Abstract
Double-stranded RNA (dsRNA) induces sequence-specific posttranscriptional gene silencing in many organisms by a process known as RNA interference (RNAi). Using a Drosophila in vitro system, we demonstrate that 21- and 22-nt RNA fragments are the sequence-specific mediators of RNAi. The short interfering RNAs (siRNAs) are generated by an RNase III–like processing reaction from long dsRNA. Chemically synthesized siRNA duplexes with overhanging 3′ ends mediate efficient target RNA cleavage in the lysate, and the cleavage site is located near the center of the region spanned by the guiding siRNA. Furthermore, we provide evidence that the direction of dsRNA processing determines whether sense or antisense target RNA can be cleaved by the siRNA–protein complex.
Keywords: RNAi, posttranscriptional gene silencing, dsRNA, siRNA
The term RNA interference (RNAi) was coined after the discovery that injection of dsRNA into the nematode Caenorhabditis elegans leads to specific silencing of genes highly homologous in sequence to the delivered dsRNA (Fire et al. 1998). RNAi was also observed subsequently in insects (Kennerdell and Carthew 1998), frog (Oelgeschlager et al. 2000), and other animals including mice (Svoboda et al. 2000; Wianny and Zernicka-Goetz 2000) and is likely to also exist in human. RNAi is closely linked to the posttranscriptional gene-silencing (PTGS) mechanism of cosuppression in plants and quelling in fungi (Cogoni and Macino 1999; Catalanotto et al. 2000; Dalmay et al. 2000; Ketting and Plasterk 2000; Mourrain et al. 2000; Smardon et al. 2000), and some components of the RNAi machinery are also necessary for posttranscriptional silencing by cosuppression (Catalanotto et al. 2000; Dernburg et al. 2000; Ketting and Plasterk 2000). The topic has been reviewed recently (Fire 1999; Sharp 1999; Bass 2000; Bosher and Labouesse 2000; Plasterk and Ketting 2000; Sijen and Kooter 2000; see also the entire issue of Plant Molecular Biology, Vol. 43, issue 2/3, 2000).
The natural function of RNAi and cosuppression appears to be protection of the genome against invasion by mobile genetic elements such as transposons and viruses, which produce aberrant RNA or dsRNA in the host cell when they become active (Jensen et al. 1999; Ketting et al. 1999; Ratcliff et al. 1999; Tabara et al. 1999; Malinsky et al. 2000). Specific mRNA degradation prevents transposon and virus replication, although some viruses are able to overcome or prevent this process by expressing proteins that suppress PTGS (Anandalakshmi et al. 2000; Lucy et al. 2000; Voinnet et al. 2000).
DsRNA triggers the specific degradation of homologous RNAs only within the region of identity with the dsRNA (Zamore et al. 2000). The dsRNA is processed to 21–23-nt RNA fragments (Zamore et al. 2000). These short fragments were also detected in extracts prepared from Drosophila melanogaster Schneider 2 cells that were transfected with dsRNA before cell lysis (Hammond et al. 2000) or after injection of radiolabeled dsRNA into D. melanogaster embryos (Yang et al. 2000) or C. elegans adults (Parrish et al. 2000). RNA molecules of similar size also accumulate in plant tissue that exhibits PTGS (Hamilton and Baulcombe 1999). It has been suggested that the 21–23-nt fragments are the guide RNAs for target recognition (Hamilton and Baulcombe 1999; Hammond et al. 2000), which is supported by the finding that the target mRNA is cleaved in 21–23-nt intervals (Zamore et al. 2000).
Here, we use the established Drosophila in vitro system (Tuschl et al. 1999; Zamore et al. 2000) to explore further the mechanism of RNAi. It is demonstrated that synthetic 21- and 22-nt RNAs, when base paired with 3′ overhanging ends, act as the guide RNAs for sequence-specific mRNA degradation. Short 30-bp dsRNAs are inefficiently processed to 21- and 22-nt RNAs, which may explain why they are ineffective in mediating RNAi. Furthermore, we define the target RNA cleavage sites relative to the 21- and 22-nt short interfering RNAs (siRNAs) and provide evidence that the direction of dsRNA processing determines whether a sense or an antisense target RNA can be cleaved by the siRNP endonuclease complex.
Length requirements for processing of dsRNA to 21- and 22-nt RNA fragments
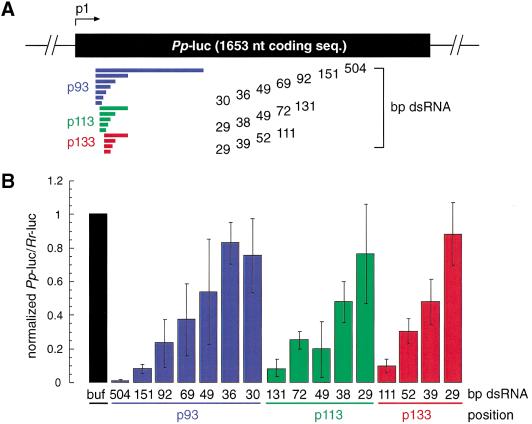
Lysate prepared from D. melanogaster syncytial embryos recapitulates RNAi in vitro, providing a tool for biochemical analysis of the mechanism of RNAi (Tuschl et al. 1999; Zamore et al. 2000). In vitro and in vivo analysis of the length requirements of dsRNA for RNAi has revealed that short dsRNA (<150 bp) are less effective than longer dsRNAs in degrading target mRNA (Ngo et al. 1998; Tuschl et al. 1999; Caplen et al. 2000; Hammond et al. 2000). The reasons for reduction in mRNA degrading efficiency are not understood. We therefore examined the precise length requirement of dsRNA for target RNA degradation under optimized conditions in the Drosophila lysate. Three series of dsRNAs were synthesized and directed against firefly luciferase (Pp-luc) reporter RNA. The dual luciferase assay was used to monitor specific suppression of target RNA expression (Tuschl et al. 1999; (Fig. 1A,B). Specific inhibition of target RNA expression was detected for dsRNAs as short as 38 bp, but dsRNAs of 29–36 bp were not effective in this process. The effect was independent of the target position and the degree of inhibition of Pp-luc mRNA expression correlated with the length of the dsRNA; that is, long dsRNAs were more effective than short dsRNAs.
Figure 1.
Double-stranded RNA as short as 38 bp can mediate RNAi. (A) Graphic representation of dsRNAs used for targeting Pp-luc mRNA. Three series of blunt-ended dsRNAs covering a range of 29–504 bp were prepared. The position of the first nucleotide of the sense strand of the dsRNA is indicated relative to the start codon of Pp-luc mRNA (p1). (B) RNA interference assay (Tuschl et al. 1999). Ratios of target Pp-luc to control Rr-luc activity were normalized to a buffer control (black bar). DsRNAs (5 nM) were preincubated in Drosophila lysate at 25°C for 15 min before the addition of 7-methyl-guanosine-capped Pp-luc and Rr-luc mRNAs (∼50 pM). The incubation was continued for another hour and then analyzed by the dual luciferase assay (Promega). The data are the average from at least four independent experiments ±S.D.
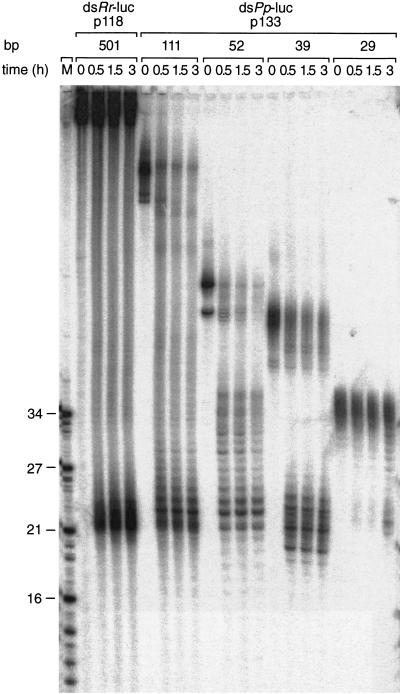
It has been suggested that the 21–23-nt RNA fragments generated by processing of dsRNAs are the mediators of RNA interference and cosuppression (Hamilton and Baulcombe 1999; Hammond et al. 2000; Zamore et al. 2000). We therefore analyzed the rate of 21–23-nt fragment formation for a subset of dsRNAs ranging in size from 501 to 29 bp. Formation of 21–23-nt fragments in Drosophila lysate (Fig. 2) was readily detectable for 39–501 bp dsRNAs but was significantly delayed for the 29-bp dsRNA. This observation is consistent with a role of 21–23-nt fragments in guiding mRNA cleavage and provides an explanation for the lack of RNAi by 30-bp dsRNAs. The length dependence of 21–23 mer formation is likely to reflect a mechanism to prevent the undesired activation of RNAi by short intramolecular base-paired structures of cellular RNAs.
Figure 2.
A 29-bp dsRNA is only slowly processed to 21–23-nt fragments. Time course of 21–23-mer formation from processing of internally 32P-labeled dsRNAs (5 nM) in the Drosophila lysate. The length and source of the dsRNA are indicated. An RNA size marker (M) has been loaded in the left lane, and the fragment sizes are indicated. Double bands at time zero are caused by incompletely denatured dsRNA.
Mapping of the cleavage sites on sense and antisense target RNAs
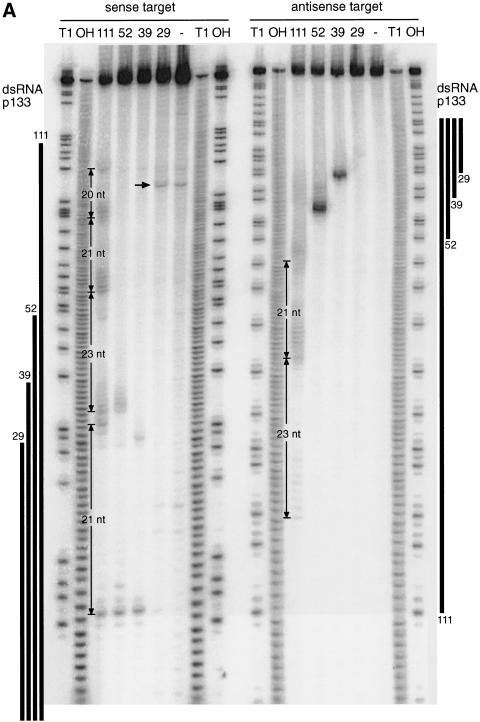
Addition of dsRNA and 5′-capped target RNA to the Drosophila lysate results in sequence-specific degradation of the target RNA (Tuschl et al. 1999). The target mRNA is only cleaved within the region of identity with the dsRNA, and many of the target cleavage sites are separated by 21–23 nt (Zamore et al. 2000). Thus, the number of cleavage sites for a given dsRNA was expected to roughly correspond to the length of the dsRNA divided by 21. We mapped the target cleavage sites on a sense and an antisense target RNA that was 5′ radiolabeled at the cap (Zamore et al. 2000; Fig. 3A,B). Stable 5′ cleavage products were separated on a sequencing gel, and the position of cleavage was determined by comparison with a partial RNase T1 and an alkaline hydrolysis ladder from the target RNA.
Figure 3.
Mapping of sense and antisense target RNA cleavage sites. (A) Denaturing gel electrophoresis of the stable 5′ cleavage products produced by 1 h incubation of 10 nM sense or antisense RNA 32P-labeled at the cap with 10 nM dsRNAs of the p133 series in Drosophila lysate. Length markers were generated by partial nuclease T1 digestion and partial alkaline hydrolysis (OH) of the cap-labeled target RNA. The regions targeted by the dsRNAs are indicated as black bars on both sides. The 20–23-nt spacing between the predominant cleavage sites for the 111-bp dsRNA is shown. The horizontal arrow indicates unspecific cleavage not caused by RNAi. (B) Position of the cleavage sites on sense and antisense target RNAs. The sequences of the capped 177-nt sense and 180-nt antisense target RNAs are represented in antiparallel orientation such that complementary sequence are opposing each other. The region targeted by the different dsRNAs are indicate by differently colored bars positioned between sense and antisense target sequences. Cleavage sites are indicated by circles (large circle for strong cleavage, small circle for weak cleavage). The 32P-radiolabeled phosphate group is marked by an asterisk.
Consistent with the previous observation (Zamore et al. 2000), all target RNA cleavage sites were located within the region of identity to the dsRNA. The 39-bp dsRNA produced a strong and a weak (often hardly detectable) cleavage site in the sense target RNA separated by 19 nt. The antisense target was only cleaved once, by the 39-bp dsRNA. The predominant cleavage site of the sense strand and the cleavage site of the antisense strand are located 10 nt from the 5′ end of the region covered by the dsRNA (Fig. 3B). The 52-bp dsRNA, which shares the same 5′ end as the 39-bp dsRNA, produces the same strong cleavage site on the sense target, located 10 nt from the 5′ end of the region of identity with the dsRNA in addition to two weaker cleavage sites 23 and 24 nt downstream of the first site. The antisense target was only cleaved once, again 10 nt from the 5′ end of the region covered by its respective dsRNA. Mapping of the cleavage sites for the 38–49-bp dsRNAs shown in Figure 1 revealed that the first and predominant cleavage site was always located 7–10 nt downstream from the 5′ boundary of the region covered by the dsRNA (data not shown). This suggests that the point-of-target RNA cleavage can be determined by the end of the dsRNA and could imply that processing to 21–23mers starts from the ends of the duplex.
Cleavage sites on sense and antisense targets for the longer 111-bp dsRNA were much more frequent than anticipated, and most of them appear in clusters separated by 20–23 nt (Fig. 3A,B). As for the shorter dsRNAs, the first cleavage site on the sense target is 10 nt from the 5′ end of the region spanned by the dsRNA, and the first cleavage site on the antisense target is located 9 nt from the 5′ end of region covered by the dsRNA. It is unclear what causes this disordered cleavage, but one possibility could be that longer dsRNAs may not only get processed from the ends but also internally, or there are some specificity determinants for dsRNA processing that we do not yet understand. Some irregularities to the 21–23 nt spacing were also noted previously (Zamore et al. 2000).
dsRNA is processed to 21- and 22-nt RNAs by an RNase III–like mechanism
To understand better the molecular basis of dsRNA processing and target RNA recognition, we decided to analyze the sequences of the 21–23-nt fragments generated by processing of 39-, 52-, and 111-bp dsRNAs in the Drosophila lysate. We first examined the 5′ and 3′ termini of the RNA fragments. Periodate oxidation of gel-purified 21–23-nt RNAs followed by β-elimination indicated the presence of a terminal 2′ and 3′ hydroxyl (data not shown). The 21–23mers were also responsive to alkaline phosphatase treatment, implying the presence of a 5′ terminal phosphate (data not shown). The presence of 5′ phosphate and 3′ hydroxyl termini suggests that the dsRNA could be processed by an enzymatic activity similar to Escherichia coli RNase III (for reviews, see Dunn 1982; Nicholson 1999; Robertson 1982, 1990).
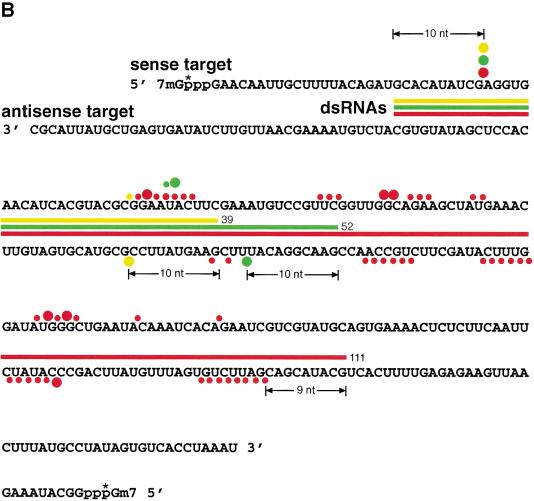
To directionally clone the 21–23-nt RNA fragments, 3′ and 5′ adapter oligonucleotides were ligated to the purified 21–23 mers using T4 RNA ligase. The ligation products were reverse transcribed, PCR-amplified, concatamerized, cloned, and sequenced. Over 220 short RNAs were sequenced from dsRNA processing reactions of the 39-, 52-, and 111-bp dsRNAs (Fig. 4A). We found the following length distribution: 1% 18 nt, 5% 19 nt, 12% 20 nt, 45% 21 nt, 28% 22 nt, 6% 23 nt, and 2% 24 nt. Sequence analysis of the 5′ terminal nucleotide of the processed fragments indicated that oligonucleotides with a 5′ guanosine were underrepresented. This bias was most likely introduced by T4 RNA ligase, which discriminates against 5′ phosphorylated guanosine as donor oligonucleotide (Romaniuk et al. 1982); no significant sequence bias was seen at the 3′ end. Many of the ∼21-nt fragments originating from the 3′ ends of the sense or antisense strand of the duplexes include 3′ nucleotides that are derived from untemplated addition of nucleotides during RNA synthesis using T7 RNA polymerase. Interestingly, a significant number of endogenous Drosophila ∼21-nt RNAs were also cloned, some of them from LTR and non-LTR retrotransposons (data not shown). This is consistent with a possible role for RNAi in transposon silencing (Ketting et al. 1999; Tabara et al. 1999).
Figure 4.
21- and 22-nt RNA fragments are generated by an RNase III–like mechanism. (A) Sequences of ∼21-nt RNAs after dsRNA processing. The ∼21-nt RNA fragments generated by dsRNA processing were directionally cloned and sequenced. Oligoribonucleotides originating from the sense strand of the dsRNA are indicated as blue lines; those originating from the antisense strand are red lines. Thick bars are used if the same sequence was present in multiple clones, the number at the right indicating the frequency. The target RNA cleavage sites mediated by the dsRNA are indicated as orange circles: large circle for strong cleavage, small circle for weak cleavage (see Fig. 3B). Circles on top of the sense strand indicated cleavage sites within the sense target, and circles at the bottom of the dsRNA indicate cleavage site in the antisense target. Up to five additional nucleotides were identified in ∼21-nt fragments derived from the 3′ ends of the dsRNA. These nucleotides are random combinations of predominantly C, G, or A residues and were most likely added in an untemplated fashion during T7 transcription of the dsRNA-constituting strands. (B) Two-dimensional TLC analysis of the nucleotide composition of ∼21-nt RNAs. The ∼21-nt RNAs were generated by incubation of internally radiolabeled 504-bp Pp-luc dsRNA in Drosophila lysate, gel purified, and then digested to mononucleotides with nuclease P1 (top row) or ribonuclease T2 (bottom row). The dsRNA was internally radiolabeled by transcription in the presence of one of the indicated α-32P nucleoside triphosphates. Radioactivity was detected by phosphorimaging. Nucleoside 5′-monophosphates, nucleoside 3′-monophosphates, nucleoside 5′,3′-diphosphates, and inorganic phosphate are indicated as pN, Np, pNp, and pi, respectively. Black circles indicate UV-absorbing spots from nonradioactive carrier nucleotides. The 3′,5′-diphosphates (red circles) were identified by comigration with radiolabeled standards prepared by 5′-phosphorylation of nucleoside 3′-monophosphates with T4 polynucleotide kinase and γ-32P-ATP (data not shown).
The ∼21-nt RNAs appear in clustered groups (Fig. 4A) that cover the entire dsRNA sequences. For the 39-bp dsRNA, two clusters of ∼21-nt RNAs were found from each dsRNA-constituting strand (including overhanging 3′ ends). Only one of the clusters from each strand can be correlated with a strong cleavage hot spot on the target sense or antisense RNA (Fig. 3A,B), indicating that dsRNA processing produced primarily two functional small RNAs originating from the 3′ ends of the duplex. Perhaps the ∼21-nt RNAs are present in double-stranded form in the endonuclease complex, but only one of the strands can be used for target RNA recognition and cleavage.
The ∼21-mer clusters for the 52- and 111-bp dsRNA are less well defined when compared to the 39-bp dsRNA. The clusters are spread over regions of 25–30 nt most likely representing several distinct subpopulations of ∼21-nt duplexes and, therefore, guiding target cleavage at several nearby sites. These cleavage regions are still predominantly separated by 20–23-nt intervals. The rules determining how dsRNA can be processed to ∼21-nt fragments are not yet understood, but it was observed previously that the ∼21–23-nt spacing of cleavage sites could be altered by a run of uridines (Zamore et al. 2000). The specificity of dsRNA cleavage by E. coli RNase III appears to be mainly controlled by antideterminants, that is, excluding some specific base pairs at given positions relative to the cleavage site (Zhang and Nicholson 1997). The sequence dependence of dsRNA processing and target RNA cleavage in RNAi needs to be examined further.
To test whether sugar-, base-, or cap-modification were present in processed ∼21-nt RNA fragments, we incubated radiolabeled 505-bp Pp-luc dsRNA in lysate for 1 h, isolated the ∼21-nt products, and digested it with P1 or T2 nuclease to mononucleotides. The nucleotide mixture was then analyzed by two-dimensional thin-layer chromatography (Fig. 4B). None of the four ribonucleotides were modified, as indicated by P1 or T2 digestion. We have previously analyzed adenosine to inosine conversion in the ∼21-nt fragments (after a 2 h incubation) and detected a small extent (<0.7%) deamination (Zamore et al. 2000); shorter incubation in lysate (1 h) reduced this inosine fraction to barely detectable levels. RNase T2, which cleaves 3′ of the phosphodiester linkage, produced nucleoside 3′-phosphate and nucleoside 3′,5′-diphosphate, thereby indicating the presence of a 5′-terminal monophosphate. All four nucleoside 3′,5′-diphosphates were detected and indicate that the internucleotidic linkage was cleaved with little or no sequence specificity for the residue 3′ to the cleavage site, and according to the sequence analysis of the cloned ∼21-nt fragments, no significant sequence bias was observed for the residue 5′ of the cleavage site. In summary, the ∼21-nt fragments are unmodified and were generated from dsRNA such that 5′-monophosphates and 3′-hydroxyls were present at the 5′-ends. Analysis of the products of dsRNA processing indicated that the ∼21-nt fragments are generated by a reaction with all the characteristics of an RNase III cleavage reaction (Dunn 1982; Robertson 1982, 1990; Nicholson 1999).
Synthetic 21- and 22-nt RNAs mediate target RNA cleavage
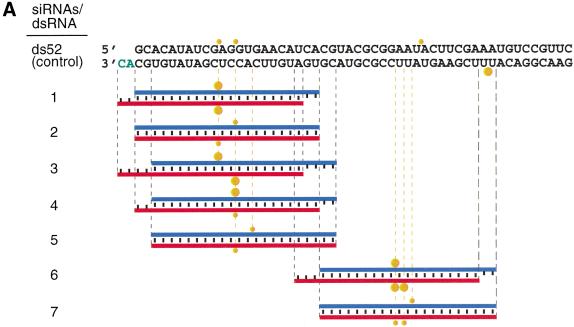
We chemically synthesized 21- and 22-nt RNAs, identical in sequence to some of the cloned ∼21-nt fragments, and tested them for their ability to mediate target RNA degradation (Fig. 5A–C). The 21- and 22-nt RNA duplexes were incubated at 100 nM concentrations in the lysate, a 10- to 20-fold higher concentration than the 52-bp control dsRNA. Under these conditions, target RNA cleavage was readily detectable. Tenfold reduced concentrations of 21- and 22-nt duplexes (10 nM) still caused target RNA cleavage but to a smaller extent (data not shown). Increasing the duplex concentration from 100 to 1000 nM, however, did not further increase target degradation (data not shown), perhaps because of a limiting protein factor within the lysate. Single-stranded sense or antisense 21- and 22-nt RNAs at 100 nM concentration did not affect target RNA expression, most likely because single-stranded RNAs are not stable in the lysate and degraded to mononucleotides within minutes (data not shown). We also found that preannealing of the short antisense RNAs to the target mRNA before the addition of lysate had no effect on target RNA expression (data not shown).
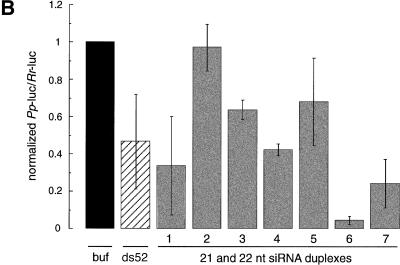
Figure 5.
Synthetic 21- and 22-nt RNAs mediate target RNA cleavage. (A) Graphic representation of control 52-bp dsRNA and synthetic 21- and 22-nt dsRNAs. The sense strand of 21- and 22-nt short interfering RNAs (siRNAs) is shown in blue, the antisense strand in red. The sequences of the siRNAs were derived from the cloned fragments of 52- and 111-bp dsRNAs (Fig. 4A), except for the 22-nt antisense strand of duplex 5. The siRNAs in duplexex 6 and 7 were unique to the 111-bp dsRNA-processing reaction. The two 3′ overhanging nucleotides indicated in green are present in the sequence of the synthetic antisense strand of duplexes 1 and 3. Both strands of the control 52-bp dsRNA were prepared by in vitro transcription, and a fraction of transcripts may contain untemplated 3′ nucleotide addition. The target RNA cleavage sites directed by the siRNA duplexes are indicated as orange circles (see legend to Fig. 4A) and were determined as shown in Figure 5B. (B) RNA interference assay. To evaluate the efficiency of target RNA degradation, control 52-bp dsRNA (5 nM) or 21- and 22-nt RNA duplexes 1–7 (100 nM) targeting full-length Pp-luc mRNA were tested in the translation-based RNAi assay as described in Figure 1B. The relative luminescence of target to control luciferase normalized to a buffer control (buf) is blotted; error bars indicate standard deviations calculated from at least two independent experiments. (C) Position of the cleavage sites on sense and antisense target RNAs. The target RNA sequences are as described in Figure 3B. Control 52-bp dsRNA (10 nM) or 21- and 22-nt RNA duplexes 1–7 (100 nM) were incubated with target RNA at 25°C for 2.5 h in Drosophila lysate. The stable 5′ cleavage products were resolved on the gel. The cleavage sites are indicated in Figure 5A. The region targeted by the 52-bp dsRNA or the sense (s) or antisense (as) strands are indicated by the black bars to the side of the gel. The cleavage sites are all located within the region of identity of the dsRNAs. For precise determination of the cleavage sites of the antisense strand, a lower percentage gel was used (data not shown).
RNase III makes two staggered cuts in both strands of the dsRNA, leaving a 3′ overhang of 2 nt. The 21- and 22-nt RNA duplexes with 2- or 3-nt overhanging 3′ ends (duplexes 1, 4, 6) were more efficient in reducing the target RNA expression than the corresponding blunt-ended dsRNAs (duplexes 2, 5, 7) or the dsRNA with 4 nt overhang (duplex 3). Duplexes 6 and 7 are generally more effective for RNAi than duplexes 1–5, probably as a consequence of target RNA accessibility (because of RNA self-structure or RNA-coating proteins) or because of sequence-specific effects in the reconstitution of the RNA-degrading complexes. The interference effects determined in the translation-based assay (Fig. 5B) correlate well with the intensity of the cleavage bands observed by targeting 5′ radiolabeled model substrates with the 21- and 22-nt RNA duplexes (Fig. 5C). Together, these data suggest that 2–3 nt of overhanging 3′ ends are beneficial for reconstitution of the RNAi nuclease complex and may be required for high-affinity binding of the short RNA duplex to the protein components. A 5′ terminal phosphate, although present after dsRNA processing, was not required to mediate target RNA cleavage and was absent from the short synthetic RNAs.
The synthetic 21- and 22-nt duplexes guided cleavage of sense as well as antisense targets within the region covered by the short duplex. This is interesting, considering that the presumably base-paired clusters of ∼21-nt fragments derived from the 39-bp dsRNA (Fig. 2) can only be correlated to a predominant cleavage site on either the sense or the antisense target but not both. We interpret this result by suggesting that only one of two strands present in the ∼21-nt duplex is able to guide target RNA cleavage and that the orientation of the ∼21-nt duplex in the nuclease complex is determined by the initial direction of dsRNA processing. It also implies that the processed short RNAs are present in a tight ribonucleoprotein complex and do not dissociate and rebind during the time scale of the experiment. The presentation of an already perfectly processed ∼21-nt duplex to the in vitro system, however, does allow formation of the active sequence-specific nuclease complex with two possible orientations of the symmetric RNA duplex. This results in cleavage of sense as well as antisense targets within the region of identity with the ∼21-nt RNA duplex.
The target cleavage site is located near center of the region covered by the 21- or 22-nt RNAs, 11 or 12 nt downstream of the first nucleotide that is complementary to the 21- or 22-nt guide sequence (Fig. 4A,B). Displacing the sense strand of a 22-nt duplex by two nucleotides (cf. duplexes 1 and 3 in Fig. 5A) displaced the cleavage site of only the antisense target by two nucleotides. Displacing both sense and antisense strands by two nucleotides shifted both cleavage sites by two nucleotides (cf. duplexes 1 and 4). We predict that it will be possible to design a pair of 21- or 22-nt RNAs to cleave a target RNA at almost any given position.
The specificity of target RNA cleavage guided by 21- and 22-nt RNAs appears exquisite, as no cleavage sites are detected outside of the region of complementarity to the 21- and 22-nt RNAs (Fig. 5C). It should, however, be noted that the nucleotides present in the 3′ overhang of the 21- and 22-nt RNA duplex may contribute less to substrate recognition than the nucleotides near the cleavage site. This is based on the observation that the 3′-most nucleotide of the antisense strand of active duplexes 1 or 3 (Fig. 5A) is not complementary to the target. A detailed analysis of the specificity of RNAi can now be readily undertaken using synthetic 21- and 22-nt RNAs.
On the basis of the evidence that synthetic 21- and 22-nt RNAs with overhanging 3′ ends mediate RNA interference, we propose to name the ∼21-nt RNAs short interfering RNAs, or siRNAs, and the respective RNA-protein complex a small interfering ribonucleoprotein particle, or siRNP.
3′ overhangs of ∼20 nt on short dsRNAs inhibit RNAi
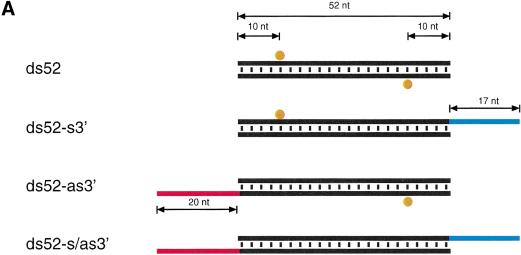
We have also analyzed dsRNAs with 17–20 nt overhanging 3′ ends that were less potent than blunt-ended dsRNAs (data not shown). The inhibitory effect of long 3′ ends was particularly pronounced for dsRNAs <100 bp. The effect was not caused by imperfect dsRNA formation, based on native gel analysis (data not shown). We tested if the inhibitory effect of long overhanging 3′ ends could be used as a tool to initiate dsRNA processing at only one of the two ends of a short RNA duplex.
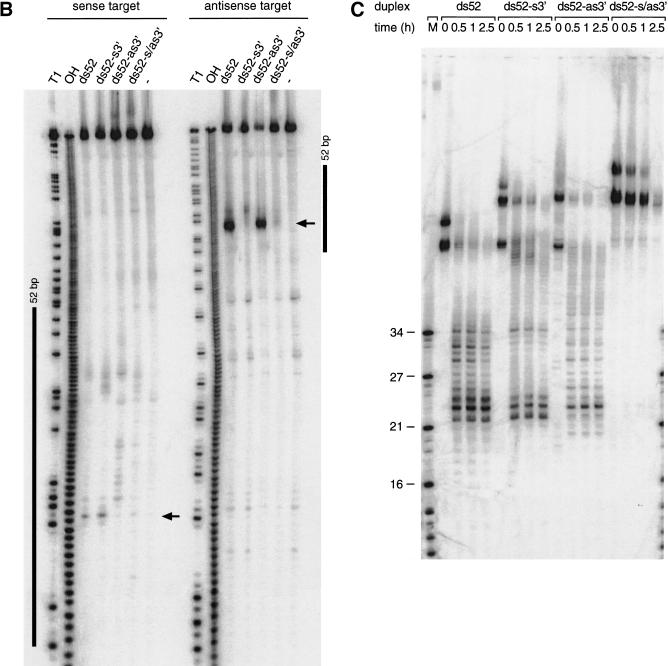
We synthesized four combinations of the 52-bp model dsRNA, blunt-ended, 3′ extension only on the sense strand, the 3′ extension only on the antisense strand, and the double 3′ extension on both strands and mapped the target RNA cleavage sites and monitored ∼21-nt formation after incubation in lysate (Fig. 6A–C). The first and predominant cleavage site of the sense target was lost when the 3′ end of the antisense strand of the duplex was extended, and the strong cleavage site of the antisense target was lost when the 3′ end of sense strand of the duplex was extended. Extending the 3′ ends on both strands rendered the 52-bp dsRNA virtually inactive. These observations correlate with the formation of ∼21-nt fragments from blunt-ended dsRNAs or dsRNAs with only one 3′ extension and the absence of ∼21-nt fragments when both 3′ ends of the duplex are extended (Fig. 6C). One explanation for the dsRNA inactivation by ∼20-nt 3′ extensions could be the association of single-stranded RNA-binding proteins that could interfere with the association of one of the dsRNA-processing factors at this end. This is supported by the significantly longer persistence of the double 3′ extended dsRNA in the lysate (Fig. 6C). Together, these results are consistent with our model where only one of the strands of the siRNA duplex in the assembled siRNP is able to guide target RNA cleavage. The orientation of the strand that guides RNA cleavage is defined by the direction of the dsRNA processing reaction. A block at the 3′ end of the sense strand will only permit dsRNA processing from the opposing 3′ end of the antisense strand. This, in turn, generates siRNPs in which only the antisense strand of the siRNA duplex is able to guide sense target RNA cleavage. The same is true for the reciprocal situation. The less pronounced inhibitory effect of long 3′ extensions in the case of longer dsRNAs (≥500 bp, data not shown) suggests that long dsRNAs may also contain internal dsRNA-processing signals or may get processed cooperatively because of the association of multiple cleavage factors.
Figure 6.
Long 3′ overhangs on short dsRNAs inhibit RNAi. (A) Graphic representation of 52-bp dsRNA constructs. The 3′ extensions of sense and antisense strand are indicated in blue and red, respectively. The observed cleavage sites on the target RNAs are represented as orange circles analogous to Figure 4A and were determined as shown in B. (B) Position of the cleavage sites on sense and antisense target RNAs. The target RNA sequences are as described in Figure 3B. DsRNA (10 nM) was incubated with target RNA at 25°C for 2.5 h in Drosophila lysate. The stable 5′ cleavage products were resolved on the gel. The major cleavage sites are indicated with a horizontal arrow and are also represented in A. The region targeted by the 52-bp dsRNA is represented as a black bar at both sides of the gel. (C) Processing of 52-bp dsRNAs with different 3′ extensions. Internally 32P-labeled dsRNAs (5 nM) were incubated in Drosophila lysate, and reaction aliquots were analyzed at the indicated time points. An RNA size marker (M) has been loaded in the left lane, and the fragment sizes are indicated. Double bands at time zero are caused by incompletely denatured dsRNA.
A model for dsRNA-directed mRNA cleavage
The new biochemical data update the model for how dsRNA targets mRNA for destruction (Fig. 7). Based on the 21–23-nt length of the processed RNA fragments, it had already been speculated that an RNase III–like activity may be involved in RNAi (Bass 2000). Double-stranded RNA is first processed to short RNA duplexes of predominantly 21 and 22 nt in length and with staggered 3′ ends similar to an RNase III–like reaction (Dunn 1982; Robertson 1982; Nicholson 1999). This hypothesis is further supported by the presence of 5′ phosphates and 3′ hydroxyls at the termini of the siRNAs (Fig. 4B) as observed in RNase III reaction products (Dunn 1982; Nicholson 1999).
Figure 7.
Proposed model for RNAi. RNAi is predicted to begin with processing of dsRNA (sense strand in black, antisense strand in red) to predominantly 21- and 22-nt short interfering RNAs (siRNAs). Short overhanging 3′ nucleotides, if present on the dsRNA, may be beneficial for processing of short dsRNAs. The dsRNA-processing proteins, which remain to be characterized, are represented as green and blue ovals and assemble on the dsRNA in asymmetric fashion. In our model, this is illustrated by binding of a hypothetical blue protein or protein domain with the siRNA strand in 3′ to 5′ direction while the hypothetical green protein or protein domain is always bound to the opposing siRNA strand. These proteins or a subset remain associated with the siRNA duplex and preserve its orientation, as determined by the direction of the dsRNA processing reaction. Only the siRNA sequence associated with the blue protein is able to guide target RNA cleavage. The endonuclease complex is referred to as small interfering ribonucleoprotein complex or siRNP. It is presumed here that the endonuclease that cleaves the dsRNA may also cleave the target RNA, probably by temporarily displacing the passive siRNA strand not used for target recognition. The target RNA is then cleaved in the center of the region recognized by the sequence-complementary guide siRNA. Because the cleavage site is displaced by 10–12 nt relative to the dsRNA processing site, a conformational rearrangement or a change in the composition of an siRNP must occur before target RNA cleavage.
Bacterial RNase III and the eukaryotic homologs Rnt1p in Saccharomyces cerevisiae and Pac1p in Schizosaccharomyces pombe have been shown to function in processing of ribosomal RNA as well as snRNA and snoRNAs (see, for example, Chanfreau et al. 2000). Little is known about the biochemistry of RNase III homologs from plants, animals, or human. Two families of RNase III enzymes have been identified predominantly by database-guided sequence analysis or cloning of cDNAs. The first RNase III family is represented by the 1327-amino-acid D. melanogaster protein drosha (accession no. AF116572). The carboxyl terminus is composed of two RNase III domains and one dsRNA-binding domain, and the amino terminus is of unknown function. Close homologs are also found in C. elegans (accession no. AF160248) and human (accession no. AF189011; Filippov et al. 2000; Wu et al. 2000). The drosha-like human RNase III was recently cloned and characterized (Wu et al. 2000). The gene is ubiquitously expressed in human tissues and cell lines, and the protein is localized in the nucleus and the nucleolus of the cell. Based on results inferred from antisense inhibition studies, a role of this protein for rRNA processing was suggested. The second class is represented by the C. elegans gene K12H4.8 (accession no. S44849) coding for an 1822-amino-acid protein. This protein has an amino-terminal RNA helicase motif, which is followed by two RNase III catalytic domains and a dsRNA-binding motif, similar to the drosha RNase III family. There are close homologs in S. pombe (accession no. Q09884), Arabidopsis thaliana (accession no. AF187317), D. melanogaster (accession no. AE003740), and human (accession no. AB028449) (Jacobsen et al. 1999; Filippov et al. 2000; Matsuda et al. 2000). It is tempting to speculate that the K12H4.8 RNase III/helicase is the likely candidate to be involved in RNAi.
Genetic screens in C. elegans identified rde-1 and rde-4 as essential for activation of RNAi, without an effect on transposon mobilization or cosuppression (Tabara et al. 1999; Dernburg et al. 2000; Grishok et al. 2000; Ketting and Plasterk 2000). This led to the hypothesis that these genes are important for dsRNA processing but are not involved in mRNA target degradation. The function of both genes is as yet unknown, the rde-1 gene product is a member of a family of proteins similar to the rabbit protein eIF2C (Tabara et al. 1999), and the sequence of rde-4 has not yet been described. Future biochemical characterization of these proteins should reveal their molecular function.
Processing of dsRNA to siRNA duplexes appears to start from the ends of both blunt-ended dsRNAs or dsRNAs with short (1–5 nt) 3′ overhangs and proceeds in ∼21–23-nt steps. Long (∼20 nt) 3′ staggered ends on short dsRNAs suppress RNAi, possibly through interaction with single-stranded RNA-binding proteins. The suppression of RNAi by single-stranded regions flanking short dsRNA and the reduced rate of siRNA formation from short 30-bp dsRNAs may explain why structured regions within mRNAs do not lead to activation of RNAi. In C. elegans, it was observed recently that injection of a 26-bp dsRNA could trigger RNAi of the unc-22 gene; however, a >250-fold higher concentration of 26-bp dsRNA was necessary compared to an 81-bp dsRNA control (Parrish et al. 2000). It is conceivable that siRNA production from the 26-bp dsRNA was rate limiting and was compensated for by increasing the concentration of 26-bp dsRNA.
In our model, we presume that the dsRNA-processing proteins or a subset of them remain associated with the siRNA duplex after the processing reaction. The orientation of the siRNA duplex relative to these proteins determines which of the two complementary strands functions in guiding target RNA degradation. Chemically synthesized siRNA duplexes guide cleavage of sense as well as antisense target RNA, as they are able to associate with the protein components in either of the two possible orientations. A distinct role of the two strands of an siRNP is consistent with the recent observation in C. elegans that certain chemical modifications (e.g., 2′-aminouridine, 2′-deoxythymidine, or 5-iodouridine) incorporated into dsRNA are well tolerated at the sense, but not the cleavage-guiding antisense, strand (Parrish et al. 2000).
The finding that synthetic 21- and 22-nt siRNA duplexes can be used for efficient mRNA degradation demonstrates that the targeting step can be uncoupled from the dsRNA-processing step. This raises the prospects of using siRNA duplexes as new tools for sequence-specific regulation of gene expression in functional genomics as well as biomedical studies. The siRNAs may be effective in mammalian systems, where long dsRNAs can not be used because they activate the dsRNA-dependent protein kinase (PKR) response (Clemens 1997). As such, the siRNA duplexes may represent a new alternative to antisense or ribozyme therapeutics.
Materials and methods
In vitro RNAi
In vitro RNAi and lysate preparations were performed as described previously (Tuschl et al. 1999; Zamore et al. 2000) using a final concentration of 0.03 mg/mL creatine kinase in the RNAi reaction. It is critical to use freshly dissolved creatine kinase (Roche or Sigma) for optimal ATP regeneration. The RNAi translation assays (Fig. 1) were performed with dsRNA concentrations of 5 nM and an extended preincubation period at 25°C for 15 min before the addition of in vitro transcribed, capped, and polyadenylated Pp-luc and Rr-luc reporter mRNAs. The incubation was continued for 1 h, and the relative amount of Pp-luc and Rr-luc protein was analyzed using the dual luciferase assay (Promega) and a Monolight 3010C luminometer (PharMingen).
RNA synthesis
Standard procedures were used for in vitro transcription of RNA from PCR templates carrying T7 or SP6 promoter sequences (see, for example, Tuschl et al. 1998). Synthetic RNA was prepared using Expedite RNA phosphoramidites (Proligo). The 3′ adapter oligonucleotide was synthesized using dimethoxytrityl-1,4-benzenedimethanol-succinyl-aminopropyl-CPG, a generous gift from B. Sproat (Catholic University Leuven, Belgium). The oligoribonucleotides were deprotected in 3 mL of 32% ammonia/ethanol (3:1) at 55°C for 4 h (Expedite RNA) or at 55°C for 16 h (3′ and 5′ adapter DNA/RNA chimeric oligonucleotides) and then desilylated and gel purified as described previously (Tuschl et al. 1993). RNA transcripts for dsRNA preparation including long 3′ overhangs were generated from PCR templates that contained a T7 promoter in sense and an SP6 promoter in antisense direction. The transcription template for sense and antisense target RNA was PCR amplified with GCGTAATACGACTCAC TATAGAACAATTGCTTTTACAG (underlined, T7 promoter) as 5′ primer, ATTTAGGTGACACTATAGGCATAAAGAATT GAAGA (underlined, SP6 promoter), as 3′ primer and the linearized Pp-luc plasmid (pGEM-luc sequence; Tuschl et al. 1999) as template; the T7-transcribed sense RNA was 177 nt long with the Pp-luc sequence between positions 113–273 relative to the start codon and followed by 17 nt of the complement of the SP6 promoter sequence at the 3′ end. Transcripts for blunt-ended dsRNA formation were prepared by transcription from two different PCR products that only contained a single promoter sequence.
DsRNA annealing was carried out using a phenol/chloroform extraction. Equimolar concentration of sense and antisense RNA (50 nM to 10 μM, depending on the length and amount available) in 0.3 M NaOAc (pH 6) were incubated at 90°C for 30 sec and then extracted at room temperature with an equal volume of phenol/chloroform and followed by a chloroform extraction to remove residual phenol. The resulting dsRNA was precipitated by addition of 2.5–3 volumes of ethanol. The pellet was dissolved in lysis buffer (100 mM KCl, 30 mM HEPES-KOH at pH 7.4, 2 mM Mg(OAc)2), and the quality of the dsRNA was verified by standard agarose gel electrophoresis in 1× TAE-buffer. The 52-bp dsRNAs with the 17-nt and 20-nt 3′ overhangs (Fig. 6) were annealed by incubating at 95°C for 1 min and then were rapidly cooled to 70°C and followed by slow cooling to room temperature over a 3-h period (50 μL annealing reaction, 1 μM strand concentration, 300 mM NaCl, 10 mM Tris-HCl at pH 7.5). The dsRNAs were then phenol/chloroform extracted, ethanol precipitated, and dissolved in lysis buffer.
Transcription of internally 32P-radiolabeled RNA used for dsRNA preparation (Figs. 2,4) was performed using 1 mM ATP, CTP, GTP; 0.1 or 0.2 mM UTP; and 0.2–0.3 μM [α-32P]UTP (3000 Ci/mmol) or the respective ratio for radiolabeled nucleoside triphosphates other than UTP. Labeling of the cap of the target RNAs was performed as described previously (Zamore et al. 2000). The target RNAs were gel purified after cap labeling.
Cleavage site mapping
Standard RNAi reactions were performed by preincubating 10 nM dsRNA for 15 min followed by addition of 10 nM cap-labeled target RNA. The reaction was stopped after a further 2-h (Fig. 3A) or 2.5-h incubation (Figs. 5C,6B) by proteinase K treatment (Tuschl et al. 1999). The samples were then analyzed on 8% or 10% sequencing gels. The 21- and 22-nt synthetic RNA duplexes were used at 100 nM final concentration (Fig. 5B,C).
Cloning of ∼21-nt RNAs
The 21-nt RNAs were produced by incubation of radiolabeled dsRNA in Drosophila lysate in absence of target RNA (200 μL reaction, 1 h incubation, 50 nM dsP111, or 100 nM dsP52 or dsP39). The reaction mixture was subsequently treated with proteinase K (Tuschl et al. 1999), and the dsRNA-processing products were separated on a denaturing 15% polyacrylamide gel. A band, including a size range of at least 18–24 nt, was excised and then eluted into 0.3 M NaCl overnight at 4°C in siliconized tubes. The RNA was recovered by ethanol precipitation and then dephosphorylated (30 μL reaction, 50°C, 30 min, 10 U alkaline phosphatase; Roche). The reaction was stopped by phenol/chloroform extraction, and the RNA was ethanol precipitated. The 3′ adapter oligonucleotide (pUUUaaccg catccttctcx: uppercase, RNA; lowercase, DNA; p, phosphate; x, 4-hydroxymethylbenzyl) was then ligated to the dephosphorylated ∼21-nt RNA (20 μL reaction, 37°C, 30 min, 5 μM 3′ adapter, 50 mM Tris-HCl at pH 7.6, 10 mM MgCl2, 0.2 mM ATP, 0.1 mg/mL acetylated BSA, 15% DMSO, 25 U T4 RNA ligase; Amersham-Pharmacia) (Pan and Uhlenbeck 1992). The ligation reaction was stopped by the addition of an equal volume of 8 M urea/50 mM EDTA stopmix and directly loaded on a 15% gel. Ligation yields were >50%. The ligation product was recovered from the gel and 5′ phosphorylated (20 μL reaction, 37°C, 30 min, 2 mM ATP, 5 U T4 polynucleotide kinase; NEB). The phosphorylation reaction was stopped by phenol/chloroform extraction, and RNA was recovered by ethanol precipitation. Next, the 5′ adapter (tactaatacgactcactAAA: uppercase, RNA; lowercase, DNA) was ligated to the phosphorylated ligation product as described above. The new ligation product was gel purified and eluted from the gel slice in the presence of reverse transcription primer (GACTAGCTGGAATTCAAG GATGCGGTTAAA: bold, EcoRI site), used as carrier. Reverse transcription (15 μL reaction, 42°C, 30 min, 150 U Superscript II reverse transcriptase; Life Technologies) was followed by PCR using a 5′ primer CAGCCAACGGAATTCATACGACTCAC TAAA (bold, EcoRI site) and the 3′ RT primer. The PCR product was purified by phenol/chloroform extraction and ethanol precipitated. The PCR product was then digested with EcoRI (NEB) and concatamerized using T4 DNA ligase (high concentration; NEB). Concatamers of a size range of 200–800 bp were separated on a low-melt agarose gel, recovered from the gel by a standard melting and phenol extraction procedure, and ethanol precipitated. The unpaired ends were filled in by incubation with Taq polymerase under standard conditions at 72°C for 15 min, and the DNA product was directly ligated into the pCR2.1-TOPO vector using the TOPO TA cloning kit (Invitrogen). Colonies were screened using PCR and M13–20 and M13 Reverse sequencing primers. PCR products were directly submitted for custom sequencing (Sequence Laboratories Göttingen). On average, four to five ∼21-mer sequences were obtained per clone.
2D-TLC analysis
Nuclease P1 digestion of radiolabeled, gel-purified siRNAs and 2D-TLC was carried out as described (Zamore et al. 2000). Nuclease T2 digestion was performed in 10 μL reactions at 50°C for 3 h in 10 mM ammonium acetate (pH 4.5) using 2 μg/μL carrier tRNA and 30 U ribonuclease T2 (Life Technologies). The migration of nonradioactive standards was determined by UV shadowing. The identity of nucleoside-3′,5′-diphosphates was confirmed by comigration of the T2 digestion products with standards prepared by 5′-32P-phosphorylation of commercial nucleoside 3′-monophosphates using [γ-32P]ATP and T4 polynucleotide kinase (data not shown).
Acknowledgments
We acknowledge Heike Taubner for assistance with fly work; Uschi Kutzke for chemical RNA synthesis; Gordon Dowe for some of the sequencing; and H. Jäckle, R. Lührmann, and F. Eckstein for support. We thank N.J. Watkins, T. Achsel, J. Ludwig, P.D. Zamore, D.P. Bartel, and P.A. Sharp for advice and comments on the manuscript. I would also like to thank P.D.Z and P.A.S. for the suggestion of naming the short interfering 21- and 22-nt RNAs siRNAs. This work was supported by BMBF Biofuture grant number 0311856.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ttuschl@mpibpc.gwdg.de; FAX 49-551-201-1197.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.862301.
References
- Anandalakshmi R, Marathe R, Ge X, Herr JM, Jr, Mau C, Mallory A, Pruss G, Bowman L, Vance VB. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science. 2000;290:142–144. doi: 10.1126/science.290.5489.142. [DOI] [PubMed] [Google Scholar]
- Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- Bosher JM, Labouesse M. RNA interference: Genetic wand and genetic watchdog. Nat Cell Biol. 2000;2:E31–E36. doi: 10.1038/35000102. [DOI] [PubMed] [Google Scholar]
- Caplen NJ, Fleenor J, Fire A, Morgan RA. dsRNA-mediated gene silencing in cultured Drosophila cells: A tissue culture model for the analysis of RNA interference. Gene. 2000;252:95–105. doi: 10.1016/s0378-1119(00)00224-9. [DOI] [PubMed] [Google Scholar]
- Catalanotto C, Azzalin G, Macino G, Cogoni C. Gene silencing in worms and fungi. Nature. 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Buckle M, Jacquier A. Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc Natl Acad Sci. 2000;97:3142–3147. doi: 10.1073/pnas.070043997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens MJ. PKR—A protein kinase regulated by double-stranded RNA. Int J Biochem Cell Biol. 1997;29:945–949. doi: 10.1016/s1357-2725(96)00169-0. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Homology-dependent gene silencing in plants and fungi: A number of variations on the same theme. Curr Opin Microbiol. 1999;2:657–662. doi: 10.1016/s1369-5274(99)00041-7. [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Dernburg AF, Zalevsky J, Colaiacovo MP, Villeneuve AM. Transgene-mediated cosuppression in the C. elegans germ line. Genes & Dev. 2000;14:1578–1583. [PMC free article] [PubMed] [Google Scholar]
- Dunn JJ. Ribonuclease III. In: Boyer PD, editor. The enzymes. 15, part B. New York: Academic Press; 1982. pp. 485–499. [Google Scholar]
- Filippov V, Solovyev V, Filippova M, Gill SS. A novel type of RNase III family proteins in eukaryotes. Gene. 2000;245:213–221. doi: 10.1016/s0378-1119(99)00571-5. [DOI] [PubMed] [Google Scholar]
- Fire A. RNA-triggered gene silencing. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz ME. Disruption of an RNA helicase/RNase III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- Jensen S, Gassama MP, Heidmann T. Taming of transposable elements by homology-dependent gene silencing. Nat Genet. 1999;21:209–212. doi: 10.1038/5997. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Plasterk RH. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Lucy AP, Guo HS, Li WX, Ding SW. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 2000;19:1672–1680. doi: 10.1093/emboj/19.7.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky S, Bucheton A, Busseau I. New insights on homology-dependent silencing of I factor activity by transgenes containing ORF1 in Drosophila melanogaster. Genetics. 2000;156:1147–1155. doi: 10.1093/genetics/156.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Ichigotani Y, Okuda T, Irimura T, Nakatsugawa S, Hamaguchi M. Molecular cloning and characterization of a novel human gene (HERNA) which encodes a putative RNA-helicase. Biochim Biophys Acta. 2000;1490:163–169. doi: 10.1016/s0167-4781(99)00221-3. [DOI] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson AW. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Oelgeschlager M, Larrain J, Geissert D, De Robertis EM. The evolutionarily conserved BMP-binding protein Twisted gastrulation promotes BMP signalling. Nature. 2000;405:757–763. doi: 10.1038/35015500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Uhlenbeck OC. In vitro selection of RNAs that undergo autolytic cleavage with Pb2+ Biochemistry. 1992;31:3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- Parrish S, Fleenor J, Xu S, Mello C, Fire A. Functional anatomy of a dsRNA trigger: Differential requirement for the two trigger strands in RNA Interference. Mol Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Plasterk RH, Ketting RF. The silence of the genes. Curr Opin Genet Dev. 2000;10:562–567. doi: 10.1016/s0959-437x(00)00128-3. [DOI] [PubMed] [Google Scholar]
- Ratcliff FG, MacFarlane SA, Baulcombe DC. Gene silencing without DNA: RNA-mediated cross-protection between viruses. Plant Cell. 1999;11:1207–1216. doi: 10.1105/tpc.11.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HD. Escherichia coli ribonuclease III cleavage sites. Cell. 1982;30:669–672. doi: 10.1016/0092-8674(82)90270-7. [DOI] [PubMed] [Google Scholar]
- ————— Escherichia coli ribonuclease III. Methods Enzymol. 1990;181:189–202. doi: 10.1016/0076-6879(90)81121-a. [DOI] [PubMed] [Google Scholar]
- Romaniuk E, McLaughlin LW, Neilson T, Romaniuk PJ. The effect of acceptor oligoribonucleotide sequence on the T4 RNA ligase reaction. Eur J Biochem. 1982;125:639–643. doi: 10.1111/j.1432-1033.1982.tb06730.x. [DOI] [PubMed] [Google Scholar]
- Sharp PA. RNAi and double-strand RNA. Genes & Dev. 1999;13:139–141. [PubMed] [Google Scholar]
- Sijen T, Kooter JM. Post-transcriptional gene-silencing: RNAs on the attack or on the defense? Bioessays. 2000;22:520–531. doi: 10.1002/(SICI)1521-1878(200006)22:6<520::AID-BIES5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke J, Stacey S, Klein M, Mackin N, Maine E. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tuschl T, Ng MM, Pieken W, Benseler F, Eckstein F. Importance of exocyclic base functional groups of central core guanosines for hammerhead ribozyme activity. Biochemistry. 1993;32:11658–11668. doi: 10.1021/bi00094a023. [DOI] [PubMed] [Google Scholar]
- Tuschl T, Sharp PA, Bartel DP. Selection in vitro of novel ribozymes from a partially randomized U2 and U6 snRNA library. EMBO J. 1998;17:2637–2650. doi: 10.1093/emboj/17.9.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell. 2000;103:157–167. doi: 10.1016/s0092-8674(00)00095-7. [DOI] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III is a 160 kDa protein involved in preribosomal RNA processing. J Biol Chem. 2000;17:17. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- Yang D, Lu H, Erickson JW. Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr Biol. 2000;10:1191–1200. doi: 10.1016/s0960-9822(00)00732-6. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zhang K, Nicholson AW. Regulation of ribonuclease III processing by double-helical sequence antideterminants. Proc Natl Acad Sci. 1997;94:13437–13441. doi: 10.1073/pnas.94.25.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]