Abstract
Although several tools for the analysis of ChIP-seq data have been published recently, there is a growing demand, in particular in the plant research community, for computational resources with which such data can be processed, analyzed, stored, visualized and integrated within a single, user-friendly environment. To accommodate this demand, we have developed PRI-CAT (Plant Research International ChIP-seq analysis tool), a web-based workflow tool for the management and analysis of ChIP-seq experiments. PRI-CAT is currently focused on Arabidopsis, but will be extended with other plant species in the near future. Users can directly submit their sequencing data to PRI-CAT for automated analysis. A QuickLoad server compatible with genome browsers is implemented for the storage and visualization of DNA-binding maps. Submitted datasets and results can be made publicly available through PRI-CAT, a feature that will enable community-based integrative analysis and visualization of ChIP-seq experiments. Secondary analysis of data can be performed with the aid of GALAXY, an external framework for tool and data integration. PRI-CAT is freely available at http://www.ab.wur.nl/pricat. No login is required.
INTRODUCTION
Chromatin immunoprecipitation in combination with high-throughput sequencing (ChIP-seq) enables the detection of in vivo protein–DNA binding events at a genome-wide scale and with unprecedented resolution and statistical power (1). One requirement for ChIP-seq analysis is availability of a complete genome sequence of the organism under study (2). As the number of sequenced plant genomes has increased significantly in the last couple of years (3), the number of species amenable to ChIP-seq experiments has increased accordingly and the new methodology will soon facilitate studies of in vivo protein–DNA interaction across a wide range of plant species.
One major bottleneck in ChIP-seq experiments is the primary analysis of the data, which is time-consuming and computationally expensive. In addition, publicly available software is often command-line based and not user-friendly (4). Primary analysis of ChIP-seq data consists of alignment or ‘mapping’ of the short sequences (reads) to the genome of the species under study, followed by detection of genomic regions with a significant enrichment of mapped reads. Candidate protein–DNA binding events are inferred from this enrichment.
Subsequent to the inference of protein–DNA binding events, analysis of ChIP-seq experiments can be extended to the integration of other types of genomic information, including data on DNA methylation, histone marks, DNA conservation, etc. This allows for a better understanding of transcriptional regulation and function underlying binding events in general. To facilitate data integration, it is important to develop a central resource for the storage of DNA binding maps obtained from ChIP-seq experiments and any other useful genomic data.
In contrast to the relatively large number of web-tools currently available for ChIP-chip analysis (5–8), we are aware of only one freely available web-tool for ChIP-seq analysis, W-ChIPeaks (7). However, W-ChIPeaks functionality is confined to a method for the detection of enriched regions. W-ChIPeak is not able to perform read mapping, which is, as mentioned above, a major computational bottleneck. Furthermore, W-ChIPeaks is focused on human and mouse ChIP-seq experiments and does not include any plant genome.
Here, we present PRI-CAT (Plant Research International ChIP-seq analysis tool), our proposed solution to the growing demand for a user-friendly web-based tool for plant ChIP-seq analysis. PRI-CAT is structured into three modules that reflect our design objectives for its main functionality: (i) primary ChIP-seq analysis; (ii) storage and visualization of DNA binding maps; and (iii) data integration through compatibility with GALAXY (9).
PRIMARY ANALYSIS
PRI-CAT offers a complete solution for the primary analysis of plant ChIP-seq experiments. The methodology implemented has been used successfully in our analyses of MADS transcription factor (TF) binding sites (1,10) and is described in detail in our published protocol (11). In brief, sets of short sequences uploaded in FASTA or FASTQ format are mapped independently against the reference genome of a selected species with the program SOAPv2 (12). Currently only Arabidopsis thaliana is available in PRI-CAT. Reads that map either to more than one genomic location or to the mitochondrial or plastid genome are not considered for further analysis. Subsequently, the R/Bioconductor (13) package CSAR (http://bioconductor.org/help/bioc-views/release/bioc/html/CSAR.html) is used to detect regions with a significant enrichment in mapped reads. Briefly, CSAR virtually extends the mapped reads to match the average DNA fragment size reported by the user. Experimental (IP) and control samples are normalized to obtain the same read-coverage distribution. A ratio-based score is used to test significance, using a permutation test to obtain false discovery rate (FDR) control. CSAR was designed to be robust against PCR-artifacts. PCR-artifacts in ChIP-seq can arise from differential amplification of DNA fragments (14,15). Over-amplification increases the number of reads that map to particular genomic locations. Such reads are revealed as duplicated sequences and when left uncorrected, will increase the probability of false discovery of binding sites. CSAR has been shown to be specially robust against PCR-artifacts (16). Cairns et al. (17) presented a new peak-calling algorithm and compared its performance with existing algorithms in their Supplementary Material section. In the particular case that they studied, CSAR showed the lowest number of false positives.
PRI-CAT uses the default settings of CSAR and reports score values per nucleotide position which are based on the ratio between the number of mapped reads in experimental and control data sets. These ratios allow a more fair comparison across experiments than commonly used ‘Poisson-based scores’, because they are less dependent on the different number of reads sequenced in each experiment.
Although this somewhat reduces the flexibility offered to the user, the fact that data sets are analyzed with the same method makes the comparison and integration of different experiments more robust, which is one main objective of the PRI-CAT web tool.
To safeguard efficient use of internet bandwidth, PRI-CAT only allows upload of FASTA or FASTAQ compressed data files. For each submitted data set several quality controls are reported and can be visualized graphically. The total number of mapped reads and the number of mapped reads with non-duplicated sequences are provided to indicate possible problems caused by PCR over-amplification. In our experience, libraries with <40% of mapped reads with non-duplicated sequences may indicate biased amplification, and users should be cautious with the interpretation of results from such experiments. A more general quality control provided is the distribution of regions showing an enrichment in mapped reads relative to gene positions. The binding event of a specific type of protein may be expected at a particular type of location, for example TFs are expected to bind in promoter regions, while the histone mark H3K27me3 seems to be usually localized within gene sequences in A. thaliana (18). Such prior knowledge can help to detect problems in particular data sets.
Users can choose to make their data sets publically available through PRI-CAT. In this way, other users can reanalyze and integrate prior data sets in the context of their own experiments. For example, control libraries are essential for proper ChIP-seq analysis and because these are often not specific for the protein studied, in a first approach, users can reuse a publicly available control data set from PRI-CAT for their own analysis, provided that the experimental conditions used are similar. These datasets will be not deleted from our server. The default option of PRI-CAT is that uploaded data sets are not made publicly available. Such data sets are however deleted from our server after one month.
The output from primary analysis of ChIP-seq data with PRI-CAT consists of (i) a list of genomic regions that show a significant enrichment in mapped reads at different FDR control levels; (ii) a list of genes at a maximum distance (3-kb upstream and 1-kb downstream of the gene) of the inferred binding events, as potential target genes of the TF; (iii) a DNA binding map in Wiggle format representing the level of enrichment per nucleotide position. The map can be visualized in any Wiggle-compatible genome browser.
PRI-CAT is running in a queue system on a high-end cluster with a total of 56 dedicated CPU cores and 88 GB total RAM with one terabyte of storage space. An additional server with 256 GB RAM is available as backup to guarantee availability of enough resources for the most demanding jobs. The mapping process of a typical data set (4 million reads) takes 15 min, and the CSAR analysis around 20 min. Therefore, our server can handle an average of 42 jobs consisting of two IP data sets compared to two control data sets per hour.
STORAGE AND VISUALIZATION
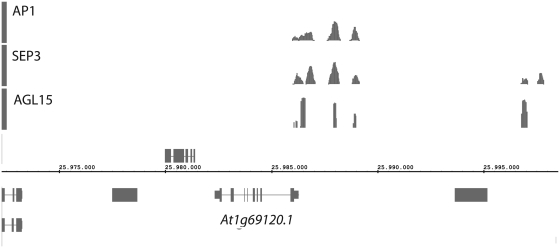
The high resolution of DNA binding maps obtained with ChIP-seq experiments makes them well suited for visualization in a genome browser. Simultaneous visualization of multiple binding maps can provide very useful information (Figure 1). All publically available DNA binding maps generated by PRI-CAT are therefore transferred to our QuickLoad server, which can be connected to a compatible genome browser for visualization. We advise the use of the Integrated Genome Browser (19) because it allows for secondary analyses of data, for example searching for DNA motifs. All PRI-CAT maps have a unique internal ID for future reference.
Figure 1.
DNA binding maps of three MADS-box TFs in the promoter region of the AP1 gene (indicated below, At1g69120). From top to bottom: AP1, SEP3 and AGL15. The visualization of multiple binding maps can provide useful information; in the current display, for example, it appears that the binding sites of the three TFs map to the same genomic positions. This can be explained by the fact that they form dimers with each other (1).
In order to take benefit of previous experiments, we have re-analyzed most of the plant ChIP-seq (seven data sets) and ChIP-chip (33 data sets) experiments with a control data set available at the moment of publication. In addition, we generated in silico maps representing DNA conservation among A. thaliana ecotypes and closely related species (Supplementary Table S1). ChIP-seq experiments were re-analyzed with PRI-CAT. For ChIP-chip analysis, probe sequences were remapped to the TAIR9 Arabidopsis genome with the Starr package (20). Only probes that mapped to unique locations were retained. Subsequently, CisGenome (21) was used to detect potential binding site regions, using the hidden Markov model to combine intensities of neighboring probes. Alignments needed to calculate DNA conservation between A. thaliana with A. lyrata and Carica papaya were downloaded from the VISTA web server (http://genome.lbl.gov/vista); the percentage of identical A. thaliana nucleotides in the alignment is reported in windows of 25-bp. Alignments of 80 Arabidopsis ecotype genomes were downloaded from the 1001 genome project (http://www.1001genomes.org/), and the percentages of ecotypes with a different nucleotide or insertion/deletion relative to the Col-0 ecotype are reported per nucleotide position. Only positions where the sequences of at least 40 ecotypes are known were considered. To be conservative, nucleotide positions with <10% of ecotypes showing differences were discarded.
DATA INTEGRATION WITH GALAXY
GALAXY is an external framework for tool and data integration that focuses on the analysis of genomic data (9). New tools for it are being developed by a large and growing bioinformatics community. To interactively download data sets from our QuickLoad server to GALAXY, users need to install the tool PRICAT4GALAXY (available at http://www.ab.wur.nl/pricat) in their local GALAXY instance. Data sets currently available consist of lists of DNA binding events in BED format for each public data set stored in PRI-CAT (Supplementary Table S1). With Galaxy, users can easily combine several data sets and discover new patterns in their data, for example the co-localization of binding events from different proteins, which may indicate possible protein–protein interactions.
CONCLUSIONS
PRI-CAT was developed with the aim to evolve into a central hub for the analysis, storage and visualization of plant ChIP-seq experiments. The first step towards this goal is achieved with the current release of the tools needed for complete primary analysis of ChIP-seq experiments in A. thaliana, the most important model organism in plants. Other species will be implemented in future versions. Secondary analysis of ChIP-seq data is facilitated through compatibility of PRI-CAT with GALAXY. GALAXY offers a graphical interface for the analysis and integration of different genomic data sets, including the ones available in PRI-CAT. More than 35 DNA binding maps are now available in PRI-CAT (Supplementary Table S1), including maps for particular TFs binding events, histone marks, DNA methylation and DNA conservation. All maps can be easily visualized with a genome browser. Integration of the available data sets with newly submitted ChIP-seq data sets will greatly help in obtaining better insight in the mechanisms of protein–DNA binding and gene regulation.
SUPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Netherlands Genomics Initiative’s Horizon (grant 93519015 to A.D.J.v.D.). Funding for open access charge: The Netherlands Genomics Initiative’s Horizon.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Jan van Haarst, Henri van de Geest and Bas te Lintel Hekkert for technical support and Dr Hailiang Mei for advice regarding GALAXY.
REFERENCES
- 1.Kaufmann K, Muino JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009;7:e1000090. doi: 10.1371/journal.pbio.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buisine N, Sachs L. Impact of genome assembly status on ChIP-Seq and ChIP-PET data mapping. BMC Res. Notes. 2009;2:257. doi: 10.1186/1756-0500-2-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feuillet C, Leach JE, Rogers J, Schnable PS, Eversole K. Crop genome sequencing: lessons and rationales. Trends Plant Sci. 2011;16:77–88. doi: 10.1016/j.tplants.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cesaroni M, Cittaro D, Brozzi A, Pelicci PG, Luzi L. CARPET: a web-based package for the analysis of ChIP-chip and expression tiling data. Bioinformatics (Oxford, England) 2008;24:2918–2920. doi: 10.1093/bioinformatics/btn542. [DOI] [PubMed] [Google Scholar]
- 6.Gibbons FD, Proft M, Struhl K, Roth FP. Chipper: discovering transcription-factor targets from chromatin immunoprecipitation microarrays using variance stabilization. Genome Biol. 2005;6:R96. doi: 10.1186/gb-2005-6-11-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan X, Bonneville R, Apostolos J, Wu W, Jin VX. W-ChIPeaks: a comprehensive web application tool for processing ChIP-chip and ChIP-seq data. Bioinformatics (Oxford, England) 2011;27:428–430. doi: 10.1093/bioinformatics/btq669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang ZD, Rozowsky J, Lam HY, Du J, Snyder M, Gerstein M. Tilescope: online analysis pipeline for high-density tiling microarray data. Genome Biol. 2007;8:R81. doi: 10.1186/gb-2007-8-5-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufmann K, Wellmer F, Muino JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueno F, Krajewski P, Meyerowitz EM, et al. Orchestration of floral initiation by APETALA1. Science (New York, N.Y.) 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- 11.Kaufmann K, Muino JM, Osteras M, Farinelli L, Krajewski P, Angenent GC. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP) Nat. Protoc. 2010;5:457–472. doi: 10.1038/nprot.2009.244. [DOI] [PubMed] [Google Scholar]
- 12.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics (Oxford, England) 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 13.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat. Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G + C)-biased genomes. Nat. Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muiño JM, Kaufmann K, van Ham RCHJ, Angenent GC, Krajewski P. ChIP-seq Analysis in R (CSAR): An R package for the statistical detection of protein-bound genomic regions. Plant Methods. 2011;7:11. doi: 10.1186/1746-4811-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cairns J, Spyrou C, Stark R, Smith ML, Lynch AG, Tavare S. BayesPeak–an R package for analysing ChIP-seq data. Bioinformatics (Oxford, England) 2011;27:713–714. doi: 10.1093/bioinformatics/btq685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics (Oxford, England) 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zacher B, Kuan PF, Tresch A. Starr: Simple Tiling ARRay analysis of Affymetrix ChIP-chip data. BMC Bioinformatics. 2010;11:194. doi: 10.1186/1471-2105-11-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]