Abstract
The cell wall is an important determinant of plant cell form. Here we define a class of Arabidopsis root hair mutants with defective cell walls. Plants homozygous for kojak (kjk) mutations initiate root hairs that rupture at their tip soon after initiation. The KJK gene was isolated by positional cloning, and its identity was confirmed by the molecular complementation of the Kjk− phenotype and the sequence of three kjk mutant alleles. KOJAK encodes a cellulose synthase-like protein, AtCSLD3. KOJAK/AtCSLD3 is the first member of this subfamily of proteins to be shown to have a function in cell growth. Subcellular localization of the KOJAK/AtCSLD3 protein using a GFP fusion shows that KOJAK/AtCSLD3 is located on the endoplasmic reticulum, indicating that KOJAK/AtCSLD3 is required for the synthesis of a noncellulosic wall polysaccharide. Consistent with the cell specific defect in the roots of kjk mutants, KOJAK/AtCSDL3 is preferentially expressed in hair cells of the epidermis. The Kjk− phenotype and the pattern of KOJAK/AtCSLD3 expression suggest that this gene acts early in the process of root hair outgrowth. These results suggest that KOJAK/AtCSLD3 is involved in the biosynthesis of β-glucan-containing polysaccharides that are required during root hair elongation.
Keywords: Arabidopsis, cell wall, cellulose synthase-like gene, β-glucan, root hair, morphogenesis
Cell morphogenesis is the process by which postmitotic cells attain their final form and is responsible for the diversity of cell morphologies found in plants. Spatially controlled and cell-specific cell wall deposition is a major determinant of plant cell form. Few genes involved in determining plant cell shape have been characterized at the molecular level. It is clear the cytoskeleton is a major determinant of cell shape (Mathur and Chua 2000), and genetic analysis of the ZWICHEL (ZWI) gene has defined a role for a class of kinesin-related proteins in the formation of the stellate trichome of the shoot epidermis of Arabidopsis (Oppenheimer et al. 1997). The swollen cell phenotype of plants homozygous for loss of function mutations in the RADIAL SWELLING1 (RSW1) gene illustrates the role of cell wall cellulose in the establishment and maintenance of cell shape (Arioli et al. 1998). Cellulose, which is a major component of the cell wall, is synthesized at the plasma membrane. Matrix polysaccharides that constitute the noncellulosic wall fraction, such as xyloglucan and pectin, are synthesized in the endomembrane system of the cell and are delivered to the cell surface in Golgi-derived vesicles. Little is known about the enzymes responsible for the biosynthesis of wall polysaccharide components. None of the enzymes associated with the synthesis of xyloglucan backbone, xylan, or callose has yet been isolated. To understand the molecular mechanism underpinning wall formation during cell growth in plants, we have identified a set of root hair mutants in Arabidopsis thaliana with defective cell walls.
The Arabidopsis root hair has been used to genetically dissect the process of cellular morphogenesis in plants. Root hairs are tubular extensions that form on the outer surface of specialized epidermal cells called trichoblasts (Schiefelbein and Somerville 1990; Dolan et al. 1994). Hairs play important roles in a diverse array of processes including water and ion uptake and anchorage and are the site of interaction with a range of symbiotic microorganisms. The first stage of root hair morphogenesis involves a change in the polarity of cell growth leading to the formation of a bulge at the distal end of the trichoblast. Bulge formation is accompanied by local cell wall thinning and a decrease in apoplastic pH that may be required for the modification of cell wall polymers (Bibikova et al. 1998). A polarized cytoplasm organizes in this protrusion with the formation of a characteristic tip growth morphology when the hair is 20–40 μm in length. The tip is a site of localized cell wall synthesis through an accumulation of secretory vesicles containing protein and polysaccharide polymers. Once tip growth is established, the hair continues to grow, generating a long tubular-shaped cell. Physiological studies have shown that a tip-focused calcium influx generates a cytosolic Ca2+ gradient necessary for root hair growth (Schiefelbein et al. 1992; Wymer et al. 1997). Growth ceases on vacuolation of the hair tip.
The genetic analysis of root hair development has identified several genes that are required for the initiation and growth of the root hair. RHL1, RHL2, and RHL3 genes are active during the formation of a bulge early in root hair growth. RHL1 encodes a nuclear protein of unknown function that is required for the formation of the polarized outgrowth (Schneider et al. 1998). RHD6 activity is necessary to localize the site of hair initiation in the trichoblast. RHD6 acts through an auxin/ethylene pathway, as the rhd6 mutant phenotype can be rescued by the application of either auxin or ethylene (Masucci and Schiefelbein 1994). RHD1 strengthens the cell wall in the vicinity of the bulge (Schiefelbein and Somerville 1990). RHD2 is necessary for hair outgrowth, as plants homozygous for recessive loss of function alleles stop growing soon after the formation of a bulge (Schiefelbein and Somerville 1990). Later-acting genes such as COW1 (Grierson et al. 1997), TIP1 (Ryan et al. 1998), and RHD3 (Galway et al. 1997), and RHD4 are active in the maintenance of hair elongation and polarity (Schiefelbein and Somerville 1990). RHD3 encodes a protein with GTP-binding motifs that may be involved in cell signaling during hair formation, its precise role has not been characterized (Wang et al. 1997). While genetic analysis has defined a set of genes required for root hair morphogenesis, the molecular basis of hair outgrowth is still far from being understood.
To identify genes required for cell wall biosynthesis during cell growth, we screened our root hair mutant collection for phenotypes that resemble root hairs treated with an inhibitor of cellulose synthesis, 2,6-dichlorobenzonitrile (DCB; Delmer et al. 1987). DCB-treated root hairs rupture at their tip because of the uncoupling of cellulose biosynthesis and protoplast growth (L. Dolan, unpubl). Here we report the identification of an A. thaliana cellulose synthase-like protein, AtCSLD3, encoded by the KOJAK gene that is required for the formation of the cell wall during root hair morphogenesis.
Results
Root hairs of kjk mutants rupture soon after initiation
Arabidopsis root hairs grown in the presence of the cellulose synthesis inhibitor DCB (2,6-dichlorobenzonitrile) rupture at their tips (data not shown). To identify mutants with cell wall defects, we screened our collection of Arabidopsis root hair mutants for ruptured root hair phenotypes. In this screen, one line (D4225) was isolated from a Ds transposon-mutagenized Landsberg erecta (Ler) population (Bancroft et al. 1992), and two lines, E1025 and E1100, were isolated from an EMS-mutagenized Columbia (Col) population. The progeny of pairwise combinations of crosses between these mutants had mutant phenotypes indicating that the mutations in these lines are alleles of a single gene. This gene was named KOJAK (KJK) because of the bald phenotype of the mutant roots. Lines D4225, E1025, and E1100 were designated kjk-1, kjk-2, and kjk-3, respectively. Backcrosses of each of the kjk mutants to their respective wild type produced F1 plants with wild-type phenotypes. The 3:1 segregation of Kjk+ and Kjk− plants in the F2 generation of these backcrosses indicated that the mutations segregate as single recessive alleles. Linkage analysis showed that the kjk-1 mutation was not caused by the insertion of a Ds transposon. All F1 plants derived from the cross between plants homozygous for kjk and plants homozygous for previously published mutations in root hair growth (rhd1, rhd2, rhd3, rhd4, rhd6, tip1, and cow1) had wild-type root hair phenotypes indicating that kjk defines a new gene required for root hair growth.
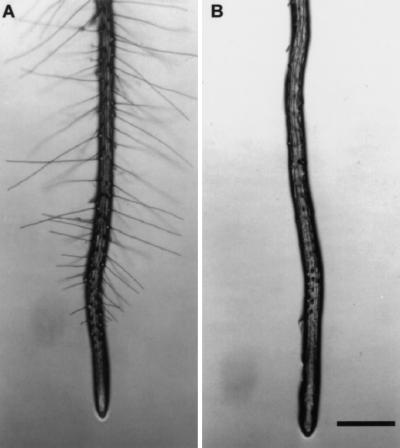
Plants homozygous for any kjk allele formed no hairs (Fig. 1), although deformed root hairs were occasionally observed. During normal root hair development, the trichoblast produced a small swelling (bulge) at the end of the cell nearest the meristem, from which a tip growing hair emerged. kjk mutants produced the initial bulge but failed to develop a root hair. Instead, the bulge continued to swell spherically until it burst eventually, resulting in the extrusion of the protoplast through a fracture in the wall (Fig. 2A–D). The rupture always formed near the dome of the bulge. The dead cells were visualized because of the staining of their cytoplasmic constituents with propidium iodide, which is normally restricted to the cell wall of living cells (Fig. 2E,F). Files of stained cells indicated that every hair cell in the file undergoes lysis (Fig. 2F). These files were located over the anticlinal cell walls of underlying cortical cells (the location in which hair cells normally develop), indicating that KJK played no role in the specification of root epidermal cell identity. The gross morphology of the shoot system of kjk mutants was indistinguishable from wild type.
Figure 1.
Plants homozygous for the kjk mutation do not form root hairs. (A) Elongated root hairs are visible on the wild-type (Ler ecotype) root. (B) Plants homozygous for the kjk-1 mutation do not form root hairs. The characteristic bulges are visible in the early differentiation zone. Scale bar, 100 μm.
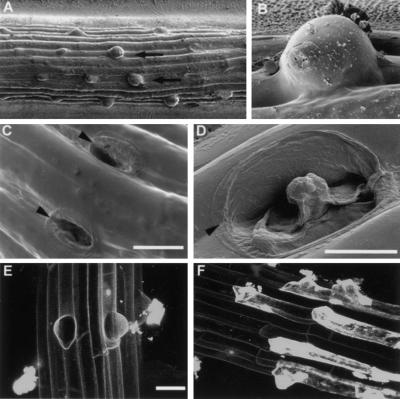
Figure 2.
Root hairs swell and burst soon after initiation in plants homozygous for the kjk mutation. (A–D) Cryo-Scanning electron micrographs of roots of plants homozygous for the kjk-1 mutation show that the outer wall of the trichoblast swells (arrows) excessively (A). The bulge continues to swell until it bursts (B), killing the trichoblasts (arrowheads) (C,D). (E,F) Confocal images of the root homozygous for the kjk mutation stained with propidium iodide, which stains the intercellular spaces between living cells and the contents of dead cells (E). Stripes of compressed dead trichoblasts between nonhair epidermal cell files (F). Scale bars, 10 mm.
kjk is epistatic to cow1
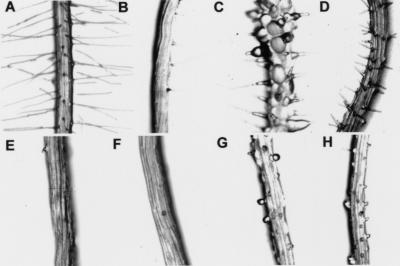
To characterize the role of KJK in root hair morphogenesis, double mutants comprising kjk and other mutants with defects in root hair elongation were constructed. Double mutants were identified in the F2 populations of crosses between kjk and the respective root hair mutant and verified by backcrossing to both parents (test cross; Fig. 3). kjk and rhd2 single mutants had identical phenotypes. The phenotype of the kjk rhd2 double mutant was indistinguishable from either parent, indicating that two genes may act at the same stage of development. rhd6 mutants develop few hairs that originate in variable positions along the trichoblast. In roots of seedlings homozygous for both kjk and rhd6, small bulges were produced in place of hairs as in the kjk mutant, but they form in variable positions along the trichoblast as in the rhd6 single mutant. This additive phenotype indicates that KJK activity is required for the growth of root hairs in the rhd6 background (Fig. 3B–F). While the rhd1 mutant produced root hairs with excessive bulges at the base (Fig. 3C; Schiefelbein and Somerville 1990), the entire outer cell wall of the kjk rhd1 trichoblast was swollen, and no root hairs were formed (Fig. 3G). tip1 root hairs were shorter and more branched than wild type, whereas kjk tip1 double mutants developed short, thickened root hairs (Fig. 3D–H). Hairs on rhd3 mutants were wavy, and hairs on rhd4 plants formed bulges and constrictions along their length. kjk rhd3 double mutants had very short, slightly wavy root hairs, and kjk rhd4 had shortened root hairs with bulges and constrictions along their length (data not shown). The double mutants with rhd1, tip1, rhd3, and rhd4 had intermediate phenotypes with characteristics of each single mutant, indicating that KJK acts independent of these genes. Interestingly, the cell-rupture phenotype of kjk mutants is suppressed by tip1, rhd1, rhd3, and rhd4. cow1 root hairs were shorter and branch more often than wild type. kjk is epistatic to cow1; that is, double mutant displayed a Kjk− phenotype, indicating that these genes act in the same pathway, perhaps with KJK acting earlier in hair growth.
Figure 3.
Root phenotype of double mutants. (A) Root hairs of the wild-type root. (B–E) Single mutant phenotypes and (F–H) double mutant phenotypes. Roots of plants homozygous for rhd6-1 (B) are largely hairless. Plants homozygous for the rhd1-1 mutation (C) produce hairs with excessive swellings at its base. Trichoblasts of plants homozygous for tip1 (D) can produce up to three short, branched root hairs. Characteristic bulges on the root of a plant homozygous for the kjk-2 mutation (E). Those hairs that form on roots of plants homozygous for both kjk-2 and rhd6 are short (F). The entire outer cell wall of the trichoblast of a plant homozygous for both kjk-2 and rhd11 is swollen (G), but trichoblasts neither rupture nor form a hair. Roots of plants homozygous for both kjk-1 and tip1-2 (H) produce a few abnormal root hairs.
Molecular cloning of the KJK gene
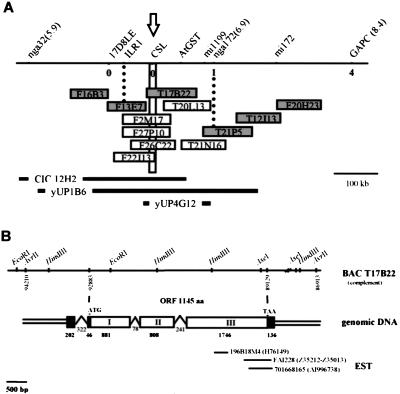
The KJK gene was cloned using a map-based cloning strategy. A segregating F2 family was made from a cross between plants homozygous for the kjk-1 mutation in the Ler background and wild-type Col plants. kjk-1 was initially mapped to the top of chromosome 3 between SSLP markers nga32 and nga172 (Bell and Ecker 1994) using a population of 80 chromosomes (Fig. 4A). Analysis of a further 200 chromosomes identified a single recombination event between nga172 and kjk-1 whereas no recombinants were identified between kjk-1 and CAPS marker 17D8LE (Bartel and Fink 1995). Examination of BAC sequences in this region identified a gene encoding a cellulose synthase-like (CSL) protein, AtCSLD3. Using a CAPS marker in the AtCSLD3 gene (CSL, see Material and Methods) that identified a polymorphism between Ler and Col, we found no recombinants between CSL and kjk-1 in the population of 220 chromosomes.
Figure 4.
Mapping of the kjk mutation and organization of the KJK/AtCSLD3 gene. (A) KJK is located on the top of chromosome 3. Adjacent SSLP and CAPS markers with genetic distances are indicated. The number of recombinants identified from a population of 200 mutant chromosomes is given above the respective loci. The vertical bar indicates the region of chromosome 3 containing the AtCSLD3 gene. The positions of BAC and YAC clones spanning this region are indicated. The sequenced BACs are represented by filled rectangles. (B) The restriction map of the KJK/AtCSLD3 gene is aligned with a graphical representation of the structure of the KJK/AtCSLD3 gene. Open boxes are exons (I to III, determinated from cDNA) and the gap between is introns. Shaded boxes indicate untranslated sequences. ATG (start codon), TGA (stop codon) and Arabidopsis ESTs showing identities with the gene are indicated.
To show that AtCSLD3 encodes the gene that is defective in the kjk-1 mutant, we transformed plants homozygous for the kjk-1 mutation with a 7.2-kb AvrII genomic fragment that includes the entire AtCSLD3 gene (Fig. 4B). Twelve independent primary transformants selected on kanamycin had a wild-type root hair phenotype (data not shown). Examination of T2 plants showed the cosegregation of the kanamycin resistance with the wild-type phenotype. This result provides firm genetic evidence that the Kjk− phenotype is caused by mutation in the AtCSLD3 gene. This gene is hereafter designated KOJAK/Arabidopsis thaliana cellulose synthase-like protein D3 (KJK/AtCSLD3). Using the first exon of KJK/AtCSLD3 as a probe on a Southern blot of Ler genomic DNA, we showed that there is a single copy of the gene per haploid genome (data not shown).
The KJK/AtCSLD3 cDNA was cloned by RACE PCR (Frohman et al. 1988). One transcript was detected. The KJK/AtCSLD3 cDNA is 3819 nucleotides long with an open reading frame (ORF) of 1145 amino acids (Fig. 4B). Translation was assumed to begin at nucleotide 249, the first ATG codon of the open reading frame. Blast searching (Altschul et al. 1997) the Stanford A. thaliana database identified similar (99% identical at nucleotide level) expressed sequence tags from A. thaliana (Fig. 4B). Comparison of cDNA and genomic sequences revealed that the KJK/AtCSLD3 gene has three introns, including one in the 5′UTR (Fig. 4B).
KJK encodes a cellulose synthase-like protein
Analysis of the deduced amino acid sequence of KJK/AtCSLD3 showed that it is a member of the D subfamily of cellulose synthase-like (CSL) genes identified in A. thaliana (Cutler and Somerville 1997). The KJK/AtCSLD3 amino acid sequence shares 64%–68% identity and 76%–81% similarity with the other four CSLD members (CSLD1, D2, D4, and D5; Fig. 5A). The KJK/AtCSLD3 protein is similar in size to several cellulose synthase catalytic subunit proteins (CESA). These include the Arabidopsis RSW1, ATHA, ATHB (Arioli et al. 1998), IRX3 (Taylor et al. 1999), and ARAXCELA (Wu et al. 1998); the cotton CELA1 (GhCESA1; Pear et al. 1996); and PtCESA2 from Populus tremuloides. The CSLD family constitutes a distinct lineage among the CESA and AtCSL proteins (Fig. 5B).
Figure 5.
KJK encodes a cellulose synthase-like protein. (A) Amino acids sequence of the KJK/AtCSLD3 protein. Conserved residues (white letters) between Arabidopsis cellulose synthase-like AtCSLD (AtCSLD1, AtCSLD4, AtCSLD5) and CESA members from Arabidopsis thaliana (ATHA, ATHB, RSW1, IRX3, ARAXCELA), Populus tremuloides (PtCESA2) and Gossypium hirsutum (GhCESA1). Additional amino acids present in AtCSLD subfamily are underlined and italicized. The position of the cysteine-rich region (C in red), the eight presumed transmembrane segments (green upperlining), the conserved subdomains U1 to U4 (red upperlining) that contain the three D residues and the QxxRW (in red) signature are indicated. The position of the three kjk mutations (amino acids Q121, R889, substituted by stop codon in kjk-2 and kjk-1 alleles, respectively, and E830–K substitution in kjk-3) are indicated by an asterisk. (B) Schematic representation of KJK/AtCSLD3 structure. Shaded boxes represent conserved sequences. The cysteine-rich region, the U1 to U2 subdomains, the plant-specific and conserved region (P-CR), and the plant hypervariable region (HVR) are indicated. (C) Relationship tree of CESA and AtCSL subfamilies (A, B, D, and G) products. Agrobacterium tumefaciens CELA (AgtCESA1) was used to root the tree.
Alignment of sequences shows that several regions in the KJK/AtCSLD3 protein are conserved in the plant CESA and Arabidopsis CSLD proteins (Fig. 5A–C). KJK/AtCSLD3 contains the four highly conserved subdomains (U-1 through U-4) that characterize the processive β-glycosyl transferases in plants and bacteria (Saxena et al. 1995; Pear et al. 1996). These regions contain the three conserved aspartate (D) residues and the QxxRW motif that have been proposed to be involved in substrate (UDP-glucose) binding and/or catalysis (Fig. 5A–C). Surrounding these motifs, KJK/AtCSLD3 shares one plant-specific, conserved region (P-CR) and one hypervariable region (HVR) with plant CESA proteins (Fig. 5C; Pear et al. 1996; Arioli et al. 1998; Delmer 1999; Taylor et al. 1999). However, KJK/AtCSLD3 has additional amino acids in the P-CR domain (Fig. 5A). These additional amino acids are only found in the AtCSLD protein subfamily. The region close to the N terminus contains a cysteine-rich region (Fig. 5A–C), which has been suggested to form a zinc finger–binding domain involved in protein–protein interactions (Delmer 1999). This motif is also present in CESA cellulose synthases (Pear et al. 1996; Arioli et al. 1998; Taylor et al. 1999) but is located closer to the N-terminal region. Therefore, KJK/AtCSLD3 is likely to be involved in the synthesis of cellulose or other related β-glycans (Cutler and Somerville 1997).
Molecular analysis of the kjk mutant alleles
To define the molecular basis for the kjk mutations, the genomic DNA sequence of the three kjk mutant alleles was determined. We identified a single mutation in each of the mutant alleles. kjk-1 has a C to T transition, which results in the introduction of a stop codon in place of the arginine (R) at position 889 in the conserved QxxRW motif (Fig. 5A–C). kjk-2 has a stop codon in place of the glutamine (Q) at position 121, which is predicted to cause premature termination of translation. kjk-2 is a putative complete loss of function mutation. kjk-3 has a G to A transition, resulting in a change of a glutamic acid (E) to a lysine (K) at position 830 close to the conserved U3 segment (Fig. 5A–C).
KJK encodes a membrane protein located in the endoplasmic reticulum
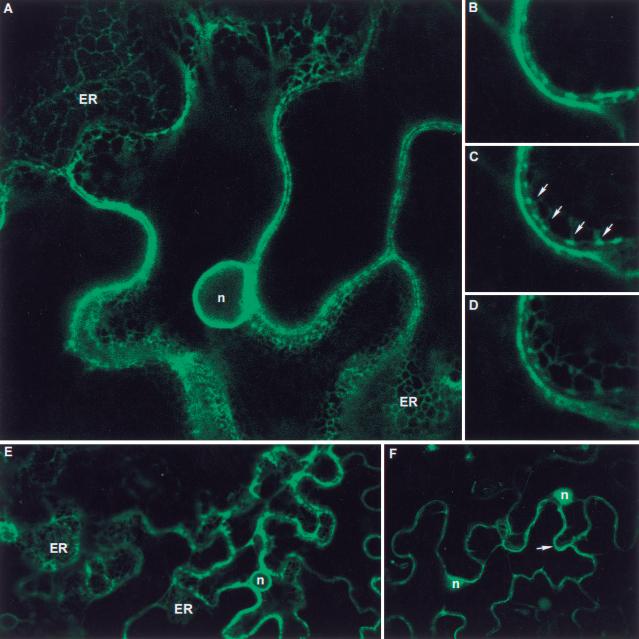
Analysis of peptide structure and hydrophobicity predicted that KJK/AtCSLD3 has eight membrane-spanning domains. Two transmembrane segments are located between residues 292–312 and 319–335, and six further transmembrane segments are detected in the C-terminal region between residues 926 and 1129 (Fig. 5A–C). KJK/AtCSLD3 has no obvious N-terminal signal sequence. These data suggest that KJK/AtCSLD3 is a membrane protein anchored to the plasma membrane, as predicted for a cellulose synthase catalytic subunit, or to a membrane in the endomembrane system. Subcellular localization of KJK/AtCSLD3 protein was determined in vivo using a translational fusion between KJK/AtCSLD3 and the green fluorescent protein 4 (GFP4). The KJK/AtCSLD3::mGFP4 construct under the control of the constitutive 35S promoter was delivered into Nicotiana benthamiana leaf cells by a transient Agrobacterium infiltration assay. All infiltrated epidermal cells showed that the KJK/AtCSLD3–GFP4 fusion protein was present in the endoplasmic reticulum that is located in the thin layer of cytoplasm between the plasma membrane and the vacuole (Fig. 6A–D). No KJK/AtCSLD3–GFP4 was detected in the plasma membrane, but it is possible that small amounts were present that we were unable to detect. The KJK/AtCSLD3–GFP4 fusion protein was excluded from vacuoles, organelles, and nucleus, and no free cytoplasmic fluorescence was detected. A similar subcellular distribution was observed in cells that were transformed with a GFP4 that is targeted specifically to the endoplasmic reticulum (ER-GFP4; Fig. 6E), as described previously (Haseloff et al. 1995). Cells transformed with GFP4 alone, which is located in the cytoplasm and the nucleus, are shown for comparison (Fig. 6F). These data suggest that KJK/AtCSLD3 is located in the ER membrane.
Figure 6.
Subcellular localization of the KJK/AtCSLD3 protein using GFP. Tobacco epidermis was infiltrated with Agrobacterium tumefaciens containing a binary vector with the 35S promoter and GFP fusion. (A–D) Confocal sections of transformed epidermal cells. KJK/AtCSLD3-GFP fusion protein is located in the endoplasmic reticulum (ER) (A). Successive 1-μm confocal sections (B,C,D) show that GFP fluorescence is located in the endoplasmic reticulum (arrows). A similar subcellular distribution was observed with the control ER-targeted GFP (E). The cytoplasmic GFP is distributed throughout the cytoplasm (arrow) restricted between the vacuole and the cell wall (F). (ER) Endoplasmic reticulum; (n) nucleus.
Expression pattern of KJK
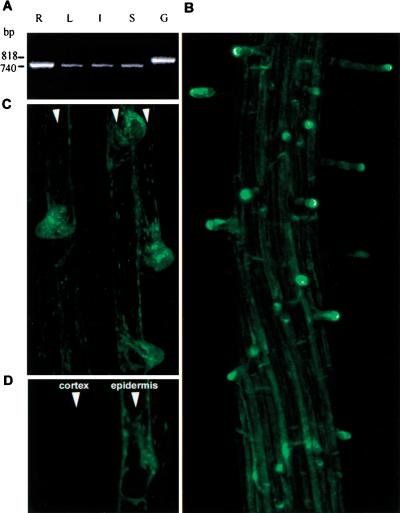
Steady-state levels of KJK/AtCSLD3 mRNA were examined by Northern blot and RT–PCR analysis. Northern blot analysis failed to detect KJK/AtCSLD3 transcripts. In contrast, RT–PCR showed that this gene is expressed throughout the wild-type plant. Signal was detected in leaves, roots, stems, and inflorescences (Fig. 7A). KJK/AtCSLD3 gene expression was also characterized in mutants homozygous for the three alleles to examine the effect of these mutations on KJK/AtCSLD3 mRNA accumulation. A dramatic reduction in the abundance of the transcript was detected in seedlings homozygous for the kjk-2 allele (data not shown). Wild-type levels of KJK/AtCSLD3 mRNA were observed in seedlings homozygous for kjk-1 and kjk-3, which implies that these mutations alter KJK/AtCSLD3 protein function but not KJK/AtCSLD3 transcript synthesis or stability.
Figure 7.
Analysis of KJK/AtCSLD3 expression. (A) RT–PCR analysis of KJK/AtCSLD3. Total RNA was isolated from roots (R), leaves (L), inflorescences (I), and inflorescence stems (S) of wild-type plants. Wild-type genomic DNA (G) was used as control. (B–D) Projections of confocal images of a 5-d-old wild-type seedling transformed with KJK/AtCSLD3::GFP. GFP fluorescence is detected preferentially in root hair cells (arrowheads) (B). Weak KJK/AtCSLD3 expression is visualized in the nonhair epidermal cells (C). No expression was detected in cortical cells (D).
To determine the cells in which KJK/AtCSLD3 is expressed, the KJK/AtCSLD3 gene and its native promoter were fused to the endoplasmic reticulum targeted-GFP2.5 (ERGFP2.5). Images of roots from transformed wild-type plants showed that KJK/AtCSLD3–ERGFP2.5 fusion is preferentially expressed in the root hair cells (Fig. 7B,C). KJK/AtCSLD3 expression is first detected in cells at the early stages of root hair growth—during the formation of the bulge—and continues to be expressed in growing root hairs. KJK/AtCSLD3 is occasionally expressed at much lower levels in non–root hair epidermal cells (Fig. 7C) and in lateral root cap cells (data not shown). No expression was observed in the cortical or endodermal cells (Fig. 7D) or vascular cylinder (data not shown). No GFP fluorescence was detected in the shoot meristem, cotyledons, or leaves of young seedlings, indicating that KJK/AtCSLD3 is not expressed at this stage of shoot development (infloresencences were not examined in this study). Thus, KJK/AtCSLD3 is preferentially expressed in growing root hair cells in the root, consistent with its requirement in root hair cell expansion. The KJK/AtCSLD3–ERGFP2.5 construct did not complement the Kjk− phenotype when transformed into plants homozygous for the kjk mutation. This is not unexpected, as the GFP2.5 is itself targeted to the ER and may therefore have altered the topology or function of the KJK/AtCSLD3 protein.
Discussion
A screen for cell wall mutants in A. thaliana using a root hair rupture phenotype led to the isolation of three recessive alleles in the KJK gene. KJK was cloned and found to encode a cellulose synthase-like protein, KJK/AtCSLD3, required for the formation of an intact cell wall during root hair morphogenesis. This is the first report of the isolation and characterization of a cellulose synthase-like protein involved in wall formation during root hair morphogenesis. KJK/AtCSLD3 is one of a battery of effector genes that are likely to be targets of the transcription factor cascade that has been shown to be involved in the specification of cell identity in the root epidermis (Lee and Schiefelbein 1999). Other targets of these transcriptional regulators will be involved in the cellular functions required for the growth and shaping of the root hair cell.
KJK gene is required for root hair cell morphogenesis
One of the earliest stages of root hair growth is the formation of a bulge on the outer face of the trichoblast from which a tip growing root hair emerges. In plants homozygous for each of the three kjk mutations, root hairs initiate in the correct position on the trichoblast but tip growth is not established. Instead, the bulge continues to expand slowly and eventually ruptures. DCB, a cellulose synthase inhibitor, which also causes hair cell rupture, phenocopies this mutation. This suggests that hairs of plants homozygous for kjk mutations have a localized cell wall defect resulting in the formation of a weakened wall. KJK/AtCSLD3 is required at or before the establishment of tip growth for the maintenance of the integrity of the cell wall, and in the absence of KJK/AtCSLD3 activity, the cell wall cannot withstand the internal pressure of the growing protoplast.
KJK acts early in the temporal sequence of hair growth
A number of other genes required for root hair morphogenesis have been described in Arabidopsis. Only RHL1 and RHD3 have been cloned. RHL1 encodes a nuclear protein of unknown function (Schneider et al. 1998), and RHD3 encodes a G-protein of unknown function (Wang et al. 1997). The Kjk− phenotype indicates that KJK/AtCSLD3 acts early in the temporal sequence of hair growth. kjk is epistatic to cow1, suggesting that these genes act in the same pathway. Alternatively, KJK/AtCSLD3 activity may be needed before COW1 activity is required during hair growth. KJK/AtCSLD3 and RHD2 genes are both required at the same stage of development for the transition from swelling formation to tip-growth. RHD2 is required for the formation of the Ca2+ gradient, although it is as yet unclear what RHD2 encodes (Bibikova et al. 1998). The intermediate phenotype of the double mutants with root hair morphogenesis mutants rhd1, rhd3, rhd4, and tip1 suggests that KJK/AtCSLD3 acts independently of these genes. It is probable that these genes act independently at the same stage of hair elongation. Interestingly, these mutations suppress cell rupture of root hairs in double mutants. The slow growth rate of these mutant root hairs may decrease the expansive pressure on the cell wall of kjk hairs and thereby suppress the rupturing. It is nevertheless formally possible that the respective gene products interact directly in the developing root hair cell.
KJK is a member of the cellulose synthase-like family
The sequence of KJK/AtCSLD3 indicates that it encodes a protein similar to processive β-glycosyltransferases that catalyze the transfer of multiple sugar residues from an activated donor substrate to an acceptor molecule. KJK/AtCSLD3 is a member of a multigene family in Arabidopsis that is similar to the catalytic subunit responsible for the synthesis of cellulose (CESA; Cutler and Somerville 1997). More than 40 members have been identified in the Arabidopsis genome. These cellulose synthase-like (CSL) genes were classified into five subfamilies: AtCSLA, AtCSLB, AtCSLD, AtCSLE, and AtCSLG. The hydrophobicity plots of CESA and CSL members predict a structure that is generally similar for each with a number of membrane-spanning domains and highly conserved subdomains U-1 through U-4, which are proposed to be involved in substrate (UDP-glucose) binding and/or catalysis.
We identified mutations in the KJK/AtCSLD3 sequence in each of the different alleles. Plants homozygous for each of the kjk alleles have indistinguishable root hair phenotypes. kjk-1 and kjk-2 are predicted to encode truncated proteins. kjk-2 has a stop codon near the N terminus and low levels of expression indicating that it is a complete loss of function allele. The U-4 conserved subdomain and the six C terminus transmembrane domains that are missing in kjk-1 are therefore essential for function. The missense mutation representing change in one amino acid was found in kjk-3, demonstrating a functional role for this residue (E830).
Plant CESA homologs were first identified from developing cotton fibers (Pear et al. 1996). However, the proof of function in vivo of plant CESA genes was established recently by the characterization of two Arabidopsis mutants defective in cellulose deposition. The temperature-sensitive mutant radial swelling1 (rsw1) has a reduction in cellulose content, an accumulation of noncrystalline β-1,4-glucan, and morphological abnormalities (Arioli et al. 1998). The irregular xylem3 (irx3) mutant has collapsed xylem vessels and decreased cellulose content in inflorescence stems (Turner and Somerville 1997; Taylor et al. 1999).
KJK/AtCSLD3 is located in the endoplasmic reticulum
The primary structure of KJK/AtCSLD3 predicts that it is a membrane protein located in the endomembrane system or plasma membrane. Expression of KJK/AtCSLD3 fused to GFP indicated that KJK/AtCSLD3 is located in the endoplasmic reticulum. The endoplasmic reticulum plays a central role in the biosynthesis of the macromolecules. Many lipids, glycoproteins, and polysaccharides originate in the endoplasmic reticulum and are progressively modified as they move via the Golgi to the plasma membrane. However, we cannot entirely rule out the possibility that low levels of KJK/AtCSLD3 are also present in the plasma membrane or in the Golgi apparatus, which is known to contain glycosyltransferase activities. We are currently generating antibodies to the KJK/AtCSLD3 protein to further characterize it's subcellular distribution.
Role of KJK/AtCSLD3 in wall biosynthesis of the root hair cell
KJK/AtCSLD3 mRNA preferentially accumulates in growing root hair cells and consequently may be involved in cell type–specific β-glucan synthesis. This early and localized expression in the root hair cell is consistent with the mutant phenotype and with the proposed role for KJK/AtCSLD3 in the establishment of the growth of the hair. The plant primary cell wall is composed of a network cellulose microfibrils embedded in a Golgi-derived matrix of hemicellulose, pectins, and proteins. Hemicellulose, pectin, and proteins are processed in the endomembrane system secreted into incipient cell plate and subsequently into the cell wall proper. In contrast, cellulose microfibrils are synthesized in terminal complexes at the plasma membrane (Brown et al. 1996; Kimura et al. 1999).
Whereas KJK/AtCSLD3 is similar to CESA and therefore may be involved in cellulose biosynthesis, its subcellular localization in the endoplasmic reticulum suggests that AtCSLD3 functions in the biosynthesis of other polysaccharides. These include β-xylans, mannans, or the backbone of xyloglucan (Cutler and Somerville 1997; Carpita and Vergara 1998; Delmer 1999). The biochemical analysis of the cell wall of kjk roots will allow the elucidation of the precise function of KJK/AtCSLD3 in the biosynthesis of the cell wall. It is to be anticipated that a defect in KJK/AtCSLD3 will result in a wall with an altered composition, structure, and rheological properties. Unfortunately, the cell-specific phenotype of plants homozygous for kjk mutations makes the analysis of the biochemical defect in the wall of mutants difficult. Future experiments in which KJK/AtCSLD3 is mis- or overexpressed may obviate this problem and identify the precise biochemical function of KJK/AtCSLD3.
Does KJK/AtCSLD3 play a role in morphogenesis of other cell types?
The visible kjk phenotype is restricted to the root hairs. No pleiotropic defects in plant stature have been observed in kjk mutants as described previously for a number of other mutants with defective root hair growth. Plants homozygous for tip1 and rhd3 mutations are smaller than wild type, and tip1 pollen growth is defective (Ryan et al. 1998; Schneider et al. 1998). This suggests that KJK, unlike many of the genes involved in root hair growth, is not required for the growth of other cells throughout the plant. However, its expression in the shoot suggests that it is likely to be involved in the formation of complex polysaccharides in other cell types. The absence of shoot phenotypes in kjk mutants may be because of a compensation of its function in regular cell growth by other pathways or by redundancy of gene function.
Materials and methods
Plant materials and growth conditions
Seeds were grown on a 1:1 potting compost (John Innes No. 1):peat moss mix at 20°C with a 12-h day. For root hair observations, seeds were surface-sterilized for 5 min in 5% sodium hypochlorite and thoroughly rinsed in sterile distilled water. Seeds were placed on the surface of 0.5% phytagel (Sigma) solid medium containing 1% sucrose and MS salts at pH 5.8. Seeds were stratified at 4°C for 48 h in darkness and incubated at 24°C under continuous illumination. Plates were inclined at an angle of 60° to allow the roots to grow along the surface.
Mutant screening and genetic analysis
Arabidopsis thaliana (L.) Heyn. lines used in these experiments were derived from the Landsberg erecta (Ler) and Columbia (Col) backgrounds. Seeds from a Ds transposon-mutagenized M2 population (Bancroft et al. 1992) and an EMS M2 population were plated on solid medium in Petri dishes, grown for 3–4 d, and screened for root hair mutant phenotypes. Putative mutants were transplanted and grown to maturity. M3 seeds were plated on solid media in Petri dishes, seedlings were rescreened, and true breeding lines were maintained. Mutants were backcrossed to the wild type for four generations to eliminate other mutations from the background. For complementation tests and double mutant analysis, kjk was crossed to homozygous root hair mutants (rhd1-1, rhd2-1, rhd3-1, rhd4-1, rhd6-1, cow1-1, and tip1-2). The heterozygous F1 families from each cross were scored and selfed and the F2 populations were examined for putative double-mutant phenotypes. Putative double mutants were verified by in a backcross to both parents (testcross). rhd1, rhd2, rhd3, rhd4, and rhd6 lines were kindly provided by John Schiefelbein, University of Michigan, Ann Arbor.
Analysis of root morphology and GFP imaging
Photographs of plants grown on phytagel medium were made using an Olympus SZH10 and a Wild M10 stereomicroscope. For cryo-scanning electron microscopy, plants were grown on phytagel medium and 3–5-d-old seedlings were placed on moist nitrocellulose paper mounted on a stub and immersed in liquid nitrogen slush. Roots were transferred to a cold stage. After removal of water by sublimation, roots were sputter coated with gold and observed using a JEOL Scanning Electron Microscope at −147°C. Roots were stained with 10 mg/mL propidium iodide and observed with a microscope Bio-Rad MRC 1024 or Leica TCS SP confocal laser scanning. A Leica TCS SP confocal laser scanning microscope was used to image GFP in transformed Arabidopsis and tobacco plants.
Genetic mapping
The kjk-1 in the Landsberg erecta background was mapped in a cross to the wild-type Columbia strain. The F2 population produced by selfing F1 individuals was screened for Kjk− phenotype. Simple sequence-length polymorphism (SSLP) (Bell and Ecker 1994) and cleaved amplified polymorphic sequences (CAPS) (Konieczny and Ausubel 1993) markers were used for mapping. DNA samples for the SSLP and CAPS mapping were prepared from single leaves of mutant F2 plants (Doyle and Doyle 1990). All amplifications and restriction enzyme digestion of the resulting PCR products were performed as described previously (Konieczny and Ausubel 1993; Bell and Ecker 1994). The following primers and restriction enzymes were used: nga172, forward primer 5′-AGCTGCTTCCTTATAGCGTCC-3′ and reverse primer 5′-CATCCGAATGCCATTGTTC-3′; 17D8LE, forward primer 5′-CTCCTTTGTCATCTCCCGAATC-3′ and reverse primer 5′-CCAACAACATGCATGATAGTTCAG-3′ and polymorphism revealed by HincII; GAPC, forward primer 5′-CTGT TATCGTTAGGATTCGG-3′ reverse primer 5′-ACGGAAA GACATTCCAGTC-3′, and polymorphism revealed by EcoRV; CSL, forward primer 5′-GAGACTAGTGGGACTTACGGTT TC-3′ and reverse primer 5′-CTCACGCTTCACCCGTCTT CG-3′ and polymorphism revealed by TaqI.
Sequencing of mutant alleles
To sequence the kjk alleles, we amplified the KJK/AtCSLD3 coding regions from wild-type and mutant plant tissues by PCR using a mix of Taq (GIBCO BRL) and Pwo polymerase (Boehringer Mannheim). The amplification condition were as follows: 3 min at 94°C, followed by 25 cycles of 30 sec at 94°C, 30 sec at 55°C, 1 min at 72°C, and a final extension at 72°C for 10 min. PCR products were used directly for sequencing. The alterations in sequence were verified by independent PCR amplifications. Nucleotide sequences were determined using the Ready Reaction ABI sequencing kit mix (Perkin Elmer).
Southern blot and Northern blot analysis
For Southern blot analysis, genomic DNA was isolated from 4-wk-old plants grown in vitro (Doyle and Doyle 1990). Southern blots were done using 5 μg of genomic DNA (Sambrook et al. 1989). For RNA analysis on wild-type and kjk mutants, total RNAs from leaves, roots, stems, and inflorescences were isolated using RNeasy Plant Minikit (Qiagen). For Northern blot analysis, RNA samples were separated in a formaldehyde containing agarose gel, blotted, and hybridized (Sambrook et al. 1989).
RACE and RT–PCR analysis
The 5′ and 3′ end of the AtCSLD3 cDNA was obtained by RACE PCR (GIBCO BRL) with total RNA from the wild-type A. thaliana ecotype Ler (Frohman et al. 1988). The primers CS4 5′-CACGGTCTGCTCATCAGATCCTGC-3′ and nested CS2 5′-CATCACCTTGACATCACAACCAG-3′ were used for 5′ RACE and primers CS9 5′-CGTTGGAATCTACCCGTTCA CATC-3′ and CS11 5′-GTGTGATTCCGCAGTGGAGTAA GT-3′ for 3′ RACE. The cDNA was also amplified with CS1 5′-GCTACAAAGTCCGGTGGATAGTGTT-3′ and CS12 5′CTTGGCCAATCTCTGTCTCCATCTT-3′. PCR products were cloned in pGEM-T vector (Promega) and sequenced. For RT–PCR analysis, total RNAs were reverse transcribed with the oligo(dT) primer and were used as templates for PCR amplification. PCR reactions used CSL forward and reverse primers (see above) according to the following temperature profile (30 cycles): 94°C, 30 sec; 55°C, 30 sec; 72°C, 1 min. These primers produce a 740-bp fragment when cDNA is used as template and an 818-bp fragment when the genomic DNA is used.
Constructs and plant transformation
For the complementation of kjk mutation, a 7.2-kb AvrII fragment of the genomic DNA containing the KJK/AtCSLD3 gene, its promoter, and its terminator was cloned into the pGreen0029 vector (Roger et al. 2000). DNA was isolated from BAC clone T17B22 (accession no. ATAC012328) by the alkaline method and its integrity confirmed by Southern blot analysis with probes from the KJK/AtCSLD3 interval. This construct was introduced into Agrobacterium tumefaciens strain GV3101. The resulting strain was used to transform homozygous kjk mutant or Ler plants by vacuum infiltration (Bechtold et al. 1993). Primary transformants were selected on MS medium containing 50 mg/L kanamycin and transferred to soil. Plants were grown to maturity and allowed to self.
Expression pattern of KJK
To identify in which cells KJK/AtCSLD3 is expressed, the 5.0-kb AvrII–AseI fragment containing the KJK/AtCSLD3 gene and its promoter was fused in frame to an ER targeted GFP2.5–nopaline synthase terminator cassette in pGreen0229 (Roger et al. 2000). The construct was introduced into wild-type Arabidopsis as described above. The endoplasmic reticulum targeted GFP2.5 facilitates imaging cell-specific patterns, which can be problematic when using cytoplasmic GFPs. As a negative control, the promoterless GFP construct in pGreen 0229 was used.
Subcellular localization and Agrobacterium infiltration
For the subcellular localization of the KJK/AtCSLD3 protein, a 3.8-kb SspI–AseI fragment of the KJK/AtCSLD3 gene was fused in frame to a cytosolic mGFP4 (Haseloff et al. 1995)—35S terminator cassette under the control of the 35S promoter in pGreen029 vector. We used the 35S CaMV promoter to control expression of the KJK/AtCSLD3::GFP4 fusion protein because it allows examination of the subcellular localization of the fusion in the large epidermal cells of tobacco. As controls, a cytosolic and an ER-targeted GFP4 under the control of the CaMV35S promoter were used. These constructs were introduced into A. tumefaciens strain GV3101. Infiltration of Agrobacterium into N. benthamiana was carried out as described (Voinnet and Baulcombe 1997).
Sequence analysis
The BLAST search program (Altschul et al. 1997) was used for sequence analysis and comparisons in the Genbank, EMBL, and swissProt databases at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and in the Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/blast). For protein structure prediction, the following servers were used: TMHMM 1.0 (http://www.cbs.dtu.dk/services/TMHMM-1.0), TMDAS Transmembrane Prediction Server (http://www.biokemi.su.se/server/DAS/; Cserzo et al. 1997), TopPred (http://www.sbc.su.se/∼erikw/toppred2; von Heijne 1992), PSort (http://psort.nibb.ac.jp), and DNA Strider 1.2 software. Multiple sequence alignments and the relationship tree were done with CLUSTAL W. All the sequences of the cellulose synthase homologs were retrieved from C. Somerville and T. Richmond's server (http://cellwall.stanford.edu/cellwall).
Accession number
The sequence data of the A. thaliana cellulose synthase-like KJK/AtCSLD3 cDNA has been submitted to the DDBJ/EMBL/Genbank databases under accession no. AF232907.
Acknowledgments
We thank Silvia Costa for assistance with confocal microscopy, Todd Richmond and Chris Somerville for the cell wall carbohydrate analysis, Isabelle Malcuit and Patrick Laufs for help with GFP fusions, and Keith Roberts and Pierre Abad for critical reading of the manuscript. We acknowledge the input of three anonymous referees who helped us to highlight the importance of KJK. L.D. is grateful to Molly and Lew Tilney for help with preliminary experiments and valuable thoughts about how plant cells grow. We thank the Nottingham and Ohio Stock Arabidopsis stock centers for providing mutant seeds. BACs containing DNA from the Columbia accession of Arabidopsis were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University. This work was supported by BBSRC, INRA, and the EU.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL liam.dolan@bbsrc.ac.uk; FAX 44-1603-450022.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.188801.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Bancroft I, Bhatt AM, Sjodin C, Scofield S, Jones JDG, Dean C. Development of an efficient two-element transposon tagging system in Arabidopsis thaliana. Mol Gen Genet. 1992;233:449–461. doi: 10.1007/BF00265443. [DOI] [PubMed] [Google Scholar]
- Bartel B, Fink GR. ILR1, an amidohydrolase that release active indole-3-acetic acid from conjugates. Science. 1995;268:1745–1748. doi: 10.1126/science.7792599. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Elis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana. CR Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bibikova TN, Jacob T, Dahse I, Gilroy S. Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development. 1998;125:2925–2934. doi: 10.1242/dev.125.15.2925. [DOI] [PubMed] [Google Scholar]
- Brown RM, Saxena IM, Kudlicja K. Cellulose biosynthesis in higher plants. Trends Plant Sci. 1996;1:149–156. [Google Scholar]
- Carpita N, Vergara C. A recipe for cellulose. Science. 1998;279:672–673. doi: 10.1126/science.279.5351.672. [DOI] [PubMed] [Google Scholar]
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane α-helices in procaryotic membrane proteins: The Dense Alignment Surface method. Prot Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- Cutler S, Somerville C. Cellulose synthesis: Cloning in silico. Curr Genet. 1997;7:R108–R111. doi: 10.1016/s0960-9822(06)00050-9. [DOI] [PubMed] [Google Scholar]
- Delmer DP. Cellulose biosynthesis: Exciting times for a difficult field of study. Annu Rev Plant Physiol Mol Biol. 1999;50:245–277. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- Delmer DP, Reed SM, Cooper G. Identification of a receptor protein in cotton fibers for the herbicide 2,6-dichlorobenzonitrile. Plant Physiol. 1987;84:415–420. doi: 10.1104/pp.84.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Duckett C, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K. Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development. 1994;120:2465–2474. [Google Scholar]
- Doyle JJ, Doyle DJ. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway ME, Heckman JJW, Schiefelbein JW. Growth and ultrastructure of Arabidopsis root hairs: The rhd3 mutation alters vacuole enlargement and tip growth. Planta. 1997;201:209–218. doi: 10.1007/BF01007706. [DOI] [PubMed] [Google Scholar]
- Grierson CS, Roberts K, Feldmann KA, Dolan L. The COW1 locus of Arabidopsis acts after RHD2 and in parallel with RHD3 and TIP1, to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. Plant Physiol. 1997;115:981–990. doi: 10.1104/pp.115.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci. 1995;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Laosinchain W, Itoh T, Cui X, Linder CR, Brown RM. Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell. 1999;11:2075–2085. doi: 10.1105/tpc.11.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 1994;106:1335–1346. doi: 10.1104/pp.106.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Chua N-H. Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell. 2000;12:465–478. doi: 10.1105/tpc.12.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, Szymanski DB, Ericson B, Feldmann K, Marks MD. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc Natl Acad Sci. 1997;94:6261–6266. doi: 10.1073/pnas.94.12.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger P, Hellens E, Edwards A, Leyland NR, Bean S, Mullineaux PM. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Ryan E, Grierson CS, Cavell A, Steer M, Dolan L. TIP1 is required for both tip growth and non-tip growth in Arabidopsis. New Phytol. 1998;138:49–58. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saxena IM, Brown RM, Fevre M, Geremia RA, Henrissat B. Multidomain architecture of β-glycosyl transferases. J Bact. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C. Genetic control of root hair development in Arabidopsis thaliana. Plant Cell. 1990;2:235–243. doi: 10.1105/tpc.2.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Shipley A, Rowse P. Calcium influx at the tip of growing root-hair cells of Arabidopsis thaliana. Plant J. 1992;187:455–459. doi: 10.1007/BF00199963. [DOI] [PubMed] [Google Scholar]
- Schneider K, Mathur J, Boudonck K, Wells B, Dolan L, Roberts K. The ROOT HAIRLESS 1 gene encodes a nuclear protein required for root hair initiation in Arabidopsis. Genes & Dev. 1998;12:2013–2021. doi: 10.1101/gad.12.13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Scheible W-R, Cutler S, Somerville C, Turner SR. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell. 1999;11:769–779. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Somerville CR. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Baulcombe DC. Systemic signaling in gene silencing. Nature. 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction, hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW. The ROOT HAIR DEFECTIVE 3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes & Dev. 1997;11:799–811. doi: 10.1101/gad.11.6.799. [DOI] [PubMed] [Google Scholar]
- Wu L, Joshi CP, Chiang VL. AraxCelA, a new member of the cellulose synthase gene family from Arabidopsis. Plant Physiol. 1998;117:1125. [Google Scholar]
- Wymer CL, Bibikova TN, Gilroy S. Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J. 1997;12:427–439. doi: 10.1046/j.1365-313x.1997.12020427.x. [DOI] [PubMed] [Google Scholar]