remains [52] may eventually permit the analysis of sufficient samples to describe the population structure of the observed diversity.
We suggest that while the direct and indirect fitness benefits that humans derive from between-group competition have probably been important in the evolution of human cooperation, our results imply that additional factors should be considered to explain why cooperation is so different in humans than in other animals, like chimpanzees, who also gain fitness benefits from between-group competition. In his original work on this topic, Bowles [15] argued that unlike non-human primates, where reproduction is skewed towards dominant individuals, humans possess distinctive practices that limit the ability of selfish individuals to outcompete altruists within groups, including culturally mandated resource and information sharing, consensus decision making, collective restraints on potential aggrandizers and monogamy. However, as some critics have noted [53,54], these ‘reproductive levelling’ mechanisms may rest on exactly the same altruistic behaviour that the model purports to explain, and Bowles' [16] subsequent model did not include a reproductive levelling term. While it is possible that variance in lifetime reproductive success is lower in humans than in chimpanzees for reasons that do not themselves rely on altruism (i.e. ecological constraints that limit the ability of particular individuals to monopolize fitness-limiting resources), the extremely slow life history of chimpanzees means that the data necessary to make the comparisons are currently unavailable.
Contemporary humans and chimpanzees differ in their cognitive abilities and capacity for language, and such factors may also have played a role in facilitating the evolution of altruistic cooperation in humans. Humans are noteworthy in the extent to which socially learned, culturally transmitted information leads to between-group variation in adaptive behaviour. A number of factors, including the tendency of emigrants to adopt the cultural traits of their new group, can lead to more cultural than genetic differentiation between human groups [54,55]. In this regard, our results leave open the possibility that both genetic and cultural differentiation between groups played a role in the evolution of altruistic cooperation [1–3,54,55].
Acknowledgements
All research on chimpanzees complied with the guidelines of local authorities and of the relevant research institutions.
We thank S. Pääbo for comments on an earlier version of the manuscript, R. Mundry for statistical assistance, S. Tüpke for assistance with the figures, C. Boesch for discussion and for facilitating collection of Taï chimpanzee samples, J. Mitani, D. Watts, J. Lwanga, J. Lloyd, the Uganda Wildlife Authority and the Uganda National Council of Science and Technology for facilitating collection of Ngogo and Kanyanchu chimpanzee samples, and J. Friedlaender, F. Friedlaender, P. Verdu, J. Buckleton and S. Walsh for providing detailed human genetic data. The manuscript was greatly improved by the comments of two anonymous referees and R. Bshary. This research was supported by the Alexander von Humboldt Foundation, the Max Planck Society, the National Science Foundation (USA), the Leakey Foundation, the Wenner-Gren Foundation and the Cleveland Metroparks Zoo.
References
- 1.Boyd R., Richerson P. J. 1996. Culture and the evolution of human social instincts. In Roots of human sociality: culture, cognition, and interaction (eds Enfield N. J., Levinson S. C.), pp. 453–477 New York, NY: Berg [Google Scholar]
- 2.Henrich J. 2004. Cultural group selection, coevolutionary processes and large-scale cooperation. J. Econ. Behav. Organ. 53, 3–35 [Google Scholar]
- 3.Henrich J., Henrich N. 2004. Culture, evolution and the puzzle of human cooperation. Cogn. Syst. Res. 7, 220–245 10.1016/j.cogsys.2005.11.010 (doi:10.1016/j.cogsys.2005.11.010) [DOI] [Google Scholar]
- 4.Boyd R., Richerson P. J. 1988. The evolution of reciprocity in sizable groups. J. Theor. Biol. 132, 337–356 [DOI] [PubMed] [Google Scholar]
- 5.Van der Dennen J. 1999. Human evolution and the origin of war: a Darwinian heritage. In The Darwinian heritage and sociobiology (eds Van der Dennen J., Smillie D., Wilson D.), pp. 163–186 Westport, CT: Greenwood Press [Google Scholar]
- 6.Darwin C. 1871. The descent of man, and selection in relation to sex. Princeton, NJ: Princeton University Press [Google Scholar]
- 7.Wilson E. O. 1975. Sociobiology. Cambridge, MA: Belknap Press of Harvard University Press [Google Scholar]
- 8.Alexander R. D., Borgia G. 1978. Group selection, altruism, and levels of organization of life. Annu. Rev. Ecol. Syst. 9, 449–474 [Google Scholar]
- 9.Bernhard H., Fischbacher U., Fehr E. 2006. Parochial altruism in humans. Nature 442, 912–915 10.1038/nature04981 (doi:10.1038/nature04981) [DOI] [PubMed] [Google Scholar]
- 10.Efferson C., Lalive R., Fehr E. 2008. The coevolution of cultural groups and ingroup favoritism. Science 321, 1844–1849 10.1126/science.1155805 (doi:10.1126/science.1155805) [DOI] [PubMed] [Google Scholar]
- 11.Sherif M., Harvey O. J., White B. J., Hood W. R., Sherif C. W. 1988. The robbers cave experiment: intergroup conflict and cooperation. Middletown, CT: Wesleyan University Press [Google Scholar]
- 12.Wilson D. S. 1997. Evolution: human groups as units of selection. Science 276, 1816–1817 10.1126/science.276.5320.1816 (doi:10.1126/science.276.5320.1816) [DOI] [PubMed] [Google Scholar]
- 13.Maynard Smith J. 1964. Group selection and kin selection. Nature 201, 145–147 [Google Scholar]
- 14.Williams G. C. 1966. Adaptation and natural selection. Princeton, NJ: Princeton University Press [Google Scholar]
- 15.Bowles S. 2006. Group competition, reproductive leveling, and the evolution of human altruism. Science 314, 1569–1572 10.1126/science.1134829 (doi:10.1126/science.1134829) [DOI] [PubMed] [Google Scholar]
- 16.Bowles S. 2009. Did warfare among ancestral hunter-gatherers affect the evolution of human social behaviors? Science 324, 1293–1298 10.1126/science.1168112 (doi:10.1126/science.1168112) [DOI] [PubMed] [Google Scholar]
- 17.Mesa N. R., et al. 2000. Autosomal, mtDNA, and Y-chromosome diversity in Amerinds: pre- and post-Columbian patterns of gene flow in South America. Am. J. Hum. Genet. 67, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murdoch G. P. 1981. Atlas of world cultures. Pittsburgh, PA: University of Pittsburgh Press [Google Scholar]
- 19.Heyer E., Balaresque P., Jobling M. A., Quintana-Murci L., Chaix R., Segurel L., Aldashev A., Hegay T. 2009. Genetic diversity and the emergence of ethnic groups in Central Asia. BMC Genet. 10, 49. 10.1186/1471-2156-10-49 (doi:10.1186/1471-2156-10-49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manica A., Prugnolle F., Balloux F. 2005. Geography is a better determinant of human genetic differentiation than ethnicity. Hum. Genet. 118, 366–371 10.1007/s00439-005-0039-3 (doi:10.1007/s00439-005-0039-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sokal R. R. 1988. Genetic, geographic, and linguistic distances in Europe. Proc. Natl Acad. Sci. USA 85, 1722–1726 10.1073/pnas.85.5.1722 (doi:10.1073/pnas.85.5.1722) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langergraber K. E., Mitani J. C., Vigilant L. 2007. The limited impact of kinship on cooperation in wild chimpanzees. Proc. Natl Acad. Sci. USA 104, 7786–7790 10.1073/pnas.0611449104 (doi:10.1073/pnas.0611449104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller M. N., Mitani J. C. 2005. Conflict and cooperation in wild chimpanzees. Adv. Study Behav. 35, 275–331 [Google Scholar]
- 24.Crofoot M. C., Wrangham R. W. 2010. Intergroup aggression in primates and humans: the case for a unified theory. In Mind the gap: tracing the origins of human universals (eds Kappeler P. M., Silk J. B.), pp. 171–195 Berlin, Germany: Springer [Google Scholar]
- 25.Wrangham R. W. 1999. Evolution of coalitionary killing. Yearb. Phys. Anthropol. 42, 1–30 [DOI] [PubMed] [Google Scholar]
- 26.Wrangham R. W., Wilson M. L., Muller M. N. 2006. Comparative rates of violence in chimpanzees and humans. Primates 47, 14–26 10.1007/s10329-005-0140-1 (doi:10.1007/s10329-005-0140-1) [DOI] [PubMed] [Google Scholar]
- 27.Mitani J. C., Watts D. P., Amsler S. J. 2010. Lethal intergroup aggression leads to territorial expansion in wild chimpanzees. Curr. Biol. 20, R507–R508 [DOI] [PubMed] [Google Scholar]
- 28.Becquet C., Patterson N., Stone A. C., Przeworski M., Reich D. 2007. Genetic structure of chimpanzee populations. PLoS Genet. 3, e66. 10.1371/journal.pgen.0030066 (doi:10.1371/journal.pgen.0030066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer A., Pollack J., Thalmann O., Nickel B., Pääbo S. 2006. Demographic history and genetic differentiation in apes. Curr. Biol. 16, 1133–1138 [DOI] [PubMed] [Google Scholar]
- 30.Langergraber K. E., Siedel H., Mitani J. C., Wrangham R. W., Reynolds V., Hunt K. D., Vigilant L. 2007. The genetic signature of sex-biased migration in patrilocal chimpanzees and humans. PLoS ONE 10, e973. 10.1371/journal.pone.0000973 (doi:10.1371/journal.pone.0000973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonder M. K., Disotell T. R., Oates J. F. 2006. New genetic evidence on the evolution of chimpanzee populations and implications for taxonomy. Int. J. Primatol. 27, 1103–1127 10.1007/s10764-006-9063-y (doi:10.1007/s10764-006-9063-y) [DOI] [Google Scholar]
- 32.Rodseth L., Wrangham R. W., Harrigan A. M., Smuts B. B. 1991. The human community as a primate society. Curr. Anthropol. 32, 221–254 [Google Scholar]
- 33.Fox R. F. 1967. Kinship and marriage. Middlesex, UK: Penguin Books [Google Scholar]
- 34.Di Fiore A. 2003. Molecular genetic approaches to the study of primate behavior, social organization, and reproduction. Yearb. Phys. Anthropol. 46, 62–99 10.1002/ajpa.10382 (doi:10.1002/ajpa.10382) [DOI] [PubMed] [Google Scholar]
- 35.Goodall J. 1986. The chimpanzees of Gombe: patterns of behavior. Cambridge, MA: Harvard University Press [Google Scholar]
- 36.Nishida T., Hiraiwa-Hasegawa M., Hasegawa K., Takahata Y. 1985. Group extinction and female transfer in wild chimpanzees in the Mahale Mountains National Park, Tanzania. Z. Tierpsychol. 67, 281–301 [Google Scholar]
- 37.MacDonald D. H., Hewlett B. S. 1999. Reproductive interests and forager mobility. Curr. Anthropol. 40, 501–523 [Google Scholar]
- 38.Marlowe F. W. 2005. Hunter-gatherers and human evolution. Evol. Anthropol. 14, 54–67 10.1002/evan.20046 (doi:10.1002/evan.20046) [DOI] [PubMed] [Google Scholar]
- 39.Nsubuga A. M., Robbins M. M., Roeder A. D., Morin P. A., Boesch C., Vigilant L. 2004. Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Mol. Ecol. 13, 2089–2094 10.1111/j.1365-294X.2004.02207.x (doi:10.1111/j.1365-294X.2004.02207.x) [DOI] [PubMed] [Google Scholar]
- 40.Langergraber K., Mitani J., Vigilant L. 2009. Kinship and social bonds in female chimpanzees (Pan troglodytes). Am. J. Primatol. 71, 840–851 10.1002/ajp.20711 (doi:10.1002/ajp.20711) [DOI] [PubMed] [Google Scholar]
- 41.Lukas D., Reynolds V., Boesch C., Vigilant L. 2005. To what extent does living in a group mean living with kin? Mol. Ecol. 14, 2181–2196 10.1111/j.1365-294X.2005.02560.x (doi:10.1111/j.1365-294X.2005.02560.x) [DOI] [PubMed] [Google Scholar]
- 42.Schubert G. 2010. Genetic differentiation, gene flow and kinship in bonobos, chimpanzees and humans. PhD thesis, University of Leipzig, Germany [Google Scholar]
- 43.Arandjelovic M., Guschanski K., Schubert G., Harris T. R., Thalmann O., Siedel H., Vigilant L. 2009. Two-step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Mol. Ecol. Res. 9, 28–36 10.1111/j.1755-0998.2008.02387.x (doi:10.1111/j.1755-0998.2008.02387.x) [DOI] [PubMed] [Google Scholar]
- 44.Tishkoff S. A., et al. 2009. The genetic structure and history of Africans and African Americans. Science 324, 1035–1044 10.1126/science.1172257 (doi:10.1126/science.1172257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verdu P., et al. 2009. Origins and genetic diversity of Pygmy hunter-gatherers from western central Africa. Curr. Biol. 19, 312–318 10.1016/j.cub.2008.12.049 (doi:10.1016/j.cub.2008.12.049) [DOI] [PubMed] [Google Scholar]
- 46.Walsh S. J., Mitchell R. J., Watson N., Buckleton J. S. 2007. A comprehensive analysis of microsatellite diversity in Aboriginal Australians. J. Hum. Genet. 52, 712–728 10.1007/s10038-007-0172-z (doi:10.1007/s10038-007-0172-z) [DOI] [PubMed] [Google Scholar]
- 47.Friedlaender J. S., et al. 2008. The genetic structure of Pacific islanders. PLoS Genet. 4, e19. 10.1371/journal.pgen.0040019 (doi:10.1371/journal.pgen.0040019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S., et al. 2007. Genetic variation and population structure in Native Americans. PLoS Genet. 3, 2049–2067 10.1371/journal.pgen.0030185 (doi:10.1371/journal.pgen.0030185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jost L. 2008. G(ST) and its relatives do not measure differentiation. Mol. Ecol. 17, 4015–4026 10.1111/j.1365-294X.2008.03887.x (doi:10.1111/j.1365-294X.2008.03887.x) [DOI] [PubMed] [Google Scholar]
- 50.Nei M. 1973. Analysis of gene diversity in subdivided populations. Proc. Natl Acad. Sci. USA 70, 3321–3323 10.1073/pnas.70.12.3321 (doi:10.1073/pnas.70.12.3321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grinker R. R. 1994. Houses in the rainforest: ethnicity and inequality among farmers and foragers in Central Africa. Berkeley, CA: University of California Press [Google Scholar]
- 52.Krause J., Briggs A. W., Kirchner M., Maricic T., Zwyns N., Derevianko A., Pääbo S. 2010. A complete mtDNA genome of an early modern human from Kostenki, Russia. Curr. Biol. 20, 231–236 10.1016/j.cub.2009.11.068 (doi:10.1016/j.cub.2009.11.068) [DOI] [PubMed] [Google Scholar]
- 53.Boyd R. 2006. The puzzle of human sociality. Science 314, 1555–1556 10.1126/science.1136841 (doi:10.1126/science.1136841) [DOI] [PubMed] [Google Scholar]
- 54.Bell A. V., Richerson P. J., McElreath R. 2009. Culture rather than genes provides greater scope for the evolution of large-scale human prosociality. Proc. Natl Acad. Sci. USA 106, 17 671–17 674 10.1073/pnas.0903232106 (doi:10.1073/pnas.0903232106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richerson P. J., Boyd R. P. D. 2005. Not by genes alone: how culture transformed human evolution. Chicago, IL: University of Chicago Press [Google Scholar]