Abstract
DNA polymerase η carries out translesion synthesis past UV photoproducts and is deficient in xeroderma pigmentosum (XP) variants. We report that polη is mostly localized uniformly in the nucleus but is associated with replication foci during S phase. Following treatment of cells with UV irradiation or carcinogens, it accumulates at replication foci stalled at DNA damage. The C-terminal third of polη is not required for polymerase activity. However, the C-terminal 70 aa are needed for nuclear localization and a further 50 aa for relocalization into foci. Polη truncations lacking these domains fail to correct the defects in XP-variant cells. Furthermore, we have identified mutations in two XP variant patients that leave the polymerase motifs intact but cause loss of the localization domains.
Keywords: DNA polymerase, DNA replication, foci, translesion synthesis, UV radiation, xeroderma pigmentosum
Ultraviolet (UV) light produces photoproducts in cellular DNA, which in normal individuals are repaired by nucleotide excision repair (NER). Most patients with the highly cancer-prone disorder xeroderma pigmentosum (XP) are defective in NER (Berneburg and Lehmann 2000; de Laat et al. 1999). Cells from one group of XP individuals, the XP variants (XP-V), are, however, not deficient in NER. Instead, they are deficient in postreplication repair (PRR), which enables cells to synthesize intact daughter DNA strands, despite the presence of persisting damage in the template strands (Lehmann et al. 1975). XP-V cell lines are highly mutable by UV light (McGregor et al. 1999), and the patients have a dramatically increased incidence of skin cancer, just like the NER-defective XPs. Thus, the ability to carry out PRR is crucial for cancer avoidance.
Until very recently, little was known about the mechanisms of PRR and the defect in XP-V cells, but a series of studies showed that cell-free extracts of XP-V cells were unable to replicate past DNA damage (Cordeiro-Stone et al. 1997; Ensch-Simon et al. 1998; Svoboda et al. 1998), pointing to a defect in translesion synthesis (TLS; Cordonnier et al. 1999). These studies culminated in the cloning of the gene defective in XP-V patients (Johnson et al. 1999a; Masutani et al. 1999a,b). The gene encodes a DNA polymerase, designated polη, that is a member of a recently discovered superfamily of novel DNA polymerases (Woodgate 1999) that are related in structure to each other but are unrelated to classical DNA polymerases. They have in common a highly distributive, rather than processive, mode of synthesis on undamaged templates and a relatively low stringency. This endows them with a high error rate when replicating undamaged templates (Maor-Shoshani et al. 2000; Matsuda et al., 2000; Tissier et al., 2000) but facilitates TLS past different types of DNA damage. This distinguishes them from the classical polymerases, which typically have a high processivity and high fidelity but are blocked by most DNA lesions.
Both yeast and human polη are able to replicate past T–T cyclobutane photodimers and undamaged T–T residues with the same efficiency (Johnson et al. 1999b; Masutani et al. 1999b), and in the majority of cases they insert the complementary bases (adenines) opposite the T–T photodimer (Johnson et al. 2000; Masutani et al. 2000). There have been several reports on the enzymology of human polη describing its error proneness (Johnson et al. 2000; Masutani et al. 1999a, 2000; Matsuda et al. 2000), its mode of synthesis on damaged and undamaged templates, and its ability to carry out TLS on templates carrying different types of damage (Masutani et al. 2000; Yuan et al. 2000). Although these findings are of importance for understanding the mode of action of polη, they give only limited insight into the events that occur inside the cell when the replication fork encounters DNA damage. In this article, we show that following UV irradiation of human fibroblasts, polη is relocalized into nuclear foci, we identify the sequences required for this relocalization, and we show that relocalization of polη is vital for its biological function.
Results
Polη is localized in the nucleus and relocalizes after UVC irradiation
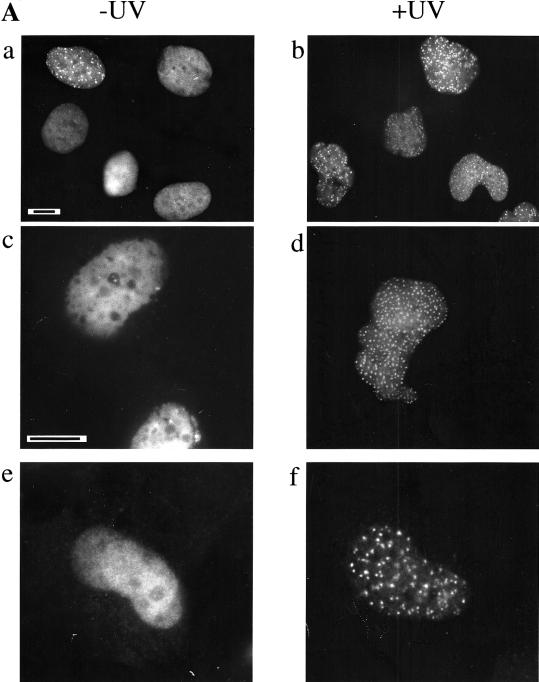
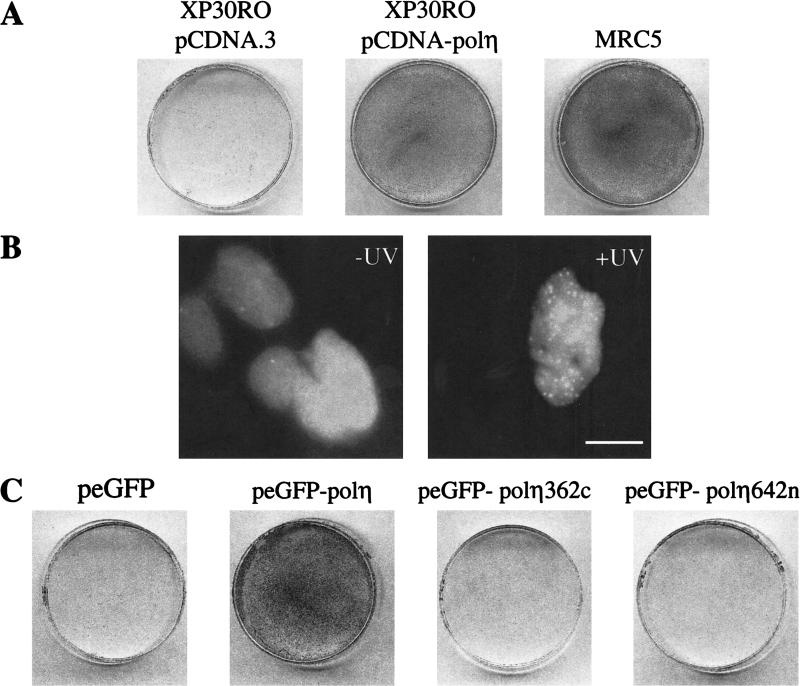
To study the cellular localization of polη, the cDNA-encoding enhanced green fluorescent protein (eGFP) was fused in-frame to the amino-terminus of polη (eGFP-polη), and this construct was transfected into SV40-transformed MRC5 fibroblasts. In most cells, the tagged protein was distributed homogeneously within the nucleus (Fig. 1A, panel a). In 10%–15% of transfected cells, however, eGFP-polη was localized in many brightly fluorescent spots, suggesting the presence of high local concentrations of eGFP-polη. To determine whether the distribution of polη changes after DNA damage, we irradiated the transfected cells with 7 J/m2 UVC and analyzed the pattern of eGFP-polη 8 h later (Fig. 1A, panel b). Strikingly, in >60% of transfected cells, eGFP-polη was concentrated into foci throughout the nucleoplasm, the number varying from ∼50 to several hundred per cell. This focal distribution of eGFP-polη was also observed in transformed XP-V fibroblasts (see below). Similar localization was detected in living cells or after paraformaldehyde fixation, ruling out the possibility of artefacts caused by the fixation (Fig. 1A, panels c,d). It is unlikely that the eGFP-polη foci were artefacts resulting from overexpression of the protein, both because we also observed them in stable transfected clones with low expression levels (see below) and because the number of cells with foci was much higher after UVC irradiation, whereas the protein level remained similar to that in unirradiated cells (data not shown). To verify that the distribution of eGFP-polη was not caused by tagging artefacts, we examined the distribution of untagged polη using a pCDNA-polη construct and anti-polη antibodies. Similar foci were observed (Fig. 1A, panels e,f). We conclude that this localization reflects the endogenous nuclear distribution of polη and indicates that the protein is relocalized into intranuclear foci after UVC irradiation.
Figure 1.
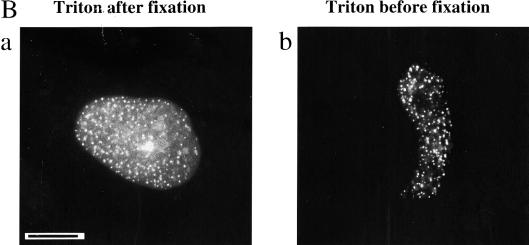
Polη relocalizes into intranuclear foci after UVC-irradiation. (A) MRC5 cells were transfected with plasmids encoding either eGFP-polη (a–d) or untagged polη (e–f). Twenty hours after transfection, cells were irradiated with 7 J/m2 (b,d,f). Eight hours later, the distribution of eGFP-polη was examined after paraformaldehyde fixation (a,b) or in living cells (c,d). For cells transfected with pCDNA-polη, polη distribution was revealed with anti-polη pAb and FITC-conjugated secondary antibody (e,f). Bar = 10 μm. (B) Twenty hours after transfection of MRC5 cells with plasmid encoding eGFP-polη, cells were irradiated with 7 J/m2 UV and then incubated for 8 h. Distribution of eGFP-polη was examined either after paraformaldehyde fixation followed by Triton X-100 permeabilization (a) or with permeabilization carried out before fixation (b). The diffuse staining of eGFP-polη disappeared when cells were preextracted in 1% Triton X-100 before fixation, whereas eGFP-polη foci remain.
To investigate if the polη foci are attached to nuclear structures, we treated the transfected cells with 1% Triton X-100 before fixation. In irradiated cells, eGFP-polη foci were resistant to Triton X-100 extraction (Fig. 1B), whereas the uniform staining in both irradiated (Fig. 1B) and unirradiated cells (not shown) was greatly reduced. This suggests that the foci were tightly associated with nuclear substructures.
Relocalization of polη is a specific response to unrepaired DNA lesions
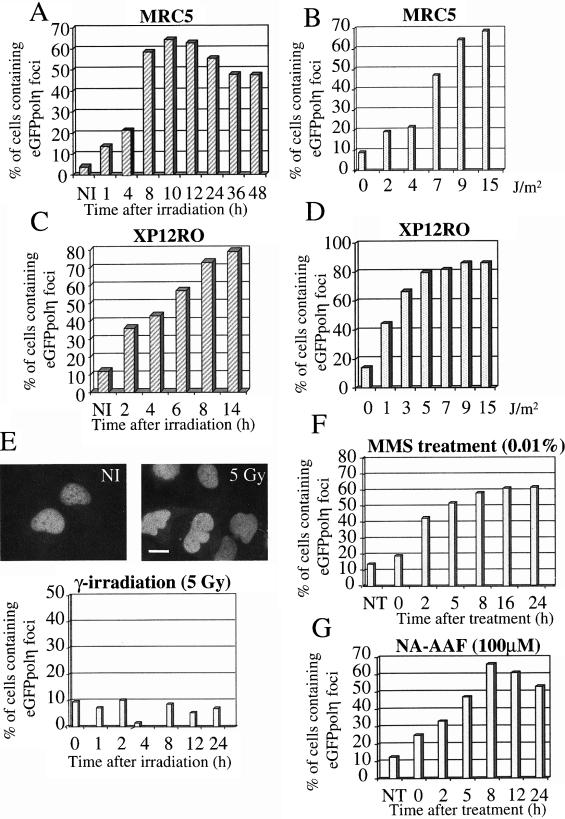
To determine the kinetics of the relocalization of polη after UV-irradiation, transfected MRC5 cells were irradiated with 7 J/m2 and then cultured for various times before analysis of foci formation. The maximal response (>60% of cells with foci) was obtained 8–12 h after UVC irradiation and declined slowly thereafter (Fig. 2A). The localization of polη into foci 8 h after irradiation was dose dependent (Fig. 2B).
Figure 2.
The relocalization of polη is specific for certain types of DNA lesions. MRC5 (A,B,E–G) or XP12RO (XP-A) (C,D) cells were transfected with eGFP-polη and incubated for 20 h. Cells were then UV irradiated with 7 J/m2 (A,C) and incubated for different times or (B,D) irradiated with different UV doses and incubated for 8 h. The proportion of eGFPpolη-containing cells in which the polη was localized in foci was determined. (E–G) Transfected cells were treated with 5-Gy γ rays, 0.01% MMS, or 100 μM N-AAAF and then incubated for the indicated times before analysis for foci. Top of panel E shows an example of lack of foci after γ irradiation. All panels show representatives of three experiments.
We postulated that the relocalization of polη into intranuclear foci after UV irradiation reflects its ability to function in the bypass of unrepaired DNA lesions during DNA replication. To examine if the relocalization of eGFP-polη was dependent on unrepaired damage, we used XP-A cells (XP12RO), which are defective in NER and, thus, fail to remove UV lesions. The percentage of cells with intranuclear foci was significantly higher in XP-A cells than in normal cells (Fig. 2C), with the fraction of cells with eGFP-polη foci reaching a maximum at a UVC dose of 5 J/m2 in XP-A cells compared with 15 J/m2 in normal cells (Fig. 2D). Taken together, these data strongly suggest that in vivo polη relocalizes to unrepaired UV damage.
To rule out the possibility that the relocalization could be a nonspecific cellular response to DNA damage, we analyzed the distribution of eGFP-polη after γ irradiation. Transfected cells were irradiated with 5 Gy, and the distribution of eGFP-polη was examined after various times. We did not observe any relocalization of eGFP-polη after γ irradiation (Fig. 2E). This is consistent with the sensitivity of XP-V cells to UV but not to γ irradiation (Arlett and Harcourt 1980). These results indicate that polη-foci formation is not part of a nonspecific global response to DNA damage but is specific to certain classes of DNA lesions.
In vitro polη is also able to bypass other DNA lesions such as acetylaminofluorene (AAF)-guanine adducts and abasic sites (Masutani et al. 2000). We therefore tested the distribution of eGFP-polη after NA-AAF and MMS (monofunctional DNA-alkylating agent that generates AP sites) treatment. Both carcinogens resulted in formation of polη-foci (Fig. 2F,G). These observations are in agreement with the biochemical data and consistent with the hypothesis that polη foci colocalize with sites of replication forks blocked by several but not all types of DNA lesions.
Polη foci result from relocalization rather than de novo synthesis
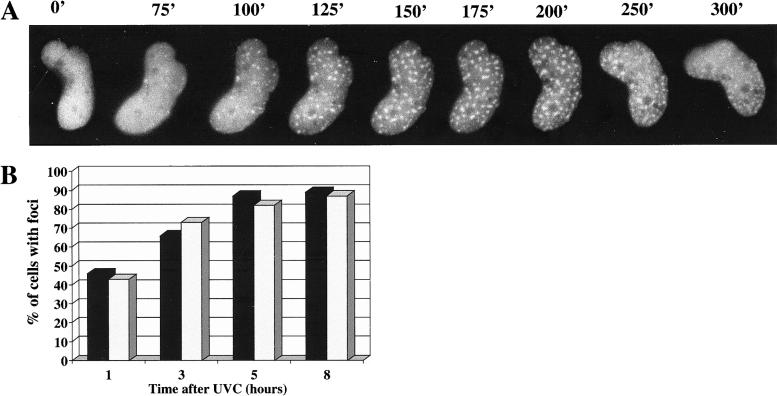
We have analyzed the formation of foci in living cells following UV irradiation using time-lapse microscopy. Figure 3A shows a single MRC5 cell at various times after UV irradiation with a dose of 10 J/m2. In this cell, foci appeared 2 h after irradiation; their intensity was maximum at 3 h and then subsided over the following 2 h. The formation of polη foci was accompanied by a marked decrease in intensity of the uniformly distributed polη. Quantification of the intensity of the polη image over the whole nucleus indicated that the total amount of nuclear polη did not change significantly (data not shown). This result suggests that the foci result from relocalization of polη rather than de novo synthesis. Consistent with these observations, we found that incubation of cells after UV irradiation with the protein synthesis inhibitor, cycloheximide, did not affect foci formation (Fig. 3B). (We used XP12RO cells for this experiment because foci appear in a shorter time than in normal cells [see Fig. 2C]. In this way we were able to minimize the time that the cells spent in cycloheximide, thereby reducing any secondary effects of this general inhibitor.)
Figure 3.
Foci result from relocalization of existing Polη. (A) MRC5 cells were transfected with peGFP-polη and UV irradiated, and individual living cells were examined by time-lapse fluorescence microscopy. Accumulation of polη into foci at 120–200 min is associated with a decrease in intensity of uniform staining throughout the nucleus. (B) XP12RO cells transfected with peGFP-polη were UV irradiated (7 J/m2) and incubated for different times with or without 20 μg/mL cycloheximide. The proportion of cells with foci was determined. (Black bars) Without cycloheximide; (gray bars) with cycloheximide.
Polη colocalizes with PCNA
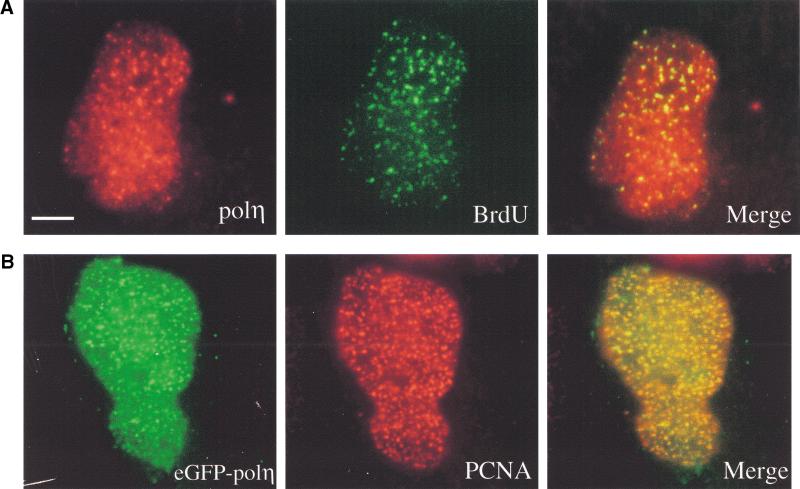
In eukaryotes, DNA replication takes place in discrete replication foci that vary in morphology during S phase and contain proteins involved in replication such as PCNA (Bravo and Macdonald-Bravo 1987), RPA, and DNA ligase I (Montecucco et al. 1995). We have reported that eGFP-polη is focally concentrated in 10%–15% of unirradiated cells. To determine if these foci might be coupled to the replication machinery, we have examined whether nuclear spots of polη localize to replication foci. Unirradiated cells were transfected with pCDNA-polη, incubated for 20 h to allow expression of exogenous polη, and then pulse labeled for 15 min with BrdU. Replication foci and polη were visualized using antibodies against BrdU (Fig. 4A, green staining) and against polη (red staining; note that the endogenous levels of polη are insufficient to be detected by our antibody). In the 10%–15% of cells with polη foci, a substantial proportion of the foci colocalized with BrdU. In these cells, nearly all replication foci staining with anti-BrdU also contained polη (Fig. 4A). Likewise, in cells transfected with peGFP-polη (green staining, Fig. 4B), autofluorescent foci colocalized with PCNA (red staining). The overlapping of polη with either BrdU or PCNA is visualized as yellow foci (Fig. 4A,B, right panels) and suggests that in 10%–15% of untreated cells, polη localizes in replication foci. As only a proportion of cells are transfected, we cannot make conclusive statements, but our data are consistent with the suggestion that polη is located in replication foci during DNA replication. Likewise, we found that after UV irradiation, in the much greater number of cells with eGFP-polη foci, all the foci colocalized with PCNA (results not shown). These findings suggest that during replication in UV-irradiated cells, polη is localized at replication forks that may be at the sites of unrepaired UV damage.
Figure 4.
Polη is associated with the replication machinery and partially colocalizes with Rad51. (A) MRC5 cells transfected with pcDNA-polη and pulse labeled with BrdU for 15 min before fixation were double stained with anti-BrdU mAb (green staining) and anti-polη pAb (red staining). (B) Cells transfected with eGFP-polη were fixed and stained with anti-PCNA mAb and TRITC-conjugated secondary antibody. The staining pattern of PCNA (red staining) and the autofluorescent signal of GFP (green staining) in the same cell are shown. Colocalization between BrdU and polη (A) or PCNA and eGFP-polη (B) is indicated by a yellow pattern (right panels). Bar = 5 μm. (C,D) Partial colocalization between polη and Rad51 proteins after UVC irradiation. Twenty hours after transfection with peGFP-polη, cells were UV irradiated with 15 J/m2. Eight hours later, they were fixed and stained with anti-Rad51 pAb and TRITC-conjugated secondary antibody (red staining). The distribution of polη was detected by autofluorescence of the GFP (green staining). In ∼20% of transfected cells, complete colocalization between Rad51 and eGFP-polη foci was observed as shown by yellow staining where red and green signals overlap (C, right panel). In most of the cells, there was a significant but partial colocalization of Rad51 and eGFP-polη foci, as shown by arrows in D. Bar = 5 μm.
Rad51 partially colocalizes with polη after UVC irradiation
Human Rad51, a structural homolog of the bacterial recombination repair protein RecA, relocalizes into multiple subnuclear foci after various DNA damaging agents such as UVC and γ irradiation, MMS, or hydroxyurea (Haaf et al. 1995; Scully et al., 1997). We asked whether polη and Rad51 colocalize in the same complex. Twenty hours after transfection by peGFP-polη, cells were irradiated with 15 J/m2 and fixed for immunostaining 8 h later. Rad51 foci were visualized by anti-Rad51 antibodies (Fig. 4C,D, red staining), and polη spots were detected by autofluorescence of eGFP (Fig. 4C,D, green staining). In some cells, there was almost complete colocalization of Rad51 and polη foci (e.g., Fig. 4C) but in the majority, there was partial but not complete colocalization (Fig. 4D). This suggests that unrepaired UV damage elicits the relocalization of Rad51 and polη in the same areas. The distribution of Rad51 into foci in polη-deficient XP30RO cells was similar to that observed in normal cells, indicating that the relocalization of Rad51 after UV irradiation is not dependent on functional polη (not shown).
Polη localizes at sites of unrepaired DNA damage
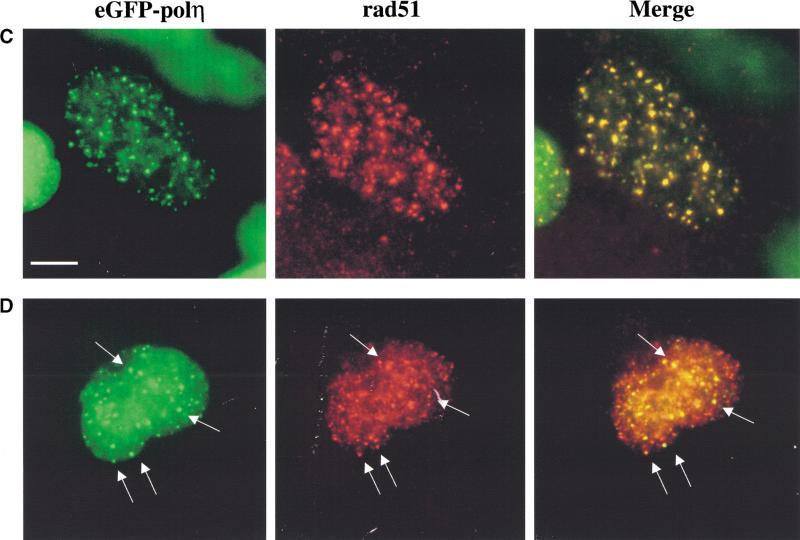
The data presented so far indicate that polη accumulates at replication forks stalled at damaged sites. However, it is well documented that UV damage results in a decreased rate of traverse through S phase, with a resulting substantial increase in the number of S-phase cells several hours after irradiation (Domon and Rauth 1969; Lehmann et al. 1979). The increase in number of cells with foci might, therefore, merely reflect the increase in the number of S-phase cells. To exclude this possibility, we used a filter to induce local UV damage in the nucleus in cells transfected with pCDNA-polη. After incubation for 12 h and fixation, the sites of irradiation were visualized by immunofluorescence, using an antibody that recognized UV-induced cyclobutane pyrimidine dimers (CPDs). (Note that even in normal cells, which are able to remove UV photoproducts, >50% of the CPDs remain after 12 h [van Hoffen et al. 1993].) The localization of polη was then visualized by means of the anti-polη antibodies. Strikingly, polη (Fig. 5, left panel) accumulates almost exclusively in the areas of the nucleus that have been exposed to UV (middle panel), as confirmed by the yellow staining in the right panel. These results strongly support the idea that polη accumulates at replication forks located at the sites of damage and argues against the idea that the foci solely reflect an increased proportion of S-phase cells.
Figure 5.
Recruitment of polη to DNA lesions after UV irradiation. Twenty hours after transfection of MRC5 cells with pCDNA-polη, cells were locally irradiated using a filter. Twelve hours later, polη was detected using anti-polη pAb and FITC-conjugated secondary antibody (green staining), and UV-induced damage in the same cell was revealed using anti-TMD2 mAb and TRITC-conjugated secondary antibody (red staining). Most of the polη foci colocalize with UV lesions, as shown in the right panel (yellow staining). Bar = 5 μm.
Analysis of mutations in different XP-V cell lines
We have sequenced the Polη cDNA in many XP-V cell lines in our collection. Details of the mutations that we have found will be presented elsewhere. We were particularly intrigued by two mutations resulting in truncations close to the C terminus, both in XP37BR—a frameshift mutation at codon 556—and in XP1AB—a nonsense mutation in codon 548 (Fig. 6A). Polη was originally isolated by Masutani et al. (1999a) on the basis of its activity, as a 511-aa C-terminally truncated protein whose activity was comparable with the full-length recombinant protein. Thus, the C-terminal 200 aa are entirely dispensable for polymerase activity and we anticipate that polη will be fully active in XP37BR and XP1AB. The mutation in these patients must therefore affect some other aspect of the enzyme's function, defined by the C-terminal 200 aa.
Figure 6.
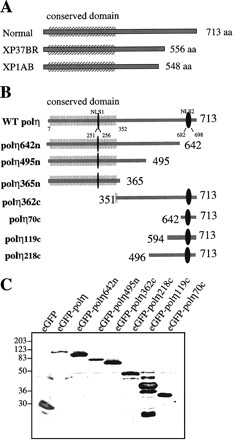
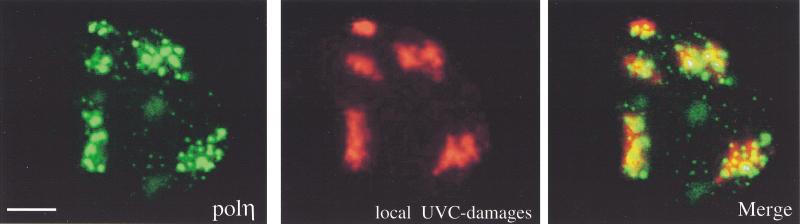
The C-terminal domain of polη is required for the nuclear localization and relocalization after UVC irradiation. (A) Predicted proteins encoded by mutated polη gene in two XP-V cell lines, XP37BR and XP1AB. (B) GFP-tagged deletion mutants are depicted schematically, and the name of each truncated protein is indicated on the left. The conserved domain (aa 1–352) is indicated as well as two putative nuclear localization signals deduced from the amino acid sequence of polη at positions 251–256 and 682–698. (C) Western blots of MRC5 cells transfected with plasmids encoding truncated proteins listed above the panel. Numbers on the left indicate apparent molecular weights in kilodaltons. (D) MRC5 cells were transiently transfected with plasmids expressing GFP-polη642n (a,b), GFP-polη362c (c,d), GFP-polη70c (e,f), and GFP-polη119c (g,h). Twenty hours later, cells were irradiated with 7 J/m2 (b,d,f,h). After 8 h incubation, the localization of the truncated polη proteins was determined by the autofluorescent signal of GFP. Bar = 10 μm.
Deletion analysis of polη
Two putative nuclear localization signals (NLS) have been described in polη at positions 251–256 and 682–698 (Masutani et al. 1999b). To identify the sequences that are required for nuclear localization and UV-relocalization, a series of GFP-tagged deletion mutants were constructed as shown in Figure 6B. All fusion proteins were tested for stability in vivo: MRC5 cells were transfected with expression constructs, and protein extracts were analyzed by Western blotting (Fig. 6C). All constructs gave stable fusion products whose apparent molecular weights are consistent with values deduced from the amino acid sequences. The mutation deleting amino acids 643–713 demonstrated unambiguously that the first putative NLS at codons 251–256 is inactive because the fusion protein eGFP-polη642n was distributed diffusely in the whole cell (Fig. 6D, panels a, b). Two larger deletions (of amino acids 496–713 or 367–713) led to the same localization as eGFP-polη642n (Fig. 7), demonstrating that this distribution is not caused by a cytoplasmic retention signal, as has been described for other proteins (e.g., Schmidt-Zachmann et al. 1993). Moreover, even though some of the eGFP-polη642n was localized in the nucleus, it did not relocalize into nuclear foci after UV irradiation, showing that the catalytic domain (amino acids 1–352) by itself does not localize into foci (Fig. 6D, panel b). In striking contrast, GFP-tagged polη362c, in which the catalytic N-terminal domain is deleted, displayed total nuclear localization, relocalized into multiple nuclear foci after UVC irradiation (Fig. 6D, panels c,d), and colocalized with PCNA after UVC treatment (not shown). These results indicate that the C-terminal half of polη, which is truncated in two XP-V patients, contains an active NLS and the domain required for UV relocalization. Consequently, we assume that the nuclear localization of polη is caused by the bipartite NLS KRNPKSPLACTNKRPRP at position 682–698.
Figure 7.
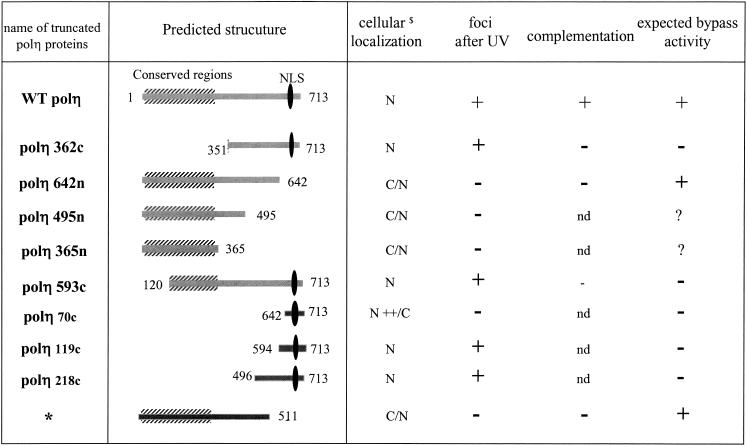
Summary of polη domains involved in its cellular localization and bypass activity. (*) Expected results from original truncated protein isolated by Masutani et al. (1999a) with bypass activity comparable to the wild-type protein. (C) Cytoplasmic; (N) nuclear; (C/N) both cytoplasmic and nuclear. (nd) Not determined.
We then tested if the C-terminal 70 amino acids containing the NLS were sufficient to direct the fusion protein (eGFP-polη70c) into the nucleus in unirradiated cells and into foci after UVC irradiation. As shown in Figure 6D, panel e, the last 70 residues of polη were sufficient to target most but not all of the protein into the nucleus. A small fraction of eGFP-polη70c remained in the cytoplasm, suggesting that total nuclear localization requires sequences upstream of the NLS. More strikingly, however, no nuclear foci were seen in any unirradiated cells, and GFP-polη70c localization was not affected by UVC (Fig. 6D, panel f), demonstrating that the motif involved in UV-relocalization is distinct from the NLS. Localization analysis of two other mutant constructs in which aa 1–495 and 1–593 were deleted (mutants named eGFP-polη218c and eGFP-polη119c, respectively) showed that they were not only localized to the nucleus but also formed foci after UVC irradiation (Fig. 6D, panels g,h; Fig. 7). These findings demonstrate that the domain required for UVC relocalization is contained within residues 594–713.
Biological significance of polη localization
All the data corresponding to the localization of polη were obtained with transiently transfected cells. To confirm these observations, we made a series of stable clones and verified the localization of the ectopic protein as well as its capacity to complement XP-V cells. We used SV40-transformed XP30RO fibroblasts, in which the polη gene is mutated and encodes a truncated protein containing only 42 residues and can, therefore, be considered as a knock-out cell line (Masutani et al. 1999b; B.C. Broughton and A.R. Lehmann, unpubl.). XP-V cells are only slightly sensitive to UV irradiation, but this sensitivity can be dramatically increased by postirradiation incubation in caffeine (UV + caffeine; Arlett et al. 1975). Stable XP30RO transfectants expressing polη protein were much more resistant to UV + caffeine compared with XP30RO cells transfected with vector alone (Fig. 8A). UV resistance was not, however, totally restored to that of the normal MRC5 cells. Next, we checked the complementation and localization of GFP-tagged polη protein in stable clones. The fusion cDNA (eGFP-polη) was stably transfected into XP30RO cells. Autofluorescence analysis showed that these clones expressed low levels of the fusion protein. Nevertheless, eGFP-polη relocalized into intranuclear foci after UVC irradiation, confirming that the pattern observed after transient transfection is not caused by overexpression of polη (Fig. 8B). Moreover, eGFP-polη cDNA was able to correct the UV + caffeine sensitivity of XP-V cells (Fig. 8C), indicating that addition of the eGFP tag did not detectably interfere with polη function in vivo. In contrast, as anticipated, no complementation was observed with clones stably transfected by peGFP-polη362c (truncated in the N-terminal conserved domain of polη; see Fig. 6B), although the fusion protein was localized in the nucleus and formed foci after UVC irradiation (not shown). This result demonstrates that the catalytic N-terminal domain of polη is required for its UV-damage bypass activity.
Figure 8.
eGFP-polη is functional in vivo and complements XP-variant cells. (A) XP30RO cells were transfected with pCDNA.3 (left) or pCDNA3-polη and incubated for 30 h. The plates were then exposed to 7 J/m2 UV irradiation and incubated in the presence of 75 μg/mL caffeine. After 4 d, the cells were stained with methylene blue. Right panel: MRC5 cells mock transfected and then treated in the same way. (B) XP30RO cells were transfected with peGFP-polη, and stable clones were isolated. UV irradiation results in relocalization of the eGFP-polη like in normal cells. (C) XP30RO cells were transfected with plasmids containing the indicated inserts and then treated as in A. Only the full-length plasmid corrects the UV + caffeine sensitivity.
We also stably transfected XP30RO with peGFP-polη642n, which expressed polη lacking the C-terminal 70 aa required for nuclear localization. As after transient transfection, the fusion protein displayed diffuse staining throughout the cell and did not relocalize after UVC irradiation (not shown). The complementation assay showed that cells remained sensitive to UVC irradiation like the parental XP30RO fibroblasts (Fig. 8C).
Altogether, these results indicate that not only is the catalytic N-terminal domain of polη required for its enzymatic activity but also that the correct localization of polη is indispensable for complementation of UV + caffeine sensitivity. These observations suggest that in the two XP-V patients in which polη protein is truncated in the C-terminal region, the XP phenotype is probably caused by the protein not being correctly localized in the cell. Therefore, in vitro, this protein will appear functional, whereas in vivo, it will not.
Discussion
The dramatic proneness to skin cancer of XP-V individuals, who are defective in polη, testifies to the importance of this novel DNA polymerase in cancer avoidance. The deficiency in XP-V cells in the ability to synthesize intact daughter strands after UV irradiation in vivo (Lehmann et al. 1975) is readily explained by the ability of polη to carry out TLS past T–T cyclobutane dimers in vitro (Masutani et al. 1999b, 2000). The polymerase activity of polη resides entirely in the first 511 aa of the 713-aa protein (Masutani et al. 1999a). Two important questions are posed by these findings: What is the function of the C-terminal 200 aa? What is the nature of the events inside the cell that result in TLS and other possible damage avoidance processes being integrated into the DNA replication process in an orderly manner? In this article, we have provided answers to the first question and begun to shed light on the second. Our results are summarized in Figure 7.
Localization of polη is determined by sequences close to the C terminus
We have shown that DNA polymerase η has a dynamic cellular organization, that the C-terminal 70 aa containing a bipartite NLS are required for nuclear localization of polη, and that an additional 50 aa are needed for relocalization of polη into multiple intranuclear foci following UV irradiation (see Fig. 6). These C-terminal 120 aa are necessary and sufficient to target the protein into foci after DNA damage and are also required for complementation of the defect in XP30RO cells (Fig. 8C).
The C-terminal 100 aa encompass a single postulated C2H2 zinc finger. In vitro, this motif is clearly not required for polymerase activity, but in vivo, when replication forks are blocked by UV lesions, it might play an important role by increasing the capacity of polη to bind to UV-damaged DNA. This domain is moderately well conserved from yeast to man (Fig. 9A). In the human, mouse, Drosophila, Schizosaccharomyces pombe, and Saccharomyces cerevisiae orthologs, the amino acid sequence encompassing the putative zinc finger motif is very well conserved. However, although the two cysteines are separated from the histidines by 11 residues in human, mouse, Drosophila and S. pombe, in S. cerevisiae, the two cysteines are immediately adjacent to each other and separated from the two histidines by 14 residues. Furthermore, there is only one histidine and no cysteines in the Caenorhabditis elegans ortholog (not shown). More work is required to understand the function of this motif. Apart from this putative zinc finger and the NLS, the C-terminal 100 aa do not show any sequence homology to other proteins in the sequence databases.
Figure 9.
Alignment of the C-terminal sequences of polη and Rad30 orthologs. (A) The C-terminal 100 aa of human polη (hpolη) are aligned with those of orthologs from mouse (mpolη), Drosophila (dmpolη), Schizosaccharomyces pombe (Eso1), and Saccharomyces cerevisiae (scrad30). Identical aa are indicated in black, conserved aa in gray. The NLS and putative C2H2 zinc finger motifs are indicated by bars above the alignment. (B) Putative PCNA binding motifs in polη are aligned with those from other PCNA-binding proteins (Montecucco et al. 1998).
Polη is localized at unrepaired DNA-damage sites during DNA replication
We have shown that in untreated cells, polη is distributed uniformly in the nucleoplasm except in 10%–15% of cells, in which it is concentrated into intranuclear foci and colocalizes with BrdU and PCNA, suggesting that the foci occur exclusively at the sites of replication. This suggests that polη is associated with the replication machinery, as also proposed by Limoli et al. (2000). Nevertheless, it does not appear to play a role in the replication process in undamaged cells, as XP-V cells replicate their DNA with the same efficiency as normal cells (Lehmann et al. 1975).
UV irradiation results in a marked time- and dose-dependent increase in the number of cells containing polη in foci. These foci must represent a substantial number of polη molecules to be visible as microscopic dots. We propose that the foci represent replication factories in undamaged cells and that the increased number of cells displaying polη foci results from replication forks stalled at UV photoproducts before effecting TLS. Several lines of evidence support this hypothesis. First, the polη foci colocalize with PCNA, showing that they are at the sites of replication forks. Second, foci were found after treatments with UV, MMS, and AAAF, which introduce lesions that result in stalling of replication forks. Third, they were not observed after γ irradiation, which inhibits DNA synthesis almost exclusively by inhibition of DNA initiation by trans acting factors (Lamb et al. 1989). There is minimal effect on chain elongation. Fourth, when cells were UV irradiated at localized sites within the nuclei, most of the foci were localized at the sites of UV damage.
There are several possible explanations for the specific involvement of polη in DNA replication after UV irradiation. First, during normal replication, polδ (and possibly polη also) synthesizes the new DNA strands in a highly processive manner. Because of this high processivity, polη may not be able to gain access to unimpeded replication forks and may only be loaded when the fork is stalled by damage. Second, polη may not be located right at the replication fork but may be sequestered close by in the replication factories. On this model, stalling of the replication fork might send a local signal to activate polη at the fork. Third, loading polη might require the degradation of stalled polδ following ubiquitination by the Rad6–Rad18 complex (Bailly et al. 1994; Tateishi et al. 2000). Rad6p and Rad18p are essential for PRR in UV-damaged S. cerevisiae. This degradation might be necessary to allow access of polη at the replication forks, possibly assisted by PCNA. Goodman has proposed a model for DNA polymerase V-catalyzed error-prone translesion synthesis in Escherichia coli (Goodman 2000) in which β clamp is involved in loading polV at stalled replication forks in E. coli. By analogy, in mammalian cells, PCNA might be involved directly in loading polη when replication forks encounter UV-lesions but not during normal replication. A PCNA-binding motif has been postulated in several proteins involved in DNA replication, DNA repair, or cell cycle control, such as DNA-cytosine-5-methyl-transferase (MCMT; Chuang et al. 1997) or DNA ligase I (Montecucco et al. 1998). There are three putative PCNA-binding motifs in the C-terminal half of polη (Fig. 9B).
Relationship between translesion synthesis and homologous recombination
Numerous proteins involved in DNA damage responses, such as Rad51 (Haaf et al. 1995), BRCA1 (Scully et al. 1997), UNG2 (Otterlei et al. 1999), and XRCC1 (Taylor et al. 2000), are localized in replication sites and/or recruited into these foci after DNA damage. We have observed that polη foci partly colocalized with Rad51, indicating that Rad51 relocalized to unrepaired DNA lesions during the replication, as has been proposed previously by Scully et al. (1997). The simplest explanation is that the Rad51 protein, involved in homologous recombination, might be recruited at the arrested replication forks that are recombination substrates, while polη is recruited at the same sites for its bypass activity.
Biological relevance of the localization of polη
We have shown that the localization of polη is crucial for its function inside the cell. Constructs lacking the C-terminal localization signals failed to correct the UV + caffeine sensitivity of XP-V cells, and in two of the XP-V patients that we have analyzed, polη was truncated upstream of the localization region. Polη activity should be normal in these individuals, as the 511 aa sufficient for activity are intact, but it cannot be recruited to replication forks stalled at DNA photoproducts because it lacks localization signals. These data emphasize that not only the activity of polη but also its localization is vital for its function.
Materials and methods
Cell lines and culture conditions
SV40-transformed human fibroblasts were used in all experiments. MRC5V1 (normal), XP30RO(sv) (XP-V; also designated GM3617; Cleaver et al. 1999) and XP12ROSV40 (XP-A) cells were grown in Eagle's MEM supplemented with 10% fetal calf serum (FCS). The designations of the cell lines are abbreviated to MRC5, XP30RO, and XP12RO in this article.
DNA transfection and selection of stable transfectants
Plasmids were transfected into transformed fibroblasts using Fugene 6 according to the manufacturer's protocol (Roche). Cells were processed after 20–30 h incubation.
For stable transfectants, XP30RO cells were transfected with plasmids derived from pCDNA.3zeo (Invitrogen) or peGFP-C3 (Clontech). Forty hours after transfection, cells were incubated in selection medium containing either 150 μg/mL zeocin or 600 μg/mL G418. Selection was continued for 2 wk, and stable transfectants were isolated. Complementation of the defect in XP-V (XP30RO) cells was determined for UV resistance by exposing cells to a UVC dose of 7 J/m2 and incubating for 4 d in medium containing 75 μg/mL caffeine. The plates were then stained with methylene blue.
Irradiation and drug treatments
UV irradiation at 254 nm was performed with a germicidal lamp at a fluence rate of 0.5 J/m2/sec. Localized UV irradiation within the nucleus was achieved with a filter, using a technique that will be described in detail elsewhere (M. Volker, L.H.F. Mullenders, and R. van Driel, in prep.). The dose of irradiation was 30J/m2.
Cells in PBS were γ irradiated from a 60Co source at a dose rate of 1 Gy/min. For drug treatments, MMS was added to the culture medium for 50 min at a final concentration of 0.01%; cells were then washed twice with PBS and incubated in drug-free medium for various times. For NA–AAF treatment, cells were first incubated with 10−8M paraoxon to inhibit deacetylase activity (thereby preventing the formation of unwanted nonacetylated metabolites of AAAF) for 15 min before addition of NA–AAF for 30 min at 37°C in complete medium. After incubation, cells were washed twice with PBS and fresh medium was added. Cells were fixed at the indicated time points.
Construction of XPV expression vector
Full-length cDNA encoding polη was obtained by PCR using pET21a-polη as a template, with Pfu DNA polymerase and primers: 5′-gaattcATGGCTACTGGACAGGATCGA-3′ and 5′-ggatccCTAATGTGTTAATGGCTTAAAAAATGA-3′. The PCR product was digested with EcoRI and BamHI and inserted into the EcoRI-BamHI sites of pCDNA3.zeo downstream of the CMV promoter (Invitrogen), producing the plasmid pCDNA-polη. The sequence of polη cDNA was confirmed by DNA sequencing.
Construction of GFP fusion proteins
A series of GFP-tagged deletion mutants of polη was generated to study the cellular organization of polη. All polη deletion mutants were cloned in-frame downstream of the eGFP-cDNA in peGFP-C3. We modified polη cDNA by deleting the first ATG to avoid undesirable translation products starting at the first methionine of polη. The modified polη cDNA was generated by PCR using pCDNA-polη as a template, with Pfu DNA polymerase and the primers: 5′-ctcgagctcgagGCTACTGGA CAGGATCGA-3′ and 5′-ggatccCTAATGTGTTAATGGCT TAAAAAATGA-3′. The product was digested with XhoI and BamHI and inserted into the XhoI-BamHI sites of the vector to produce the plasmid peGFP-polη. peGFP-polη642n and peGFP-polη365n were obtained by subcloning the 1927-bp XhoI-KpnI fragment or 1489-bp XhoI-HindIII fragment from peGFP-polη. To generate peGFP-polη495n, peGFP-polη was digested with BstXI and BamHI to remove the 3′-terminal 700 bp of polη cDNA. To produce peGFP-polη70c, peGFP-polη119c and peGFPpolη218c, peGFP-polη 3′-terminal 215-bp KpnI-BamHI, 366-bp PstI-BamHI, or 653-bp HindIII-BamHI fragments were respectively subcloned into the vector. To generate peGFP-polη362c, the 3′ 1086-bp fragment of polη cDNA was obtained by PCR using pCDNA-polη plasmid as template, with Pfu DNA polymerase and the following primers: 5′-ctcgagAGACTGAC TAAAGACCGAAATGAT-3′ and 5′-ggatccCTAATGTGT TAATGGCTTAAAAAATGA-3′. The PCR product was digested with XhoI and BamHI and inserted into the XhoI-BamHI sites of peGFP-C3.
Sequencing
Mutations in XP-V patients were identified using RT–PCR with RNA extracted from XP-V fibroblasts. The gene was amplified from cDNA using gene-specific primers, and the PCR products were sequenced directly, using procedures described previously (Broughton et al. 1994).
Immunofluorescence microscopy
For visualization of eGFP-tagged proteins, cells were rinsed twice in PBS and examined without further fixation. Alternatively, cells grown on coverslips were rinsed in PBS and fixed with 3% paraformaldehyde for 10 min. To visualize polη or simultaneously detect Rad51 and eGFP-polη proteins, cells were fixed with 3% paraformaldehyde for 30 min at room temperature, then permeabilized with 0.5% Triton X-100 for 15 min. For detection of PCNA and eGFP-polη proteins, cells were fixed in cold methanol for 20 min at −20°C and then incubated for 30 sec with cold acetone to extract the soluble PCNA fraction. The detection of eGFP-polη foci was not affected by this latter fixation condition. Cells were subsequently washed twice with PBS and incubated for 1 h at room temperature with the primary antibody. Then, cells were washed with PBS and incubated for 45 min with TRITC-conjugated swine antirabbit IgG (Dako), FITC-conjugated goat antimouse IgG (Sigma), or FITC-conjugated goat antirabbit IgG (Sigma). After washing three times for 5 min with PBS, cells were mounted with Glycergel (Dako). No significant signal attributable to secondary antibody alone was detected. For the simultaneous visualization of polη protein and sites of DNA synthesis, cells were incubated in 50 μM BrdU for 15 min before fixation. Immunostaining for polη was performed as described above. After the final wash, antigen-antibody complexes were fixed with 2% formaldehyde in PBS for 10 min. Cells were treated with 2 M HCl at 37°C for 40 min, then neutralized in 0.1 M borate buffer (pH 8.5). After washing in PBS, cells were incubated for 1 h at room temperature with the FITC-conjugated anti-BrdU mAb (Roche). For the simultaneous detection of polη protein and UV-induced damage, cells locally irradiated were fixed with 3% formaldehyde containing 0.5% Triton X-100 for 30 min on ice. Immunostaining for polη was performed as described above. After the final wash, cells were again fixed with 2% formaldehyde in PBS for 10 min and treated with 2 M HCl at 37°C for 10 min. After extensive washes in PBS + 0.5% BSA, cells were incubated for 1 h at room temperature with antithymidine dimers antibody. After three washes in PBS + 0.5% BSA, cells were incubated for 45 min at room temperature with Texas-red conjugated affiniPure goat antimouse IgG (Jackson ImmunoResearch laboratories), washed three times in PBS + 0.5% BSA, and mounted in Vectashield Mounting Medium (Vector Laboratories) containing DAPI.
Cells were photographed with a Zeiss Axiophot2 Photomicroscope. Images were captured using a CCD camera (Princeton Instruments) and colored using MetaMorph software (MetaMorph Imaging System). A minimum of 200 nuclei were analyzed for each cell line and treatment.
Time-lapse microscopy
For observations with living cells, cells were plated onto dishes with coverslips (MatTek), transfected with peGFP-pol, and UV irradiated as described above. The dishes were then assembled into a live-cell microscopy chamber set to 37°C, that was mounted onto a Zeiss Axiovert 125 inverted microscope. At 1-min intervals, images were captured with a CCD camera.
Antibodies
Polη protein was detected using a rabbit polyclonal antibody directed against a peptide corresponding to polη residues 19–34 VQVEQRQNPHLRNKPC diluted 1 : 200. Anti-Rad51 protein, from Oncogene Research Products, was diluted 1 : 100; PC10 monoclonal anti-PCNA protein was from Santa Cruz Biotechnology. Thymidine dimers were detected using TDM2 mouse monoclonal antibody diluted 1 : 2000 (Mori et al. 1991). All antibodies were diluted in PBS containing 3% BSA.
Western blot analysis
Cells scraped from culture dishes were washed with PBS, centrifuged and directly lysed in Laemmli buffer, boiled for 10 min, and separated on a 10% SDS-polyacrylamide gel. Proteins were transferred onto PVDF membranes (Hybond P, Amersham), and probed for 1 h with rabbit anti-GFP antibodies diluted 1 : 200 (Clontech). After washing, the membrane was incubated with horseradish peroxidase-conjugated antirabbit-IgG (1 : 5000; Dako), and signals were visualized using enhanced chemiluminescence (ECL, Amersham).
Acknowledgments
We are grateful to O. Nikaido for the antithymidine dimer antibody; J.E. Cleaver for the XP30ROsv cell line; Heather Fawcett for assistance with cell culture; and Elaine Taylor, Bryn Bridges, and Penny Jeggo for helpful criticism of the manuscript. We are particularly indebted to Daniel Zicha and Colin Gray, ICRF, London, for enabling us to carry out the time-lapse microscopy experiments of Figure 3A. This work was supported by grants from AICR (99–063), EU FP5 (QLG1-CT1999–0181), and EU Concerted Action (BMH4-CT98–3045).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL a.r.lehmann@sussex.ac.uk; FAX 44-1273-678121.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.187501
References
- Arlett CF, Harcourt SA. Survey of radiosensitivity in a variety of human cell strains. Cancer Res. 1980;40:926–932. [PubMed] [Google Scholar]
- Arlett CF, Harcourt SA, Broughton BC. The influence of caffeine on cell survival in excision-proficient and excision-deficient xeroderma pigmentosum and normal human cell strains following ultraviolet light irradiation. Mutation Res. 1975;33:341–346. doi: 10.1016/0027-5107(75)90209-2. [DOI] [PubMed] [Google Scholar]
- Bailly V, Lamb J, Sung P, Prakash S, Prakash L. Specific complex formation between yeast RAD6 and RAD18 proteins: A potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes & Dev. 1994;8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- Berneburg M, Lehmann AR. Advances in Genetics. Vol. 43. San Diego, CA: Academic Press; 2001. Xeroderma pigmentosum and related disorders: Defects in DNA repair and transcription; pp. 71–102. [DOI] [PubMed] [Google Scholar]
- Bravo R, Macdonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: Association with DNA replication sites. J Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton BC, Steingrimsdottir H, Weber C, Lehmann AR. Mutations in the xeroderma pigmentosum group D DNA repair gene in patients with trichothiodystrophy. Nat Genet. 1994;7:189–194. doi: 10.1038/ng0694-189. [DOI] [PubMed] [Google Scholar]
- Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- Cleaver JE, Afzal V, Feeney L, McDowell M, Sadinski W, Volpe JP, Busch DB, Coleman DM, Ziffer DW, Yu Y, et al. Increased ultraviolet sensitivity and chromosomal instability related to p53 function in the xeroderma pigmentosum variant. Cancer Res. 1999;59:1102–1108. [PubMed] [Google Scholar]
- Cordeiro-Stone M, Zaritskaya LS, Price LK, Kaufmann WK. Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J Biol Chem. 1997;272:13945–13954. doi: 10.1074/jbc.272.21.13945. [DOI] [PubMed] [Google Scholar]
- Cordonnier AM, Lehmann AR, Fuchs RPP. Impaired translesion synthesis in xeroderma pigmentosum extracts. Mol Cell Biol. 1999;19:2206–2211. doi: 10.1128/mcb.19.3.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes & Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- Domon M, Rauth AM. Ultraviolet-light irradiation of mouse L cells: Effects on cells in the DNA synthesis phase. Radiat Res. 1969;40:414–429. [PubMed] [Google Scholar]
- Ensch-Simon I, Burgers PM, Taylor JS. Bypass of a site-specific cis-syn thymine dimer in an SV40 vector during in vitro replication by HeLa and XPV cell-free extracts. Biochemistry. 1998;37:8218–8226. doi: 10.1021/bi972460j. [DOI] [PubMed] [Google Scholar]
- Goodman MF. Coping with replication “train wrecks” in Escherichia coli using Pol V, Pol II and RecA proteins. Trends Biochem Sci. 2000;25:189–195. doi: 10.1016/s0968-0004(00)01564-4. [DOI] [PubMed] [Google Scholar]
- Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999a;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Polη. Science. 1999b;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- Lamb JR, Petit-Frère C, Broughton BC, Lehmann AR, Green MHL. Inhibition of DNA replication by ionizing radiation is mediated by a trans-acting factor. Int J Radiat Biol. 1989;56:125–130. doi: 10.1080/09553008914551271. [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Kirk-Bell S, Arlett CF, Paterson MC, Lohman PHM, de Weerd-Kastelein EA, Bootsma D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc Natl Acad Sci. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, Kirk-Bell S, Mayne L. Abnormal kinetics of DNA synthesis in ultraviolet light–irradiated cells from patients with Cockayne's syndrome. Cancer Res. 1979;39:4237–4241. [PubMed] [Google Scholar]
- Limoli CL, Giedzinski E, Morgan WF, Cleaver JE. Inaugural article: Polymerase η deficiency in the xeroderma pigmentosum variant uncovers an overlap between the S phase checkpoint and double-strand break repair. Proc Natl Acad Sci. 2000;97:7939–7946. doi: 10.1073/pnas.130182897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor-Shoshani A, Reuven NB, Tomer G, Livneh Z. Highly mutagenic replication by DNA polymerase V (UmuC) provides a mechanistic basis for SOS untargeted mutagenesis. Proc Natl Acad Sci. 2000;97:565–570. doi: 10.1073/pnas.97.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999a;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The human XPV (xeroderma pigmentosum variant) gene encodes human polymerase η. Nature. 1999b;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Iwai S, Hanaoka F. Accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel T. Low fidelity DNA synthesis by human DNA polymerase-η. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- McGregor WG, Wei D, Maher VM, McCormick JJ. Abnormal, error-prone bypass of photoproducts by xeroderma pigmentosum variant cell extracts results in extreme strand bias for the kinds of mutations induced by UV light. Mol Cell Biol. 1999;19:147–154. doi: 10.1128/mcb.19.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A, Savini E, Weighardt F, Rossi R, Ciarrocchi G, Villa A, Biamonti G. The N-terminal domain of human DNA ligase I contains the nuclear localization signal and directs the enzyme to sites of DNA replication. EMBO J. 1995;14:5379–5386. doi: 10.1002/j.1460-2075.1995.tb00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, Tomkinson AE. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6′-4′) photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, Krokan HE. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Dargemont C, Kuhn LC, Nigg EA. Nuclear export of proteins: The role of nuclear retention. Cell. 1993;74:493–504. doi: 10.1016/0092-8674(93)80051-f. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Svoboda DL, Briley LP, Vos JM. Defective bypass replication of a leading strand cyclobutane thymine dimer in xeroderma pigmentosum variant cell extracts. Cancer Res. 1998;58:2445–2448. [PubMed] [Google Scholar]
- Tateishi S, Sakuraba Y, Masuyama S, Inoue H, Yamaizumi M. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc Natl Acad Sci. 2000;97:7927–7932. doi: 10.1073/pnas.97.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RM, Moore DJ, Whitehouse J, Johnson P, Caldecott KW. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol Cell Biol. 2000;20:735–740. doi: 10.1128/mcb.20.2.735-740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A, McDonald JP, Frank EG, Woodgate R. Polι, a remarkably error-prone human DNA polymerase. Genes & Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- van Hoffen A, Natarajan AT, Mayne LV, van Zeeland AA, Mullenders LHF, Venema J. Deficient repair of the transcribed strand of active genes in Cockayne's syndrome cells. Nucleic Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R. A plethora of lesion-replicating DNA polymerases. Genes & Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- Yuan F, Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, Taylor JS, Wang Z. Specificity of DNA lesion bypass by the yeast DNA polymerase η. J Biol Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]