Abstract
In archaea and bacteria, the late steps in adenosylcobalamin (AdoCbl) biosynthesis are collectively known as the nucleotide loop assembly (NLA) pathway. In the archaeal and bacterial NLA pathways, two different guanylyltransferases catalyze the activation of the corrinoid. Structural and functional studies of the bifunctional bacterial guanylyltransferase that catalyze both ATP dependent corrinoid phosphorylation and GTP dependent guanylylation are available, but similar studies of the monofunctional archaeal enzyme that catalyzes only GTP dependent guanylylation are not. Herein, the three-dimensional crystal structure of the guanylyltransferase (CobY) enzyme from the archaeon Methanocaldococcus jannaschii (MjCobY)1 in complex with GTP is reported. The model identifies the location of the active site. An extensive mutational analysis was performed, and the functionality of the variant proteins was assessed in vivo and in vitro. Substitutions of residues Gly8, Gly153, or Asn177 resulted in ≥94% loss of catalytic activity, thus variant proteins failed to support AdoCbl synthesis in vivo. Results from isothermal titration calorimetry experiments showed that MjCobYG153D had 10-fold higher affinity for GTP than MjCobTWT, but failed to bind the corrinoid substrate. Results from Western blot analyses suggested that the above-mentioned substitutions render the protein unstable and prone to degradation; possible explanations for the observed instability of the variants are discussed within the framework of the three-dimensional crystal structure of MjCobYG153D in complex with GTP. The fold of MjCobY is strikingly similar to that of the N-terminal domain of Mycobacterium tuberculosis GlmU (MtbGlmU), a bifunctional acetyltransferase/uridyltransferase that catalyses the formation of uridine-diphosphate-N-acetylglucosamine (UDP-GlcNAc).
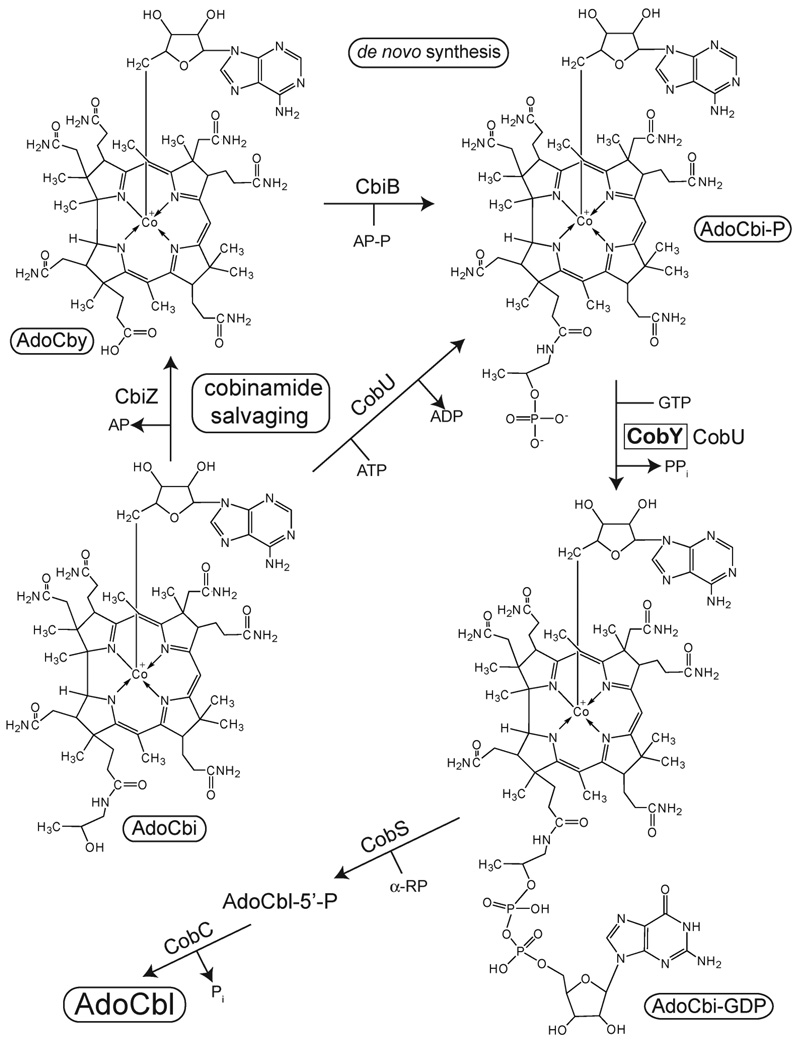
Many prokaryotes and eukaryotes use adenosylcobalamin (AdoCbl, coenzyme B12, Figure 1) in diverse catabolic and anabolic processes, such as the generation of carbon and energy from fatty acids and other small molecules, and the biosynthesis of lipids, methionine, or deoxyribonucleotides (1–5). Although the use of AdoCbl is broadly distributed in all domains of life, its biosynthesis is restricted to some bacteria and archaea (6). Some AdoCbl producers assemble AdoCbl de novo, while others can only synthesize it if incomplete corrinoids (e.g., cobinamide (Cbi), cobyric acid (Cby), Figure 1) are present in the environment (a process known as corrinoid salvaging) (6).
Figure 1.
De novo synthesis of the nucleotide loop, and cobinamide salvaging in bacteria and archaea
Bacterial and archaeal AdoCbl producers use different strategies to salvage cobinamide (Figure 1). Bacteria use a bifunctional enzyme (ATP:AdoCbi kinase, EC 2.7.1.156; GTP:AdoCbi-P guanylyltransferase, EC 2.7.7.62) to convert AdoCbi into AdoCbi-GDP via an AdoCbi-P intermediate (7–11). Two additional reactions convert AdoCbi-GDP to AdoCbl. The first one yields AdoCbl-5’-P (catalyzed by AdoCbl-5’-P synthase, EC 2.7.8.26), and the second one yields AdoCbl (catalyzed by AdoCbl-5’-P phosphatase, EC 3.1.3.73) (12–15). In contrast, archaea use an amidohydrolase (EC 3.5.1.90) to convert AdoCbi into AdoCby (16– 19), which is condensed with 1-aminopropan-2-yl-phosphate yielding AdoCbi-P in a single step (catalyzed by AdoCbi-P synthetase EC 4.1.1.81) (20). As in bacteria, the next step in the archaeal pathway also involves a GTP:AdoCbi-P guanylyltransferase, but unlike the bacterial enzyme, the archaeal enzyme lacks AdoCbi kinase activity (21). Results from early studies of the gene encoding the archaeal GTP:AdoCbi-P guanylyltransferase (CobY) established that cobY is a non-orthologous replacement of the gene encoding the bifunctional enzyme found in bacteria (21, 22). More recently, the initial biochemical characterization of the CobY enzyme from the hyperthermophilic methanogenic archaeon Methanocaldoccocus jannaschii was reported (23). These studies revealed that MjCobY binds GTP before AdoCbi-P, that the enzyme does not bind AdoCbi-P in the absence of GTP, that the upper ligand is important for binding the corrinoid substrate, and that the enzyme would not bind GDP, nor the GTP analogues guanosine-5’-[β,γ-imido]-triphosphate (GMP-PNP) and guanylyl-5’-[β,γ-methylene]-diphosphonate (GMP-PCP). There is no sequence similarity between the bacterial and archaeal enzymes. The bacterial enzymes show a clear sequence signature for a classic P-loop, whereas the archaeal enzymes lack this motif, which suggests a different structural architecture in both classes of protein. Herein we report the three-dimensional crystal structure of MjCobY in complex with GTP at 2.8-Å resolution. Results from bioinformatics studies revealed striking similarities in the fold of MjCobY to the N-terminal domain of Mycobacterium tuberculosis GlmU (MtbGlmU), a bifunctional acetyltransferase/uridyltransferase, which catalyses the formation of uridine-diphosphate-N-acetylglucosamine (UDP-GlcNAc).
EXPERIMENTAL PROCEDURES
Microbiological Techniques. Bacteria, Culture Media, and Growth Conditions
Bacterial strains and plasmids used in these studies are listed in Table S1 (supporting information). Salmonella enterica strains were cultured in nutrient broth (NB; Difco); E. coli strains were cultured in lysogeny broth (LB) (24, 25). No-carbon essential (NCE) medium (26, 27) supplemented with glycerol (30 mM) or ethanolamine (30 mM,) MgSO4 (1 mM,) and trace minerals (28) was used to grow cells under nutrient-defined conditions. Plasmids were maintained by the addition of ampicillin and/or chloramphenicol to rich medium at 100 µg/mL and 20 µg/mL, respectively, and to minimal medium at 25 µg/mL and 5 µg/mL, respectively. When added, the concentration of corrinoid [cyanocobyric acid (CNCby), dicyanocobinamide [(CN)2Cbi], or cyanocobalamin (CNCbl)] in the medium was 15 nM. (CN)Cby was a gift from Paul Renz (Institut für Biologische Chemie und Ernahrungswissenschaft, Universität-Hohenheim, Stuttgart, Germany); (CN)2Cbi and CNCbl were purchased from Sigma.
Growth of Strains for Western Blot Analysis
Two mL of nutrient broth (NB) were inoculated with isolated colonies of strains tested and grown at 37°C with shaking for approximately 16 h. The overnight NB cultures were diluted 1:50 into 5 mL of NCE minimal medium, which was supplemented with glycerol (22 mM), MgSO4 (1 mM), trace minerals, NH4Cl (30 mM), L-methionine (300 µM), and cyanocobyric acid (CNCby, 15 nM). Cultures were incubated for approximately 20 h at 37°C with shaking. The overnight NCE cultures were normalized by calculating the volume of each culture required to make 1 mL of culture with a final OD600 of ~ 3.0. The appropriate volume of each overnight NCE culture was centrifuged, and the cell pellet was resuspended in 50 µL of Tris buffer (100 mM, pH 7.9), containing BugBuster™ Protein Extraction Reagent (Novagen.) The suspension was incubated at room temperature for 15 min and centrifuged at 18,000 × g in an Eppendorf microfuge for 2 min. The soluble portion was placed in a thin-walled PCR tube. Two hundred µL of cracking buffer [Tris-HCl buffer (60 mM, pH 6.8) + SDS (1%, w/v) + glycerol (10%, v/v), bromophenol blue (0.1%, w/v) + DTT (200mM)] were added to the tube, and the solution was heated to 99°C for 5 min. Ten µL of the heated solution were loaded into a PAGEr® pre-cast 15% Tris-glycine gel (Lonza). Purified, wild-type MjCobY protein was loaded as positive control.
Genetic and Recombinant DNA techniques. Cloning, Mutagenesis, and In Vivo Assessment of CobY Function
Method A. M. jannaschii cobY from plasmid pCobY14 (23) was mutated using the QuikChange XL kit (Stratagene) according to the manufacturer’s instructions. Method B. Multiple-mutant CobY alleles L38K/K41D, L38D/K41D, and I144D/F146D/Y163R were constructed in single reactions in which the mutagenic primers were combined with the plasmid pCobY14. Primer sequences are shown in Table S2 (supplemental information). All mutations were verified by DNA sequencing using BigDye® protocols (ABI PRISM). Reaction mixtures were resolved by the UW-Madison Biotechnology Center. The pT7-7 (29) plasmid carrying the wild-type cobY allele was used as a template for polymerase chain reaction (PCR)-based site directed mutagenesis. The functionality of CobY variant proteins was assessed in vivo using S. enterica strain JE8268 (Δcob ΔycfN, Table S1), which lacks NTP:adenosylcobinamide-phosphate nucleotidyl transferase activity (30).
Biochemical Techniques. Expression and Purification of MjCobY Variants
MjCobY proteins were overproduced and purified as described (23). Briefly, wild type and mutant cobY alleles were overexpressed in E. coli strain BL21-CodonPlus (DE3)-RIL after induction with isopropyl-β-D-1-thiogalactoside (IPTG). Cells were lysed by sonication, and recombinant proteins were purified by thermal denaturation (15 min at 75°C), ammonium sulfate precipitation, hydrophobic interaction chromatography, anion exchange chromatography, and size-exclusion chromatography prior to flash freezing into liquid N2, and storage at −80°C (30).
Expression and Purification of Selenomethionine-CobY G153D Protein
A selenomethionine (L-Se-Met) derivative of the CobYG153D variant protein was overproduced in M9 medium using metabolic inhibition (31). A 500-mL starter culture was used to inoculate 15.5 L of M9 minimal medium supplemented with ampicillin (100 µg/mL) and chloramphenicol (20 µg/mL). Cells were grown in a 16-L (working volume) stainless steel bioreactor at 37°C, with agitation (300 RPM), and airflow (5 SLPM). After 16 h of growth, the culture was supplemented with an amino acid cocktail containing L-Se-Met (50 mg/L). Thirty minutes after the addition of L-Se-Met, expression of allele cobY1381 (encodes CobYG153D) was induced by the addition of IPTG to a final concentration of 0.5 mM. The culture was grown for an additional 12 h at 37°C, and cells were harvested by centrifugation and re-suspended in 300 mL of HEPES buffer (50 mM, pH 7.5) containing MgCl2 (10 mM), DTT (5 mM), lysozyme (0.2 mg/mL), and DNase I (0.2 mg/mL). Cells were frozen and stored at −80°C. Subsequent extraction and purification steps were performed as described elsewhere without modifications (23).
Synthesis of AdoCbi and AdoCbi-P
AdoCbi-P, the corrinoid substrate of CobY is not commercially available and was synthesized from AdoCbi, which was also synthesized in-house. Protocols for the enzymic synthesis and isolation of AdoCbi and AdoCbi-P have been described and were used without modifications (30, 32). Syntheses and purification steps were performed under red light to minimize cleavage of the Co-C bond. AdoCbi-P was resolved from contaminants by reverse phase high-performance liquid chromatography (RP-HPLC) using a System Gold HPLC system (Beckman/Coulter) equipped with an Alltima HP C18 AQ 5-µm column (150 by 4.6 mm) (Alltech). The System II mobile phase described by Blanche et al for the separation of corrinoids was used as described, except that cyanide was omitted (33).
In Vitro Synthesis of AdoCbi-GDP by MjCobY
In vitro conditions for assaying the GTP:AdoCbi-P guanylyltransferase activity of MjCobY were recently described (23). Briefly, reaction mixtures contained 2-amino-2-hydroxymethyl-propane-1,3-diol (Tris-HCl buffer; 50 mM, pH 7.9), GTP (2 mM), NaCl (50 mM), MgCl2 (10 mM), dithiothreitol (DTT, 10 mM), MjCobY protein (~2 pmol), and AdoCbi-P (200 µM) in a final volume of 40 µL, and were incubated at 37°C. Reaction mixtures were incubated under dim, red light for 15 min, after which corrinoids were converted to their cyano forms by the addition of 3 µL of KCN (100 mM), heating at 80 °C for 10 min, and irradiation with a 60 W incandescent light at a distance of 6 cm for 15 min. Cyanocorrinoids were resolved by RP-HPLC (see above). The column was equilibrated with a buffer system of 77%A:23% B (see below). A linear gradient (~14 min, 1 mL min−1) was applied until the composition of the buffer system was 67% A:33% B. The composition of buffers were: buffer A [100 mM potassium phosphate buffer (pH 6.5) + KCN (10 mM)]; buffer B [100 mM potassium phosphate buffer (pH 8.0) + 10 mM KCN]:CH3CN (1:1). Elution of cyanocorrinoids from the column was monitored at 367 nm with a photodiode array detector. (CN)2Cbi was used as a standard to establish the limit of detection of photodiode array detector; the response was linear between 1 pmol and 3 nmol (r2 = 0.9984).
Assessment of Protein Purity
Protein concentration was determined by the Bradford method (34) using a kit from Bio-Rad Labs. To assess protein purity, proteins were first resolved by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (35), and stained with Coomassie brilliant blue (36). Protein purity was established by densitometry using a Fotodyne imaging system (Foto/Analyst version 5.00 software, Fotodyne, Inc) for image acquisition, and TotalLab software (version 2005, Nonlinear Dynamics) for analysis.
Generation of Rabbit Polyclonal Antibodies Against the MjCobY Enzyme
Polyclonal antibodies were elicited by subcutaneous injection of purified MjCobY protein (23) into a New Zealand White (HsdOkd:NZW) rabbit (Harlan Bioproducts for Science, Inc).
Pre-clearing Rabbit α-MjCobY Antiserum
Fifty mL of lysogeny broth were inoculated with Salmonella enterica strain JE8268 (ΔcobU ΔycfN, Table S1), and incubated at 37°C with shaking until the culture reached OD650 ~ 1.0. Cells were harvested by centrifugation at 4°C at 10,000 × g for 15 min using a JA-25.50 rotor in an Avanti J-25I Beckman/Coulter refrigerated centrifuge. The cell pellet was resuspended in 1 mL of Tris-HCl buffer (50 mM, pH 7.5) containing 2% (w/v) SDS, boiled for 5 min, and immediately placed on ice. Nine mL of Tris-HCl buffer (50 mM, pH 7.5) were added to the cooled cell suspension. One hundred µL of rabbit α-MjCobY antiserum were added, and the cell suspension was incubated at 4°C for 24–36 h. The cell suspension was centrifuged at 10,000 × g for 15 min, and the supernatant (pre-cleared α-MjCobY antiserum) was collected. Pre-cleared α-MjCobY antiserum was stored at 4°C for up to 2 weeks.
Western Blot Analysis
After electrophoresis at 125V for 45 min, proteins were transferred from the SDS-PA gel to polyvinylidene fluoride (PVDF) membrane using the iBlot™ Dry Blotting System (Invitrogen) for 7 min at a Power Level 3 setting. α-MjCobY Western blots were performed using protocols available online (http://www.millipore.com/catalogue/module.do;jsessionid=1857AE2A539F78DCD3F93AE91E50ABD6?id=C3117 ). These protocols were performed with a 1:100 dilution of pre-cleared rabbit a-MjCobY antibody (total 1:10,000 dilution of rabbit serum) as primary antibody, and a 1:25,000 dilution of goat a-rabbit immunoglobulin G (IgG, heavy and light chains) conjugated to calf intestinal alkaline phosphatase (Pierce) as secondary antibody. Signal was detected using nitro-blue-tetrazolium chloride and 5-bromo-4-chloro-3'-indolyphosphate p-toluidine salt (NBT-BCIP) as described (37). An α-MjCobY standard curve was obtained to determine the limit of detection of MjCobY by Western blot analysis; as little as 32 pg of MjCobY were reliably detected when the blot was developed with NBT-BCIP. Alternatively, a 1:250,000 dilution of horseradish peroxidase-conjugated donkey α-rabbit antibody (Pierce) was used as secondary antibody. In this case, signal was detected using the SuperSignal West Femto Maximum Sensitivity Substrate (Pierce) according to the manufacturer’s instructions on a computer-controlled Typhoon Trio phosphorimager (GE Healthcare) equipped with ImageQuant v.5.2 imaging software (Molecular Dynamics.)
Isothermal Titration Calorimetry (ITC)
MjCobYG153D and MjCobYG8D proteins were dialyzed against Tris-HCl buffer, pH 8.0 (50 mM, 4 °C), containing NaCl (50 mM), MgCl2 (10 mM), and tris(2-carboxyethyl)phosphine hydrochloride (4 mM); GTP was dissolved in spent dialysis buffer. ITC experiments were conducted at 27 °C and consisted of 27 10-µL injections of a 1 mM stock of ligand into a 1.45-mL sample cell containing an 18 µM solution of MjCobYG153D or MjCobYG8D dimers. Injections were made over a period of 20 s with a 3 min interval between injections. The sample cell was stirred at 307 RPM. Data were acquired on a VP-ITC microcalorimeter (Microcal, Northhampton, MA) and analyzed with the ORIGIN v.7.0383 software provided with the microcalorimeter.
Crystallization, Data Collection, and Structure Determination of the MjCobYG153D Protein
Crystallization conditions were surveyed at 25°C and 4°C by vapor diffusion with a 144-condition sparse matrix screen developed in the Rayment laboratory. Crystals of both the native and Se-Met substituted proteins MjCobYG153D protein in complex with GTP were grown by mixing equal volumes of a solution containing 10 mg/mL MjCobYG153D, GTP (2.5 mM), Tris (10 mM, pH 8.0), NaCl (50 mM) with reservoir solution containing 60% (w/v) 2-methyl-2,4-pentanediol and 2-(N-morpholino)ethanesulfonic acid buffer (100 mM, pH 6.0). The mixture was centrifuged at 16,000 × g for 5 min to remove nuclei. Four µL of the mixture were suspended over the reservoir solution in Linbro plates at 4°C. The crystallization experiments were nucleated after 24 h with a fine cat whisker from crushed crystals obtained from the earlier crystallization screens. Crystals appeared 24 −36 h after nucleation and continued to grow for an additional 7 days. The crystals were flash frozen in liquid N2 directly from the crystallization drop. The crystals belong to the space group P31 with 4 subunits in the asymmetric unit. X-ray data were collected at the Advanced Photon Source in Argonne, IL utilizing the Structural Biology Center beamline 19BM. Diffraction data were integrated and scaled with HKL3000 (38). Data collection statistics are summarized in Table 1. The structure of the Se-Met substituted protein is reported since these crystals diffracted better than the native protein.
Table 1.
Data Collection and Refinement Statistics
| Data collection | |
| space group | P31 |
| unit-cell parameters (Å,°) | 63.6, 63.6, 222.5 |
| Wavelength (Å) | 0.9791 |
| resolution range (Å) | 50.0–2.8 (2.85–2.80)a |
| reflections: measured | 177049 |
| reflections: unique | 24528 |
| redundancy | 7.2 (7.3) |
| completeness (%) | 98.8 (98.6) |
| average I/ σ | 31.6 (3.75) |
| Rmerge (%) | 9.9 (57.5) |
| Refinement | |
| Rcryst | 0.231 (0.270) |
| Rfree | 0.276 (0.291) |
| no. protein atoms | 5642 |
| no. of ligand atoms | 128 |
| no. water molecules | 108 |
| average B factors (Å2) | 46.0 |
| Ramachandran (%) | |
| most favored | 96.4 |
| allowed | 2.7 |
| Disallowed | 0.9 |
| rms deviations | |
| bond lengths (Å) | 0.013 |
| bond angles (°) | 1.70 |
| Chiral |
Values in parentheses are those for the highest-resolution shell.
Rmerge = Σ|I(hkl) – I| ~1/Σ|I(hkl)|, where the average intensity I is taken over all symmetry equivalent measurements and I(hkl) is the measured intensity for a given reflection.
Rfactor = Σ|F(obs) – F(calc)| / Σ | F(obs)|, where Rwork refers to the Rfactor for the data utilized in the refinement and Rfree refers to the Rfactor for 5% of the data that were excluded from the refinement.
Structure Determination and Refinement
The structure of the MjCobYG153D protein was solved by single wavelength SAD phasing (λpeak). The program SOLVE was used to locate the selenium atoms and to generate initial phases (39). Solvent flattening and molecular averaging with RESOLVE resulted in an interpretable electron density at 2.8 Å resolution (40). The final model was generated with alternate cycles of manual model building and least squares refinement with the programs COOT (41) and Refmac (42). Refinement statistics are presented in Table 1.
RESULTS AND DISCUSSION
MjCobYG153D variant Monomer Structure
An extensive mutational analysis of the conserved residues in CobY was initiated prior to the structural determination. At that time, efforts to crystallize the wild-type protein were unsuccessful, however, the on-going mutational analysis lead to the identification of the CobYG153D variant as a potential target protein because of its low specific activity. For the sake of clarity, the crystal structure of MjCobYG153D is described first, so that the effects of the mutations can be discussed in the context of a three-dimensional model.
The structure of the MjCobYG153D variant in complex with GTP has been determined at 2.8-Å resolution. The protein crystallized in the hexagonal space group P31 with 4 chains in the asymmetric unit. The electron density is similar for all chains, and is well defined except for a stretch from Asp74 - Ile81, which is mostly disordered in all chains. As discussed below, this stretch is almost certainly involved in binding to the corrinoid and most likely undergoes a disorder-order transition during the enzymatic cycle. The four chains are very similar. The root mean square (rms) differences between subunit A and the three other subunits B, C, and D are 0.50, 0.60, and 0.68 Å respectively.
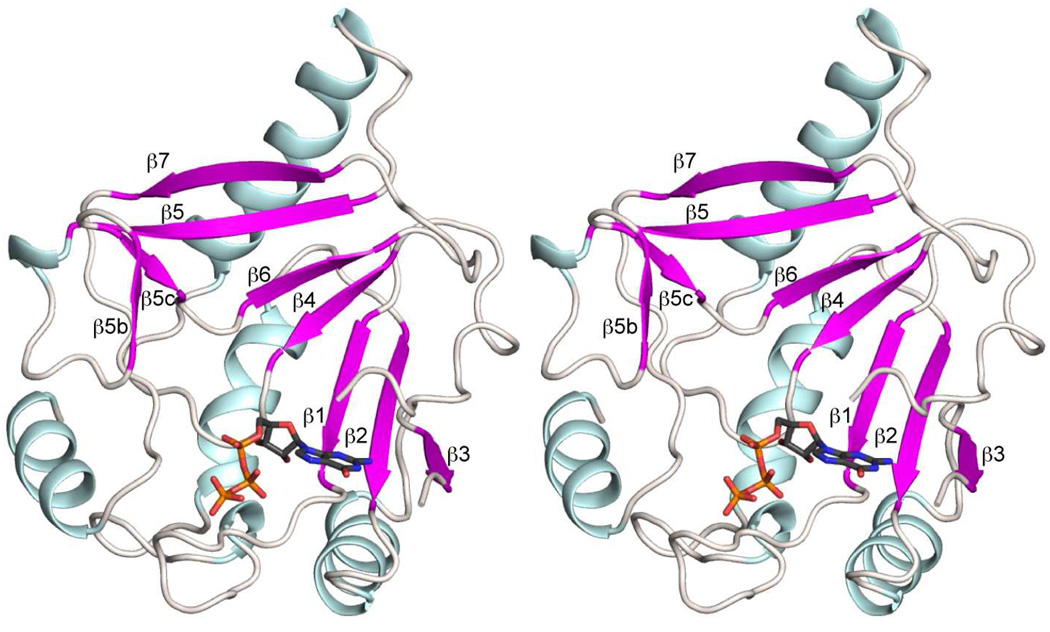
MjCobYG153D monomer consists of a single globular domain with one GTP molecule bound per subunit (Figure 2). The protein fold contains six α-helices and nine β-strands. Seven of the β-strands (β1–5, β8, β9) combine to form a large mixed β-sheet, which underlies the entire structural motif. The α-helices lie on the outside convex face of the sheet. This leaves a substantial bowl-shaped depression that is closed off in part by the loops that connect the antiparallel strands. This depression contains the nucleotide at its base and is presumably the binding site for the corrinoid. The α-helices do not make contact with the nucleotide.
Figure 2. Stereoview of a cartoon representation of CobYG153D.
The dominant feature of the protein fold is a large, mixed β-sheet that underlies the GTP and putative corrinoid binding sites. This figure corresponds to subunit D in the coordinate file. Figures 2 – 7 were prepared with Pymol (49).
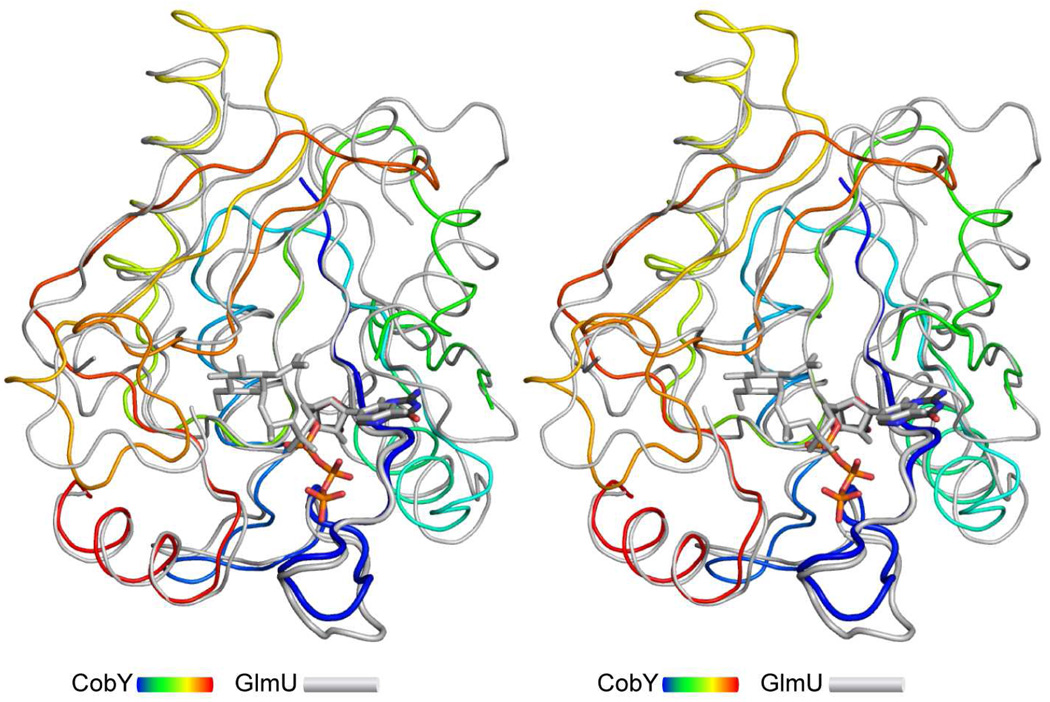
A search for structurally related proteins with the DALI server shows that MjCobY is most similar to the N-terminal domain of Mycobacterium tuberculosis GlmU (MtbGlmU), a bifunctional acetyltransferase/uridyltransferase that catalyses the formation of uridine-diphosphate-N-acetylglucosamine (UDP-GlcNAc) from glucosamine-1-phosphate (GlcN-1-P), acetyl-coenzyme A (AcCoA), and UTP (43–45). MjCobY thus belongs to the UDP-glucose pyrophosphorylase family of protein structures, which themselves exhibit the nucleotide diphosphate sugar transferase fold. The rms difference between MjCobY and MtbGlmU [RCSB 3DJ4, (44)] is only 1.84 Å for 144 structurally equivalent amino acids, which is remarkable considering the lack of significant sequence similarity except for the nucleotide-binding motif. Both MjCobY and MtbGlmU exhibit a glycine rich pyrophosphorylase consensus sequence that binds the nucleotide (46). This lies at the base of the nucleotide-binding site (Figure 3) as described below.
Figure 3. Structural comparison of CobY with the uridyltransferase domain of MtbGlmU.
MjCobY is depicted in a rainbow coloration scheme (N-terminus blue), whereas the uridyltransferase domain is colored in gray. The wide diameter ribbon indicates the consensus pyrophosphorylase motif. The structures are remarkably similar considering the lack of sequence similarity. Indeed, the rms difference between 144 structurally similar α-carbons is only 1.84 Å. The coordinates for MtbGlmU were obtained from the RCSB, accession number 3DJ4 (44). Several surface loops (151–176 and 197–222) were removed from GlmU to emphasize the similarity between these proteins.
MjCobYG153D Ligand Binding
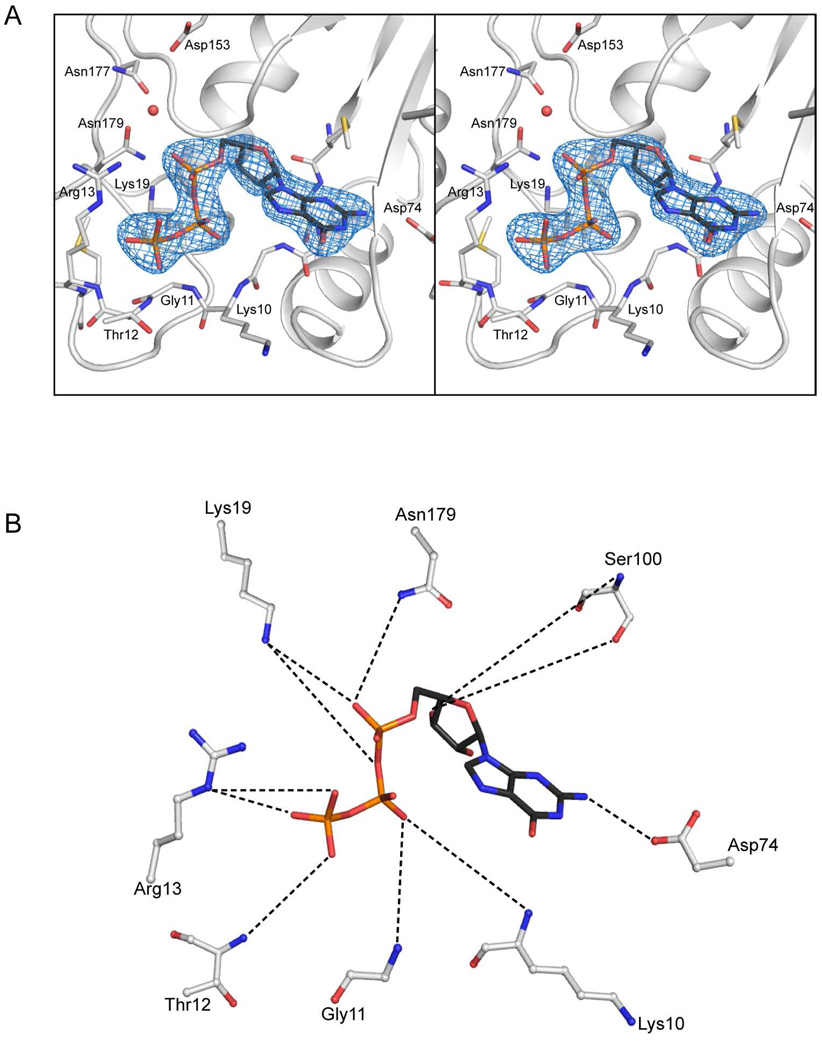
As noted above, the structure of the MjCobYG153D variant was determined in the presence of GTP. The nucleotide and its associated binding site are well defined in all subunits, though the electron density is most clear in subunit D as shown in Figure 4A. The conformation of the nucleotide is essentially the same in all subunits. The residues involved in ligand binding are described in Figure 4B. At 2.8 Å resolution it is inappropriate to discuss the details of the hydrogen bonding, but the residues that are most likely involved in nucleotide binding are readily identifiable based on their proximity to the GTP and structural similarity with MtbGmlU. It is difficult to identify all of the interactions specific to the guanine moiety of the nucleotide given the disordered loop between Asp74 -Ile81, which is presumed to abut O6 and N7 of the guanine ring. However, there is apparent shape complementarity throughout the nucleotide-binding site. In particular, the glycine rich random coil that constitutes the pyrophosphorylase binding motif [MAGGKGTRMGGVEKP(20)] extends under the entire length of GTP where it interacts with the guanine base, the ribose and the β- and γ-phosphoryl moieties. The presence of numerous glycine residues allows the amide hydrogens of Lys10, Gly11, and Thr12 to be in a position to putatively interact with the β- and γ-phosphoryl moieties. Charge neutralization appears to be contributed by Arg13, which is in proximity to the γ-phosphate of the GTP. Lys19 also lies in proximity to both the β- and γ-phosphoryl moieties and might form an interaction with the bridging oxygen. If this were the case it would explain why neither GMP-PNP or GMP-PCP were observed to bind to CobY (23). The solvent structure associated with the nucleotide is not well defined, however there does not appear to be a metal ion associated with the phosphoryl moieties as observed in the complex of MtbGlmU with diphosphate-N-acetylglucosamine (44). The lack of a metal ion may be due to the absence of the corrinoid substrate.
Figure 4. Stereoview of the electron density for GTP in complex with CobYG153D.
(A) The map was calculated from coefficients of the form Fo-Fc where the ligand was omitted from the phase calculation and refinement. (B) The ligand interaction diagram was derived from LIGPLOT (50) and displayed using Pymol. The dashed lines indicate putative hydrogen bonding interactions.
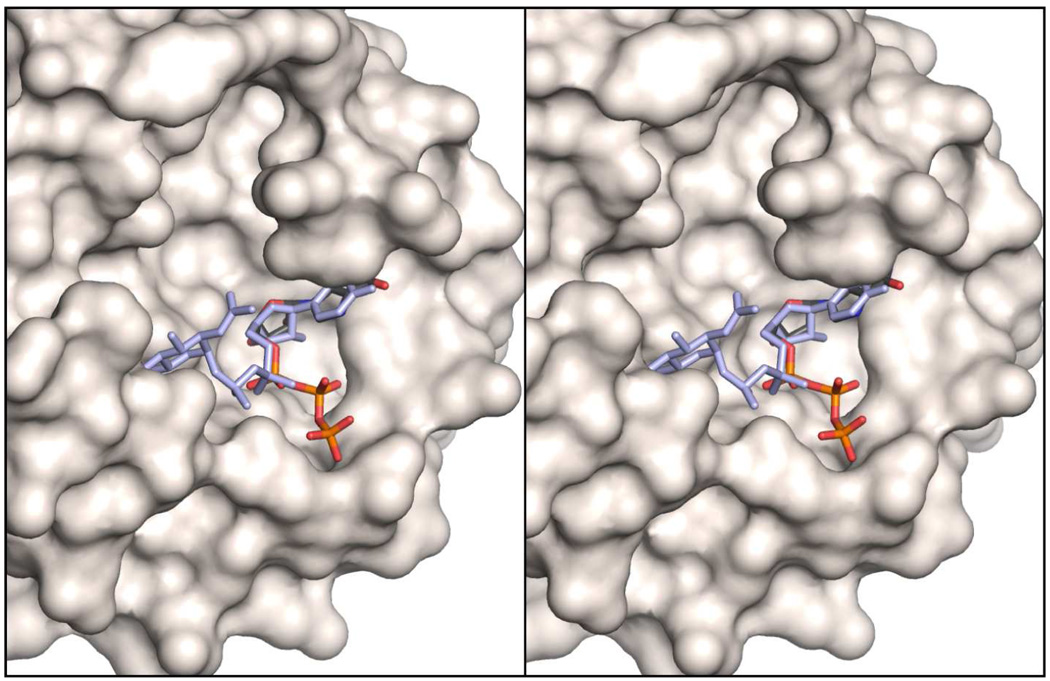
Superposition of the MjCobY monomer onto the N-terminal domain of MtbGlmU with UDP-GlcNAc bound in active site shows that the GMP and UMP components lie in a very similar location. The orientation of the β- and γ- phosphoryl moieties of GTP adopt a different orientation compared to the N-acetylglucosamine phosphate, but this is to be expected because GTP is a substrate whereas UDP-GlcNAc is a product. Both enzymes generate pyrophosphate. The GlcNAc-1-P moiety provides insight into the location of the second substrate for MjCobY, whereas the β- and γ- phosphates indicate the orientation of pyrophosphate as it leaves the active site. As can be seen from a surface representation, when MjCobY and MtbGlmU are superimposed, the GlcNAc moiety lies in a large pocket that must constitute the binding site for AdoCbi-P (Figure 5).
Figure 5. Surface representation of the CobY active site superimposed on N-acetylglucosamine phosphate from MtbGlmU.
The GTP bound to CobY lies at the base of a deep pocket that is assumed to be the binding site for AdoCbi-P. Superposition of the uridyltransferase domain of MtbGlmU (RCSB accession number 3DJ4) shows that N-acetylglucosamine phosphate lies within the pocket and thus defines the location that the phosphoryl moiety of AdoCbi-P must adopt for guanylyl transfer. The cavity that extends away from the α-phosphate appears to have a magnitude that is sufficient to accommodate an adenosylated corrinoid.
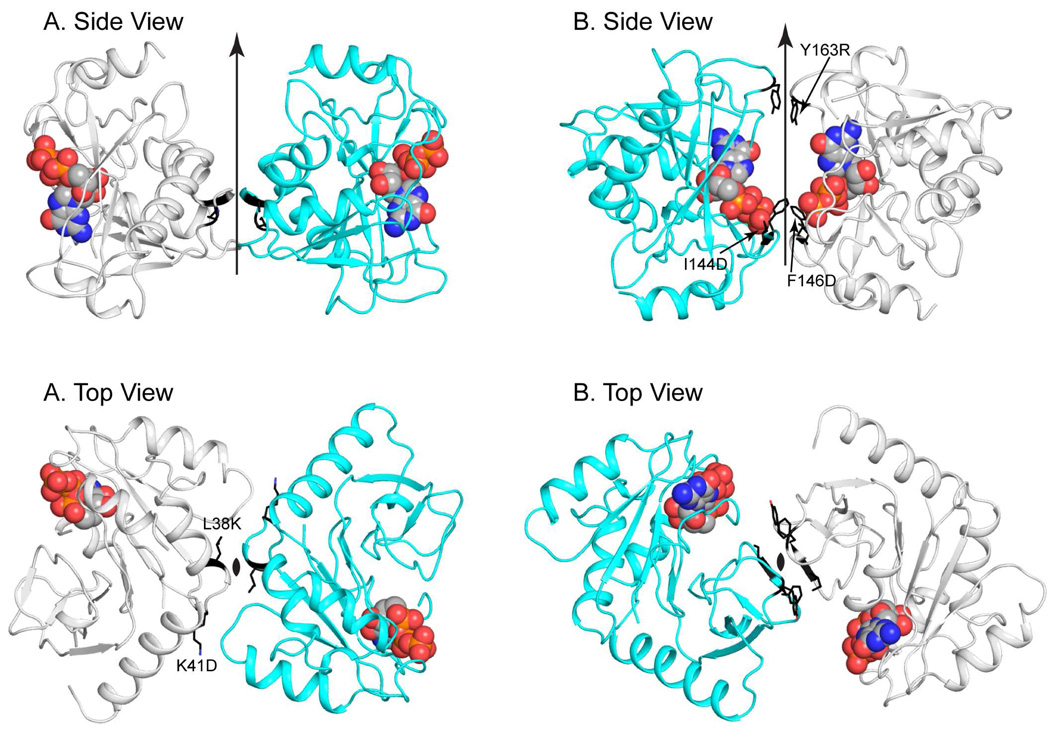
Quaternary Structure of CobY
The MjCobYG153D variant crystallized with four monomers in the asymmetric unit. Examination of the crystal lattice reveals that there are two plausible dimeric arrangements. These are shown in Figure 6. Because of the pseudosymmetric packing, all four subunits participate in both types of dimers. Consequently the crystal packing alone cannot distinguish which dimer is observed in solution. In interface A the active sites are positioned distal from one another with apparent dimer contacts along the loop that connects α1 with β2. Numerous hydrophobic contacts are observed along this interface among residues L37, L38 and K41. In interface B the active sites are positioned inward and β6 from both subunits combine to form an antiparallel β sheet across this interface. Interfaces A and B bury 525 and 434 Å2 accessible surface area per subunit, respectively, as calculated with the software Surfrace (47). These are both at the lower end of stable interfaces (48). These small interfaces are consistent with the observation of a mixed population of monomers and dimers in solution. (23). As might be expected, based on the small interfaces, structural analysis software such as the EBI PISA server failed to identify a stable molecular dimer.
Figure 6. Dimeric assemblies of CobY observed in the crystal lattice.
Two types of dimers are observed in the crystal lattice (A and B). In A the active sites are distal to the dimer interface, whereas in B the active sites face each other. Both types of interface are observed for four subunits in the asymmetric unit on account of pseudocrystallographic symmetry. There are thus two examples of each type interface in the crystal lattice, which suggests that the interactions are not a packing artifact. The dimers shown here correspond to the interactions between subunit B (cyan) and D (gray). GTP is depicted in a space-filling representation.
To experimentally distinguish which interface was the biologically relevant dimer, substitutions were introduced at L38K/D, K41D to probe the A interface, and I144D, F146D and Y163R to probe the B interface (Table 2). These substitutions were designed to introduce destabilizing charge repulsions at the putative interfaces. Analysis of the mutant enzymes by gel filtration revealed that mutations along interface A had little effect on the monomer: dimer ratio while mutations along interface B resulted in an enrichment of the monomer pool (77.2% monomer for mutant versus 58.6% WT). This appears to suggest that interface B is the biologically relevant assembly, though the effects are not dramatic, which is consistent with the small interface. Interestingly, the specific activity measurements on the separate pools for the mutant enzymes showed that CobY is active in both the monomeric and dimeric forms. Together, these results suggest that dimerization is not important for function. This conclusion is supported by results of the mutational analysis of conserved residues described below, which also shows no correlation between the monomer:dimer ratio and the specific activity of the variants (Table 3).
Table 2.
Mutational analysis of MjCobY dimerization interfaces.
| Protein tested | Functional in vivo? |
Doubling time (min) |
Monomer:Dimer: Aggregate Ratio |
Specific Activity (pmol/min/µg protein) |
|||
|---|---|---|---|---|---|---|---|
| Mixture | Monomer | Dimer | Aggregate | ||||
| MjCobYWT | Yes | 79 ± 3 | 59: 41: 0 | 103 ± 5a | ND | ND | NA |
| MjCobYL38D, K41D | Yes | 78 ± 1 | 52 : 26 : 22 | ND | 124 ± 6 | 119 ± 2 | 85 ± 7 |
| MjCobYL38K, K41D | Yes | 79 ± 1 | 53 : 25 : 22 | ND | 125 ± 2 | 120 ± 10 | 78 ± 3 |
| MjCobYI144D, F146D, Y163R | Yes | 83 ± 4 | 77 : 23 : 0 | ND | 156 ± 23 | 49 ± 10 | NA |
MjCobYWT specific activity of a mixture of monomers and dimers. Doubling time was determined during growth in minimal medium containing glycerol (30 mM) + CNCby (15 nM).
ND, not determined; NA, not applicable.
Table 3.
Functionality of MjCobY variants.
| Protein Encoded |
Functional In vivo? |
Doubling Time (min) |
% in vitro activity (relative to CobYWT) |
Monomer: Dimer Ratio |
Specific Activity (pmol/min/µg protein) |
|---|---|---|---|---|---|
| WT | Yes | 63 ± 4 | 100 | 59 : 41 | 116 ± 3 |
| G8D | No | NA | 3 | 33 : 67 | 4 ± 0 |
| R13A | Yes | 187 ± 30 | 15 | 14 : 86 | 18 ± 1 |
| E18D | Yes | 61 ± 4 | 56 | 40 : 60 | 66 ± 2 |
| K19A | Yes | 39 ± 11 | 16 | 36 : 64 | 19 ± 1 |
| P20A | Yes | 61 ± 5 | 93 | 63 : 37 | 102 ± 4 |
| S50A | Yes | 64 ± 6 | 100 | ND | 112 ± 7 |
| T53S | Yes | 58 ± 3 | 84 | 76 : 24 | 97 ± 4 |
| P54A | Yes | 58 ± 4 | 62 | 50 : 50 | 73 ± 3 |
| T56A | Yes | 119 ± 12 | 102 | ND | 107 ± 3 |
| Y80F | Yes | 60 ± 4 | 65 | 74 : 26 | 69 ± 4 |
| D83G | Yes | 60 ± 5 | 100 | 82 : 18 | 110 ± 5 |
| P94A | Yes | 68 ± 5 | 43 | 52 : 48 | 71 ± 3 |
| S100A | Yes | 117 ± 7 | 100 | ND | 101 ± 3 |
| D101G | Yes | 64 ± 5 | 71 | 34 : 66 | 79 ± 5 |
| G153D | No | NA | 2 | 54 : 46 | 3 ± 0 |
| N177R | No | NA | 5 | ND | 6 ± 0 |
| N179D | Yes | 67 ± 11 | 95 | ND | 93 ± 2 |
| T180A | Yes | 98 ± 6 | 73 | ND | 106 ± 5 |
The ability of the above-mentioned variants to restore AdoCbl synthesis from CNCby in the S. enterica ΔcobU ΔycfN strain was determined as described under Experimental Procedures. Doubling times of cultures grown in NCE glycerol medium supplemented with CNCby (15 nM) were calculated graphically from semi-log plots of OD595 as a function of time and are shown as the arithmetic mean ± standard error of the mean (n=3), using the Prism® v4 program (GraphPad Software). Monomer:dimer ratios and specific activities were calculated as described elsewhere (23); NA, not applicable; ND, not determined.
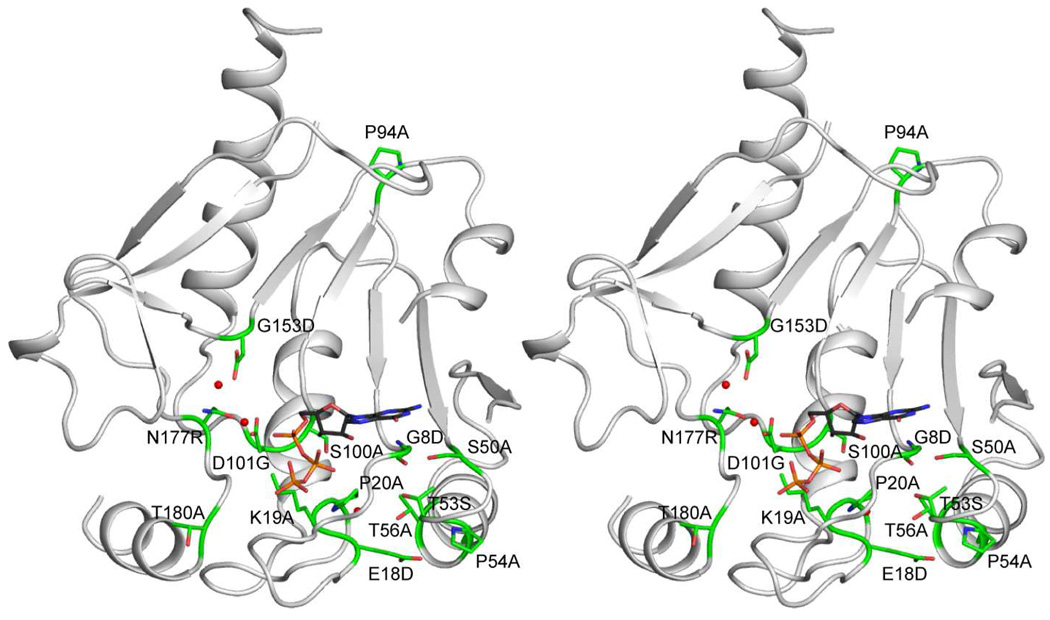
Mutational Analysis of Conserved Residues
Prior to the structural determination we investigated the functional role of conserved residues observed in 12 orthologues of CobY (Fig. S1). Site-directed mutagenesis was performed on 14 of the residues that are conserved in 12/12 CobY proteins, 3 residues that were conserved in 11/12, and 1 residue that was present in 9/12 in order to identify residues critical to MjCobY function (Figure 7). Each of these variants was overproduced and purified where possible to determine the effect of the substitutions on the activity in vitro and in vivo. The in vitro synthesis of AdoCbi-GDP was measured by RP-HPLC as described previously (23). Specific activities of each functional variant varied from 56–94% of wild-type activity, and are reported as pmol min−1 µg protein−1 in Table 3. The results of complementation tests indicated that all but three of the variants tested retained sufficient activity to support the conversion of AdoCby to AdoCbl, and their remaining activity supported Cbl-dependent growth as efficiently as the wild-type MjCobT protein as indicated by their doubling time (Table 3). Notably, cells that synthesized MjCobYT56A, MjCobYS100A, or MjCobYT180A grew at half the rate of cells expressing wild-type MjCobY. However, the specific activity of the proteins was similar to that of wild-type MjCobY, suggesting that substitutions at these locations do not compromise the activity of the enzyme, but affect AdoCbl synthesis through some other mechanism. Three non-functional mutants were identified that had substitutions at locations Gly8, Gly153, or Asn177.
Figure 7. Location of mutations introduced into CobY.
Conserved residues in 12 orthologues of CobY were targeted for mutagenesis. The targeted residues are primarily associated with the active site, where most are in close proximity to the pyrophosphorylase consensus sequence which is involved in nucleotide binding. There is no satisfactory explanation for the conservation of Pro94. Interestingly most of the conserved residues can be changed without loss of function.
The enzymatic activity of the MjCobYG8D, MjCobYG153D, and MjCobYN177R variants was determined in vitro by assessing the synthesis of AdoCbi-GDP by RP-HPLC. In all cases, the specific activities were dramatically decreased with ≥95% loss of activity relative to that of the wild-type enzyme. These data explain why the MjCobYG8D, MjCobYG153D, and the MjCobYN177R proteins failed to support AdoCbl biosynthesis in vivo, but did not shed light on whether the variant proteins were not functional solely because of the reduced enzymatic activity or whether they were also compromised by changes in the protein levels in vivo due to reduced protein stability.
The MjCobYG8D, MjCobYG153D, and the MjCobYN177R variants are unstable. Results from Western blot analyses using rabbit polyclonal antibodies against MjCobY show that the structural stability of these MjCobY variants is also compromised during constitutive expression. Strains JE8335, JE9293, JE9299, JE9306, and JE8269 (ΔcobU ΔycfN strains harboring plasmids encoding wild-type or null cobY alleles or the empty vector control; see Table S1) were grown under conditions that demanded GTP:AdoCbi-P guanylyltransferase activity for growth. These cultures of strains were examined by Western blot analysis that employed polyclonal rabbit antibodies raised against wild-type MjCobY. Detection of MjCobY variant proteins would suggest that the substitution likely affected MjCobY activity, rather than its stability in vivo, while absence of protein could suggest that the substitution affected its stability against degradation. While both the wild-type protein and the pure-protein control were observed, MjCobYG8D, MjCobYG153D, and the MjCobYN177R variants were not detected in the crude cell-free extracts (data not shown). The yield of overproduced MjCobYG8D, MjCobYG153D and MjCobYN177R in E. coli was similar to that of MjCobYWT. However, the stability of the proteins was greatly diminished. The MjCobYWT protein, and all of the other variants tested, were stable at 4°C for longer than two weeks without additional treatment. In spite of their reduced stability, sufficient amounts of MjCobG8D and MjCpobYG153D were isolated, and isothermal titration calorimetry experiments were performed. The MjCobYN177R variant, however, degraded rapidly after purification (within two days), making it necessary to complete in vitro studies of its function immediately after purification. This suggests that the observed instability of the MjCobYG8D, MjCobYG153D, and in particular MjCobYN177R is the reason why these variants failed to restore AdoCbl biosynthesis from CNCby in the S. enterica ΔcobU ΔycfN strain.
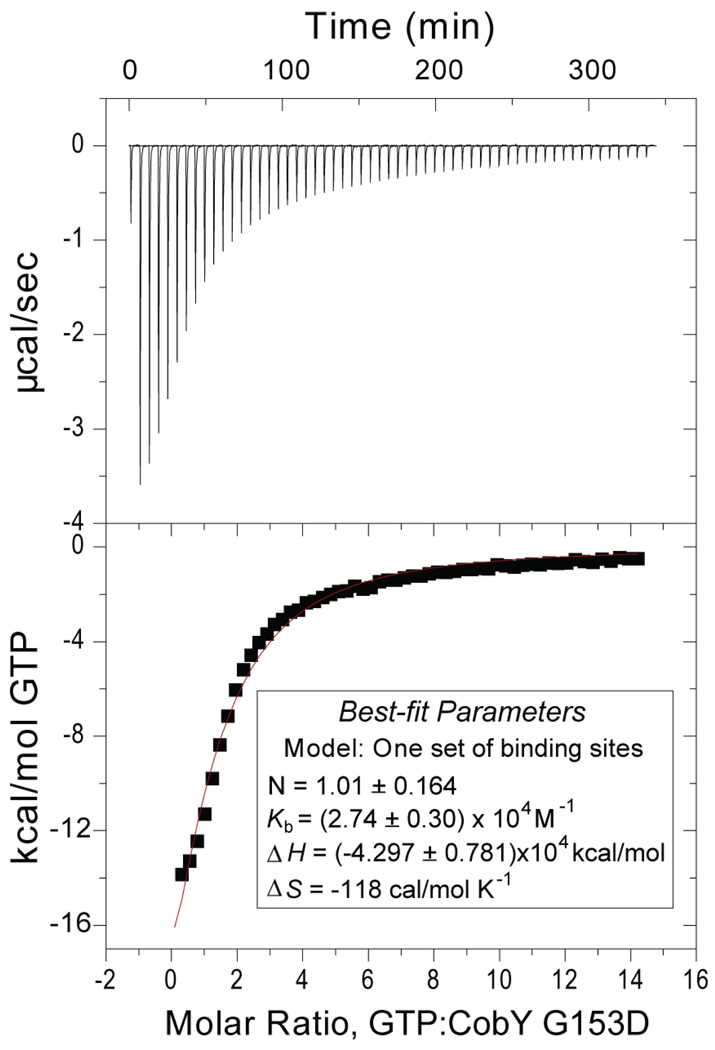
The G153D substitution increases the affinity of the enzyme for GTP and blocks corrinoid binding
Results from ITC experiments revealed that MjCobYG153D dimers bound GTP with approximately 10-fold higher affinity for the substrate than did the wild-type enzyme (Figure 8, Table 4). As seen in figure 7, the introduced G153D substitution is located at the back of the substrate-binding pocket where it interacts indirectly with residues that are in contact with GTP. This substitution serves to increase the number of hydrogen bonding partners in the active site and thus is expected to alter the water structure and the anticipated metal ion binding. It is seems likely this is the reason for the higher GTP affinity. Similarly, inclusion of an additional negative charge in the back of the substrate-binding site would be expected to interfere with corrinoid binding. The corrinoid ring carries numerous hydrophobic and polar groups on its periphery (Figure 1). Any modification in the way that these interact with the enzyme is expected to lower the affinity. Results from ITC experiments show that AdoCbi does not bind to the CobYG153D/GTP complex (data not shown), which is consistent with this hypothesis.
Figure 8. CobYG153D binds GTP.
A. Twenty-seven injections of 1 mM GTP were added to 18 µM CobYG153D dimers in the ITC cell. B. The area under each injection peak (top panel) equals the total heat released for that injection. The heat of dilution of GTP into buffer was subtracted. A binding isotherm for the interaction between CobYG153D and GTP was obtained from the integrated heat (bottom panel). Shown is the enthalpy per mole of GTP injected as a function of the molar ratio of CobYG153D dimers to GTP. The best-fit values for stoichiometry (N), binding constant (Kb), enthalpy (ΔH), and entropy (ΔS) are shown in the inset, and a comparison to previously published data for the wild-type enzyme is presented in Table 4.
Table 4.
Thermodynamic parameters for association of MjCobYWT and MjCobYG153D with GTP
| Protein | Kb (M−1) |
Kd (µM) |
ΔS (kcal mol−1 K−1) |
ΔH (kcal /mol) |
|---|---|---|---|---|
| MjCobYa | (2.00 ± 0.11) × 105 | 5.0 ± .02 | −32.5 | (−1.7 ± 0.02) × 104 |
| MjCobYG153D | (2.74 ± 0.30) × 104 | 0.4 ± .04 | −118.0 | (−4.3 ± 0.78) × 104 |
Previously reported (23).
The effect of the G8D substitution is caused by a perturbation in nucleotide binding
Residue Gly8 lies within a glycine-rich region that is part of the nucleotide-binding site. As described earlier, this residue is a component of the pyrophosphorylase consensus sequence LXXGXGTXMXXXXPK where Gly8 is underlined. In MjCobY the sequence of the pyrophosphorylase loop is MAGGKGTRMGGVEKP, where the order of the terminal proline and lysine are interchanged, but the penultimate lysine in the MjCobY motif still is able to interact with the β-phosphoryl moiety as does the terminal lysine in the canonical motif. Gly8 lies against the purine ring of the guanine, and as such, there is no space for a side chain. Replacement of Gly8 with any other amino acid is predicted to prevent binding of GTP and destabilize the interactions of the consensus motif with its neighboring residues. This prediction is consistent with the ITC measurements that show that the CobYG8D variant protein does not bind GTP (data not shown) and the observed instability of the protein.
Why Does the N177R Substitution Destabilize the Protein?
The structure of the CobYG153D variant shows that Asn177 lies at the base of the substrate-binding pocket in close proximity to the bound GTP (Figure 7). The amide side chain of Asn177 is involved in several close, potential hydrogen bonding interactions with Asn179, Tyr139, and an ordered water molecule. These interactions would be lost on replacement of Asn177 with an arginine. Introduction of the three methylene groups associated with the arginine side chain would eliminate the ability of Asn179 and Tyr139 to fulfill their hydrogen bonding potential and thus would be very destabilizing. Interestingly, Asp153 in the crystal structure (MjCobYG153D) lies close to Asn177. This suggests that the integrity of this region is critical for both enzyme stability and activity.
CONCLUSIONS
The structure of the archaeal MjCobYG153D variant protein provides valuable insights into the oligomeric structure of the wild-type enzyme, the placement of GTP in the active site, and the architecture of the active site. Although we know that the Ado moiety of AdoCbi-P is critical for binding (23), we can only speculate its location in the active site. We suggest that AdoCbi-P binds to the active site in an orientation that places the phosphoryl moiety close to residue G153. If that were the case, binding of AdoCbi-P to the active site of MjCobG153D would be prevented by strong repulsions between Asp and the Pi moiety of AdoCbi-P. This idea is supported by results of ITC studies that attempted to quantify the interactions between the MjCobYG153D/GTP complex and AdoCbi-P. Under the conditions used, AdoCbi-P did not bind to MjCobYG153D/GTP.
The model also revealed that the MjCobYG153D variant crystallized as a dimer, and that two putative dimerization interfaces exist, both of which are small. Experimental evidence favors a dimerization arrangement in which the active sites are proximal. The existence of small dimerization interfaces explains why homogeneous MjCobYWT is comprised of a population of dimers and monomers. We suggest that MjCobYWT functions as a dimer in vivo.
MjCobY contains a pyrophosphorylase-binding motif common to the nucleotide diphosphate sugar transferase superfamily. The fold seen in MjCobY is most similar to the one found in the uridyltransferase domain of the Mycobacterium tuberculosis GlmU protein. The structure shows that the pyrophosphorylase motif of MjCobY is involved in GTP binding. The structural similarities between MjCobY and MtbGlmU will prove valuable to future mechanistic studies of MjCobY function.
We suggest caution on the interpretation of the apparent absence of any effects of substitutions of many of the conserved residues on MjCobY function, especially because MjCobY function was assessed using a heterologous bacterial system (i.e., S. enterica) that employs a vastly different enzyme to convert AdoCbi-P to AdoCbi-GDP. Although there is insufficient information to propose a mechanism of catalysis for MjCobY, at this point there is no evidence to suggest that the MjCobY reaction proceeds via an enzyme bound GMP intermediate as with the bacterial enzyme (10). However, on the basis of the structural and mutational analyses of MjCobY we conclude that residues G8 and G153 are important to the binding of GTP and the corrinoid substrate, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kathy Krasny for technical assistance. We also thank Paul Renz for the gift of cyanocobyric acid.
Footnotes
This work was supported by NIH grants GM086351 to I.R. and GM40313 to J.C.E.-S. Use of the SBC BM19 beamline at the Argonne National Laboratory Advanced Photon Source was supported by the U.S. Department of Energy, Office of Energy Research, under Contract No. W-31-109-ENG-38.
The atomic coordinates and structure factors (code 3RSB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
ABBREVIATIONS: MjCobY, Methanocaldococcus jannaschii CobY; Ado, adenosyl; MtbGlmU, Mycobacterium tuberculosis GlmU; AdoCbl, adenosylcobalamin or coenzyme B12; Cbi, cobinamide; (CN)2Cbi, dicyanocobinamide; AdoCbi, adenosylcobinamide; Cby, cobyric acid; CNCby, cyanocobyric acid; AdoCby, adenosylcobyric acid; CNCby, cyanocobyric acid; GMP-PNP, guanosine-5’-[β,γ-imido]-triphosphate; GMP-PCP, guanylyl-5’-[β,γ-methylene]-diphosphonate; IPTG, isopropyl-β-D-1-thiogalactoside; NBT-BCIP, nitro-blue tetrazolium chloride and 5-bromo-4-chloro-3'-indolyphosphate p-toluidine salt; NCE, No-carbon essential medium; NLA, nucleotide loop assembly; RP-HPLC, reverse phase high-performance liquid chromatography; SDS-PAGE, sodium dodecylsulfate-polyacrylamide gel electrophoresis; DTT, dithiothreitol; HEPES, 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid; Tris, 2-amino-2-hydroxymethyl-propane-1,3-diol; UDP-GlcNAc, uridine-diphosphate-N-acetylglucosamine; GlcN-1-P, glucosamine-1-phosphate; Ac-CoA, acetyl-coenzyme A; RPM, revolutions per minute; SLPM, standard liters per minute; NBT-BCIP, nitro-blue-tetrazolium chloride and 5-bromo-4-chloro-3'-indolyphosphate p-toluidine salt; PVDF, polyvinylidene fluoride.
Supporting Information
Lists of bacterial strains, experimental plasmids, and PCR primers together with a sequence analysis are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Banerjee R. Radical carbon skeleton rearrangements: catalysis by coenzyme B12-dependent mutases. Chem. Rev. 2003;103:2083–2094. doi: 10.1021/cr0204395. [DOI] [PubMed] [Google Scholar]
- 2.Fontecave M, Mulliez E. Ribonucleotide Reductases. In: Banerjee R, editor. Chemistry and Biochemistry of B12. New York: John Wiley & Sons, Inc; 1999. pp. 731–756. [Google Scholar]
- 3.Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. How a protein binds B12: A 3.0Å X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 4.Shibata N, Tamagaki H, Hieda N, Akita K, Komori H, Shomura Y, Terawaki SI, Mori K, Yasuoka N, Higuchi Y, Toraya T. Crystal structures of ethanolamine ammonia-lyase complexed with coenzyme B12 analogs and substrates. J. Biol. Chem. 2010;285:26484–26493. doi: 10.1074/jbc.M110.125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toraya T. Radical catalysis in coenzyme B12-dependent isomerization (eliminating) reactions. Chem. Rev. 2003;103:2095–2127. doi: 10.1021/cr020428b. [DOI] [PubMed] [Google Scholar]
- 6.Escalante-Semerena JC, Warren MJ. Biosynthesis and Use of Cobalamin (B12) In: Böck A III, Curtiss R, Kaper JB, Karp PD, Neidhardt FC, Nyström T, Slauch JM, Squires CL, editors. EcoSal - Escherichia coli and Salmonella: cellular and molecular biology. Washington, D. C: ASM Press; 2008. module 3.6.3.8. [Google Scholar]
- 7.Ronzio RA, Barker HA. Enzymic synthesis of guanosine diphosphate cobinamide by extracts of propionic acid bacteria. Biochemistry. 1967;6:2344–2354. doi: 10.1021/bi00860a009. [DOI] [PubMed] [Google Scholar]
- 8.Thomas MG, Thompson TB, Rayment I, Escalante-Semerena JC. Analysis of the adenosylcobinamide kinase / adenosylcobinamide phosphate guanylyltransferase (CobU) enzyme of Salmonella typhimurium LT2. Identification of residue H46 as the site of guanylylation. J. Biol. Chem. 2000;275:27376–27386. doi: 10.1074/jbc.M000977200. [DOI] [PubMed] [Google Scholar]
- 9.Thompson TB, Thomas MG, Escalante-Semerena JC, Rayment I. Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase from Salmonella typhimurium determined to 2.3 Å resolution. Biochemistry. 1998;37:7686–7695. doi: 10.1021/bi973178f. [DOI] [PubMed] [Google Scholar]
- 10.Thompson TB, Thomas MG, Escalante-Semerena JC, Rayment I. Three-dimensional structure of adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase (CobU) complexed with GMP: evidence for a substrate-induced transferase active site. Biochemistry. 1999;38:12995–13005. doi: 10.1021/bi990910x. [DOI] [PubMed] [Google Scholar]
- 11.Blanche F, Debussche L, Famechon A, Thibaut D, Cameron B, Crouzet J. A bifunctional protein from Pseudomonas denitrificans carries cobinamide kinase and cobinamide phosphate guanylyltransferase activities. J. Bacteriol. 1991;173:6052–6057. doi: 10.1128/jb.173.19.6052-6057.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maggio-Hall LA, Escalante-Semerena JC. In vitro synthesis of the nucleotide loop of cobalamin by Salmonella typhimurium enzymes. Proc. Natl. Acad. Sci. U S A. 1999;96:11798–11803. doi: 10.1073/pnas.96.21.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maggio-Hall LA, Claas KR, Escalante-Semerena JC. The last step in coenzyme B12 synthesis is localized to the cell membrane in bacteria and archaea. Microbiology. 2004;150:1385–1395. doi: 10.1099/mic.0.26952-0. [DOI] [PubMed] [Google Scholar]
- 14.Cameron B, Blanche F, Rouyez MC, Bisch D, Famechon A, Couder M, Cauchois L, Thibaut D, Debussche L, Crouzet J. Genetic analysis, nucleotide sequence, and products of two Pseudomonas denitrificans cob genes encoding nicotinate-nucleotide: dimethylbenzimidazole phosphoribosyltransferase and cobalamin (5'-phosphate) synthase. J. Bacteriol. 1991;173:6066–6073. doi: 10.1128/jb.173.19.6066-6073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zayas CL, Escalante-Semerena JC. Reassessment of the late steps of coenzyme B12 synthesis in Salmonella enterica: Evidence that dephosphorylation of adenosylcobalamin-5'-phosphate by the CobC phosphatase is the last step of the pathway. J. Bacteriol. 2007;189:2210–2218. doi: 10.1128/JB.01665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodson JD, Escalante-Semerena JC. CbiZ, an amidohydrolase enzyme required for salvaging the coenzyme B12 precursor cobinamide in archaea. Proc. Natl. Acad. Sci. USA. 2004;101:3591–3596. doi: 10.1073/pnas.0305939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodson JD, Zayas CL, Escalante-Semerena JC. A new pathway for salvaging the coenzyme B12 precursor cobinamide in archaea requires cobinamide-phosphate synthase (CbiB) enzyme activity. J. Bacteriol. 2003;185:7193–7201. doi: 10.1128/JB.185.24.7193-7201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray MJ, Escalante-Semerena JC. In vivo analysis of cobinamide salvaging in Rhodobacter sphaeroides strain 2.4.1. J. Bacteriol. 2009;191:3842–3851. doi: 10.1128/JB.00230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray MJ, Escalante-Semerena JC. The cobinamide amidohydrolase (cobyric acid-forming) CbiZ enzyme: a critical activity of the cobamide remodelling system of Rhodobacter sphaeroides. Mol. Microbiol. 2009;74:1198–1210. doi: 10.1111/j.1365-2958.2009.06928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zayas CL, Claas K, Escalante-Semerena JC. The CbiB protein of Salmonella enterica is an integral membrane protein involved in the last step of the de novo corrin ring biosynthetic pathway. J. Bacteriol. 2007;189:7697–7708. doi: 10.1128/JB.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas MG, Escalante-Semerena JC. Identification of an alternative nucleoside triphosphate: 5'- deoxyadenosylcobinamide phosphate nucleotidyltransferase in Methanobacterium thermoautotrophicum ΔH. J. Bacteriol. 2000;182:4227–4233. doi: 10.1128/jb.182.15.4227-4233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodson JD, Peck RF, Krebs MP, Escalante-Semerena JC. The cobY gene of the archaeon Halobacterium sp. strain NRC-1 is required for de novo cobamide synthesis. J. Bacteriol. 2003;185:311–316. doi: 10.1128/JB.185.1.311-316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otte MM, Escalante-Semerena JC. Biochemical characterization of the GTP:adenosylcobinamide-phosphate guanylyltransferase (CobY) enzyme of the hyperthermophilic archaeon Methanocaldococcus jannaschii. Biochemistry. 2009;48:5882–5889. doi: 10.1021/bi8023114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertani G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 2004;186:595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkowitz D, Hushon JM, Whitfield HJ, Jr, Roth J, Ames BN. Procedure for identifying nonsense mutations. J. Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel HJ, Bonner DM. Acetylornithase of Escherichia coli: partial purification, and some properties. J. Biol. Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 28.Balch WE, Wolfe RS. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 1976;32:781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabor S. Expression using the T7 RNA polymerase/promoter system. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1990. 16.12.11. [Google Scholar]
- 30.Otte MM, Woodson JD, Escalante-Semerena JC. The thiamine kinase (YcfN) enzyme plays a minor but significant role in cobinamide salvaging in Salmonella enterica. J. Bacteriol. 2007;189:7310–7315. doi: 10.1128/JB.00822-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 32.O'Toole GA, Escalante-Semerena JC. Purification and characterization of the bifunctional CobU enzyme of Salmonella typhimurium LT2. Evidence for a CobU~GMP intermediate. J. Biol. Chem. 1995;270:23560–23569. doi: 10.1074/jbc.270.40.23560. [DOI] [PubMed] [Google Scholar]
- 33.Blanche F, Thibaut D, Couder M, Muller JC. Identification and quantitation of corrinoid precursors of cobalamin from Pseudomonas denitrificans by high-performance liquid chromatography. Anal. Biochem. 1990;189:24–29. doi: 10.1016/0003-2697(90)90038-b. [DOI] [PubMed] [Google Scholar]
- 34.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Sasse J. Detection of proteins. In: Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1991. 10.16.11–10.16.18. [Google Scholar]
- 37.Blake MS, Johnston KH, Russell-Jones GJ, Gotschlich EC. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal. Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the Maximum-Likelihood Method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Kostrewa D, D'Arcy A, Takacs B, Kamber M. Crystal structures of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, in apo form at 2.33 Å resolution and in complex with UDP-N-acetylglucosamine and Mg(2+) at 1.96 Å resolution. J. Mol. Biol. 2001;305:279–289. doi: 10.1006/jmbi.2000.4296. [DOI] [PubMed] [Google Scholar]
- 44.Parikh A, Verma SK, Khan S, Prakash B, Nandicoori VK. PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J. Mol. Biol. 2009;386:451–464. doi: 10.1016/j.jmb.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z, Bulloch EM, Bunker RD, Baker EN, Squire CJ. Structure and function of GlmU from Mycobacterium tuberculosis. Acta Crystallogr. D Biol. Crystallogr. 2009;65:275–283. doi: 10.1107/S0907444909001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mio T, Yabe T, Arisawa M, Yamada-Okabe H. The eukaryotic UDP-N-acetylglucosamine pyrophosphorylases. Gene cloning, protein expression, and catalytic mechanism. J. Biol. Chem. 1998;273:14392–14397. doi: 10.1074/jbc.273.23.14392. [DOI] [PubMed] [Google Scholar]
- 47.Tsodikov OV, Record MT, Jr, Sergeev YV. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 48.Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Q. Rev. Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 49.Delano WL. The Pymol Molecular Graphics System. 2002 on http://www.pymol.org.
- 50.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.