Abstract
Studies of viral pathogenesis have relied heavily on analyses of specific clones and their genetic determinants of virulence. It is sometimes difficult to apply this reductionist approach to the study of RNA viruses, which by virtue of their very high mutation rates, exist as a complex mixture of mutants. While quasispecies theory has provided an intellectual framework for exploring the relationship between the viral population structure and phenotype, experimental studies have been limited by the relatively poor resolution of traditional sequencing-based approaches. We have addressed this problem by developing a molecular barcoding strategy in which viral subpopulations are tagged with unique 20-nucleotide sequences. The behavior of these subpopulations can be monitored using a universal barcode microarray. We demonstrate the performance of our barcode microarray platform using poliovirus, a model RNA virus. Using this platform, we explored the fitness landscape occupied by an artificial quasispecies consisting of 48 randomly mutagenized clones. We were able to rapidly derive precise fitness measurements for a majority of these clones and identified a neutral space surrounding the wild type. The experimental paradigm presented here is readily adaptable to other viral systems and can potentially be used to track thousands of variants in a cost-effective manner.
INTRODUCTION
The evolution of RNA viruses has been the subject of intense interest over the last 30 years (14, 16, 41). The ongoing HIV and influenza pandemics and the emergence of new infectious agents highlight the importance of these viruses as human pathogens. The study of RNA viruses has also increased our general understanding of adaptive evolution (40). The RNA-dependent RNA polymerases of riboviruses exhibit characteristically low fidelity with measured mutation rates of 10−3 to 10−5 mutations per nucleotide copied per replication cycle (3, 17, 61). These mutation rates are orders of magnitude greater than those of nearly all double-stranded-DNA-based viruses and organisms. Given the short generation times and large population sizes observed in both experimental and natural infections, RNA viruses exist as rapidly evolving populations of related mutants (18, 22). Because their evolution can be monitored in “real time,” RNA viruses have become important model systems for evolutionary biologists.
Quasispecies theory is a mathematical framework that has been used to describe the dynamics of viral populations (6, 20). It builds on classical population genetics but seeks to explore the consequences of rapid, error-prone replication and near-infinite population sizes for genome evolution. A viral quasispecies is a cloud of diverse variants that are genetically linked through mutation and collectively contribute to the characteristics of the population. Understanding the contributions of genetically distinct variants to a given viral phenotype is therefore of critical importance to pathogenesis in infected hosts. In contrast, much of our understanding of viral pathogenesis derives from studies of single viral clones, which may not reveal many of the most important aspects of natural infections, particularly for RNA viruses. Indeed, work in our laboratory and others has shown that changes in the overall structures of viral populations can have a profound influence on viral pathogenesis that is not attributable to individual variants (46, 56, 64). Further characterization of these effects will require new experimental methods, capable of measuring changes in population structure.
Experimental approaches to viral evolution seek to characterize how evolutionary forces, such as mutation, selection, and genetic drift, impact the population (41). Techniques currently in use draw on traditional population genetics and rely on markers linked to a given genotype (29). An ideal marker is selectively neutral and easy to score. Many early studies measured the relative frequencies of two variants by scoring monoclonal antibody resistance in standard plaque assays (31, 37). Other assays have used restriction digests or real-time PCR to quantify the frequencies of sequence polymorphisms in a population (8, 63). While these methods have been widely applicable to studies of viral fitness and population bottlenecks, they are limited by the paucity of suitable markers. As a result, inferences must be made about the population based on analyses of a few clones, and they are relatively insensitive to smaller differences in population structure.
Advances in molecular biology and laboratory instrumentation now permit the simultaneous measurement of hundreds or thousands of markers. In a process known as genetic footprinting, unique DNA sequence tags serve as molecular “barcode” identifiers for specific variants in a mixed population (59). These tags are easily detected using spotted microarrays, making it a parallel-screening strategy. Molecular barcoding is now used in signature-tagged mutagenesis of bacteria and in the construction of libraries for RNA interference screens (38). Here, we apply a similar approach to studies of viral evolution. By marking viral clones with 20-nucleotide sequence tags, we can follow a large number of subpopulations within an evolving quasispecies. In contrast to other systems, our strategy does not rely on polymorphic viral sequences (35). We describe its utility for quantifying viral fitness and measuring the segregation of populations in vivo. We applied our platform to characterize the fitness landscape occupied by a mutagenized poliovirus population and documented the mutational robustness predicted to play a critical role in the dynamics of an RNA virus population in which a minority of the mutants exhibited significant fitness defects.
MATERIALS AND METHODS
Viruses and cells.
HeLaS3 cells (ATCC CCL-2.2) were kindly provided by R. Geller and J. Frydmann (Stanford University). The cells were maintained in 50% Dulbecco's modified Eagle medium (DMEM)/50% F-12 medium supplemented with 10% newborn calf serum and 2 mM glutamine (Invitrogen). All viruses were derived from two Mahoney type I poliovirus clones: prib(+)XpAlong and pMoV2.115UTR RE (2, 30). The latter was engineered to contain unique restriction sites in the 5′ untranslated region (UTR) (BssHII and SacI) and the 2A cleavage site (EcoRI and XhoI) to facilitate cloning of exogenous sequences.
Microarray printing and postprocessing.
Array oligonucleotides (see Table S1 in the supplemental material) were synthesized (Invitrogen), resuspended in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) to a final concentration of 20 μM, and spotted with S75 printing tips (Parallel Synthesis) onto polylysine-coated slides (ThermoScientific), using a DeRisi Linear Servo Microarray and Arraymaker software. Arrays were printed in tandem so that each slide had two replicate microarrays. The slides were postprocessed in groups of 20 as follows. After being UV cross-linked, the slides were soaked in 3× SSC, 0.2% SDS at 65°C for 5 min, rinsed in water and 100% ethanol, and dried. The slides were then immersed for 15 min in a solution containing 5.5 g succinic anhydride, 335 ml 1-methyl-2-pyrolidinone, and 15 ml 1 M sodium borate, pH 8.0. They were then rinsed in water and ethanol, dried, and stored for subsequent use.
Molecular biology.
All 5′ barcoded viruses were generated by ligation of double-stranded oligonucleotides into the BssHII and SacI sites of pMoV2.115UTR RE. Complementary oligonucleotides with the appropriate overhangs (Invitrogen) were mixed in annealing buffer (40 mM Tris, pH 7.5, 20 mM MgCl2), heated for 5 min at 95°C, and cooled slowly to room temperature.
The oligonucleotide pairs are listed in Table S2 in the supplemental material. For clones 5′ 285, 5′ 286, 5′ 287, and 5′ 288, the double-stranded oligonucleotides were ligated individually, and plasmids were prepared from single bacterial colonies. For the WT96 library, equal quantities of the remaining 96 double-stranded oligonucleotides were pooled, ligated, and transformed together. Approximately 100,000 transformants were pooled, and the plasmid library was prepared directly from the colony pool. Given the large number of transformants pooled, we assumed that each barcode was equally represented in the library.
The 3′-barcoded clone, 3′ 288, was generated by overlap extension PCR, using pMoV2.115UTR RE as the initial template (51). The barcode sequence was inserted 3′ of the stop codons for the viral polyprotein. An additional three guanines 3′ of the barcode were also inserted to generate a unique ApaI site for subsequent cloning. In the first-round reactions, a 1.8-kb fragment upstream of the insertion site was amplified using primers F.5654 (5′-TGTGATCGATGGCAAAGAAGTGG-3′) and re7451APA288 (5′-CCCTATGCAAATTCGTGCGTGTGTTACTAAAATGAGTCAAGCCAACGG-3′), and a 0.7-kb fragment downstream of the insertion site was amplified using primers F.7494APA288 (5′-CCCCACACGCACGAATTTGCATAGTAAATTTTTCTTTAATTCGGAGAAAAA-3′) and re.8199 (5′-CCTTCGGTCCTCCGATCGTTG-3′). These primers contain complementary sequences encoding the insertion. The PCR products were purified and mixed together in a second-round PCR supplemented with primers F.5654 and re.8199. The 2.5-kb product was digested with BglII and MluI and ligated into pMoV2.115UTR RE using the same sites.
To generate the double-barcoded clone, 5′ 285/3′ 288, an MluI/BglII fragment containing the 5′ half of 5′ 285 was ligated to a BglII/MluI fragment containing the 3′ half of 3′ 288.
A wild-type clone, pAL-WT, containing the 3′ ApaI restriction site, was generated by replacing the NheI/BglII fragment of 3′ 288 with the corresponding fragment from prib(+)XpAlong. Note that this intermediate clone retains the 3′ 288 barcode. A guanidine-resistant clone, pAL-GR, encoding an asparagine-to-glycine substitution at amino acid 179 of the 2C protein, was generated by overlap extension PCR using pAL-WT as a template. In the first-round reaction, a 0.4-kb fragment upstream of the mutation was amplified using primers for 4379 (5′-CAGTCTAAGAGGTTTGCCCCTCTTTACGCAGTGGAA-3′) and rev4727GR (5′-CTGGGTTTTGCCCCAGGTCGTCCATAATCACCACTCCCTGTTGTT-3′), and a 0.9-kb fragment downstream of the mutation was amplified using primers for4727GR (5′-GGACGACCTGGGGCAAAACCCAGATGGTGCGGACATGAAGCTGTTCTG-3′) and rev5696 (5′-CGCTTTGGCATCCAAGATCTC-3′). These PCR products were purified and mixed together in a second reaction mixture supplemented with primers for4379 and rev5696. The 1.3-kb product was digested with BmgBI and BglII and ligated into pMoV2.115UTR RE using the same sites.
Double-barcoded spike-in and guanidine-resistant clones were generated by PCR using pAL-WT and pAL-GR as templates, respectively. The primers in these reaction mixtures contained barcode sequences at their 5′ termini (see Table S3 in the supplemental material). The WT40 library was also generated by PCR using Herculase II (Stratagene). Forty individual PCR products, each generated with a distinct pair of barcode primers, were pooled and digested with BssHII and ApaI. This pool was ligated to a 5′ DNA fragment spanning the T7 promoter and poliovirus 5′ untranslated region and a 3′ DNA fragment spanning the poliovirus 3′ untranslated region. This WT40 ligation reaction served as a template for subsequent in vitro transcription of the corresponding viral RNA. A similar protocol was used to generate the MUT40 library. In this case, wild-type poliovirus was passaged twice in 400 μM ribavirin. The titer of this stock was determined, and single clones were isolated by endpoint dilution. RNA was prepared from 48 of these clones, and full-length viral cDNA was generated using Thermoscript (Invitrogen). This cDNA was amplified using Herculase II (Stratagene).
Preparation of viral stocks.
Viral genomic RNA was generated by in vitro transcription of the plasmids and libraries described above. For each virus or viral library, 20 μg of RNA was electroporated into 4 × 106 HeLaS3 cells in a 4-mm cuvette with the following pulse: 300 V, 24 Ω, and 1,000 μF. The resulting passage 0 (P0) stock was passed once on HeLaS3 cells at high multiplicity (multiplicity of infection [MOI], 1 to 10) to generate a P1 stock used for subsequent experiments. All viral stocks and experimental supernatants were harvested following three successive freeze-thaw cycles of the cell culture dish and clarified by centrifugation at 1,000 rpm for 5 min in a benchtop centrifuge (Sorvall Legend T, rotor 75006434).
One-step growth curves.
HeLaS3 cells were plated in 12-well dishes at 5 × 105 cells per well on the day prior to infection. Three wells were used for each virus and time point. On the day of infection, the medium was removed and the cells were washed once with phosphate-buffered saline (PBS). The cells were infected with 106 PFU of wild-type, 5′-barcode (285), 3′-barcode (288), or 5′/3′-barcode (285/288) virus. The inocula were removed at 60 min (time 0 [t0]), and each well was washed twice with PBS to remove residual virus. Samples were removed at various time points (t = 0, 2, 4, 6, 8, 10, 12, and 24 h), and the titer was determined by standard plaque assay on HeLaS3 cells. Triplicate values for each virus and time point were averaged.
RNA preparation and reverse transcription (RT) single-stranded PCR.
RNA was isolated from cell-free viral supernatants using Trizol (Invitrogen) according to the manufacturer's instructions. For experiments with spike-in control viruses, 450 μl of each supernatant was mixed with 50 μl of the spike-in pool prior to adding Trizol. RNA from 500 μl supernatant was precipitated with 20 μg glycogen and resuspended in 8 μl nuclease-free water. cDNA was synthesized from 7 μl total RNA using Superscript III reverse transcriptase (Invitrogen), 100 ng random hexamers, and 50 pmol oligo(dT) in a 20-μl reaction mixture. Short, ∼60-base single-stranded fragments containing either the 5′ or 3′ barcode were amplified and labeled in two separate single-stranded PCRs (ssPCRs). Each 25-μl reaction mixture contained 1 μl cDNA, 250 μM each deoxynucleoside triphosphate (dNTP), 2.5 U Taq polymerase (Denville Scientific), 50 nM forward primer (Elim Biopharmaceuticals), 1 μM Cy-labeled reverse primer (Integrated DNA Technologies). Five prime barcodes were amplified using primers F.661 (5′-CTGTTTGCTGGATCCGCGCGC-3′) and Cy(3 or 5)-rev5BC (5′-GTACTTAGAGTAAACACACTCAATG-3′). Three prime barcodes were amplified using primers F.7430.1 (5′-CCGCCGTTGGCTTGACTCATTTTAGT-3′) and Cy(3 or 5)-rev3BC (5′-CAATTCGACTGAGGTAGGG-3′). Reaction mixtures were heated to 94°C for 2 min and subjected to 40 cycles of 94°C for 15 s, 55°C for 15 s, and 72°C for 15 s.
Infection of susceptible mice.
The University of California, San Francisco, Institutional Animal Care and Use Committee approved the protocols for the mouse studies described here. In these experiments, we used 6- to 8-week-old cPVR mice, which express the poliovirus receptor under the control of the beta-actin promoter (11). Mice were infected under anesthesia with 100 μl of viral supernatant via the intraperitoneal or intravenous route (i.e., the tail vein). The mice were monitored daily for the onset of paralysis and euthanized when total paralysis was imminent. Whole organs were isolated from all mice and homogenized in 2 ml PBS with a T1 Basic S1 disperser (Ika Works). Viral supernatants were recovered from the tissue homogenates following three freeze-thaw cycles and centrifugation at 1,000 rpm for 5 min in a benchtop clinical centrifuge (Sorvall Legend T, rotor 75006434).
Microrray hybridization, washing, and scanning.
Four PCR products were pooled in equal quantities for each microarray—5′ and 3′ reactions for each sample. The pools were precipitated, resuspended in 20 μl hybridization buffer (3× SSC, 25 mM HEPES, pH 7.0, 0.25% SDS), denatured at 95°C for 5 min, and allowed to cool slowly to ambient temperature. The hybridization solution was injected between the microarray slide and the coverslip, and the arrays were incubated overnight at 44°C in sealed hybridization chambers (DieTech). The slides were washed twice in 0.2× SSC-0.3% SDS at 44°C, once in 0.2× SSC at 44°C, and once in 0.2× SSC at ambient temperature. The arrays were dried by brief centrifugation using a benchtop minicentrifuge (Labnet) and scanned at 10-μm resolution using an Axon 4000B scanner (Molecular Devices).
Data analysis and statistics.
Array images (16-bit TIFF files) were analyzed using GenePix Pro 6.1 (Molecular Devices). Grids were applied using the appropriate.gal file. Poor-quality spots were identified based on the following criteria: a signal-to-noise ratio of <2 in either channel, fewer than 50% of pixels with signals greater than 2 standard deviations above background in either channel, or greater than 10% saturated pixels in either channel. The associated data were excluded from subsequent analysis.
Statistical analyses were performed with Limma and R (60). Data were normalized within each array by fitting a loess curve to the control spots. Replicate spots within the same array were then averaged. For experiments comparing the guanidine-resistant mutant to the wild type, arrays with P0 and P2 references were analyzed separately using common reference design. Dye swap and biological-replicate effects (n = 5) were controlled by including them as fixed covariates, while the technical-replicate effect was included as a random effect. For the mutant passage experiment, analyses were done separately for Cy3 and Cy5 channels using common reference design. Biological replicates (n = 5) were controlled as described above.
RESULTS
Overview of the barcode microarray platform.
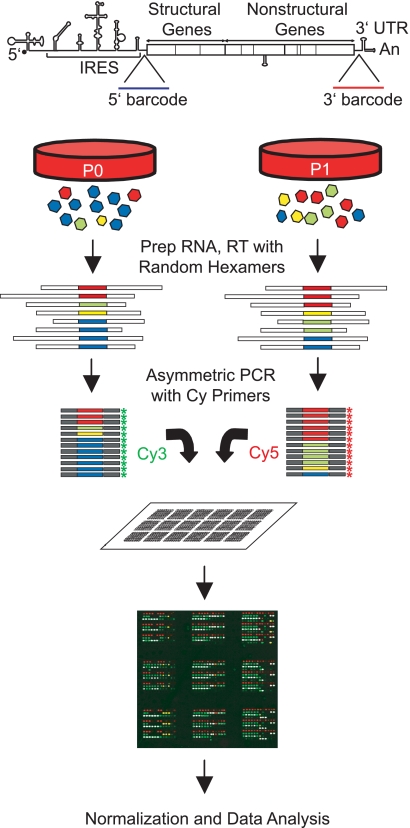
Our assay couples molecular barcoding and microarray-based quantitative phenotypic analysis, an approach used successfully in signature-tagged mutagenesis screens of bacteria and yeast. In such screens, individual mutants are generated and labeled with a distinguishing molecular tag, usually a 20- to 40-nucleotide sequence (Fig. 1). These “barcoded” strains are then pooled and grown as a mixed population. The population is sampled at defined time points, and the level of each strain under a given competitive growth condition is quantified using a microarray that contains oligonucleotides complementary to each molecular tag. The advantage of this approach is that it is highly quantitative and massively parallel, providing information on thousands of strains replicating in parallel. The general schema of our assay is similar, but we have had to adapt our platform to satisfy the constraints of the poliovirus system and perform additional controls required for experiments with a highly mutable RNA virus.
Fig. 1.
Overview of the barcode microarray platform. A library of distinct viral subpopulations is generated by insertion of 20-nucleotide sequence tags into the poliovirus genome. These “barcodes” are unique identifiers of that viral subpopulation. The population of barcoded variants is passaged on susceptible cells. The fitter variants, and their corresponding barcodes, increase in frequency, while the less fit variants decrease. Viral genomic RNA is recovered from each population, and the barcode tags are amplified by reverse transcription and asymmetric PCR using Cy-labeled primers. In each competitive hybridization, one population is labeled with Cy3 and the other with Cy5. The labeled barcodes are hybridized to a microarray that contains oligonucleotides complementary to each barcode. The fluorescence intensity of each element on the microarray reflects the frequency of the corresponding viral variant in the population. IRES, internal ribosome entry site.
Fundamental to any high-throughput assay based on signature tagging is obtaining a library of molecular barcodes that reliably distinguish individual variants within a complex population. We used the barcode sequences published by the Yeast Genome Deletion Project. This set of computationally optimized barcodes offers several advantages: (i) The 20-nucleotide barcodes can be readily inserted using standard molecular biology techniques. (ii) Each 20-mer differs from every other barcode at 5 positions, allowing substantial discriminatory power. (iii) All barcodes are predicted to be devoid of RNA secondary structures and have similar melting temperatures, simplifying competitive hybridization experiments. Because our marker strategy uses an available library and does not rely on virus-encoded polymorphisms, our system is applicable to a broad range of viruses.
Locations of barcodes and genetic neutrality.
All marker strategies for distinguishing viral variants require that the genetic marker be selectively neutral (29, 31, 37). If this assumption is violated, it is difficult to discern whether an observed viral phenotype is due to viral polymorphisms or confounding effects of the associated marker. Our assay relies on the insertion of exogenous sequences at two distinct sites within the poliovirus genome. As such, there are three issues relating to the neutrality of these markers. (i) Do the insertion sites tolerate exogenous sequence? (ii) Do the sequences themselves affect viral fitness? (iii) Do the sequences exert epistatic effects on the distribution of mutants within a viral population? RNA viruses are notorious for their compact genomes; the 7,440-nucleotide poliovirus RNA codes for 11 viral proteins and regulates their efficient expression throughout its replication cycle (25). To minimize direct and epistatic effects of sequence insertion on the viral phenotype, we focused on noncoding sequences that are also predicted to be devoid of RNA secondary structure. We inserted 5′ barcodes between the structural elements of the internal ribosome entry site and the start codon for the viral polyprotein (Fig. 1). The 3′ barcodes were inserted between the polyprotein stop codon and the regulatory stem-loop structures in the 3′ untranslated region.
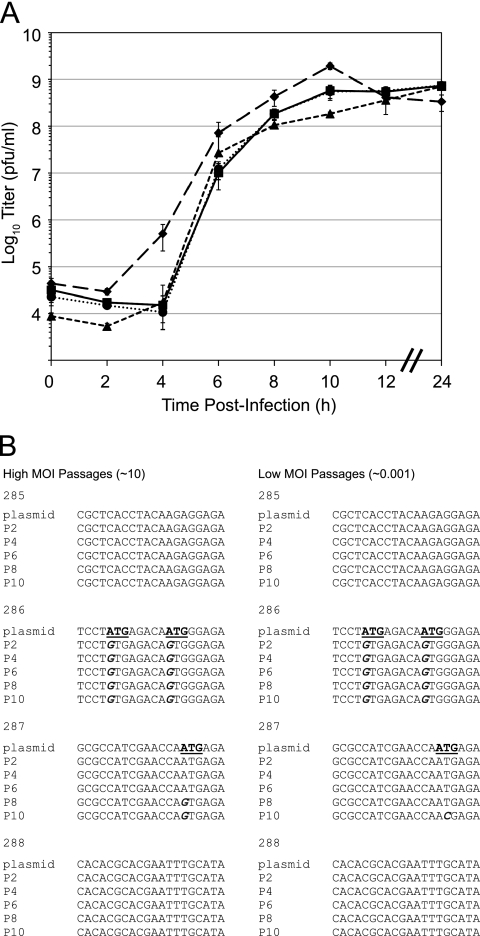
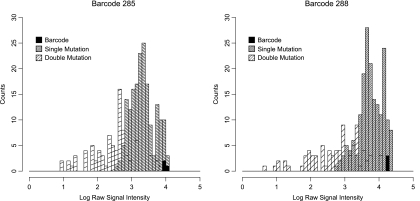
We assayed the effects of 5′ and 3′ barcodes on viral fitness using one-step growth curves, which measure the kinetics of viral replication through a single cycle of infection. While there were slight differences at individual time points (Fig. 2A, 5′/3′ barcodes at t of 4 and 10 h and 3′ barcode at t of 10 h), the overall growth rates were similar. While growth curves are insufficiently sensitive to exclude small effects on fitness, our data suggest that the locations of the barcodes do not significantly affect viral replication. We further characterized the selective neutrality of the barcodes by examining their genetic stability over repeated passages. We and others have generally observed rapid deletion of exogenous sequences by RNA viruses over several passages due to pleiotropic effects on viral fitness. Mutations within the barcodes are also a potential complication of sequence tagging a ribovirus, and approximately 0.5% of barcodes will carry a mutation at any time (assuming a constant mutation rate across the genome, two 20-nucleotide barcodes, 7,481 total bases, and one mutation per genome per replication cycle). However, without a fitness effect, these barcode variants will achieve a mutation-selection balance and remain at a low frequency in the population. Thus, we carried out population sequencing of individual viruses after repeated passages. We detected only an occasional substitution at a late passage, suggesting that most barcodes do not exert negative selective pressure on the virus (Fig. 2B). Interestingly, all of the mutations identified changed candidate AUG codons within the barcode sequence. Because these barcode-encoded codons could serve as alternative translation start sites, they are an illustrative exception to the selective-neutrality hypothesis and were excluded from further experiments. We also found no evidence of insertions or deletions within the barcodes by sequence (see Fig. S1 in the supplemental material) or sensitive RT-PCR analysis (see Fig. 5A). This result further argues for the relative neutrality of the insertion site and the exogenous sequence. Of note, we found that the low observed level of mutation in neutral barcodes did not significantly affect the microarray hybridization efficiency or discriminatory power (Fig. 3) (68). Together, our data suggest that barcode insertions at the selected sites have only minor effects on viral fitness. Because of the large number of barcode sequences used in these experiments, it is not feasible to quantify the effect of each barcode by competition with a wild-type reference over extended passages. However, small barcode-mediated effects should not significantly confound fitness measurements in short-term experiments (<10 passages), particularly if multiple barcodes are used for each tagged variant. In the experiments described below, we further controlled for the effects of the barcode sequences on a given variant by using a barcoded wild-type reference.
Fig. 2.
Wild-type and barcoded polioviruses replicate with similar kinetics. (A) Replication kinetics of wild-type (solid line; squares), 5′-barcoded (dotted line; circle), 3′-barcoded (dashed line; triangle), and double-barcoded (large dashed line; diamond) viruses. HeLa cells were infected in triplicate at an MOI of 1. Samples were taken at the indicated time points (x axis), and the titer of progeny virus was determined by plaque assay. The viral titers (y axis) represent the means ± standard deviations (SD) of three replicates. (B) Genetic stability of barcode sequences after repeated passages. Viruses containing the indicated barcodes (285 to 288) at the 5′ insertion site were passaged 10 times at either high (MOI = 10) or low (MOI = 0.001) multiplicity. Virus was recovered for population sequencing at every other passage. The population consensus sequences of the barcodes are shown compared to the plasmid clone, with mutations indicated in italics.
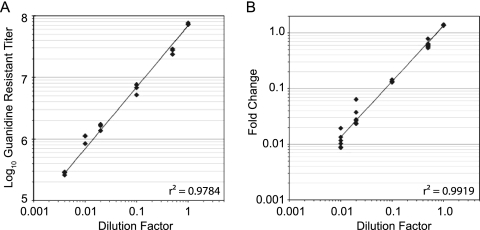
Fig. 5.
Dynamic range and accuracy of the barcode microarray platform. A double-barcoded guanidine-resistant virus (5′ U178/3′ U196) was serially diluted in a stock of the WT96 library. (A) Titer of guanidine-resistant virus, measured by plaque assay (y axis, log scale), as a function of the dilution factor (x axis, log scale). (B) Fold change of guanidine-resistant virus, measured by microarray (y axis, log scale) as a function of the dilution factor (x axis, log scale). Measurements derived from triplicate array elements for each of the barcodes are shown.
Fig. 3.
Effect of barcode mutation on hybridization efficiency. Viral genomic RNA from a double-barcoded virus (5′285/3′288) was reverse transcribed, Cy labeled by asymmetric PCR, and hybridized to a custom array tiled with all possible point mutants and a subset of double mutants for each barcode. The raw signal intensity on each spot reflects the hybridization efficiency of each barcode against a mismatched partner and is shown as a histogram with wild-type barcodes, single-mutant barcodes, and double-mutant barcodes indicated.
Amplification of barcodes by RT-ssPCR.
We examined a number of protocols for preparing fluorescently labeled template from viral genomic RNA. Direct labeling of the barcodes during cDNA synthesis resulted in long probes with relatively low specific activity and cross hybridization of labeled viral sequences. We therefore pursued a PCR strategy in which cyanine dye (Cy)-labeled primers were used to amplify the barcode sequences. Because amplification converted long cDNA samples into short, ∼60-base amplicons, it reduced the complexity of the hybridized sample, thereby improving assay specificity. We also boosted the amount of nucleic acid available for hybridization by using single-stranded rather than conventional PCR.
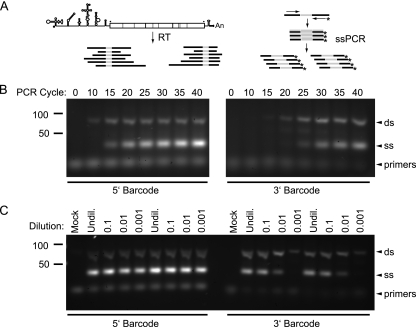
Random priming in a reverse transcription reaction was used to create an unbiased representation of cDNA from the corresponding viral genomic RNA (Fig. 4A). The 5′ and 3′ barcode identifiers were then amplified from this template by asymmetric PCR (47, 52). In our protocol, the reverse primer is end labeled with either Cy3 or Cy5 and present in 20-fold molar excess over the forward primer. The forward primer is exhausted after approximately 10 cycles, and further cycling generates an excess of antisense, Cy-labeled, single-stranded DNA (Fig. 4B). Each reaction mixture has the same quantity of limiting primer, which ensures that the yields of labeled product are similar over a wide range of template concentrations (Fig. 4C). Because this step effectively normalizes the total Cy3 and Cy5 signal in each viral population, the fluorescence of any given barcode measures the relative amount of that subpopulation within the larger sample. The amplification step also allows analysis of relatively small populations.
Fig. 4.
Amplification and labeling of barcode tags by RT-ssPCR. (A) Protocol schematic. Barcode sequences are shown in gray. Random hexamers and oligo(dT) are used to prime reverse transcription of short cDNA from the viral RNA. This cDNA is used as a template in two separate PCRs, one for each barcode. In this asymmetric PCR, the Cy-labeled primer (*) is present at 20-fold molar excess relative to the forward, sense primer. A double-stranded DNA product is produced for a limited number of cycles until the forward primer is used up. The remaining cycles result in linear amplification of single-stranded, antisense, Cy-labeled product. An, poly(A) tail of the viral genome RNA. (B) ssPCR reactions were run in octuplicate for each barcode, indicated below, and a sample was removed every 5 cycles. The amplified product was electrophoresed through a 4% agarose gel and stained with SYBR gold. Positions of markers (in bp), double-stranded (ds) and single-stranded (ss) DNA, and primers on the gel are indicated. (C) Genomic RNA was isolated from serial 10-fold dilutions of a viral stock (∼109 PFU/ml) and subjected to RT-ssPCR as in panel B.
Precision and accuracy of array measurements.
We next tested the dynamic range and accuracy of our platform for detecting changes in the frequency of a single barcoded subpopulation within a complex pool. We generated a barcoded poliovirus clone bearing a single point mutation, C4605U, which renders the virus resistant to the antiviral drug guanidine (49). This clone contained three distinct genetic markers, which allowed detection by independent techniques. The relative frequency of barcodes 178 and 196 were measured by the microarray, and the relative titer of guanidine-resistant virus was measured by plaque assay in the presence and absence of drug. We serially diluted a stock of the 5′ 178/3′ 196-C4605U virus into a stock of a barcoded wild-type library and measured the relative frequency of this variant by microarray and plaque assay. We found that the microarray platform is comparable in precision and accuracy to plaque assay, and both protocols are linear over a 100- to 250-fold dynamic range (Fig. 5). Below this threshold, accurate microarray quantitation is problematic because of background fluorescence and a low signal-to-noise ratio. In plaque assays, this background signal reflects the presence of preexisting guanidine-resistant variants in “wild-type” quasispecies (12). Nevertheless, these data indicate that the barcode microarray provides a robust platform for accurate measurements of genome frequency within a complex population.
Normalization of microarray data and statistical analyses.
Data normalization allows comparisons within and between experiments and is a key element of microarray data analysis (33). It corrects for nonbiological sources of error, including unequal quantities of RNA or ssPCR-amplified product, bias in labeling or detection efficiency, and differences in loading. Many standard techniques available in commercial platforms rely on total-intensity-based normalization, in which one assumes that the average fold change across all samples is equal to 1. This assumption is not valid in many viral-evolution experiments, where bottlenecks, population expansions, and selective events may drastically alter the total population size. Because our PCR amplification step equalizes the total numbers of barcoded genomes sampled from the two different populations, competitive hybridization on our barcode microarray reveals changes in the relative rather than the absolute frequency of a given subpopulation within the larger quasispecies. As a result, a high fitness variant that increases its absolute number from one passage to the next will exhibit no change in our assay if the total population increases by a corresponding amount.
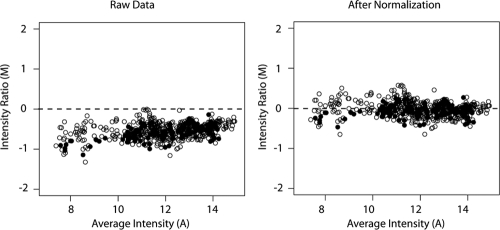
We addressed this issue by designing a normalization protocol based on the addition of external “spike-in” controls (23, 43). Changes in the relative frequency of these controls can be used to correct for total population size, thereby converting measurements of relative frequency to estimates of changes in absolute frequency. The spike-in controls are a set of 12 wild-type poliovirus clones that each have a unique pair of barcodes in the locations defined above. The titers of viruses derived from these plasmids were determined, and the viruses were diluted and pooled so that each of the 12 stocks was represented in a 2-fold serial dilution series spanning a 4,096-fold dynamic range (e.g., 212). Defined volumes of this pool were added to each viral supernatant prior to sample processing. Because the absolute numbers of barcodes amplified from the two samples are essentially equal, the relative frequency of spike-in barcodes is inversely proportional to the starting population size. In each experiment, we derived a standard curve from the signal on the spike-in control spots, which was used to normalize the fluorescence signal on the experimental spots (Fig. 6). Pilot experiments and dye flips with technical replicates from the same sample indicate that this normalization protocol was reproducible and accurate.
Fig. 6.
Normalization of microarray data using spike-in pools. Equal volumes of a spike-in pool of 12 barcoded viruses were mixed with two aliquots of the same barcoded viral stock. The two samples were labeled with either Cy3 or Cy5 as described in the legend to Fig. 5 and hybridized to the same microarray. The intensity ratio (M; log2Cy3 − log2Cy5) for each array element is plotted against its average intensity [A; (log2Cy3 + log2Cy5)/2]. Spots corresponding to spike-in viruses are shown as filled circles, and spots corresponding to experimental samples are shown as open circles.
Measuring viral fitness by competitive hybridization.
Experimental approaches to viral evolution rely heavily on fitness measurements for inferences about the adaptive process and its molecular correlates. Direct-competition assays, in which a given variant is competed against a marked reference strain, measure relative fitness, and serial passage allows precise measurement of small fitness differences (31, 37). We used fitness assays as an approach to validate the performance of our microarray platform. As in our initial pilot studies, we used a barcoded guanidine-resistant strain to distinguish this variant by both plaque assay and microarray. We competed the strain, 5′ 182/3′ 202-C4605U, against a library of 40 barcoded wild-type variants. The competitions were carried out in the absence of drug. By measuring the decline of the C4605U variant relative to the wild-type competitors, we were able to determine the fitness cost of the drug resistance mutation.
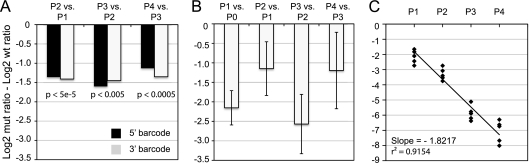
We mixed the 5′ 182/3′ 202-C4605U mutant with the WT40 library at a 5:1 starting ratio and infected five replicate dishes of HeLa cells at a multiplicity of 1. As in other competition experiments, an excess of the less fit mutant strain was used to ensure its presence in the population over multiple passages (31, 37). We attempted to limit coinfection and associated recombination within the population by using an MOI of ≤1 and limiting each of the four passages to the time required for one infectious cycle. Given the burst size of poliovirus, the number of progeny far exceeds the remaining input virus (10, 24, 26). For each sample, we measured the relative frequencies of barcodes 182 and 202 by microarray. We also compared our array measurements of fitness to titer-based determinations by plaque assay in the presence and absence of drug. In both assays, we found a steady and consistent decline in the C4605U drug-resistant variant relative to the wild type. A linear regression of microarray data from the five biological replicates measured the relative fitness of this variant as 0.38 to 0.39 over the first two passages with good concordance between the data from the 5′ and 3′ barcodes (Fig. 7A). There was more variability at later passages, with fitness estimates of 0.32 to 0.45. Close inspection of the data suggests that some of this variation is attributable to differences in the reference population used for each experiment. Because the decline of the guanidine-resistant variant after two passages exceeded the dynamic range of the array, populations at passages 3 and 4 were compared to that at passage 2, as opposed to the more uniform P0 population. As before, we found that there was good agreement between the microarray measurements and plaque titers. On a per-passage basis, we measured the relative fitness of the guanidine-resistant variants by plaque assay as 0.45, 0.17, and 0.44 at passages 2,3, and 4, respectively (Fig. 7B). Because of the larger variability in titer across biological replicates, we also estimated fitness as a function of passage by fitting a standard curve through all data points (Fig. 7C). This estimate, 0.28, is consistent with the per-passage data and is in good agreement with the microarray data. These data confirm the reliability of the microarray platform, especially given the inherent ambiguities of comparing relative frequencies of viral genomes to measurements of infectious particles.
Fig. 7.
Measurements of viral fitness in a competition assay. A double-barcoded guanidine-resistant virus (5′ U182/3′ U202) was mixed with the WT40 library and serially passaged on HeLa cells in quintuplicate at an MOI of 1. Samples from each passage and replicate were recovered for titering by plaque assay and microarray analysis. Fitness was measured as a change in the ratio of the mutant (mut) in the population over time relative to the change in the ratio of the wild type (wt). This concept is expressed mathematically as log [NM(t)/NM(t0)] = w × log [NW(t)/NW(t0)], where NM(t) and NW(t0) represent the titer of the mutant and wild type, respectively, at the beginning of the infection (t0) and time in passage postinfection. Therefore, fitness corresponds to the difference between the log2 mutant ratio and the log2 wild-type ratio, which is shown on the y axis. (A) Microarray analysis of the fold change in the guanidine-resistant mutant relative to the median of measurements derived from 40 wild-type viruses. The data are derived from linear regression across all five biological replicates at each passage. P values, corrected for multiple-hypothesis testing, are shown. Measurements derived from the 5′ and 3′ barcodes are shown in black and light gray, respectively. (B) Measurement of fitness by plaque assay. Titers of the guanidine-resistant variant and wild-type reference were measured by plaque assay in 2 mM guanidine or no drug, respectively. The data are expressed at each passage with means ± standard deviations for the five biological replicates. For comparison to the microarray, the data are expressed in log2 scale. (C) As in panel B, except the trend line is fitted through data from all five biological replicates and all passages. The slope of this regression reflects the relative fitness according to the equation above.
Suitability for in vivo studies and demonstration of a stochastic CNS bottleneck.
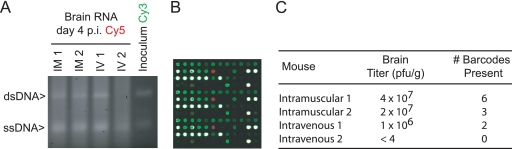
The population dynamics of viral infection in infected hosts are complex, and recent studies suggest that bottleneck events and population segregation may play a significant role in viral evolution and pathogenesis (4, 35, 45, 50). Tagging strategies are useful for these studies, as the barcodes function as lineage markers for each subpopulation. We tested the utility of our platform for in vivo studies of viral pathogenesis using a murine model of poliovirus infection. Work in this system suggests that multiple host bottleneck events restrict population diversity prior to central nervous system (CNS) invasion (35, 45). For our in vivo studies, we used a wild-type population consisting of equal quantities of 96 identical subpopulations, each with a unique 5′ barcode (WT96 library; see Materials and Methods). Mice were infected either intramuscularly or intravenously with 108 PFU of the library, the median lethal dose. High titers were achieved in a majority of the mice (Fig. 8), and the RT-ssPCR protocol was successful at amplifying barcode sequences from a variety of tissues for subsequent microarray analysis. We used the array as a semiquantitative tool to identify the origins of the viral populations in each tissue. Subpopulations were identified as present in the tissue if they increased in relative frequency compared to the inoculum and absent if they decreased. In all cases, the signal for present and absent barcodes differed by several orders of magnitude. Using this method, we observed a stringent CNS bottleneck with at most 6 out of the 96 subpopulations able to access the brain. Importantly, the specific barcodes identified in the brain were different for each mouse. This stochastic result is expected given that each subpopulation of 106 PFU in the library is essentially isogenic, except the selectively neutral barcode. These results highlight the power of our platform for quantifying host bottlenecks and its utility for in vivo studies of viral evolution.
Fig. 8.
Analysis of population segregation in vivo. Mice were infected in pairs, either intramuscularly (IM) or intravenously (IV), with the 5′-barcoded library WT96 at the median lethal dose. They were euthanized 4 days postinfection. (A) RT-ssPCR with Cy-labeled primers of total RNA from brain homogenates and the inoculum. (B) Sample microarray showing a single-barcoded variant (red spots; in triplicate) present in brain tissue compared to other barcodes in the inoculum, which were not isolated from the brain (green). Also shown are control spots, which are saturated in this image (white). (C) Viral titer and number of barcodes present in the brain of each mouse. No signs of infection were observed in the IV 2 mouse.
Tracking subpopulations within an evolving quasispecies.
Quasispecies theory suggests that the genetic architecture of the viral population determines its phenotype. We examined the ability of our system to measure such changes by generating an artificial quasispecies composed of 48 distinct subpopulations. We derived a highly diverse population by passaging a wild-type population in 400 μM ribavirin, an RNA mutagen that increases the viral mutation rate to roughly 8 mutations per genome (10). The vast majority of variants in this population harbored at least one lethal mutation, as we observed a 100-fold reduction in titer after a single passage. Array-based resequencing of this population further suggested that the viable fraction also had an increased mutational load compared to the wild type (65). We cloned unique, viable variants from the population by endpoint dilution after a single drug passage and barcoded them by RT-PCR. Forty-eight of these barcoded clones were pooled, and the corresponding viral population was passaged on HeLa cells. Sequence analysis of the P1 population of 48 clones indicate that our sample of the ribavirin-treated population has an increased mutational load compared to the wild type, with a strong statistical trend (Table 1 ). These barcoded variants represent a region of sequence space more distant from the master sequence than variants in standard wild-type populations.
Table 1.
Mutational loads of populations
| Population | No. of clones sequenced | No. of unique clonesa | No. of nucleotides sequenced | Total no. of mutations | No. of mutations per genomeb |
|---|---|---|---|---|---|
| Mutant pool | 62 | 32 | 73,822 | 67 | 6.75 |
| Wild type | 35 | 17 | 41,474 | 26 | 4.66 |
Only clones with unambiguous sequences and ≥1 mutation over a 1,240-nucleotide region (capsid region; positions 1353 to 2592).
Based on a 7,440-nucleotide genome. P = 0.1210 for all clones, and P = 0.1239 for unique clones by the Mann-Whitney U test.
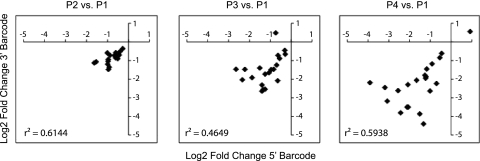
We used the microarray to measure changes in frequency of each barcoded subpopulation at every passage. As mentioned above, linear regression of microarray data from the five biological replicates measured was used to estimate the relative fitness of each variant. We measured a statistically significant change in the relative frequencies of 23 out of the 48 variants based on analyses of both barcodes (Fig. 9A; see Table S4 in the supplemental material). Among these 23 mutants, the vast majority encoded changes with deleterious effects. After two passages, the relative fitness ranged from 0.34 to 0.83. The relative fitness of the deleterious variants over four passages ranged from 0.09 to 0.73. Although we were able to distinguish minor fitness differences after just one passage, the larger effects observed after multiple passages resulted in more statistical power. We found reasonable concordance in fitness measurements obtained by the 5′ and 3′ barcodes for each variant (Fig. 10). This was generally poorer than expected. In most cases, the differences were small and could be due to small differences in the discriminatory power or detection threshold for individual barcodes. Similar effects have been documented in yeast, where in some cases only one of two barcodes provides usable data (67). Although we tried to limit each passage to one replication cycle, we cannot exclude the possibility of recombination and breakdown of barcode linkage, particularly at later passages.
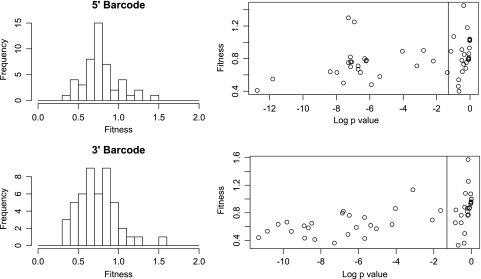
Fig. 9.
Fitness distribution of a randomly mutagenized viral population. Fitness was quantified as the slope of the linear regression of the relative barcode frequency over multiple passages, as in Fig. 7. The 5′ and 3′ barcodes were analyzed independently, and data are presented for all barcodes, regardless of the statistical significance. The histograms (left) show the distribution of fitness values for all 48 barcoded variants derived from a mutagenized population. The scatterplots (right) show the fitness values for each variant plotted against the log of the P value. The vertical lines at −1.3 correspond to a P value of 0.05; values to the left of these lines were statistically significant after correction for multiple-hypothesis testing.
Fig. 10.
Mutant passage. Shown is the correlation between the fold changes measured by the 5′ barcode (x axis) and 3′ barcode (y axis) for each barcode pair. The passages in each analysis are shown above each plot. The data are from Table 1 and were analyzed as in Fig. 9.
Fitness values for the remaining 25 barcoded variants in the population were closer to wild type and did not achieve statistical significance. While discordant results across the five biological replicates did play a role, many of these 25 variants exhibited only minor fitness defects (relative fitness, 0.85 to 1) that were difficult to resolve from a wild-type reference (Fig. 9B). This result suggests that poliovirus inhabits a neutral region of sequence space where the majority of mutations do not lead to significant fitness effects. These data extend previous studies from our laboratory, where we found that many of the viable mutants isolated following ribavirin passage encode synonymous mutations with minor fitness effects (65). Together, our data demonstrate the power of our microarray system for tracking viral subpopulations within an evolving quasispecies.
DISCUSSION
The field of population genetics has long relied on various marker-based strategies to make inferences about fitness and genetic variation within complex populations (29). Experimental approaches to RNA virus evolution have generally been limited by the paucity of neutral genetic markers for given variants or subpopulations. We applied a signature-tagging approach to this problem and developed a high-throughput microarray platform capable of tracking distinct subpopulations within a larger viral quasispecies. Because existing libraries of computationally optimized barcodes have over 9,000 such sequences, our scalable assay is well suited to studies of RNA virus population structure. The barcode amplification and hybridization protocols described here provide accurate and precise measurements over a 100-fold dynamic range. By using spike-in controls and loess-based normalization, we are able to determine relative frequency in populations that differ in absolute titer (60). We found that array-based measurements of viral fitness closely mirrored those derived from the viral titer and quantitated the fitness of a large number of mutants replicating as a mixed population. Experimental infections in a transgenic-mouse model demonstrate the utility of our platform for in vivo studies.
A system for studying viral populations.
The primary objective of this project was to develop an experimental platform for characterizing the complex genetic architecture of RNA virus populations. The application of quasispecies theory to the study of pathogenesis and antiviral resistance has been limited by the relatively poor sensitivity of sequence-based approaches for population level changes (discussed in reference 65). Conventional sequence analysis of a viral quasispecies will give a consensus sequence, which is essentially a weighted average of all the sequences present in the population and is not necessarily present as a single genome (15). While newer “deep-sequencing” technologies have revolutionized experimental approaches to complex systems, the reported error rate of 0.5 to 1% means that they can only reliably discern the sequence of variants present at frequencies of greater than 1% in a population that may number up to a billion (34). The short read length of many next-generation sequencing platforms also provides little information on linkage among mutations or overall haplotype. Finally, the current cost of such studies makes it difficult to perform replicate experiments on the scale proposed here.
Our platform addresses many of these issues and could be a valuable complement to sequencing-based approaches. Whereas sequencing can describe the mutational spectrum of a population, the barcode microarray defines the relative frequencies of variants within the larger quasispecies. Each barcode identifier serves as a lineage marker for a given subpopulation and provides information on the associated genome, or haplotype. The barcodes may also function as molecular handles for subsequent PCR amplification and sequencing. Because thousands of markers are available, the array can theoretically track haplotypes present at a low frequency within the population. However, data from signature-tagged-mutagenesis studies suggests that pools with large numbers of marked strains (>1,000) may suffer from poorer sensitivity for rarer subpopulations that escape amplification and detection. Our system could also be used to track recombination between two genomes if markers are placed on opposite ends of the RNA. Although our pilot studies were insufficiently powered for measurements of recombination, the 5′ and 3′ barcodes maintained partial linkage. A similar multiple-marker approach could be used to characterize reassortment in segmented viruses or cross-packaging in retroviruses. Our system also has significant advantages in terms of overall cost. Because most institutions are equipped with basic microarray functionality, our assay is accessible to many small academic laboratories. Oligonucleotide synthesis is the main cost limitation. After setup, the recurring cost is mainly consumables related to RT-PCR amplification.
Application to experiments on viral evolution.
Evolutionary theory seeks to define how mutation, selection, recombination, and drift affect the adaptive process. The challenge for the experimentalist is to observe and measure these phenomena in vitro and in vivo. As illustrated here, our microarray platform is well suited to such experiments. Fitness measurements are common in experimental evolution, and most published assays are able to compare only a few candidates. In contrast, we were able to determine the fitness of a large number of variants in an artificial quasispecies, and the assay has the capacity to track hundreds or even thousands using the same protocol. This expanded scope and resolution should be useful in studies of RNA viruses where the degree to which minority variants influence the fitness and adaptability of a population is a largely unanswered question (13, 64). Our data also show that the barcode microarray assay is sufficiently sensitive for in vivo studies of population segregation and migration. Work from our laboratory suggests that virulence is related to quasispecies diversity and that positive interactions among members of a population can influence the infectious outcome (64). The spatial and temporal nature of these interactions is unclear, and the host environment is known to present a number of stringent bottlenecks that segregate viral populations. As in other systems, the size of a genetic bottleneck can be measured by applying a simple probabilistic model to microarray data from animal infections with heterogeneously barcoded wild-type viruses (4, 35, 45, 50). After several passages, each of these tagged wild-type viruses will give rise to a tagged population. Assuming similar mutant distributions among these barcoded populations, stochastic events will govern which barcoded variants access a given tissue. The number of distinct barcodes reflects the number of founders that transited the bottleneck. Data on population segregation and migration can also be used to construct models of in vivo population dynamics, as has been described for Salmonella spp. (28). Unfortunately, our assay was insufficiently quantitative to provide precise information about the frequency of viral recombination in vivo, and therefore, we relied on a single-marker strategy for these studies.
Poliovirus populations occupy a neutral space.
The characteristically low replicative fidelity of RNA viruses ensures that almost every genome contains at least one mutation compared to its parent (3, 17, 61). Viral populations have the capacity to explore vast regions of sequence space, and many have focused on the relationship between the mutation rate and adaptability. However, it is also clear that mutation presents a double-edged sword (18, 22). Because RNA viruses tend to have small and tightly organized genomes, many mutations have deleterious effects on viral fitness. For example, Sanjuán and colleagues have shown that the vast majority of point mutations in vesicular stomatitis virus negatively impact viral fitness (54, 55). Recent studies suggest that many RNA viruses have adapted to their low replicative fidelity by becoming “robust” to the effects of mutation (9, 21, 39, 53). In evolutionary models, mutational robustness refers to constancy of phenotype despite the occurrence of deleterious mutations. Although the mechanisms underlying viral mutational robustness are unclear, a viral population can be intrinsically robust if it occupies a neutral region of sequence space. Because this neutral space corresponds to flatter regions of the corresponding fitness landscape, quasispecies theory predicts that RNA viruses exhibit a phenomenon termed “survival of the flattest” (9, 53, 62, 66).
Our results with a mutagenized population of polioviruses are consistent with this model. Ribavirin is a potent viral mutagen and increases the basal mutation rate 4-fold at the concentrations used here (10). Each of our randomly selected variants would therefore be expected to encode several mutations compared to consensus sequence, and our sequence analysis of individual clones supports this assumption. Despite this mutational load, we found that only a minority exhibited significant fitness defects. Many of those that achieved statistical significance had relatively modest effects. These data complement recent studies from our laboratory, where we found that most of the accumulated mutations in this population are synonymous and therefore more likely to be selectively neutral (65). The size of the neutral space may also be strongly influenced by epistatic effects among individual mutations (21, 54). We are cautious, however, in the interpretation of our results. Because our experimental system does not provide information on lethal mutations, our description of the poliovirus fitness landscape is incomplete. Furthermore, the cloning and library construction process may have been biased against more highly mutated variants with low relative fitness.
The scope and complexity of neutral space has practical implications for the lethal mutagenesis of RNA viruses. A number of studies suggest that viral populations can be extinguished by treatment with mutagenic drugs (10, 12, 32, 36, 58). These efforts have been inspired by quasispecies theory, which predicts a mutational meltdown with loss of meaningful genetic information at supranormal mutation rates (5, 19). Experimental demonstration of a classic error catastrophe with loss of the master sequence has proven difficult, and many have argued for a distinction between lethal mutagenesis and error catastrophe (7). Indeed, careful studies of lethal mutagenesis by Grande-Pérez and colleagues have shown imperfect correlation between the mutational load and population extinction (27). Preextinction populations exhibit marked heterogeneity in both the locations and numbers of mutations per genome. Although our fitness measurements are derived from a relatively small population recovered after a single drug passage, they do suggest that heterogeneity in the mutational load within a population could complicate efforts at extinction through lethal mutagenesis. Without a true mutational meltdown, a less mutagenized preextinction population could serve as a reservoir for novel mutants that mediate antiviral resistance or immune escape (1, 42, 44, 48, 57). Larger-scale studies using our platform with poliovirus and other RNA viruses may shed more light on this important problem.
A scalable and versatile platform.
In conclusion, we have designed a barcode microarray platform suitable for a range of studies in RNA virus evolution. The experimental and analytical protocols presented here allow a significant amount of assay flexibility, as they work independently of the number of markers. Because our system relies on exogenous sequence tags rather than viral polymorphisms, it can easily be adapted to other viral systems. While the barcodes can exert small fitness effects and confound certain experiments in a general or cell-type-specific manner, these problems can be minimized by choosing a selectively neutral insertion site. Finally, the cost and ease of setup should make this approach widely accessible to the academic virology community.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Institute of Allergy and Infectious Disease to R.A. (R01 AI36178 and R01 AI40085) and from the NIAID (K08 AI081754-01) and the American Heart Association (0875026N) to A.S.L.
We thank Dwight Barnes for assistance with the mouse experiments, Dale Webster for assistance with microarray printing, Armin Hekele for helpful discussions, and members of the Andino laboratory for technical assistance and critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 2 February 2011.
REFERENCES
- 1. Arnold J. J., Vignuzzi M., Stone J. K., Andino R., Cameron C. E. 2005. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J. Biol. Chem. 280:25706–25716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnes D., Kunitomi M., Vignuzzi M., Saksela K., Andino R. 2008. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe 4:239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Batschelet E., Domingo E., Weissmann C. 1976. The proportion of revertant and mutant phage in a growing population, as a function of mutation and growth rate. Gene 1:27–32 [DOI] [PubMed] [Google Scholar]
- 4. Bergstrom C. T., McElhany P., Real L. A. 1999. Transmission bottlenecks as determinants of virulence in rapidly evolving pathogens. Proc. Natl. Acad. Sci. U. S. A. 96:5095–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biebricher C. K., Eigen M. 2005. The error threshold. Virus Res. 107:117–127 [DOI] [PubMed] [Google Scholar]
- 6. Biebricher C. K., Eigen M. 2006. What is a quasispecies? Curr. Top. Microbiol. Immunol. 299:1–31 [DOI] [PubMed] [Google Scholar]
- 7. Bull J. J., Sanjuán R., Wilke C. O. 2007. Theory of lethal mutagenesis for viruses. J. Virol. 81:2930–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carrasco P., Daròs J. A., Agudelo-Romero P., Elena S. F. 2007. A real-time RT-PCR assay for quantifying the fitness of tobacco etch virus in competition experiments. J. Virol. Methods 139:181–188 [DOI] [PubMed] [Google Scholar]
- 9. Codoñer F. M., Daros J. A., Sole R. V., Elena S. F. 2006. The fittest versus the flattest: experimental confirmation of the quasispecies effect with subviral pathogens. PLoS Pathog. 2:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crotty S., Cameron C. E., Andino R. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. U. S. A. 98:6895–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crotty S., Hix L., Sigal L. J., Andino R. 2002. Poliovirus pathogenesis in a new poliovirus receptor transgenic mouse model: age-dependent paralysis and a mucosal route of infection. J. Gen. Virol. 83:1707–1720 [DOI] [PubMed] [Google Scholar]
- 12. Crotty S., et al. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375–1379 [DOI] [PubMed] [Google Scholar]
- 13. de la Torre J. C., Holland J. J. 1990. RNA virus quasispecies populations can suppress vastly superior mutant progeny. J. Virol. 64:6278–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Domingo E., et al. 2006. Viruses as quasispecies: biological implications. Curr. Top. Microbiol. Immunol. 299:51–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Domingo E., Sabo D., Taniguchi T., Weissmann C. 1978. Nucleotide sequence heterogeneity of an RNA phage population. Cell 13:735–744 [DOI] [PubMed] [Google Scholar]
- 16. Domingo E., Wain-Hobson S. 2009. The 30th anniversary of quasispecies. Meeting on ‘Quasispecies: past, present and future’. EMBO Rep. 10:444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Drake J. W., Holland J. J. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 96:13910–13913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Duffy S., Shackelton L. A., Holmes E. C. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9:267–276 [DOI] [PubMed] [Google Scholar]
- 19. Eigen M. 2002. Error catastrophe and antiviral strategy. Proc. Natl. Acad. Sci. U. S. A. 99:13374–13376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eigen M. 1971. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 58:465–523 [DOI] [PubMed] [Google Scholar]
- 21. Elena S. F., Carrasco P., Daros J. A., Sanjuan R. 2006. Mechanisms of genetic robustness in RNA viruses. EMBO Rep. 7:168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elena S. F., Sanjuán R. 2005. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 79:11555–11558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fardin P., et al. 2007. Normalization of low-density microarray using external spike-in controls: analysis of macrophage cell lines expression profile. BMC Genomics 8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fields B. N., Knipe D. M., Howley P. M. 2007. Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 25. Fields B. N., Mahan Knipe D., Howley P. M. 2007. Fields virology, 5th ed., vol. 1, p. 3177 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 26. Geller R., Vignuzzi M., Andino R., Frydman J. 2007. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 21:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grande-Pérez A., Lázaro E., Lowenstein P., Domingo E., Manrubia S. C. 2005. Suppression of viral infectivity through lethal defection. Proc. Natl. Acad. Sci. U. S. A. 102:4448–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grant A. J., et al. 2008. Modelling within-host spatiotemporal dynamics of invasive bacterial disease. PLoS Biol. 6:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hartl D. L. 2000. A primer of population genetics, p. 305 Sinauer Associates, Sunderland, MA [Google Scholar]
- 30. Herold J., Andino R. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holland J. J., de la Torre J. C., Clarke D. K., Duarte E. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holland J. J., Domingo E., de la Torre J. C., Steinhauer D. A. 1990. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J. Virol. 64:3960–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holloway A. J., van Laar R. K., Tothill R. W., Bowtell D. D. 2002. Options available—from start to finish—for obtaining data from DNA microarrays II. Nat. Genet. 32(Suppl.):481–489 [DOI] [PubMed] [Google Scholar]
- 34. Holt R. A., Jones S. J. 2008. The new paradigm of flow cell sequencing. Genome Res. 18:839–846 [DOI] [PubMed] [Google Scholar]
- 35. Kuss S. K., Etheredge C. A., Pfeiffer J. K. 2008. Multiple host barriers restrict poliovirus trafficking in mice. PLoS Pathog. 4:e1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee C. H., et al. 1997. Negative effects of chemical mutagenesis on the adaptive behavior of vesicular stomatitis virus. J. Virol. 71:3636–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martínez M. A., et al. 1991. Fitness alteration of foot-and-mouth disease virus mutants: measurement of adaptability of viral quasispecies. J. Virol. 65:3954–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mazurkiewicz P., Tang C. M., Boone C., Holden D. W. 2006. Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nat. Rev. Genet. 7:929–939 [DOI] [PubMed] [Google Scholar]
- 39. Montville R., Froissart R., Remold S. K., Tenaillon O., Turner P. E. 2005. Evolution of mutational robustness in an RNA virus. PLoS Biol. 3:e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moya A., Elena S. F., Bracho A., Miralles R., Barrio E. 2000. The evolution of RNA viruses: a population genetics view. Proc. Natl. Acad. Sci. U. S. A. 97:6967–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moya A., Holmes E. C., González-Candelas F. 2004. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2:279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mulder L. C., Harari A., Simon V. 2008. Cytidine deamination induced HIV-1 drug resistance. Proc. Natl. Acad. Sci. U. S. A. 105:5501–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oshlack A., Emslie D., Corcoran L. M., Smyth G. K. 2007. Normalization of boutique two-color microarrays with a high proportion of differentially expressed probes. Genome Biol. 8:R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pfeiffer J. K., Kirkegaard K. 2003. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. U. S. A. 100:7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pfeiffer J. K., Kirkegaard K. 2006. Bottleneck-mediated quasispecies restriction during spread of an RNA virus from inoculation site to brain. Proc. Natl. Acad. Sci. U. S. A. 103:5520–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pfeiffer J. K., Kirkegaard K. 2005. Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog. 1:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pierce K. E., Sanchez J. A., Rice J. E., Wangh L. J. 2005. Linear-After-The-Exponential (LATE)-PCR: primer design criteria for high yields of specific single-stranded DNA and improved real-time detection. Proc. Natl. Acad. Sci. U. S. A. 102:8609–8614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pillai S. K., Wong J. K., Barbour J. D. 2008. Turning up the volume on mutational pressure: is more of a good thing always better? (A case study of HIV-1 Vif and APOBEC3). Retrovirology 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pincus S. E., Diamond D. C., Emini E. A., Wimmer E. 1986. Guanidine-selected mutants of poliovirus: mapping of point mutations to polypeptide 2C. J. Virol. 57:638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sacristán S., Malpica J. M., Fraile A., García-Arenal F. 2003. Estimation of population bottlenecks during systemic movement of tobacco mosaic virus in tobacco plants. J. Virol. 77:9906–9911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 52. Sanchez J. A., Pierce K. E., Rice J. E., Wangh L. J. 2004. Linear-after-the-exponential (LATE)-PCR: an advanced method of asymmetric PCR and its uses in quantitative real-time analysis. Proc. Natl. Acad. Sci. U. S. A. 101:1933–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sanjuán R., Cuevas J. M., Furio V., Holmes E. C., Moya A. 2007. Selection for robustness in mutagenized RNA viruses. PLoS Genet. 3:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sanjuán R., Moya A., Elena S. F. 2004. The contribution of epistasis to the architecture of fitness in an RNA virus. Proc. Natl. Acad. Sci. U. S. A. 101:15376–15379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sanjuán R., Moya A., Elena S. F. 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc. Natl. Acad. Sci. U. S. A. 101:8396–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sanz-Ramos M., Diaz-San Segundo F., Escarmis C., Domingo E., Sevilla N. 2008. Hidden virulence determinants in a viral quasispecies in vivo. J. Virol. 82:10465–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sierra M., et al. 2007. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J. Virol. 81:2012–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sierra S., Davila M., Lowenstein P. R., Domingo E. 2000. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J. Virol. 74:8316–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smith V., Botstein D., Brown P. O. 1995. Genetic footprinting: a genomic strategy for determining a gene's function given its sequence. Proc. Natl. Acad. Sci. U. S. A. 92:6479–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smyth G. K. 2005. Limma: linear models for microarray data, p. 397–420In Gentleman R., et al. (ed.), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY [Google Scholar]
- 61. Steinhauer D. A., de la Torre J. C., Meier E., Holland J. J. 1989. Extreme heterogeneity in populations of vesicular stomatitis virus. J. Virol. 63:2072–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Swetina J., Schuster P. 1982. Self-replication with errors. A model for polynucleotide replication. Biophys. Chem. 16:329–345 [DOI] [PubMed] [Google Scholar]
- 63. van Maarseveen N. M., et al. 2006. A novel real-time PCR assay to determine relative replication capacity for HIV-1 protease variants and/or reverse transcriptase variants. J. Virol. Methods 133:185–194 [DOI] [PubMed] [Google Scholar]
- 64. Vignuzzi M., Stone J. K., Arnold J. J., Cameron C. E., Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Webster D. R., et al. 2009. An enhanced single base extension technique for the analysis of complex viral populations. PLoS One 4:e7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wilke C. O., Wang J. L., Ofria C., Lenski R. E., Adami C. 2001. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature 412:331–333 [DOI] [PubMed] [Google Scholar]
- 67. Winzeler E. A., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906 [DOI] [PubMed] [Google Scholar]
- 68. Yuan D. S., et al. 2005. Improved microarray methods for profiling the Yeast Knockout strain collection. Nucleic Acids Res. 33:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.