Abstract
The proteasome is well known for its regulation of the cell cycle and degradation of mis-folded proteins, yet many of its functions are still unknown. We show that RPN11, a gene encoding a subunit of the regulatory cap of the proteasome, is required for UV-stimulated activation of Gcn4p target genes, but is dispensable for their activation by the general control pathway. We provide evidence that RPN11 functions downstream of RAS2, and show that mutation of two additional proteasome subunits results in identical phenotypes. Our analysis defines a novel function of the proteasome: regulation of the RAS- and AP-1 transcription factor-dependent UV resistance pathway.
Keywords: Proteasome, GCN4, AP-1, RAS, UV resistance
Ultraviolet (UV) radiation and other environmental stresses continuously damage DNA and other cellular components. Eukaryotes have evolved highly regulated systems to combat these assaults, and an essential component of all known resistance pathways is the ability to alter the patterns of gene expression in response to stress. At least two independent pathways exist in eukaryotes to resist damage caused by UV radiation. The best characterized of the two is the DNA damage–cell cycle checkpoint pathway that responds to DNA lesions, resulting in cell cycle arrest and the increased expression of DNA repair genes (Elledge 1996; Weinert 1998). The second, less characterized, pathway responds to the non-genomic effects of UV irradiation, and its signals are generated by cytoplasmic determinants (Devary et al. 1993; Radler-Pohl et al. 1993). In mammalian cells, this second response is dependent upon the membrane-bound Ha-Ras molecule, which in turn activates c-Jun N-terminal kinase (JNK), a component of a mitogen-activated protein (MAP) kinase cascade (Davis 1994; Dérijard et al. 1994; Karin 1998). Phosphorylation of c-Jun and ATF-2 increases their ability to activate transcription of various stress resistance genes (for review, see Davis 1994; Karin 1998).
As in higher eukaryotes, UV irradiation of the budding yeast Saccharomyces cerevisiae activates the AP-1-like transcription factor Gcn4p through a RAS2-dependent pathway (Engelberg et al. 1994). UV-mediated activation of Gcn4p occurs in the absence of GCN2 function and in cells expressing a constitutive version of GCN4 that escapes translational control (Engelberg et al. 1994); thus it is separate from the well characterized amino acid starvation response (Hinnebusch 1988; Dever et al. 1992). The only genes that are known to be required for this response are RAS2 and GCN4. Unfortunately, genes functioning downstream of RAS2 and upstream of GCN4 have not yet been identified.
A connection between protein degradation and the AP-1 transcription factor pathway has emerged with the identification of genes required for the function of AP-1 transcription factors in mediating stress responses. The Schizosaccharomyces pombe gene pad1+ (pap-dependent transcriptional activator 1) and its human homolog POH1 confer multidrug and UV resistance when overexpressed in their native organisms (Shimanuki et al. 1995; Spataro et al. 1997). Pad1+ is a positive regulator of pap1+ (pombe AP-1)-dependent transcription, and although its mechanism is unknown, it clearly does not do so by regulating the levels of Pap1 or its DNA-binding activity (Shimanuki et al. 1995). Biochemical analyses of Pad1+ and Poh1 indicate that they are regulatory subunits of the 26S proteasome (Spataro et al. 1997; Penney et al. 1998). Both pad1+ and POH1 are related to the S. cerevisiae gene RPN11/MPR1. Rpn11p is likewise a bona-fide subunit of the 26S proteasome (Glickman et al. 1998), and its gene was initially cloned for its role in maintaining mitochondrial function and cell cycle progression through the G2/M checkpoint (Rinaldi et al. 1998).
Here, we describe the characterization of RPN11, the gene of a regulatory subunit of the proteasome. Using mutant alleles of RPN11 and other proteasome subunit genes, we show that the proteasome is required for the UV- and methyl methanesulfonate (MMS)-induced activation of Gcn4p target genes, but not for their activation through the amino acid starvation pathway. We provide evidence that RPN11 acts downstream of RAS2 and upstream of GCN4. Our analysis has identified a novel function for the proteasome: the regulation of AP-1-like transcription factors in stress-mediated pathways.
Results and Discussion
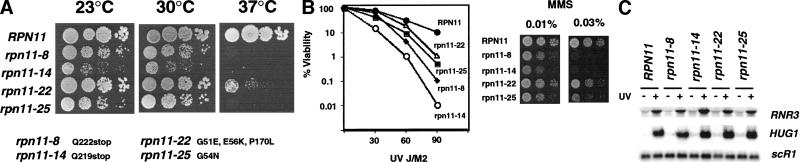
RPN11/TSG6/MPR1 was isolated as a gene-dosage suppressor of a TATA box binding protein associated factor (TAF) mutant, taf68-9 (Reese et al. 2000; J. Reese and M. Green, in prep). Recently, Rpn11p was verified to be a non-ATPase subunit of the 19S regulatory “cap” of the proteasome (Glickman et al. 1998) and has been implicated in controlling mitochondrial function and G2/M progression (Rinaldi et al. 1998). From this point on, we will refer to RPN11/TSG6/MPR1 as RPN11. In an attempt to characterize the potential transcriptional regulatory functions of RPN11, a screen for temperature-sensitive (ts-) mutants was performed and four mutants were chosen for characterization. Figure 1A shows the growth phenotypes of these mutants at 23°C, 30°C, and 37°C. Three of the four mutants (rpn11-8, rpn1-14, and rpn11-25) displayed growth defects at permissive temperatures and arrested growth at 37°C, whereas the rpn1-22 allele grew well at 23°C and 30°C and displayed a weaker ts- phenotype at 37°C.
Figure 1.
RPN11 is required for normal resistance to UV radiation (A). Isolation of temperature-sensitive mutants. Tenfold serial dilutions of cultures of YJR124-0 (RPN11), YJR124-8 (rpn11-8), YJR124-14 (rpn11-14), YJR124-22 (rpn11-22), YJR124-25 (rpn11-25) were spotted onto YPAD plates and incubated at 23°C, 30°C, or 37°C for 3 d. The amino acid substitutions in each allele are indicated below. (B) UV (left) and MMS (right) sensitivity of RPN11 mutants. For UV sensitivity studies, cells (YJR124) were grown in YPAD at 30°C until mid-log, and then appropriate dilutions were spread onto pre-warmed YPAD plates in triplicate. The plates were treated with the doses of UV radiation indicated in the figure and then incubated at 30°C in the dark. Sensitivity to MMS was measured on plates containing 0.01% and 0.03% MMS incubated at 30°C for 2 and 4 d, respectively. (C) Expression of RNR3 and HUG1 in RPN11 mutants. Cells were treated as described in Fig. 2A. RNR3 and HUG1 mRNA was detected by Northern blotting. ScR1 served as a loading control.
Because a previously identified mutation in RPN11 caused cell cycle arrest in G2/M at the restrictive temperature (mpr1-1, described in Rinaldi et al. 1998), the cell cycle phenotype of the mutants described here was examined by flow cytometry. Even at the permissive temperature (30°C), all four mutants displayed a significant accumulation of cells with a 2N DNA content, suggesting a delay in G2/M (data not shown). Consistent with a delay in G2/M, microscopic examination of the cells revealed that most contained a single large bud (not shown).
Overexpression of POH1 resulted in a measurable increase in the resistance of tissue culture cells to UV radiation (Spataro et al. 1997), indicating that it may regulate UV resistance pathways in mammals. We therefore examined the sensitivity of the RPN11 mutants to UV radiation. The results presented in Figure 1B show that all four mutants were significantly more sensitive to UV radiation compared to the wild-type strain. Moreover, the sensitivity of the mutants to UV radiation correlated with the severity of their growth defects (Fig. 1, cf. A and B). The two mutants that are truncated within the C terminus, rpn11–8 and rpn11–14, were approximately 100- and 1000-fold more sensitive than the wild type to UV, respectively. In contrast, the rpn11-22 and rpn11-25 alleles were less sensitive, displaying a roughly 10-fold increase in sensitivity. We next tested the sensitivity of the mutants to MMS, a DNA alkylating agent. Figure 1B shows that all strains grew at 30°C on plates containing 0.01% MMS, and that the rpn11-8 and rpn11-14 alleles failed to grow on medium containing 0.03% MMS. Neither the rpn11-22 nor the rpn11-25 allele was more sensitive than the wild-type strain, even at the maximal permissive concentration of MMS (0.05%, not shown). Moreover, despite showing some sensitivity to MMS, the rpn11-8 and rpn11-14 mutants were only threefold more sensitive to MMS; thus, they are significantly less sensitive to MMS compared to UV radiation.
To ascertain the functions of RPN11 in mediating the resistance to UV radiation, we examined the integrity of the two known UV resistance pathways in yeast. The first pathway is dependent upon the DNA damage cell cycle checkpoint genes and regulates the expression of DNA repair genes (Elledge et al. 1993; Kiser and Weinert 1996; Basrai et al. 1999). We examined the integrity of the DNA damage checkpoint pathway in the RPN11 mutants by monitoring the induction of two prototypical target genes whose expression is dependent upon checkpoint function, namely RNR3 and HUG1 (Elledge et al. 1993; Kiser and Weinert 1996; Basrai et al. 1999). The Northern blot presented in Figure 1C shows that the induction of RNR3 and HUG1 mRNA was not affected by mutations in RPN11, indicating that the DNA damage checkpoint pathways are not dependent upon RPN11.
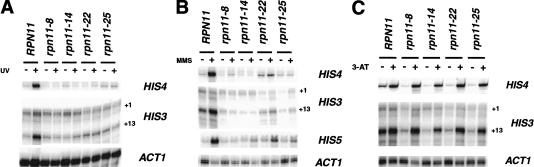
The second resistance pathway does not require the DNA damage checkpoint genes in yeast (J. Reese, unpubl.) or nuclear function in mammals (Devary et al. 1993; Radler-Pohl et al. 1993) and is mediated by RAS2 and the AP-1-like transcription factor Gcn4p (Engelberg et al. 1994). Given the genetic evidence implicating RPN11 in the regulation of AP-1-like transcription factors (Shiminuki et al. 1995), we examined the UV-induced expression of Gcn4p target genes in the RPN11 mutants. As reported in a previous study (Engelberg et al. 1994), the treatment of wild-type cells with UV radiation increased the expression of two well-characterized Gcn4p target genes, HIS3 and HIS4 (Fig. 2A). In contrast, UV-induced activation of both genes was abolished in all four RPN11 mutants even when the cells were maintained at a permissive temperature for growth, 30°C (Fig. 2A, lanes 4,6,8,10). Moreover, mutation of RPN11 resulted in the selective reduction in the transcript initiating at +13 (Fig. 2A), which is preferentially utilized by Gcn4p (Collart and Struhl 1993).
Figure 2.
RPN11 is required for the UV- and MMS-induced expression of Gcn4p target genes. (A) UV-induced expression of HIS3 and HIS4. Wild-type and mutant cells (JR493) were treated on plates with 70 J/m2 UV and returned to prewarmed YPAD for 75 min. HIS3 and ACT1 mRNAs were detected by S1 nuclease protection, and HIS4 mRNA by primer extension. +1 and +13 indicate the two major transcription start sites of HIS3. (B) MMS-induced induction of HIS3, HIS4, and HIS5. Cells were treated with (+) or without (−) 0.1% MMS for 1 h at 30°C. HIS5 mRNA was detected by Northern blotting. (C) Examination of HIS3 and HIS4 expression under amino acid starvation. Cells were grown at 30°C in liquid SC media (−) or in media lacking histidine and treated with 15 mM 3-AT for 60 min (+). RNA was analyzed as described in A.
A genome-wide analysis of transcription revealed that most Gcn4p-dependent genes are induced by MMS treatment (Jelinsky and Samson 1999). Therefore, we examined the induction of three representative genes using MMS as an inducing agent. Similar to what was observed upon UV irradiation, treatment of wild-type cells with 0.1% MMS caused a strong induction of HIS3 (+13) and HIS4 mRNA (Fig. 2B). Also consistent with the results shown in Figure 2A, MMS-induced expression of these genes was eliminated in all four mutants (Fig. 2B). In this experiment, the RPN11 mutants displayed reduced levels of the +13 transcript of HIS3, even in the absence of MMS treatment. This difference is likely caused by residual Gcn4p activity in the untreated wild-type cells, which is reduced in the mutants.
GCN4 was initially identified by its regulation of the amino acid starvation response, and most studies carried out to date have focused on this function (for review, see Hinnebusch 1988; Grant et al. 1998). Gcn4p is also activated by UV irradiation through a RAS2-dependent, but GCN2- and amino acid starvation–independent pathway (Engelberg et al. 1994). To determine whether RPN11 specifically regulates the UV response pathway, and whether it broadly affects GCN4 function, we examined the activation of HIS3 and HIS4 by inducing amino acid starvation conditions using 3-amino-1H-1,2,4-triazole (3-AT), a competitive inhibitor of the HIS3 gene product (Kanazawa et al. 1988). The data shown in Figure 2C show that amino acid starvation strongly induced HIS3 and HIS4 expression in the mutant strains; thus, the amino acid starvation pathway is intact in RPN11 mutants. These results clearly demonstrate that RPN11 is specifically required for the GCN4-dependent UV resistance pathway, and that it does not generally control GCN4 function.
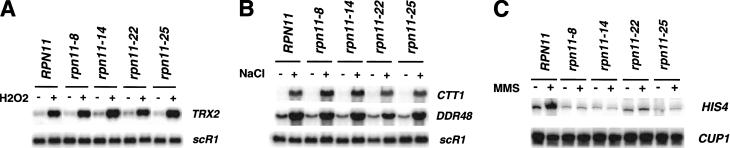
The activation of Gcn4p by UV irradiation requires RAS2 (Engelberg et al. 1994). To rule out the possibility that RPN11 simply regulates RAS activity, rather than specifically affecting GCN4 function, we examined the expression of three RAS2-dependent genes. Overexpression of RAS2 suppresses the transcription of CTT1, DDR48, TRX2 and heat shock genes, and conversely, mutation of RAS2 causes their constitutive expression (Stanhill et al. 1999). Therefore, if RPN11 controls RAS activity, changes in the expression of these genes would be evident in the mutants. The results shown in Figure 3 A and B demonstrate that the uninduced and stress-induced levels of transcription of TRX2, DDR48, and CTT1 were not affected in the RPN11 mutants. In addition to the data presented here, we also verified that the induction of CUP1 by copper sulfate and that of SSA4 by heat shock were unaffected in all four mutants (data not shown). These results indicate that RPN11 is not generally required for stress-induced transcription, and that it does not affect the activity of RAS2.
Figure 3.
RPN11 functions downstream of RAS2. (A,B) The expression of stress-induced genes is not affected by RPN11 mutations. Wild-type (JR124-0) and mutant strains were grown in YPAD at 30°C to an OD of 0.4 and then treated with 1 mM hydrogen peroxide (H2O2, A) or 1 M NaCl (B) for 90 min. RNA levels were measured by Northern blotting. (C) MMS-induced expression of HIS4 in strains overexpressing Ras2val19. Wild-type and mutant strains containing a high-copy number plasmid expressing the activated form of RAS2 (Whistler and Rine 1997) were grown in selective media and treated with 0.1% MMS for 1 h (+). RNA was analyzed as described in Fig. 2B.
Expression of an activated form of Ras2p, Ras2val19p, increased the expression of Gcn4-target genes (Engelberg et al. 1994). We used this observation to more definitively place RPN11 downstream of RAS2 by examining the expression of HIS4 in cells transformed with a high copy plasmid containing Ras2val19. Enhanced RAS activity did not suppress the ts- growth defects or UV sensitivity of the mutant strains (not shown). In addition, the data shown in Figure 3C clearly indicate that overexpression of this hyperactive form of RAS2 did not suppress the defects in HIS4 expression observed in the mutants. Collectively, the results presented in Figure 3 indicate that RPN11 functions downstream of RAS2, and that it is not generally required for stress-induced transcription.
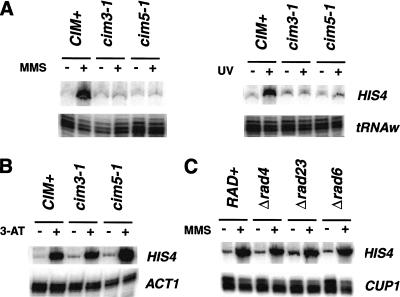
Next, we investigated whether RPN11 specifically regulates the RAS/AP-1-dependent UV resistance pathway, and whether the regulatory cap of the proteasome is required for this function. The 19S regulatory cap of the proteasome is composed of at least 11 subunits, each separated into one of two categories: ATPase and non-ATPase subunits (Glickman et al. 1998). RPN11 is a representative of the latter category. We examined the requirement of two ATPase domain-containing subunits, Cim3/Sug1/Rpt6 and Cim5/Rpt1 (Glickman et al. 1998) for the UV-mediated activation of Gcn4p. The cim3-1 and cim5-1 mutants are moderately sensitive to UV radiation (Schauber et al. 1998; data not shown) and accumulate in the G2/M phase of the cell cycle (Ghislain et al. 1993); thus, they are good candidates to analyze. As the results in Figure 4A show activation of HIS4 by either MMS or UV treatment is severely compromised in both the cim3-1 and cim5-1 strains. Moreover, similar to the RPN11 mutants, the cim3-1 and cim5-1 mutations had no effect on the starvation-induced activation of HIS4 expression (Fig. 4B). In fact, we reproducibly observed a slight increase in the accumulation of HIS4 mRNA in 3-AT-treated cim5-1 cells (see below). These results indicate that the regulatory cap of the proteasome, and not RPN11 specifically, is required for this function.
Figure 4.
The regulatory cap of the proteasome regulates Gcn4p activity independent of its nucleotide excision repair functions. (A). UV- and MMS-induced expression of HIS4. Wild-type (CIM+), cim3-1, and cim5-1 strains were treated with 0.1% MMS (left) or with 70 J/m2 UV (right) at 30°C and then processed. HIS4 mRNA was analyzed as described in Figure 2. (B) Starvation-induced expression of HIS4. Cells were grown to mid-log phase growth in SC medium at 30°C, collected by centrifugation and resuspended in SC (−) or SC minus histidine plus 15 mM 3-AT (+). After 60 min at 30°C the cultures were harvested. (C). Expression of HIS4 in RAD4, RAD6, and RAD23 mutants. Cells were treated as described in Fig. 2B.
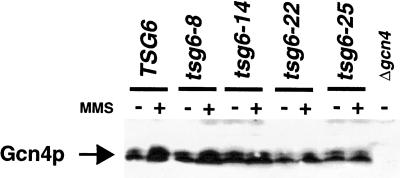
Rad23p and Rad4p, two components of the nucleotide excision repair system, associate with the regulatory cap of the proteasome (Schauber et al. 1998; Russell et al. 1999). It is possible that the defects in HIS4 transcription caused by mutations in proteasome subunits result from altered nucleotide excision repair function. To rule out this possibility, we analyzed the expression of HIS4 in strains containing a deletion of RAD23 and RAD4. The results shown in Figure 4C demonstrate that the stimulation of HIS4 by MMS is unaffected in these mutants; thus, the proteasome, and not the nucleotide excision repair system, is required for this function. In addition, we examined the induction of HIS4 transcription in a RAD6 mutant, a ubiquitin ligase required for normal resistance to UV and for the degradation of Gcn4p (Kornitzer et al. 1994). Because deletion of RAD6 did not affect HIS4 expression, the effects we observe in the RPN11 mutants are unlikely to result from the alteration of Gcn4p half-life. To directly demonstrate that the defects in HIS4 expression are not the result of altered Gcn4p levels, we immunoblotted for Gcn4p protein in the RPN11 mutants. Figure 5 shows that Gcn4p levels were not significantly different in the mutants versus the wild type, either prior to or after 60 min of MMS treatment.
Figure 5.
RPN11 mutations do not affect Gcn4p levels. Cells were grown under nonstarvation conditions in SC complete medium and treated (+) or not treated (−), with 0.1% MMS for 60 min at 30°C. Gcn4p was detected by immunoblotting using anti-Gcn4 antibodies. An extract from a Δgcn4 strain was loaded as a negative control.
RPN11 is specially required for the UV-mediated activation of Gcn4p, but not for general control. These defects are not explained by trivial changes in Gcn4p levels or turnover. We did not find significant differences in the levels of Gcn4p in the mutants compared to a wild-type strain. If protein turnover is the predominant mechanism, we predict that the expression of Gcn4p target genes would increase, and that the starvation response would likewise be affected in a similar fashion (Kornitzer et al. 1994; Meimoun et al. 2000). In fact, the half-life of Gcn4p is extended in a CIM5/RPT1 mutant (Kornitzer et al. 1994), and consistent with this, we observed a slight increase in HIS4 transcription in 3-AT-stimulated cim5-1 cells. In contrast, UV-stimulated expression is completely abolished in this mutant. Secondly, deletion of RAD6 (Fig. 3C), a gene that regulates Gcn4p half-life (Kornitzer et al. 1994), does not affect the MMS-induced activation of HIS4. Thus the defects we observed are not simply the result of changes in Gcn4p levels or turnover rates.
What is the function of the proteasome in this pathway? One possibility is that UV irradiation stimulates the RAS pathway, which in turn triggers the destruction of a negative regulator of Gcn4p through the proteasome. A negative regulator of GCN4 function has not been discovered; however, most analyses have centered on its translational regulation by GCN2 (for review, see Hinnebusch 1988). Our data, and those of Engelberg et al. (1994), clearly indicate that two distinct pathways activate Gcn4p. The advantage of an alternate mechanism is that the activation of Gcn4p target genes can occur rapidly to repair damaged cellular compartments prior to the next cell division and can function independently of amino acid availability. The mechanism may be analogous to the regulation of NF-κB in mammalian cells. Like AP-1 transcription factors, NF-κB is stimulated by UV irradiation through the RAS pathway in mammalian cells (Devary et al. 1993). Ubiquitination and proteasome-mediated destruction of IκB, an inhibitor of NF-κB, are required to activate NF-κB-dependent genes (Winston et al. 1999). Alternatively, the proteasome degrades an inhibitor of the MAP kinase pathway that activates Gcn4p under UV stress. Treatment of mammalian cells with proteasome inhibitors prevents the degradation of MAP kinase phosphatase (Brondello et al. 1999), an antagonist of the MAP kinase pathway. Likewise in yeast, the proteasome may degrade a phosphatase (or other type of inhibitor) that antagonizes the actions of the MAP kinase pathway. At this time, the MAP kinase genes that activate Gcn4p in S. cerevisiae are not known, but future studies aimed in this direction will help illuminate the functions of the proteasome in the RAS-dependent UV resistance pathway.
RPN11 certainly has functions in UV resistance in addition to regulating the RAS-AP-1 response. This is demonstrated by the effect of each allele on the expression of Gcn4p-dependent genes versus their degrees of sensitivity to UV. All four mutants displayed strong effects on the expression of HIS3 and HIS4, and yet the UV sensitivities varied as much as 100-fold among the alleles. Our observations cannot be explained by the reduced expression of Gcn4p-dependent genes not tested, because gcn4 or ras2 mutants are only threefold more sensitive to UV radiation compared to wild-type strains (Engelberg et al. 1994), whereas mutation of RPN11 causes as much as a 1000-fold increase. The proteasome associates with Rad23 and Rad4, two components of the nucleotide excision repair system (Schauber et al. 1998; Russell et al. 1999). Mutation of RPN11 may affect the association of the Rad23p/Rad4 complex with the proteasome, independent of its function in regulating the UV response. An alternate, but not mutually exclusive, explanation for the more severe phenotype of the mutants is that RPN11 controls the activity of other transcription factors involved in stress resistance. Genetic evidence suggests that it is another AP-1-like transcription factor (Shimanuki et al. 1995). S. cerevisiae have at least 8 bZip transcription factors (YAPs) that have a high degree of sequence homology to Gcn4p (Fernandes et al. 1997), and the functions of these genes have yet to be elucidated. Additional studies such as genome-wide microarray expression experiments and genetic screens using RPN11 mutants will hopefully identify additional targets of the proteasome that will increase our understanding of its functions in UV and drug resistance.
Materials and methods
Isolation of temperature-sensitive mutants of RPN11
Temperature sensitive mutants of RPN11 were isolated by the plasmid shuffle technique by introducing a pool of hydroxylamine-treated pRS414-RPN11 (Guthrie and Fink 1991) into YJR124–0 (Mat a; ΔRPN11:hisg; ade2-11; his3-11,15; leu2-3,112; ura3-1; trp1-1; can1-100; [RS426-RPN11], a W303 background). Tryptophan prototrophs were replica-plated onto 5-fluoroorotic acid-containing media and placed at 23°C and 37°C, and mutants that supported growth at 23°C, but not 37°C, were isolated for further analysis. Mutants that ceased growth at ≤2 doubling times were sequenced and subjected to additional analysis. The YJR493 background (HIS3+) was constructed by transforming the YJR124 strains with NsiI-digested pRS403. The cim3-1 and cim5-1 temperature-sensitive mutants (Ghislain et al. 1993) were described in previous publications. The RAD deletion strains were purchased from Research Genetics.
Growth and phenotypic analysis of yeast strains
Cells were typically grown in YPD supplemented with 20 μg/mL adenine sulfate (YPAD) or in the appropriate synthetic medium (Guthrie and Fink 1991) at 30°C unless indicated otherwise. The temperature and drug sensitivity phenotypes of RPN11 mutants were verified by spotting 10-fold serial dilutions of growing cells onto YPAD agar plates supplemented with the agents indicated in the figure legends. For UV sensitivity studies, 200 μL of the appropriate serial dilutions made from log-phase cultures were spread in triplicate onto YPAD agar plates. Each of the five plates was irradiated with 0 J/m2, 30 J/m2, 60 J/m2, or 90 J/m2 of ultraviolet radiation, respectively, and immediately placed in the dark at 30°C. Colonies were counted on each plate, and percent viability was calculated by dividing the number of colonies on irradiated plates by the number of colonies on the untreated plates.
RNA isolation and analysis
Strains were typically grown in YPAD at 30°C until an OD of 0.4, treated in culture with the agents indicated in the figures legends, collected by centrifugation and used for RNA isolation as described previously (Apone et al. 1996; Walker et al. 1996). The detection of specific messages was performed by S1 nuclease protection (Cormack and Struhl 1992), primer extension analysis (Engelberg et al. 1994) or Northern blotting (Sambrook et al. 1989). The 3-amino-1H-1,2,4-triazole (3-AT) induction experiments were conducted as described in Natarajan et al. (1998). Cells were treated with UV on solid media as described previously (Kiser and Wienert 1996) and returned to pre-warmed YPAD for 60–90 min prior to collection and RNA isolation.
Acknowledgments
The authors are grateful to Carl Mann, Linda Breeden, Shelley Berger and Fred Winston for yeast strains used in these studies and those not shown. We also thank Chris Norbury for sequencing the RPN11 mutants and comments on this paper, and Jasper Rhine for the plasmids expressing the RAS derivatives. We acknowledge Alan Hinnebusch for antibodies and plasmids used in this study and Jerry Workman and Patrick Grant for advice on SAGA purification and comments on this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jcr8@psu.edu; FAX (814) 863-7024.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.863801.
References
- Apone LM, Virbasius CM, Reese JC, Green MR. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes & Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- Basrai MA, Velculescu VE, Kinzler KW, Hieter P. NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7041–7049. doi: 10.1128/mcb.19.10.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondello JM, Pouyssegur J, McKenzie FR. Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science. 1999;286:2514–2517. doi: 10.1126/science.286.5449.2514. [DOI] [PubMed] [Google Scholar]
- Collart MA, Struhl K. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 1993;12:177–186. doi: 10.1002/j.1460-2075.1993.tb05643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- Davis RJ. MAPKs: New JNK expands the group. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Dérijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ. JNK1: A protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- Devary Y, Rosette C, DiDonato JA, Karin M. NF-κB activation by ultraviolet light is not dependent on a nuclear signal. Science. 1993;261:1442–1445. doi: 10.1126/science.8367725. [DOI] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translation control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB, Navas TA. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: Preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Engelberg DE, Klein C, Martinetto H, Struhl K, Karin M. The UV response involving the Ras signalling pathway and AP-1 transcription factors is conserved between yeast and mammals. Cell. 1994;77:381–390. doi: 10.1016/0092-8674(94)90153-8. [DOI] [PubMed] [Google Scholar]
- Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Sterner DE, Duggan LJ, Workman JL, Berger SL. The SAGA unfolds: Convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. New York, NY: Academic Press; 1991. [Google Scholar]
- Hinnebusch AG. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988;52:248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa S, Driscoll M, Struhl K. ATR1, a Saccharomyces cerevisiae gene encoding a transmembrane protein required for aminotriazole resistance. Mol Cell Biol. 1988;8:664–673. doi: 10.1128/mcb.8.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. Mitogen-activated protein kinase cascades as regulators of stress responses. Annals NY Acad Sci. 1998;851:139–146. doi: 10.1111/j.1749-6632.1998.tb08987.x. [DOI] [PubMed] [Google Scholar]
- Kiser GL, Weinert TA. Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol Biol Cell. 1996;7:703–718. doi: 10.1091/mbc.7.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornitzer D, Raboy B, Kulka RG, Fink GR. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meimoun A, Holtzman T, Weissman Z, McBride HJ, Stillman DJ, Fink GR, Kornitzer D. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCFCDC4 ubiquitin–ligase complex. Mol Biol Cell. 2000;11:915–927. doi: 10.1091/mbc.11.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Jackson BM, Rhee E, Hinnebusch AG. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol Cell. 1998;2:683–692. doi: 10.1016/s1097-2765(00)80166-5. [DOI] [PubMed] [Google Scholar]
- Penney M, Wilkinson C, Wallace M, Javerzat JP, Ferrell K, Seeger M, Dubiel W, McKay S, Allshire R, Gordon C. The Pad1+ gene encodes a subunit of the 26S proteasome in fission yeast. J Biol Chem. 1998;273:23938–23945. doi: 10.1074/jbc.273.37.23938. [DOI] [PubMed] [Google Scholar]
- Radler-Pohl A, Sachsenmaier C, Gebel S, Auer H-P, Bruder JT, Rapp U, Angel P, Rahmsdorf HJ, Herrlich P. UV-induced activation of AP-1 involves obligatory extranuclear steps including Raf-1 kinase. EMBO J. 1993;12:1005–1012. doi: 10.1002/j.1460-2075.1993.tb05741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese JC, Zhang ZJ, Kurpad H. Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J Biol Chem. 2000;275:17391–17398. doi: 10.1074/jbc.M001635200. [DOI] [PubMed] [Google Scholar]
- Rinaldi T, Ricci C, Porro D, Bolotin-Fukuhara M, Frontali L. A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces a cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol Biol Cell. 1998;9:2917–2931. doi: 10.1091/mbc.9.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Reed SH, Huang W, Friedberg EC, Johnston SA. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol Cell. 1999;3:687–695. doi: 10.1016/s1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- Shimanuki M, Saka Y, Yanagida M, Toda T. A novel essential fission yeast gene pad1+ positively regulates pap1+-dependent transcription and is implicated in the maintenance of chromosome structure. J Cell Sci. 1995;108:569–579. doi: 10.1242/jcs.108.2.569. [DOI] [PubMed] [Google Scholar]
- Spataro V, Toda T, Craig R, Seeger M, Dubiel W, Harris AL, Norbury C. Resistance to diverse drugs and ultraviolet light conferred by overexpression of a novel human 26S proteasome subunit. J Biol Chem. 1997;272:30470–30475. doi: 10.1074/jbc.272.48.30470. [DOI] [PubMed] [Google Scholar]
- Stanhill A, Schick N, Engelberg D. The yeast ras/cyclic AMP pathway induces invasive growth by suppressing the cellular stress response. Mol Cell Biol. 1999;19:7529–7538. doi: 10.1128/mcb.19.11.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SS, Reese JC, Apone LM, Green MR. Transcription activation in cells lacking TAFIIS. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- Weinert T. DNA damage checkpoints update: Getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- Whistler JL, Rine J. Ras2 and Ras1 protein phosphorylation in Saccharomyces cerevisiae. J. Biol. Chem. 1997;272:18790–18800. doi: 10.1074/jbc.272.30.18790. [DOI] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCF beta–TRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in I kappa B alpha and beta-catenin and stimulates I kappa B alpha ubiquitination in vitro. Genes & Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]