Abstract
Herpesvirus DNA replication proceeds via concatemeric replicative intermediates that are comprised of head-to-tail linked genomes. Genome maturation is carried out by the terminase, an enzyme complex that mediates both the insertion of concatemer DNA into capsids and its subsequent cleavage to release genomes within these capsids. This cleavage is sequence specific, but the governing cis-acting DNA sequences are only partially characterized. Two highly conserved motifs, the pac1 and pac2 motifs, lie near the ends of herpesvirus genomes and are known to be critical for genome maturation. In murine cytomegalovirus, poorly conserved sequences distal to the pac2 motif up to 150 bp from the point of cleavage are also important for cleavage. Here, we sought to identify the cleavage/packaging signals of human cytomegalovirus. Our results show that a previously proposed pac2-like poly(A) tract is dispensable for cleavage/packaging function and suggest that human cytomegalovirus may utilize a cryptic pac2 motif that lacks a poly(A) tract characteristic of pac2 motifs in other herpesviruses. Additional distal sequences 47 to 100 bp from the point of cleavage were found to enhance cleavage efficiency. These results should facilitate the identification of trans-acting factors that bind to these cis elements and elucidation of their functions. Such information will be critical for understanding the molecular basis of this complex process.
INTRODUCTION
Herpesviruses have large (130- to 235-kb) linear double-stranded DNA genomes that replicate via concatemeric intermediates consisting of head-to-tail linked genomes. An enzyme complex called terminase functions to package the concatemers into preformed capsids and cleave the DNA at precise locations to release unit-length genomes within the capsids (3). This process is thought to be governed in part by two cis-acting sequences, designated the pac1 and pac2 sequences, which were first identified as herpesvirus-conserved motifs located approximately 30 bp from opposite ends of herpesvirus genomes (7). Typically, pac1 motifs consist of 3- to 7-bp A- or T-rich regions flanked on each side by poly(C) tracts (7, 32), whereas pac2 motifs consist of 5- to 10-bp A-rich regions (7). Subsequent studies of several viral systems suggest that both pac1 and pac2 motifs play important roles in genome maturation (5–8, 11, 12, 21, 26, 31, 34, 38, 40).
In the simplest model, the pac1 motif-containing end of one genome is fused to the pac2 motif-containing end of the adjacent genome in the concatemer. Hence, pac1 and pac2 motifs are about 60 bp apart, and cleavage occurs at a precise nucleotide position in between. The situation is more complex for viruses with class E genomes (27), such as herpes simplex virus 1 (HSV-1) and human cytomegalovirus (HCMV). In HSV-1, the pac1 and pac2 motifs lie near opposite ends of the terminal repeat, or a sequence, that is present as a single copy adjacent to the c sequence at the short (S) segment terminus (ca), as one or more reiterated copies adjacent to the b sequence at the long (L) segment terminus (anb), and as one or more copies in inverted orientation (relative to the terminal a sequences) at the junction between the L and S segments (b′a′mc′) (28). For HSV-1, the observation that all genomes have at least one a sequence at each end, yet are cleaved from concatemers that predominantly contain junctions with single a sequences, suggests that a sequences must first be duplicated by an unknown mechanism such that cleavage can occur between two adjacent a sequences.
Earlier efforts to identify cis cleavage/packaging sequences by insertion of a sequence subfragments into the HSV-1 genome were problematic due to recombination between the subfragments and native a sequences present in inverted orientation at L/S junctions (31). To avoid this problem, we chose to study murine cytomegalovirus (MCMV), which has a simple genome structure that lacks internal repeats and invertible elements (9, 24). We developed a system based on the premise that a second or “ectopic” cleavage site in the genome will function as an alternate to the natural cleavage site. However, because cleavage sites must be spaced nearly a genome length apart, genome packaging will occur in two frames: a natural frame in which cleavage occurs at natural cleavage sites and intervening ectopic sites are skipped over, and an ectopic frame in which cleavage occurs at ectopic cleavage sites and intervening natural sites are skipped over. Consequently, virions will contain a mixture of two genome types: those cleaved at natural sites, having natural termini and uncleaved internal ectopic junctions, and those cleaved at ectopic sites, having ectopic termini and uncleaved internal natural junctions. If the ectopic sites function with the same efficiency as the natural sites, the ratio of the two genome types will be roughly equal, while mutations that reduce the efficiency of ectopic sites will shift the ratio to favor genomes cleaved at natural sites. Therefore, the ratio can be used to quantitate the efficiency of ectopic site function (22). By using this system, it was shown that for MCMV, both pac1 and pac2 motifs are critically important for the full process of producing genomes encapsidated in extracellular virions, whereas a CGCGGCG motif (found distal to the pac2 motif in some herpesviruses) (7) serves to augment cleavage site efficiency (22). Subsequent studies revealed that sequences distal to the pac2 motif (up to 150 bp from the terminus) contribute significantly to the efficiency of cleavage site function (39).
While these studies did much to probe the boundaries of herpesvirus genome maturation signals, HCMV did not seem to fit the paradigm. HCMV has a class E genome structure similar to that of HSV-1 (16, 18), but unlike HSV-1, a significant number of HCMV genomes have S termini that lack an a sequence and instead end with a c sequence (32). This suggests that in HCMV, both c-a and a-a junctions represent functional cleavage sites. While both contain a typical pac1 element at one a sequence end, neither the c sequence end nor the other a sequence end (which opposes the pac1 motif across the point of cleavage) contains an A-rich region characteristic of pac2 motifs. In 1989, Kemble and Mocarski suggested that an ATAAAA motif might represent the HCMV pac2 motif despite its unusual location 644 bp distal from the predicted pac2 terminus (Fig. 1A) (15). Subsequently, however, a proposed “cryptic pac2” element, which functions in DNA maturation and occupies the expected pac2 position yet lacks an A-rich region, was identified for guinea pig cytomegalovirus (21). This suggested that HCMV may have similar cryptic pac2 elements located near the c and a sequence ends. Partial homologies between a and c sequences located an appropriate distance from their respective genomic termini were used to identify putative cryptic pac2 elements that were designated pac2a and pac2c elements (Fig. 1A) (23).
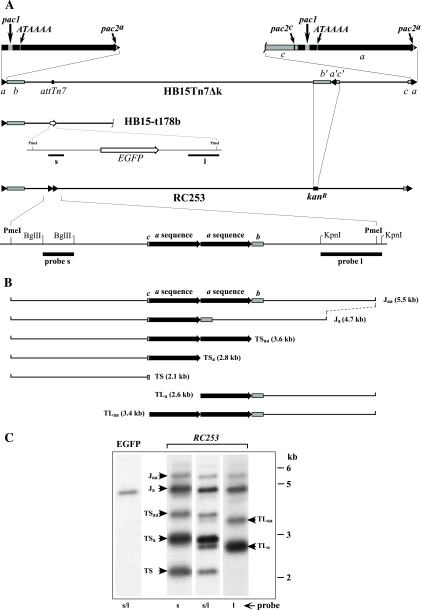
Fig. 1.
The HCMV ectopic cleavage site system. (A) The genome of wild-type BAC-derived virus HB15Tn7Δk is shown at the top with terminal a sequences (triangles) expanded above (black arrows) to illustrate the relative positions of putative cis elements. Below this are shown the genomes of virus HB15-t178b, illustrating the insertion of an EGFP expression cassette (open arrow) at the attTn7 site, and RC253, illustrating the replacement of the b′a′c′ segment with the Kanr gene (kanR) and ectopic insertion of two a sequences at the attTn7 site. The RC253 ectopic cleavage site is expanded below to show the two a sequences (black arrows) with flanking partial b and c sequences (gray boxes). Fragments used as hybridization probes s and l are indicated by black bars. (B) Schematics illustrate restriction fragments predicted from PmeI digestion of RC253 DNA representing junctions (J) and terminal fragments (TS and TL) predicted to arise from cleavage at the ectopic locus. Predicted fragment sizes (kb) are in parentheses. (C) Virion DNAs from viruses HB15-t178b (EGFP) and RC253 were digested with PmeI, separated by agarose electrophoresis, transferred to a nylon membrane, and sequentially hybridized with probe s, probe l, or a mixture of s and l (s/l) as indicated below each lane. Fragment identifications (arrows) are based on molecular weights and probe hybridizations. The positions of DNA size standards are shown on the right.
With the advent of infectious bacterial artificial chromosome (BAC) clones of the HCMV genome, it became theoretically possible to delete the entire internal b′a′c′ L/S junction region, producing an HCMV genome that lacks the internal inverted a sequences that were problematic for HSV-1 studies. This was achieved through BAC mutagenesis with Escherichia coli, and the resulting BAC gave rise to an HCMV mutant, HB15ΔL/S, that replicates with near wild-type efficiency in vitro but has a simple genome that does not undergo segment inversion (29). Here, we show that ectopic cleavage sites inserted into the HB15ΔL/S genome are cleaved efficiently and that the ATAAAA motif identified by Kemble and Mocarski (15) is dispensable for efficient cleavage and therefore does not represent the HCMV pac2 motif. We further show that the region opposite the pac1 motif predicted to contain a cryptic pac2 motif (23) is critically important for HCMV genome maturation.
MATERIALS AND METHODS
Recombinant virus construction and propagation.
Construction of bacterial artificial chromosome (BAC) HB15ΔL/S from parental BAC HB15Tn7Δk been previously described (29). Ectopic cleavage sites were inserted into plasmid pFastBac1 (Invitrogen) and shuttled into the attTn7 site of HB15ΔL/S by Tn7-mediated site-specific transposition as described previously (10). The control virus HB15-t178b was derived by transposition of an expression cassette for enhanced green fluorescent protein (EGFP) into BAC HB15Tn7Δk. Clones were screened for correct transposition as described previously (10). Human MRC-5 fibroblast cells (ATCC CCL-171) were propagated in Eagle's minimal essential medium supplemented with 10% fetal calf serum (HyClone Laboratories), 10,000 IU/liter penicillin, and 10 mg/liter streptomycin (Gibco-BRL). Recombinant viruses were reconstituted by transfection of 2 to 4 μg of BAC DNA into subconfluent MRC-5s grown in 6-well plates as described previously (10). Viruses were named by adding the prefix “RC” to the number from the plasmid that was used for transposition (e.g., virus RC253 contains the ectopic cleavage site from plasmid pMA253). The correct sequence for each ectopic site was confirmed by Sanger dideoxy sequencing of DNA isolated from virions as previously described (22).
Plasmids.
Plasmid pMA253 was constructed by excision of a 2.7-kb XhoI fragment from pON232 (25) and ligation into XhoI-restricted pFastBac1. Plasmid pMA258 was constructed by restricting pMA253 with SgrAI, blunt ending the resulting ends with T4 DNA polymerase plus all four deoxynucleoside triphosphates, and ligating the construct into StuI-restricted pFastBac1. Plasmid pON265 was constructed by restricting pMA253 with MluI/ApaI, blunt ending as described above, and ligating into StuI-restricted pFastBac1. Plasmid pMA273 was constructed by restricting pMA265 with BsiBI/BssHII, blunt ending as described above, and ligating into StuI-restricted pFastBac1. Plasmid pMA315 was constructed by restricting pMA273 with PleI/BseYI, blunt ending as described above, and ligating into StuI-digested pFastBac1. Plasmids pMA297, pMA298, pMA302, pMA312, and pMA337 were constructed by PCR amplification of pMA273 DNA using the primers shown in Table 1, cloning the PCR products into pCR8/GW/TOPO (Invitrogen), and then moving the resulting inserts, contained in EcoRI fragments, into EcoRI-restricted pFastBac1.
Table 1.
Oligonucleotides used for plasmid construction
| Plasmid | Oligonucleotide | Sequence (5′ to 3′) |
|---|---|---|
| pMA297 | MOL233 | ACCGACACACCCGACACACG |
| MOL235 | CAACGCGACTCCAAGACTCC | |
| pMA298 | MOL233 | ACCGACACACCCGACACACG |
| MOL241 | ATTTCACCCCCCCGCTAAAA | |
| pMA302 | MOL245C | ACCCCCCCGCTAAAAACA |
| MOL246 | CACACCCAACCCGACGCC | |
| pMA312 | MOL241 | ATTTCACCCCCCCGCTAAAA |
| MOL265 | GACACACCCGGCACGCA | |
| pMA337 | MOL241 | ATTTCACCCCCCCGCTAAAA |
| MOL266 | ACGCACGCACCCACCCA |
Analysis of ectopic cleavage site function.
Cells were infected at a multiplicity of infection (MOI) of ∼0.1. Ten days after infection, virion DNAs were purified from extracellular viral particles, restricted with PmeI, separated by agarose electrophoresis, transferred to nylon membranes, and hybridized using 32P-labeled probes as described previously (22). The pFastBac1 probe consisted of NotI-digested pFastBac1 DNA. Probes s and l were BglII (s) or KpnI (l) fragments gel-purified from BglII- or KpnI-restricted pMA253 DNA (Fig. 1A). Autoradiographs of hybridized membranes were scanned as transparencies, and fragment intensities were quantitated as previously described (39). Percent cleavage efficiencies were calculated using the formula (TS − background)/(J − background) × 100, where TS is a terminal fragment and J is a junction.
RESULTS
Ectopically inserted tandem a sequences are cleaved efficiently.
Previous studies have shown that L termini and L/S junctions of HCMV genomes can contain one to several a sequences in tandem arrangement, while S termini generally contain one or, in some cases, no copies of the a sequence (20, 33). It was therefore uncertain whether one a sequence or two in tandem would be needed for ectopic cleavage. A fragment containing two tandemly repeated a sequences derived from HCMV strain Towne was transposed into the attTn7 site of HB15ΔL/S to generate virus RC253. The ectopic Towne a sequences differed somewhat from that of the native AD169 a sequences of HB15ΔL/S, but regions of partial homology and the location of the pac1 motif indicated that the ectopic a sequences were inserted in direct orientation relative to the native a sequences, as illustrated in Fig. 1A. Virus HB15-t178b, which has an expression cassette for EGFP transposed into the attTn7 site, as illustrated in Fig. 1A, was used as a control. Virion DNAs from viruses RC253 and HB15-t178b were digested with the restriction enzyme PmeI and analyzed by Southern hybridization. Figure 1B shows the ectopic cleavage site of RC253 as well as restriction fragments predicted from PmeI digestion of RC253 DNA representing ectopic junctions that retain two a sequences (Jaa) or one a sequence (Ja), ectopic S termini having zero (TS), one (TSa), or two terminal a sequences (TSaa), and ectopic L termini having one (TLa) or two terminal a sequences (TLaa). L ends lacking an a sequence were not predicted, as they are not observed at the native ends of normal HCMV genomes (20, 33).
The membrane was initially hybridized with a mixture of two probes, s and l, containing sequences derived from both sides of the ectopic cleavage site (probe locations are shown in Fig. 1A). As expected, the probe mixture hybridized with only one fragment in the control virus HB15-t178b, and its 5-kb size was consistent with the PmeI fragment predicted from this virus (Fig. 1C). In contrast, seven fragments were detected in RC253 DNA (Fig. 1C), and their sizes matched the sizes of the predicted terminal and junction fragments illustrated in Fig. 1B. To confirm their identities, the membrane was stripped and sequentially rehybridized with probe s and then probe l. The results precisely matched the predictions for each fragment. Both probes hybridized to the 4.7- and 5.5-kb fragments predicted to be junction fragments Ja and Jaa; probe l hybridized to the 2.6- and 3.4-kb fragments predicted to be TLa and TLaa and failed to detect the three TS fragments; and probe s hybridized to the 2.1-, 2.8-, and 3.6-kb fragments predicted to be TS, TSa, and TSaa but failed to detect either of the TL fragments (Fig. 1C). From these results, we conclude that during replication of RC253, most of the Jaa junctions present in BAC DNA were recombined to give rise to Ja junctions. One or both of these junctions were able to serve as cleavage packaging sites, giving rise to ectopic termini having various numbers of terminal a sequences, precisely as observed previously for wild-type HCMV (20, 33).
Fragments spanning the junction between two a sequences are cleaved efficiently.
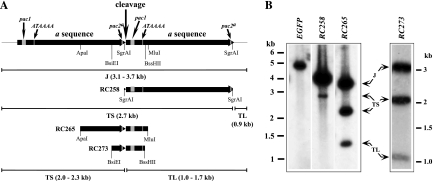
We next predicted that sequences spanning the junction between two a sequences would be sufficient to mediate cleavage and would lack the complexity arising from multiple a sequences. Various restriction fragments, shown in Fig. 2A, were transposed into the attTn7 site of BAC HB15ΔL/S, and the resulting viruses were assayed for cleavage by detection of TL and TS fragments. In virus RC265, an ApaI-MluI fragment, in which pac1 and pac2a motifs flank the predicted point of cleavage and the ATAAAA motif is distal to the pac1 sequence relative to the point of cleavage (Fig. 2A), gave rise to TL and TS fragments with precisely the predicted sizes (Fig. 2B). In virus RC273, a smaller BsiEI-BssHII fragment, again containing all three cis elements in the same orientations, was similarly cleaved (Fig. 2B). In contrast, in virus RC258, a SgrAI fragment containing the same putative cis elements but with pac1 and pac2a motifs at opposite ends of the fragment (Fig. 2A) was not cleaved near the pac1 end, as predicted 2.0- and 1.7-kb fragments were not detected (Fig. 1B). A small amount of 2.7-kb fragment was observed, consistent with the size predicted for a TS fragment resulting from cleavage at the right end of the SgrAI fragment adjacent to the pac2a motif (Fig. 2A); however, no signal was detected for the corresponding 911-bp TL fragment (Fig. 2B) even after extended exposure (not shown). These results indicate that (i) the BsiEI-BssHII fragment is sufficient to mediate efficient cleavage and packaging, (ii) in the absence of the pac1 motif, inefficient cleavage and packaging can occur via cleavage adjacent to the pac2a motif, and (iii) efficient cleavage and packaging requires the pac1 motif counterposed across the point of cleavage from the pac2a motif (or adjacent sequences).
Fig. 2.
Analysis of fragments spanning the junction between two a sequences. (A) Two adjacent a sequences are shown to illustrate the positions of putative cis elements, the pac2a, pac1, and ATAAAA elements, relative to the predicted point of cleavage. Restriction fragments shown below were tested as ectopic cleavage sites. Lines with bars illustrate PmeI junctions (J) and terminal fragments (TS and TL) predicted to arise from cleavage at the ectopic loci. Sizes (kb) predicted for these fragments are shown in parentheses and vary slightly between sites. (B) Virion DNAs from viruses HB15-t178b (EGFP), RC258, RC265, and RC273 were digested with PmeI, separated by agarose electrophoresis, transferred to a nylon membrane, and hybridized with a probe consisting of radiolabeled pFastBac1 sequences. Arrows indicate J, TL, and TS fragments, and the positions of DNA size standards are shown to the left or right of each panel.
The minimal functional cleavage site retains the pac1 and pac2a motifs but not the ATAAAA motif.
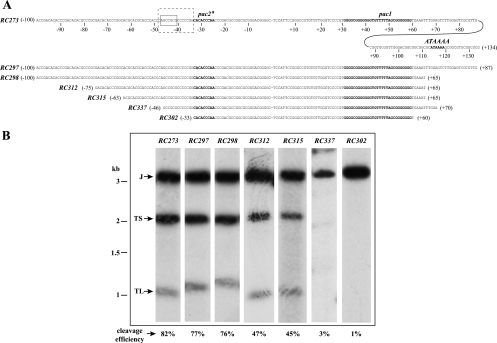
The BsiEI-BssHII fragment in virus RC273 consists of 234 nucleotides. Assigning the zero position to the point of cleavage, we numbered the nucleotides on the pac2a side from −1 to −100 and those on the pac1 side from +1 to +134 (Fig. 3A). Nested deletions, shown in Fig. 3A, were constructed and assayed for cleavage by Southern hybridization (Fig. 3B) as described above. To quantify the efficiency of ectopic cleavage, hybridization signals for J and TS were measured densitometrically and corrected for background. Cleavage efficiencies were then calculated as the ratios of TS to J signal intensities. This analysis revealed an 82% cleavage efficiency for the BsiEI-BssHII cleavage site in virus RC273. This high efficiency remained essentially unchanged in viruses RC297 and RC298 (77% and 76%, respectively), which lack sequences to the right of the pac1 motif (Fig. 3A). These results demonstrate that virtually all sequences distal to the pac1 motif (nucleotides +65 to +134) are not important for HCMV DNA cleavage and packaging. As the ATAAAA motif is contained within this region, we conclude that the ATAAAA motif is not involved in HCMV DNA cleavage and packaging and hence cannot represent the HCMV pac2 motif.
Fig. 3.
Nested deletions of the ectopic cleavage site. (A) The full sequence of the BsiEI-BssHII fragment is shown with a dash (–) indicating the point of cleavage. Nucleotides to the left are numbered −1 to −100, and nucleotides to the right are numbered +1 to +134. Putative cis packaging elements, the pac2a, pac1, and ATAAAA elements, are shown in bold. A conserved block (dashed box) and sequences similar to CGCGGCG motifs (on the complementary strand; solid box) were identified in a prior study (23). Sequences below represent cleavage sites introduced into recombinant viruses. The nucleotide positions for left and right ends of each construct are in parentheses. (B) Virion DNAs from the indicated viruses were digested with PmeI, separated by agarose electrophoresis, transferred to a nylon membrane, and hybridized with a probe consisting of radiolabeled pFastBac1 sequences. Arrows indicate J, TS, and TL fragments. Below each lane are the ectopic cleavage site efficiencies that were calculated from densitometric measurements of J and TS fragment intensities of each virus. The positions of DNA size standards are shown on the left.
Ectopic cleavage efficiency dropped precipitously as sequences were deleted from the pac2a side of the cleavage site. The 76% cleavage efficiency of virus RC298 decreased to 47% when nucleotides −100 to −76 were deleted in virus RC312. No further decrease was observed when additional nucleotides, from −75 to −64, were deleted in virus RC315, but deletion of distal sequences through −47 in virus RC337 and through −32 in RC302 fully eliminated cleavage. These results suggest that (i) sequences in the −100 to −76 region, though not essential for cleavage, play a modest role in augmenting cleavage efficiency, (ii) sequences from −75 to −64 are not important for cleavage, and (iii) sequences in the region from −63 to −47 are crucial for cleavage site function.
DISCUSSION
Herpesvirus DNA maturation, including DNA translocation and site-specific cleavage, are critical for viral replication but are of no importance for cellular processes. Hence, they represent attractive targets for antiviral development. Several compounds that inhibit DNA maturation have been identified (4, 13, 17, 35–37), and one, active against HCMV, has entered clinical trials (19). However, our understanding of herpesvirus DNA packaging at the molecular level remains fragmentary. Terminase clearly recognizes specific DNA sequences that direct both where to cleave and which end to insert into empty procapsids to initiate packaging. Evidence to date suggests that multiple cis-acting elements are involved, but these elements have not been precisely defined and it remains uncertain how or if different elements interact with different terminase subunits to mediate different functions (e.g., initiation of packaging versus terminal cleavage).
The pac1 and pac2 cis-acting elements are well conserved near the termini of herpesvirus genomes (7, 21). However, the pac2 element of HCMV has remained an enigma, as regions predicted to contain pac2 elements lack the poly(A) tract that is characteristic of pac2 elements from other viruses. While the ATAAAA motif identified by Kemble and Mocarski (15) has a plausible pac2-like sequence, its location distal to the pac1 sequence is not consistent with the location of pac2 elements (opposite the point of cleavage from the pac1 element) seen for other viruses. This led us to propose that, like guinea pig cytomegalovirus (21), HCMV uses cryptic pac2 elements that carry out the function(s) of pac2 elements but lack the characteristic poly(A) tract (23). Moreover, because HCMV forms two types of short-arm ends (one ending with ca and one ending with c) we postulated the existence of two distinct cryptic pac2 elements and proposed putative cis elements, the pac2a and pac2c elements, based on position and sequence conservation between the two ends (23).
The work presented here clearly eliminates the ATAAAA motif as having any involvement in DNA maturation. Indeed, there appear to be no cis-acting sequences distal to the pac1 element/+65 that are important to this process. This finding is fully consistent with our results from MCMV as well as those from HSV-1, where deletions, insertions, or substitutions of sequences distal to the pac1 motif had little or no impact (12, 22, 34, 39). On the proposed cryptic pac2 side of the cleavage site, we found that sequences greater than 100 nucleotides from the point of cleavage are dispensable. Within the end-proximal 100 nucleotides, there are at least two regions of importance. The first, between −100 and −76, is nonessential but augments cleavage site efficiency nearly 2-fold. This is consistent with findings for other herpesviruses, including indirect evidence for the existence of augmenting sequences at positions −101 to −78 of HSV-1 (12) and mutagenic evidence defining augmenting sequences within two regions for MCMV (−150 to −127 and −117 to −101) (39) and at −89/88 of varicella-zoster virus (14) (in each virus, nucleotides are numbered negatively from the pac2 end).
The second region, from −63 to −47, contains sequences essential for cleavage site function. Although these results neither confirm nor preclude the existence of a cryptic pac2 (e.g., pac2a) element within sequences proximal to −47, they do affirm the presence of critical cis-acting sequences on this side of the cleavage site. Indeed, the fact that conserved sequences that include a CGCGGCG-like motif (CGCGGCT) lie adjacent/distal to the pac2a motif (Fig. 3A, boxed regions) suggests the possibility that the HCMV pac2 motif encompasses the pac2a motif and these additional distal sequences, perhaps extending into the −63 to −47 region. This hypothesis is supported by the observation that a 6-bp insertion 1 bp distal to the MCMV pac2 poly(A) eliminates cleavage site function (22). In contrast, pac2-adjacent sequences are not important in HSV-1, as substitution mutants of its CGCGGCG motif or sequences immediately adjacent to the pac2 poly(A) tract had no apparent impact on DNA cleavage or packaging (12, 34). Mutants with more detailed substitutions will be needed to directly demonstrate the importance of HCMV sequences proximal to position −47.
Curiously, the presence of TS fragments within virions (Fig. 1B) would seem to contradict the above results derived using a-a junctions. The latter clearly demonstrate the importance of sequences greater than 46 nucleotides from the point cleavage, while the fact that only 34 bp of c sequence are present in the RC253 cleavage site suggests that cleavage and packaging of genomes with TS ends does not require the pac2c element or paralogs of the distal cis elements identified above. Perhaps the key to this contradiction is the context: one experiment involves full a sequences, while the other involves small fragments spanning a-a junctions. Thus, cis elements required for cleavage at c-a or a-a junctions may be different from those required for cleavage when a full a sequence is present and a sequence duplication is presumably occurring.
As our studies examined the contents of extracellular virions, they reveal only the cis elements essential for the complete process and cannot differentiate distinct functions. However, other studies support models in which the pac1 and pac2 sequences govern formation of their respective termini, while pac2 and/or associated sequences direct the insertion of concatemer ends into procapsids to initiate packaging (12, 23, 30, 34, 38). Consistent with this, virus RC258 produced a small number of TS fragments in the absence of detectable TL fragments, suggesting that cleavage can occur adjacent to the pac2a element in the absence of a counterposing pac1 element, but of the two ends that are formed, only those containing the pac2a element are packaged into virions. This is highly reminiscent of results described by Tong and Stow, in which cells infected by HSV-1 mutants lacking the pac1 motif formed both genome ends, but only the pac2-containing ends were protected from nuclease and hence presumably packaged (34). If indeed cis sequences are required for insertion of concatemer ends into procapsids, the detection of TS ends with no a sequence in virions suggests that such sequences lie within the terminal 34 bp of the c sequence. It is interesting to note that analogous HSV-1 sequences (−1 to −20) comprising the DR1 repeat were recently shown to be crucial for genome maturation (34). This suggests the intriguing possibility that while the pac2 motif may mediate formation of concatemer ends, the extreme terminal sequences of these ends may govern concatemer/procapsid docking and/or initiation of DNA packaging. Future studies examining packaged and unpackaged intracellular viral DNA species formed by our mutants may yield additional clues to the unique functions of specific cis-acting sequences.
An accurate delineation of the cis elements required for DNA maturation should facilitate identification of functional interactions between cis elements and terminase, its subunits, or other proteins involved in genome maturation. While pac1 binding has been reported for terminase subunits UL28 of HSV-1 (1) and UL56 of HCMV (2), protein interactions with pac2 elements have been difficult to demonstrate. Prior reports showing binding to the ATAAAA motif by proteins in HCMV-infected cells (2), HCMV UL56 (2), or an unidentified cellular protein (15) must now be considered irrelevant to DNA packaging in light of our finding that the ATAAAA motif is dispensable. Nevertheless, future efforts to demonstrate sequence-specific DNA cleavage or relevant DNA-protein interactions will benefit greatly from a detailed understanding of the functional cis elements required for this process.
ACKNOWLEDGMENTS
We are grateful to Gabriella Hahn for providing BAC HB15Tn7Δk and to Edward Mocarski for plasmid pON232.
This work was supported by Public Health Services grants R01AI46668 and R21AI43527 (to M.A.M.).
Footnotes
Published ahead of print on 23 February 2011.
REFERENCES
- 1. Adelman K., Salmon B., Baines J. D. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. U. S. A. 98:3086–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bogner E., Radsak K., Stinski M. F. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72:2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown J. C., McVoy M. A., Homa F. L. 2002. Packaging DNA into herpesvirus capsids.p. 111–153 In Bogner E., Holzenburg A. (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum Publishers, London, United Kingdom [Google Scholar]
- 4. Buerger I., et al. 2001. A novel nonnucleoside inhibitor specifically targets cytomegalovirus DNA maturation via the UL89 and UL56 gene products. J. Virol. 75:9077–9086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chowdhury S. I., Buhk H. J., Ludwig H., Hammerschmidt W. 1990. Genomic termini of equine herpesvirus 1. J. Virol. 64:873–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davison A. J. 1984. Structure of the genome termini of varicella-zoster virus. J. Gen. Virol. 65:1969–1977 [DOI] [PubMed] [Google Scholar]
- 7. Deiss L. P., Chou J., Frenkel N. 1986. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J. Virol. 59:605–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deiss L. P., Frenkel N. 1986. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J. Virol. 57:933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ebeling A., Keil G. M., Knust E., Koszinowski U. H. 1983. Molecular cloning and physical mapping of murine cytomegalovirus DNA. J. Virol. 47:421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hahn G., Jarosch M., Wang J. B., Berbes C., McVoy M. A. 2003. Tn7-mediated introduction of DNA sequences into bacmid-cloned cytomegalovirus genomes for rapid recombinant virus construction. J. Virol. Methods 107:185–194 [DOI] [PubMed] [Google Scholar]
- 11. Hammerschmidt W., Ludwig H., Buhk H. J. 1988. Specificity of cleavage in replicative-form DNA of bovine herpesvirus 1. J. Virol. 62:1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hodge P. D., Stow N. D. 2001. Effects of mutations within the herpes simplex virus type 1 DNA encapsidation signal on packaging efficiency. J. Virol. 75:8977–8986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hwang J. S., et al. 2007. Identification of acetylated, tetrahalogenated benzimidazole d-ribonucleosides with enhanced activity against human cytomegalovirus. J. Virol. 81:11604–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaufer B. B., Smejkal B., Osterrieder N. 2010. The varicella-zoster virus ORFS/L (ORF0) gene is required for efficient viral replication and contains an element involved in DNA cleavage. J. Virol. 84:11661–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kemble G. W., Mocarski E. S. 1989. A host cell protein binds to a highly conserved sequence element (pac-2) within the cytomegalovirus a sequence. J. Virol. 63:4715–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kilpatrick B. A., Huang E. S. 1977. Human cytomegalovirus genome: partial denaturation map and organization of genome sequences. J. Virol. 24:261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krosky P. M., et al. 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. LaFemina R. L., Hayward G. S. 1980. Structural organization of the DNA molecules from human cytomegalovirus, p. 39–55 In Fields B. N., Jaenisch R. (ed.), Animal virus genetics. Academic Press, New York, NY [Google Scholar]
- 19. Lischka P., Zimmermann H. 2008. Antiviral strategies to combat cytomegalovirus infections in transplant recipients. Curr. Opin. Pharmacol. 8:541–548 [DOI] [PubMed] [Google Scholar]
- 20. McVoy M. A., Nixon D. E. 2005. Impact of 2-bromo-5,6-dichloro-1-beta-d-ribofuranosyl benzimidazole riboside and inhibitors of DNA, RNA, and protein synthesis on human cytomegalovirus genome maturation. J. Virol. 79:11115–11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McVoy M. A., Nixon D. E., Adler S. P. 1997. Circularization and cleavage of guinea pig cytomegalovirus genomes. J. Virol. 71:4209–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McVoy M. A., Nixon D. E., Adler S. P., Mocarski E. S. 1998. Sequences within the herpesvirus-conserved pac1 and pac2 motifs are required for cleavage and packaging of the murine cytomegalovirus genome. J. Virol. 72:48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McVoy M. A., Nixon D. E., Hur J. K., Adler S. P. 2000. The ends on herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences. J. Virol. 74:1587–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mercer J. A., Marks J. R., Spector D. H. 1983. Molecular cloning and restriction endonuclease mapping of the murine cytomegalovirus genome (Smith strain). Virology 129:94–106 [DOI] [PubMed] [Google Scholar]
- 25. Mocarski E. S., Liu A. C., Spaete R. R. 1987. Structure and variability of the a sequence in the genome of human cytomegalovirus (Towne strain). J. Gen. Virol. 68:2223–2230 [DOI] [PubMed] [Google Scholar]
- 26. Nasseri M., Mocarski E. S. 1988. The cleavage recognition signal is contained within sequences surrounding an a-a junction in herpes simplex virus DNA. Virology 167:25–30 [DOI] [PubMed] [Google Scholar]
- 27. Roizman B., Pellett P. E. 2001. The family of Herpesviridae: a brief introduction, p. 2381–2397 In Knipe D. M., Howley P. M. (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 28. Roizman B., Sears A. E. 1996. Herpes simplex viruses and their replication, p. 1048–1066 In Fields B. N., et al. (ed.), Fundamental virology. Raven Press, New York, NY [Google Scholar]
- 29. Sauer A., Wang J. B., Hahn G., McVoy M. A. 2010. A human cytomegalovirus deleted of internal repeats replicates with near wild type efficiency but fails to undergo genome isomerization. Virology 401:90–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schynts F., et al. 2003. The structures of bovine herpesvirus 1 virion and concatemeric DNA: implications for cleavage and packaging of herpesvirus genomes. Virology 314:326–335 [DOI] [PubMed] [Google Scholar]
- 31. Smiley J. R., Duncan J., Howes M. 1990. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J. Virol. 64:5036–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tamashiro J. C., Filpula D., Friedmann T., Spector D. H. 1984. Structure of the heterogeneous L-S junction region of human cytomegalovirus strain AD169 DNA. J. Virol. 52:541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamashiro J. C., Spector D. H. 1986. Terminal structure and heterogeneity in human cytomegalovirus strain AD169. J. Virol. 59:591–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tong L., Stow N. D. 2010. Analysis of herpes simplex virus type 1 DNA packaging signal mutations in the context of the viral genome. J. Virol. 84:321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Underwood M. R., et al. 2004. Mechanism of action of the ribopyranoside benzimidazole GW275175X against human cytomegalovirus. Antimicrob. Agents Chemother. 48:1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Underwood M. R., et al. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Zeijl M., et al. 2000. Novel class of thiourea compounds that inhibit herpes simplex virus type 1 DNA cleavage and encapsidation: resistance maps to the UL6 gene. J. Virol. 74:9054–9061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Varmuza S. L., Smiley J. R. 1985. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell 41:793–802 [DOI] [PubMed] [Google Scholar]
- 39. Wang J. B., Nixon D. E., McVoy M. A. 2008. Definition of the minimal cis-acting sequences necessary for genome maturation of the herpesvirus murine cytomegalovirus. J. Virol. 82:2394–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zimmermann J., Hammerschmidt W. 1995. Structure and role of the terminal repeats of Epstein-Barr virus in processing and packaging of virion DNA. J. Virol. 69:3147–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]