Abstract
Xenotransplantation of porcine cells, tissues, and organs shows promise to surmount the shortage of human donor materials. Among the barriers to pig-to-human xenotransplantation are porcine endogenous retroviruses (PERV) since functional representatives of the two polytropic classes, PERV-A and PERV-B, are able to infect human embryonic kidney cells in vitro, suggesting that a xenozoonosis in vivo could occur. To assess the capacity of human and porcine cells to counteract PERV infections, we analyzed human and porcine APOBEC3 (A3) proteins. This multigene family of cytidine deaminases contributes to the cellular intrinsic immunity and act as potent inhibitors of retroviruses and retrotransposons. Our data show that the porcine A3 gene locus on chromosome 5 consists of the two single-domain genes A3Z2 and A3Z3. The evolutionary relationships of the A3Z3 genes reflect the evolutionary history of mammals. The two A3 genes encode at least four different mRNAs: A3Z2, A3Z3, A3Z2-Z3, and A3Z2-Z3 splice variant A (SVA). Porcine and human A3s have been tested toward their antiretroviral activity against PERV and murine leukemia virus (MuLV) using novel single-round reporter viruses. The porcine A3Z2, A3Z3 and A3Z2-Z3 were packaged into PERV particles and inhibited PERV replication in a dose-dependent manner. The antiretroviral effect correlated with editing by the porcine A3s with a trinucleotide preference for 5′ TGC for A3Z2 and A3Z2-Z3 and 5′ CAC for A3Z3. These results strongly imply that human and porcine A3s could inhibit PERV replication in vivo, thereby reducing the risk of infection of human cells by PERV in the context of pig-to-human xenotransplantation.
INTRODUCTION
Xenotransplantation, the transplantation of viable cells, organs, or tissues between species, has been proposed as a solution to the shortage of human organs for the treatment of organ failure. Advances in nonclinical xenotransplantation have increased the feasibility of clinical trials. The prevention of xenogeneic infections is central to the ultimate success of xenotransplantation (24, 25).
Pigs are considered a source species for clinical xenotransplantation. They are easy to breed, are good size matches for humans, and have been genetically engineered to express or suppress a variety of proteins relevant to transplantation. Even though swine pose greater immunological barriers to transplantation into primates, they can be bred to exclude many common potential human pathogens with barrier maintenance according to designated pathogen-free conditions.
Concerns have been raised regarding the retroviral safety of clinical xenotransplantation since porcine endogenous retroviruses (PERV), belonging to the genus Gammaretrovirus, have been identified in the pig genome, and infectivity of some of these viruses for human cells has been demonstrated in vitro (1, 51, 58, 59, 72, 76, 77, 99, 104). Pigs harbor three replication-competent PERV classes, termed PERV-A, PERV-B, and PERV-C (1, 51). While PERV-A and PERV-B are also able to infect human cells at least in vitro, PERV-C is restricted to pigs (99). The capacity of PERV to infect nonhuman primate cells is limited, at least partially due to differential display and efficiency of PERV receptors on these cells. No evidence of infection of human cells in vivo has been demonstrated, and no disease resulting from this family of viruses has been described in swine or humans to date (26, 38, 75, 106).
PERV mRNAs are expressed in different pig tissues and organs (12, 64); thus, any transplanted organ may potentially produce virus. Pigs are classified as transmitters or nontransmitters, depending on whether porcine peripheral blood mononuclear cells (PBMCs) transmit PERV to human cells in vitro. Null animals do not transmit PERV to either pig or human cells (72). Expression and infection may be amplified by stimulation of swine peripheral blood lymphocytes (PBLs) in vitro (104).
A number of strategies have been devised to reduce the risk of PERV infection in xenograft recipients. These include the use of nontransmitter pigs or pigs without active PERV loci as source animals, use of antiretroviral agents, such as the reverse transcriptase (RT) inhibitors zidovudine (AZT) and dideoxyinosine (ddI) in recipients (80, 81), viral vaccines, and antibodies to target PERV (22), or the reduction of viral replication in vitro using RNA interference (16).
Due to the limited genetic coding capacity of viruses, they strongly depend on a plethora of factors provided by their host cells (10, 101). The permissiveness of a given cell may be additionally determined by the presence or absence of restriction factors that evolved during host-virus coevolution (27, 28, 33, 105).
During recent years three proteins belonging to these restriction factors were characterized to be of particular importance for the replication of retroviruses: TRIM5α, tetherin, and APOBEC3 ([A3] for apolipoprotein B mRNA-editing catalytic polypeptide 3) (61, 105). The mammalian APOBEC3 genes are part of the AID/APOBEC gene family including AID, APOBEC1 (A1), APOBEC2 (A2), and APOBEC4. All family members share a structural and functional domain of zinc-dependent deaminases (reviewed in reference 13). APOBEC cytidine deaminases have the inherent enzymatic capacity to deaminate cytidine residues in single-stranded RNA and DNA molecules that are recognized by currently unknown mechanisms. Proteins of the A3 group contain one or two zinc (Z)-coordinating domains and can be classified according to the presence/absence of a Z1, Z2, or Z3 motif (47, 67). Different lineages of placental mammals show an individual A3 locus with a variable arrangement of A3 genes varying in number and types (A3Z1, -Z2, -Z3, -Z2-Z1, -Z2-Z2, -Z2-Z3) that appears to be the result of a specific host adaptation to viruses (47, 67, 114, 115). In humans, the A3 locus contains seven A3 genes; the A3 locus contains one A3 gene in rodents, two in pigs, six in horses, and four in cats (8, 40, 48, 67, 71, 114). The nomenclature for human/primate A3 follows the original proposed system (A3A to A3H), while a novel nomenclature for nonprimate A3 has been suggested based on the presence or absence of the Z domains (47).
Human immunodeficiency virus type 1 (HIV-1) mutants lacking the vif gene can package APOBEC3G (A3G) into viral particles. Incorporated A3G specifically deaminates cytidine residues to uracil in growing single-stranded DNA during reverse transcription, leading to HIV genome degradation and/or hypermutation (7, 32, 49, 55, 56, 112). More recent studies indicate that deaminase-independent mechanisms might also be involved in the antiviral activity of A3 (6, 36, 37, 39, 60, 69). The question of how retroviruses counteract or escape the A3s from their own host species is important to understand virus tropism and host-virus coevolution (84). In the case of HIV-1, the amount of cellular A3G in wild-type (wt) HIV-1-infected cells is dramatically reduced by a Vif-dependent degradation via the ubiquitination-proteasome pathway (57, 89, 110, 111). In contrast to the well-characterized A3-Vif interaction, little is known yet about A3-neutralizing strategies used by retroviruses that do not encode a Vif protein. Gammaretroviruses such as murine or feline leukemia virus (MuLV or FeLV) appear not to express accessory proteins with a function similar to that of the lentiviral Vif or to the foamy viral protein Bet (52, 67, 79, 85), both inhibiting the encapsidation of A3 proteins. Despite many studies, the debate on the mechanism of resistance of MuLVs to murine A3 (muA3) has not resulted in a generally convincing model (7, 9, 41, 45, 46, 56). However, recent data clearly show that muA3 is an important in vivo restriction factor of Friend virus complex and the Moloney murine leukemia virus (54, 87, 98) that is used in our experiments as an internal control.
In initial studies, porcine A3Z2-Z3 (poA3Z2-Z3) was found to strongly inhibit HIV-1 and to weakly restrict MuLV (42). In a second publication, it was reported that overexpressed poA3Z2-Z3 did not significantly interfere with PERV transmission, and the authors concluded that PERV is resistant to its species-own A3 protein (43).
Here, we reanalyzed the chromosomal porcine A3 locus for poA3Z2 and poA3Z3 and found evidence that pigs express four different A3 mRNAs encoding poA3Z2, poA3Z3, and by read-through transcription and alternatively splicing poA3Z2-Z3 and poA3Z2-Z3 splice variant A (SVA). We found that PERV was severely inhibited by various porcine A3s in single-round as well as in spreading-virus assays. The PERV inhibition strongly correlated with a specific cytidine deamination in viral genomes in the trinucleotide 5′ TGC (edited nucleotide underlined) for poA3Z2 as well as poA3Z2-Z3 and 5′ CAC for A3Z3.
MATERIALS AND METHODS
Cell lines.
The cell lines 293T (CRL-11268; ATCC LGC Standards GmbH, Wesel, Germany) and MAX-T (A. Saalmüller, Vienna, Austria; derived from d/d haplotype miniature swine [78]) were maintained in Dulbecco's high-glucose modified Eagle's medium (DMEM) (Biochrom, Berlin, Germany) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, penicillin (100 units/ml), and streptomycin (100 μg/ml). For 293T cells, 500 μg/ml Geneticin (G418; Invitrogen, Darmstadt, Germany) was added to the culture medium. The cell lines Dubca (CRL-2276; ATCC LGC Standards GmbH) and SCP (CRL-1700; ATCC LGC Standards GmbH) were cultivated in minimum essential medium (MEM) (Biochrom) supplemented with 10% FBS, 2 mM l-glutamine, penicillin (100 units/ml), and streptomycin (100 μg/ml). The suspension cell line L45 (ECACC 91012320) was maintained in RPMI 1640 medium (Biochrom) containing 10% FBS, 5 × 10−5 M 2-mercaptoethanol, 1 mM sodium pyruvate, and 2 mM l-glutamine. Peripheral blood mononuclear cells were isolated from heparin-treated whole blood by Histopaque-1077 (Sigma Aldrich, München, Germany) gradient centrifugation and either used directly for preparation of high-molecular-weight genomic DNA or cultured before RNA isolation after activation with phytohemagglutinin (PHA; 3 μg/ml) for 3 days in RPMI 1640 medium containing 10% FBS, 5 × 10−5 M 2-mercaptoethanol, and 2 mM l-glutamine and afterwards with human recombinant interleukin-2 (100 U/ml). All cells were maintained at 37°C and 5% CO2.
Porcine A3 mRNA expression studies.
Total RNA was isolated with TRIzol reagent (Invitrogen) according to the instructions of the manufacturer. Expression studies of porcine A3 mRNAs in MAX-T, L45, and PHA-activated PBMCs of a domestic pig were done by reverse transcription-PCR (RT-PCR) using 5 μg of total RNA to produce cDNA by SuperScript III RT (Invitrogen). cDNAs were used to perform PCRs to screen for porcine A3 expression. To check the quality of the cDNA PCR was performed amplifying the constitutively expressed porcine glutamine–fructose-6–phosphate amidotransferase (GFAT) by PCR (30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min) with the primer combination poGFAT_for (5′-AACCCAGTCCTGTCAATAGCC-3′) and poGFAT_rev (5′-ACGAGAGAGATTGCAGCTTCC-3′).
Human A3 sequences were used to search for orthologous, porcine expressed sequence tags (ESTs). Three porcine ESTs were found (BP170197, DN107667, and DN107314) that could be aligned via overlapping identical sequence regions. Within this prolonged EST sequence a putative porcine A3 open reading frame (ORF) was identified with a length of 798 bp encoding a protein sequence of 256 amino acids (aa). On this basis, primers (10f_poAPO3, 5′-ATGGATCCTCAGCGCCTGAGACAATGGCC-3′; 9r_poAPO3, 5′-TCACCACCTGGCGTGAGCAC-3′) were generated, and PCR was run with 35 cycles of 94°C for 30 s, 64°C for 30 s, and 72°C for 1 min. This led to the identification of poA3Z2. A second porcine A3 sequence was used for this study, published by Conticello et al. (13). To amplify this double-domain poA3Z2-Z3, the primers f_pA3_cDNA (5′-ATGGATCCTCAGCGCCTGAG-3′) and r_pA3_cDNArc (5′-TCATCTCGAGTCACTTCTTGATG-3′) were used. PCR was run for 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 70 s. A third porcine A3 sequence was published by LaRue et al. (48). This single-domain poA3Z3 ORF was directly amplified with specific primers to construct the porcine A3 expression plasmid (see below). The identified open reading frames served as templates for primer generation to find the untranslated regions (UTRs) of the mRNA sequences.
Porcine mRNA was isolated with an mRNA-Only Eukaryotic mRNA Isolation Kit (Epicentre Biotechnologies, Madison, WI). Following cDNA synthesis, PCRs were performed using a 5′/3′ RACE Kit, 2nd Generation (Roche Applied Science, Mannheim, Germany) and an Expand High Fidelity PCR System (Roche Applied Science) according to the manufacturer's instructions. All oligonucleotides used are listed in Table 1.
Table 1.
Primers applied to identify the UTRs of the porcine mRNA sequencesa
| mRNA region | Reaction | Primer name (reference) | 5′→3′ Sequence |
|---|---|---|---|
| cDNA | 1r_Ss3FII | CCATTGTCTCAGGCGCTGAGGATCCAT | |
| 5′ UTR A3Z2 | 1st PCR | r_E1_UTR | GTGGTTTGCTTCCTCAGGCTCAGAC |
| 2nd PCR | rev_A3Z2-Z3LR | CCTCCGAGCAGCAGGACCCAAT | |
| cDNA | r_Exon5_1 | AGCAGCCTTTGTCAAGAATGGAGTCATC | |
| 5′ UTR A3Z3 | 1st PCR | r_A3Z3_E2_b | CTGTTGTATGAATATATTTTCTTTTAGTAGATTCAT |
| 2nd PCR | r_5′ UTR_A3Z3 | CAGATTCCGGGAGTCCTGAGGC | |
| cDNA | Oligo-dT-Anchor primer | GACCACGCGTATCGATGTCGAC(T)16V | |
| 3’ UTR A3Z3 | 1st PCR | A3Z2-Z3 outer 3′ RACE (48) | CCAAGGAGCTGGTTGATTTC |
| 2nd PCR | A3Z2-Z3 poA3 inner 3′ RACE (48) | CTGGAGCAATACAGCGAGAG | |
| cDNA | Oligo-dT-Anchor primer | GACCACGCGTATCGATGTCGAC(T)16V | |
| 3′ UTR A3Z2 | 1st PCR | f_EI_8 | TCGCCTCTACTACTTCTGGAAGTCAG |
| 2nd PCR | f_Exon4_3 | CAACACTGCTGGAACAACTTCGTG |
The particular primers were used in the indicated order. The coding A3 sequences served as templates for the generation of the oligonucleotides (where V is A, C, or G).
Quantitative RT-PCR (qRT-PCR).
mRNA expression levels of porcine A3 transcripts in different porcine tissues were quantified by one-step RT-PCR using a Roche LightCycler instrument (Roche Applied Science) according to the manufacturer's protocols. Reactions were performed in 20 μl of LightCycler capillaries (Roche Applied Science) with a QuantiTect SYBR green RT-PCR Kit (Qiagen, Hilden), according to the manufacturer's instructions, containing 0.5 μM each primer and 20 ng of template. One-step RT-PCR was performed as follows: 50°C for 20 min, 95°C for 15 min, and then 40 to 50 cycles of 94°C for 15 s, 56°C for 15 s, and 72°C for 17 s. To check for the accuracy of the reactions a melting curve analysis was performed. Table 2 lists the sequences of the poA3 and beta-actin primers. The threshold cycles were calculated using the LightCycler software, LCDA, version 3.5.28. PCRs were performed in triplicates for each sample, and all of the A3 mRNA levels were normalized to beta-actin.
Table 2.
A3 qRT-PCR primers
| mRNA | NCBI accession no. | Primer name | 5′→3′ Sequence |
|---|---|---|---|
| A3Z2 | FJ716801 | A3Z2_QRT_sense | GGAGGAACCTGCAACCTATG |
| A3Z2_QRT_antise | CGTGAGCTGTGGTTAGGTCT | ||
| A3Z3 | HM347451 | A3Z3_QRT_sense | CTGGGAGGTTGCTCACAGATAA |
| A3Z3_QRT_antise | TGAGGCAGACTCCTTGTCCC | ||
| A3Z2-Z3 | EU871586 | A3Z2-Z3_Q_sense | CGCTTGGTCACAGAGCTGAAGC |
| A3Z2-Z3_Q_anti | GTAGCACAAGTAGGTCTTCCTC | ||
| A3Z2-Z3 SVA | FJ716802 | LC_17_s | CTCCTTCCACTTTCGCAACC |
| LC_15_a | GACATAGCAGATGATTCTGTAGC | ||
| Beta-actin | X00351 | beta-actin_LC_s | GACGTGGACATCCGCAAAGAC |
| beta-actin_LC_a | GTGATCTCCTTCTGCATCCTGTCG |
Porcine A3 expression plasmids.
The identified sequences were modified by adding a Kozak sequence (in italics in the forward [f] primers) in front of the start codon as well as a C-terminal hemagglutinin (HA)-tag (in italics in the reverse [r] primers) on the 3′ end of the A3 ORFs. All restriction sites are underlined. The primers f_NheI_K_APO (5′-GCTAGCACTATGGATCCTCAGCGCCTGAG-3′) and r_843_HA_XhoI (5′-CTCGAGTCAAGCATAATCTGGAACATCATATGGATAGCGGTAACAAATCCAACTAGCATC-3′) were used to amplify poA3Z2. PCR was run for 30 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min. In the case of poA3Z2-Z3 SVA and poA3Z2-Z3, PCRs were run for 30 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 60 s (poA3Z2-Z3 SVA) or 70 s (poA3Z2-Z3) using primers f_Acc65I_K_708_APO (5′-GGTACCACTATGGATCCTCAGCGCCTGAG-3′) and r_708_HA_NotI (5′-GCGGCCGCTCAAGCATAATCTGGAACATCATATGGATATCTCGAGTCACTTCTTGATG-3′). poA3Z3 was amplified by PCR (30 cycles of 98°C for 10 s, 62°C for 20 s, and 72°C for 30 s) using Phusion HF DNA polymerase (NEB GmbH, Frankfurt, Germany) with the primer combination f_Acc65I_K_A3Z3 (5′-GGTACCACTATGAATCTACTAAAAGAAAATATATTCATACAACAG-3′) and r_pA3_HA_NotI_rc (5′-GCGGCCGCTCAAGCATAATCTGGAACATCATATGGATATCTCGAGTCACTTCTTGATGATGAAGGTGATG-3′). These HA-tagged porcine A3 sequences were cloned in pcDNA3.1Zeo(+) (Invitrogen) using NheI/XhoI in the case of poA3Z2 and Acc65I/NotI restriction sites in the case of poA3Z2-Z3, poA3Z3, and poA3Z2-Z3 SVA. The expression of poA3Z2 was confirmed by the specific reverse primer r2_SV-N_poAPO3 (5′-TCAGCGGTAACAAATCCAACTAGCATCCACGAG-3′) in combination with primer 10f_poAPO3 in a PCR (35 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 1 min).
Plasmids.
For cloning of the PERV-enhanced green fluorescent protein (EGFP) reporter virus expression plasmid, the EGFP sequence was amplified (30 cycles of 94°C for 30 s, 62°C for 5 s, and 72°C for 1 min) using pLEGFP-N1 (BD Biosciences, Heidelberg, Germany) and primers KpnI_GFP_for (5′-CAGTGAGGTACCATGGTGAGCAAGGGCGAGGAG-3′) and GFP_PacI_rev (5′-TCTCTGTTAATTAATTACTTGTACAGCTCGTCCATGCC-3′). The PCR product was inserted in the env region (nucleotides [nt] 7022 to 8057) of a replication-competent PERV-B expression plasmid (pGTe[PERV-B(33)ATG]Δchrom), originating from plasmid PERV-B(33)ATG (AJ133816) (14) that lacks the chromosomal flanks. To enhance EGFP expression, a human cytomegalovirus (CMV) immediate-early promoter was inserted in front of EGFP via KpnI. CMV sequence was amplified by PCR (30 cycles of 94°C for 30 s, 58°C for 20 s, and 72°C for 1 min) using pLEGFP-N1 and primers For_KpnI_CMV (5′-GGTACCTAGTTATTAATAGTAATCAATTACGGGGTC-3′) and Rev_CMV_KpnI (5′-GGTACCGTCGACTGCAGAATTC-3′). The MuLV-EGFP reporter virus expression plasmid was constructed similarly. The CMV-EGFP-cassette of pLEGFP-N1 was amplified with the primers f_NsiI_CMV (5′-CAGTGAATGCATTAGTTATTAATAGTAATCAATTACGGGGTCATTAG-3′) and GFP_PacI_rev (5′-TCTCTGTTAATTAATTACTTGTACAGCTCGTCCATGCC-3′) using 30 PCR cycles of 94°C for 15 s, 63°C for 30 s, and 72°C for 70 s. This PCR product was inserted within the env sequence (nt 7491 to 7834) of the replication-competent MuLV plasmid pM91MS (34) that encodes Moloney MuLV with an amphotropic env gene. For the production of pseudotyped viral particles, the vesicular stomatitis virus G protein (VSV-G) encoding plasmid pMD.G2 was applied (68). The following APOBEC expression plasmids were used: muA3, pc-Mu-APOBEC3G-HA (56), human AID (huAID), pcDNA3.1-AID-V5-6xHis, huA1, pcDNA3.1-APOBEC1-V5-6xHis, and huA2, pcDNA3.1-APOBEC2-V5-6xHis (113). All human A3 sequences (A3A, -B, -C, -F, -G, and -H) are expressed as C-terminal hemagglutinin (HA)-tagged proteins in pcDNA3.1Zeo(+) (65, 103, 113).
Transfections and retrovirus infection assays.
A total of 1 × 106 293T cells were seeded in six-well plates. One day later plasmid DNA was transfected with Lipofectamine and Plus Reagent (Invitrogen) according to the manufacturer's instructions. One microgram of one reporter virus expression plasmid was cotransfected with 0.1 μg of pMD.G2 and 1 μg of an A3 expression plasmid or pcDNA3.1Zeo(+) as an empty vector control. Two days posttransfection (dpt) the supernatants were filtered (0.45-μm pore size) and analyzed for reverse transcription (RT) activity using a C-type RT activity assay (Cavidi, Uppsala, Sweden) according to the manufacturer's instructions (protocol B). RT-normalized viral supernatants (80 mU for PERV-EGFP and 50 mU for MuLV-EGFP) were used to transduce 1.5 × 105 293T cells seeded in 12-well plates. At 3 days posttransduction, the percentage of fluorescent cells (percent positive cells) was analyzed by flow cytometry (FACScan; BD Biosciences, Heidelberg, Germany).
Spreading-virus replication studies.
For studies of spreading-virus replication, stable A3-expressing (huA3B, huA3F, poA3Z2, poA3Z3, and poA3Z2-Z3) cells were constructed by transfecting 0.5 μg of each plasmid in 1 × 106 293T cells. At 2 dpt, cultures were selected using 400 μg/ml zeocin (Invitrogen). Resistant cells were transfected with 1 μg of pBS_Not[PERV-B(33)ATG] (14), and viral supernatants were collected regularly over a period of 30 days to determine the RT activity. Stable A3 expression was analyzed by immunoblotting.
Cloning and sequencing of the porcine A3 gene locus.
Based on the construct poA3Z2-Z3, primers were designed to amplify porcine genomic DNA to obtain intron sequences and to determine the exon-exon junctions. The primer pairs that successfully identified the sequence of the porcine A3 genes (between the start codon of poA3Z2 and the stop codon of poA3Z3) are listed in Table 3. For each PCR, 400 ng of genomic DNA of MAX-T cells supplemented with 0.5 M betaine (Sigma Aldrich) and KOD XL polymerase (Merck Chemicals Ltd., Nottingham, United Kingdom) were used, and 30 cycles of 94°C for 30 s, annealing for 5 s (temperature dependent on the applied primer pair), and 74°C for 1 to 6 min (depending on the length of the PCR product) were applied. All PCR products were cloned into pGEM-T Easy (Promega GmbH, Mannheim, Germany) and sequenced.
Table 3.
Primers applied to identify the porcine A3 gene locus reaching from the start codon of poA3Z2 to the stop codon of poA3Z3a
| Primer name | 5′→3′ Sequence | poA3 gene locus region (nt) |
|---|---|---|
| 9f_poAPO3 | ATGGATCCTCAGCGCCTGAGAC | 1–2370 |
| 3r_RH_poAPO3 | GGTTTTGAAAGATGCCTTGGAAGAAACAG | |
| f_Exon2_1 | CCAGAGGAGTGGTTCCATGAGTTATCTCC | 2226–6054 |
| r_EI_3 | CTGACTTCCAGAAGTAGTAGAGGCGAGC | |
| for_EI_8 | TCGCCTCTACTACTTCTGGAAGTCAG | 2294–6001 |
| 7r_poAPO3 | CCACGGCTGGAAGGGCATCC | |
| f_Exon4_3 | CAACACTGCTGGAACAACTTCGTG | 6532–14901 |
| r_Exon5_2 | GCTGGTTGCCAAACTGTTGTATG | |
| for_EI_7 | CAACCAGCCCCGGGTCTTGGC | 14893–16084 |
| r_EI_6 | AGGTGGTGCCGATTGCTGATGAAATCAACC | |
| for_EI_3 | TGATTTCATCAGCAATCGGCACCACCTG | 16058–16547 |
| 3r_Ss3F | AGAGCCGTCGGCTTATGCTCTCGC | |
| for_EI_2 | AAAGCGGGTCTCCGTGGCTGTCATG | 16163–16934 |
| 2r_Ss3F | GAACCGAGTCGCAAATCTCTGAAGGAATCC |
The generation of the oligonucleotides was based on the ORF of the porcine full-length construct poA3Z2-Z3.
Southern blot hybridization.
High-molecular-weight genomic DNA of pig, camel, human, sheep, and cow was prepared from PBMCs or cell lines according to standard protocols (86) and digested with BglII (100 U) or EcoRI (100 U) (NEB), separated in a 0.8% Tris-acetate-EDTA (TAE)-agarose gel, and alkali blotted onto a nylon membrane (Porablot NY Amp; Macherey and Nagel, Düren, Germany). The porcine A3Z3 probe (1,038 bp, ranging from nt 16407 to nt 17444 bp in poA3 gene locus, corresponding to the 5′ part of exon 3 to the stop codon in exon 5 of the poA3Z3 gene sequence) was generated by PCR with genomic DNA of MAX-T as a template using the primers for_EI_5 (5′-AGCGACATGCAGAAATTCGTTTTATTGACAAG-3′) and 1r_Ss3F (5′-TCATCTCGAGTCACTTCTTGATGATGAAGGTC-3′). PCR product was cloned into pGEM-T Easy (Promega GmbH), sequenced, and cut out with NotI. This probe was labeled with a Megaprime DNA labeling Kit (GE Healthcare, Munich, Germany) and 1.85 MBq [α-32P]dCTP (Hartmann Analytic, Braunschweig, Germany). The membrane was hybridized with 2 × 107 to 4 × 107 counts per minute (cpm) of the probe overnight at 65°C. The filter was washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at ambient temperature and 0.2× SSC–0.1% SDS at 65°C. Autoradiography was done using Hyperfilm MP (Amersham Biosciences, Freiburg, Germany).
Immunoblot analysis.
293T cells were cotransfected with plasmids encoding PERV-EGFP, VSV-G, and A3-HA. At 2 dpt cell lysates were prepared using lysis buffer (100 mM NaCl, 20 mM Tris [pH 7.5], 10 mM sodium deoxycholate, 1% Triton X-100) supplemented with a protease inhibitor mix. Protein quantification was carried out with a Bradford assay (1× Bradford dye reagent; Bio-Rad, Munich, Germany). Virions were concentrated by two ultracentrifugation steps of the filtered culture supernatants through a 20% sucrose cushion at 28,000 rpm for 1.5 h in an SW28 rotor (Beckman Coulter, Krefeld, Germany). Viruses were diluted in phosphate-buffered saline (PBS), and RT activity was measured. Twenty micrograms of each cell lysate and 400 mU of RT of viral isolates were separated by SDS-PAGE in a 5% stacking gel and a 12.5% resolving gel and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore GmbH, Schwalbach, Germany). Membranes were blocked (0.1% Tween in Tris-buffered saline [TBS-T] and 10% skim milk powder) and then incubated with either a monoclonal mouse anti-HA antibody (clone HA-7; 1:4,000; Sigma Aldrich) to detect A3 expression, mouse anti-V5 antibody (1:4,000; MCA1360; Serotec) for huAID, huA1, and huA2 expression, monoclonal mouse anti-beta-actin antibody (clone AC-15; 1:5,000; Biozol, Eching, Germany), or polyclonal rabbit anti-PERV p30 antiserum (1:3,000; Eurogentec, Köln, Germany) (23) against the capsid protein of the PERV. As secondary antibodies, horseradish peroxidase-conjugated rabbit anti-mouse or donkey anti-rabbit antibodies (each, 1:10,000; Dianova, Hamburg, Germany) were used. Detection was done using ECL chemiluminescence reagents (GE Healthcare) and hyperfilm ECL (GE Healthcare).
RH mapping.
The INRA-University of Minnesota porcine radiation hybrid panel (35, 62, 107) was received from INRA, Laboratoire de Genetique Cellulaire, BP27, Castanet-Tolosan, France. The DNAs of 118 hybrid clones of the radiation hybrid (RH) panel were used for two different PCRs. The first PCR was performed with the primers f_Intron1_1 (5′-CTGGAAAGTCTTCCAGCAGGGAATTC-3′) and r_Exon2_2 (5′-GATGCCTTGGAAGAAACAGTTCTTTCC-3′) flanking a region of 276 bp including the main part of exon 2 and the 3′ part of intron 1 of the porcine A3Z2 gene. With the second primer pair f_Exon5_1 (5′-TCTTGGCGCCCTACTATCTGAGGAAG-3′) and r_Intron5_1 (5′-TGTTCTCAGACGCTGAATGGAGGAGC-3′), a 479-bp fragment was amplified containing the 3′ part of exon 2 and the 5′ part of intron 2 of the porcine A3Z3 gene. PCRs were run for 35 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 45 s. Positive signals were scored, and the results were statistically analyzed using the IMpRH mapping tool (http://www.toulouse.inra.fr/).
Isolation of a porcine BAC clone.
For the isolation of a porcine bacterial artificial chromosome (BAC) clone containing the poA3 gene locus, the CHORI-242 porcine (Sus scrofa) BAC library (http://bacpac.chori.org/porcine242.htm) (73) was screened with the help of a high-density hybridization (HDH) filter set (number 007344), where a part of the library was overlaid on a grid of 22- by 22-cm positively charged nylon filters. The filters were hybridized with the [α-32P]dCTP-radiolabeled porcine A3Z3 probe (see “Southern blot hybridization”) and further treated according to Southern blotting conditions. One positive BAC clone (clone CH242-330G8) was chosen and used for a colony PCR using the specific primers f_Intron1_1 and r_Exon2_2 (see “RH mapping”).
Chromosome preparation and FISH.
Porcine metaphase spreads on GTG-banded chromosomes were prepared using phytohemagglutinin (PHA)-stimulated porcine blood lymphocytes. Slides were prepared using standard cytogenetic techniques. Prior to fluorescence in situ hybridization (FISH), the well-banded spread metaphase chromosomes were photographed using a highly sensitive charge-coupled device (CCD) camera and IPLab, version 2.2.3 (Scanalytics Inc., Reutlingen, Germany). Identification of chromosomes followed (31). The BAC clone containing the porcine A3 gene locus was labeled with digoxigenin by nick translation using a Nick Translation Mix (Roche Applied Science). FISH was performed using 750 ng of digoxigenin-labeled BAC DNA. In this experiment 12 μg of porcine Cot-1 DNA and 10 μg of salmon sperm were used as competitors. After hybridization overnight, signal detection was performed using a digoxigenin-fluorescein isothiocyanate (FITC) detection kit (Qbiogene, Heidelberg, Germany). The chromosomes were counterstained with 4′,6′- diamidino-2-phenylindole (DAPI) and embedded in propidium iodide-antifade solution. Thirty photographed metaphases were reexamined after hybridization with a Zeiss Axioplan 2 microscope (Zeiss, Jena, Germany) equipped for fluorescence.
Sequencing of viral transcripts.
A total of 1 × 106 293T cells were transduced with RT-normalized (60 mU) and DNase I (2,000 U/ml; addition of 1/20 volume; NEB)-treated PERV-EGFP (VSV-G). The pseudotyped viruses were produced in the absence [usage of pcDNA3.1/Zeo(+)] or presence of different A3 proteins (huA3B, huA3F, huA3G, muA3, poA3Z2-Z3, poA3Z2, poA3Z3, and poA3Z2-Z3 SVA) as described previously. After 10 h of incubation, cells were washed five times with PBS, and DNA was isolated using a DNeasy Blood and Tissue Kit (Qiagen). The EGFP sequence of the reporter virus genome was amplified using Taq DNA polymerase (NEB) and the primers KpnI_EGFP_for (5′-CAGTGAGGTACCATGGTGAGCAAGGGCGAGGAG-3′) and GFP_PacI_rev (5′-TCTCTGTTAATTAATTACTTGTACAGCTCGTCCATGCC-3′) with 35 cycles of 94°C for 30 s, 62°C for 45 s, and 72°C for 1 min. The PCR products were cloned into pGEM-T Easy (Promega). For each approach the nucleotide sequences of 10 independent clones were analyzed.
Biostatistical analyses.
To determine the nucleotide preferences of the A3 proteins, the distribution of the nucleotides in the neighborhood (±5 bp) of the C-to-T mutations compared to the nonedited EGFP sequence within the minus-strand DNA was analyzed. Evaluation was performed for each surrounding position separately with a chi-square test (significance level α of 0.05; P values adjusted according to the Bonferroni-Holm method for multiple comparisons). To separate preferred from disfavored surroundings, we further estimated the frequency of C-to-T mutations occurring at a certain NNC (mutated residue underlined) trinucleotide context by taking also the number of examined clones for each A3 protein into consideration. This accounts for the fact that mutations appear in only some of the 10 clones at a certain position (few “hot spots,” with a maximum of 6/10), and actually no mutation occurred in many identical NNC contexts at different positions of the clone. The favored surroundings for mutations were displayed graphically with 95% confidence limits based on binomial distribution (exact Clopper-Pearson confidence intervals). The statistical analysis was performed with the software SAS/STAT (version 9.2; SAS Institute Inc., Cary, NC).
Phylogenetic analysis of the porcine A3 gene locus.
To infer the phylogenetic relationships for the porcine A3 locus, the largest part of the poA3Z3 gene was amplified using genomic DNA of different pig species: German landrace (source, PK15 and PBMCs from two unrelated individuals), mini-pig (source, MAX-T), wild pig (source, PBMCs from three unrelated individuals), and Yucatan pig (source, PBMCs from two unrelated individuals). Additionally, genomic DNA of two relatives to pig were included: Pecari tajacu (source, PBMCs of two breed individuals) and Camelus dromedarius (source, cell line Dubca). PCRs were performed in the presence of 0.5 M betaine (Sigma Aldrich), genomic DNAs (100 ng each), and KOD XL polymerase (Merck Chemicals Ltd.). Two independent PCRs were run for 35 cycles of 94°C for 30 s, 62°C for 5 s, and 74°C for 1 min with the primer pair for_EI_5 (5′-AGCGACATGCAGAAATTCGTTTTATTGACAAG-3′) and 1r_Ss3F (5′-TCATCTCGAGTCACTTCTTGATGATGAAGGTC-3′), amplifying a genomic region of 1,038 nt on average (in the case of MAX-T, genomic region from nt 16407 to 17444) and the primers f_Intron4_C2 (5′-TCCTGGCCAACAAAGAAGGATGAGAC-3′) and r_Exon6_C (5′-TCTTGTCAATAAAACGAATTTCTGCATGTCGC-3′) amplifying a sequence of an average size of 1,270 bp (in the case of MAX-T, genomic region from nt 15170 to 16439). From each PCR product at least five clones were sequenced. Sequences were aligned to establish a consensus sequence, and both consensus sequences for each species were concatenated via overlapping sequence regions. These sequences were used to reconstruct the phylogenetic tree. A3Z3 sequences in the databases from mammalian species were retrieved for analysis. Within the Laurasiatheria, the following were used: Sus scrofa, A3Z2-Z3 C terminus (NM_001097446); Bos taurus, A3Z2-Z3 C terminus (NM_001077845); Ovis aries, A3Z2-Z3 C terminus (NM_001093784); Equus caballus, A3Z3 (FJ532289); Canis lupus, A3Z3 (XM_538369); and Felis silvestris, A3Z3 (NM_001112710). Within the Euarchontoglires the following were used: Rattus norvegicus, A3Z2-Z3 C terminus (NP_001028875); Mus musculus, A3Z2-Z3 C terminus (ACH89410); Homo sapiens, A3H (NP_861438); Pan troglodytes, A3H (XP_525599); Gorilla gorilla, A3H (ABD72580); Pongo pygmaeus, A3H (Q1WBT4); and Macaca mulatta, A3H (XP_001096739). Additionally, the sequences generated from Camelus dromedarius, Pecari tajacu, and the different races from Sus scrofa were also included. The sequences were aligned at the amino acid level with MUSCLE (20, 21), visualized for manual correction, and back-translated to the nucleotide level using PAL2NAL (http://www.bork.embl.de/pal2nal) (97). Maximum-likelihood (ML) phylogenetic analysis was performed with RAxML HPC (92, 93) using the GTR+Γ4 (general time-reversible with gamma-distributed rate variation across sites) model of evolution and the CAT (category amino acid site-heterogeneous mixture) approximation of rate heterogeneity (91), introducing three partitions that corresponded to each of the codon positions. Two independent runs of 5,000 bootstraps each were performed, and for each one, the final tree topology was optimized without resorting to the CAT approximation. The best of the two final trees was chosen (−ln = 5191.168; respective optimized values for the alpha parameter of the gamma function for each of the three codon positions were 1.058, 1.077, and 1.984), but there were no significant differences among the corresponding likelihood values using a Shimodaira-Hasegawa (SH) test (90). Bootstrap support values for the nodes were obtained from the combined set of 10,000 trees. Bayesian phylogenetic analysis was performed with BEAST, version 1.4.8 (http://beast.bio.ed.ac.uk) (19), with the GTR+Γ4 model of evolution, and for both strict clock and uncorrelated log normal relaxed clock, introducing three partitions that corresponded to each of the codon positions and unlinking parameters across codon positions. Two independent chains of 50 million steps were calculated and analyzed with a burn-in of 10 million steps. Compatibility of both chains was assessed by calculating the corresponding Bayes factor (96), and both chains were combined into one. The combined chains from the relaxed and the strict clock were also compared by calculating the Bayes factor on the corresponding likelihoods. There was only marginally better support for the relaxed clock calculations (log Bayes factor, 1.177). The Bayesian maximum clade credibility tree was built on the relaxed clock combined chain, and its likelihood was calculated under ML framework and compared with the best RAxML tree. The topologies of the Bayesian maximum clade credibility tree and that of the best tree under ML were identical. Fine phylogenetic analysis of sequences generated here from Camelidae, Tayassuidae, and Suidae was performed concatenating both coding and noncoding regions. Four partitions were generated, one for each of the three codon positions, and a fourth one for the concatenated introns, and ML and Bayesian analyses were performed as described above. There was no difference in the likelihood values of the trees obtained under strict clock and under relaxed clock premises (log Bayes factor, 0.034). The best ML tree (−ln = 5820.605; respective optimized values for the alpha parameter of the gamma function for each of the three codon positions of 1.958, 0.312 and 3.856; alpha parameter for the concatenated introns of 1.490) had an identical topology to that of the maximum clade credibility tree of the Bayesian analyses. The reference cladogram for the animal species included in the taxon sample was pruned out from a mammalian tree constructed according to supertree methodology (5), using TREEPRUNER (http://www.uni-oldenburg.de/molekularesystematik/33997.html). Figures were generated with FigTree, version 1.1.2 (http://tree.bio.ed.ac.uk/software/figtree).
Nucleotide sequence accession numbers.
Nucleotide sequences for the porcine cDNA sequences and for the poA3 gene locus sequence have been deposited in the GenBank database as follows (accession number): porcine A3Z2-Z3 gene (EU871587), poAZ2-Z3 mRNA (EU871586), poA3Z2-Z3 SVA mRNA (FJ716802), poA3Z2 mRNA (FJ716801) and poA3Z3 mRNA (HM347451). For the amplified main part of the A3Z3 gene sequence (ranging from the 3′ end of intron 1 to the stop codon in exon 5) used to develop a phylogenetic tree the following GenBank files were also generated (accession number): Sus scrofa_german landrace, A3Z3 (FJ716803); Sus scrofa_mini pig, A3Z3 (FJ716804); Sus scrofa_Yucatan pig, A3Z3 (FJ716805); Sus scrofa_wild pig, A3Z3 (FJ716806); Pecari tajacu, A3Z3 (FJ716807); and Camelus dromedarius, A3Z3 (FJ716808).
RESULTS
The porcine A3 gene locus encodes two genes.
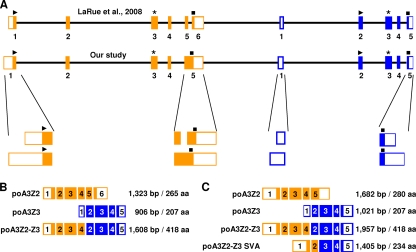
In order to identify the sequence of the porcine A3 gene locus, genomic DNA of the porcine kidney cell line MAX-T was used for PCR-based cloning. Based on the cDNA sequence of the porcine A3Z2-Z3 (13), we looked for possible exon-exon junctions (BLAST search [http://genome.ucsc.edu/]) (95, 102) and designed primers (Table 3) to amplify chromosomal DNA sequences. Using this approach, we identified the sequence of the porcine A3 exons and introns containing the region from the start codons to the stop codons without the untranslated regions (UTRs). With regard to data published by LaRue and coworkers (48), the identified gene locus consists of two single Z-domain genes, Z2 and Z3, and spans over 17 kb (Fig. 1A). Both genes have five exons, and each gene encodes the zinc-coordinating domain in exon 3. All exon/intron boundaries conform to the GT/AG rule (95, 102).
Fig. 1.
Porcine A3 gene locus and its transcriptional potential. (A) The gene locus consists of the two single domains, Z2 (orange) and Z3 (blue). The identified exon-intron composition was compared to published data by LaRue et al. (48). Regions that differ between the two studies are enlarged. The exons are shown as filled boxes, whereas the untranslated regions are indicated as open boxes. Black lines indicate introns, and stars mark the position of the zinc-coordinating domains. The start codon is plotted as a triangle, and the stop codon is shown as a square. The scheme is depicted true to scale. All mRNA transcripts identified by LaRue and coworkers (B) and by this study (C) are listed and include the nucleotide lengths in base pairs (bp) of the mRNA as well as the lengths of amino acid (aa) sequences.
Complex splicing of porcine A3 genes results in four mRNAs.
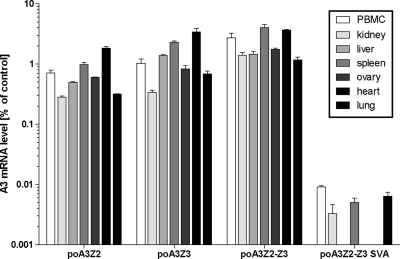
To identify porcine A3 transcripts, total RNA from the porcine cell line MAX-T, porcine activated peripheral blood mononuclear cells (PBMCs), and the porcine T-cell line L45 were used for RT-PCR. From these sources we were able to clone four different porcine cDNAs (poA3Z2, poA3Z3, poA3Z2-Z3, and poA3Z2-Z3 SVA) (Fig. 1C), whereas the last-mentioned cDNA was not found in the porcine cell lines. Using quantitative RT-PCR on primary porcine tissues (PBMCs, kidney, liver, spleen, ovary, heart, and lung, the detected A3 expression levels were between 0.5 and 4% of the beta-actin mRNA control levels for poA3Z2, poA3Z3, and poA3Z2-Z3 in all tissues, whereas the expression levels for poA3Z2-Z3 SVA were very low (maximum, 0.01% of control) in PBMCs, kidney, spleen, and lung (Fig. 2). No expression of SVA was observed in liver, ovary, and heart.
Fig. 2.
mRNA expression of the porcine A3 transcripts in porcine tissues detected by quantitative PCR. All mRNA expression levels are normalized to beta-actin mRNA expression. Mean values and standard deviations for each triplicate are depicted.
The cDNAs for poA3Z2 (open reading frame [ORF] length of 843 bp) and poA3Z3 (ORF of 624 bp) encode proteins consisting of 280 aa and 207 aa, respectively. The read-through transcript poA3Z2-Z3 (ORF of 1,257 bp) that was originally described by Conticello et al. (13) consists of exons 1 to 4 of poA3Z2 and exons 2 to 5 of poA3Z3 and codes for a protein of 418 aa (Fig. 1C). Thus, sequences of exon 5 of poA3Z2 and exon 1 of poA3Z3 are not part of the mRNA of poA3Z2-Z3. Exon 5 of poA3Z2, which possesses an additional stop codon, is used only for the poA3Z2 transcript. The additional, shorter read-through transcript, poA3Z2-Z3 SVA (ORF of 705 bp, 234 aa), consists of exons 1 and 2 of poA3Z2 and exon 3 to 5 of poA3Z3 (Fig. 1C).
To characterize the untranslated regions (UTRs) of poA3Z2 and poA3Z3 mRNAs, we applied 5′ and 3′ rapid amplification of cDNA ends (RACE) using mRNA of MAX-T as a template (Fig. 1C). The 5′ UTR of poA3Z2 is located in exon 1, which has a total maximum length of 553 bp. However, we also detected smaller RACE products in this reaction (data not shown). Exon 1 contains 473 bp of presumed untranslated sequence, followed by the start codon and 80 bp of coding region. The 3′ UTR of poA3Z2 is located in exon 5. This exon possesses a length of 583 bp (nt 1100 to 1682); the stop codon is at nt 1314 to 1316 and is terminated by the polyadenylation signal ACTAAA, a rare hexanucleotide motif found only in 0.6% of all mRNAs (4). The 5′ RACE of the poA3Z3 mRNA identified a 5′ UTR of 170 bp and various shorter lengths. The start codon for the poA3Z3 mRNA is at nt 171 to 173 in exon 2. Thus, exon 1 of poA3Z3 consists completely of a noncoding region. Exon 5 of poA3Z3 has a length of 317 bp, reaching from nt 705 to 1021. The stop codon is localized at nt 792 to 794, followed by the 3′ UTR of 227 bp. At the 3′ end of the noncoding sequence, the hexanucleotide ATTAAA was identified as the presumed polyadenylation signal (4). Including the UTRs, the four mRNAs have the following lengths: poA3Z2-Z3, 1,957 bp; poA3Z2, 1,682 bp; poA3Z3, 1,021 bp; and poA3Z2-Z3 SVA, 1,405 bp (Fig. 1C).
The porcine A3 genes are located on chromosome 5.
To verify the copy number of the porcine A3 gene locus, Southern blot analyses were performed (Fig. 3). To this end, genomic DNA of different species (human, pig, camel, sheep, and cow) was digested with two restriction endonucleases (EcoRI or BglII) and analyzed. For hybridization we used a C-terminal probe of 1,038 bp reaching from exon 3 to the stop codon in exon 5 of poA3Z3 (Fig. 3A). In the autoradiography, both lanes of porcine DNA had one detectable signal with sizes of 15 kb (EcoRI restriction) and 5 kb (BglII restriction) (Fig. 3B, lane 3 plus lane 8). Since only one signal was obtained, we concluded that the porcine A3Z3 gene locus is unique and that probably the complete poA3 locus is unique too. However, probes binding in the poA3Z2 gene did not work under our experimental conditions, and we cannot formally rule out that the swine genome contains more than one A3Z2 gene.
Fig. 3.
Southern blotting to determine the number of gene copies of the A3Z3 gene in the porcine genome. (A) Hybridization site of the probe within the A3 gene locus as well as the nucleotide positions of the restriction sites used for digestion of the genomic DNA. (B) Genomic DNA from different species was digested with the restriction enzymes EcoRI (lanes 1 to 5) and BglII (lanes 6 to 10). The membrane was hybridized with the radiolabeled C-terminal porcine A3Z3 probe. The λ-HindIII-marker served as a size standard. Lanes 1 and 10, human; lanes 2 and 9, camel; lanes 3 and 8, pig; lanes 4 and 7, sheep; lanes 5 and 6, cow.
The porcine A3Z3 probe also generated a specific signal using camel DNA digested with EcoRI (Fig. 3B, lane 2). Thus, it is likely that the distantly related camel (3, 70) carries an A3Z3 gene that has sequence homology to porcine A3Z3. No signal was found when genomic camel DNA was restricted with the enzyme BglII. A subsequent sequence analysis of the identified camel A3Z3 gene sequence (FJ716808) showed that there is a BglII restriction site within the binding region of the probe (data not shown).
In order to determine the chromosomal localization of the porcine A3 genes, genomic DNA of the INRA-University of Minnesota porcine radiation hybrid (IMpRH) panel of cell lines (35, 107) was used in combination with PCR primers. Two sequences were amplified, with one fragment located in the 5′ part of the gene poA3Z2 (crossover between intron 1 and exon 2) and the second fragment located in the 5′ part of poA3Z3 gene (crossover between exon 2 and intron 2). With both probes the two-point analysis showed a significant linkage of the porcine A3 genes with the microsatellite marker SW1482 and the gene ACO2 (logarithm of the odds [LOD] of >5) (Table 4), where both markers build a linkage group on the short arm of the porcine chromosome 5 (SSC5p) near the telomere (35, 82). We conclude that the porcine A3 genes are located on the short arm of chromosome 5. Comparing maps of human and pig chromosomes (29, 83; http://www.toulouse.inra.fr/lgc/pig/compare/compare.htm), we were able to underpin our results: the genomic region of Sus scrofa chromosome 5p14-pter (where ter is telomere) is syntenic to Homo sapiens chromosome 22q12-qter. The human A3 gene cluster is located on chromosome 22q13.1 (http://www.genome.ucsc.edu/).
Table 4.
Results of the IMpRH panel
| RH panel | Primer paira | Retention fraction (%)b | Marker | Chromosome | Distance (cR)c | LODd |
|---|---|---|---|---|---|---|
| I | f_Intron1_1 | 33 | ACO2 | 5p | 66 | 5.88 |
| r_Exon2_2 | SW1482 | 5p | 92 | 3.44 | ||
| II | f_Exon5_1 | 37 | ACO2 | 5p | 51 | 8.23 |
| r_Intron5_1 | SW1482 | 5p | 65 | 5.98 |
Two different primer pairs were used to map the porcine A3 genes in the INRA-University of Minnesota porcine radiation hybrid panel.
The retention frequency indicates the fraction of clones showing a positive signal in the PCR.
The distance gives the chromosomal interspace between the markers and the porcine A3 genes. cR, centiray.
An LOD (logarithm of the odds) score of >5 is an indicator for a gene linkage.
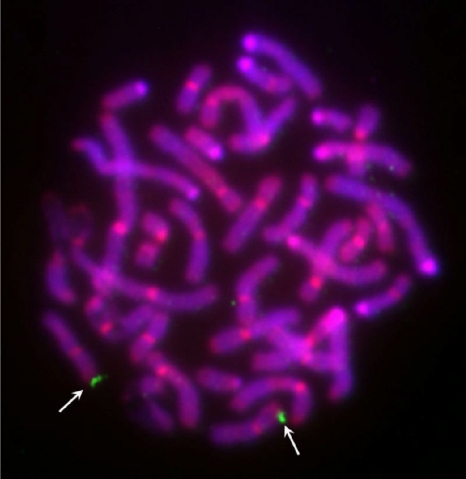
To visualize and further confirm the chromosomal location, fluorescence in situ hybridization (FISH) analysis was performed. For this purpose, a digoxigenin-labeled BAC clone harboring the complete porcine A3 locus was hybridized to porcine metaphase chromosomes. The A3 probe localized only to chromosome 5p1.5, nicely confirming the results obtained by the RH panel (Fig. 4).
Fig. 4.
Localization of the porcine A3 gene locus by fluorescence in situ hybridization. The digoxigenin-labeled BAC clone CH242-330G8 harboring the complete porcine A3 gene sequence was hybridized to GTG-banded metaphase chromosomes. Signals (marked with white arrows) are localized on the short arm of pig chromosome 5 near the telomere (SSC5p1.5). The chromosomes were counterstained with DAPI and propidium iodide.
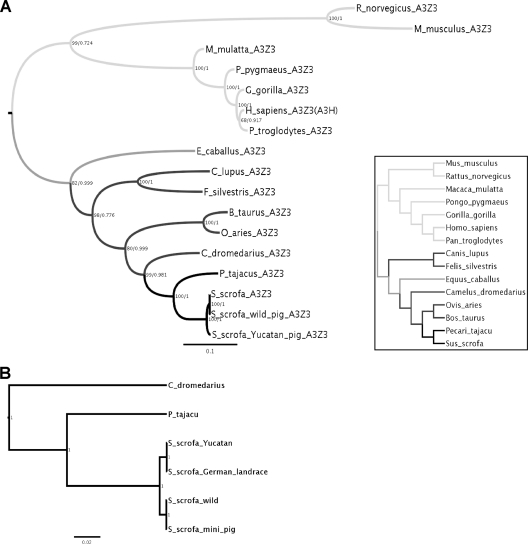
The evolution of the porcine A3Z3 gene.
To get more insight into the evolution of the porcine A3 gene locus, we amplified the main section of the A3Z3 gene (3′ end of intron 1 to stop codon in exon 5) (Fig. 1A) from different pig breeds (Sus scrofa) and the nearest relatives to pigs, peccary (Pecari tajacu) and camel (Camelus dromedarius). A comparison of the obtained sequences with the orthologous C termini of the A3 genes acquired from different species within Laurasiatheria and Euarchontoglires showed that the evolution of A3Z3 reflects the presently accepted evolutionary history of the corresponding species (Fig. 5A and inset). Topologies inferred under ML and under Bayesian approaches were identical, and there was no increased support for a relaxed clock compared to a strict clock. Thus, the fusion of the proto-A3Z3 gene with the proto-A3Z2 gene in these taxa did not modify the evolutionary pattern of the A3Z3 moiety. By analyzing the sequence data from camel, peccary, and pig breeds, we identified two different sequences within A3Z3 in Sus scrofa, which could correspond to two ancestral alleles (Fig. 5B). We interpret the minor additional differences between Yucatan and German races as well as between wild pig and “mini-pig” sequences, which are present exclusively in the noncoding region, as polymorphisms of these two alleles. The evolutionary relationships of the porcine A3Z2 gene were not analyzed because our PCRs on peccary and camel DNA failed to produce an amplification product.
Fig. 5.
Best known maximum-likelihood trees of the A3Z3 genes encoded in Laurasiatheria (dark gray) and Euarchontoglires (light gray) based on amino acid sequences (A) and in Camelidae, Tayassuidae, and Suidae based on nucleotide sequences (B). Values in nodes reflect maximum-likelihood bootstrap support values/Bayesian posterior probability values. The scale bar is given in substitutions per site. The inset in panel A shows the currently accepted phylogenetic relationships among the species included in the A3Z3 trees (5).
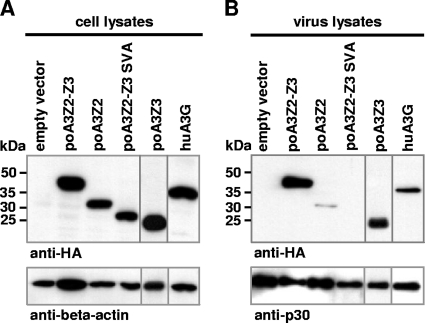
The porcine A3 proteins are packaged into PERV particles.
To find out whether porcine A3 proteins are encapsidated in PERV particles, PERV-EGFP reporter particles were generated in the presence or absence of porcine A3s and huA3G. As shown in Fig. 6A, all A3 proteins were expressed to equal amounts in the virus-producing cells but were differently incorporated into viral particles (Fig. 6B). huA3G, poA3Z3, and poA3Z2-Z3 were found to be efficiently encapsidated; in contrast, poA3Z2 was only weakly packaged in PERV particles (Fig. 6B). The virus particles did not encapsidate poA3Z2-Z3 SVA although the protein was expressed in the virus-producing cells (Fig. 6A). These results suggest that PERV does not harbor a mechanism to prevent the encapsidation of its species-own cytidine deaminases poA3Z2-Z3 and poA3Z3. It is unclear whether poA3Z2 and poA3Z2-Z3 SVA proteins are weakly packaged or not packaged because they lack active encapsidation domains or because PERV excludes these proteins by unknown means.
Fig. 6.
Packaging of the A3 proteins in PERV particles. (A) Immunoblot of 293T cells after transfection to determine A3 expression. (B) Immunoblot of the produced PERV particles to determine the packaging of the A3 proteins. The cytidine deaminases were detected with an anti-HA antibody. As a loading control either an anti-beta-actin antibody (A) or an anti-PERV p30 antibody (B) was used.
Human and porcine A3 proteins restrict PERV infection in a dose-dependent manner and inhibit spreading replication.
To investigate the effect of porcine A3 proteins on viral infectivity, VSV-G pseudotyped PERV-EGFP particles were generated in 293T cells in the absence or presence of A3 expression plasmids (muA3; huA3A, -B, -C, -F, -G, -H; and poA3Z2, -Z3, -Z2-Z3, and -Z2-Z3 SVA). Additionally, expression plasmids for huAID, huA1, and huA2 were included in this study. Because the literature regarding the A3 sensitivity of the related gammaretrovirus MuLV is conflicting (7, 9, 41, 45, 46, 56), we added MuLV reporter viruses to the analysis. Viral supernatants were harvested at 2 dpt, and the virus-producing cells were lysed to monitor intracellular A3 expression.
Altogether, the inhibitory effects of the tested A3 proteins were similar against PERV and MuLV (Fig. 7A and B). Porcine cytidine deaminases inhibited PERV replication in an effective manner by a factor of 65 for poA3Z2-Z3, by a factor of 38 for poA3Z2, and by a factor of 17 for poA3Z3 (Fig. 7A). PERV was significantly restricted by huA3B, huA3G, and muA3 by factors of 32, 38, and 19, respectively (Fig. 7A). Human AID, A1, and A2 were inactive against PERV or MuLV (Fig. 7A and B), while poA3Z2-Z3 SVA and huA3A, -C, -F, and -H reduced the infectivity of PERV moderately by a factor of 2 to 9 (Fig. 7A). The indicated significance levels do not always conform to the factor of reduction because of high values of the standard deviation of some experiments. The pattern of inhibition of PERV by human A3s was similarly found in the experiments using the MuLV reporter virus (compare Fig. 7A and B). The corresponding immunoblots show that almost all A3s were expressed (Fig. 7C). Only huA3H showed low expression. Although the protein levels for poA3Z2 were lower than those for poA3Z2-Z3, poA3Z2 effectively restricted PERV and MuLV replication. All porcine A3s inhibited MuLV, and of these poA3Z2-Z3 SVA again showed the weakest restriction activity. MuLV was resistant to its species-own muA3 (Fig. 7B), an important difference from PERV, which was sensitive to porcine A3s. We conclude that most porcine and human A3s and muA3 are PERV inhibitors.
Fig. 7.
Antiviral activity of the A3 proteins against PERV and MuLV. 293T cells were infected with VSV-G pseudotyped PERV (A) or VSV-G pseudotyped MuLV (B) that was produced in the presence or absence of the indicated A3 proteins. For each approach, two independent experiments were done, and the results were compared to get a standard deviation. Levels of significance are indicated: *, P < 0.05; **, P < 0.01; ***, P < 0.0001. (C) Immunoblotting to determine the intracellular A3 expression. The cytidine deaminases were detected with an anti-HA or an anti-V5 antibody. An anti-beta-actin antibody served as a loading control.
To determine the dose-dependent effect of A3 proteins on PERV infectivity, cotransfections with different amounts (0.2, 0.4, 0.6, 0.8, and 1 μg) of the poA3-encoding plasmids were performed. As a positive control huA3G was included. The results in Fig. 8 A show that even low plasmid concentrations below 1 μg seem to be enough to see a significant reduction in PERV infectivity. But also in this instance the indicated significance levels for huA3G do not conform to the factor of reduction because of the high values of the standard deviation. For example, an antiretroviral effect was already detectable with the lowest plasmid concentration (0.2 μg): huA3G and poA3Z2, -Z3, and -Z2-Z3 showed inhibitory effect factors of 5, 19, 7, and 6, respectively. As seen before, for poA3Z2-Z3 SVA no restriction was observed (Fig. 8A). Interestingly, poA3Z3 showed no dose-dependent increase in the antiviral activity, indicating an early saturation at an uncharacterized step. The A3 protein expression in the virus-producing cells was confirmed by immunoblot analyses (Fig. 8B).
Fig. 8.
Porcine A3 proteins inhibit PERV infectivity in a dose-dependent manner. (A) VSV-G pseudotyped PERV particles were produced in 293T cells transfected with increasing amounts of huA3G or porcine A3 expression plasmids. The concentration of the A3 expression vector plasmid transfected is shown on the x axis. Infectivity of the viruses was determined on 293T cells by intracellular EGFP fluorescence. Levels of significance are indicated: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) Immunoblots of transfected 293T cells. A3 protein expression was detected with an anti-HA antibody. An anti-beta-actin antibody served as a loading control.
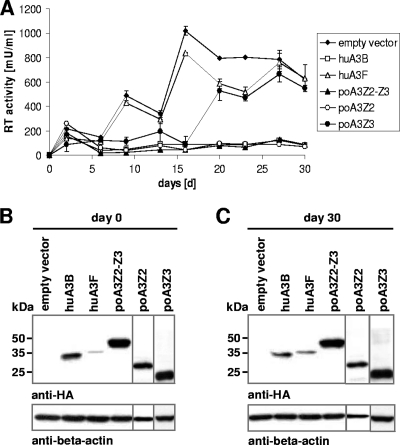
As our previous experiments were done with VSV-G pseudotyped single-round reporter viruses, we asked whether A3s could also inhibit the spreading replication of wild-type PERV during a longer observation period. Therefore, we generated different cell lines based on 293T cells that stably expressed poA3Z2-Z3, poA3Z2, poA3Z3, huA3B, and huA3F. To start PERV infection, cells were transfected with a plasmid encoding a replication-competent PERV-B (pBS_Not[PERV-B(33)ATG]) (14). PERV replication was followed for a period of 30 days and monitored by measuring the RT activity in the supernatants of the cell cultures (Fig. 9A).
Fig. 9.
Long-term infectivity of PERV in the presence of A3 proteins. (A) Stable A3-expressing 293T cells were transfected with a plasmid encoding a replication-competent PERV-B clone. Viral supernatants were harvested every 3 or 4 days, and RT activity was measured. Stable A3 expression was checked via immunoblotting at day 0 (B) and day 30 (C) of the experiment. A3 expression was detected with an anti-HA antibody. An anti-beta-actin antibody served as a loading control.
We found that huA3B, poA3Z2-Z3, and poA3Z2 suppressed PERV replication during the whole observation period, whereas huA3F and, surprisingly, poA3Z3 were not able to restrict PERV replication (Fig. 9A). The reason why huA3F and poA3Z3 were able to inhibit PERV in single-round assays (Fig. 7A) but failed to show this restrictive activity in spreading PERV infection is unclear, but it is in line with similar observations on HIV-1 and huA3F (63). Together, our results demonstrate that PERV cannot replicate over several rounds in human cells expressing poA3Z2, poA3Z2-Z3, or huA3B. Studies with a longer observation period would be required to estimate whether PERV would be able to develop adaptive mutations in vivo that could circumvent the A3 restriction (63). To confirm that the A3 proteins were expressed during the whole experiment, cells of the cultures were harvested at day 0 and day 30 and analyzed by immunoblotting (Fig. 9B and C).
PERV restriction is attributed to cytidine deamination.
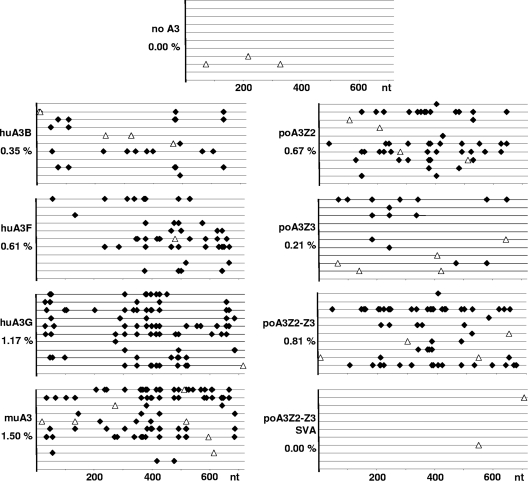
To search for an antiviral mechanism of the porcine A3 proteins, we analyzed viral cDNAs for mutations. Genomic DNA from 293T cells was prepared at 10 h postinfection and used for PCR, amplifying EGFP of the viral reporter genome. In addition to the porcine A3s (A3Z2, -Z3, -Z2-Z3, and -Z2-Z3 SVA), we included huA3B, -F, and -G and muA3 in these experiments.
As expected, murine and human A3 proteins induced G-to-A hypermutations in the PERV genome effectively: huA3B mutated 0.35%, huA3F mutated 0.61%, huA3G mutated 1.17%, and muA3 mutated 1.50% of all nucleotides (Fig. 10). Interestingly, we could not detect any G-to-A mutations in the viral cDNAs in experiments using huA3A (data not shown) although it reduced PERV transduction by a factor of 5 (Fig. 7A). In contrast to the report from Jonsson et al. (43), G-to-A hypermutations were also detected for poA3Z2-Z3 with 0.81%, poA3Z2 with 0.67%, and poA3Z3 with 0.21% of all nucleotides. Cytidine deamination activity was undetectable for the weakly antiviral poA3Z2-Z3 SVA (Fig. 7A and 10). Based on these results, we conclude that the A3 proteins mainly restrict PERV replication by hypermutating the viral genome.
Fig. 10.
A3 proteins induce G-to-A hypermutation in the PERV genome. 293T cells were transduced with pseudotyped PERV-EGFP/VSV-G particles produced in the absence or presence of huA3G, huA3F, huA3B, muA3, poA3Z2, poA3Z3, poA3Z2-Z3, and poA3Z2-Z3 SVA. At 10 h posttransduction genomic DNA was prepared and used for PCR, amplifying the 720-bp-long EGFP sequence from the PERV-EGFP genome. Ten clones were sequenced for one approach. One line represents one sequenced clone. Each G-to-A mutation is plotted (black diamond), and the mutation frequency for each A3 protein is indicated (percent G-to-A mutations compared to 100 nucleotides). Open triangle, non-G-to-A mutation.
Sequence preferences for porcine A3-mediated hypermutation.
To identify the preferred nucleotide context for poA3 cytidine deamination, we analyzed the nucleotide context five bases downstream and five bases upstream of all C-to-T mutation sites and performed a chi-square test with Bonferroni-Holm correction for each nucleotide position. From this analysis, we obtained the nucleotide distribution around the mutation sites (Fig. 11) and identified a trinucleotide preference for all A3 proteins tested (Fig. 12). huA3G favored 5′ T/CCC (mutation site is underlined), whereas the muA3 preferred 5′ TT/CC, confirming the results previously published by others (2, 41, 108, 109). For huA3F and likely for huA3B (Fig. 12), a preference for 5′ T/ATC was seen, in agreement with the report of Armitage et al. (2). Both poA3Z2-Z3 and poA3Z2 showed a nucleotide preference for 5′ TGC (42), whereas poA3Z3 seemed to favor the trinucleotide 5′ CAC (Fig. 12). These results were statistically confirmed by reanalyzing the data regarding the context of all C nucleotides, whether they were mutated or not. The diagrams of all favored trinucleotides from the most to the least preferred sequence for each A3 protein are shown in Fig. 12. The observed preferences confirmed the result of the first analysis (Fig. 11). In the experiments using huA3B and poA3Z3, no significant preference for the −2 position could be shown probably due to the small data set for these two A3 proteins based on the low mutation rates (Fig. 10).
Fig. 11.
Nucleotide preferences of the investigated A3 proteins toward PERV proviruses. The distribution of the nucleotides at each position from +5 to −5 bases around the mutation site on the proviral minus-strand DNA is depicted. Numbering starts at the site of deamination. Differences between distributions in the hypermutated EGFP sequences (proviruses produced in the presence of A3 proteins) compared to nonmutated sequences (proviruses produced in the absence of A3 proteins) were evaluated with a chi-square test with Bonferroni-Holm correction denoting significance level α: (*, P < 0.05; **, P < 0.01; ***, P < 0.0001).
Fig. 12.
Trinucleotide preferences of the investigated A3 proteins. For each A3 protein, the observed mutations within the 10 sequenced clones in a C-containing context (NNC-to-NNT mutations) were analyzed; they are listed here as proportions of mutations and are arranged from the most to the least preferred sequence. Error bars represent 95% confidence limits based on binomial distribution (exact Clopper-Pearson confidence intervals).
We noted that in addition to both nucleotides 5′ of the mutation site, the position +4 downstream also seems to be important for the deamination event (Fig. 11). In the case of huA3G a mutated A residue is found at the +4 position, and a mutated T residue is found in the cases of huA3B, muA3, and poA3Z2. The sequence preference for huA3G extends further, as nearly each nucleotide within the +5 to −5 positions around the mutation shows a significant conservation (Fig. 11).
DISCUSSION
In this study we analyzed the chromosomal A3 gene locus in pigs and its mRNAs. While we agree for the most part with previous reports (43, 48), our data disagree in a number of aspects. Importantly, we confirm that the porcine A3 gene locus consists of the two genes poA3Z2 and poA3Z3 (48). In addition to transcripts for the single genes (poA3Z2 and poA3Z3) and the read-through transcript poA3Z2-Z3, we characterized a second read-through transcript, poA3Z2-Z3 SVA, expressed at low levels in some primary tissues including PBMCs. Our data are in contrast with the previously predicted exon composition of poA3Z2. We reanalyzed the genomic sequence of the poA3Z2 gene and did not detect an intron between the previously predicted exons 5 and 6 (48); thus, we propose five instead of six exons for poA3Z2 (Fig. 1).
Comparing the mRNAs of our study with the sequences described by LaRue and coworkers (48), we identified several differences. We mapped the transcriptional start sites and found longer 5′ UTRs than previously known, expanding the lengths of the exons 1 of poA3Z2 and poA3Z3 (Fig. 1). As in the huA3G promoter, the presumed promoter regions of the porcine A3s do not display any TATA and CCAAT boxes (66). The lack of these promoter elements is often associated with multiple transcription start sites (40, 66). Accordingly, we as well as LaRue et al. (48) isolated also smaller 5′ RACE products, suggesting that the porcine A3 genes possess numerous transcription start sites, as is the case for huA3G (66).
Retroviruses evolved at least three different, not exclusive, strategies to maintain replication in their natural hosts. Some viruses, such as primate or feline lentiviruses and foamy retroviruses, express accessory proteins that directly target cellular A3s to prevent A3 encapsidation (11, 52, 67, 79). Other viruses exclude A3s by less understood mechanisms through their structural proteins. Examples for this group are the human T cell leukemia virus and Moloney MuLV (15, 17). Finally, the replication of a third group of retroviruses (e.g., equine infectious anemia virus) is restricted to cells that express very small amounts of A3 (114). The question of how PERV, a porcine endogenous gammaretrovirus of unknown pathogenicity, interacts with porcine and nonporcine A3 restriction factors was addressed here. In this study we made several important findings: (i) PERV is very sensitive to its species-own porcine A3s; (ii) porcine A3s inhibit PERV by cytidine deamination; (iii) PERV is restricted by human and murine A3 proteins. These findings are of utmost importance in estimating the risk of zoonotic PERV transmission after xenotransplantation and in understanding the PERV-pig interaction.
All porcine A3 proteins except poA3Z2-Z3 SVA were able to inhibit PERV and MuLV replication in a significant way. The novel isolated transcript poA3Z2-Z3 SVA has an unclear biological relevance. By overexpressing the corresponding protein (Fig. 7A and B), we observed a weak restrictive effect toward PERV and MuLV, but neither packaging nor cytidine deamination activity was detectable for this protein (Fig. 6B and 10). More research is needed to discover whether poA3Z2-Z3 SVA has other activities than the inhibition of PERV or shows higher expression in tissues that were not analyzed in this study. In contrast, poA3Z2-Z3, poA3Z2, and poA3Z3 effectively reduced retroviral infectivities in single-round infection assays, and at least poA3Z2 and poA3Z2-Z3 blocked PERV replication in a long-term replication study. These findings are in contrast to the previous report of Jonsson et al. (43), which found that PERV is resistant to poA3Z2-Z3. While we used human 293T cell lines stably expressing human and porcine A3 proteins, Jonsson et al. analyzed PERV replication in porcine PK15 cells expressing endogenous levels of poA3Z2-Z3 (43). It is thus possible that low expression of endogenous A3 or the detected sequence variation of A3 in PK15 cells (43) can explain the PERV replication in PK15 cells.
The probability of transmission of potentially zoonotic porcine viruses via a xenotransplant has led to a number of studies on methods to eliminate or prevent expression and transmission of these viruses. The main focus of these studies has been PERV. Our data strongly argue that humans could be fairly resistant against PERV viremia and that PERV transmission after xenotransplantations may be aborted by the human A3 defense shield. However, the recent findings of (xenotropic murine leukemia virus-related virus (XMRV) in humans raise concerns about this assumption. XMRV is a gammaretrovirus that has been detected in human patients with prostate carcinoma (88, 100) and the neurologic disease chronic fatigue syndrome (53). It is controversial how causative and widespread XMRV is (for an overview, see references 18 and 44). With the knowledge of the A3-resistant factors, the question of how XMRV survives in vivo in humans without expressing a counteractive factor is unresolved. Similarly to our findings that PERV is inhibited 9- to 38-fold by huA3F and huA3G (Fig. 7A), three studies showed that XMRV is strongly restricted by huA3G and to a lesser extent by huA3F (30, 74, 94). Xenotransmitted PERV could therefore, possibly like XMRV, replicate in vivo in human cells expressing no or only very small amounts of huA3F and huA3G or evade A3 restriction by unknown mechanisms.
It is not clear whether A3 proteins have had an important effect in the establishment of endogenous retroviruses in the porcine genome. The proviruses of human endogenous retroviruses (HERVs) and endogenous MuLVs in mice were studied to determine whether they show signs of cytidine deamination reminiscent of A3 activity. In the endogenous polytropic and modified polytropic (but not in the xenotropic) murine leukemia virus groups, mutations attributable to murine A3 constitute a large fraction of the total (41). Two variants from the HERV-K (HML2) family were also subjected to A3 hypermutation (2, 50). Information about the presence or absence of G-to-A hypermutations in endogenous PERVs is currently not available and might be clear when the swine genome is fully sequenced. Altogether, the results suggest that A3 editing immediately preceding the integration event could have led to retroviral endogenization. However, other factors contributing to inactivation of infectivity in some endogenous retroviruses must also have been involved.
Taking these results together, we could show that the human as well as the porcine A3 proteins are strong inhibitors of PERV. Based on distinct G-to-A mutations detected in the viral genomes, we suggest that the retroviral restriction of PERV by human and porcine A3s is mainly based on cytidine deamination. Further studies are needed to elucidate whether porcine A3s are functionally important to prevent PERV viremia and protect pigs from retroviral cross-species infections.
ACKNOWLEDGMENTS
We thank the Wuppertal Zoo for the gift of reagents. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program: AID, A1, and A2. We thank Dieter Häussinger for continuous support.
I.G.B. is funded by the Programa Ramón y Cajal, Ministry of Science and Innovation, Spain. C.M. is supported by the Heinz-Ansmann Foundation for AIDS Research. E.D. was supported by grant QLK2-CT-2002-70785 from the European Union, Brussels, Belgium, given to R.R.T. The study was supported by grant TO 117/1 from the Deutsche Forschungsgemeinschaft, Bonn, Germany, to R.R.T.
Footnotes
Published ahead of print on 9 February 2011.
REFERENCES
- 1. Akiyoshi D. E., et al. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 72:4503–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Armitage A. E., et al. 2008. Conserved footprints of APOBEC3G on hypermutated human immunodeficiency virus type 1 and human endogenous retrovirus HERV-K(HML2) sequences. J. Virol. 82:8743–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balmus G., et al. 2007. Cross-species chromosome painting among camel, cattle, pig and human: further insights into the putative Cetartiodactyla ancestral karyotype. Chromosome Res. 15:499–515 [DOI] [PubMed] [Google Scholar]
- 4. Beaudoing E., Freier S., Wyatt J. R., Claverie J. M., Gautheret D. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10:1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bininda-Emonds O. R., et al. 2007. The delayed rise of present-day mammals. Nature 446:507–512 [DOI] [PubMed] [Google Scholar]
- 6. Bishop K. N., Holmes R. K., Malim M. H. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450–8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bishop K. N., et al. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392–1396 [DOI] [PubMed] [Google Scholar]
- 8. Bogerd H. P., Tallmadge R. L., Oaks J. L., Carpenter S., Cullen B. R. 2008. Equine infectious anemia virus resists the antiretroviral activity of equine APOBEC3 proteins through a packaging-independent mechanism. J. Virol. 82:11889–11901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Browne E. P., Littman D. R. 2008. Species-specific restriction of Apobec3-mediated hypermutation. J. Virol. 82:1305–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bushman F. D., et al. 2009. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 5:e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chiu Y. L., Greene W. C. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317–353 [DOI] [PubMed] [Google Scholar]
- 12. Clemenceau B., Lalain S., Martignat L., Sai P. 1999. Porcine endogenous retroviral mRNAs in pancreas and a panel of tissues from specific pathogen-free pigs. Diabetes Metab. 25:518–525 [PubMed] [Google Scholar]
- 13. Conticello S. G., Thomas C. J., Petersen-Mahrt S. K., Neuberger M. S. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22:367–377 [DOI] [PubMed] [Google Scholar]
- 14. Czauderna F., Fischer N., Boller K., Kurth R., Tönjes R. R. 2000. Establishment and characterization of molecular clones of porcine endogenous retroviruses replicating on human cells. J. Virol. 74:4028–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Derse D., Hill S. A., Princler G., Lloyd P., Heidecker G. 2007. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl. Acad. Sci. U. S. A. 104:2915–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dieckhoff B., et al. 2007. Inhibition of porcine endogenous retroviruses (PERVs) in primary porcine cells by RNA interference using lentiviral vectors. Arch. Virol. 152:629–634 [DOI] [PubMed] [Google Scholar]
- 17. Doehle B. P., Schafer A., Wiegand H. L., Bogerd H. P., Cullen B. R. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 79:8201–8207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dolgin E. 2010. Chronic controversy continues over mysterious XMRV virus. Nat. Med. 16:832. [DOI] [PubMed] [Google Scholar]
- 19. Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edgar R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fiebig U., Stephan O., Kurth R., Denner J. 2003. Neutralizing antibodies against conserved domains of p15E of porcine endogenous retroviruses: basis for a vaccine for xenotransplantation? Virology 307:406–413 [DOI] [PubMed] [Google Scholar]
- 23. Fischer N., Krach U., Niebert M., Tönjes R. R. 2003. Detection of porcine endogenous retrovirus (PERV) using highly specific antisera against Gag and Env. Virology 311:222–228 [DOI] [PubMed] [Google Scholar]
- 24. Fishman J. A. 2007. Infection in solid-organ transplant recipients. N. Engl. J. Med. 357:2601–2614 [DOI] [PubMed] [Google Scholar]
- 25. Fishman J. A., Patience C. 2004. Xenotransplantation: infectious risk revisited. Am. J. Transplant. 4:1383–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garkavenko O., et al. 2004. Monitoring for presence of potentially xenotic viruses in recipients of pig islet xenotransplantation. J. Clin. Microbiol. 42:5353–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goff S. P. 2004. HIV: replication trimmed back. Nature 427:791–793 [DOI] [PubMed] [Google Scholar]
- 28. Goff S. P. 2004. Retrovirus restriction factors. Mol. Cell 16:849–859 [DOI] [PubMed] [Google Scholar]
- 29. Goureau A., et al. 1996. Human and porcine correspondence of chromosome segments using bidirectional chromosome painting. Genomics 36:252–262 [DOI] [PubMed] [Google Scholar]
- 30. Groom H. C., Yap M. W., Galao R. P., Neil S. J., Bishop K. N. 2010. Susceptibility of xenotropic murine leukemia virus-related virus (XMRV) to retroviral restriction factors. Proc. Natl. Acad. Sci. U. S. A. 107:5166–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gustavsson I. 1988. Standard karyotype of the domestic pig. Committee for the Standardized Karyotype of the Domestic Pig. Hereditas 109:151–157 [DOI] [PubMed] [Google Scholar]
- 32. Harris R. S., et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 33. Harris R. S., Liddament M. T. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868–877 [DOI] [PubMed] [Google Scholar]
- 34. Hartl I., et al. 2005. Library-based selection of retroviruses selectively spreading through matrix metalloprotease-positive cells. Gene Ther. 12:918–926 [DOI] [PubMed] [Google Scholar]
- 35. Hawken R. J., et al. 1999. A first-generation porcine whole-genome radiation hybrid map. Mamm. Genome 10:824–830 [DOI] [PubMed] [Google Scholar]
- 36. Holmes R. K., Koning F. A., Bishop K. N., Malim M. H. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 282:2587–2595 [DOI] [PubMed] [Google Scholar]
- 37. Holmes R. K., Malim M. H., Bishop K. N. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32:118–128 [DOI] [PubMed] [Google Scholar]
- 38. Irgang M., et al. 2003. Porcine endogenous retroviruses: no infection in patients treated with a bioreactor based on porcine liver cells. J. Clin. Virol. 28:141–154 [DOI] [PubMed] [Google Scholar]
- 39. Iwatani Y., et al. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 35:7096–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jarmuz A., et al. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285–296 [DOI] [PubMed] [Google Scholar]
- 41. Jern P., Stoye J. P., Coffin J. M. 2007. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS. Genet. 3:2014–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jonsson S. R., et al. 2006. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 34:5683–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jonsson S. R., et al. 2007. The restriction of zoonotic PERV transmission by human APOBEC3G. PLoS One 2:e893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kaiser J. 2011. Chronic fatigue syndrome. Studies point to possible contamination in XMRV findings. Science 331:17. [DOI] [PubMed] [Google Scholar]
- 45. Kobayashi M., et al. 2004. APOBEC3G targets specific virus species. J. Virol. 78:8238–8244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Langlois M. A., Kemmerich K., Rada C., Neuberger M. S. 2009. The AKV murine leukemia virus is restricted and hypermutated by mouse APOBEC3. J. Virol. 83:11550–11559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. LaRue R. S., et al. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 83:494–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. LaRue R. S., et al. 2008. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC. Mol. Biol. 9:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lecossier D., Bouchonnet F., Clavel F., Hance A. J. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 50. Lee Y. N., Malim M. H., Bieniasz P. D. 2008. Hypermutation of an ancient human retrovirus by APOBEC3G. J. Virol. 82:8762–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Le Tissier P., Stoye J. P., Takeuchi Y., Patience C., Weiss R. A. 1997. Two sets of human-tropic pig retrovirus. Nature 389:681–682 [DOI] [PubMed] [Google Scholar]
- 52. Löchelt M., et al. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 102:7982–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lombardi V. C., et al. 2009. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science 326:585–589 [DOI] [PubMed] [Google Scholar]
- 54. Low A., et al. 2009. Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene. Virology 385:455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mangeat B., et al. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103 [DOI] [PubMed] [Google Scholar]
- 56. Mariani R., et al. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31 [DOI] [PubMed] [Google Scholar]
- 57. Marin M., Rose K. M., Kozak S. L., Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398–1403 [DOI] [PubMed] [Google Scholar]
- 58. Martin S. I., Wilkinson R., Fishman J. A. 2006. Genomic presence of recombinant porcine endogenous retrovirus in transmitting miniature swine. Virol. J. 3:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Martin U., et al. 2000. Productive infection of primary human endothelial cells by pig endogenous retrovirus (PERV). Xenotransplantation. 7:138–142 [DOI] [PubMed] [Google Scholar]
- 60. Mbisa J. L., et al. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Meije Y., Tönjes R. R., Fishman J. A. 2010. Retroviral restriction factors and infectious risk in xenotransplantation. Am. J. Transplant. 10:1511–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Milan D., et al. 2000. IMpRH server: an RH mapping server available on the Web. Bioinformatics 16:558–559 [DOI] [PubMed] [Google Scholar]
- 63. Miyagi E., et al. 2010. Stably expressed APOBEC3F has negligible antiviral activity. J. Virol. 84:11067–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moon H. J., et al. 2009. Comparison of the age-related porcine endogenous retrovirus (PERV)expression using duplex RT-PCR. J. Vet. Sci. 10:317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Muckenfuss H., et al. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161–22172 [DOI] [PubMed] [Google Scholar]
- 66. Muckenfuss H., et al. 2007. Sp1 and Sp3 regulate basal transcription of the human APOBEC3G gene. Nucleic Acids Res. 35:3784–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Münk C., et al. 2008. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 9:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Naldini L., et al. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed] [Google Scholar]
- 69. Newman E. N., et al. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166–170 [DOI] [PubMed] [Google Scholar]
- 70. Nikaido M., Rooney A. P., Okada N. 1999. Phylogenetic relationships among cetartiodactyls based on insertions of short and long interpersed elements: hippopotamuses are the closest extant relatives of whales. Proc. Natl. Acad. Sci. U. S. A. 96:10261–10266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. OhAinle M., Kerns J. A., Malik H. S., Emerman M. 2006. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 80:3853–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Oldmixon B. A., et al. 2002. Porcine endogenous retrovirus transmission characteristics of an inbred herd of miniature swine. J. Virol. 76:3045–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Osoegawa K., et al. 1998. An improved approach for construction of bacterial artificial chromosome libraries. Genomics 52:1–8 [DOI] [PubMed] [Google Scholar]
- 74. Paprotka T., et al. 2010. Inhibition of xenotropic murine leukemia virus-related virus by APOBEC3 proteins and antiviral drugs. J. Virol. 84:5719–5729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Paradis K., et al. 1999. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with living pig tissue. The XEN 111 Study Group. Science 285:1236–1241 [DOI] [PubMed] [Google Scholar]
- 76. Patience C., et al. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 75:2771–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Patience C., Takeuchi Y., Weiss R. A. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282–286 [DOI] [PubMed] [Google Scholar]
- 78. Pauly T., et al. 1995. Classical swine fever virus-specific cytotoxic T lymphocytes and identification of a T cell epitope. J. Gen. Virol. 76:3039–3049 [DOI] [PubMed] [Google Scholar]
- 79. Perkovic M., et al. 2009. Species-specific inhibition of APOBEC3C by the prototype foamy virus protein bet. J. Biol. Chem. 284:5819–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Powell S. K., et al. 2000. Antiretroviral agents inhibit infection of human cells by porcine endogenous retroviruses. Antimicrob. Agents Chemother. 44:3432–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Qari S. H., et al. 2001. Susceptibility of the porcine endogenous retrovirus to reverse transcriptase and protease inhibitors. J. Virol. 75:1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rettenberger G., Bruch J., Fries R., Archibald A. L., Hameister H. 1996. Assignment of 19 porcine type I loci by somatic cell hybrid analysis detects new regions of conserved synteny between human and pig. Mamm. Genome 7:275–279 [DOI] [PubMed] [Google Scholar]
- 83. Rink A., et al. 2002. A first-generation EST RH comparative map of the porcine and human genome. Mamm. Genome 13:578–587 [DOI] [PubMed] [Google Scholar]
- 84. Ross S. R. 2009. Are viruses inhibited by APOBEC3 molecules from their host species? PLoS Pathog. 5:e1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Russell R. A., et al. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79:8724–8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 87. Santiago M. L., et al. 2008. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science 321:1343–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Schlaberg R., Choe D. J., Brown K. R., Thaker H. M., Singh I. R. 2009. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc. Natl. Acad. Sci. U. S. A. 106:16351–16356 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89. Sheehy A. M., Gaddis N. C., Malim M. H. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404–1407 [DOI] [PubMed] [Google Scholar]
- 90. Shimodaira H., Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to pylogenetic inference. Mol. Biol. Evol. 16:1114–1116 [Google Scholar]
- 91. Stamatakis A. 2006. Phylogenetic models of rate heterogeneity: a high performance computing perspective, poster, p. 253 Proc. 20th IEEE/ACM Int. Parallel Distributed Processing Symp., Rhodos, Greece, 25 to 29 April 2006 [Google Scholar]
- 92. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 93. Stamatakis A., Ludwig T., Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463 [DOI] [PubMed] [Google Scholar]
- 94. Stieler K., Fischer N. 2010. Apobec 3G efficiently reduces infectivity of the human exogenous gammaretrovirus XMRV. PLoS One 5:e11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Strachan T., Read A. P. 1999. Human molecular genetics, 2nd ed. Wiley-Liss, Wilmington, DE [Google Scholar]
- 96. Suchard M. A., Weiss R. E., Sinsheimer J. S. 2001. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 18:1001–1013 [DOI] [PubMed] [Google Scholar]
- 97. Suyama M., Torrents D., Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Takeda E., et al. 2008. Mouse APOBEC3 restricts friend leukemia virus infection and pathogenesis in vivo. J. Virol. 82:10998–11008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Takeuchi Y., et al. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986–9991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Urisman A., et al. 2006. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101. Watanabe T., Watanabe S., Kawaoka Y. 2010. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7:427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wessagowit V., Nalla V. K., Rogan P. K., McGrath J. A. 2005. Normal and abnormal mechanisms of gene splicing and relevance to inherited skin diseases. J. Dermatol. Sci. 40:73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wiegand H. L., Doehle B. P., Bogerd H. P., Cullen B. R. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wilson C. A., et al. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J. Virol. 72:3082–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wolf D., Goff S. P. 2008. Host restriction factors blocking retroviral replication. Annu. Rev. Genet. 42:143–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wood J. C., et al. 2004. Identification of exogenous forms of human-tropic porcine endogenous retrovirus in miniature Swine. J. Virol. 78:2494–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yerle M., et al. 1998. Construction of a whole-genome radiation hybrid panel for high-resolution gene mapping in pigs. Cytogenet. Cell Genet. 82:182–188 [DOI] [PubMed] [Google Scholar]
- 108. Yu Q., et al. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379–53386 [DOI] [PubMed] [Google Scholar]
- 109. Yu Q., et al. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435–442 [DOI] [PubMed] [Google Scholar]
- 110. Yu X., et al. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060 [DOI] [PubMed] [Google Scholar]
- 111. Yu Y., Xiao Z., Ehrlich E. S., Yu X., Yu X. F. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhang H., et al. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zheng Y. H., et al. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zielonka J., et al. 2009. Restriction of equine infectious anemia virus by equine APOBEC3 cytidine deaminases. J. Virol. 83:7547–7559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zielonka J., et al. 2010. Vif of feline immunodeficiency virus from domestic cats protects against APOBEC3 restriction factors from many felids. J. Virol. 84:7312–7324 [DOI] [PMC free article] [PubMed] [Google Scholar]