Abstract
The FALC-Loop web server provides an online interface for protein loop modeling by employing an ab initio loop modeling method called FALC (fragment assembly and analytical loop closure). The server may be used to construct loop regions in homology modeling, to refine unreliable loop regions in experimental structures or to model segments of designed sequences. The FALC method is computationally less expensive than typical ab initio methods because the conformational search space is effectively reduced by the use of fragments derived from a structure database. The analytical loop closure algorithm allows efficient search for loop conformations that fit into the protein framework starting from the fragment-assembled structures. The FALC method shows prediction accuracy comparable to other state-of-the-art loop modeling methods. Top-ranked model structures can be visualized on the web server, and an ensemble of loop structures can be downloaded for further analysis. The web server can be freely accessed at http://falc-loop.seoklab.org/.
INTRODUCTION
Protein loops are often responsible for functional specificity of a given protein by contributing to recognition of interaction partners, enzymatic reactions with substrates or conformational changes relevant to function. The special properties of protein loops originate from the variable loop structures that occur as a result of substitutions, insertions or deletions in sequence during evolution.
Many available loop modeling web servers use database search methods (1–3) that search for loops of related sequences in the structure database. When loops with reasonable sequence similarity are not found, one may have to rely on ab initio methods. However, typical ab initio methods that rely mainly on intensive energy optimizations are very time consuming and therefore may not be suitable for web-based service where predictions have to be produced in relatively short time.
In this article, we introduce FALC-Loop server, a protein loop modeling web server that implements an efficient ab initio loop modeling method, FALC (fragment assembly and analytical loop closure) (4). The FALC method is relatively faster than typical ab initio methods because the use of fragments derived from a structure database reduces conformational search space drastically and a knowledge-based potential allows fast scoring of the generated conformations. The fragment-assembled structures are not geometrically consistent with a given framework protein, but the backbone loop dihedral angles can be adjusted to fit into the framework efficiently by solving the analytical loop closure equation (5,6). The prediction accuracy of the FALC method is comparable to other ab initio methods due to the excellent loop sampling performance (4). A combination of the efficient loop sampling method with a more intensive energy optimization can improve the prediction accuracy, but with a large increase in computation time (Park, H. and Seok, C., manuscript in preparation).
FALC-LOOP METHOD
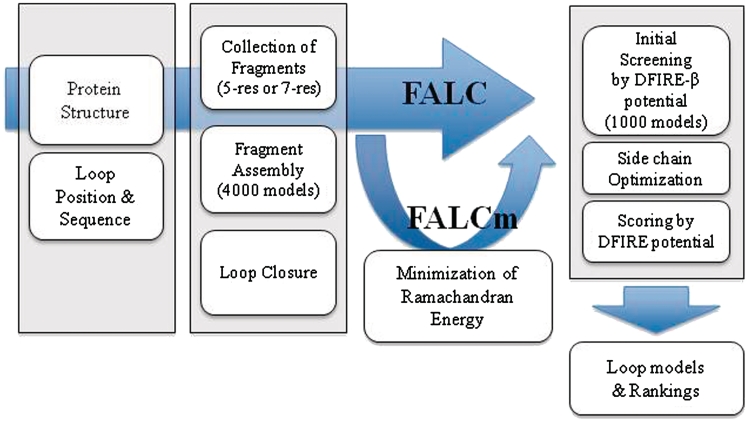
A flowchart of the FALC-Loop modeling procedure is shown in Figure 1. The FALC-Loop server employs the loop modeling method that combines fragment assembly and analytical loop closure developed in Ref. (4).
Figure 1.
A flowchart of the FALC-Loop modeling procedure.
First, 4000 candidate loop structures are generated by fragment assembly. For each residue of target loops, 200 fragment structures of length 5 (for loop length ≤ 5-residue) or length 7 (for loop length > 5-residue) with similar sequence features are collected from the ASTRAL SCOP (version 1.63) structure database (7–9), filtered to maximum pairwise sequence identity 25% (4362 chains, 905 684 residues). The collected fragments are assembled by sequentially adding randomly chosen fragments starting from the N-terminal region of the loop, requiring that the fragments have similar torsion angles at junctions. The average length of the joined segments is about two residues.
Second, the analytical loop closure algorithm (5,6) is applied to fit the candidate structures into the rest of the protein structure by rotating the six backbone torsion angles of randomly chosen three residues. In a variant method called FALCm, an additional step is taken in which an energy devised to enforce the torsion angles to lie within the allowed regions of Ramachandran map is minimized while satisfying the loop closure restraint simultaneously (4). Only the backbone conformations are generated up to this stage.
Third, 1000 backbone-only models are selected from the closed loop candidate structures for each of the model sets generated by the FALC and FALCm methods by scoring with the DFIRE-β potential (4,10). Side chain conformations are then built and optimized for the 2000 models using our in-house version of SCWRL (4). These models are scored by the DFIRE potential, and top-ranked models are reported.
Performance of the method
The FALC method was shown in Ref. (4) to outperform several of the best previous loop sampling methods. For example, it shows better performance in loop sampling than the recently published method SOS (11) when tested on 30 loops [Table I of Ref. (4)]: average of the minimum RMSD from native improves from 1.2 to 0.8 Å and 2.3 to 1.8 Å for 8- and 12-residue loops, respectively. The loop sampling method was also tested on 317 loops and gave better results than RAPPER (12) [Table III of Ref. (4)].
In Ref. (4), the FALC-Loop method provides higher accuracy loop modeling results than RAPPER combined with DFIRE scoring (13) [Table IV of (4)]. The FALC-loop server also shows better performance than the well-known loop modeling server, ModLoop (14), as shown in Table 1, for longer loops of 8- and 12-residues when tested on the 30 loops listed in Table 1 of Ref. (4). (Homologous proteins were removed from the database during fragment library generation for this comparison.) The performance of the FALC-loop method (RMSD = 3.1, 3.4 and 3.8 Å for 10, 11 and 12 residues, respectively) [Table IV of Ref. (4)] is also comparable to those of commercially available programs Prime (Schrödinger, LLC), MODELLER (Accelrys Software, Inc.), ICM (Molsoft, LLC) and Sybyl (Tripos, Inc.), 3–5 Å for 10–12 residues (15), although different benchmark sets were used. However, it may be less accurate than other loop modeling methods that employ more extensive energy optimizations such as ROSETTA (16,17).
Table 1.
The average RMSD of the loop conformations predicted by ModLoop and FALC-Loop (FALC and FALCm) when tested on the 30 loops listed in Table 1 of Ref. (4)
| Loop length (aa) | Average RMSD from native (Å) |
||
|---|---|---|---|
| ModLoop | FALC | FALCm | |
| 4 | 0.66 | 0.87 | 0.93 |
| 8 | 2.46 | 2.34 | 1.87 |
| 12 | 4.48 | 3.13 | 3.07 |
FALC-LOOP WEB SERVER
Hardware and software
The FALC-Loop server runs on a Linux server of a 2.8 GHz Intel Xeon processor that consists of two cores. The web application uses Python and the MySQL database. The loop prediction pipeline is implemented using Python by combining the fragment assembly program implemented in C and the algorithms for loop closure, side chain optimization and DIFRE scoring implemented in Fortran 90. The JMol (http://www.jmol.org) is used for visualization of predicted structures.
Input
The FALC-loop server accepts as input a protein structure and the positions and sequences of one or more loops. The maximum sizes of the protein chain and the loops are set to 1500 and 50 amino acids, respectively, for efficient service. For a protein larger than the maximum size, the user may truncate parts of the protein structure that are away from the loops of interest. Typical computation time is about 3 h for loops of 8–12 residues in protein chains of less than 500 residues.
The protein structure has to be provided in the PDB format. It is expected that the structure file contains coordinates of all residues except for those of loop regions. The server reads the SEQRES and ATOM lines in the PDB file and identifies stretches of the residues with missing ATOM lines as loops. If the PDB file does not contain SEQRES lines, a separate SEQ file must be provided in the FASTA format. After submission of a structure file and an optional sequence file, loops identified by the server are displayed. Once the loops to be modeled are selected, the job is added to the modeling queue. The modeling progress can be checked by following the link for the report page or through the Queue page.
Output
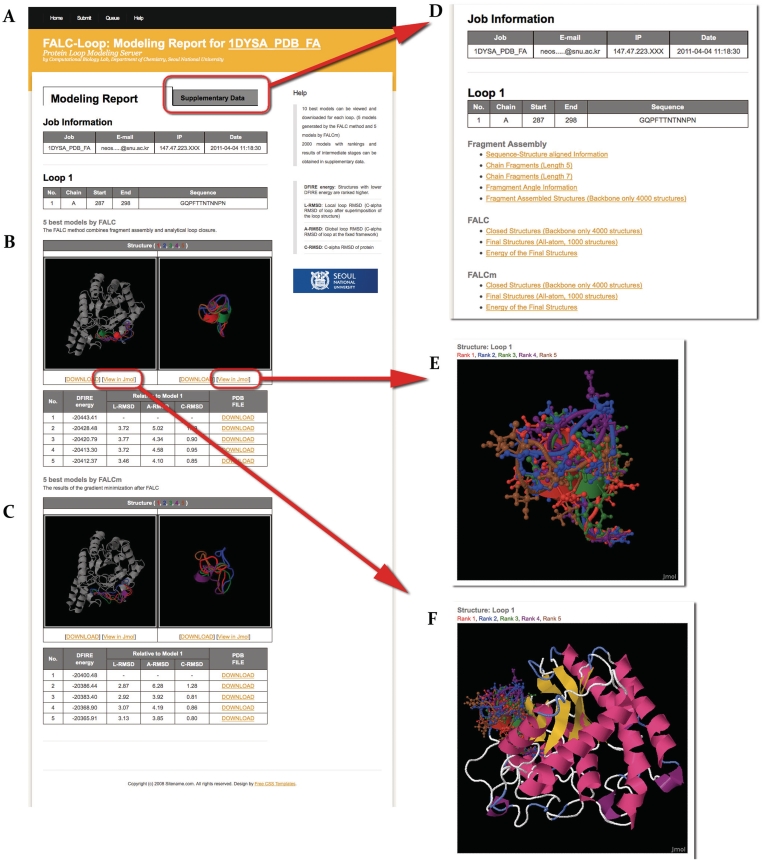
The FALC-Loop output consists of two pages, Modeling Report. On the Modeling Report page (Figure 2A–C), the top five models obtained from each of the methods FALC and FALCm are presented. Static structure images both with and without the protein framework can be viewed on the web page. Structures can also be examined using the Jmol structure viewer by clicking the ‘View in Jmol’ link. The loop structures are colored by the rank of the DFIRE potential. The PDB files used to draw the images can be downloaded from the DOWNLOAD link. Comparison of the DFIRE scores and the following RMSD measures from the first model is summarized in a table: L-RMSD (C-α RMSD of loop after superimposition of loop structures), A-RMSD (C-α RMSD of loop at the fixed framework) and C-RMSD (C-α RMSD of protein structure). The DFIRE scores can be used as a guideline if stabilities of different loop conformations need to be compared, although it is challenging to estimate the model quality from such scores in general. The RMSD measures may be used to get a quick idea on the relative differences of the models. Each loop conformation can also be downloaded from the table.
Figure 2.
FALC-Loop output page. The Modeling Report page shows (A) job information, loop information and five top-ranking loop models obtained by (B) the FALC method and (C) the FALCm method. The static images for the loop structures are shown with and without the framework structure. DFIRE scores and RMSDs from the best model are tabulated. The structures can also be viewed using the Jmol structure viewer with and without the framework (E and F) by clicking the ‘View in Jmol’ link. (D) In the page, fragment libraries, structures from the intermediate stages, as well as the final structures and DFIRE energy scores can be downloaded.
The FALC-Loop server provides additional data in the page (Figure 2D). The ensemble of the 2000 final models and the DFIRE scores can be used for analysis of alternative structures. Other data may be used for further research such as method developments for fragment assembly (fragment libraries), loop closure (fragment-assembled structures) or side chain optimization (closed backbone-only structures).
CONCLUSIONS
The FALC-Loop web server is a protein loop modeling server that employs an efficient ab initio loop modeling method that has aspects of knowledge-based methods such as the use of structure fragments derived from a structure database and scoring by a knowledge-based potential. The server does not require availability of related loops in the structure database for high accuracy prediction unlike the web servers based on database search methods. Therefore, the FALC-Loop server may also be applied to modeling designed loops, loops in multiple states, etc.
FUNDING
National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (2010-0000220 to J.L., 305-20100007 to C.S.); National Institutes of Health (R01 GM 090205-02 to E.A.C.); Center for Marine Natural Products and Drug Discovery (CMDD), one of the MarineBio21 programs funded by the Ministry of Land, Transport, and Maritime Affairs of Korea (to J.K. and H.P.). Funding for open access charge: Seoul National University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hildebrand PW, Goede A, Bauer RA, Gruening B, Ismer J, Michalsky E, Preissner R. SuperLooper-A prediction server for the modeling of loops in globular and membrane proteins. Nucleic Acids Res. 2009;37:W571–W574. doi: 10.1093/nar/gkp338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez-Fuentes N, Zhai J, Fiser A. ArchPRED: a template based loop structure prediction server. Nucleic Acids Res. 2006;34:W173–W176. doi: 10.1093/nar/gkl113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng H-P, Yang A-S. Modeling protein loops with knowledge-based prediction of sequence-structure alignment. Bioinformatics. 2007;23:2836–2842. doi: 10.1093/bioinformatics/btm456. [DOI] [PubMed] [Google Scholar]
- 4.Lee J, Lee D, Park H, Coutsias EA, Seok C. Protein loop modeling by using fragment assembly and analytical loop closure. Proteins. 2010;78:3428–3436. doi: 10.1002/prot.22849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coutsias EA, Seok C, Jacobson MP, Dill K. A kinematic view of loop closure. J. Comput. Chem. 2004;25:510–528. doi: 10.1002/jcc.10416. [DOI] [PubMed] [Google Scholar]
- 6.Coutsias EA, Seok C, Wester MJ, Dill K. Resultants and loop closure. Int. J. Quantum Chem. 2006;106:176–189. [Google Scholar]
- 7.Brenner SE, Koehl P, Levitt M. The ASTRAL compendium for protein structure and sequence analysis. Nucleic Acids Res. 2000;28:254–256. doi: 10.1093/nar/28.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sim J, Kim S-Y, Lee J. Prediction of protein solvent accessibility using fuzzy k-nearest neighbor method. Bioinformatics. 2005;21:2844–2849. doi: 10.1093/bioinformatics/bti423. [DOI] [PubMed] [Google Scholar]
- 9.Sim J, Kim S-Y, Lee J. Fuzzy k-nearest neighbor method for protein secondary structure prediction and its parallel implementation. In: Huang DS, Li K, Irwin GW, editors. Computational Intelligence and Bioinformatics. Heidelberg: Springer Berlin; 2006. pp. 444–453. [Google Scholar]
- 10.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu P, Zhu F, Rassokhin DN, Agrafiotis DK. A self-organizing algorithm for modeling protein loops. PLOS Comput. Biol. 2009;5:e1000478. doi: 10.1371/journal.pcbi.1000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePristo MA, de Bakker PIW, Lovell SC, Blundell TL. Ab initio construction of polypeptide fragments: efficient generation of accurate, representative ensembles. Proteins. 2002;51:41–55. doi: 10.1002/prot.10285. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C, Liu S, Zhou Y. Accurate and efficient loop selections by the DFIRE-based all-atom statistical potential. Protein Sci. 2004;13:391–399. doi: 10.1110/ps.03411904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 15.Rossi KA, Weigelt CA, Nayeem A, Krystek SR., Jr Loopholes and missing links in protein modeling. Protein Sci. 2007;16:1999–2012. doi: 10.1110/ps.072887807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J. Mol. Biol. 2007;373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 17.Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]