Abstract
BACKGROUND
Approximately 30% to 40% of all patients with osteosarcomas ultimately experience recurrence. The study investigated the hypothesis that the resistance of osteosarcoma to chemotherapy may be related to the expression of a pregnane xenobiotic receptor (PXR) variant protein and its role as the major inducer of P450 3A4 in these tumors.
METHODS
Polymerase chain reaction (PCR) and Western blot analysis were used to determine PXR mRNA and protein expression, respectively. Real-time PCR and CYP3A catalytic activity using 7-benzyl-trifluoromethyl coumarin (BFC) as the probe substrate were used to measure the induction of P450 3A4 or MDR1. siRNA transfections were performed for PXR and cytotoxicity determined by a colorimetric based assay or Annexin v-Fitc staining.
RESULTS
Differences were observed in the molecular size of the PXR protein expressed in sarcoma cell lines when compared with the wildtype PXR expressed in normal liver, kidney, or small intestine. A polyclonal PXR antibody raised against the N-terminus of the wildtype PXR did not detect PXR expressed in these sarcoma cell lines. In the osteosarcoma cell lines, etoposide and doxorubicin were better inducers of P450 3A4 and MDR1 than rifampin. siRNA against PXR down-regulated P450 3A4 expression only in the osteosarcoma cell line. Cytotoxicity assays showed that the resistance of the osteosarcoma cell lines to etoposide correlated with PXR protein expression levels and activation of P450 3A4 and could be prevented by ketoconazole.
CONCLUSION
The results suggest that PXR plays a critical role in the regulation of P450 3A4 expression in osteosarcoma and that its expression and activation in these tumors may influence the effect of chemotherapeutic agents on the induction of target genes implicated in drug resistance.
Keywords: PXR, P450 3A4, MDR1, osteosarcoma, drug resistance
Osteogenic sarcoma is the most common primary malignant bone tumor, occurring in approximately 1 out of every 100,000 people per year.1 Resistance to chemotherapy remains a major obstacle to the successful treatment of osteosarcoma.2,3 Delineating the clinical and biological features that could predict and/or may be responsible for treatment failures may help identify those patients who would benefit the most from tailored therapy.
Pregnane xenobiotic receptor (PXR), also known as steroid and xenobiotic receptor (SXR, PAR, and NR112),4 is an orphan nuclear receptor that regulates the transcription of genes such as cytochrome P450 3A4 (P450 3A4) and the multidrug-resistant gene (MDR1) that are involved in the metabolism and elimination of a number of xenobiotics.5,6 P450 3A4 is a major Phase I drug-metabolizing enzyme involved in the biotrans-formation of xenobiotics to active and inactive metabolites and which activates several potent chemical carcinogens.7 MDR1 is a gene whose product (P-glycoprotein, PGP) is believed to play an important role in the resistance of tumor cells to chemotherapy by acting as an extruding pump, reducing the accumulation of drugs and cytotoxic agents in these cells.8 Several reports have described the important role of PGP in the resistance of human osteosarcomas to chemotherapy.9,10 A number of clinical trials have attempted to alter PGP activity and thus improve clinical outcomes11–13; however, the majority of these studies showed no clear-cut success. We hypothesized that modulation of these target genes involved in drug metabolism and transport by PXR may play an important role in the mechanisms for drug resistance observed in some osteosarcoma patients.
In the current study we found that a variant form of PXR is expressed in osteosarcoma cells that does not respond to rifampin, but does respond to etoposide or doxorubicin to induce P450 3A4 and MDR1, correlating with drug resistance. This receptor also plays a major role in modulating the expression of P450 3A4 in osteosarcoma cell lines.
MATERIALS AND METHODS
Acquisition and Characterization of Cell Lines
Normal human liver (NL), kidney, and small intestine cells, as well as tumor samples from the Ewings and synovial sarcoma WW were derived by dissociation of tissues into single cells using a 40-µm metal mesh. Collection and handling of human samples was done using procedures approved by the Institutional Review Board of the University of Michigan Medical Center. The OS187, WOL, and COL osteosarcoma cell lines have been described previously.14 LS174T/ LS180, a colon adenocarcinoma cell line, and HepG2, a hepatoma cell line, were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Primary tumors were maintained in Dulbecco Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 1% insulin trans-sel-X 100X, 1% penicillin/streptomycin, and 1% glutamine purchased from GIBCO (Grand Island, NY). LS174T and HepG2 cells were maintained using complete DMEM components but without the 1% insulin trans-sel-X 100X. Whole cell lysates from the BT20 breast cancer cell line and the A549 lung adenocarcinoma cell line were generous gifts from Santa Cruz Biotechnology (Santa Cruz, CA). Chemotherapeutic agents were obtained from the University of Michigan Comprehensive Cancer Center Pharmacy or purchased from Sigma (St. Louis, MO). LS174T, HepG2 cells, or NL, kidney, and small intestine, all characterized for wild-type PXR, were used as positive controls whenever possible. Concentrations that inhibited cell proliferation by 50% (IC50 values) were established for all agents used in all cell lines by standard methods.15 The concentrations of the chemotherapeutic agents used for subsequent studies were less than or equal to the IC50 in all cell lines unless otherwise stated.
Determination of Expression of P450 3A4, PXR, and MDR1 mRNA by RT-PCR
Total mRNA was isolated from cells treated with 50 µM etoposide,16 25 µM doxorubicin, 200 µM ifosfamide, or 30 µM rifampin for 48 hours or from untreated cells using the TriZol method (Invitrogen, La Jolla, CA). cDNA was synthesized from RNA (1 µg) using a first strand synthesis kit for reverse-transcriptase polymerase chain reaction (RT-PCR) (Retro-script, Ambion, Austin TX, or Promega, Madison WI), or Superscript and PE Sybr Green. Quantitative RT-PCR (QT-PCR) was carried out with the following primer pairs: GAPDH, product size: 225 bp, Forward: 5′-GAA GGT GAA GGT CGG AGT C-3′, Reverse: 5′-GAA GAT GGT GAT GGG ATT TC-3′; P450 3A4, product size: 128 bp, Forward: 5′-CAT TCC TCA TCC CAA TCT TTG AAG T-3′, Reverse: 5′-CCA CTC GGT GCT TTT GTG TAT CT-3′, Probe: 5′-CGA GGC GAC TTT CTT TCA TCC TTT TTA CAG ATT TTC-3′; MDR1, product size: 183 bp, Forward: 5′-AGG AAG ACA TGA CCA GGT ATG C-3′: Reverse: 5′-CCA ACA TCG TGC ACA TCA AAC-3′ and PXR, Forward: 5′-CAA GCG GAA GAA AAG TGA ACG-3′ Reverse: 5′-CTG GTC CTC GAT GGG CAA GTC-3′. mRNA from NL or LS174T cells was extracted under identical conditions and used as positive controls. The delta/delta CT formula for real-time PCR was used to determine differences in mRNA expression compared with the liver cells or LS174T positive controls.17,18
Silencing of PXR Protein Expression With Small Interfering RNA (siRNA)
PXR siRNA duplexes that target the sequences 5′-AU-GUCAUGACAUUGAAGUGAUAGCC-3′ (siRNA-74); 5′-UUUCAUCUGAGCGUCCAUCAGCUCC-3′ (siRNA-75); 5′-AAAUGGGAGAAGGUAGUGUCAAAAG-3′ (siRNA-76); 5′-GGAGCUGAUGGACGCUCAGAUGAAA-3′(siRNA-229); 5′-UCUUGUUCCUGAAGAUCAUGGCUAU-3′ (siRNA-132); 5′-GGAGAAAUCCCUCAGAUCCCACUAA-3′ (siRNA-345) were synthesized by Invitrogen Life Technologies (Carlsbad, CA). High-purity control scramble sequence (Low GC) siRNA oligonucleotides were purchased from Invitrogen Life Technologies. This scrambled sequence does not match any human genome sequence. Transfection of siRNA was conducted with Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. Briefly, OS187 and HepG2 cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. The cells were seeded into 6-well plates at concentrations of 4 × 105 per well in triplicate for 24 hours before siRNA transfection. The cells were then transfected with 100 nM PXR siRNA and incubated with the transfection complexes for 24, 48, and 72 hours. At the end of the experiment the cells were trypsinized and the cell number was determined using the Trypan blue exclusion method for the cytotoxic assay. Fifty micrograms of the whole cell lysates from each sample were used for immunoblotting analysis as described below to determine the gene-silencing efficiency.
Western Blot Analysis of PXR and P450 3A4
Western blot analysis was performed on whole cell lysates obtained from the osteosarcoma primary cell lines, the LS174T colon cancer cells, or human tissue samples of the liver, small intestine, and kidney. Escherichia coli expressed human P450 3A4 protein samples purified in our laboratory were run in parallel as positive controls. PXR expression was determined using anti-PXR antibodies A-20 and N-19 from Santa Cruz Biotechnology or anti-PXR antibodies from Active Motif (Carlsbad, CA) at 1:1000 dilutions, all of which detected PXR at the same molecular size.19 P450 3A4 expression was determined using an anti-P450 3A4 antibody (a gift from Dr. Kan He, Bristol Meyers Squibb, Princeton, NJ). The secondary antibodies, antigoat for PXR (Santa Cruz Biotechnology), antisheep for P450 3A4 (a gift from Dr. Kan He), and antimouse for actin (Bio-Rad Laboratories, Hercules, CA) were used to probe membranes using the enhanced chemiluminescent assay (ECL) from Pierce (Rockford, IL) for the detection of horseradish peroxidase (HRP) conjugated to secondary antibodies.
Determination of CYP3A Catalytic Activity in Osteosarcoma Cell Lines Using the P450 3A4 Probe Substrate, 7-Benzyl-trifluoromethyl Coumarin (BFC)
BFC is metabolized by P450 3A4 to give the highly fluorescent 7-hydroxy-4(trifluoromethyl) coumarin that can readily be detected spectrofluorometrically.20,21 Cells (100,000/mL) were seeded in phenolred free DMEM. LS174T cells or HepG2 cells were used for positive controls. All cells were incubated with or without rifampin (30 µM), ketoconazole (10 µM), etoposide (25–150 µM), doxorubicin (25–50 µM), or ifosfamide (400 µM) for 24 or 48 hours as indicated. Cells were detached with trypsin, washed with phosphate-buffered saline (PBS), and incubated in phenol-red free DMEM containing 50 µM 7-benzyl-4(trifluoromethyl) coumarin. After 4–6 hours the supernatant was collected, centrifuged, and transferred into a clean Eppendorf tube. Stop solution (80% CH3CN + 20% 0.5 M Tris base) equal to 40% of the reaction volume was added and the samples were read on an RF-5301 PC Spectrofluorophotometer (Shimadzu Scientific Instruments, Wooddale, IL) using an excitation wavelength of 410 nm and an emission wavelength of 530 nm. Each experiment was conducted with duplicate samples. The induction of P450 3A4 activity by rifampin in the LS174T or HepG2 cells was used as positive control.
Cytotoxic Response of Cells Exposed to Etoposide Alone or in Combination With Ketoconazole
The Annexin V-FITC Apoptosis Detection Kit (Bio-Vision Research Products, Mountain View, CA) was used according to the manufacturer’s protocol to determine apoptosis of etoposide-treated COL or OS187 cells that were analyzed using a Coulter Elite ESP Cell Sorter from Beckman (Fullerton, CA). We also determined the effect of ketoconazole on the response of COL to etoposide using the Trypan blue (Gibco, Carlsbad, CA) exclusion method.22 All cytotoxicity assays were further confirmed using either a colorimetric assay based on the cleavage of the tetrazolium salt WST-1 (Roche Molecular Biochemicals, Vienna, Austria) by mitochondrial dehydrogenases in viable cells23 or the release of lactate dehydrogenases (LDH) for nonviable cells.24
RESULTS
Expression of PXR in Primary and Stable Sarcoma Cell Lines
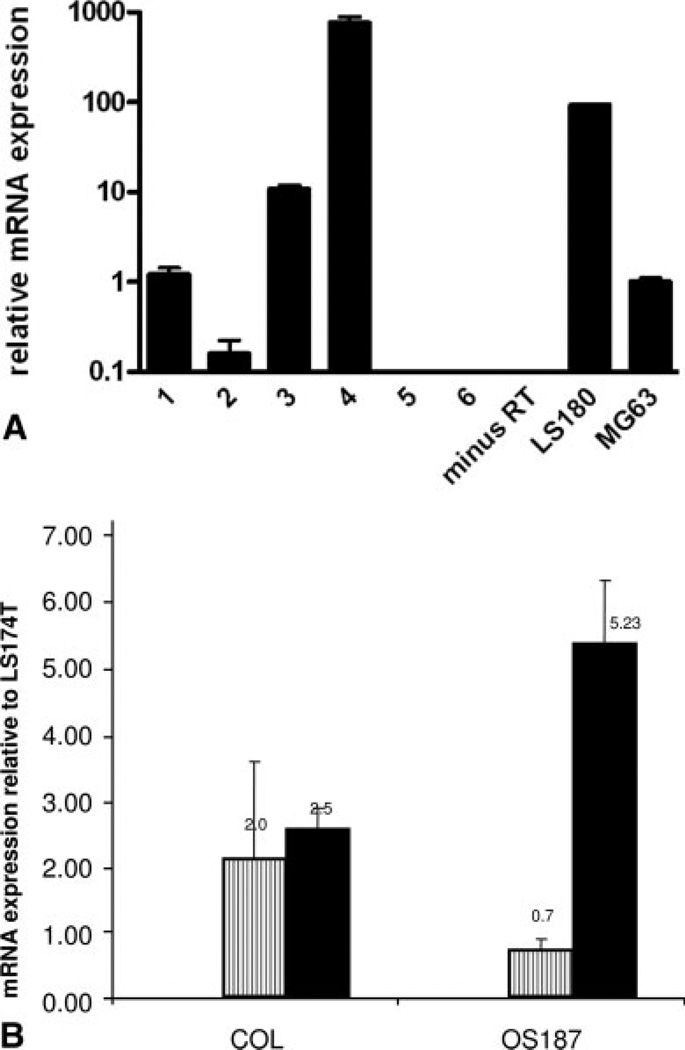
Figure 1A is a representative figure showing the differential expression of mRNA for PXR in a number of primary and stable sarcoma cell lines. Expression of PXR in the stable osteosarcoma cell line MG63 has previously been reported by Tabb et al.25 The expression level was arbitrarily set at 1 for MG63. The colon adenocarcinoma cell line LS180, known for exhibiting high expression levels of PXR, was used as a positive control. Our results suggest that there is significant variation in the expression levels of PXR mRNA (over a 1000-fold range) in the sarcoma cell lines tested, with the highest expression found in primary synovial sarcoma cells and the lowest expression determined in the liposarcoma cell line. Because there were variations in PXR expression in the various sarcoma cell lines tested, we determined if expression of target genes P450 3A4 and MDR1 levels were also different in at least 2 of the osteosarcoma cell lines chosen for further analysis. Figure 1B shows the baseline expression of P450 3A4 and MDR1 mRNA in COL and OS187 osteosarcoma cell lines relative to expression in the colon adenocarcinoma cell line LS174T. Our data indicate that osteosarcoma cell line COL expresses higher levels of P450 3A4 and less MDR1 mRNA than OS187.
FIGURE 1.
(A) PXR expression in osteosarcoma cell lines. Total RNA was isolated from a number of primary and stable sarcoma cell lines. Real-time polymerase chain reaction (PCR) was performed using the SYBR Green-PCR-Master-Mix (Applied Biosystems, Foster City, CA). cDNA was made using Superscript III from Invitrogen (La Jolla, CA) using 1 µg total RNA input. Osteosarcoma cell line MG63 was set as 1 and all expression was determined relative to MG63. The colon adenocarcinoma cell line LS180 was used as positive control and minus RT for negative control. Expression is depicted on a logarithmic scale to facilitate comparison of widely divergent expression levels. Lanes 1–6 represent: primary osteosarcoma cell line (COL); liposarcoma; osteosarcoma cell line (SAOS2); primary synovial sarcoma cells; primary embryonal rhabdomyosarcoma cells (no RNA detected); rhabdomyosarcoma cells (no RNA detected), respectively. (B) Quantitative RT-PCR for P450 3A4 and MDR1 in COL and OS187 cells. P450 3A4 or MDR1 levels were determined relative to LS174T mRNA and represented as P450 3A4 (−) and MDR1 (−). mRNA values represent the average of triplicates, standardized to the expression of GAPDH.
Expression of a Disparately Sized PXR Variant Protein in Osteosarcoma
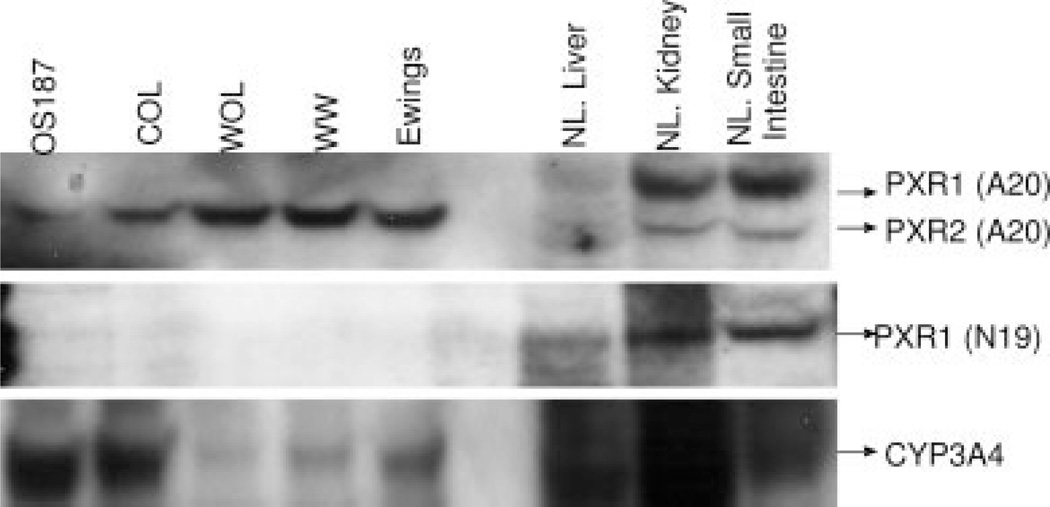
We determined the expression of the PXR protein in a number of osteosarcoma primary cell lines (OS187, COL, and WOL), Ewings sarcoma, and a synovial sarcoma, WW. Using Western blot analysis with antibodies against various epitopes of the PXR protein, we investigated the molecular size of PXR in these cell lines and compared it to PXR expressed in normal liver, kidney, or small intestine.25,26 Negative controls consisted of the lung adenocarcinoma cell line A54927 and the breast cancer cell line BT20 as previously described (data not shown).28 Higher levels of PXR protein were detected in the osteosarcoma cell line COL compared with OS187 cells (Fig. 2) (A20). The PXR protein expressed in the osteosarcoma cell lines exhibited a molecular size approximately 1000 Da lower than PXR protein variant 1 (PXR1) and approximately 1000 Da higher than PXR variant 2 (PXR2).29 This size difference was confirmed when a mixture of whole cell lysates from OS187 cells and small intestine was subjected to Western blot analysis. Electrophoretic separation of the mixture of PXR proteins expressed in small intestine and OS187 cells revealed 3 distinct bands after probing with the anti-PXR antibody (data not shown). We considered the possibility that the reduced size of the PXR protein expressed in the osteosarcoma cells might be due to a mutation or truncation in the N-terminal region of the PXR protein. In order to investigate this possibility, we used a PXR1 antibody (N19) raised against an epitope located in the N-terminal region at amino acids 1–20 of the PXR protein and found that it detected the expression of PXR1 alone, and only in the liver, small intestine, and kidney, but not in the osteosarcoma cell lines, as shown in Figure 2. Finally, we determined the protein levels of P450 3A4 in the different cell lines that also showed variable expression.
FIGURE 2.
Western blot analysis for the expression of PXR or CYP3A4 protein in nontreated cells. Total cell lysates (50 µg) were separated on a 10% SDS-PAGE gel and immunoblotted with polyclonal anti-PXR antibodies against the conserved epitope (A-20) or the n-terminal (N-19), and detected using the enhanced chemiluminiscent substrate.
PXR siRNA Significantly Down-regulates the Expression of P450 3A4 in the Osteosarcoma Cell Line OS187 Compared With HepG2 Cells
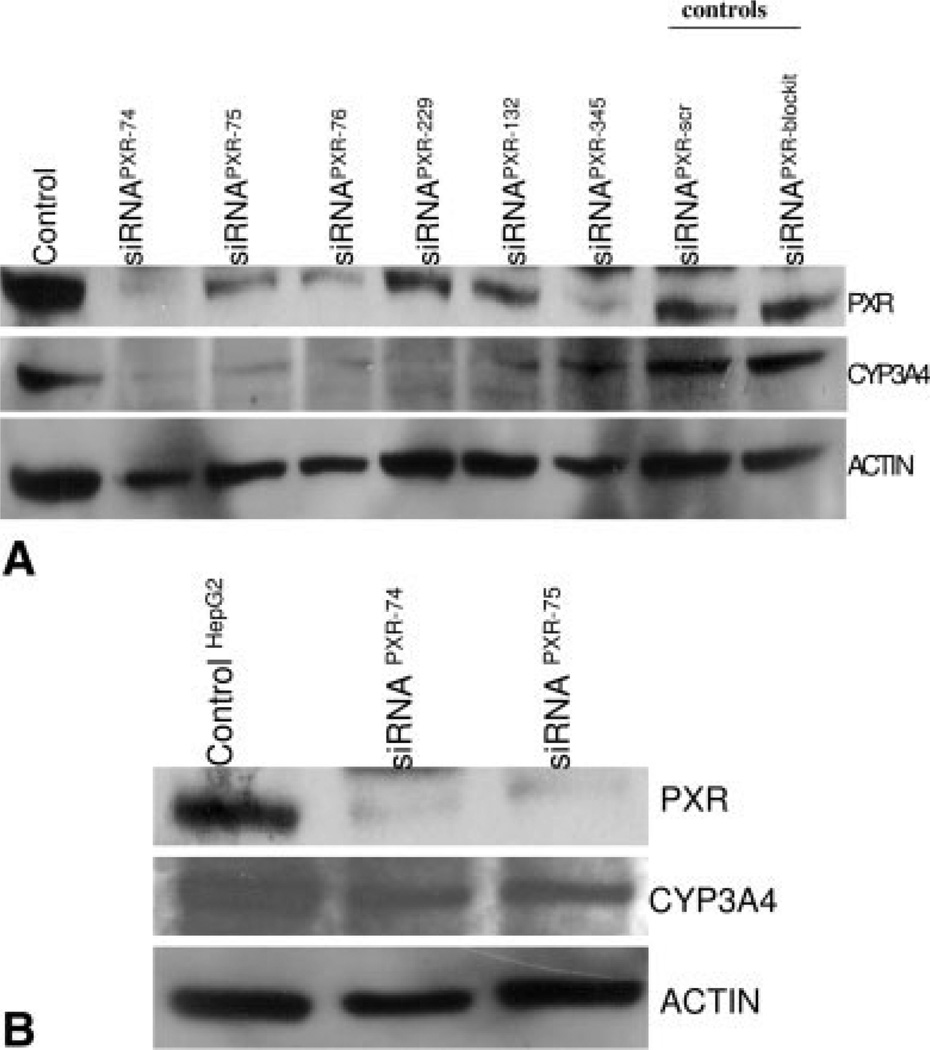
To test the hypothesis that PXR is the critical regulator of P450 3A4 in osteosarcoma cells, we analyzed the effects of inhibiting the expression of endogenous PXR on the expression of P450 3A4 protein using siRNA-mediated gene knockdown. We synthesized a number of different siRNAs corresponding to distinct regions of human PXR spanning the n-terminal, conserved region with the DNA binding domain, and the c-terminal region. When these siRNAs for PXR were transfected into OS187 cells, there was significant down-regulation of endogenous PXR expression levels. Controls consisted of siRNA scrambled oligos or siRNA block-it, a fluorescent oligo used to determine transfection efficacy (Fig. 3A). Down-regulation of PXR protein significantly lowered the expression of endogenous P450 3A4 protein in these cells. In Figure 3B we show that silencing of the PXR gene only modestly down-regulated P450 3A4 expression in HepG2 cells compared with the OS187 cell line. Thus, whereas PXR siRNA efficiently knocked down endogenous PXR levels in OSI87 and HepG2 cells, P450 3A4 levels were down-regulated only in the OS187 cell line, indicating that PXR plays a critical role as a regulator of P450 3A4 expression in the osteosarcoma cell line, but not in the HepG2 cells.
FIGURE 3.
(A) Effect of treatment with PXR siRNA on the expression of P450 3A4 in OS187 cells. Cells were transfected with siRNA for PXR, along with siRNA scramble sequence or the sequence for fluorescent oligos “block-it” used as controls and to determine transfection efficacy, respectively. Total cell extracts were prepared for Western blotting and probed with anti-PXR antibody or anti-P450 3A4 antibody. The membranes were reprobed for actin. (B) Effect of treatment with PXR siRNA on the expression of P450 3A4 in HepG2 cells. Cells were transfected with siRNA for PXR. Total cell extract was prepared for Western blotting and probed with anti-PXR antibody or anti-P450 3A4 antibody. Membranes were reprobed for actin.
Variant PXR Protein Expressed in Osteosarcoma Cell Lines Induces P450 3A4 and MDR1 in Response to Etoposide But Not Rifampin
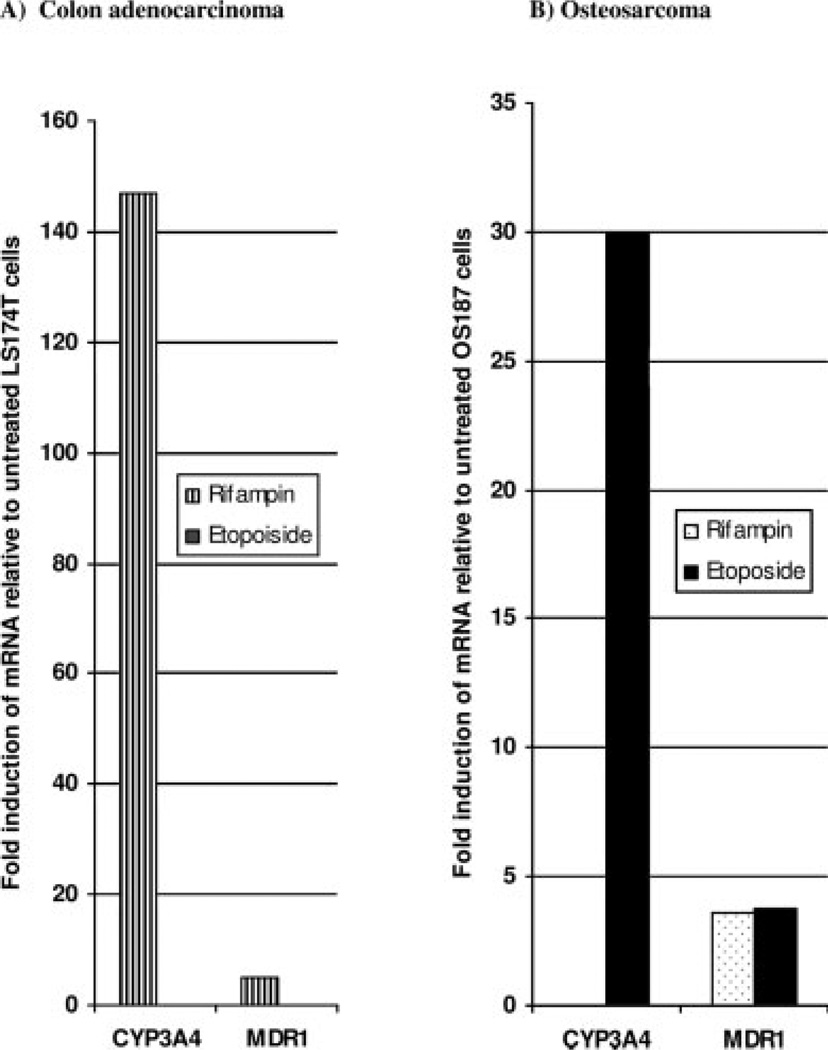
Rifampin is known to activate wildtype PXR-mediated transcription leading to increased P450 3A4 expression and enzymatic activity.4 To further test the hypothesis that the PXR variant expressed in osteosarcoma cell lines regulates the ligand-dependent transcription of target genes differently from wild-type PXR expressed in the colon adenocarcinoma cell line, LS174T, we investigated the ability of the PXR-ligands rifampin and etoposide to increase the mRNA levels of P450 3A4 and MDR1 in both cell lines using QT-PCR. Figure 4 shows that in the osteosarcoma cell line etoposide increased P450 3A4 mRNA expression by 30-fold and MDR1 expression by 3.8-fold. In these cells rifampin had no effect on P450 3A4 expression, even though it induced MDR1 expression by 3.6-fold, similar to etoposide. However, in LS174T, colon adenocarcinoma cells, rifampin increased the mRNA of P450 3A4 more than 140-fold and MDR1 by approximately 5-fold, whereas etoposide had no effect on the expression of P450 3A4 and MDR1 mRNA in these cell lines. Zhuo et al.30 demonstrated that etoposide is unable to induce P450 3A4 through wildtype PXR due to the presence of a repressor that may bind to the N-terminus and would be predicted to have no effect on a variant PXR with a mutation at the 5′ flanking sequence.16 Indeed, their report supports our findings because etoposide but not rifampin induced the expression of PXR target genes in osteosarcoma cell lines that express a PXR with a mutation in the N-terminal region of the protein.
FIGURE 4.
Effect of etoposide and rifampin on the induction of P450 3A4 and MDR1 mRNA in LS174T (A) or OS187 (B) cells. Cells were treated with 30 µM rifampin or 50 µM etoposide for 48 hours. Total mRNA was isolated and subjected to quantitative polymerase chain reaction (QT-PCR). Standard curves representing concentrations of GAPDH, 3A4, and MDR1 in pg/µL for mRNA copies were generated. Expression is represented as the fold increase in 3A4 or MDR1 relative to the untreated LS174T OS187 cells.
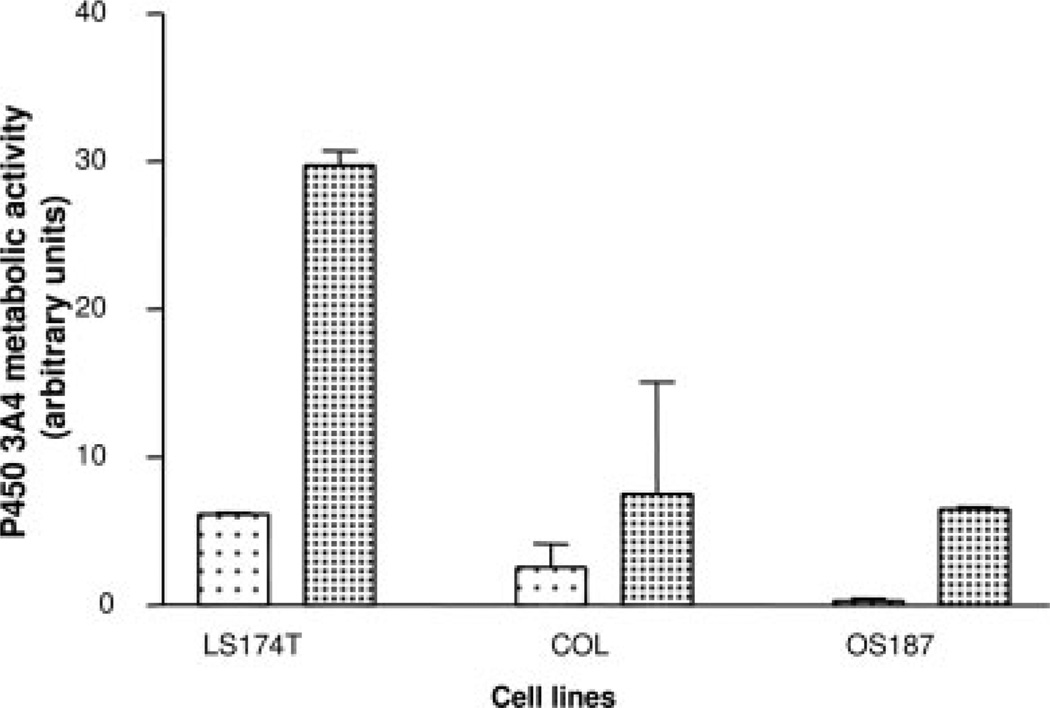
Rifampin Has No Effect on P450 3A4 Metabolic Activity in Osteosarcoma Cell Lines
We further investigated if there was a correlation between the rifampin induction of P450 3A4 mRNA and increase in P450 3A4 catalytic activity. In Figure 5 we show that rifampin is a potent inducer of P450 3A4 activity in the colon adenocarcinoma cell line LS174T consistent with an increase in mRNA levels (Fig. 4). Rifampin, however, did not induce the catalytic activity of P450 3A4 in COL cells, and only modestly induced P450 3A4 in OS187 cells.
FIGURE 5.
Induction of CYP3A activity by rifampin (–) in LS174T and primary osteosarcoma cell lines. Increased activity is represented as arbitrary fluorescent units on the Y-axis. The experimental details are described in Material and Methods. (–) represents control untreated cells.
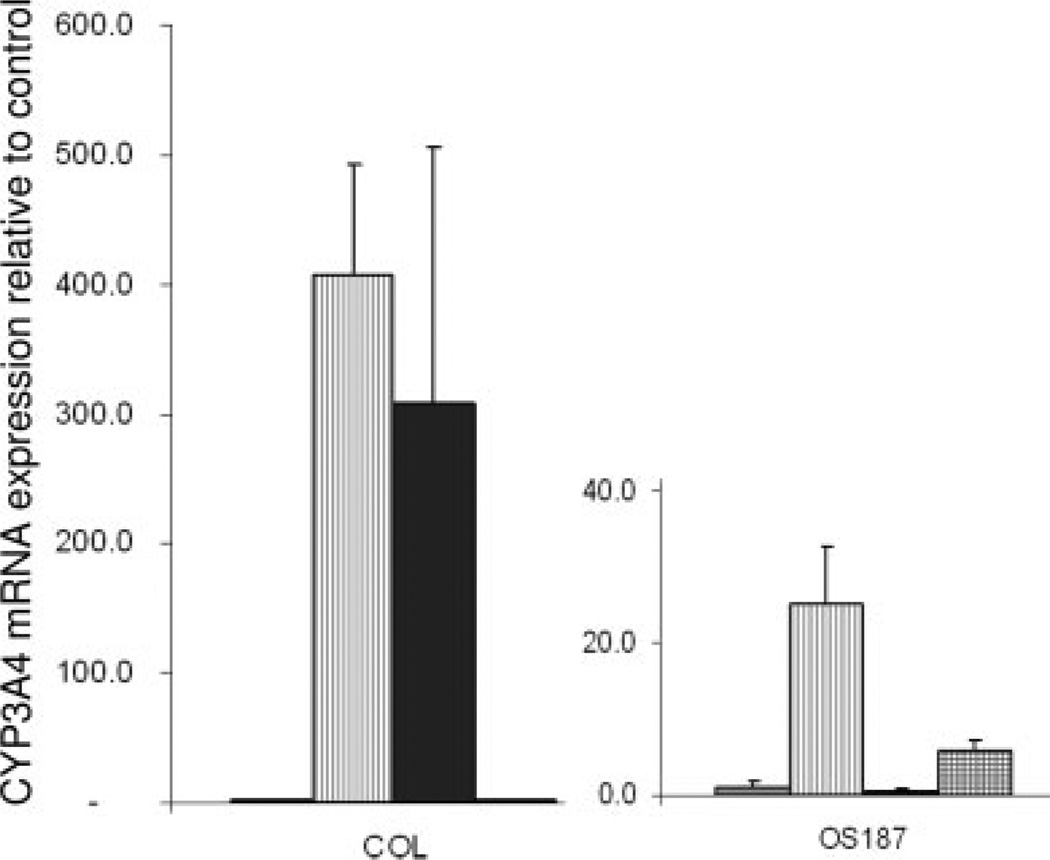
Effect of Etoposide, Doxorubicin, or Ifosfamide on the Expression of P450 3A4 mRNA in Osteosarcoma Cell Lines
We investigated the ability of the conventional chemotherapeutic agents doxorubicin and ifosfamide, which are used for the treatment of osteosarcoma, to increase P450 3A4 mRNA levels and compared them with the nonconventional drug etoposide using QT-PCR (Fig. 6). Etoposide caused significant increases in the expression of P450 3A4 mRNA in the COL osteosarcoma cell line, which expresses higher levels of the PXR protein compared with OS187, as shown in Figure 2. Additionally, doxorubicin increased the expression levels of P450 3A4 mRNA only in COL cells, whereas ifosfamide caused a modest induction of P450 3A4 mRNA only in the OS187 cells.
FIGURE 6.
Induction of P450 3A4 mRNA levels in OS187 and COL cells by etoposide (–), doxorubicin (–), and ifosfamide (–). P450 3A4 mRNA expression levels were determined by quantitative reverse-transcriptase polymerase chain reaction (RT-PCR). The values represent the average of triplicates standardized to GAPDH expression and are represented relative to untreated controls for each cell line. (–) represents control untreated cells.
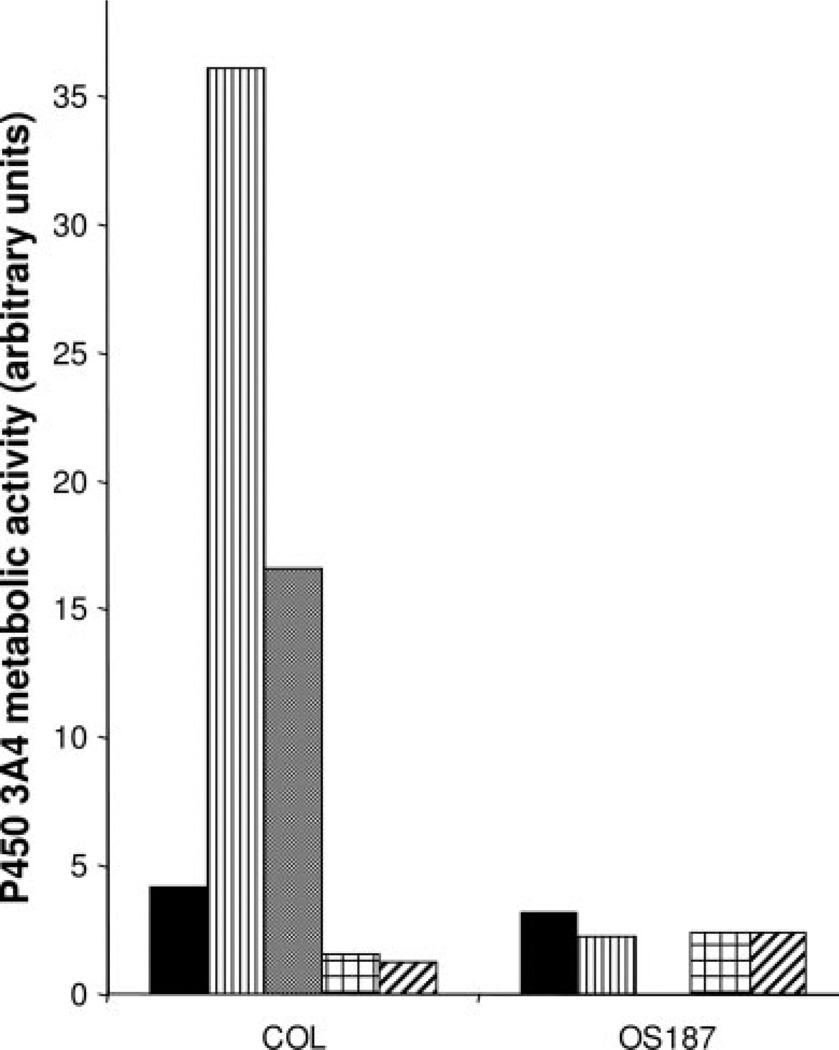
Effect of Etoposide, Doxorubicin, or Ifosfamide on P450 3A4 Enzymatic Activity in Osteosarcoma Cell Lines
We further investigated if the increase in P450 3A4 mRNA caused by etoposide in the COL cells correlated with an increase in P45 3A4 metabolic activity. Figure 7 shows that etoposide increased P450 3A4 metabolic activity only in the COL cell line, and this activity was reversed with the addition of the P450 3A4 inhibitor ketoconazole. Neither doxorubicin nor ifosfamide had an effect on P450 3A4 catalytic activity in either cell line.
FIGURE 7.
Induction of P450 3A4 activity in COL and OS187 cell lines by etoposide, doxorubicin, and ifosfamide represented by control; etoposide; etoposide + ketoconazole doxorubicin; and ifosfamide. CYP3A catalytic activity was measured using 7-benzyl-trifluoromethyl coumarin (BFC) as a probe substrate as described in Materials and Methods.
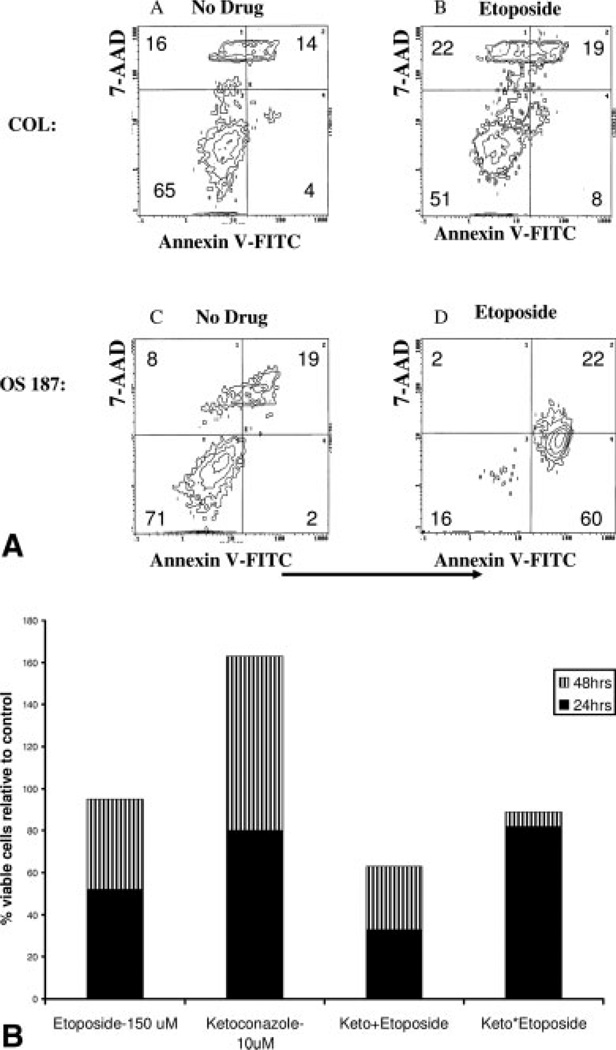
PXR Confers Resistance to Etoposide in Osteosarcoma Cells
To determine if induction of P450 3A4 enzymatic activity by etoposide correlated with resistance in COL cells, we investigated the cytotoxic effect of etoposide on COL and OS187 cell lines. As shown in Figure 8A, etoposide induced apoptosis in the majority of OS187 cells, but caused only a slight increase in the percentage of COL cells undergoing apoptosis. Because ketoconazole reversed the metabolic activity induced by etoposide, we hypothesized that resistance of COL cells to etoposide will also be reversed if cells are exposed to both agents. In Figure 8B we show that the resistance of COL cells to etoposide was decreased upon 24-hour pretreatment of cells with ketoconazole before etoposide exposure. Addition of etoposide and ketoconazole simultaneously was a less effective regime at 24 and 48 hours in the osteosarcoma cell line compared with the sequential exposure. This indicates that inhibition of P450 3A4 activity by ketoconazole indeed sensitizes the COL cells to etoposide in a schedule-dependent manner.
FIGURE 8.
(A) COL and OS187 cells were cultured with 150 µM etoposide for 24 hours and then examined by flow cytometry, using FITC-annexin V and 7-amino-actinomycin D (7AAD). Viable cells do not stain with either reagent. Staining with 7-AAD alone indicates necrotic cells. Staining with Annexin V identifies apoptotic cells and 7AAD staining separates early apoptotic (7AAD−) from late apoptotic (7AAD+) cells. (B) Viable COL cells remaining after exposure to etoposide, or ketoconazole and in combination for 24 hours (−) and 48 hours (−). (Keto+ represents simultaneous addition of both agents; Keto* represents 24 hours pretreatment with ketoconazole before the addition of etoposide.)
DISCUSSION
Approximately 40% of osteosarcoma patients do not achieve the desired results after chemotherapy. Drug resistance is believed to be the most common reason for the failure of chemotherapy in these patients.31 The contributions of PXR to resistance mechanisms remains obscure and an improved understanding of its role may reveal that it is an important determinant for clinical outcomes.
PXR is predominately expressed in the liver, small intestine, and kidney and has been shown to act as a xenobiotic sensor for the regulation of a number of genes including P450 3A4 and MDR1.32 Recent reports have shown that PXR is also expressed in endometrial and breast cancer cells19,28 and plays an important role in vitamin D and bone homeostasis.33 However, the functional implications of PXR expression in these tumors are not clear.
We have demonstrated that PXR is expressed in a number of primary and secondary sarcoma cell lines. Whereas a number of PXR variants have been reported previously, our results show for the first time that a variant form of PXR is expressed in osteosarcoma cells and is recognized differently by antibodies raised against different epitopes. Interestingly, the disparately sized PXR variant expressed in these cell lines correlates approximately with a PXR variant reported by Hustert et al.34 that is ≈1.7 kDa lower in molecular size than the wildtype. The inability of an antibody against the N-terminal region of the protein to detect PXR in these cell lines, and the reduced molecular size of the receptor compared with wild-type, suggests a splice variant with a truncated or modified N-terminal. The N-terminal region of the PXR protein contains the AF-1 region, which plays a role in ligand-mediated transcription of target genes.35,36 Our results suggest that a mutation on the 5′-flanking region of the PXR protein may have an effect on the etoposide-induced PXR-transcription of P450 3A4 in the osteosarcoma cell lines, consistent with data from Koyano et al.29
Our results also show that PXR regulates the expression of P450 3A4 in osteosarcoma and that the induction of P450 3A4 activity via this receptor may be an important mechanism for drug resistance. The regulation of MDR1 in osteosarcomas appears to involve a mechanism independent of PXR because we observed no differences in the induction of MDR1 mRNA (Fig. 3) or PGP activity (data not shown) by either etoposide or rifampin. Additionally, we have observed that CITCO, a ligand specific for the constitutive androstane receptor (CAR), which may play a role in the induction of P450 3A4,37 exhibited no effect on the induction of P450 3A4 in the osteosarcoma cell lines tested (data not shown), ruling out CAR as a possible regulator.
In conclusion, we have determined that a variant PXR protein is expressed in osteosarcoma cell lines. PXR is the major regulator of P450 3A4 protein expression in these tumors and the activity of its target genes appears to correlate with drug resistance. We determined that etoposide increased the expression and catalytic activity of P450 3A4 in the osteosarcoma cell line that expressed higher levels of the PXR protein that correlated with etoposide resistance. Pretreatment of osteosarcoma cells with ketoconazole, a PXR antagonist before exposure to etoposide, significantly increased the sensitivity of these cells to the selected chemotherapeutic agents. The use of compounds such as ketoconazole may be of value for the treatment of patients exhibiting high levels of PXR in their tumor biopsies.
Acknowledgments
Supported in part by the “Robert and Heather Urich Research and Patient Care Fund,” the Walter Cancer Institute, the University of Michigan Comprehensive Cancer Center innovation grant P/G F004721, and in part by grant from the Environmental Protection Agency STAR-R830686 (to B.B.).
Laurence Baker receives grant support from Abbott, Aventis (Sanofi), Lilly, National Institutes of Health, the Robert Urich Foundation, the Hyatt Corporation, and the Walther Foundation. He is on the advisory boards of Ascenta, Hope Foundation, Kanisa, the NCCN Guidelines Committee, and SARC.
Bruce Blumberg is a named inventor on several patents related to SXR: US 6,756,491, US 6,809,178, US 6,984,773.
REFERENCES
- 1.Bielack SS, Kempf-Bielack B, Delling G, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20:776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 2.Antman KH, Ryan L, Elias A, Sherman D, Grier HE. Response to ifosfamide and mesna: 124 previously treated patients with metastatic or unresectable sarcoma. J Clin Oncol. 1989;7:126–131. doi: 10.1200/JCO.1989.7.1.126. [DOI] [PubMed] [Google Scholar]
- 3.Elias A, Ryan L, Sulkes A, Collins J, Aisner J, Antman KH. Response to mesna, doxorubicin, ifosfamide, and dacarbazine in 108 patients with metastatic or unresectable sarcoma and no prior chemotherapy. J Clin Oncol. 1989;7:1208–1216. doi: 10.1200/JCO.1989.7.9.1208. [DOI] [PubMed] [Google Scholar]
- 4.Blumberg B, Sabbagh W, Jr, Juguilon H, et al. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 6.Pascussi JM, Gerbal-Chaloin S, Drocourt L, Maurel P, Vilarem MJ. The expression of CYP2B6, CYP2C9 and CYP3A4 genes: a tangle of networks of nuclear and steroid receptors. Biochim Biophys Acta. 2003;1619:243–253. doi: 10.1016/s0304-4165(02)00483-x. [DOI] [PubMed] [Google Scholar]
- 7.Wrighton SA, Schuetz EG, Thummel KE, Shen DD, Korzekwa KR, Watkins PB. The human CYP3A subfamily: practical considerations. Drug Metab Rev. 2000;32:339–361. doi: 10.1081/dmr-100102338. [DOI] [PubMed] [Google Scholar]
- 8.Ling V. Multidrug resistance: molecular mechanisms and clinical relevance. Cancer Chemother Pharmacol. 1997;40 Suppl:S3–S8. doi: 10.1007/s002800051053. [DOI] [PubMed] [Google Scholar]
- 9.Potter DA, Glenn J, Kinsella T, et al. Patterns of recurrence in patients with high-grade soft-tissue sarcomas. J Clin Oncol. 1985;3:353–366. doi: 10.1200/JCO.1985.3.3.353. [DOI] [PubMed] [Google Scholar]
- 10.Oda Y, Ohishi Y, Saito T, et al. Nuclear expression of Y-box-binding protein-1 correlates with P-glycoprotein and topo-isomerase II alpha expression, and with poor prognosis in synovial sarcoma. J Pathol. 2003;199:251–258. doi: 10.1002/path.1282. [DOI] [PubMed] [Google Scholar]
- 11.Dalton WS, Crowley JJ, Salmon SS, et al. A phase III randomized study of oral verapamil as a chemosensitizer to reverse drug resistance in patients with refractory myeloma. A Southwest Oncology Group study. Cancer. 1995;75:815–820. doi: 10.1002/1097-0142(19950201)75:3<815::aid-cncr2820750311>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 12.Sonneveld P, Suciu S, Weijermans P, et al. Cyclosporin A combined with vincristine, doxorubicin and dexamethasone (VAD) compared with VAD alone in patients with advanced refractory multiple myeloma: an EORTC-HOVON randomized phase III study (06914) Br J Haematol. 2001;115:895–902. doi: 10.1046/j.1365-2141.2001.03171.x. [DOI] [PubMed] [Google Scholar]
- 13.Leonard GD, Fojo T, Bates SE. The role of ABC transporters in clinical practice. Oncologist. 2003;8:411–424. doi: 10.1634/theoncologist.8-5-411. [DOI] [PubMed] [Google Scholar]
- 14.Hughes DP, Thomas DG, Giordano TJ, Baker LH, McDonagh KT. Cell surface expression of epidermal growth factor receptor and Her-2 with nuclear expression of Her-4 in primary osteosarcoma. Cancer Res. 2004;64:2047–2053. doi: 10.1158/0008-5472.can-03-3096. [DOI] [PubMed] [Google Scholar]
- 15.Allison DC, Ridolpho P. Use of a trypan blue assay to measure the deoxyribonucleic acid content and radioactive labeling of viable cells. J Histochem Cytochem. 1980;28:700–703. doi: 10.1177/28.7.6156203. [DOI] [PubMed] [Google Scholar]
- 16.Schuetz E, Lan L, Yasuda K, et al. Development of a realtime in vivo transcription assay: application reveals pregnane X receptor-mediated induction of CYP3A4 by cancer chemotherapeutic agents. Mol Pharmacol. 2002;62:439–445. doi: 10.1124/mol.62.3.439. [DOI] [PubMed] [Google Scholar]
- 17.Yao D, Ding S, Burchell B, Wolf CR, Friedberg T. Detoxication of vinca alkaloids by human P450 CYP3A4-mediated metabolism: implications for the development of drug resistance. J Pharmacol Exp Ther. 2000;294:387–395. [PubMed] [Google Scholar]
- 18.Xie W, Barwick JL, Downes M, et al. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- 19.Masuyama H, Hiramatsu Y, Kodama J, Kudo T. Expression and potential roles of pregnane X receptor in endometrial cancer. J Clin Endocrinol Metab. 2003;88:4446–4454. doi: 10.1210/jc.2003-030203. [DOI] [PubMed] [Google Scholar]
- 20.Crespi CL, Miller VP, Penman BW. Microtiter plate assays for inhibition of human, drug-metabolizing cytochromes P450. Anal Biochem. 1997;248:188–190. doi: 10.1006/abio.1997.2145. [DOI] [PubMed] [Google Scholar]
- 21.Obach RS. Nonspecific binding to microsomes: impact on scale-up of in vitro intrinsic clearance to hepatic clearance as assessed through examination of warfarin, imipramine, and propranolol. Drug Metab Dispos. 1997;25:1359–1369. [PubMed] [Google Scholar]
- 22.Gennuso F, Fernetti C, Tirolo C, et al. Bilirubin protects astrocytes from its own toxicity by inducing up-regulation and translocation of multidrug resistance-associated protein 1 (Mrp1) Proc Natl Acad Sci U S A. 2004;101:2470–2475. doi: 10.1073/pnas.0308452100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen J, Mao HQ, Li W, Lin KY, Leong KW. Biodegradable polyphosphoester micelles for gene delivery. J Pharm Sci. 2004;93:2142–2157. doi: 10.1002/jps.20121. [DOI] [PubMed] [Google Scholar]
- 24.Liu SQ, Saijo K, Todoroki T, Ohno T. Induction of human autologous cytotoxic T lymphocytes on formalin-fixed and paraffin-embedded tumour sections. Nat Med. 1995;1:267–271. doi: 10.1038/nm0395-267. [DOI] [PubMed] [Google Scholar]
- 25.Tabb MM, Sun A, Zhou C, et al. Vitamin K2 regulation of bone homeostasis is mediated by the steroid and xerobiotic receptor size. J Biol Chem. 2003;45:43919–43927. doi: 10.1074/jbc.M303136200. [DOI] [PubMed] [Google Scholar]
- 26.Honkakoski P, Negishi M. Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem J. 2000;347(Pt 2):321–337. doi: 10.1042/0264-6021:3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hukkanen J, Vaisanen T, Lassila A, et al. Regulation of CYP3A5 by glucocorticoids and cigarette smoke in human lung-derived cells. J Pharmacol Exp Ther. 2003;304:745–752. doi: 10.1124/jpet.102.038208. [DOI] [PubMed] [Google Scholar]
- 28.Dotzlaw H, Leygue E, Watson P, Murphy LC. The human orphan receptor PXR messenger RNA is expressed in both normal and neoplastic breast tissue. Clin Cancer Res. 1999;5:2103–2107. [PubMed] [Google Scholar]
- 29.Koyano S, Kurose K, Saito Y, et al. Functional characterization of four naturally occurring variants of human pregnane X receptor (PXR): one variant causes dramatic loss of both DNA binding activity and the transactivation of the CYP3A4 promoter/enhancer region. Drug Metab Dispos. 2004;32:149–154. doi: 10.1124/dmd.32.1.149. [DOI] [PubMed] [Google Scholar]
- 30.Zhuo X, Zheng N, Felix CA, Blair IA. Kinetics and regulation of cytochrome P450-mediated etoposide metabolism. Drug Metab Dispos. 2004;32:993–1000. [PubMed] [Google Scholar]
- 31.Keohan ML, Taub RN. Chemotherapy for advanced sarcoma: therapeutic decisions and modalities. Semin Oncol. 1997;24:572–579. [PubMed] [Google Scholar]
- 32.Pascussi JM, Jounaidi Y, Drocourt L, et al. Evidence for the presence of a functional pregnane X receptor response element in the CYP3A7 promoter gene. Biochem Biophys Res Commun. 1999;260:377–381. doi: 10.1006/bbrc.1999.0745. [DOI] [PubMed] [Google Scholar]
- 33.Pascussi JM, Robert A, Nguyen M, et al. Possible involvement of pregnane X receptor-enhanced CYP24 expression in drug-induced osteomalacia. J Clin Invest. 2005;115:177–186. doi: 10.1172/JCI21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hustert E, Zibat A, Presecan-Siedel E, et al. Natural protein variants of pregnane X receptor with altered transactivation activity toward CYP3A4. Drug Metab Dispos. 2001;29:1454–1459. [PubMed] [Google Scholar]
- 35.Kumar R, Thompson EB. The structure of the nuclear hormone receptors. Steroids. 1999;64:310–319. doi: 10.1016/s0039-128x(99)00014-8. [DOI] [PubMed] [Google Scholar]
- 36.Kumar R, Johnson BH, Thompson EB. Overview of the structural basis for transcription regulation by nuclear hormone receptors. Essays Biochem. 2004;40:27–39. doi: 10.1042/bse0400027. [DOI] [PubMed] [Google Scholar]
- 37.Maglich JM, Parks DJ, Moore LB, et al. Identification of a novel human constitutive androstane receptor (CAR) agonist and its use in the identification of CAR target genes. J Biol Chem. 2003;278:17277–17283. doi: 10.1074/jbc.M300138200. [DOI] [PubMed] [Google Scholar]