Abstract
Silencing of the cryptic mating-type locus HMR requires recognition of a small DNA sequence element, the HMR-E silencer, by the Sir1p, one of four Sir proteins required for the assembly of silenced chromatin domains in Saccharomyces cerevisiae. The Sir1p recognizes the silencer through interactions with the origin recognition complex (ORC), a protein complex that binds the silencer DNA directly. Sir1p was physically associated with HMR in chromatin, and this association required a Sir1p–ORC interaction, suggesting that it reflected the Sir1p silencer-recognition function required for silencing. Sir1p was not associated with nonsilencer replication origins that bind the ORC, indicating that a Sir1p–ORC interaction is confined to silencers. Significantly, the other SIR genes were required for Sir1p's association with HMR. Thus, multiple protein contacts required for and unique to silent chromatin may confine a Sir1p–ORC interaction to silencers. The Sir1p was present at extremely low concentrations in yeast cells yet was associated with HMR at all stages of the cell cycle examined. These data provide insights into the mechanisms that establish and restrict the assembly of silenced chromatin to only a few discrete chromosomal domains.
Keywords: Silencing, Sir, ORC, yeast, chromatin
Silencing is a form of transcriptional repression in yeast that requires the assembly of a repressive chromatin structure analogous to heterochromatin in multicellular eukaryotes (Hendrich and Willard 1995; Pillus and Grunstein 1995). Both heterochromatic and silenced domains contain relatively hypoacetylated histones that, in combination with specialized nonhistone chromatin-associated proteins, form a repressive chromatin structure recalcitrant to a variety of activities requiring access to the chromosomal DNA. These specialized chromatin domains can transcriptionally repress genes contained within them, but they also can influence several other aspects of DNA metabolism including DNA replication, recombination, and repair and chromosome stability (Gottlieb and Esposito 1989; Terleth et al. 1989, 1992; Freeman-Cook et al. 1999; Stevenson and Gottschling 1999; Henikoff 2000). Histone modifications and nonhistone proteins required for the assembly and maintenance of these specialized chromatin structures have been identified (Eissenberg et al. 1995; Hendrich and Willard 1995; Pillus and Grunstein 1995), but the mechanisms that target and confine the formation of repressive chromatin to specific domains within the eukaryotic genome are unclear.
Silencing of the cryptic mating-type loci, HMR and HML, in the yeast Saccharomyces cerevisiae is an example of the role repressive chromatin structures play in blocking transcription. Silencing of HMR and HML is not required for yeast cell viability but is important in regulating haploid mating types by repressing transcription of cryptic copies of mating-type genes present at HMR and HML (Herskowitz et al. 1992). Silencing at both loci requires the combined action of small DNA elements (∼150 bp) called silencers, several multifunctional DNA-binding proteins that bind silencers directly (silencer-binding proteins), and the four specialized silencing-specific Sir (Silent Information Regulator) proteins (Loo and Rine 1995). The silencer-binding proteins are proposed to nucleate the assembly of a Sir protein complex(es) that encompass ∼4-kb pairs of chromosomal DNA at the HMR and HML loci (Loo and Rine 1994; Triolo and Sternglanz 1996; Donze et al. 1999).
The recognition of the HMR-E silencer by the Sir1 protein is proposed to be a key step in targeting the assembly of silenced chromatin to HMR. The HMR-E silencer contains binding sites for three multifunctional nuclear proteins: the origin recognition complex (ORC), Rap1p, and Abf1p. The ORC must bind to its site (an ACS) within HMR-E to function in silencing HMR (McNally and Rine 1991; Foss et al. 1993; Micklem et al. 1993; Fox et al. 1995; Loo et al. 1995; Palacios DeBeer and Fox 1999). The available evidence indicates that the primary role for the ORC in silencing HMR is recruitment of the Sir1p to HMR-E. In particular, the requirement for the ORC in silencing HMR can be bypassed by tethering a Gal4-Sir1p fusion protein to the HMR-E silencer through a Gal4-binding site that replaces the silencer's ACS (Chien et al. 1993; Fox et al. 1997). This Gal4–Sir1p-tethered silencing requires the three other Sir proteins (Sir2p, Sir3p, and Sir4p), indicating that Sir1p can recruit the other Sir proteins to HMR in the absence of an intact silencer (Chien et al. 1993). In addition, Sir1p and Orc1p, the largest subunit of the six-subunit ORC (Bell and Stillman 1992), interact directly (Triolo and Sternglanz 1996), and mutations in SIR1 that specifically abolish this interaction lead to defects in silencing (Gardner et al. 1999). These and additional observations have led to a model in which an ORC bound to a silencer recruits the Sir1p that in turn helps to recruit the other Sir proteins to the silent loci where they function as structural components of silenced chromatin. In this view, Sir1p has a regulatory role in the establishment of silenced chromatin but is less important to the inherent structure of silenced chromatin or its maintenance and inheritance once established (discussed in Loo and Rine 1995).
The HMR-E silencer also contains one binding site each for the Rap1p and Abf1p proteins, two abundant nuclear proteins with additional roles in telomere structure, transcription activation, and DNA replication and repair at other positions in the genome (Marahrens and Stillman 1992; Shore 1994; Kang et al. 1995; Gailus-Durner et al. 1996; Rolfes et al. 1997; Reed et al. 1999). Like ORC, both Rap1p and Abf1p must bind their sites within HMR-E to function in silencing. However, there is no evidence that either Rap1p or Abf1p functions in recruiting the Sir1p to HMR-E.
As is true for Rap1p and Abf1p, the ORC also has a role in addition to silencing. Specifically, the ORC is required for the initiation of DNA replication at each of the hundreds of chromosomal origins scattered throughout the yeast genome (Bell and Stillman 1992; Fox et al. 1995; Liang et al. 1995). The ORC's two different roles in yeast, silencing and DNA replication, both of which require its binding to an ACS, raise questions about the factors that govern a Sir1p–ORC interaction and the role this interaction plays in defining a silencer. In contrast with the ORC, the Sir1p functions only in silencing. Because the ORC binds to hundreds of positions in the yeast genome but only four of these positions correspond to known silencers, an important question is what factors distinguish an ORC bound to a silencer from an ORC bound to a nonsilencer replication origin. A prevalent but untested hypothesis is that Sir1p binds to silencer-bound ORC molecules but not to the ORC molecules bound to nonsilencer replication origins.
In this report, we addressed the current view of the Sir1p in silencing HMR. Chromatin immunoprecipitation experiments indicated that the Sir1p specifically associated with HMR in chromatin, and that this association required the Sir1p–Orc1p interaction. Significantly, Sir1p did not associate with nonsilencer replication origins, providing direct evidence that the Sir1p–ORC interaction is a determinant that distinguishes a silencer from a nonsilencer replication origin. Interestingly, Sir1p's association with HMR required SIR2, indicating that other components of silenced chromatin may contribute to a stable Sir1p–ORC interaction at silencers. The Sir1p was present at very low concentrations in yeast, yet associated with HMR throughout the cell cycle in a manner similar to Sir3p, a known structural component of silent chromatin. Taken together, these data put constraints on models for the role of Sir1p in the establishment of a silenced chromosome domain.
Results
Sir1p was physically associated with HMR in chromatin
A current model for silencing HMR posits that Sir1p binds to the HMR-E silencer in chromatin through interactions with the ORC that is bound directly to the silencer ACS. To test directly whether Sir1p is in close physical proximity to the HMR-E silencer DNA, we performed chromatin immunoprecipitation with antibodies against the hemagglutinin epitope (α-HA) by using yeast expressing a HA-tagged version of Sir1p (Sir1p–3xHA). The synthetic HMR-E silencer (HMR-SS) was used in most experiments because it lacks the redundancy present in the natural silencer and absolutely requires both Sir1p and individual binding sites within the silencer for silencing (McNally and Rine 1991).
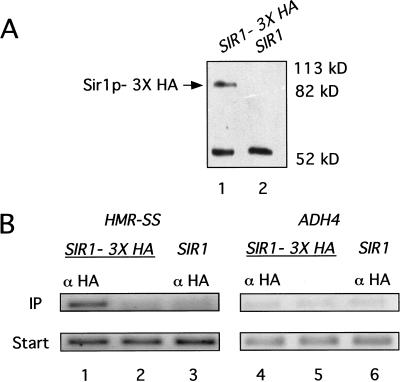
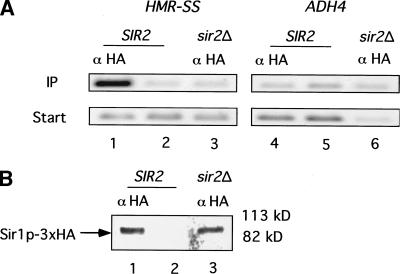
If Sir1p was associated with HMR-SS in chromatin, then HMR-SS DNA should be enriched in a protein–DNA fraction prepared by immunoprecipitation of Sir1p–3xHA from a yeast whole cell extract. Sir1p–3xHA functioned indistinguishably from wild-type Sir1p in vivo (data not shown) and was detectable with an antibody to HA (α-HA) as an ∼85-kD protein on an immunoblot (Fig. 1A). The smaller immunoreactive band visible in both lanes in this experiment was due to the immunoglobulin heavy chain from α-HA. Yeast cells harboring a chromosomal copy of SIR1–3xHA were cross-linked with formaldehyde to create covalent linkages between closely associated proteins and proteins and DNA. The cross-linked chromatin was isolated and sheared, and Sir1p–3xHA was immunoprecipitated from this mixture by using α-HA. The HMR-SS locus and the nonsilent ADH4 or ACT1 locus were detected in the starting mixtures and in the immunoprecipitates with specific primers (Table 1) and PCR.
Figure 1.
Sir1p was physically associated with HMR in chromatin. (A) Protein immunoblot analysis of Sir1p–3xHA quantitatively immunoprecipitated with α-HA. Twenty A600 cell equivalents of the immunoprecipitated fraction were analyzed in each lane. The smaller band seen in this immunoblot is due to binding of secondary antibody to the heavy chain of the α-HA. (B) Chemically cross-linked and sheared chromatin was immunoprecipitated with α-HA from strains that expressed either SIR1–3xHA (lanes 1 and 4; CFY815) or untagged SIR1 (lanes 3 and 6; CFY345). HMR-SS DNA (lanes 1–3) and ADH4 DNA (lanes 4–6) were analyzed in the crude extract (Start) and immunoprecipitated (IP) fraction by using PCR directed by specific primers (Table 1). A mock precipitation of extracts from cells expressing SIR1–3xHA was performed that was identical to the α-HA–directed immunoprecipitation except that it lacked α-HA antibody (lanes 2 and 5).
Table 1.
Oligonucleotide primer pairs used in this study
| Locus
|
Primer pair
|
|
|---|---|---|
| ADH4 | ggctactaacggtggggaaatcggagac | gcacaggcatcggtgattgggttagaggc |
| ACT1 | cggtattgtcaccaactgggacgatatgg | gcagcggtttgcatttcttgttcgaagtcc |
| ARS306 | ctgtagtttggacaaggtgaccctgccaag | gtgacttactaacgtcaacgtacaatcgcg |
| ARS1 | ggcgttattggtgttgatgtaagcggagg | gcaagaccgagaaaaggctagcaagaatcg |
| ARS1412 | gggaatagctaatcaagtggataagacgc | ccaactcctctctacttgcgtgtgtatttg |
| HMR (a) | ctaacttcaactgttgctggtgtcgcctgc | gcccatttcgaagaatgattgagcaccgtc |
| HMR (b) | caagggcctacgattactatgtactggaagg | ggtcaacataaagtggcgagaaaaacgccctg |
| HMR-SS | ggtagagttccttgttgaacgtgataaccc | gatgtctgggtttgtttggcatgcatcagc |
| HMR (d) | cttcttctgttgttacactctctggtaacttagg | cctgttctaaaaatgcccgtgcttggggtg |
| HMR (e) | caaactttgagagaaatatgtctttctactgcg | cctaccacattatcaatccttgcatccagc |
| HMR (f) | gccaacaatggaatctcattacccatcccaag | gtactcacacctttgcaagactaccaggatc |
| HMR (g) | gatatcgccactgcatcatttctgtagtcg | cagtcagacaggtcgatcaaagttgaaattgg |
Significantly, HMR-SS was enriched in the immunoprecipitate, and this enrichment required both SIR1–3xHA and α-HA (Fig. 1B, cf. lanes 1–3). Importantly, this enrichment was specific for silencer DNA; the nonsilenced control locus ADH4 was not enriched specifically in the immunoprecipitate (Fig. 1B, lanes 4–6). In separate experiments, the natural HMR-E silencer was specifically enriched in a Sir1p–3xHA-dependent chromatin immunoprecipitation experiment, indicating that Sir1p–3xHA's behavior with the HMR-SS was an accurate reflection of natural Sir1p function, consistent with the previously published genetic analysis of this silencer (McNally and Rine 1991).
Sir1p's association with HMR required the silencer ORC-binding site and a region of Sir1p required for interaction with Orc1p
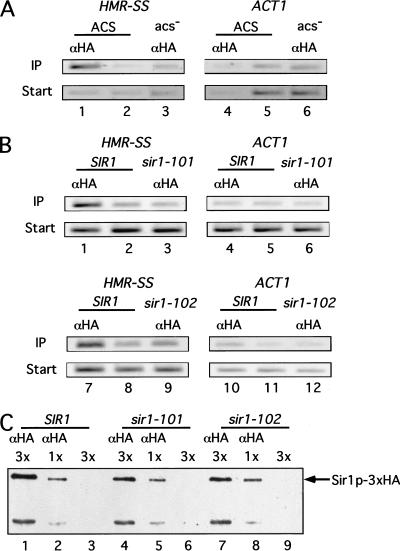
Several independent genetic experiments indicate that Sir1p's recognition of the HMR-E silencer in vivo requires the ORC bound to the silencer ORC-binding site (ACS). If Sir1p's physical association with HMR observed above reflected this recognition, then the silencer ACS should be required for enrichment of HMR in a Sir1p–3xHA-dependent chromatin immunoprecipitation experiment. Therefore, we determined whether the association between Sir1p and HMR was abolished by a mutation of this silencer's ACS (Fig. 2A). Significantly, in a strain containing a mutation of the ACS in the synthetic silencer (HMR-SS[acs-]), the silencer DNA was not enriched in the immunoprecipitate (Fig. 2A).
Figure 2.
Sir1p's association with HMR required the silencer ACS and a region of Sir1p required for interaction with Orc1p. The chromatin immunoprecipitation experiments were performed as described in Fig. 1 except that ACT1 was used as a nonsilenced control locus. (A) A mutation of the silencer ACS abolished Sir1p's association with HMR. Chromatin immunoprecipitation of HMR-SS was compared for two isogenic strains that contained either wild-type HMR-SS (ACS; CFY476; lanes 1 and 2 and 4 and 5) or HMR-SS containing a mutation in the ACS (HMR-SS [acs−]) (acs−; CFY633; lanes 3 and 6). (B) A mutation in the region of Sir1p required for interaction with Orc1p abolished Sir1p's association with HMR. Chromatin immunoprecipitation of HMR-SS was compared for three isogenic strains that contained either wild-type SIR1–3xHA (SIR1; CFY815; lanes 1 and 2, 4 and 5, 7 and 8, and 10 and 11) or one of two sir1srd–3xHA alleles (sir1–101; CFY689; lanes 3 and 6; sir1–102; CFY687; lanes 9 and 12) at the chromosomal SIR1 locus. (C) The mutant Sir1srd–3xHA proteins were expressed at the same levels as wild-type Sir1p–3xHA. Protein immunoblot analysis of wild-type and mutant Sir1p–3xHA proteins quantitatively immunoprecipitated with α-HA from the strains used in the experiment described above (B). Protein was analyzed in the immunoprecipitated fractions of extracts prepared from 45 A600 (3×) (lanes 1,4,7) or 15 A600 (1×) (lanes 2,5,8) cell equivalents. Mock precipitations were performed as described in Fig. 1 (lanes 3, 6, and 9).
The Sir1p contains a small region that is dedicated to Sir1p's recognition of a silencer and required for Sir1p's interaction with the Orc1p, the largest subunit of ORC (Gardner et al. 1999). Mutant Sir1 proteins with single amino acid substitutions in this region function normally when tethered to the silencer through a heterologous DNA-binding domain but fail to silence HMR by the normal mechanisms that presumably require Sir1p's physical association with the HMR-E silencer. These mutant Sir1 proteins are referred to as silencer-recognition-defective (Sir1psrd), and the region required for interaction with Orc1p is referred to as the SRD region of Sir1p. To test whether the SRD region of Sir1p was required for Sir1p's ability to associate with HMR in chromatin, we examined whether two different mutant Sir1psrd proteins enriched the HMR-SS DNA in a chromatin immunoprecipitation experiment (Fig. 2B). Both mutant Sir1psrd proteins were expressed from the SIR1 locus as fusions to the 3x-HA epitope (sir1–101–3xHA; sir1–102–3xHA). Notably, neither mutant Sir1psrd enriched HMR-SS DNA as efficiently as wild-type Sir1p (Fig. 2B), indicating that this region of Sir1p was required for Sir1p's association with a silencer in chromatin. Importantly, the mutant proteins were expressed at levels equivalent to wild-type Sir1p, indicating that the inability of these mutant proteins to enrich silencer DNA in the immunoprecipitate was not due to significant reductions in the levels of the mutant Sir1psrd proteins (Fig. 2C).
Taken together, these data indicate that Sir1p is physically associated with HMR in chromatin and support the hypothesis that this association is required for Sir1p's function in silencing.
The Sir1p did not associate with nonsilencer origins
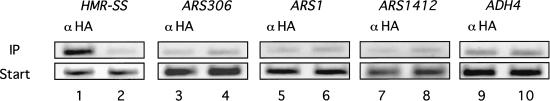
If a Sir1p–silencer interaction were a key determinant of silent chromatin assembly at HMR, then Sir1p should not associate with nonsilencer origins even though these elements bind the ORC. Therefore, we tested whether several characterized replication origins were associated with Sir1p by using chromatin immunoprecipitation (Fig. 3; data not shown). In these experiments, Sir1p associated with the HMR-E silencer within chromatin but not with other origins including an early (ARS306), middle (ARS1), and late (ARS1412) replication origin. In addition, Sir1p did not associate with several different inefficient origins on chromosome VI, nor did it associate with natural telomeric origin sequences (data not shown). Thus, although the available evidence supports only a direct interaction between Sir1p and Orc1p, other factors in addition to ORC contribute to Sir1p's association with HMR in chromatin.
Figure 3.
The Sir1p was not associated with nonsilencer origins. Chromatin immunoprecipitation with α-HA was performed using a strain expressing SIR1–3xHA (CFY815; lanes 1,3,5,7,9). The appropriate primers (Table 1) were used to examine the Sir1p's association with HMR-SS and several nonsilencer origins as indicated. A mock precipitation was performed as described in Fig. 1, and the precipitate was examined for DNA with the same primers (lanes 2,4,6,8,10).
The other SIR genes were required for the association of Sir1p with HMR
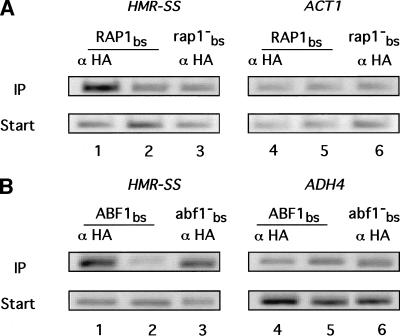
The HMR-E silencer contains binding sites for two other silencer-binding proteins in addition to the ORC, specifically the Rap1p and the Abf1p, that could contribute to Sir1p's preference for a silencer over nonsilencer origins. Therefore, we tested whether the Rap1p or Abf1p binding sites were required for enrichment of the HMR-SS (Fig. 4). Significantly, in a strain containing a mutation of the Rap1p-binding site in the synthetic silencer (HMR-SS[rap1−]), Sir1p–3xHA did not associate with the HMR-SS (Fig. 4A). In contrast, an analogous experiment demonstrated that the Abf1p-binding site was not required for Sir1p–3xHA's association with HMR-SS in chromatin (Fig. 4B).
Figure 4.
The silencer Rap1p-binding site, but not the Abf1p-binding site, was required for Sir1p's association with HMR. Chromatin immunoprecipitation was performed as described in Fig. 1. (A) A mutation of the silencer Rap1p-binding site abolished Sir1p's association with HMR. Chromatin immunoprecipitation of HMR-SS was compared for two isogenic strains that contained either wild-type HMR-SS (RAP1bs; CFY476; lanes 1and 2 and 4 and 5) or HMR-SS containing a mutation in the RAP1bs (HMR-SS[rap1−bs]) (rap1−bs; CFY632; lanes 3 and 6). (B) A mutation of the silencer Abf1p-binding site did not abolish Sir1p's association with HMR. Chromatin immunoprecipitation of HMR-SS was compared for two isogenic strains that contained either wild-type HMR-SS (ABF1bs; CFY476; lanes 1 and 2 and 4 and 5) or HMR-SS containing a mutation in the ABF1bs (HMR-SS[abf1−bs]) (abf1−bs; CFY625; lanes 3 and 6).
A crucial role of the Rap1p in silencing at both telomeres and the HM loci is to recruit the Sir2/Sir3/Sir4 protein complex that functions in the assembly and maintenance of silent chromatin (Moretti et al. 1994; Grunstein 1997). Thus, the requirement for the silencer's Rap1p-binding site in Sir1p–3xHA's association with HMR-SS could actually reflect a requirement for the other Sir proteins. Therefore, we tested whether Sir1p–3xHA associated with HMR-SS in yeast cells harboring a deletion of SIR2 (Fig. 5). Significantly, in strains lacking SIR2, Sir1p–3xHA failed to enrich for HMR-SS (Fig. 5A). The nuclear concentration of Sir1p–3xHA was not reduced in sir2Δ mutant strains, indicating that the lack of a Sir1p–3xHA–silencer interaction in chromatin in this mutant was not due to a lack of nuclear Sir1p–3xHA (Fig. 5B). In separate experiments, a sir3Δ mutant behaved similarly (data not shown). Thus, the other SIR genes are required for a stable association between the Sir1p and HMR in chromatin.
Figure 5.
SIR2 was required for Sir1p's association with HMR in chromatin. Chromatin immunoprecipitations were performed as described in Fig. 1. (A) A sir2Δ mutation abolished Sir1p's association with HMR. Chromatin immunoprecipitation of HMR-SS was compared in two isogenic strains that contained SIR1–3xHA and either a wild-type copy of SIR2 (CFY815; SIR2; lanes 1 and 2 and 4 and 5) or a deletion of SIR2 (sir2Δ; CFY105; lanes 3 and 6). (B) The nuclear concentration of Sir1p–3xHA was not reduced by a mutation in SIR2. Sir1p was immunoprecipitated from nuclear extracts prepared from isogenic strains that each contained SIR1–3xHA and either wild-type SIR2 (CFY815; lanes 1 and 2 and lanes 4 and 5) or sir2Δ (CFY105; lanes 3 and 6).
The Sir1p and Sir3p associated similarly with HMR
A prevalent view of Sir1p in silencing HMR differs considerably from the view of Sir3p. In particular, the Sir1p, a proposed regulator of silenced chromatin assembly (Pillus and Rine 1989; Loo and Rine 1995; Triolo and Sternglanz 1996), might be expected to associate with HMR differently from Sir3p. Therefore, we compared the association of Sir1p and Sir3p with HMR in a chromatin immunoprecipitation experiment (Fig. 6).
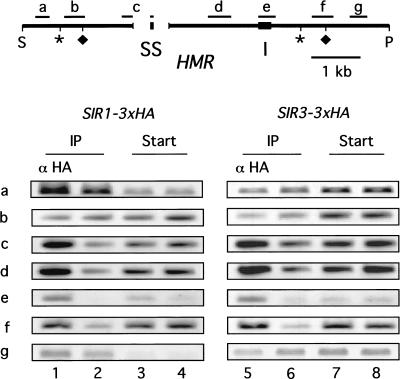
Figure 6.
The Sir1p and Sir3p associated similarly with HMR. Chromatin immunoprecipitations were performed as described in Fig. 1 by using isogenic strains expressing either SIR1–3xHA (CFY762 containing pRS316–SIR1–3xHA [pCF448]; lanes 1 and 4) or SIR3–3xHA (CFY914 containing pRS316–SIR3–3xHA [Ansari and Gartenberg 1999]; lanes 5 and 8). The presence of particular fragments within and surrounding HMR in the immunoprecipitates was determined using specific primers diagrammed in the figure and listed in Table 1. (S) Sph1 site on chromosome III at Saccharomyces Genome Database (SGD) coordinate 289759; (P) Pvu1 site at SGD coordinate 297608. The gap in the line represents a deletion of ∼800 bp that includes the HMR-E silencer and flanking sequences; these deleted sequences were replaced with the HMR-SS (McNally and Rine 1991). One primer of the pair used to analyze fragment c within HMR annealed to sequences within HMR-SS, and the second primer annealed to sequences flanking the 800-bp deletion, which is why fragment c is depicted as “interrupted.” (Asterisks and diamonds) Boundaries of silenced chromatin as determined by Loo and Rine (1994) and Donze et al. (1999), respectively. The sequences of all primers used to generate the fragments indicated in the figure are listed in Table 1.
To compare the association of Sir1p and Sir3p with HMR, chromatin immunoprecipitation experiments were performed with two isogenic strains that differed in the form of Sir protein that was expressed as an HA-tagged protein. Specifically, one strain expressed Sir1p–3xHA and the second strain expressed Sir3p–3xHA. After chromatin isolation, the sheared chromatin was immunoprecipitated from each strain, and different regions of HMR were examined using the appropriate PCR primers as diagrammed in Figure 6 and listed in Table 1.
In three independent experiments, one of which is shown in Figure 6, the association of Sir1p and Sir3p with HMR was virtually identical. Although the enrichment of the most outlying HMR fragments varied somewhat in individual experiments, the behavior of Sir1p–3xHA and Sir3p–3xHA was comparable. For example, in the experiment shown, fragment b was not associated with either Sir1p–3xHA or Sir3p–3xHA based on chromatin immunoprecipitation. In other experiments, fragment b was associated with both Sir1p–3xHA and Sir3p–3xHA. Thus, at the low resolution offered by these experiments, Sir1p–3xHA and Sir3p–3xHA associated similarly with HMR.
The Sir1p was expressed at low levels in yeast cells and was associated exclusively with a chromatin-containing fraction
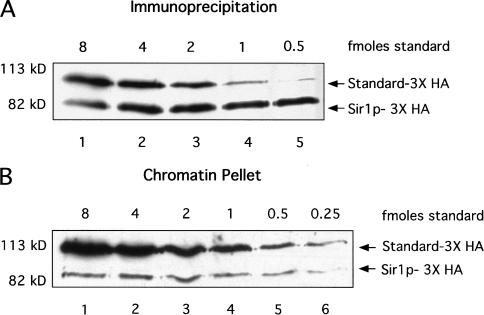
The data described above supported the view that a Sir1p–ORC interaction is confined to silenced regions such as HMR. Assuming that a Sir1p–ORC interaction is stable, one factor that could help confine it to silenced regions is a low concentration of Sir1p in yeast. Therefore, we determined the levels of Sir1p–3xHA expressed per yeast cell by comparing the amount of Sir1p–3xHA in a known number of yeast cells to a 3xHA-containing standard protein of known concentration (Fig. 7). In one approach, Sir1p–3xHA was quantitatively immunoprecipitated from 30 A600 cell equivalents of a denatured crude yeast cell extract, and known amounts of a purified standard protein (GST–Ssq1p–3xHA; Voisine et al. 2000) were added to the immunoprecipitate before gel electrophoresis (Fig. 7A). Importantly, Sir1p–3xHA could not be detected in a second immunoprecipitation of the supernatant that remained after the first immunoprecipitation, suggesting that the Sir1p–3xHA had been quantitatively immunoprecipitated (K.A. Gardner, data not shown). Moreover, digestion of the yeast whole cell extract with DNAse and/or treatment with denaturing detergents did not change the amount of Sir1p–3xHA that could be immunoprecipitated (K.A. Gardner, data not shown). These experiments indicated that the Sir1p–3xHA was present at a concentration of ∼10– 30 molecules per cell, with the precise number varying between individual experiments.
Figure 7.
The Sir1p was expressed at low levels and was associated exclusively with a chromatin-containing fraction. (A) Quantitative immunoprecipitation of Sir1p–3xHA from yeast containing SIR1–3xHA at the chromosomal SIR1 locus (CFY815) indicated that Sir1p–3xHA concentration was ∼20 molecules per yeast cell. Sir1p–3xHA was quantitatively immunoprecipitated from crude yeast extracts and 5.5 A600 cell equivalents of the immunoprecipitated fraction were mixed with decreasing amounts of a standard protein (standard–3xHA; purified and quantified Ssq1p–3xHA [Voisine et al. 2000]). (B) Sir1p–3xHA was present exclusively in a chromatin-containing fraction; 0.85 A600 cell equivalents of the chromatin-containing fraction were mixed with decreasing amounts of a standard protein (Ssq1–3xHA) as described in (A). Sir1p–3xHA could not be detected in the soluble fraction even after immunoprecipitation and protein immunoblot analysis.
As a second independent method for measuring the concentration of Sir1p–3xHA that did not rely on an immunoprecipitation step, a crude yeast extract was separated into chromatin and soluble fractions, and the level of Sir1p–3xHA in these fractions was determined (Fig. 7B). Most of the Sir1p–3xHA was present in the chromatin pellet; Sir1p–3xHA could not be detected in the soluble fraction even after immunoprecipitation before protein immunoblot analysis (data not shown). The concentration of Sir1p–3xHA in a yeast cell based on the amount present in the chromatin fraction was ∼20 molecules, a value close to that determined by quantitative immunoprecipitation of Sir1p–3xHA from a denatured crude extract (Fig. 7A). Therefore, only a few Sir1p molecules are present in a yeast cell, and most of these molecules are associated with a chromatin-containing fraction.
The Sir1p was associated with HMR at the G1, S, and M phases of the cell cycle
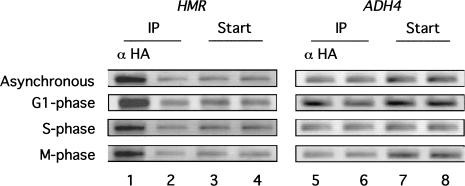
Sir1p has been postulated to function in the S-phase requirement for the establishment of silenced chromatin at the HM loci (Miller and Nasmyth 1984; Pillus and Rine 1989, Loo and Rine 1995; Triolo and Sternglanz 1996; Fox et al. 1997). One intriguing possibility raised by this postulate is that an S-phase requirement for establishment could be accomplished, at least in part, by confining an ORC–Sir1p interaction at HMR to the S phase of the cell cycle (Triolo and Sternglanz 1996). Therefore, we tested whether Sir1p–3xHA was associated with HMR in chromatin at three different stages of the cell cycle by performing Sir1p–3xHA-dependent chromatin immunoprecipitation experiments in yeast cells arrested in either the G1, S, or M phase of the cell cycle (Fig. 8). Notably, the Sir1p–3xHA associated with HMR at each stage of the cell cycle as efficiently as it associated with HMR in the same yeast strain growing asynchronously (Fig. 8). Therefore, if Sir1p's role in silencing is confined to S phase, this cell cycle regulation is not exerted at the level of Sir1p's physical association with HMR in chromatin.
Figure 8.
The Sir1p was associated with HMR at the G1, S, and M phases of the cell cycle. Chemically cross-linked and sheared chromatin was immunoprecipitated with α-HA (lanes 1 and 5) from a MATa HMRa strain that expressed SIR1–3xHA (CFY416) that was growing asynchronously or arrested with α-factor (G1 phase), hydroxyurea (S phase), or nocodazole (M phase). The natural HMR locus (lanes 1–4) and ADH4 DNA (lanes 5–8) were analyzed in the crude extract (Start) and immunoprecipitated (IP) fractions by using PCR directed by specific primers. A mock precipitation of extracts was performed that was identical to the α-HA–directed immunoprecipitation except that it lacked α-HA antibody (lanes 2 and 6).
Discussion
A Sir1p–ORC interaction in defining a silencer
Several independent studies support the conclusion that a Sir1p–ORC interaction is critical for Sir1p's function in silencing HMR (Chien et al. 1993; Triolo and Sternglanz 1996; Fox et al. 1997; Gardner et al. 1999). The work presented here extends this conclusion by demonstrating directly that the natural Sir1p expressed from its normal locus physically associates with HMR in chromatin in a manner that requires a Sir1p–ORC interaction. Moreover, this study provides evidence that a Sir1p–ORC interaction is a molecular feature that distinguishes a silencer from a nonsilencer replication origin because a Sir1p–ORC interaction could not be detected at several different nonsilencer origins.
Two specific and interdependent findings reported here contribute insights into the mechanisms that could confine a Sir1p–ORC interaction to silenced chromatin. First, the low concentration of Sir1p and its association with HMR at various stages of the cell cycle are consistent with a stable Sir1p–ORC interaction within silenced chromatin domains. Such stability would limit the opportunity for Sir1p to interact with nonsilencer ORC molecules within the genome, a possibly important limitation considering Sir1p's role in assembling repressive chromatin and the discovery that at least one Sir protein can modulate the activity of replication origins (Stevenson and Gottschling 1999). Second, the requirement for the other SIR genes in Sir1p's association with HMR indicates that the other Sir proteins, known structural components of silenced chromatin, may stabilize a Sir1p–ORC interaction that by itself may be inherently weak. Notably, a Sir1p–Sir4p interaction has been observed (Triolo and Sternglanz 1996), indicating that Sir1p does indeed contact at least one other Sir protein directly that could function in this way. Thus, multiple, and possibly individually relatively weak, interactions between Sir proteins and between Sir proteins and silencer-binding proteins may contribute to the ultimate stability of a Sir1p–ORC interaction in silenced chromatin, and this interaction in turn could enhance the stability of the silenced chromatin of which it is a part. The inference that additional proteins that comprise silent chromatin contribute to the stability and specificity of a Sir1p–ORC interaction underscores the probability that the assembly of large chromosomal regions into heritable transcriptional states involves a high degree of molecular cooperativity.
The establishment and maintenance of a silenced chromatin domain
Analysis of silencing during the cell cycle (Miller and Nasmyth 1984), and at the single-cell level (Pillus and Rine 1989; Mahoney et al. 1991), helped formulate the hypothesis that the Sir1p and silencers play regulatory roles in silencing. According to this hypothesis, the Sir1p is required for the establishment of silenced chromatin during the S phase of the cell cycle but not the subsequent maintenance or propagation of the silenced state (discussed in Fox et al. 1993; Loo and Rine 1995). In addition, this Sir1p/S-phase–dependent establishment is required in only a few cell divisions to switch a cell from the nonsilenced to silenced state. The demonstration that Sir1p mediates its role in silencing through the silencers refined the view of Sir1p-mediated establishment (Chien et al. 1993). Specifically, establishment of silencing at HMR often is depicted as the Sir1p binding to silencers in S phase through interactions with the ORC and nucleating the assembly of silenced chromatin by recruiting a complex consisting of the other Sir proteins (Triolo and Strenglanz 1996; Gardner et al. 1999). If establishment is required only rarely, Sir1p would need to “nucleate” silencing only once every several S phases because, once assembled, the silenced state self-propagates efficiently (Fox et al. 1993). In contrast with this view of Sir1p, the other Sir proteins are viewed as being important for maintaining the inherent structure of silenced chromatin throughout the cell cycle. Indeed, this view for the Sir2/3/4 proteins is supported by the important observation that these proteins are structural components of silenced chromatin (Hecht et al. 1995, 1996; Strahl-Bolsinger et al. 1997).
The data presented here put constraints on the mechanisms by which Sir1p could function in establishing silenced chromatin and highlight the possibility that Sir1p may have an equally important role in maintaining silenced chromatin throughout the cell cycle. In particular, the demonstration that the Sir1p is associated with HMR throughout the cell cycle makes it improbable that a potential S-phase role for Sir1p in silencing is due to confining a Sir1p–ORC interaction to S phase (Triolo and Sternglanz 1996). Moreover, the cell cycle data presented here are equally consistent with Sir1p having a “structural” role in silent chromatin at HMR, similar to the other Sir proteins. It is possible, for example, that one or more Sir1p molecules recognize ORC(s) within previously assembled, and therefore probably condensed, silenced chromatin and, by binding to these ORC molecules and other Sir proteins, structurally stabilize this chromatin state. If Sir1p does recognize and stabilize previously assembled and condensed silenced chromatin at HMR, only a few Sir1 protein molecules would be necessary to “contact” the entire domain. Thus, a molecular component of SIR1-dependent establishment may be Sir1p's recognition and stabilization of previously assembled silenced chromatin. In this view, the establishment and stable propagation of silenced chromatin in a wild-type yeast population are not mechanistically distinct events. Rather than nucleating a new silenced state, Sir1p simply enhances the already relatively efficient process of self-propagation of a previously assembled silenced state. This view, although different from models for Sir1p function proposed previously, is entirely consistent with genetic data concerning the role of SIR1 and silencers in the establishment of silencing at the HM loci (Pillus and Rine 1989; Mahoney et al. 1991). Because Sir1p functions, at least in part, through the silencers, this view of Sir1p is also consistent with recent elegant molecular experiments that indicate that the silencers are required to maintain silencing throughout the cell cycle (Holmes and Broach 1996; Cheng and Gartenberg 2000).
There are many possible mechanisms by which a Sir1p–ORC interaction could enhance the stability of silenced chromatin and restrict it to a few discrete domains in the genome. As discussed above, the Sir1p–ORC interaction could prevent the inherent instability of Sir2/3/4 silencing (Pillus and Rine 1989; Gottschling et al. 1990; Aparicio et al. 1991) by a direct structural mechanism. Alternatively, Sir1p could function as an enzyme that modifies a component of silenced chromatin, perhaps ORC itself, and this modification in turn could play a stabilizing structural role. The low concentration of Sir1p and the recent exciting data demonstrating that yeast Sir2p is an enzyme make this possibility worth considering (Tanney et al. 1999; Imai et al. 2000; Landry et al. 2000; Smith et al. 2000).
Although there are no known homologs of SIR1, it is probable that interactions between ORC and heterochromatin-associated proteins with roles analogous to yeast Sir1p will be important for regulating silencing in all eukaryotic organisms. In particular, the association of the Drosophila HP1 protein with heterochromatin requires an HP1–ORC interaction reminiscent of the Sir1p–ORC interaction in yeast (Pak et al. 1997; Huang et al. 1998), and, significantly, a human homolog of HP1 has been proposed to silence genes involved in breast cancer metastasis (Kirschmann et al. 2000). Given that ORC homologs have been identified in a variety of metazoans including humans (Quintana and Dutta 1999), it will be interesting to learn whether the function of this human HP1 homolog is mediated through interactions with the human ORC. Regardless of the exact mechanism by which Sir1p contributes to silencing, a comprehensive analysis of the Sir1p–ORC interaction in yeast undoubtedly will provide mechanistic insights relevant to the role of the ORC in chromosome structure and gene silencing in all eukaryotes.
Materials and methods
Yeast strains
The yeast strains used in this study are listed in Table 2 and were constructed using standard yeast molecular genetic techniques (Guthrie and Fink 1991). Chromosomal SIR1–3xHA was constructed using a PCR-mediated epitope-tagging method (Schneider et al. 1995). pRS316-containing SIR1–3xHA was constructed using a C-terminal SIR1–3xHA fragment derived from the chromosomal SIR1–3xHA. pRS316-containing SIR3–3xHA was provided by Marc Gartenberg (Ansari and Gartenberg 1999).
Table 2.
Strains used in this study
| Strain
|
Genotype
|
|---|---|
| JRY3009 | MATαade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 (W303-1B) |
| CFY345 | JRY3009 HMR-SS |
| CFY815 | JRY3009 HMR-SS SIR1-3xHA |
| CFY416 | JRY3009 MATa SIR1-3xHA |
| CFY434 | JRY3009 MATa HMR-SS |
| CFY419 | JRY3009 MATa HMR-SS(acs−) |
| CFY423 | JRY3009 MATa HMR-SS(rap1−) |
| CFY425 | JRY3009 MATa HMR-SS(abf1−) |
| CFY474 | JRY3009 hmrΔ::URA3 SIR1-3xHA ADE2 lys2Δ |
| CFY476 | CFY474xCFY434 |
| CFY629 | CFY474xCFY416 |
| CFY633 | CFY474xCFY419 |
| CFY632 | CFY474xCFY423 |
| CFY625 | CFY474xCFY425 |
| CFY687 | JRY3009 HMR-SS sir1-102-3xHA |
| CFY689 | JRY3009 HMR-SS sir1-101-3xHA |
| CFY105 | JRY3009 MATa HMR-SS SIR1-3xHA sir2Δ::LEU2 |
| CFY762 | JRY3009 HMR-SS sir1Δ::LEU2 |
| CFY914 | JRY3009 HMR-SS sir3Δ::LEU2 |
Immunoprecipitation and immunoblotting
To prepare extracts for analysis of Sir1p, the appropriate yeast cells were grown to a concentration of 2 × 107 cells/mL. The cells were harvested by centrifugation, washed twice with TBS (20 mM Tris-HCl at pH 7.5, 200 mM NaCl), and resuspended in 400 μL FA-LB (50 mM Hepes-KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 0.1% sodium deoxycholate, and 1:500 dilution of CalBiochem protease inhibitor cocktail). The resuspended cells were mechanically broken by bead beating. To immunoprecipitate Sir1p–3xHA, we incubated anti-HA antibodies (BAbCo) with the extract overnight at 4°C, and the extract/antibody mixture then was incubated an additional 2–6 h with 200 μ 50:50 slurry of protein A–Sepharose CL-4B beads (Pharmacia). Immunoprecipitates were washed with 1 mL of each of the following: FA-LB, FA-LB/0.5M NaCl, wash 3 (10 mM Tris-HCl at pH 8.0, 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA), and TE (20 mM Tris-HCl at pH 8.0, 1 mM EDTA), separated by SDS–polyacrylamide gel electrophoresis (8%), and transferred to nitrocellulose. Sir1p–3xHA was detected with anti-HA antibodies. Protein–antibody complexes were visualized by enhanced chemiluminescence (Pierce SuperSignal DURA Detection System). To test whether Sir1p–3xHA had been quantitatively immunoprecipitated, we passed the supernatant fraction from an immunoprecipitation experiment through the immunoprecipitation protocol a second time, and the resulting immunoprecipitate was analyzed for Sir1p–3xHA.
To quantify the levels of Sir1p–3xHA in yeast, we compared the amount of Sir1p–3xHA immunoprecipitated from a crude extract or present in a chromatin fraction (see below) with a known amount of purified Ssq1–3xHA (Voisine et al. 2000).
Chromatin immunoprecipitation and analysis of immunoprecipitated DNA
The chromatin immunoprecipitation protocol used here was as described (Strahl-Bolsinger et al. 1997) except that 200 mL of yeast cells was grown in minimal media to a concentration of 2.0 × 107cells/mL before cross-linking, and, except for the experiments shown in Figures 6 and 8, cross-linking was performed for 60 min. In addition, 125 mM glycine and 2% ammonium hydroxide were added to quench the cross-linking reaction. The cells were harvested and broken as described above. The purified DNA from the immunoprecipitate was resuspended in 25 μL TE. The purified DNA in the starting material (whole cell extract before immunoprecipitation) was resuspended in 1.0 mL TE.
To analyze for the presence of specific DNA fragments in the immunoprecipitate, we amplified the immunoprecipitated DNA by PCR using specific primers (Table 1). The PCRs were performed in 50 μL with one-sixth of the immunoprecipitated material and 1/375th of the starting material. The number of PCR cycles yielding product within the linear range was determined empirically by analysis of twofold serial dilutions of the starting material. PCR products (15 μL) were separated on 1% agarose gels and visualized with ethidium bromide.
To measure the association between Sir1p and HMR at different stages of the cell cycle, we performed chromatin immunoprecipitation experiments as described above except that 400 mL of yeast cells was grown to a concentration of 5 × 106 cells/mL in rich media (YPD). The cells then were concentrated to 200 mL fresh YPD and allowed to continue growing asynchronously, or incubated with 0.2 M hydroxyurea (S phase) or 10 μg/mL nocodazole (M phase) for 3 h. To arrest cells in G1, we concentrated the cells to 40 mL and incubated them with 6 μM α-factor (G1 phase) for 3 h. When arrest was complete, the cells were diluted to 400 mL YPD and then incubated with formaldehyde. The yeast cells arrested with >90% efficiency at all three stages as judged by cell morphology and cross-linking was performed for 15 min.
Nuclear and crude chromatin isolation
To determine the approximate nuclear concentration of Sir1p–3xHA in sir2Δ and sir3Δ mutant strains, we isolated nuclei as described (Lue and Kornberg 1987) except the spheroplasts were washed on a sucrose cushion as described (Bell and Stillman 1992). Purified nuclei were resuspended in phosphate-buffered saline. Fifty A600 cell equivalents of isolated nuclei were used for nuclear immunoprecipitations.
Chromatin was isolated as described (Donovan et al. 1997) except that spheroplasts were prepared as described in the nuclear isolation procedure (Bell and Stillman 1992). The spheroplasts were harvested and washed two times with ice-cold wash buffer (50 mM Tris at pH 7.5, 20 mM MgCl2, 100 mM NaCl). The cells were lysed on ice 20 min at a concentration of 250 A600 cell equivalents/mL in lysis buffer (wash buffer + 0.5% Triton X-100), and chromatin was isolated by centrifugation (15,000 rpm, 4°C for 5 min).
Acknowledgments
We thank Marc Gartenberg for providing pRS316–SIR3–3xHA and Rohinton Kamakaka for α-HA used to begin this study and valuable advice and consultation during the course of this work. We also thank Jaerek Marszalek for purified and quantified Ssq1–3xHA protein. We are grateful to Michael Sheets and members of the Fox and Sheets laboratories for constructive criticism. This work was supported by a grant from the National Institutes of Health (GM56890 to C.A.F.). K.A.G. was supported by a Biotechnology Training Grant to the University of Wisconsin–Madison (NIH 5T32 GM08349). C.A.F. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cfox@facstaff.wisc.edu; FAX (608) 262-5253.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.852801.
References
- Ansari A, Gartenberg MR. Persistence of an alternate chromatin structure at silenced loci in vitro. Proc Natl Acad Sci. 1999;96:343–348. doi: 10.1073/pnas.96.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Cheng TH, Gartenberg MR. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes & Dev. 2000;14:452–463. [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes & Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin SR, Paro R. Epigenetic regulation in Drosophila: A conspiracy of silence. In: Elgin SCR, editor. Chromatin structure and gene expression. New York: Oxford University Press Inc.; 1995. pp. 147–171. [Google Scholar]
- Foss M, McNally FJ, Laurenson P, Rine J. A role of the origin recognition complex (ORC) in transcriptional silencing and DNA replication in Saccharomyces cerevisiae. Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- Fox CA, Loo S, Rivier DH, Foss M, Rine J. A transcriptional silencer as a specialized origin of replication that establishes functional domains of chromatin. Cold Spring Harb Symp Quant Biol. 1993;58:443–455. doi: 10.1101/sqb.1993.058.01.051. [DOI] [PubMed] [Google Scholar]
- Fox CA, Loo S, Dillin A, Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes & Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- Fox CA, Ehrenhofer-Murray AE, Loo S, Rine J. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science. 1997;276:1547–1551. doi: 10.1126/science.276.5318.1547. [DOI] [PubMed] [Google Scholar]
- Freeman-Cook LL, Sherman JM, Brachmann CB, Allshire RC, Boeke JD, Pillus L. The Schizosaccharomyces pombe hst4(+) gene is a SIR2 homologue with silencing and centromeric functions. Mol Biol Cell. 1999;10:3171–3186. doi: 10.1091/mbc.10.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailus-Durner V, Xie J, Chintamaneni C, Vershon AK. Participation of the yeast activator Abf1 in meiosis-specific expression of the HOP1 gene. Mol Cell Biol. 1996;16:2777–2786. doi: 10.1128/mcb.16.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner KA, Rine J, Fox CA. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics. 1999;151:31–44. doi: 10.1093/genetics/151.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast trasncriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Hecht A, Laroche T, Strahl BS, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Hendrich BD, Willard HF. Epigenetic regulation of gene expression; the effect of altered chromatin structure from yeast to mammals. Hum Mol Gen. 1995;4:1765–1777. doi: 10.1093/hmg/4.suppl_1.1765. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Heterochromatin function in complex genomes. Biochim Biophy Acta. 2000;1470:1–8. doi: 10.1016/s0304-419x(99)00034-7. [DOI] [PubMed] [Google Scholar]
- Herskowitz I, Rine J, Strathern J. Mating-type determination and mating-type interconversion in Saccharomyces cerevisiae. In: Jones E, Pringle JR, Broach JR, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae: Gene expression. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1992. pp. 583–656. [Google Scholar]
- Holmes SG, Broach JR. Silencers are required for inheritance of the repressed state in yeast. Genes & Dev. 1996;10:1021–1032. doi: 10.1101/gad.10.8.1021. [DOI] [PubMed] [Google Scholar]
- Huang DW, Fanti L, Pak DTS, Botchan MR, Pimpinelli S, Kellum R. Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: Their phosphorylation levels and associations with origin recognition complex proteins. J Cell Biol. 1998;142:307–318. doi: 10.1083/jcb.142.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is a NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Kang JJ, Yokoi TJ, Holland MJ. Binding sites for abundant nuclear factors modulate RNA polymerase I-dependent enhancer function in Saccharomyces cerevisiae. J Biol Chem. 1995;270:28723–28732. doi: 10.1074/jbc.270.48.28723. [DOI] [PubMed] [Google Scholar]
- Kirschmann DA, Lininger RA, Gardner LM, Seftor EA, Odero VA, Ainszten AM, Earnshaw WC, Wallrath LL, Hendrix MJ. Down-regulation of HP1Hsalpha expression is associated with the metastatic phenotype in breast cancer. Cancer Res. 2000;60:3359–3363. [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- ————— Silencing and domains of heritable gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- Loo S, Fox CA, Rine J, Kobayashi R, Stillman B, Bell S. The origin recognition complex in silencing, cell-cycle progression, and DNA replication. Mol Biol Cell. 1995;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue NF, Kornberg RD. Accurate initiation at RNA polymerase II promoters in extracts from Saccharomyces cerevisiae. Proc Natl Acad Sci. 1987;84:8839–8843. doi: 10.1073/pnas.84.24.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney DJ, Marquardt R, Shei GJ, Rose AB, Broach JR. Mutations in the HML-E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes & Dev. 1991;5:605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- McNally FJ, Rine J. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5648–5659. doi: 10.1128/mcb.11.11.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem G, Rowley A, Harwood J, Nasmyth K, Diffley JF. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature. 1993;366:87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- Miller AM, Nasmyth KA. Role of DNA replication in the repression of silent mating type loci in yeast. Nature. 1984;312:247–251. doi: 10.1038/312247a0. [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes & Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Pak DTS, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Palacios DeBeer MA, Fox CA. A role for a replicator dominance mechanism in silencing. EMBO J. 1999;18:3808–3819. doi: 10.1093/emboj/18.13.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- Pillus L, Grunstein M. Chromatin structure and epigenetic regulation in yeast. In: Elgin SCR, editor. Chromatin structure and gene expression. New York: Oxford University Press; 1995. pp. 123–146. [Google Scholar]
- Quintana DG, Dutta A. The metazoan origin recognition complex. Front. Biosci. 1999;4:805–815. doi: 10.2741/quintana. [DOI] [PubMed] [Google Scholar]
- Reed SH, Akiyama M, Stillman B, Friedberg EC. Yeast autonomously replicating sequence binding protein is involved in nucleotide excision repair. Genes & Dev. 1999;13:3052–3058. doi: 10.1101/gad.13.23.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfes RJ, Zhang F, Hinnebusch AG. The transcriptional activators BAS1, BAS2, and ABF1 bind positive regulatory sites as the critical elements for adenine regulation of ADE5,7. J Biol Chem. 1997;272:13343–13354. doi: 10.1074/jbc.272.20.13343. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Shore D. RAP1: A protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+ dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JB, Gottschling DE. Telomeric chromatin modulates replication timing near chromosome ends. Genes & Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. Sir2 and Sir4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Terleth C, van Sluis CA, van de Putte P. Differential repair of UV damage in Saccharomyces cerevisiae. Nucleic Acids Res. 1989;17:4433–4439. doi: 10.1093/nar/17.12.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleth C, Schenk P, Poot R, Brouwer J, van de Putte P. Differential repair of UV damage in rad mutants of Saccharomyces cerevisiae: A possible function of G2 arrest upon UV irradiation. Mol Cell Biol. 1992;10:4678–4684. doi: 10.1128/mcb.10.9.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and Sir1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- Voisine C, Schilke B, Ohlson M, Beinert H, Marszalek J, Craig EA. Role of the mitochondrial Hsp70s, Ssc1 and Ssq1, in the maturation of Yfh1. Mol Cell Biol. 2000;20:3677–3684. doi: 10.1128/mcb.20.10.3677-3684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]