Abstract
Trichomonas vaginalis, which causes the most common nonviral sexually transmitted disease worldwide, is itself commonly infected by nonsegmented double-stranded RNA (dsRNA) viruses from the genus Trichomonasvirus, family Totiviridae. To date, cDNA sequences of one or more strains of each of three trichomonasvirus species have been reported, and gel electrophoresis showing several different dsRNA molecules obtained from a few T. vaginalis isolates has suggested that more than one virus strain might concurrently infect the same parasite cell. Here, we report the complete cDNA sequences of 3 trichomonasvirus strains, one from each of the 3 known species, infecting a single, agar-cloned clinical isolate of T. vaginalis, confirming the natural capacity for concurrent (in this case, triple) infections in this system. We furthermore report the complete cDNA sequences of 11 additional trichomonasvirus strains, from 4 other clinical isolates of T. vaginalis. These additional strains represent the three known trichomonasvirus species, as well as a newly identified fourth species. Moreover, 2 of these other T. vaginalis isolates are concurrently infected by strains of all 4 trichomonasvirus species (i.e., quadruple infections). In sum, the full-length cDNA sequences of these 14 new trichomonasviruses greatly expand the existing data set for members of this genus and substantiate our understanding of their genome organizations, protein-coding and replication signals, diversity, and phylogenetics. The complexity of this virus-host system is greater than has been previously well recognized and suggests a number of important questions relating to the pathogenesis and disease outcomes of T. vaginalis infections of the human genital mucosa.
INTRODUCTION
Trichomonas vaginalis is a flagellated protozoan parasite that colonizes the human genitourinary tract and is the causative agent of trichomoniasis, the most common nonviral sexually transmitted disease (STD) in the world (50). T. vaginalis infections are a cause of considerable morbidity worldwide, with about 180 million new infections each year, the vast majority of which occur in resource-poor settings in both developed and developing countries (13, 29). In addition to causing symptomatic vaginitis, infections by T. vaginalis are increasingly recognized as being associated with other serious clinical outcomes, including premature delivery, low birth weight, cervical cancer, and prostate cancer, as well as an increased risk of infection by HIV and human papillomavirus (4, 8, 13, 35). There is additional concern because resistance to metronidazole, the first-line treatment for trichomoniasis, is on the rise (38).
As an extracellular parasite, T. vaginalis colonizes the mucosal surfaces of the human genitourinary tract without direct invasion (parasite biology recently reviewed in reference 13). Despite its extracellular life style, however, T. vaginalis can effect mucosal damage through an apparent variety of mechanisms, some of which are dependent on epithelial cell contact (e.g., adherence-based cytotoxicity) and others of which are not (e.g., release of soluble factors such as proteinases) (reviewed in reference 13). Moreover, in many cases, T. vaginalis infection results in an exuberant host immune response that leads to inflammation and further mucosal damage (13, 14, 31, 36). Notwithstanding the isolation and characterization of several virulence factors in recent years, the pathogenesis of trichomoniasis, as well as that of the other serious clinical associations of T. vaginalis infection, remains poorly understood.
Many isolates of T. vaginalis, including fresh clinical isolates, are infected with one or more double-stranded RNA (dsRNA) viruses (1, 15, 17, 37, 42, 43, 47, 48). These T. vaginalis viruses (TVVs) form isometric viral particles (4–6, 15, 41–43) that contain a nonsegmented, 4,500- to 5,000-bp dsRNA genome encoding a viral capsid protein (CP) and a viral RNA-dependent RNA polymerase (RdRp) in 2 overlapping open reading frames (ORFs) (5, 6, 41). The CP ORF is expressed as an independent protein, whereas the RdRp ORF is expressed only as part of a CP/RdRp fusion protein following either a −1 or a +1 ribosomal frameshifting mechanism before termination of the CP ORF (5, 6, 26, 41). This fusion protein is also incorporated into viral particles at low levels (5, 26) and permits these particles to mediate viral RNA synthesis, both transcription and replication (24). Based on these characteristics, the TVVs have for some time been tentatively assigned to the family Totiviridae of nonsegmented dsRNA viruses (19, 49).
The biological significance of T. vaginalis infection by TVVs remains poorly understood. It appears that TVV infection is generally a noncytopathic phenomenon that leads to stable, persistent infections of the protozoan host without an extracellular transmission phase (44). Transmission of TVV instead occurs upon cell division and possibly also during mating (27). Despite their nonlytic life style, infection by certain TVVs has been shown to alter the surface expression of a highly immunogenic T. vaginalis protein, P270 (6, 23, 45), as well as the expression profiles of cysteine proteinases that are known virulence factors (32). Thus, infection of T. vaginalis by TVVs could very conceivably modulate the pathogenicity of T. vaginalis infections of the human genital mucosa.
To date, the near-complete cDNA sequences of only 5 TVV strains (5, 6, 25, 40, 41), plus the protein-encoding portions of a sixth strain (51), have been reported to GenBank. Based on these sequences, phylogenetic comparisons have contributed to the recent creation of a new genus in the family Totiviridae, designated Trichomonasvirus, to accommodate these viruses (20). Accompanying the creation of this new genus has been the formal recognition of 3 distinct trichomonasvirus species, designated Trichomonas vaginalis virus 1, Trichomonas vaginalis virus 2, and Trichomonas vaginalis virus 3 (5, 6, 20, 41). Only single strains of the second and third species have been reported to date (5, 6).
The recognition of 3 distinct trichomonasvirus species, coupled with the visualization by electrophoresis of several dsRNA segments of similar sizes in TVV-infected T. vaginalis isolates, has suggested the possibility of concurrent infection by more than one TVV strain in the same parasite cell (5, 16, 22). In this report, we identify a cloned clinical isolate of T. vaginalis and determine the full-length cDNA sequences of 3 distinct TVV strains that are present in that isolate, one from each of the 3 known trichomonasvirus species, thereby confirming the capacity for concurrent (in this case, triple) infections of this protozoan host. In addition, we report the full-length cDNA sequences of an additional 11 TVVs obtained from 4 other clinical isolates of T. vaginalis that we show to be each stably infected with 1, 2, or 4 distinct TVVs. Included among these additional sequences are those of 3 TVV strains representing a new, fourth trichomonasvirus species, designated Trichomonas vaginalis virus 4. The results presented here greatly expand the existing data set for members of this genus and substantiate our understanding of their genome organizations, protein coding, and replication signals.
MATERIALS AND METHODS
Ethics statement.
Vaginal swab samples were obtained from women enrolled for this study, with written informed consent, at the Onondaga County Department of Health STD Clinic in Syracuse, NY. The collection of these samples occurred in agreement with the Declaration of Helsinki principles of human research ethics and under protocols approved by the Institutional Review Boards for human subject research at the SUNY Upstate Medical University and Brigham and Women's Hospital.
T. vaginalis isolates and culture.
All T. vaginalis isolates were originally obtained from symptomatic patients. Isolate UR1 (36, 39) was obtained from the Monroe County STD clinic in Rochester, NY, in June 1999. Isolate UH9 was obtained from Upstate University Hospital in Syracuse, NY, in September 2002 but has not been described in a previous report. Isolates OC3, OC4, and OC5 were all newly obtained from the Onondaga County STD Clinic in Syracuse, NY, between November 2009 and January 2010.
T. vaginalis isolates were cultured in Diamond's modified medium (pH 6.0) with iron and 10% heat-inactivated horse serum (HyClone) at 37°C, as reported previously (9). For RNA isolation, parasite cells were harvested in late log phase (∼24 h) by centrifugation at 1,000 × g for 10 min at 4°C. The cells were then washed once with phosphate-buffered saline (pH 7.4) and pelleted at 5,000 × g for 10 min at 4°C. The pellets were lysed immediately or in some cases frozen at −70°C until use. For the experiment described in Results and Discussion, we also reisolated T. vaginalis UR1 clones as agar-embedded colonies (10), followed by regrowth into stocks.
dsRNA extraction.
Upon thawing if previously frozen, ∼1 × 107 pelleted T. vaginalis cells were disrupted in lysis buffer (0.1 M sodium acetate, pH 5.0; 1% sodium dodecyl sulfate) at room temperature, extracted with TRIzol reagent and chloroform to remove protein and DNA, precipitated with isopropanol, washed with 70% ethanol, and dissolved in water. Contaminating single-stranded RNA was next selectively precipitated from the solution by incubation with 2 M LiCl at −20°C for 30 min, followed by centrifugation at 10,000 × g for 30 min. The remaining supernatant was enriched for dsRNA, which was lastly precipitated from the solution by incubation with 4 M LiCl at −20°C for 30 min, followed by centrifugation at 10,000 × g for 30 min. This pellet was dissolved in water to create a working solution of dsRNA.
Gel electrophoresis.
For routine examination of dsRNA and reverse transcription-PCR (RT-PCR) products, electrophoresis was performed on 0.8% agarose gels in TAE buffer (40 mM Tris, 40 mM acetic acid, 1 mM EDTA, pH 8.0) and 0.5 mg/ml ethidium bromide at 4 V/cm for 45 min at room temperature. For visualization of individual dsRNA genomes from a single isolate, electrophoresis was performed on 1.0% agarose gels in TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA, pH 8.0) at 1 V/cm for 24 h at 4°C, and gels were stained in TBE containing 1 μg/ml ethidium bromide after electrophoresis.
RT-PCR and nucleotide sequencing.
To screen T. vaginalis isolates for trichomonasviruses, dsRNA extracts from cultured T. vaginalis cells were used to carry out RT-PCR using the Qiagen one-step RT-PCR kit with species-selective screening primers (see Table S1 in the supplemental material), designed from previously reported TVV sequences (5, 6, 25, 40, 41, 51). The presence of an ∼500-bp band, which when sequenced showed homology to a known trichomonasvirus sequence, was taken as confirmation for the presence of that species in the T. vaginalis isolate.
For full-length sequencing of cDNA strands for trichomonasviruses from T. vaginalis isolates UR1 and UH9, various internal fragments of each viral genome were amplified by reverse transcription followed by PCR, using other forward and reverse primers designed from the previously reported TVV sequences (5, 6, 25, 40, 41, 51). The sequences from both strands of each amplified fragment were then determined in full directly from the PCR products. For determining the sequences from near the genome termini of TVV1-UR1 and TVV1-UH9, a poly(A) tail was added to both 3′ ends of each genomic dsRNA molecule. First-strand cDNA synthesis was then performed with reverse transcriptase, a dT17 primer that additionally ended with the reverse complement of the 5′- or 3′-terminal 10-nucleotide (nt) sequence from published TVV1 strains, and an internal primer designed from the preceding sequencing results for these strains. For determining the sequences from near the genome termini of TVV2-UR1 and TVV3-UR1, a poly(A) tail was added to both 3′ ends of each genomic dsRNA molecule. First-strand cDNA synthesis was then performed with reverse transcriptase, a dT17 primer that additionally began with a 10-nt adapter sequence, and an internal primer designed from the preceding sequencing results for these strains. For confirming the sequences from the genome segment termini of TVV2-UR1, a poly(G) tail was added to both 3′ ends of each genomic dsRNA molecule. First-strand cDNA synthesis was then performed with reverse transcriptase, a dC19 primer that additionally ended with the reverse complement of the single 5′- or 3′-terminal nucleotide of published TVV1 strains, and an internal primer designed from the preceding sequencing results for these strains. For each of these amplifications of genome termini performed after 3′ tailing, sequences from one strand of each amplified fragment (reading toward the respective genome terminus) were then determined in full directly from the respective PCR product. Sequences from near the genome segment termini of TVV1-UR1, TVV1-UH9, TVV2-UR1, and TVV3-UR1 were also redetermined and confirmed by using the protocol described in the next paragraph for the trichomonasviruses from T. vaginalis isolates OC3, OC4, and OC5.
For full-length sequencing of cDNA strands for trichomonasviruses from T. vaginalis isolates OC3, OC4, and OC5, purified dsRNA was first denatured in 30 mM methylmercury hydroxide for 10 min at room temperature, followed by addition of a poly(A) tail to both 3′ ends of each dsRNA molecule using yeast poly(A) polymerase (Epicentre Technologies). First-strand cDNA synthesis was then conducted using SuperScript III reverse transcriptase (Invitrogen) and a dT20 primer with the 5′ 20-nt extension GCGATAGTCTCCGTTCAGGA. A primer directed toward the latter sequence was then used, along with internal TVV-directed primers for added specificity, to amplify and then sequence both entire cDNA strands. All DNA sequencing was performed by GeneWiz, Inc.
Nucleotide sequence accession numbers.
Full-length cDNA sequences of TVV strains described in this paper were deposited in GenBank with the accession numbers indicated: TVV1-UR1, HQ607513; TVV2-UR1, HQ607514; TVV3-UR1, HQ607515; TVV1-UH9, HQ607516; TVV1-OC3, HQ607517; TVV2-OC3, HQ607518; TVV3-OC3, HQ607519; TVV4-OC3, HQ607520; TVV1-OC4, HQ607521; TVV4-OC4 (renamed TVV4-1 as prototype of the new species), HQ607522; TVV1-OC5, HQ607523; TVV2-OC5, HQ607524; TVV3-OC5, HQ607525; and TVV4-OC5, HQ607526.
RESULTS AND DISCUSSION
Strains of 3 trichomonasvirus species identified in a T. vaginalis clinical isolate, UR1.
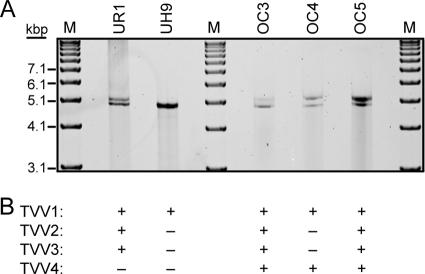
Extracts of previously characterized T. vaginalis clinical isolate UR1 (36, 39) were analyzed by agarose gel electrophoresis for the presence of dsRNAs. An ethidium-stainable band was found to migrate in the 4,000- to 5,000-bp range relative to DNA markers. Upon further analysis by extended agarose gel electrophoresis, this band was found to separate into at least 2 constituent bands (Fig. 1A). Published evidence that TVV genome lengths are consistently near this size and that T. vaginalis isolates might often be concurrently infected with more than one TVV strain led us to suspect that these bands represent the dsRNA genomes of at least 2 different TVV strains that are coinfecting T. vaginalis UR1.
Fig. 1.
dsRNA gel and TVV detection summary. (A) Preparations of dsRNA extracted from the five T. vaginalis isolates listed at top were run on a 1% agarose gel overnight, followed by staining with ethidium bromide. M, DNA markers (TrackIt 1-kb ladder from Invitrogen). (B) Summary of PCR screening and TVV detection results confirmed by determination of full-length cDNA sequences of the TVV genomes.
We next used database sequences for the TVV2 prototype, strain 1 (GenBank accession no. AF127178) (5, 20), and the TVV3 prototype, strain 1 (GenBank accession no. AF325840) (6, 20), to design forward and reverse primers for reverse transcription (RT), PCR amplification, and sequencing of any TVV strains that may be associated with T. vaginalis UR1. The primers were designed to amplify an ∼500-bp fragment from the same, CP-encoding region of the TVV2 and TVV3 genomes but in a species-selective manner (see Table S1 in the supplemental material). Each pair of primers (that designed from TVV2-1 and that designed from TVV3-1) gave rise to an ∼500-bp PCR product. Upon sequencing, these 2 PCR products were found to be distinct. The product from the primers for TVV2-1 showed the strongest sequence identity to TVV2-1 in BLAST searches: 82% at the nucleotide level and 89% identity at the amino acid level across the full length of the PCR product. The product from the primers for TVV3-1, in contrast, showed the strongest sequence identity to TVV3-1 in BLAST searches: 81% at the nucleotide level and 92% identity at the amino acid level across the full length of the PCR product. Sequence identity between the 2 PCR products was much lower: only 32% identity at the amino acid level. These results indicate that T. vaginalis UR1 is coinfected with at least 2 different TVV strains, one TVV2-like (designated TVV2-UR1) and one TVV3-like (designated TVV3-UR1).
We also used database sequences for 4 different TVV1 strains—the prototype, strain 1 (TVV1-1) (GenBank accession no. TVU08999) (20, 41), and 3 other strains designated TVV1-T5, TVV1-Ch (Ch for Changchun), and TVV1-IH-2 (GenBank accession no. TVU57898, DQ270032, and DQ528812, respectively) (25, 40, 51)—to design consensus forward and reverse species-selective primers (see Table S1 in the supplemental material) for RT, PCR amplification, and sequencing of any TVV1-like strain(s) that may also be infecting T. vaginalis UR1. The primers were designed to amplify an ∼500-bp fragment from a CP-encoding region of the TVV1 genome, which partially overlapped the region that was analyzed above for TVV2 and TVV3. Using each pair of these TVV1-specific primers, an ∼500-bp PCR product was again obtained. Upon sequencing, each of these PCR products was found to show the strongest sequence identity to the 4 TVV1 strains in BLAST searches: 76 to 80% at the nucleotide level and 79 to 86% identity at the amino acid level across the full length of each PCR product. Pairwise sequence identities between these TVV1-specific PCR products and the preceding TVV2- and TVV3-specific PCR products were much lower: ≤20% identity at the amino acid level. These results indicate that T. vaginalis UR1 is also infected with a TVV1-like strain (designated TVV1-UR1) (see summary of PCR screening results in Fig. 1B).
To determine the remaining sequences from the 3 TVV strains infecting T. vaginalis isolate UR1, we used a strategy for RT-PCR with species-selective primers (see Table S1 in the supplemental material) to obtain several other, overlapping amplicons, which in combination spanned all but the termini of the TVV genomes. We then used these and other primers to determine sequences directly from the purified PCR products. In this manner, we were able to determine fully overlapping sequences from each genomic strand (plus or minus) of each virus. To complete these sequencing efforts, we modified a well-established protocol (18) for amplifying regions proximal to and including the genome termini to generate PCR products for direct sequencing. This protocol involved 3′ poly(A) or poly(G) tailing of both genomic RNA strands, which were previously untailed, followed by tail-specific priming for RT and PCR. For PCR, a second primer was matched to an internal region of each TVV genome to provide strain specificity for each amplification. Using this protocol, we were able to complete determination of the full-length sequence of each viral genome. Analyses of the sequences are described below.
Confirmation of TVV triple infection of T. vaginalis UR1.
Having gained evidence for 3 different TVV strains in T. vaginalis UR1, we recognized that this parasite isolate had been passaged, but not clonally purified, from the original patient sample (36, 39). The UR1 stock used in the preceding experiments could thus conceivably represent a mixture of T. vaginalis strains, each containing different viruses. To address this possibility, we reisolated T. vaginalis UR1 clones as agar-embedded colonies (10) and repeated the preceding analyses on amplified stocks derived from the clonally purified parasite. The results showed that the cloned parasite contains the same 3 TVV strains as identified in the preceding section. Thus, T. vaginalis UR1 is indeed concurrently infected with at least 3 different TVV strains, one from each of the 3 known trichomonasvirus species (20).
Strains of 4 trichomonasvirus species identified in 4 other T. vaginalis clinical isolates.
We used similar strategies for gel detection of dsRNA, RT-PCR amplification, and direct sequencing from the PCR products to determine full-length sequences for the TVV strains associated with 4 other T. vaginalis clinical isolates: UH9, which was obtained in 2002 from Upstate University Hospital in Syracuse, NY, and OC3, OC4, and OC5, which were obtained in 2009 to 2010 from the Onondaga County Health Department's STD Clinic in Syracuse, NY. For T. vaginalis UH9, RNA gels showed a single band near the 5,000-bp marker, comigrating with the lower RNA band from T. vaginalis UR1 (Fig. 1A). RT-PCR screening and initial sequencing then identified the presence of only a TVV1 strain (designated TVV1-UH9) (Fig. 1B). For T. vaginalis isolates OC3, OC4, and OC5, on the other hand, RNA gels showed a double band near the 5,000-bp marker, comparable to those from T. vaginalis UR1 (Fig. 1A). RT-PCR screening and initial sequencing then identified the following: for T. vaginalis OC3, the presence of a TVV1, a TVV2, and a TVV3 strain (designated TVV1-OC3, TVV2-OC3, and TVV3-OC3, respectively); for T. vaginalis OC4, the presence of a TVV1 strain (designated TVV1-OC4) as well as a more divergent strain only distantly related to TVV1, -2, or -3; and for T. vaginalis OC5, the presence of a TVV1, a TVV2, and a TVV3 strain (designated TVV1-OC5, TVV2-OC5, and TVV3-OC5, respectively) (Fig. 1B). In subsequent work, we were able to determine the full-length sequence of each of these 9 viral genomes, which combined with those determined from T. vaginalis UR1 represent a total of 12 new full-length sequences: ones from 5 new TVV1 strains, 3 new TVV2 strains, and 3 new TVV3 strains and one new divergent strain that we consider to be the prototype of a new species, Trichomonas vaginalis virus 4 (prototype strain designated TVV4-1).
Having identified the divergent strain TVV4-1 in T. vaginalis OC4, we designed new screening primers (see Table S1 in the supplemental material) based on the TVV4-1 sequence in an effort to detect additional TVV4 strains that might be present in the other 4 T. vaginalis isolates examined in this study. In this manner, a new TVV4-related RT-PCR amplicon was generated from T. vaginalis OC5, although not from UR1, UH9, or OC3. Upon sequencing, this amplicon was found to exhibit strong sequence identity to TVV4-1 OC4, confirming it as a second TVV4 strain, TVV4-OC5. Notably, this result combines with preceding data in this report to indicate that T. vaginalis OC5 harbors 4 different TVV strains, one from each of the four known species (Fig. 1B). In subsequent work, we determined the full-length sequence of TVV4-OC5, bringing the total of new full-length TVV sequences determined in this study to 13.
After obtaining full-length sequences from the 13 new TVV strains, we recognized that our initial sets of species-selective screening primers, which were based on the 5 full-length TVV sequences that had been reported to GenBank by the time that this study was begun, bound to sequence regions that were not absolutely conserved among the different new strains of each trichomonasvirus species. To ensure that no TVV2, -3, or -4 strains had been missed in our initial screens due to poor priming, we used all 18 now-available full-length TVV sequences to identify absolutely conserved regions of 17 nt or longer within each of these species (see Table S2 in the supplemental material). We then redesigned new sets of screening primers based on these regions (Table S1) and rescreened T. vaginalis isolates UR1, UH9, OC3, and OC4 for viruses that may have been previously missed. In this manner, a TVV4-related RT-PCR amplicon was newly obtained from T. vaginalis OC3. Upon sequencing, this amplicon was found to exhibit strong sequence identity to the other TVV4 strains, confirming the presence of a new virus, TVV4-OC3. In contrast, no RT-PCR amplicons were yet obtained for a TVV2-, TVV3-, or TVV4-like strain in T. vaginalis UH9; for a TVV4-like strain in T. vaginalis UR1; or for a TVV2- or TVV3-like strain in T. vaginalis OC4. Combined with preceding data in this report, these new results indicate that T. vaginalis OC3 is another parasite isolate like T. vaginalis OC5 that harbors 4 different TVV strains, one from each of the four known trichomonasvirus species (Fig. 1B). In subsequent work, we were able to determine the full-length sequence of TVV4-OC3, bringing the total of new full-length TVV sequences determined in this study to 14, including the 3 new TVV4 strains. These additional sequences are included in the analyses and comparisons described below.
Phylogenetics.
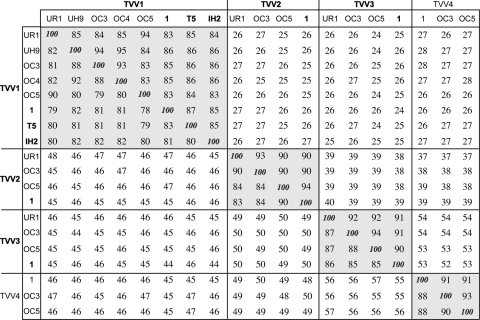
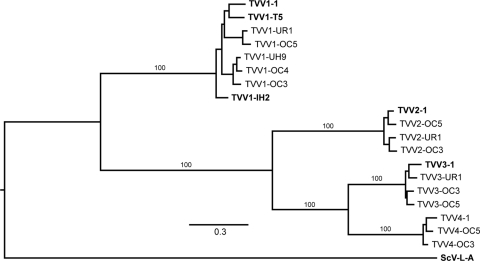
The 14 new TVV strains identified and sequenced in this study were examined for assignment to one of the 3 known trichomonasvirus species (5, 6, 20, 41), first based on amplification by species-selective primers (see Table S1 in the supplemental material) and later based on comparisons of relative overall sequence similarities with other strains of each species (Fig. 2). Assignments were then further corroborated by more formal phylogenetic analyses (Fig. 3). The illustrated phylogram is a representative one that employs the prototype of the family Totiviridae, Saccharomyces cerevisiae virus L-A (ScV-L-A) (21), as an outgroup and includes the other 5 TVV strains for which putatively full-length cDNA sequences have been deposited in GenBank to date. The 19 total TVV strains cluster into 3 well-defined clades reflecting species Trichomonas vaginalis virus 1, Trichomonas vaginalis virus 2, and Trichomonas vaginalis virus 3, with strains TVV4-1, TVV4-OC3, and TVV4-OC5 huddling outside these 3 other clusters and thus proposed to represent a previously unrecognized fourth species, Trichomonas vaginalis virus 4. As recently noted (6, 20), TVV2 and TVV3 strains are more closely related to each other than either is to TVV1 strains, and the TVV4 strains are now seen to fall within that supercluster as well, more closely related to TVV3 than to TVV2 strains (Fig. 2 and 3). Based on current results, we conclude that a whole-genome (nucleotide) or CP/RdRp (amino acid) pairwise identity score of <60% is an appropriate cutoff for assigning strains to different trichomonasvirus species (Fig. 2).
Fig. 2.
Pairwise sequence comparisons of the 19 TVV sequences. Percent identities for pairwise comparisons of the full-length genomic nucleotide sequences are shown below and left of the 100% diagonal, and those of the predicted CP/RdRp amino acid sequences are shown above and right of that diagonal. Pairwise alignments were performed by the program EMBOSS::needle(global), with default settings, at http://www.ebi.ac.uk/Tools/emboss/align. Clusters of percent identity scores of ≥78%, reflecting the four trichomonasvirus species (TVV1, -2, -3, and -4), are highlighted by background shading. Previously reported species and strains are indicated by boldface.
Fig. 3.
Phylogenetic tree for the 19 aligned TVV CP/RdRp sequences. Multiple sequence alignment was performed by the program MUSCLE v3.7, without subsequent curation. Bayesian phylogenetic analysis was then performed by the program MrBayes v3.1.2 with 10,000 iterations, a sampling interval of 100, and a burn-in period of 100. The tree was drawn with TreeDyn v198.3. All of the preceding steps were done online at http://www.phylogeny.fr (11), with the default setting unless otherwise indicated. The tree was lastly refined for publication using the program FigTree v1.3.1. Outgroup: ScV-L-A, Saccharomyces cerevisiae virus L-A (GenBank accession no. J04692), type species of the family Totiviridae.
How many different trichomonasviruses are present in each T. vaginalis clinical isolate?
To date, we have obtained agar-embeded clones for only one T. vaginalis isolate, UR1, and thereby formally shown it to be concurrently infected by strains of 3 different trichomonasvirus species. For the other parasite isolates in which strains of multiple trichomonasvirus species were found (OC3, OC4, and OC5), though it remains possible that these isolate stocks contain mixtures of parasites infected with different single TVV strains, we consider it highly likely that these other parasites are also concurrently infected, including in the case of isolates OC3 and OC5, which were found to contain strains of all 4 trichomonasvirus species. Another important point is that the number of TVV strains that we have so far identified in each parasite isolate should be viewed as only the minimum number that may be present, potentially limited in this study by our capacity to design appropriate primers to amplify each viral genome. Thus, it remains possible that additional TVV strains will be found in these same parasite isolates in future studies. Regarding the latter point, future studies involving next-generation sequencing to survey more completely the TVV contents of different T. vaginalis isolates seem especially warranted.
Important biological considerations relating to concurrent infections by multiple trichomonasviruses.
The apparent commonness of stable concurrent infections by strains of 2, 3, or 4 different trichomonasvirus species in T. vaginalis clinical isolates raises a number of interesting questions with regard to the biological basis and consequences of such concurrent infections. Do the strains of different trichomonasvirus species interact in some manner that promotes concurrent infections? Can the proteins of one trichomonasvirus species package and replicate the RNAs of the other trichomonasvirus species? Can the CPs or CP/RdRps of different trichomonasvirus species coassemble into functionally chimeric capsids? Do the strains of different trichomonasvirus species replicate and assemble in different regions of the parasite cell so as to reduce such interactions suggested in previous questions? Are important effects on the host parasite restricted to particular trichomonasvirus species or to particular combinations of those species? Are potential effects on human clinical illness restricted to T. vaginalis strains infected with strains of particular trichomonasvirus species or with particular combinations of those species? These and other important questions should be the topics of future studies.
Concurrent stable infections of host cells with 2 distinct viruses are known to occur for members of the genera Totivirus (e.g., ScV-L-A and ScV-L-BC; Ustilago maydis virus H1 [UmV-H1] and UmV-H2) and Victorivirus (e.g., Sphaeropsis sapinea RNA virus 1 [SsRV1] and SsRV2) in the family Totiviridae (19, 49). No examples of heterologous encapsidation have been reported in these cases. Concurrent infections may be of common occurrence among fungal viruses, as well as protozoan viruses, because of the intracellular means by which they are transmitted in nature and their commonly nonlytic life style. The identification of not just double but even triple and quadruple infections of host cells in this study is unusual if not unprecedented.
Diagnostic implications.
The substantial expansion of known full-length TVV sequences presented in this report, along with the identification of a fourth, previously unknown trichomonasvirus species, offers greatly improved opportunities for detection of trichomonasvirus infections in clinical isolates of T. vaginalis. In addition to use for designing primers for PCR-based detection methods as in this study, the expanded repertoire of TVV sequences should improve the quality of nucleic acid microarrays by allowing detection of TVV RNA based on strongly conserved sequences within each trichomonasvirus species (2).
Similar lengths of the new TVV genomes.
Some basic characteristics of the newly sequenced TVV genomes are listed in Table 1. The genome lengths of all 14 of these TVV strains vary across only a narrow range from 4,670 to 4,944 bp. These values are consistent with the electrophoretic mobilities of the genomic dsRNA molecules on agarose gels (Fig. 1). For new strains within each species, the range is even much narrower: 4,678 to 4,684 bp for TVV1, 4,671 to 4,674 bp for TVV2, 4,842 to 4,846 bp for TVV3, and 4,942 to 4,944 bp for TVV4.
Table 1.
Basic characteristics of TVV genome sequences
| TVV strain | Genome length (bp) | Mass (MDa)a | G:C content (%)a |
|---|---|---|---|
| TVV1-UR1 | 4,684 | 3.01 | 46 |
| TVV1-UH9 | 4,678 | 3.00 | 45 |
| TVV1-OC3 | 4,684 | 3.01 | 45 |
| TVV1-OC4 | 4,680 | 3.01 | 45 |
| TVV1-OC5 | 4,680 | 3.01 | 45 |
| TVV1-1b | 4,647 | 2.98 | 45 |
| TVV1-T5 | 4,648 | 2.98 | 45 |
| TVV1-IH2 | 4,647 | 2.98 | 45 |
| TVV2-UR1 | 4,674 | 3.00 | 45 |
| TVV2-OC3 | 4,674 | 3.00 | 45 |
| TVV2-OC5 | 4,671 | 3.00 | 45 |
| TVV2-1 | 4,674 | 3.00 | 46 |
| TVV3-UR1 | 4,845 | 3.11 | 47 |
| TVV3-OC3 | 4,846 | 3.11 | 48 |
| TVV3-OC5 | 4,842 | 3.11 | 48 |
| TVV3-1 | 4,844 | 3.11 | 48 |
| TVV4-1 | 4,943 | 3.18 | 49 |
| TVV4-OC3 | 4,944 | 3.18 | 49 |
| TVV4-OC5 | 4,942 | 3.18 | 49 |
As calculated at http://mbcf.dfci.harvard.edu/docs/oligocalc.html.
Sequences previously reported to GenBank are shown in bold.
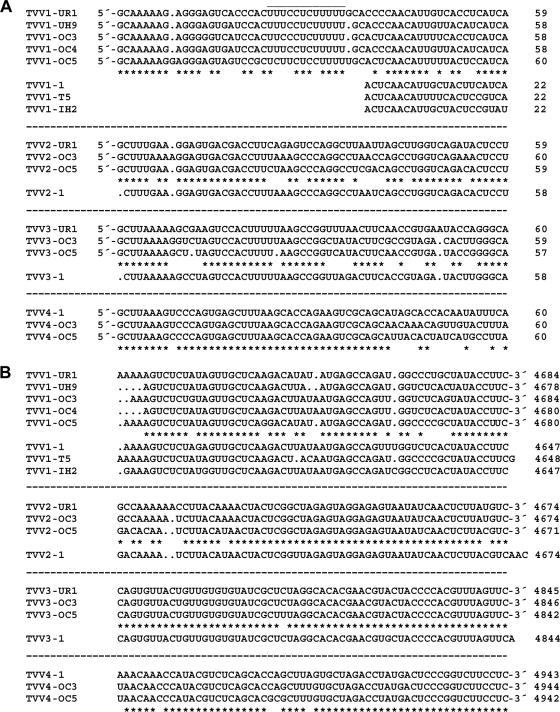
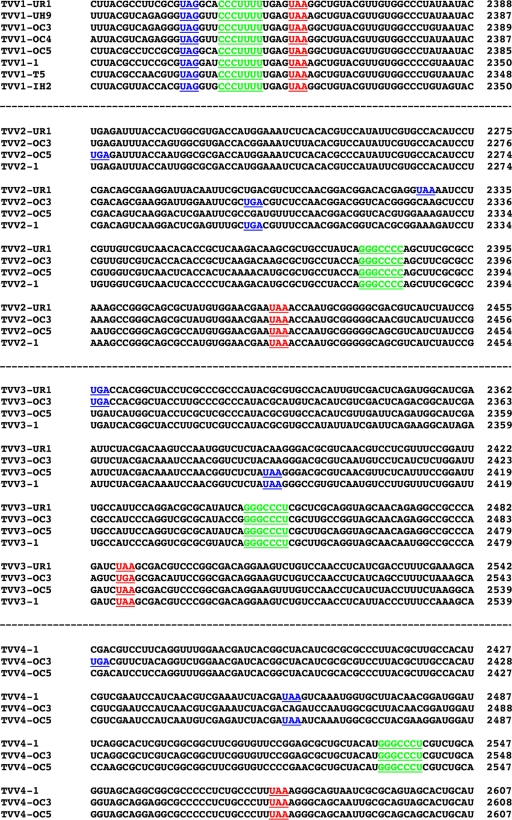
Among the putatively full-length sequences of TVV strains previously deposited in GenBank, the one TVV2 sequence and the one TVV3 sequence each fall within the respective range of genome lengths for those trichomonasvirus species observed in the current study (Table 1). Notably, however, the 3 previously deposited TVV1 sequences fall outside this range and are instead consistently 30 to 40 bp shorter than the TVV1 sequences newly determined here. Upon aligning all 8 TVV1 sequences, it is clear that each of the previously deposited TVV1 sequences is missing 30 to 40 bp from the 5′ end of the genomic plus strand (Fig. 4A). In other words, the previously deposited TVV1 sequences appear to have been truncated at that end. Although it is conceivable that the previous TVV1 sequences represent a distinct clade in which these terminal sequences are indeed absent, phylogenetic trees do not support that conclusion, since the old and new TVV1 sequences are intermixed (Fig. 3). Thus, we conclude that the previously deposited TVV1 sequences were likely truncated due to sequencing errors. In fact, in our new TVV1 sequences, the sequence aligning with the end of the previous sequences is immediately preceded by a long U-rich tract (Fig. 4A), which may have been confused with a homopolymer tail in the previous studies.
Fig. 4.
Terminal sequences of TVV plus strands. Sequences at the 5′ (A) and 3′ (B) termini are shown. Multiple sequence alignments for the viruses within each species (TVV1, -2, -3, and -4) were performed by the program Clustal 2.0.12, with default settings, at http://www.ebi.ac.uk/Tools/clustalw2. Gaps within each sequence are indicated by periods. Conserved positions in the consensus are indicated by asterisks below each set of newly determined sequences. Previously reported TVV sequences are shown below the consensus in each set. The positions of the rightmost nucleotide in each line of sequence are indicated at right.
RNA gels.
For the T. vaginalis strains showing 2 dsRNA bands in Fig. 1B, we interpret these 2 bands as an upper band representing the TVV3 and/or TVV4 genome and a lower band representing the TVV1 genome with or without a comigrating TVV2 genome. Thus, T. vaginalis UH9, which we have identified to contain only a TVV1 strain, shows only the lower band, and T. vaginalis OC4, which we have shown to contain only a TVV1 and a TVV4 strain, still shows both bands. The lower mobility of the TVV3 and TVV4 genomes according to this interpretation is consistent with their lengths being greater than those of TVV1 and TVV2 (Table 1). Moreover, the comigration of the TVV1 and TVV2 genomes according to this interpretation is consistent with their similar lengths (Table 1). We continue to investigate methods to provide better electrophoretic separation of all four viral genomes.
Terminal sequences.
Sequences at the extreme ends of viral dsRNA genome segments, though outside the protein-coding region(s), are commonly conserved among related strains because they include important signals for RNA synthesis and/or packaging. For example, the 3′ end of each strand must be able to be recognized, or at least well accommodated, by the viral RdRp for proper initiation of plus- and minus-strand synthesis. For the strains within each trichomonasvirus species, we observe that there are indeed fully conserved sequences at both ends, with the lengths and identities of these sequences varying between species (Fig. 4). At the 5′ end of the plus strand (Fig. 4A), there is the 8-nt sequence 5′-GCAAAAAG in our 5 new TVV1 strains, the 5-nt sequence 5′-GCUUU in our 3 new TVV2 strains, the 8-nt sequence 5′-GCUUAAAA in our 3 new TVV3 strains, and the 8-nt sequence 5′-GCUUAAAG in our 3 new TVV4 strains. At the 3′ end of the plus strand (Fig. 4B), there is the 9-nt sequence UAUACCUUC-3′ in our 5 new TVV1 strains, the 3-nt sequence GUC-3′ in our 3 new TVV2 strains, the 34-nt sequence UAGGCACACGAACGUACUACCCCACGUUUAGUUC-3′ in our 3 new TVV3 strains, and the 30-nt sequence GUGCUAGACCUAUGACUCCCGGUCUUCCUC-3′ in our 3 new TVV4 strains. Of additional note in these comparisons is that all of our new TVV sequences contain a conserved dinucleotide at each end, 5′-GC and UC-3′, which we therefore propose to be characteristic of all members of the genus Trichomonasvirus.
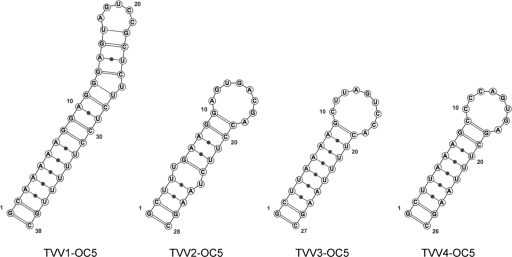
RNA folding predictions can be useful for identifying structures important for packaging, RNA synthesis, or translation (9). With regard to the 5′-proximal plus-strand sequences of the 14 new TVV strains described in this study, an interesting observation is that all exhibit the capacity to form a 26- to 38-nt-long stem-loop structure that begins with the 5′-terminal G residue and ends with a downstream C (or in one case U) residue that base pairs with the 5′-terminal G at the base of the predicted stem (Fig. 5). For all but 1 of the 14 new TVV strains, this stem-loop is present in the predicted minimum-free-energy (MFE) structure for the whole plus strand. We propose that this 5′-terminal stem-loop is involved in protecting that end of each TVV plus strand from degradation in infected cells and that it may play other roles as well in packaging, RNA synthesis, or translation. A similar structure is not consistently predicted for the 3′-proximal plus-strand sequences of the 14 new TVV strains, and indeed in all of the predicted centroid structures and all but 2 of the predicted MFE structures for the whole plus strand, four residues at the 3′ terminus are unpaired, which may be important for RdRp recognition.
Fig. 5.
Predicted 5′ stem-loop structures in full-length TVV plus-strand sequences. Structures were predicted by the program RNAfold, with default settings, at http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi. Results for the four TVV strains from T. vaginalis isolate OC5 are shown as representative examples. Note that the stem encompasses the 5′ terminus, nucleotide G1, in each virus.
Another interesting observation from RNA folding predictions is that the plus-strand RNAs of all 14 new TVV strains described in this study exhibit a propensity for 5′-3′ association through sequences at or near the 2 termini. Even though base pairing between the terminal regions does not usually appear in the MFE structure predicted for each TVV plus strand, the propensity for 5′-3′ association is consistently seen in the base-pairing probability matrix (28) of each RNA (see Fig. S1 in the supplemental material). In other viruses, 5′-3′ association has also been proposed to be important for packaging, RNA synthesis, or translation (7, 30).
Terminal UTRs.
The plus-strand 5′ untranslated regions (UTRs) of all 14 new TVV strains are quite long, ranging from 295 to 363 nt. For the strains within each species, the range is even narrower and also distinct for each trichomonasvirus species: 322 to 326 nt for TVV1 strains, 295 to 297 nt for TVV2 strains, 359 to 363 nt for TVV3 strains, and 337 to 338 nt for TVV4 strains (Table 2; Fig. 6). The putatively full-length sequences of TVV2 and TVV3 strains previously deposited in GenBank include 5′ UTRs consistent with these respective ranges. The putatively full-length sequences of TVV1 strains previously deposited in GenBank include 5′ UTRs that are consistently 30 to 40 nt shorter than those reported here, due to the putative sequencing-based truncations suggested above. For all of the TVV strains, the long 5′ UTRs are consistent with containing signals not only for RNA synthesis and packaging but also for regulating translation, such as in the form of an internal ribosome entry site. Indeed, among the 14 newly determined TVV sequences, the 5′ UTRs include as many as 8 AUG codons preceding the conserved AUG near the beginning of the CP ORF, suggesting that some mechanism must be in operation to allow sufficient bypass of these upstream potential start codons for ribosomes to gain access to the proper downstream start codon for CP translation.
Table 2.
Basic plus-strand coding characteristics of TVV genome sequences
| TVV strain | ORF (nucleotide positions)a |
Length (aa), mass (kDa) |
Lengths (nt) of 5′, 3′ UTRsd | ||
|---|---|---|---|---|---|
| CP | RdRp | CPb | CP/RdRpc | ||
| TVV1-UR1 | 271–2359 | 2346–4613 | 678, 74.0 | 1,429, 159 | 325, 68 |
| TVV1-UH9 | 319–2358 | 2345–4612 | 678, 74.6 | 1,429, 160 | 324, 63 |
| TVV1-OC3 | 321–2360 | 2347–4614 | 678, 74.5 | 1,429, 160 | 326, 67 |
| TVV1-OC4 | 316–2358 | 2345–4612 | 678, 74.6 | 1,429, 160 | 324, 65 |
| TVV1-OC5 | 317–2356 | 2343–4610 | 678, 74.1 | 1,429, 160 | 322, 67 |
| TVV1-1e | 282–2321 | 2308–4575 | 678, 74.9 | 1,429, 160 | 287, 69 |
| TVV1-T5 | 280–2319 | 2306–4573 | 678, 74.4 | 1,429, 160 | 285, 72 |
| TVV1-IH2 | 282–2321 | 2308–4575 | 678, 74.6 | 1,429, 160 | 287, 69 |
| TVV2-UR1 | 204–2423 | 2330–4603 | 709, 78.3 | 1,436, 162 | 296, 68 |
| TVV2-OC3 | 205–2424 | 2304–4604 | 709, 78.2 | 1,436, 162 | 297, 67 |
| TVV2-OC5 | 236–2422 | 2218–4602 | 709, 78.4 | 1,436, 162 | 295, 66 |
| TVV2-1 | 236–2422 | 2302–4602 | 709, 78.5 | 1,436, 162 | 295, 69 |
| TVV3-UR1 | 345–2486 | 2306–4690 | 708, 79.3 | 1,443, 162 | 362, 152 |
| TVV3-OC3 | 346–2487 | 2307–4691 | 708, 79.0 | 1,443, 162 | 363, 152 |
| TVV3-OC5 | 342–2483 | 2390–4687 | 708, 79.2 | 1,443, 162 | 359, 152 |
| TVV3-1 | 342–2483 | 2390–4687 | 708, 79.4 | 1,443, 162 | 359, 154 |
| TVV4-1 | 311–2575 | 2461–4779 | 746, 81.5 | 1,481, 165 | 337, 161 |
| TVV4-OC3 | 312–2576 | 2372–4780 | 746, 81.5 | 1,481, 165 | 338, 161 |
| TVV4-OC5 | 311–2575 | 2461–4779 | 746, 81.7 | 1,481, 165 | 337, 160 |
Open reading frame as defined by bracketing stop codons.
First conserved Met codon within the CP ORF has been interpreted as the start codon.
A +1 or −1 frameshift has been included within the second conserved trinucleotide of the proposed slippery sequence to allow interpretation of CP/RdRp length.
Untranslated regions at the genomic plus-strand termini. The 3′ UTR has been defined as excluding the RdRp stop codon.
Sequences previously reported to GenBank are shown in bold.
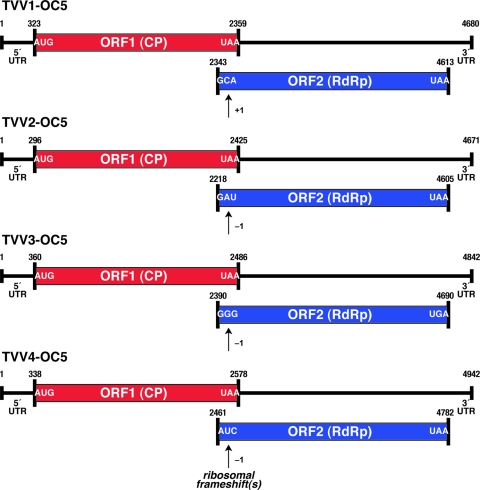
Fig. 6.
Coding diagrams for full-length TVV plus-strand sequences. ORF1 (CP) and ORF2 (RdRp) are diagrammed for TVV1, -2, -3, and -4 strains. The first and last codons for each ORF are indicated. In each virus, the RdRp is thought to be expressed as a CP/RdRp fusion following ribosomal frameshifting as indicated. 5′ and 3′ untranslated regions (UTRs) are also labeled, along with the position numbers of the first and last nucleotides of each genome and ORF. The specific nucleotides and nucleotide position numbers in this figure are shown for the four TVV strains from T. vaginalis isolate OC5 as representative examples.
The plus-strand 3′ UTRs of all 14 new TVV strains are much shorter than the 5′ UTRs, varying across only a narrow range from 63 to 161 nt (Table 2; Fig. 6). For the strains within each trichomonasvirus species, the range is even narrower: 63 to 68 nt for TVV1 strains, 66 to 68 nt for TVV2 strains, 152 nt for TVV3 strains, and 160 to 161 nt for TVV4 strains. The putatively full-length sequences of TVV1, TVV2, and TVV3 strains previously deposited in GenBank include 3′ UTRs consistent with these respective ranges.
Coding strategies for CP and RdRp.
The plus-strand sequences of all 14 new TVV strains in this study contain 2 long, partially overlapping ORFs (Table 2; Fig. 6). In each case, the upstream ORF encoding CP overlaps the downstream ORF encoding RdRp by 14 to 205 nt. In all 8 full-length TVV1 sequences determined to date, this overlap region is consistently only 14 nt, bracketed by an upstream UAG stop codon that immediately precedes the RdRp ORF and a downstream UAA stop codon that terminates the CP ORF (Fig. 7). In TVV2, -3, and -4 strains, the overlap region is larger and more variably sized: 94 to 205 nt in TVV2 strains, 94 to 181 nt in TVV3 strains, and 115 to 205 nt in TVV4 strains. Inside the overlap region in each virus, regardless of species, is a putative “slippery sequence” (XXXYYYZ heptanucleotide) that promotes ribosomal frameshifting during translation to allow bypass of the impending CP stop codon and consequent extension of translation to yield the CP/RdRp fusion protein. This putative slippery sequence is CCCUUUU in all TVV1 strains, GGGCCCC in all TVV2 strains, and GGGCCCU in all TVV3 and TVV4 strains. In TVV1 strains, the RdRp ORF is in the +1 frame relative to CP, and thus, the productive frameshifting event must be +1, or perhaps −2. In TVV2, -3, and -4 strains, the RdRp ORF is in the −1 frame relative to CP. This divergent property of TVV1 versus TVV2, -3, and -4 strains, +1 versus −1 frameshifting, correlates with their relative separations in phylogenetic analyses (Fig. 1). Expression of RdRp as part of a CP/RdRp fusion protein consequent to ribosomal frameshifting (usually −1) has been demonstrated for several other members of the family Totiviridae (19, 49), and in the case of TVV1-1 and TVV2-1, the CP/RdRp fusion product has been demonstrated by immunoblotting (5, 26). Further dissection of frameshifting mechanisms in the different TVV strains is a subject of current investigation, including analysis of potential RNA structure elements (e.g., pseudoknots) that may have important regulatory functions as seen in some other members of the family Totiviridae (12, 46). One observation of potential interest is that Y = C is disfavored in the slippery sequences of −1 frameshifting signals from a wide range of animal, plant, and fungal viruses (3), and thus, its presence in TVV2, -3, and -4 strains represents a deviation that might relate to divergent ribosomal mechanisms in the more primitive eukaryotic host T. vaginalis.
Fig. 7.
Frameshifting region of TVV plus-strand sequences. Multiple sequence alignments for the viruses within each trichomonasvirus species (TVV1, -2, -3, and -4) were performed by the program Clustal 2.0.12, with default settings, at http://www.ebi.ac.uk/Tools/clustalw2. No gaps are present in the illustrated region. Previously reported TVV sequences are shown at the bottom of each set. The positions of the rightmost nucleotide in each line of sequence are indicated at right. The proposed slippery sequence (XXXYYYZ heptanucleotide) in each virus is underlined and colored green. The stop codon defining the 5′ end of the RdRP ORF in each virus is underlined and colored blue. The stop codon defining the 3′ end of the CP ORF in each virus is underlined and colored red.
Conserved regions of CP and RdRp.
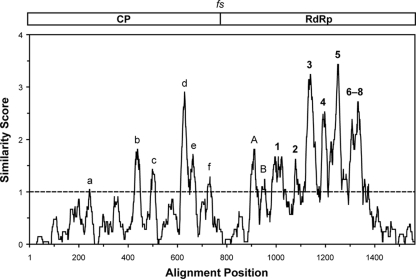
Multiple sequence alignments of the 19 predicted CP/RdRp fusion protein sequences (14 new and 5 old) used in Fig. 2 and 3 revealed only limited conservation in the CP region. This finding is consistent with low pairwise identity scores between the CPs of strains from different trichomonasvirus species (see Fig. S2 in the supplemental material) and is also well illustrated in a moving-average similarity plot obtained from the consensus of the 19 aligned CP/RdRp sequences (Fig. 8). Thus, although the 3-dimensional CP structures of the different TVV strains might be expected to be relatively similar, such structural conservation is not well reflected in the primary sequences. Of potential interest, however, are the few small regions of conserved CP sequences that are in fact seen, e.g., those labeled a to f in Fig. 8 (for aligned sequences, see Data Set S1 in the supplemental material). Whether these conserved regions have primarily structural or functional significance remains to be determined.
Fig. 8.
Similarity plot for the consensus of the 19 aligned TVV CP/RdRp sequences. The multiple sequence alignment was the same as that for Fig. 3. Similar scores were averaged over running windows of 21 amino acid positions. Scoring was as follows: identical in all 19 sequences, 4; very similar in all 19 sequences, 2; similar in all 19 sequences, 1. Averaging and graphing were performed with Microsoft Excel 2004 v11.6. High-scoring peaks in the CP region are labeled a to f. RdRp motifs are labeled 1 to 8, and other high-scoring peaks in the RdRp region are labeled A and B.
Multiple sequence alignments of the 19 predicted CP/RdRp sequences revealed higher levels of conservation in the RdRp region than in the CP region. This finding is consistent with the higher pairwise identity scores between the RdRp sequences of strains from different trichomonasvirus species (see Fig. S2 in the supplemental material) and is also well illustrated in the moving-average similarity plot described above (Fig. 8). In the RdRp region of that plot, several peaks of conservation are observed, mostly corresponding to the 8 RdRp motifs known to be conserved among dsRNA viruses (34), as labeled 1 to 8 in Fig. 8 (aligned sequences shown in Data Set S1). Also of interest are the additional small regions of conserved sequences seen near the N-terminal end of the RdRp region, as labeled A and B in Fig. 8 (aligned sequences shown in Data Set S1). Region A is especially interesting in that it aligns with a region of the ScV-L-A RdRp that has been previously shown to be required for plus-strand RNA binding in vitro and packaging into virus particles in vivo (33). Perhaps not surprisingly, due to its expected role as a flexible hinge between the CP and RdRp regions, the frameshift region of the CP/RdRp fusion proteins is one of especially low conservation in the moving-average plot (Fig. 8).
Additional plus-strand ORFs.
Viral genome sequences often contain additional, moderately sized ORFs (encoding ≥100 amino acids [aa]) of unknown translatability or significance. In the TVV2-1 plus-strand sequence, for example, Bessarab et al. (5) noted the presence of 2 such ORFs: ORF3 at nucleotide positions 936 to 1775, encoding 219 aa from the first AUG codon, and ORF4 at nucleotide positions 1794 to 2495, encoding 233 aa from the first AUG start codon. Efforts failed, however, to detect these potential translation products by immunoblotting with peptide-directed antisera. All of the newly sequenced TVV strains in this report, except for TVV3-OC3 and TVV3-OC5, include a few additional ORFs of ≥100 aa in length in their plus-strand sequences. A total of 23 such ORFs are present in these 12 other strains, and they could potentially encode proteins between 101 and 271 aa in length. Among these ORFs, the only ones that appear worthy of some consideration, in that they are found in all strains to date of a particular species, are those in TVV2 strains corresponding to the previously identified ORF3 of TVV2-1 (5). Even in that case, however, the corresponding ORFs show variable start and stop positions and lengths (e.g., encoding 136 to 271 aa) among the one old and three new TVV2 strains. We therefore consider it unlikely that any of these additional ORFs in the TVV plus-strand sequences are functionally significant.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the contribution of Shelley Gilroy and her clinical staff for providing vaginal swab samples from women enrolled at the Onondaga County Department of Health STD Clinic in Syracuse, NY. We also thank Elaine Freimont for technical assistance.
This work was supported by a Harvard Catalyst Pilot Grant (R.N.F. and M.L.N.) funded through award number UL1-RR025758, Harvard Clinical and Translational Science Center, from the National Center of Research Resources, as well as by NIH grants RC1-AI086788 (R.N.F., B.N.S., and M.L.N.), R21-HD054451 (R.N.F. and B.N.S.), and R01-AI079085 (R.N.F. and B.N.S.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Alderete J. F., et al. 2003. Trichomonas vaginalis: evaluating capsid proteins of dsRNA viruses and the dsRNA virus within patients attending a sexually transmitted disease clinic. Exp. Parasitol. 103:4450. [DOI] [PubMed] [Google Scholar]
- 2. Baptista C. S., Wu X., Munroe D. J. April 2007. International patent WO/2007/130519. Viral nucleic acid microarray and method of use. World Intellectual Property Organization, Geneva, Switzerland [Google Scholar]
- 3. Bekaert M., et al. 2003. Towards a computational model for −1 eukaryotic frameshifting sites. Bioinformatics 19:327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benchimol M., Monteiro S., Chang T. H., Alderete J. F. 2002. Virus in Trichomonas—an ultrastructural study. Parasitol. Int. 51:293–298 [DOI] [PubMed] [Google Scholar]
- 5. Bessarab I. N., Liu H. W., Ip C. F., Tai J. H. 2000. The complete cDNA sequence of a type II Trichomonas vaginalis virus. Virology 267:350–359 [DOI] [PubMed] [Google Scholar]
- 6. Bessarab I. N., Nakajima R., Liu H. W., Tai J. H. 26 November 2010. Identification and characterization of a type III Trichomonas vaginalis virus in the protozoan pathogen Trichomonas vaginalis. Arch. Virol. [Epub ahead of print.] doi:10.1007/s00705-010-0858-y [DOI] [PubMed] [Google Scholar]
- 7. Chen D., Patton J. T. 1998. Rotavirus RNA replication requires a single-stranded 3′ end for efficient minus-strand synthesis. J. Virol. 72:7387–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cotch M. F., et al. 1997. Trichomonas vaginalis associated with low birth weight and preterm delivery. Sex. Transm. Dis. 24:1–8 [DOI] [PubMed] [Google Scholar]
- 9. Davis M., Sagan S. M., Pezacki J. P., Evans D. J., Simmonds P. 2008. Bioinformatic and physical characterizations of genome-scale ordered RNA structure in mammalian RNA viruses. J. Virol. 82:11824–11836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delgadillo M. G., Liston D. R., Niazi K., Johnson P. J. 1997. Transient and selectable transformation of the parasitic protist Trichomonas vaginalis. Proc. Natl. Acad. Sci. U. S. A. 94:4716–4720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dereeper A., et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dinman J. D., Icho T., Wickner R. B. 1991. A −1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. U. S. A. 88:174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fichorova R. N. 2009. Impact of T. vaginalis infection on innate immune responses and reproductive outcome. J. Reprod. Immunol. 83:185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fichorova R. N., et al. 2006. Trichomonas vaginalis lipophosphoglycan triggers a selective upregulation of cytokines by human female reproductive tract epithelial cells. Infect. Immun. 74:5773–5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flegr J., Cerkasov J., Kulda J., Tachezy J., Stokrová J. 1987. The dsRNA of Trichomonas vaginalis is associated with virus-like particles and does not correlate with metronidazole resistance. Folia Microbiol. (Praha) 32:345–348 [DOI] [PubMed] [Google Scholar]
- 16. Flegr J., Cerkasov J., Stokrová J. 1988. Multiple populations of double-stranded RNA in two virus-harbouring strains of Trichomonas vaginalis. Folia Microbiol. (Praha) 33:462–465 [DOI] [PubMed] [Google Scholar]
- 17. Fraga J., Rojas L., Sariego I., Fernández-Calienes A. 2005. Double-stranded RNA viral infection in Cuban Trichomonas vaginalis isolates. Braz. J. Infect. Dis. 9:521–524 [DOI] [PubMed] [Google Scholar]
- 18. Frohman M. A. 1993. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 218:340–356 [DOI] [PubMed] [Google Scholar]
- 19. Ghabrial S. A. 2008. Totiviruses, p. 163–174 In Mahy B. W. J., Van Regenmortel M. H. V. (ed.), Encyclopedia of virology, 3rd ed., vol. 5 Elsevier Academic Press, San Diego, CA [Google Scholar]
- 20. Goodman R. P., Ghabrial S. A., Fichorova R. N., Nibert M. L. 2011. Trichomonasvirus: a new genus of protozoan viruses in the family Totiviridae. Arch. Virol. 156:171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Icho T., Wickner R. B. 1989. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J. Biol. Chem. 264:6717–6723 [PubMed] [Google Scholar]
- 22. Khoshnan A., Alderete J. F. 1993. Multiple double-stranded RNA segments are associated with virus particles infecting Trichomonas vaginalis. J. Virol. 67:6950–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khoshnan A., Alderete J. F. 1994. Trichomonas vaginalis with a double-stranded RNA virus has upregulated levels of phenotypically variable immunogen mRNA. J. Virol. 68:4035–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khoshnan A., Provenzano D., Alderete J. F. 1994. Unique double-stranded RNAs associated with the Trichomonas vaginalis virus are synthesized by viral RNA-dependent RNA polymerase. J. Virol. 68:7108–7114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim J. W., Chung P. R., Hwang M. K., Choi E. Y. 2007. Double-stranded RNA virus in Korean isolate IH-2 of Trichomonas vaginalis. Korean J. Parasitol. 45:87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu H. W., Chu Y. D., Tai J. H. 1998. Characterization of Trichomonas vaginalis virus proteins in the pathogenic protozoan T. vaginalis. Arch. Virol. 143:963–970 [DOI] [PubMed] [Google Scholar]
- 27. Malik S. B., Pightling A. W., Stefaniak L. M., Schurko A. M., Logsdon J. M. 2008. An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS One 3:e2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McCaskill J. S. 1990. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 29:1105–1119 [DOI] [PubMed] [Google Scholar]
- 29. McClelland R. S. 2008. Trichomonas vaginalis infection: can we afford to do nothing? J. Infect. Dis. 197:487–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mir M. A., Brown B., Hjelle B., Duran W. A., Panganiban A. T. 2006. Hantavirus N protein exhibits genus-specific recognition of the viral RNA panhandle. J. Virol. 80:11283–11292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petrin D., Delgaty K., Bhatt R., Garber G. 1998. Clinical and microbiological aspects of Trichomonas vaginalis. Clin. Microbiol. Rev. 11:300–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Provenzano D., Khoshnan A., Alderete J. F. 1997. Involvement of dsRNA virus in the protein composition and growth kinetics of host Trichomonas vaginalis. Arch. Virol. 142:939–952 [DOI] [PubMed] [Google Scholar]
- 33. Ribas J. C., Fujimura T., Wickner R. B. 1994. Essential RNA binding and packaging domains of the Gag-Pol fusion protein of the L-A double-stranded RNA virus of Saccharomyces cerevisiae. J. Biol. Chem. 269:28420–28428 [PubMed] [Google Scholar]
- 34. Routhier E., Bruenn J. A. 1998. Functions of conserved motifs in the RNA-dependent RNA polymerase of a yeast double-stranded RNA virus. J. Virol. 72:4427–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwebke J. R., Burgess D. 2004. Trichomoniasis. Clin. Microbiol. Rev. 17:794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singh B. N., et al. 2009. Structural details and composition of Trichomonas vaginalis lipophosphoglycan in relevance to the epithelial immune function. Glycoconj. J. 26:3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Snipes L. J., et al. 2000. Molecular epidemiology of metronidazole resistance in a population of Trichomonas vaginalis clinical isolates. J. Clin. Microbiol. 38:3004–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sobel J. D., Nagappan V., Nyirjesy P. 1999. Metronidazole-resistant vaginal trichomoniasis—an emerging problem. N. Engl. J. Med. 341:292–293 [DOI] [PubMed] [Google Scholar]
- 39. Sommer U., et al. 2005. Identification of Trichomonas vaginalis cysteine proteases that induce apoptosis in human vaginal epithelial cells. J. Biol. Chem. 280:23853–23860 [DOI] [PubMed] [Google Scholar]
- 40. Su H. M., Tai J. H. 1996. Genomic organization and sequence conservation in type I Trichomonas vaginalis viruses. Virology 222:470–473 [DOI] [PubMed] [Google Scholar]
- 41. Tai J. H., Chui-Fun I. P. 1995. The cDNA sequence of Trichomonas vaginalis virus-T1 double-stranded RNA. Virology 206:773–777 [DOI] [PubMed] [Google Scholar]
- 42. Wang A. L., Wang C. C. 1985. A linear double-stranded RNA in Trichomonas vaginalis. J. Biol. Chem. 260:3697–3702 [PubMed] [Google Scholar]
- 43. Wang A. L., Wang C. C. 1986. The double-stranded RNA in Trichomonas vaginalis may originate from virus-like particles. Proc. Natl. Acad. Sci. U. S. A. 83:7956–7960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang A. L., Wang C. C. 1991. Viruses of parasitic protozoa. Parasitol. Today 7:76–80 [DOI] [PubMed] [Google Scholar]
- 45. Wang A., Wang C. C., Alderete J. F. 1987. Trichomonas vaginalis phenotypic variation occurs only among trichomonads infected with the double-stranded RNA virus. J. Exp. Med. 166:142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang A. L., Yang H. M., Shen K. A., Wang C. C. 1993. Giardiavirus double-stranded RNA genome encodes a capsid polypeptide and a gag-pol-like fusion protein by a translation frameshift. Proc. Natl. Acad. Sci. U. S. A. 90:8595–8599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weber B., Mapeka T. M., Maahlo M. A., Hoosen A. A. 2003. Double stranded RNA virus in South African Trichomonas vaginalis isolates. J. Clin. Pathol. 56:542–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wendel K., Rompalo A., Erbelding E., Chang T. H., Alderete J. F. 2002. Double-stranded RNA viral infection of Trichomonas vaginalis infecting patients attending a sexually transmitted diseases clinic. J. Infect. Dis. 186:558–561 [DOI] [PubMed] [Google Scholar]
- 49. Wickner R. B., Wang C. C., Patterson J. L. 2005. Family Totiviridae, p. 571–580 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 50. World Health Organization 2001. Global programme of AIDS, p. 27–29 World Health Organization, Geneva, Switzerland [Google Scholar]
- 51. Zhao Y. P., et al. 2006. Cloning and sequence analysis of a partial gene of Trichomonas vaginalis dsRNA virus. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 34:389–390 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.